Several types of viruses encode viral miRNAs which regulate host and/or viral gene expression via posttranscriptional gene silencing, leading to efficient viral infection. Adenovirus (Ad) expresses miRNAs derived from VA-RNAs (mivaRNAI and -II); however, recent studies have revealed that processing of VA-RNAI into mivaRNAI inhibits Ad replication. Conversely, we demonstrate here that mivaRNAII significantly promotes Ad replication and that mivaRNAII-mediated suppression of CUL4A expression via posttranscriptional gene silencing induces accumulation of c-Jun, leading to promotion of Ad infection. These results exhibited the significance of VA-RNAII for supporting Ad infection through a mechanism complementary to that of VA-RNAI. These observations could provide important clues toward a new perspective on host-virus interaction. Moreover, Ad is widely used as a basic framework for viral vectors and oncolytic viruses. Our findings will help to regulate Ad infection and will promote the development of novel Ad vectors and oncolytic Ad.
KEYWORDS: JNK signaling, adenoviruses, cullin 4A, microRNA, posttranscriptional gene silencing
ABSTRACT
The adenovirus (Ad) serotype 5 genome encodes two noncoding small RNAs (virus-associated RNAs I and II [VA-RNAI and -II]), which are approximately 160-nucleotide (nt) RNAs transcribed by RNA polymerase III. It is well known that VA-RNAI supports Ad infection via the inhibition of double-stranded RNA-dependent protein kinase (PKR), which recognizes double-stranded RNA and acts as an antiviral system. Recent studies revealed that VA-RNAs are processed into VA-RNA-derived microRNAs (miRNAs) (mivaRNAI and -II); however, we and another group recently demonstrated that mivaRNAI does not promote Ad replication. On the other hand, the roles of VA-RNAII and mivaRNAII in Ad replication have remained to be clarified. In this study, we demonstrated mivaRNAII-mediated promotion of Ad replication. Transfection with chemically synthesized 3′-mivaRNAII-138, one of the most abundant forms of mivaRNAII, significantly enhanced Ad replication, while the other species of mivaRNAII did not. We identified 8 putative target genes of 3′-mivaRNAII-138 by microarray analysis and in silico analysis. Among the 8 candidates, knockdown of the cullin 4A (CUL4A) gene, which encodes a component of the ubiquitin ligase complex, most significantly enhanced Ad replication. CUL4A expression was significantly suppressed by 3′-mivaRNAII-138 via posttranscriptional gene silencing, indicating that CUL4A is a target gene of 3′-mivaRNAII-138 and mivaRNAII functions as a viral miRNA promoting Ad infection. It has been reported that CUL4A is involved in degradation of c-Jun, which acts as a transcription factor in the Jun-N-terminal kinase (JNK) signaling cascade. Treatment with JNK inhibitors dramatically suppressed Ad replication, suggesting that mivaRNAII-mediated downregulation of CUL4A enhanced JNK signaling and thereby promoted Ad infection.
IMPORTANCE Several types of viruses encode viral miRNAs which regulate host and/or viral gene expression via posttranscriptional gene silencing, leading to efficient viral infection. Adenovirus (Ad) expresses miRNAs derived from VA-RNAs (mivaRNAI and -II); however, recent studies have revealed that processing of VA-RNAI into mivaRNAI inhibits Ad replication. Conversely, we demonstrate here that mivaRNAII significantly promotes Ad replication and that mivaRNAII-mediated suppression of CUL4A expression via posttranscriptional gene silencing induces accumulation of c-Jun, leading to promotion of Ad infection. These results exhibited the significance of VA-RNAII for supporting Ad infection through a mechanism complementary to that of VA-RNAI. These observations could provide important clues toward a new perspective on host-virus interaction. Moreover, Ad is widely used as a basic framework for viral vectors and oncolytic viruses. Our findings will help to regulate Ad infection and will promote the development of novel Ad vectors and oncolytic Ad.
INTRODUCTION
MicroRNAs (miRNAs) are a class of noncoding RNAs that modulate gene expression at the posttranscriptional level. miRNAs are transcribed from the genome as precursor miRNAs (pre-miRNAs) and transported to the cytoplasm by exportin-5. The pre-miRNAs are then processed into approximately 22-nucleotide (nt) miRNA duplexes by dicer, followed by incorporation into the RNA-induced silencing complex (RISC). RISC-loaded miRNAs usually bind to target sites found within the 3′ untranslated region (3′ UTR) of targeted mRNAs, inducing the silencing of targeted genes. Numerous miRNAs have been found to play key roles in regulating a variety of cellular processes, including differentiation, proliferation, and apoptosis (1, 2).
As well as host cells, several viruses encode viral miRNAs which regulate host and/or viral gene expressions in the same manner as endogenous miRNAs (3–7). For example, simian virus 40 (SV40) encodes miR-S1, which regulates the SV40 viral T antigen expression, leading to a reduction in host cytotoxic T lymphocyte responses (8). Kaposi’s sarcoma herpesvirus miRNAs target caspase 3 and regulate apoptosis (9).
Adenoviruses (Ad) are known not only as a common cause of diseases such as colds but also as the basic framework of viral vectors and oncolytic viruses, which are expected to be useful in innovative therapies to treat cancer and other refractory diseases (10–15). It has been well established that the Ad genome encodes one or two small noncoding RNAs (virus-associated RNAs I and II [VA-RNAI and -II]), which are approximately 160-nt noncoding RNAs transcribed by RNA polymerase III (16, 17). VA-RNAs are rapidly transcribed at high levels soon after infection with Ad. VA-RNAs strongly support Ad infection, but the molecular mechanism underlying their enhancement of Ad infection has only been partly uncovered. A well-studied function of VA-RNAI is its inhibition of double-stranded RNA-dependent protein kinase (PKR), which acts as an antiviral system through the repression of viral protein synthesis, resulting in the inhibition of Ad infection (18, 19). Recently, several studies have demonstrated that VA-RNAs are processed in a manner similar to that for miRNAs, generating VA-RNA-derived miRNAs (mivaRNAs), including mivaRNAI and mivaRNAII (20, 21). 3′-mivaRNAI, a mivaRNA that is produced by dicer-mediated processing of VA-RNAI and is expressed at the highest level among the mivaRNAs in Ad-infected cells, has been shown to be efficiently incorporated into RISC (22, 23). These facts led us to hypothesize that 3′-mivaRNAI suppresses endogenous gene expression, leading to efficient infection; however, we and another group recently demonstrated that mivaRNAI did not support Ad replication and that a truncated form of VA-RNAI did not inhibit PKR activation, suggesting that when cleaved, VA-RNAI no longer supports Ad infection (24, 25).
Previous studies have demonstrated that VA-RNAII promotes Ad infection. A mutant Ad lacking only VA-RNAI expression (Sub722) exhibits approximately 6-fold-higher levels of replication than a mutant lacking both VA-RNAI and -II (26, 27). In addition, expression levels of VA-RNAII are significantly upregulated in the cells following infection with an Ad lacking VA-RNAI expression (28), although VA-RNAII inhibits PKR less efficiently than VA-RNAI (29). As described above, VA-RNAII is cleaved by dicer, producing mivaRNAII. Although the impacts of mivaRNAII on Ad replication remain to be clarified, it is known that mivaRNAII is incorporated into the RISC more efficiently than mivaRNAI, even though the copy numbers of VA-RNAII are more than 100-fold lower than those of VA-RNAI (17, 30). In addition, higher copy numbers of mivaRNAII have been detected in persistently Ad-infected lymphoid cells (31). These observations led us to hypothesize that mivaRNAII acts as a viral miRNA supporting Ad infection.
In this study, we demonstrated that mivaRNAII promotes Ad replication via posttranscriptional gene silencing, unlike mivaRNAI. Moreover, we globally searched for target genes of mivaRNAII by microarray analysis and in silico analysis, identifying the cullin 4A (CUL4A) gene as a promising target gene involved in Ad replication. CUL4A is a member of the cullin family and functions as a component of the ubiquitin ligase complex (32, 33). Suppression of CUL4A expression by mivaRNAII led to the upregulation of JNK signaling, resulting in enhancement of Ad replication. These results indicate that VA-RNAII supports Ad infection via a mechanism different from that of VA-RNAI.
RESULTS
Ad replication is upregulated by overexpression of VA-RNAII.
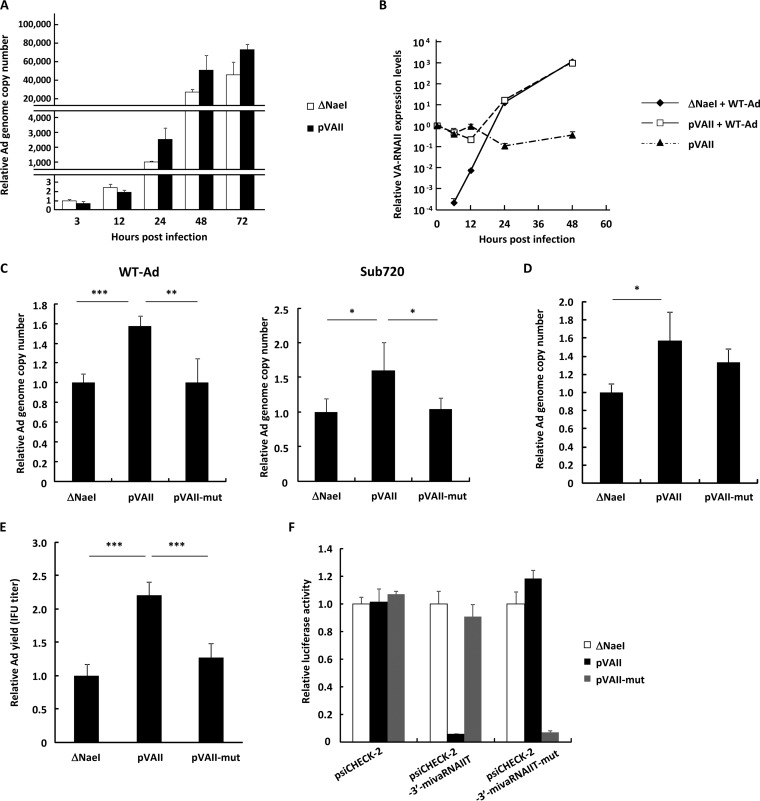
First, in order to examine whether VA-RNAII upregulated Ad replication, HeLa cells were transfected with a VA-RNAII-expressing plasmid (pVAII), followed by infection with wild-type Ad (WT-Ad; Ad serotype 5). Although there were no statistically significant differences in the Ad genome copy numbers at 3 and 12 h after infection in the cells transfected with pVAII and a control plasmid (ΔNaeI), significantly higher copy numbers of the WT-Ad genomes were found in the cells transfected with pVAII than in those transfected with ΔNaeI at 24 and 48 h postinfection (Fig. 1A). Real-time reverse transcription-PCR (RT-PCR) analysis of VA-RNAII demonstrated that transfection with pVAII resulted in much higher levels of VA-RNAII than treatment with the combination of ΔNaeI and WT-Ad during the early time points (6 and 12 h after treatment) (Fig. 1B). Subsequently, VA-RNAII copy numbers in WT-Ad-infected cells gradually increased after infection. Copy numbers of VA-RNAII 24 h after treatment with ΔNaeI and WT-Ad were approximately 120-fold higher than those in the cells after treatment with pVAII alone. In addition, VA-RNAII copy numbers in the cells treated with ΔNaeI and WT-Ad and those treated with pVAII and WT-Ad were comparable 48 h after treatment. These data indicated that WT-Ad-mediated VA-RNAII expression levels were significantly higher than those by pVAII as the Ad infection proceeded.
FIG 1.
Ad replication is upregulated by overexpression of VA-RNAII. (A) HeLa cells were transfected with a control plasmid (pAdVAntage-ΔNaeI; ΔNaeI) or a VA-RNAII-expressing plasmid (pVAII) for 48 h, followed by infection with WT-Ad at 100 VPs/cell. After 3, 12, 24, 48, and 72 h of incubation, the Ad genome copy numbers were determined by qPCR analysis and expressed as relative values (ΔNaeI, 3 h postinfection = 1). (B) HeLa cells were transfected with ΔNaeI or pVAII for 48 h, followed by infection with WT-Ad at 100 VPs/cell. After 0, 6, 12, 24, and 48 h of incubation following Ad infection, the copy numbers of VA-RNAII were determined by qRT-PCR analysis and expressed as relative values (pVAII, 0 h postinfection = 1). (C) HeLa cells were transfected with ΔNaeI, pVAII, or pVAII-mut for 48 h, followed by infection with Ad (WT-Ad or Ad lacking VA-RNAI, II; Sub720) at 100 VPs/cell. After 24 h of incubation, the Ad genome copy numbers were determined and expressed as relative values (ΔNaeI = 1). (D) A549 cells were transfected with VA-RNAII-expressing plasmids (ΔNaeI, pVAII, and pVAII-mut) for 48 h, followed by infection with WT-Ad at 100 VPs/cell. Ad genome copy numbers were determined 24 h after infection and expressed as relative values (ΔNaeI = 1). (E) HeLa cells were transfected with VA-RNAII-expressing plasmids (ΔNaeI, pVAII, and pVAII-mut) for 48 h, followed by infection with WT-Ad at 100 VPs/cell. After 24 h of incubation, infectious unit (IFU) titers of progeny WT-Ad in the cells were determined and expressed as relative values (ΔNaeI = 1). (F) HeLa cells were cotransfected with VA-RNAII-expressing plasmids (ΔNaeI, pVAII, and pVAII-mut) and reporter plasmids (psiCHECK-2, siCHECK-2-3′-mivaRNAIIT, and psiCHECK-2-3′-mivaRNAIIT-mut). After 48 h of incubation, luciferase activities were determined. RLuc activities were normalized to Fluc activities. These data are expressed as means ± SD (n = 4). *, P < 0.05; **, P < 0.01; ***, P < 0.001 (Student’s t test).
Several previous studies demonstrated that VA-RNAs are processed to mivaRNAs by dicer and that the 3′ strand of mivaRNAII (3′-mivaRNAII) is preferentially loaded into RISC compared with the other mivaRNAs (30). In order to examine whether a 3′-mivaRNAII-encoding sequence is crucial for Ad replication, we constructed pVAII-mut, a plasmid with mutations in a 3′-mivaRNAII-encoding sequence. Transfection with pVAII significantly promoted the replication of not only WT-Ad but also an Ad with a VA-RNA deletion (Sub720) at 24 h after infection, while transfection with pVAII-mut did not enhance replication of WT-Ad or Sub720 (Fig. 1C). Similar trends were found in A549 cells (Fig. 1D). Infectious Ad titers were also upregulated by overexpression of VA-RNAII (Fig. 1E). A reporter gene assay demonstrated that each of wild-type and mutated VA-RNAII suppressed the expression of the reporter gene containing the sequence complementary to the corresponding wild-type and mutated 3′-mivaRNAII in the 3′ UTR (Fig. 1F), indicating that each VA-RNAII was truncated by dicer and suppressed the expression of the reporter gene containing its target sequences via posttranscriptional gene silencing. These results indicated that VA-RNAII promoted Ad replication and that a sequence encoding 3′-mivaRNAII in VA-RNAII is crucial for VA-RNAII-mediated upregulation of Ad infection, strongly suggesting that 3′-mivaRNAII promotes Ad replication.
3′-mivaRNAII-138 promotes Ad replication.
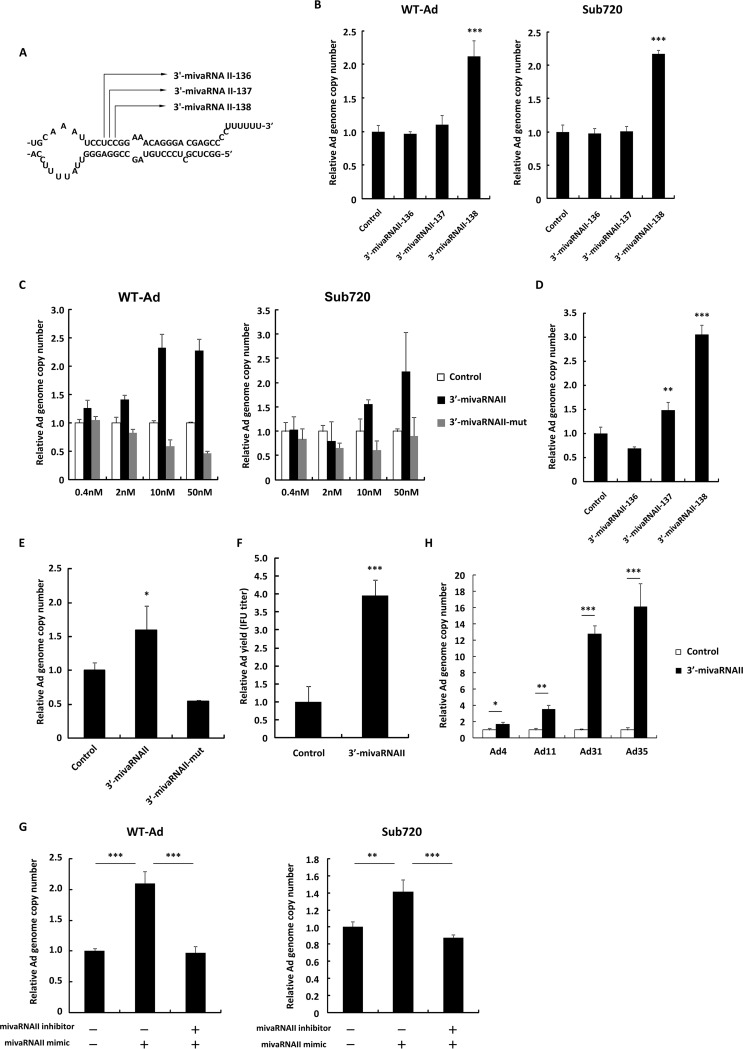
Dicer cleaves VA-RNAII at the multiple positions, producing the several 3′-mivaRNAII isoforms (23, 30) (Fig. 2A). In order to examine which isoforms of 3′-mivaRNAII promote Ad replication, HeLa cells were transfected with 3′-mivaRNAII-136, -137, and -138 mimics, followed by infection with Ad. Only the 3′-mivaRNAII-138 mimic, one of the most abundant mivaRNAs (22, 23), significantly increased Ad genome copy numbers (2.1-fold) compared with a control mimic; the other species of 3′-mivaRNAII did not (Fig. 2B). In the following experiments, we used the 3′-mivaRNAII-138 mimic as 3′-mivaRNAII, unless otherwise noted. The 3′-mivaRNAII mimic showed dose-dependent promotion of WT-Ad and Sub720 replication, while the 3′-mivaRNAII-mut mimic, which contains mutations in the seed sequence of 3′-mivaRNAII, did not (Fig. 2C). Similar results were obtained with A549 cells (Fig. 2D and E). In addition, transfection with 3′-mivaRNAII also increased infectious titers of progeny Ad (Fig. 2F). Furthermore, cotransfection with an inhibitor against 3′-mivaRNAII canceled the 3′-mivaRNAII-mediated promotion of Ad replication (Fig. 2G). Promotion of Ad replication following transfection with a 3′-mivaRNAII mimic was also found for the other human Ad serotypes (Ad31, species A; Ad11 and Ad35, species B; and Ad4, species E), although they have different sequences of mivaRNAs (16, 30) and species B Ad do not possess the VA-RNAII gene (16, 30) (Fig. 2H). These results demonstrated that 3′-mivaRNAII significantly promoted Ad replication and that the seed sequence of 3′-mivaRNAII was crucial for upregulation of Ad infection.
FIG 2.
3′-mivaRNAII promotes Ad replication. (A) A schematic diagram of the processing of VA-RNA II by dicer, producing 3′-mivaRNAII-136, -137, and -138. (B) HeLa cells were transfected with 3′-mivaRNAII-136, -137, or -138 mimics at 50 nM for 48 h, followed by infection with Ad (WT-Ad and Sub720) at 100 VPs/cell. Ad genome copy numbers were determined 24 h after infection and expressed as relative values (control = 1). (C) HeLa cells were transfected with 3′-mivaRNAII (3′-mivaRNAII-138) and 3′-mivaRNAII-mut mimics at the indicated concentrations for 48 h, followed by infection with Ad (WT-Ad and Sub720) at 100 VPs/cell. Ad genome copy numbers were determined 24 h after infection and expressed as relative values (control = 1 at the respective concentration). (D) A549 cells were transfected with 3′-mivaRNAII-136, -137, or -138 mimics at 50 nM for 48 h, followed by infection with WT-Ad at 100 VPs/cell. Ad genome copy numbers were determined 24 h after infection and expressed as relative values (control = 1). (E) A549 cells were transfected with 3′-mivaRNAII-138 and -mut mimics at 50 nM for 48 h, followed by infection with WT-Ad at 100 VPs/cell. Ad genome copy numbers were determined 24 h after infection and expressed as relative values (control = 1). (F) HeLa cells were transfected with 3′-mivaRNAII (3′-mivaRNAII-138) mimic at 50 nM and incubated for 48 h, followed by infection with WT-Ad at 100 VPs/cell. After 24 h of incubation, IFU titers of progeny WT-Ad in the cells were determined and expressed as relative values (control = 1). (G) HeLa cells were cotransfected with 3′-mivaRNAII (3′-mivaRNAII-138) mimic and negative control inhibitor or 3′-mivaRNAII inhibitor at 30 nM each for 48 h, followed by infection with WT-Ad at 100 VPs/cell. Ad genome copy numbers were determined 24 h after infection and expressed as relative values (control = 1). (H) HeLa cells were transfected with 3′-mivaRNAII (3′-mivaRNAII-138) mimic at 50 nM for 48 h, followed by infection with Ad4, Ad11, Ad31, or Ad35 at 100 VPs/cell. Ad genome copy numbers were determined 24 h after infection and expressed as relative values (control = 1). These data are expressed as means ± SD (n = 4).
Identification of 3′-mivaRNAII target genes.
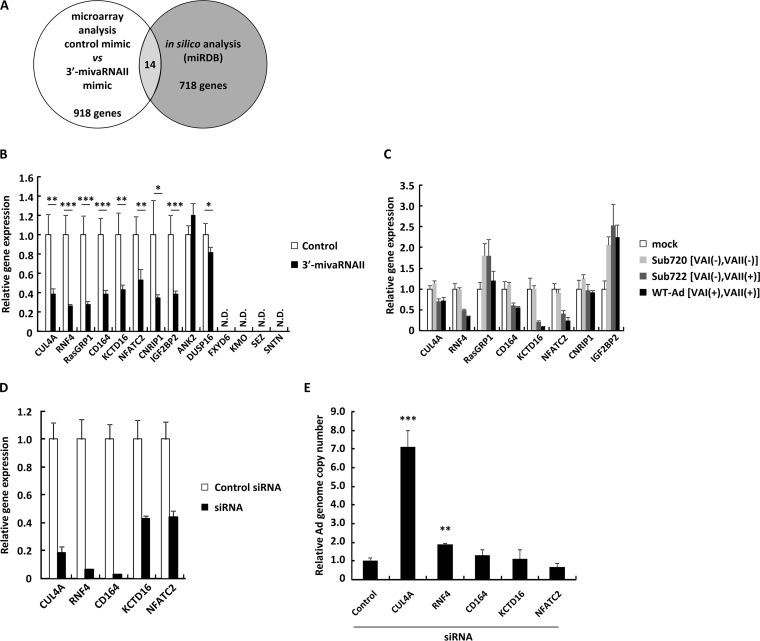
3′-mivaRNAII showed sequence-specific promotion of Ad replication in Fig. 1 and 2, indicating that 3′-mivaRNAII suppressed certain target genes in the same way as cellular miRNAs do in order to enhance Ad replication. We performed cDNA microarray gene expression analysis on HeLa cells to identify the target genes of 3′-mivaRNAII. We found that the expression levels of 918 genes in the cells transfected with the 3′-mivaRNAII mimic were reduced to less than half of those transfected with a control mimic (Fig. 3A). Among this pool of candidate genes, we selected 14 genes which have the sequence partly complementary to 3′-mivaRNAII in the 3′ UTR of their mRNA using an online database for miRNA target prediction (miRDB) (34). In order to narrow down the candidates, we determined the levels of the 14 candidate genes in the cells transfected with the 3′-mivaRNAII or infected with Ad by quantitative RT-PCR (qRT-PCR) analysis. Expression of 4 genes (FXYD6, KMO, SEZ, and SNTN) among the 14 candidates was undetectable by qRT-PCR analysis. The expression levels of the 4 genes were also very low in the cDNA microarray analysis. Among the 10 genes which were expressed in HeLa cells, the expression of 8 was significantly suppressed, more than 2-fold, in the cells by transfection with a 3′-mivaRNAII mimic, while the expression of the remaining 2 genes (ANK2 and DUSP16) was not suppressed (Fig. 3B). However, expression of 3 of the 8 genes (RasGRP1, CNRIP1, and IGF2BP2) was not significantly suppressed in WT-Ad-infected cells and Sub722-infected cells compared with either uninfected or Sub720-infected cells, while the expressions of the other 5 genes were (Fig. 3C). These data suggested that the 5 genes could be target genes of 3′-mivaRNAII.
FIG 3.
Identification of 3′-mivaRNA II target genes. (A) A schematic diagram of analysis for identification of mivaRNAII target genes. (B) HeLa cells were transfected with 3′-mivaRNAII (3′-mivaRNAII-138) mimic at 50 nM. After 48 h of incubation, mRNA levels of the indicated genes were determined by qRT-PCR analysis. (C) HeLa cells were infected with Ad (WT-Ad, Sub722, and Sub720) at 1,000 VPs/cell. After 48 h of incubation, mRNA levels of the indicated genes were determined by qRT-PCR analysis. (D and E) HeLa cells were transfected with siRNAs targeting the indicated genes at 50 nM. (D) After 48 h of incubation, mRNA levels of the indicated genes were determined by qRT-PCR analysis. (E) After 48 h of incubation, cells were infected with WT-Ad at a multiplicity of infection (MOI) of 5. Ad genome copy numbers in the cells were determined 24 h after infection and expressed as relative values (control = 1). These data are expressed as means ± SD (A to C, F, and G, n = 4; D and E, n = 3).
In order to examine which of the 5 genes were involved in Ad replication, HeLa cells were transfected with small interfering RNAs (siRNAs) against the 5 genes, followed by infection with WT-Ad. Transfection with the siRNAs induced significant knockdown of the respective target genes (Fig. 3D). Knockdown of CUL4A and RNF4 significantly enhanced Ad replication. In particular, approximately 7-fold-higher copy numbers of the Ad genome were found in the cells transfected with an siRNA against CUL4A (Fig. 3E). These results suggested that 3′-mivaRNAII suppressed the expression of the CUL4A and RNF4 genes in order to promote Ad replication and that suppression of CUL4A expression by 3′-mivaRNAII mediated the highest impact for enhancement of Ad infection among the target genes.
3′-mivaRNAII suppresses CUL4A expression via posttranscriptional gene silencing.
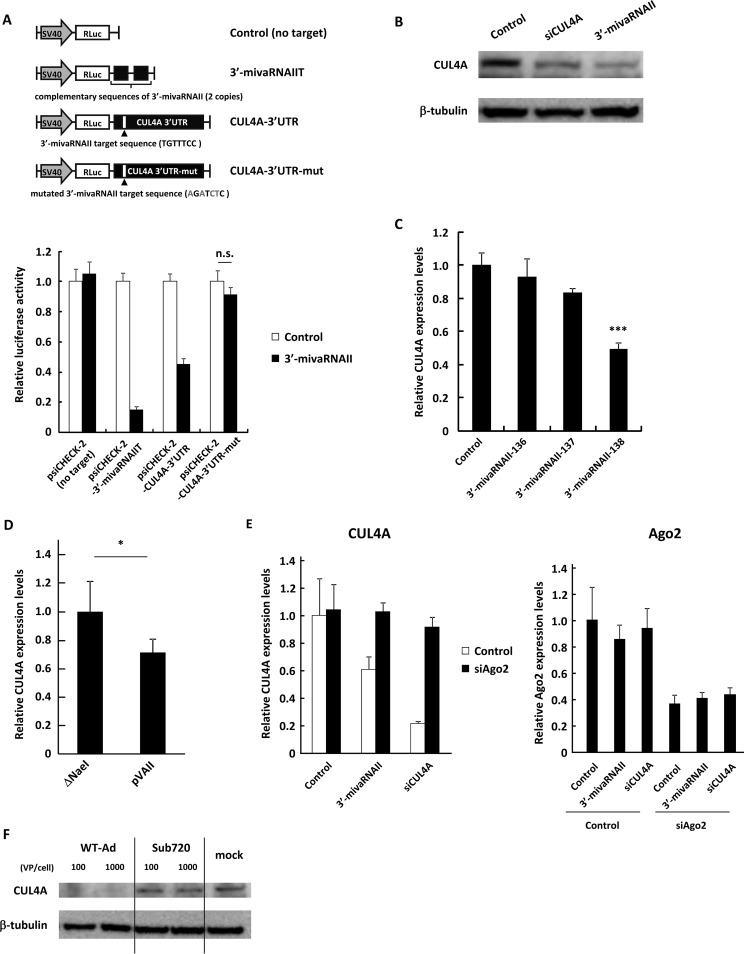
In order to examine whether 3′-mivaRNAII suppressed CUL4A expression via posttranscriptional gene silencing, we performed a reporter assay using reporter plasmids containing the wild-type or mutated 3′ UTR sequences of the CUL4A gene in the region downstream of the Renilla luciferase (RLuc) gene. The expression levels of the RLuc gene containing the wild-type 3′ UTR of the CUL4A gene and those containing the sequence perfectly complementary to 3′-mivaRNAII were significantly suppressed by transfection of the 3′-mivaRNAII mimic, while those containing the mutated 3′ UTR sequence were not (Fig. 4A). CUL4A protein levels were significantly suppressed in the cells transfected with the 3′-mivaRNAII mimic (Fig. 4B). The suppression of CUL4A expression was exclusively found in the 3′-mivaRNAII-138 mimic (Fig. 4C), as well as in pVAII (Fig. 4D). CUL4A expression was significantly restored by cotransfection with an siRNA against Ago2, a main component of RISC (Fig. 4E). In addition, infection with WT-Ad, but not Sub720, resulted in the suppression of CUL4A expression at the protein level (Fig. 4F). These results demonstrated that 3′-mivaRNAII suppressed CUL4A expression via posttranscriptional gene silencing.
FIG 4.
3′-mivaRNAII suppresses CUL4A expression via posttranscriptional gene silencing. (A) HeLa cells were cotransfected with 3′-mivaRNAII (3′-mivaRNAII-138) mimic and the indicated reporter plasmids, described in the upper portion. Luciferase activities were determined 48 h after transfection. RLuc activities were normalized to Fluc activities. The mutated nucleotides in the sequences complementary to the seed sequences of 3′-mivaRNAII are shown in gray. SV40, SV40 promoter; n.s., not significant. (B) HeLa cells were transfected with 3′-mivaRNAII (3′-mivaRNAII-138) mimic, or siCUL4A at 50 nM. After 48 h of incubation, protein levels of CUL4A were evaluated by Western blotting analysis. (C) HeLa cells were transfected with 3′-mivaRNAII-136, -137, or -138 mimics at 50 nM. After 48 h of incubation, mRNA levels of CUL4A were determined by qRT-PCR analysis. (D) HeLa cells were transfected with ΔNaeI or pVAII. After 48 h of incubation, the mRNA levels of CUL4A were determined by qRT-PCR analysis. (E) HeLa cells were cotransfected with a 3′-mivaRNAII mimic (3′-mivaRNAII-138), siCUL4A, and siAgo2 at 50 nM. After 48 h of incubation, the mRNA levels of CUL4A and Ago2 were determined by qRT-PCR analysis. (F) HeLa cells were infected with Ad (WT-Ad and Sub720) at 100 or 1,000 VPs/cell. After 48 h of incubation, protein levels of CUL4A were evaluated by Western blotting analysis. These data are expressed as means ± SD (A and C, n = 4).
Suppression of CUL4A gene expression promotes Ad infection.
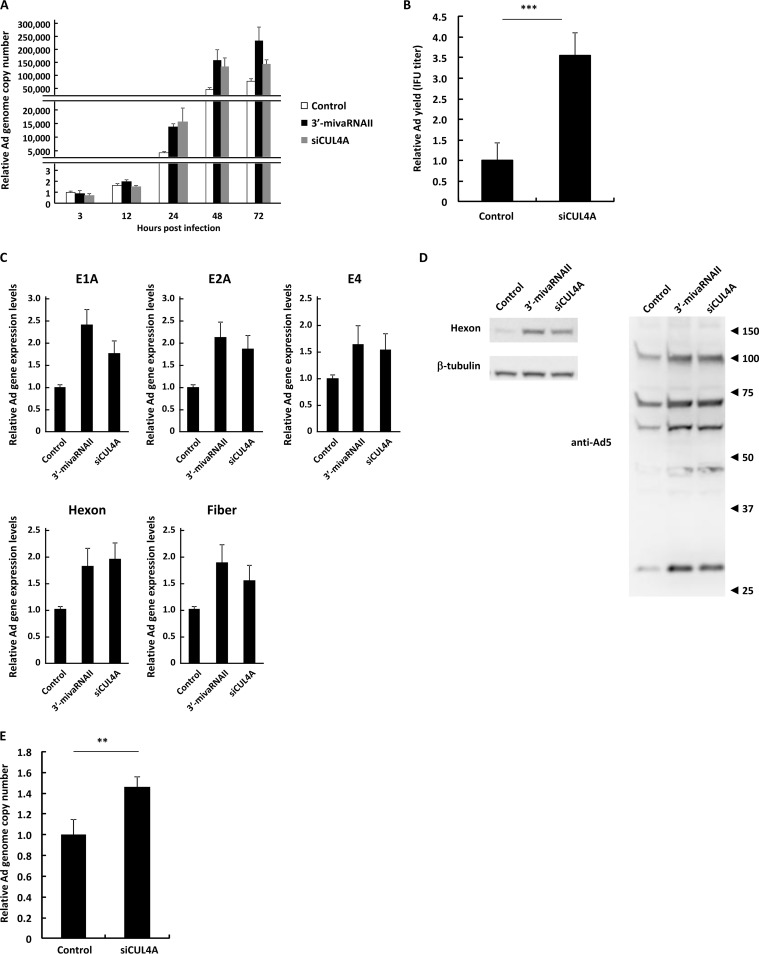
In order to further examine the effects of 3′-mivaRNAII-mediated suppression of CUL4A gene expression on Ad infection, we evaluated Ad replication, progeny virus yields, and mRNA and protein expression levels of Ad genes in the cells transfected with 3′-mivaRNAII or siCUL4A. Ad genome copy numbers in the CUL4A knockdown cells were significantly higher than those in control cells at 24, 48, and 72 h postinfection, although transfection with 3′-mivaRNAII or siCUL4A did not mediate significant differences in the Ad genome copy numbers at 3 and 12 h postinfection (Fig. 5A). Knockdown of CUL4A significantly increased the infectious Ad titers (Fig. 5B), Ad gene mRNA levels (Fig. 5C), and Ad protein levels (Fig. 5D). Knockdown of CUL4A also increased the Ad genome copy numbers in A549 cells (Fig. 5E). The 3′-mivaRNAII mimic mediated not only slightly larger increases in the virus genome copy numbers and progeny virus titers than for pVAII (Fig. 1A and C) but also significant upregulation of Ad gene expression. pVAII did not mediate any apparent upregulation of Ad gene expression (data not shown), probably because transfection with the 3′-mivaRNAII mimic introduced higher copy numbers of mivaRNAII than transfection with pVAII. These data strongly suggested that the suppression of CUL4A gene expression promoted Ad infection.
FIG 5.
Suppression of CUL4A gene expression promotes Ad infection. (A) HeLa cells were transfected with a 3′-mivaRNAII mimic (3′-mivaRNAII-138) or siCUL4A at 50 nM for 48 h, followed by infection with WT-Ad at 100 VPs/cell. After 3, 12, 24, 48, or 72 h of incubation, the Ad genome copy numbers were determined and expressed as relative values (control, 3 h postinfection = 1). (B) HeLa cells were transfected with siCUL4A at 50 nM for 48 h, followed by infection with WT-Ad at 100 VPs/cell. After 24 h of incubation, IFU titers of progeny WT-Ad were determined and expressed as relative values (control = 1). (C) HeLa cells were transfected with a 3′-mivaRNAII mimic (3′-mivaRNAII-138) or siCUL4A at 50 nM for 48 h, followed by infection with WT-Ad at 100 VPs/cell. mRNA levels of the Ad genes were determined by qRT-PCR analysis at 12 h (E1A, E2A, and E4) or 24 h (hexon and fiber) after infection. (D) HeLa cells were transfected with a 3′-mivaRNAII mimic (3′-mivaRNAII-138) or siCUL4A at 50 nM for 48 h, followed by infection with WT-Ad at 100 VPs/cell. After 24 h of incubation, the protein levels of the Ad genes were evaluated by Western blotting analysis. Numbers at the right of the membranes indicate the protein sizes in kilodaltons. Note that the sizes of the Ad major capsid proteins, hexon, penton base, and fiber, are 108 kDa, 63 kDa, and 61 kDa, respectively. (E) A549 cells were transfected with siCUL4A at 50 nM for 48h, followed by infection with WT-Ad at 100 VPs/cell. After 24 h of incubation, Ad genome copy numbers were determined and expressed as relative values (control = 1). These data are expressed as means ± SD (n = 4).
JNK signaling is involved in mivaRNAII-mediated promotion of Ad infection.
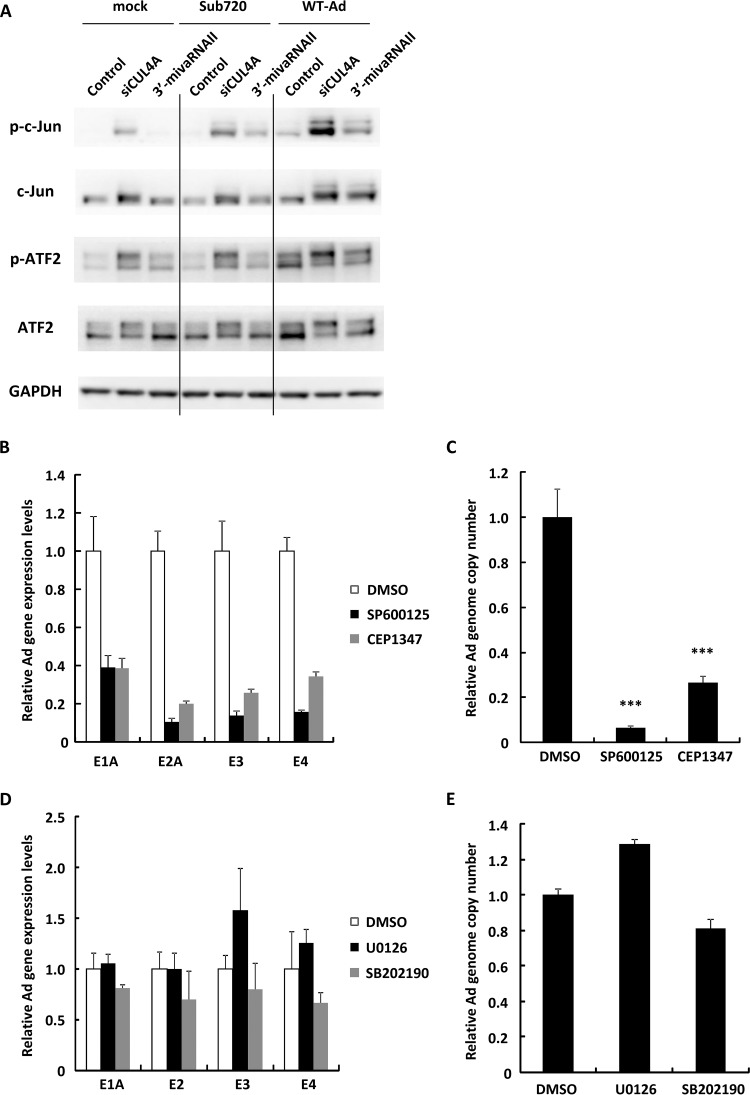
A previous study reported that CUL4A regulates Jun N-terminal kinase (JNK) signaling via proteasomal degradation of c-Jun, a proto-oncogenic transcription factor which contributes to transduction of JNK signaling (35). JNK is a member of the mitogen-activated protein kinase (MAPK) family and is activated by a variety of environmental stresses, inflammatory cytokines, growth factors, and G protein-coupled receptor agonists (36, 37). We hypothesized that JNK signaling is involved in Ad replication and that 3′-mivaRNAII promotes JNK signaling via suppression of CUL4A expression in order to promote viral growth. Suppression of CUL4A expression by transfection with siCUL4A significantly increased the amount of c-Jun and phosphorylated c-Jun (Fig. 6A). Treatment with both WT-Ad and siCUL4A or 3′-mivaRNAII mediated further increases in the protein levels of c-Jun and phosphorylated c-Jun. In addition, the expression of ATF2, which heterodimerizes with c-Jun and plays an important role in JNK signaling, significantly increased, suggesting that knockdown of CUL4A upregulated JNK signaling (Fig. 6A). In order to examine whether JNK signaling contributes to Ad replication, the JNK inhibitors SP600125 and CEP1347 were added to HeLa cells, followed by infection with WT-Ad. Inhibition of JNK signaling significantly decreased the Ad gene expression and Ad genome copy numbers (Fig. 6B and C), while treatment with other MAPK inhibitors U0126 (an extracellular signal-regulated kinase [ERK] inhibitor) and SB202190 (a p38 inhibitor) did not (Fig. 6D and E), suggesting that JNK signaling was crucial for Ad infection. These results indicated that mivaRNAII-mediated suppression of CUL4A expression resulted in the upregulation of JNK signaling, leading to an increase in Ad replication levels (Fig. 7).
FIG 6.
JNK signaling is involved in mivaRNAII-mediated promotion of Ad infection. (A) HeLa cells were transfected with 3′-mivaRNAII (3′-mivaRNAII-138) mimic or siCUL4A at 50 nM for 48 h, followed by infection with Ad (WT-Ad, Sub720) at 100 VPs/cell. After 24 h of incubation, protein levels of c-Jun and ATF2 were evaluated by Western blotting analysis. (B and C) HeLa cells were treated with JNK inhibitors (10 mM SP600125 or 20 mM CEP1347), followed by infection with WT-Ad at 100 VPs/cell. (B) After 12 h of incubation, mRNA levels of the Ad genes were determined by qRT-PCR analysis. (C) After 24 h of incubation, Ad genome copy numbers were determined and expressed as relative values (dimethyl sulfoxide [DMSO] = 1). (D and E) HeLa cells were treated with 10 mM U0126 (an ERK inhibitor) or 20 mM SB202190 (a p38 inhibitor) and infected with WT-Ad at 100 VPs/cell. (D) After 12 h of incubation, mRNA levels of the Ad genes were determined by qRT-PCR analysis. (E) After 24 h of incubation, Ad genome copy numbers were determined. These data are expressed as means ± SD (B and C, n = 4; D and E, n = 3).
FIG 7.
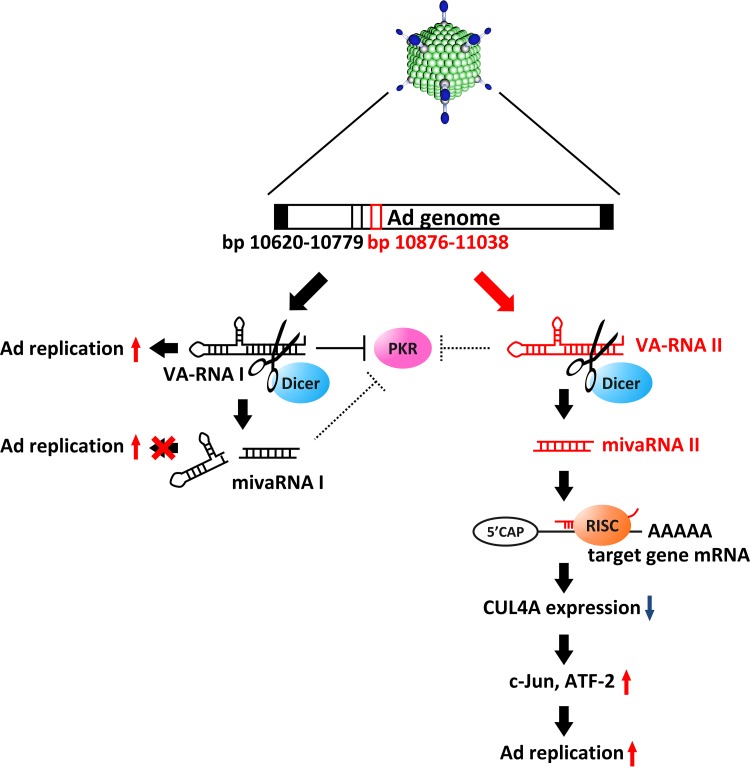
Model of VA-RNAII-mediated promotion of Ad replication via posttranscriptional gene silencing of CUL4A. After Ad infection, VA-RNAI and -II are rapidly transcribed and promote Ad infection in different ways. VA-RNAI inhibits the activation of PKR, which plays a key role in antiviral responses. VA-RNAII functions as a precursor of mivaRNAII, which promotes Ad infection, while mivaRNAI does not support Ad infection. Suppression of CUL4A, the highest-potential target of mivaRNAII, amplifies JNK signaling via stabilization of c-Jun and ATF2, leading to promotion of Ad replication.
DISCUSSION
In addition to several other types of viruses, Ad also expresses viral miRNAs (mivaRNAs), which are the truncated form of VA-RNAs; however, it is unknown whether mivaRNAs, especially miviRNAII, support Ad replication. In this study, we demonstrated that full-length VA-RNAII and 3′-mivaRNAII-138, one of the most abundant mivaRNAII isoforms in the RISC, promoted Ad replication (Fig. 1 and 2), suggesting that VA-RNAII supports Ad replication via processing into mivaRNAII (Fig. 7). These properties of VA-RNAII are contrastive to those of VA-RNAI found in our recent study. We demonstrated that only full-length VA-RNAI promoted Ad replication, while dicer-mediated cleavage of VA-RNAI inactivated VA-RNAI function (25). In addition, it has been reported that VA-RNAI antagonizes the key components of the miRNA maturation pathway, including exportin-5, dicer, and Ago2, and inhibits dicer expression, leading to a decrease in the copy numbers of endogenous mature miRNAs as well as an increase in functional full-length VA-RNAI (38–40). On the other hand, we revealed that VA-RNAII did not inhibit dicer expression (data not shown). Moreover, it has been reported that VA-RNAII inhibited PKR less efficiently than VA-RNAI (29). VA-RNAII has properties that are the opposite those of VA-RNAI, suggesting that VA-RNAII might play a role complementary to that of VA-RNAI in supporting Ad replication.
We demonstrated that only a specific isoform of 3′-mivaRNAII (3′-mivaRNAII-138) promoted Ad replication, while the other isoforms, which have different seed sequences, did not (Fig. 2A, B, and D). On the other hand, several viruses are well known to encode miRNAs which restrict viral gene expression in order to promote persistent infection (3). We found 4 copies of mivaRNAII target sequences (TGTTTCC; bp 11014 to 11020, 25140 to 25146, 32215 to 32221, and 32661 to 32667), which are perfectly complementary to the 8-nt seed sequences of mivaRNAII, on the negative strand of the Ad genome. In particular, bp 32215 to 32221 and 32661 to 32667 are located downstream of the E4 protein-coding region. However, we considered it unlikely that mivaRNAII negatively regulates expression of the E4 gene. Indeed, we demonstrated that mivaRNAII did not suppress, but rather enhanced, the expression of not only the E4 gene but also the other viral genes examined (Fig. 5C), indicating that mivaRNAII did not negatively regulate viral gene expression, at least in the case of infection for 12 h in HeLa cells. These results led us to hypothesize that 3′-mivaRNAII suppresses the expression of its specific target genes in the same manner as endogenous miRNAs, in order to support Ad replication. We performed microarray analysis and in silico analysis in order to find the target genes of 3′-mivaRNAII, identifying CUL4A as a target gene involved in Ad replication (Fig. 3 and 4).
CUL4A is a member of the cullin family (CUL1, -2, -3, -4A, -4B, and -5), which provides a scaffold for E3 ubiquitin ligases. Cullin family-mediated ubiquitination regulates diverse aspects of cellular physiology, including the cell cycle, cell signaling, tumor suppression, the DNA damage response, and chromatin remodeling, usually by inducing the proteasomal degradation of the substrates (32, 33, 41). Several viruses inhibit or recruit this machinery in order to make the profiles of cellular protein expression suitable for virus infection. Epstein-Barr virus (EBV) inhibits CUL1/4A activities in order to increase Cdt1 accumulation, promoting EBV genome replication (42). Human papillomavirus type 16 employs CUL2, resulting in aberrant degradation of the retinoblastoma tumor suppressor (43). Ad5 E1B-55k and E4orf6 proteins mediate CUL2/5-dependent protein degradation, leading to efficient infection (44). However, whether CUL4A regulates Ad infection has not previously been examined. In the present study, we found that mivaRNAII suppressed CUL4A expression in a posttranscriptional manner and that CUL4A was involved in Ad replication in this study.
CUL4A contributes to the proteasomal degradation of c-Jun (35), which is a component of transcription factor AP-1 involved in the transduction of JNK signaling. Consistently with the previous report (35), we found that infection with an Ad resulted in the suppression of CUL4A expression and thereby in an increase in the amounts of c-Jun. The JNK inhibitors SP600125 and CEP1347 significantly decreased the Ad gene expression and Ad replication, while the other MAPK inhibitors did not, suggesting that JNK regulates Ad infection (Fig. 6). These findings indicate that following Ad infection, mivaRNAII mediates downregulation of CUL4A via posttranscriptional silencing, leading to an increase in c-Jun levels and upregulation of JNK signaling. Muller et al. reported that expression of an AP-1 transcriptional factor was elevated following Ad infection and that the AP-1 transcriptional factor might bind to the AP-1 recognition site on the Ad genome (45). Several viruses activate and require the activation of JNK signaling for their efficient replication (46–48). For example, inhibition of JNK signaling suppresses human cytomegalovirus (CMV) replication. Activation of JNK signaling results in upregulation of transgene expression driven by the CMV immediate/early promoter (49, 50). These observations suggest that c-Jun binds the viral genome and directly regulates viral gene transcription. However, it remains unclear whether c-Jun regulates Ad gene expression via direct binding to the Ad genome. Further analysis will be needed to investigate this possibility.
In summary, we have demonstrated that a truncated form of VA-RNAII, mivaRNAII, promotes Ad replication in a posttranscriptional silencing manner and have identified CUL4A as a mivaRNAII target gene. Furthermore, we found that JNK signaling is involved in the regulation of Ad infection. The present findings will help to elucidate the function of VA-RNAs and will promote the development of novel Ad vectors and oncolytic Ad.
MATERIALS AND METHODS
Cells and viruses.
HeLa (a human epithelial carcinoma cell line; RCB0007), HEK293 (a transformed embryonic kidney cell line), 293T (a transformed embryonic kidney cell line expressing SV40 large T antigen), and A549 (a human alveolar adenocarcinoma cell line) cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum (FBS), streptomycin (100 μg/ml), and penicillin (100 U/ml). HeLa cells were obtained from the JCRB Cell Bank (Tokyo, Japan). The other cells were obtained from the American Type Culture Collection (ATCC; Manassas, VA).
Wild-type Ad serotypes 4 (Ad4), 5 (WT-Ad), 11 (Ad11), 31 (Ad31), and 35 (Ad35) were obtained from the ATCC. These Ad were propagated in HEK293 cells as previously described (51). Sub720, a mutant Ad serotype 5 lacking the expression of both VA-RNAI and -II, and Sub722, an Ad serotype 5 mutant from which VA-RNAI was deleted so that it expresses only VA-RNAII, were propagated in 293T cells. Sub720 and Sub722 were kindly provided by Goran Akusjarvi (Uppsala University, Uppsala, Sweden). The virus particles (VPs) were determined using a spectrophotometric method (52).
Plasmids.
A control plasmid lacking VA-RNA-expression (pAdVAntage-ΔNaeI; ΔNaeI) and a plasmid expressing only VA-RNAI (pVAI) or VA-RNAII (pVAII) were previously constructed using pAdVAntage (Promega, Madison, WI), which encodes VA-RNAI and -II of Ad serotype 2 (Ad2) (25). The VA-RNAII sequences outside the mivaRNAII coding region on Ad2 are different by 2 nt from those outside the mivaRNAII coding region on Ad5. pVAII-mut, a plasmid which expresses a VA-RNAII containing mutations in the mivaRNAII-encoding sequence, was constructed using pVAII, a QuikChange Lightning site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA), and the primers pVAII-mut-3′-F and pVAII-mut-3′-R. The primer sequences are shown in Table 1.
TABLE 1.
Oligonucleotides and primers used in this study
| No. | Name | Sequence (5′–3′)a |
|---|---|---|
| 1 | pVAIImut3′F | CCCCGCTTGCAAATTCCTCGAGCCACGGGTGCGAGCCCCTTTTTTGC |
| 2 | pVAIImut3′R | GCAAAAAAGGGGCTCGCACCCGTGGCTCGAGGAATTTGCAAGCGGGG |
| 3 | Ad5-F (Ad5-E4-F) | GGGATCGTCTACCTCCTTTTGA |
| 4 | Ad5-R (Ad5-E4-R) | GGGCAGCAGCGGATGAT |
| 5 | Ad5-probe (Ad5-E4-probe) | FAM-ACAGAAACCCGCGCTACCATACTGGAG-TAMRA |
| 6 | GAPDH-F | GGTGGTCTCCTCTGACTTCAACA |
| 7 | GAPDH-R | GTGGTCGTTGAGGGCAATG |
| 8 | GAPDH-probe | FAM-CACTCCTCCACCTTTGACGCTGGG-TAMRA |
| 9 | Ad5-VA-RNAII-F | GGCTCGCTCCCTGTAGCCGG |
| 10 | Ad5-VA-RNAII-R | AGGGGCTCGTCCCTGTTTCC |
| 11 | Ad4-F | AGACAGCGACTCTTCACTGC |
| 12 | Ad4-R | TCGTCCTCATCATCGCTTGG |
| 13 | Ad11-F | GCACTGCTATGAAGACGGGT |
| 14 | Ad11-R | TCCGGGCAATCCAACTGAAA |
| 15 | Ad31-F | ATTGATGTGGAGTCTGCCGG |
| 16 | Ad31-R | ACAGGGGGCTCCGGTAATAT |
| 17 | Ad35-F | TCCGTGGACTGTGATTTGCA |
| 18 | Ad35-R | CCAACATTGGCAGCCTTCAC |
| 19 | Ad5-E1A-F | TCCGGTCCTTCTAACACACCTC |
| 20 | Ad5-E1A-R | ACGGCAACTGGTTTAATGGG |
| 21 | Ad5-E2A-F | CACTACGGTGCGAGTGCAA |
| 22 | Ad5-E2A-R | GGTAGCTGCCTTCCCAAAAAG |
| 23 | Ad5-E3-F | AACACCTGGTCCACTGTCGC |
| 24 | Ad5-E3-R | AGCTCGGAGAGGTTCTCTCGTAG |
| 25 | Ad5-hexon-F | ACGATGACAACGAAGACGAAGTAG |
| 26 | Ad5-hexon-R | GGCGCCTGCCCAAATAC |
| 27 | Ad5-fiber-F | GCGCCTATCCGAACCTCTAGT |
| 28 | Ad5-fiber-R | AGAGGCCGTTGCCCATTT |
| 29 | mivaRNAIIT-S | TCGAGAAGGGGCTCGTCCCTGTTTCCGGACAGCAAGGGGCTCGTCCCTGTTTCCGGATTAATTAAGCGC |
| 30 | mivaRNAIIT-AS | GGCCGCGCTTAATTAATCCGGAAACAGGGACGAGCCCCTTGCTGTCCGGAAACAGGGACGAGCCCCTTC |
| 31 | mivaRNAIIT-mut-S | TCGAGAAGGGGCTCGCACCCGTGGCTCGACAGCAAGGGGCTCGCACCCGTGGCTCGATTAATTAAGCGC |
| 32 | mivaRNAIIT-mut-AS | GGCCGCGCTTAATTAATCGAGCCACGGGTGCGAGCCCCTTGCTGTCGAGCCACGGGTGCGAGCCCCTTC |
| 33 | CUL4A-F | ACCTCGCACAGATGTACCAG |
| 34 | CUL4A-R | AGGTTGACGAACCGCTCATTC |
| 35 | RNF4-F | ATGAGTACAAGAAAGCGTCGTG |
| 36 | RNF4-R | CACAAGTGAGGTCCACAATTTCA |
| 37 | RasGRP1-F | ACATCACCCAGTTCCGAATGA |
| 38 | RasGRP1-R | GCTGTCAATGAGATCGTCCAG |
| 39 | CD164-F | ACCCGAACGTGACGACTTTAG |
| 40 | CD164-R | CGTGTTCCCCACTTGACAATC |
| 41 | KCTD16-F | ATGGCTCTGAGTGGAAACTGT |
| 42 | KCTD16-R | TCAATGTGGAATGGCGAGTAAA |
| 43 | NFATC2-F | GAGCCGAATGCACATAAGGTC |
| 44 | NFATC2-R | CCAGAGAGACTAGCAAGGGG |
| 45 | CNRIP1-F | TAATGACGGCCCGGTCTTTTA |
| 46 | CNRIP1-R | TGCAGCGTGCTGGGTTTAAT |
| 47 | IGF2BP2-F | AGTGGAATTGCATGGGAAAATCA |
| 48 | IGF2BP2-R | CAACGGCGGTTTCTGTGTC |
| 49 | ANK2-F | ACCTGCAATCAGAATGGACTCA |
| 50 | ANK2-R | TGCAATGTGAAGAGCGGTATT |
| 51 | DUSP16-F | GCCCATGAGATGATTGGAACTC |
| 52 | DUSP16-R | CGGCTATCAATTAGCAGCACTTT |
| 53 | FXYD6-F | ACCCTGAGGATTGGGGGAC |
| 54 | FXYD6-R | CATTGGCGGTGATGAGGTT |
| 55 | KMO-F | TAGCCCTTTCTCATAGAGGACG |
| 56 | KMO-R | CTCTCATGGGAATACCTTGGGA |
| 57 | SEZ-F | CTGGCTCACGGACTCTCTTTA |
| 58 | SEZ-R | CTGTTGTGACAAAGTGGACGC |
| 59 | SNTN-F | TGTATGCACAGTACCCAGGAC |
| 60 | SNTN-R | AGCAGTGGTGGCAATAGCTTT |
| 61 | CUL4A-3′UTR-F | TTCTCGAGCGCATCTGCAGACGGTTC |
| 62 | CUL4A-3′UTR-R | GGGTTTAAACCACTGTTCAACCTC |
| 63 | QC-CUL4A-3′UTR-F | GGGGCTAGTGTGTTTGAGATCTCATTCTAAGATTGAGTCTGGCAG |
| 64 | QC-CUL4A-3′UTR-R | CTGCCAGACTCAATCTTAGAATGAGATCTCAAACACACTAGCCCC |
| 65 | Ago2-F | CGCGTCCGAAGGCTGCTCTA |
| 66 | Ago2-R | TGGCTGTGCCTTGTAAAACGCT |
FAM, 6-carboxyfluorescein; TAMRA, 6-carboxytetramethylrhodamine.
siRNAs, mivaRNA mimics, and mivaRNA inhibitors.
A control siRNA (Allstars negative-control siRNA), mivaRNA mimics (3′-mivaRNAII-136, -137, -138, and -mut), a control miRNA inhibitor, 3′-mivaRNAII inhibitors, and siRNAs against ANK2 (siANK2), DUSP16 (siDUSP16), and KCTD16 (siKCTD16) were purchased from Qiagen (Hilden, Germany). 3′-mivaRNAII inhibitor is an antisense oligonucleotide that is entirely composed of 2′-O-methylated RNA and is perfectly complementary to 3′-mivaRNAII. Other siRNAs were purchased from Gene Design (Osaka, Japan). The sequences of siRNAs, mivaRNA mimics, and mivaRNA inhibitors are shown in Table 2. The reagents described above were transfected into HeLa and A549 cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions.
TABLE 2.
miRNA mimics, inhibitors, and siRNAs used in this study
| No. | Name | Sequence (5′–3′) |
|---|---|---|
| 1 | 3′-mivaRNAII-136 mimic | UCCGGAAACAGGGACGAGCCCC |
| 2 | 3′-mivaRNAII-137 mimic | CCGGAAACAGGGACGAGCCCC |
| 3 | 3′-mivaRNAII-138 mimic | CGGAAACAGGGACGAGCCCC |
| 4 | 3′-mivaRNAII-mut mimic | CAAAGAUAGGGACGAGCCCC |
| 5 | 3′-mivaRNAII inhibitor | CGGAAACAGGGACGAGCCCC |
| 6 | siCUL4A | AAGCAUGAGUGCGGUGCAGCC |
| 7 | siRNF4 | GAAUGGACGUCUCAUCGUUUU |
| 8 | siCD164 | GGACUGGUGAUUCAUUUGU |
| 9 | siNFATC2 | CCAUUAAACAGGAGCAGAA |
| 10 | siAgo2 | GCACGGAAGUCCAUCUGAA |
Determination of Ad genome copy numbers.
Cells were infected with Ad, and then total DNA, including Ad genomic DNA, was isolated from the cells using a DNeasy blood and tissue kit (Qiagen). After isolation, Ad genome copy numbers were quantified using StepOnePlus real-time PCR systems (Applied Biosystems, Foster City, CA) as previously described (25). For determination of the Ad genome copy numbers in the cells, cells were treated with trypsin to remove the virus particles on the cell surface, followed by isolation of total DNA.
Infectious titer assay.
Following infection with WT-Ad, cells were recovered and subjected to 3 cycles of freezing and thawing. After centrifugation, the supernatants were added to HEK293 cells. After 48 h of incubation, the numbers of cells infected with WT-Ad were analyzed using an Adeno-X Rapid Titer kit (Clontech, Mountain View, CA).
Reporter plasmids and reporter assay.
A reporter plasmid, psiCHECK-2-mivaRNAIIT, containing two copies of sequences complementary to mivaRNAII in the 3′ UTR of the Renilla luciferase (RLuc) gene was constructed as follows. An XhoI/NotI fragment of psiCHECK-2 (Promega) was ligated with oligonucleotides encoding 2 copies of mivaRNAII-complementary sequence, mivaRNAIIT-S and mivaRNAIIT-AS, resulting in psiCHECK-2-mivaRNAIIT. psiCHECK-2-mivaRMAIIT-mut was similarly constructed using the corresponding oligonucleotides. psiCHECK-2-CUL4A-3′UTR, which contains the sequences of the wild-type 3′ UTR of the CUL4A gene in the region downstream of the RLuc gene, was constructed as follows. The fragment containing the sequence of the 3′ UTR of the CUL4A gene was amplified by PCR using cDNA prepared from HeLa cells. The PCR fragment was ligated with an XhoI/PmeI fragment of psiCHECK-2, resulting in psiCHECK-2-CUL4A-3′UTR. psiCHECK-2-CUL4A-3′UTR-mut was produced by insertion of mutations into the sequence complementary to the seed sequence of 3′-mivaRNAII using a QuikChange Lightning site-directed mutagenesis kit and the corresponding primers. The sequences of the oligonucleotides are shown in Table 1. Firefly luciferase (FLuc) and RLuc activities in the cells were determined using a dual-luciferase reporter assay system (Promega) following transfection with the reporter plasmids.
Antibodies and Western blotting.
Rabbit anti-human β-tubulin and anti-Ad5 antibodies were purchased from Abcam (Cambridge, UK). Rabbit anti-human c-Jun (60A8), phosphorylated c-Jun, ATF2 (20F1), and phosphorylated ATF2 antibodies were purchased from Cell Signaling Technology (Danvers, MA). Goat anti-human CUL4A antibody was purchased from Santa Cruz Biotechnology (Dallas, TX). Rabbit anti-human glyceraldehyde-3-phosphate dehydrogenase (anti-GAPDH) antibody was purchased from Trevigen (Gaithersburg, MD). Mouse anti-hexon antibody (65H6) was purchased from Abnova (Taipei, Taiwan). Horseradish peroxidase (HRP)-labeled anti-mouse, anti-rabbit, and anti-goat IgG antibodies were purchased from Cell Signaling Technology. Western blotting analysis was performed as previously described (25). Briefly, whole-cell extracts were prepared and electrophoresed on 10% sodium dodecyl sulfate (SDS)-polyacrylamide gels under reducing conditions, followed by electrotransfer to polyvinylidene difluoride membranes (Millipore, Bedford, MA). After blocking with 5% skim milk or 5% bovine serum albumin (BSA) prepared in TBS-T (Tris-buffered saline with Tween 20), the membrane was incubated with primary antibodies, followed by incubation in the presence of secondary antibodies. The protein bands were visualized with a chemiluminescence kit (ECL Plus Western blotting detection system; Amersham Biosciences, Piscataway, NJ).
Microarray gene expression analysis.
HeLa cells were transfected with 3′-mivaRNAII (3′-mivaRNAII-138) mimic at 50 nM using Lipofectamine 2000. After 48 h of incubation, total RNA was isolated. Microarray gene expression analysis was performed by TaKaRa Bio (Shiga, Japan). Briefly, the quality of RNA samples was checked using an Agilent 2100 bioanalyzer platform (Agilent Technologies, Santa Clara, CA). One hundred nanograms of each total RNA sample was amplified and labeled using an Agilent Low Input Quick Amp labeling kit (Agilent Technologies) according to the manufacturer’s protocol. The labeled samples were hybridized and washed using an Agilent gene expression hybridization kit and a wash buffer kit (Agilent Technologies). Fluorescence signals of the hybridized Agilent microarrays were detected using an Agilent SureScan microarray scanner (Agilent Technologies). The Agilent Feature Extraction software was used to read out and process the microarray image files.
Quantitative RT-PCR analysis.
Total RNA was isolated from cells using ISOGEN or ISOGEN II (Nippon Gene, Tokyo, Japan). cDNA was synthesized using 500 ng of total RNA with a Superscript VILO cDNA synthesis kit (Life Technologies). Quantitative RT-PCR (qRT-PCR) analysis was performed using Fast SYBR Green Master Mix (Life Technologies) or THUNDERBIRD SYBR qPCR mix (Toyobo, Osaka, Japan) and StepOnePlus real-time PCR systems (Life Technologies). The sequences of the primers used in this study are shown in Table 1.
Drugs.
SP600125 was purchased from Wako (Osaka, Japan). CEP1347 was purchased from Tocris Bioscience (Bristol, UK). SB202190 and U-0126 were purchased from Cayman Chemical (Ann Arbor, MI).
Statistical analysis.
Statistical significance was determined using Student’s t test. Data are presented as means ± standard deviations (SD).
Accession number(s).
The data for gene expression profiles have been submitted to the Gene Expression Omnibus (GEO) under accession number GSE96531.
ACKNOWLEDGMENTS
We thank Eri Hosoyamada (Graduate School of Pharmaceutical Sciences, Osaka University, Osaka, Japan) and Tomoko Yamaguchi (National Institutes of Biomedical Innovation, Health and Nutrition, Osaka, Japan) for their help.
This work was supported by grants-in-aid for Scientific Research (A) and (B) from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) of Japan. K. Wakabayashi and M. Machitani are Research Fellows of the Japan Society for the Promotion of Science.
REFERENCES
- 1.Ambros V. 2004. The functions of animal microRNAs. Nature 431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 2.Macfarlane L-AA, Murphy PR. 2010. MicroRNA: biogenesis, function and role in cancer. Curr Genomics 11:537–561. doi: 10.2174/138920210793175895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kincaid RP, Sullivan CS. 2012. Virus-encoded microRNAs: an overview and a look to the future. PLoS Pathog 8:e1003018. doi: 10.1371/journal.ppat.1003018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grundhoff A, Sullivan CS. 2011. Virus-encoded microRNAs. Virology 411:325–343. doi: 10.1016/j.virol.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gupta A, Gartner JJ, Sethupathy P, Hatzigeorgiou AG, Fraser NW. 2006. Anti-apoptotic function of a microRNA encoded by the HSV-1 latency-associated transcript. Nature 442:82–85. doi: 10.1038/nature04836. [DOI] [PubMed] [Google Scholar]
- 6.Shinozaki-Ushiku A, Kunita A, Isogai M, Hibiya T, Ushiku T, Takada K, Fukayama M. 2015. Profiling of virus-encoded microRNAs in Epstein-Barr virus-associated gastric carcinoma and their roles in gastric carcinogenesis. J Virol 89:5581–5591. doi: 10.1128/JVI.03639-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yao Y, Nair V. 2014. Role of virus-encoded microRNAs in avian viral diseases. Viruses 6:1379–1394. doi: 10.3390/v6031379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sullivan CS, Grundhoff AT, Tevethia S, Pipas JM, Ganem D. 2005. SV40-encoded microRNAs regulate viral gene expression and reduce susceptibility to cytotoxic T cells. Nature 435:682–686. doi: 10.1038/nature03576. [DOI] [PubMed] [Google Scholar]
- 9.Suffert G, Malterer G, Hausser J, Viiliainen J, Fender A, Contrant M, Ivacevic T, Benes V, Gros F, Voinnet O, Zavolan M, Ojala PM, Haas JG, Pfeffer S. 2011. Kaposi’s sarcoma herpesvirus microRNAs target caspase 3 and regulate apoptosis. PLoS Pathog 7:e1002405. doi: 10.1371/journal.ppat.1002405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feldman ER, Kara M, Coleman CB, Grau KR, Oko LM, Krueger BJ, Renne R, van Dyk LF, Tibbetts SA. 2014. Virus-encoded microRNAs facilitate gammaherpesvirus latency and pathogenesis in vivo. mBio 5:e00981-14. doi: 10.1128/mBio.00981-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blaese RM, Culver KW, Miller AD, Carter CS, Fleisher T. 1995. T lymphocyte-directed gene therapy for ADA− SCID: initial trial results after 4 years. Science 270:475–480. doi: 10.1126/science.270.5235.475. [DOI] [PubMed] [Google Scholar]
- 12.Swisher SG, Roth JA, Nemunaitis J. 1999. Adenovirus-mediated p53 gene transfer in advanced non-small-cell lung cancer. J Natl Cancer Inst 91:763–771. doi: 10.1093/jnci/91.9.763. [DOI] [PubMed] [Google Scholar]
- 13.Bischoff JR, Kirn DH, Williams A, Heise C, Horn S. 1996. An adenovirus mutant that replicates selectively in p53-deficient human tumor cells. Science 274:373–376. doi: 10.1126/science.274.5286.373. [DOI] [PubMed] [Google Scholar]
- 14.Kawashima T, Kagawa S, Kobayashi N. 2004. Telomerase-specific replication-selective virotherapy for human cancer. Clin Cancer Res 10(1 Part 1):285–292. doi: 10.1158/1078-0432.CCR-1075-3. [DOI] [PubMed] [Google Scholar]
- 15.Miest TS, Cattaneo R. 2014. New viruses for cancer therapy: meeting clinical needs. Nat Rev Microbiol 12:23–34. doi: 10.1038/nrmicro3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma Y, Mathews MB. 1996. Structure, function, and evolution of adenovirus-associated RNA: a phylogenetic approach. J Virol 70:5083–5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Söderlund H, Pettersson U, Vennström B, Philipson L, Mathews MB. 1976. A new species of virus-coded low molecular weight RNA from cells infected with adenovirus type 2. Cell 7:585–593. doi: 10.1016/0092-8674(76)90209-9. [DOI] [PubMed] [Google Scholar]
- 18.Mathews MB, Shenk T. 1991. Adenovirus virus-associated RNA and translation control. J Virol 65:5657–5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O’Malley RP, Mariano TM, Siekierka J, Mathews MB. 1986. A mechanism for the control of protein synthesis by adenovirus VA RNA I. Cell 44:391–400. doi: 10.1016/0092-8674(86)90460-5. [DOI] [PubMed] [Google Scholar]
- 20.Aparicio O, Razquin N, Zaratiegui M, Narvaiza I, Fortes P. 2006. Adenovirus virus-associated RNA is processed to functional interfering RNAs involved in virus production. J Virol 80:1376–1384. doi: 10.1128/JVI.80.3.1376-1384.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao H, Chen M, Pettersson U. 2013. Identification of adenovirus-encoded small RNAs by deep RNA sequencing. Virology 442:148–155. doi: 10.1016/j.virol.2013.04.006. [DOI] [PubMed] [Google Scholar]
- 22.Xu N, Segerman B, Zhou X, Akusjärvi G. 2007. Adenovirus virus-associated RNAII-derived small RNAs are efficiently incorporated into the RNA-induced silencing complex and associate with polyribosomes. J Virol 81:10540–10549. doi: 10.1128/JVI.00885-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bellutti F, Kauer M, Kneidinger D, Lion T. 2015. Identification of RISC-associated adenoviral microRNAs, a subset of their direct targets, and global changes in the targetome upon lytic adenovirus 5 infection. J Virol 89:1608–1627. doi: 10.1128/JVI.02336-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kamel W, Segerman B, Öberg D, Punga T, Akusjärvi G. 2013. The adenovirus VA RNA-derived miRNAs are not essential for lytic virus growth in tissue culture cells. Nucleic Acids Res 41:4802–4812. doi: 10.1093/nar/gkt172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Machitani M, Sakurai F, Wakabayashi K, Tomita K, Tachibana M, Mizuguchi H. 2016. Dicer functions as an antiviral system against human adenoviruses via cleavage of adenovirus-encoded noncoding RNA. Sci Rep 6:27598. doi: 10.1038/srep27598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bhat RA, Thimmappaya B. 1983. Two small RNAs encoded by Epstein-Barr virus can functionally substitute for the virus-associated RNAs in the lytic growth of adenovirus 5. Proc Natl Acad Sci U S A 80:4789–4793. doi: 10.1073/pnas.80.15.4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bhat RA, Domer PH, Thimmappaya B. 1985. Structural requirements of adenovirus VAI RNA for its translation enhancement function. Mol Cell Biol 5:187–196. doi: 10.1128/MCB.5.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bhat RA, Thimmappaya B. 1984. Adenovirus mutants with DNA sequence perturbations in the intragenic promoter of VAI RNA gene allow the enhanced transcription of VAII RNA gene in HeLa cells. Nucleic Acids Res 12:7377–7388. doi: 10.1093/nar/12.19.7377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma Y, Mathews MB. 1993. Comparative analysis of the structure and function of adenovirus virus-associated RNAs. J Virol 67:6605–6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kamel W, Segerman B, Punga T, Akusjärvi G. 2014. Small RNA sequence analysis of adenovirus VA RNA-derived miRNAs reveals an unexpected serotype-specific difference in structure and abundance. PLoS One 9:e105746. doi: 10.1371/journal.pone.0105746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Furuse Y, Ornelles DA, Cullen BR. 2013. Persistently adenovirus-infected lymphoid cells express microRNAs derived from the viral VAI and especially VAII RNA. Virology 447:140–145. doi: 10.1016/j.virol.2013.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sharma P, Nag A. 2014. CUL4A ubiquitin ligase: a promising drug target for cancer and other human diseases. Open Biol 4:130217. doi: 10.1098/rsob.130217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jackson S, Xiong Y. 2009. CRL4s: the CUL4-RING E3 ubiquitin ligases. Trends Biochem Sci 34:562–570. doi: 10.1016/j.tibs.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wong N, Wang X. 2015. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res 43:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wertz IE, O'Rourke KM, Zhang Z, Dornan D, Arnott D, Deshaies RJ, Dixit VM. 2004. Human de-etiolated-1 regulates c-Jun by assembling a CUL4A ubiquitin ligase. Science 303:1371–1374. doi: 10.1126/science.1093549. [DOI] [PubMed] [Google Scholar]
- 36.Chang L, Karin M. 2001. Mammalian MAP kinase signalling cascades. Nature 410:37–40. doi: 10.1038/35065000. [DOI] [PubMed] [Google Scholar]
- 37.Davis RJ. 2000. Signal transduction by the JNK group of MAP kinases. Cell 103:239–252. doi: 10.1016/S0092-8674(00)00116-1. [DOI] [PubMed] [Google Scholar]
- 38.Bennasser Y, Chable-Bessia C, Triboulet R, Gibbings D, Gwizdek C, Dargemont C, Kremer EJ, Voinnet O, Benkirane M. 2011. Competition for XPO5 binding between Dicer mRNA, pre-miRNA and viral RNA regulates human Dicer levels. Nat Struct Mol Biol 18:323–327. doi: 10.1038/nsmb.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pei Z, Shi G, Kondo S, Ito M, Maekawa A, Suzuki M, Saito I, Suzuki T, Kanegae Y. 2013. Adenovirus vectors lacking virus-associated RNA expression enhance shRNA activity to suppress hepatitis C virus replication. Sci Rep 3:3575. doi: 10.1038/srep03575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Machitani M, Katayama K, Sakurai F, Matsui H, Yamaguchi T, Suzuki T, Miyoshi H, Kawabata K, Mizuguchi H. 2011. Development of an adenovirus vector lacking the expression of virus-associated RNAs. J Control Release 154:285–299. doi: 10.1016/j.jconrel.2011.06.020. [DOI] [PubMed] [Google Scholar]
- 41.Petroski MD, Deshaies RJ. 2005. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol 6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 42.Gastaldello S, Hildebrand S, Faridani O, Callegari S, Palmkvist M, Di Guglielmo C, Masucci MG. 2010. A deneddylase encoded by Epstein-Barr virus promotes viral DNA replication by regulating the activity of cullin-RING ligases. Nat Cell Biol 12:351–361. doi: 10.1038/ncb2035. [DOI] [PubMed] [Google Scholar]
- 43.Huh K, Zhou X, Hayakawa H, Cho J-Y, Libermann TA, Jin J, Harper WJ, Munger K. 2007. Human papillomavirus type 16 E7 oncoprotein associates with the cullin 2 ubiquitin ligase complex, which contributes to degradation of the retinoblastoma tumor suppressor. J Virol 81:9737–9747. doi: 10.1128/JVI.00881-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schreiner S, Wimmer P, Dobner T. 2012. Adenovirus degradation of cellular proteins. Future Microbiol 7:211–225. doi: 10.2217/fmb.11.153. [DOI] [PubMed] [Google Scholar]
- 45.Muller U, Roberts MP, Engel DA, Doerfler W, Shenk T. 1989. Induction of transcription factor AP-1 by adenovirus E1A protein and cAMP. Genes Dev 3:1991–2002. doi: 10.1101/gad.3.12a.1991. [DOI] [PubMed] [Google Scholar]
- 46.Ludwig S, Ehrhardt C, Neumeier ER, Kracht M, Rapp UR, Pleschka S. 2001. Influenza virus-induced AP-1-dependent gene expression requires activation of the JNK signaling pathway. J Biol Chem 276:10990–10998. doi: 10.1074/jbc.M009902200. [DOI] [PubMed] [Google Scholar]
- 47.McLean TI, Bachenheimer SL. 1999. Activation of cJUN N-terminal kinase by herpes simplex virus type 1 enhances viral replication. J Virol 73:8415–8426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ceballos-Olvera I, Chavez-Salinas S, Medina F, Ludert JE, del Angel RM. 2010. JNK phosphorylation, induced during dengue virus infection, is important for viral infection and requires the presence of cholesterol. Virology 396:30–36. doi: 10.1016/j.virol.2009.10.019. [DOI] [PubMed] [Google Scholar]
- 49.Zhang H, Niu X, Qian Z, Qian J, Xuan B. 2015. The c-Jun N-terminal kinase inhibitor SP600125 inhibits human cytomegalovirus replication. J Med Virol 87:2135–2144. doi: 10.1002/jmv.24286. [DOI] [PubMed] [Google Scholar]
- 50.Bruening W, Giasson B, Mushynski W, Durham HD. 1998. Activation of stress-activated MAP protein kinases up-regulates expression of transgenes driven by the cytomegalovirus immediate/early promoter. Nucleic Acids Res 26:486–489. doi: 10.1093/nar/26.2.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mizuguchi H, Kay MA. 1998. Efficient construction of a recombinant adenovirus vector by an improved in vitro ligation method. Hum Gene Ther 9:2577–2583. doi: 10.1089/hum.1998.9.17-2577. [DOI] [PubMed] [Google Scholar]
- 52.Maizel JV Jr, White DO, Scharff MD. 1968. The polypeptides of adenovirus. I. Evidence for multiple protein components in the virion and a comparison of types 2, 7A, and 12. Virology 36:115–125. doi: 10.1016/0042-6822(68)90121-9. [DOI] [PubMed] [Google Scholar]