MLV glycoGag not only enhances MLV replication but also increases HIV-1 infectivity similarly as Nef. Recent studies have discovered that both glycoGag and Nef antagonize a novel host restriction factor Ser5 and promote viral replication. Compared to Nef, the glycoGag antagonism of Ser5 is still poorly understood. MLV glycoGag is a transmembrane version of the structural Gag protein with an extra 88-amino-acid leader region that determines its activity. We now show that glycoGag interacts with Ser5 in live cells and internalizes Ser5 via receptor-mediated endocytosis. Ser5 is polyubiquitinated and relocalized to endosomes and lysosomes for massive destruction. In addition to the previously identified tyrosine-based sorting signal, we find two more important residues for Ser5 relocalization and downregulation. We also find that the Ser5 sensitivity to glycoGag is conserved in the SERINC family. Together, our findings highlight the important role of endosome/lysosome pathway in the enhancement of viral replication by viral proteins.
KEYWORDS: HIV, MLV, Nef, SERINC5, glycoGag, restriction factor
ABSTRACT
Glycosylated Gag (glycoGag) is an accessory protein expressed by most gammaretroviruses, including murine leukemia virus (MLV). MLV glycoGag not only enhances MLV replication and disease progression but also increases human immunodeficiency virus type 1 (HIV-1) infectivity as Nef does. Recently, SERINC5 (Ser5) was identified as the target for Nef, and the glycoGag Nef-like activity has been attributed to the Ser5 antagonism. Here, we investigated how glycoGag antagonizes Ser5 using MLV glycoMA and murine Ser5 proteins. We confirm previous observations that glycoMA relocalizes Ser5 from plasma membrane to perinuclear punctated compartments and the important role of its Y36XXL39 motif in this process. We find that glycoMA decreases Ser5 expression at steady-state levels and identify two other glycoGag crucial residues, P31 and R63, for the Ser5 downregulation. The glycoMA and Ser5 interaction is detected in live cells using a bimolecular fluorescence complementation assay. Ser5 is internalized via receptor-mediated endocytosis and relocalized to Rab5+ early, Rab7+ late, and Rab11+ recycling endosomes by glycoMA. Although glycoMA is not polyubiquitinated, the Ser5 downregulation requires Ser5 polyubiquitination via the K48- and K63-linkage, resulting in Ser5 destruction in lysosomes. Although P31, Y36, L39, and R63 are not required for glycoMA interaction with Ser5, they are required for Ser5 relocalization to lysosomes for destruction. In addition, although murine Ser1, Ser2, and Ser3 exhibit very poor antiviral activity, they are also targeted by glycoMA for lysosomal destruction. We conclude that glycoGag has a broad activity to downregulate SERINC proteins via the cellular endosome/lysosome pathway, which promotes viral replication.
IMPORTANCE MLV glycoGag not only enhances MLV replication but also increases HIV-1 infectivity similarly as Nef. Recent studies have discovered that both glycoGag and Nef antagonize a novel host restriction factor Ser5 and promote viral replication. Compared to Nef, the glycoGag antagonism of Ser5 is still poorly understood. MLV glycoGag is a transmembrane version of the structural Gag protein with an extra 88-amino-acid leader region that determines its activity. We now show that glycoGag interacts with Ser5 in live cells and internalizes Ser5 via receptor-mediated endocytosis. Ser5 is polyubiquitinated and relocalized to endosomes and lysosomes for massive destruction. In addition to the previously identified tyrosine-based sorting signal, we find two more important residues for Ser5 relocalization and downregulation. We also find that the Ser5 sensitivity to glycoGag is conserved in the SERINC family. Together, our findings highlight the important role of endosome/lysosome pathway in the enhancement of viral replication by viral proteins.
INTRODUCTION
Serine incorporator (SERINC) proteins consist of five members (1 to 5), which are type III integral membrane proteins with 10 to 11 transmembrane domains. Although these proteins were proposed to play a role in lipid biosynthesis by transporting serine molecules into the hydrophobic membrane lipid bilayers, this activity has not been demonstrated, so their physiological function is still unclear (1). Recently, SERINC3 (Ser3) and SERINC5 (Ser5) were identified as novel host restriction factors that inhibit retrovirus replication (2, 3). They were discovered as the target for the HIV-1 accessory protein Nef that has an activity to increase HIV-1 particle infectivity (4–6). Nonetheless, Ser5 has a much stronger antiviral activity than Ser3. Unlike mice that express only one Ser5 isoform, humans express five Ser5 alternatively spliced isoforms with 9 or 10 transmembrane domains, but only the longest isoform is stably expressed and exhibits the antiviral activity (7). In the absence of Nef, Ser5 is incorporated into HIV-1 particles and inhibits viral replication at the entry step (2, 3, 8). Nef effectively counteracts Ser5 and restores viral infectivity by downregulating Ser5 from the cell surface and reducing its virion incorporation (2, 3). The downregulation by Nef is mediated by the cellular endocytic pathway, which targets Ser5 to lysosomes for degradation (9). The Nef antagonism plays an important role in the prevalence of primate lentiviruses in their hosts, indicating that Ser5 is an important host restriction factor for these viruses (10).
In addition to Nef, Ser5 is counteracted by murine leukemia virus (MLV) accessory protein glycosylated Gag (glycoGag) (2, 3) and equine infectious anemia virus accessory protein S2 (11, 12). The glycoGag protein is expressed from most gammaretroviruses, including MLV, which is also known as gPr80 (13–16). This nonstructural and nonenzymatic viral protein contains the entire Gag (Pr65) plus a N-terminal leader of 88 amino acids in the case of MLV via an alternative CUG initiation codon (17). This leader includes a signal peptide that directs the glycoGag precursor to the endoplasmic reticulum for N-glycosylation. Mature gPr80 proteins are proteolytically cleaved into an ∼55-kDa membrane-associated N-terminal protein, and a secreted ∼40-kDa C-terminal product (18). The N-terminal segments are either a type I integral membrane protein, with the N terminus on the exterior side of the plasma membrane; or a type II protein, with the N terminus on the cytoplasmic side (18–20). Although glycoGag is not strictly required for virus replication in cell culture, it enhances MLV replication and disease progression in vivo (21, 22). Notably, MLV glycoGag is able to replace Nef and enhance HIV-1 replication in vitro (23). Not surprisingly, glycoGag reportedly counteracts the Ser5 restriction and increases HIV-1 infectivity (2, 3).
Here, we used MLV glycoGag and murine Ser5 to investigate how glycoGag antagonizes Ser5. It was reported that glycoGag downregulates Ser5 from cell surface and relocalizes Ser5 to intracellular compartments (2, 3). However, how Ser5 is internalized and where it is finally targeted by glycoGag are still unknown. We report that Ser5 is endocytosed by the adaptor protein complex-2 (AP-2) pathway and targeted to lysosomes for destruction.
RESULTS
glycoMA counteracts Ser5 restriction via relocalizing Ser5 from plasma membrane to cytoplasmic compartments.
A minimal glycoGag fragment that only retains its N-terminal 189 amino acids was found to exhibit the full-length glycoGag activity (23). This region includes the 88-amino-acid leader and the N-terminal 101 residues of the Gag matrix (MA) protein that has 131 amino acids (23). Accordingly, we used a truncated glycoGag that consists of its 2 to 190 residues, named glycoMA (24), to study how MLV glycoGag counteracts murine Ser5 proteins.
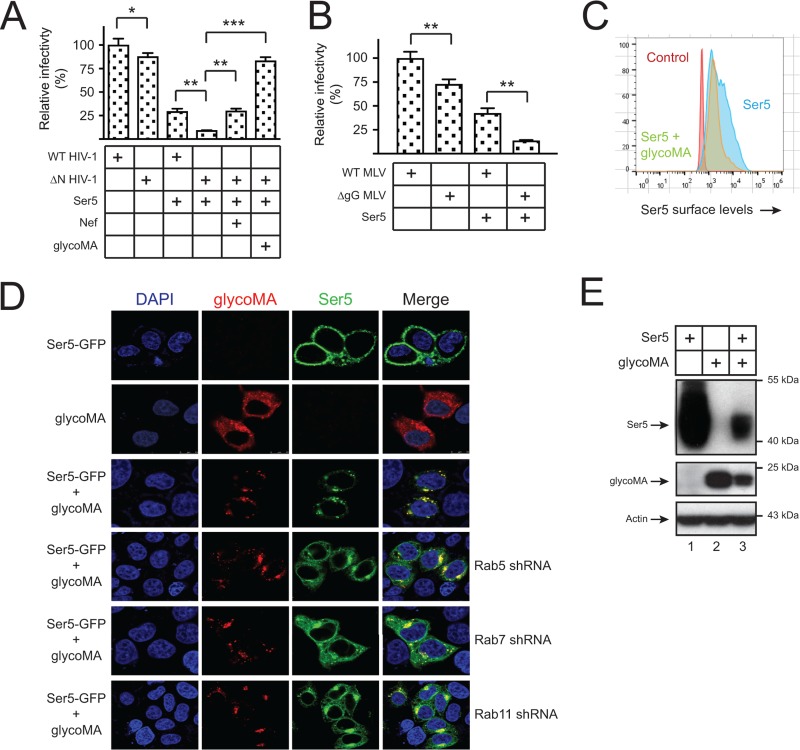
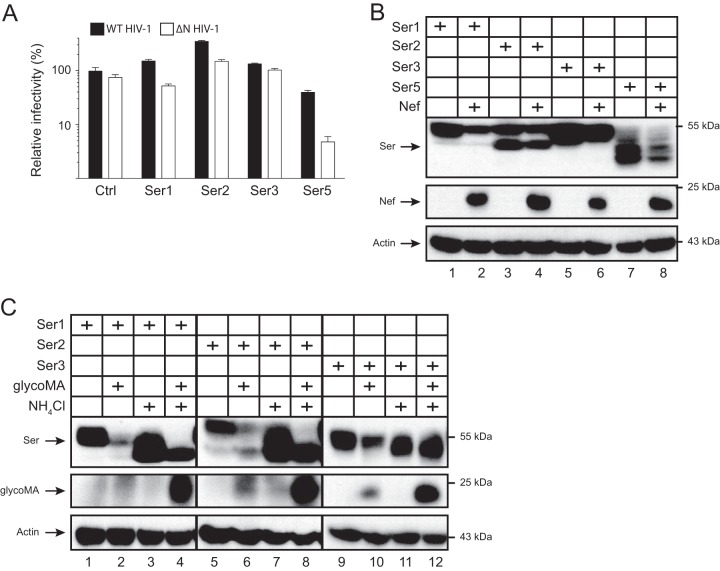
Wild-type (WT) and Nef-deficient (ΔN) HIV-1 pseudoviruses were produced from 293T cells that expressed ectopic Ser5 and glycoMA, and viral infectivity was analyzed after infection of the HIV-1 luciferase reporter TZM-bI cells. Although both WT and ΔN virus infection were inhibited by Ser5, the ΔN virus infectivity was reduced much more significantly, which was rescued by Nef. Notably, glycoMA rescued the ΔN virus infectivity much more effectively than Nef (Fig. 1A). Next, WT and glycoGag-deficient (ΔgG) MLV luciferase viruses were similarly produced from 293T cells in the presence of Ser5, and viral infectivity was analyzed after infection of mouse NIH 3T3 cells. Similarly, Ser5 inhibited the ΔgG virus replication much more strongly than the WT virus (Fig. 1B). These results confirm that Ser5 restricts HIV-1 and MLV replication, which is counteracted by glycoGag.
FIG 1.
GlycoMA counteracts Ser5 restriction via relocalizing Ser5 from plasma membrane to cytoplasmic compartments. (A) WT and ΔN HIV-1 pseudoviruses were produced from 293T cells in the presence of pBJ5-mSer5-HA (1 μg) and pcDNA3.1-glycoMA-HA or pcDNA3.1-SF2Nef-HA (2 μg). Viruses were normalized by p24Gag ELISA, and viral infectivity was determined in TZM-bI cells by measuring the luciferase activities. The infectivity of WT viruses produced in the absence of Ser5 was set as 100%. Error bars indicate the SEMs from three independent experiments. (B) WT and glycoGag-deficient (ΔgG) MLV luciferase reporter viruses were produced from 293T cells in the presence of pBJ5-mSer5-HA (1 μg). Viruses were normalized by the Gag protein levels via Western blotting, and viral infectivity was determined in NIH 3T3 cells by measuring the luciferase activities. The infectivity of WT viruses produced in the absence of Ser5 was set as 100%. Error bars indicate the SEMs from three independent experiments. (C) pBJ5-iFLAG-mSer5 (1 μg) was expressed with pcDNA3.1-glycoMA-HA (2 μg) in 293T cells, and the Ser5 expression on the cell surface was determined by flow cytometry using an anti-FLAG antibody. (D) pEGFP-N1-mSer5-FLAG (1 μg) was expressed with pcDNA3.1-glycoMA-HA (2 μg) in HeLa cells in the absence or presence of Rab5, Rab7, or Rab11 shRNA expression vectors. glycoMA was stained with a mouse anti-HA, followed by Alexa Fluor 647-conjugated goat anti-mouse antibody, and Ser5 and glycoMA expression was observed by confocal microscopy. (E) pCMV6-mSer5-FLAG (0.1μg) was expressed with pcDNA3.1-glycoMA-HA (3 μg) in 293T cells, and protein expression was analyzed by Western blotting with anti-HA, anti-FLAG, and anti-actin antibodies.
To explore the counteractive mechanism, we determined how glycoMA affects Ser5 expression. Ser5 was expressed with glycoMA, and Ser5 surface expression was analyzed by flow cytometry. The Ser5 expression was significantly reduced by glycoMA (Fig. 1C). To understand why Ser5 is downregulated, Ser5 was fused to GFP (Ser5-GFP) and expressed with glycoMA, and their subcellular localization was determined by confocal microscopy. When Ser5 and glycoMA were expressed individually, Ser5 was largely found on plasma membrane, and glycoMA was found on both plasma membrane and cytoplasm (Fig. 1D). When they were expressed together, they were both targeted to the same punctated perinuclear compartments. We reported that Ser5 is targeted to the Rab5+ early, Rab7+ late, and Rab11+ recycling endosomes by Nef (9). When cells were treated with short-hairpin RNAs (shRNAs) that specifically silence Rab5, Rab7, or Rab11 expression (9), Ser5 was less effectively compartmentalized by glycoMA (Fig. 1D). Moreover, when the Ser5 expression at steady-state levels was determined by Western blotting, the Ser5 expression was strongly reduced by glycoMA (Fig. 1E). Collectively, we confirm the previous observation that glycoGag internalizes Ser5 from cell surface and relocalizes Ser5 to intracellular compartments (2, 3), and importantly, we uncover that glycoGag reduces the Ser5 expression at steady-state levels.
Identification of crucial glycoGag residues for Ser5 downregulation.
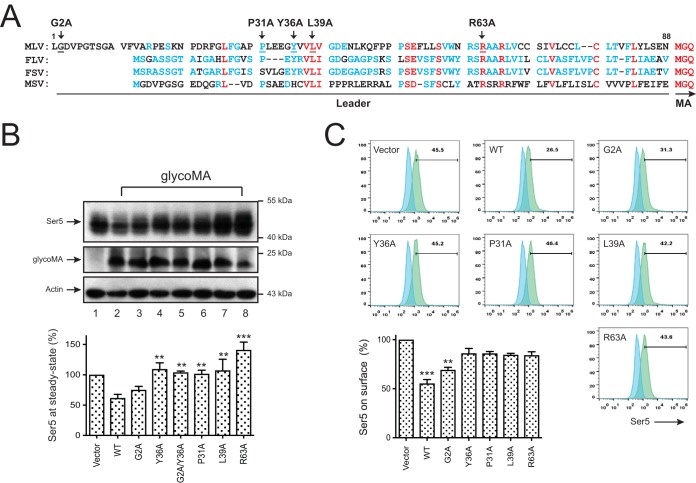
The N-terminal 88-amino-acid leader was sufficient to transfer the glycoGag activity to an unrelated type II transmembrane protein, indicating that this leader domain is responsible for the activity (24). In addition, a tyrosine-based Y36XXL39 motif plays a critical role in this domain (24). When the leader domain sequences from MLV, feline leukemia virus, feline sarcoma virus, and monkey sarcoma virus were aligned, a number of conserved residues were identified, including the two in the Y36XXL39 motif (Fig. 2A).
FIG 2.
Identification of critical glycoGag residues for Ser5 downregulation. (A) The N-terminal glycoGag protein sequences of MLV (accession number J02255), feline leukemia virus (FLV; AAA43055), feline sarcoma virus (FSV; P0DP83), and monkey sarcoma virus (MSV; ALV83311.1) are aligned. Red, blue, and dashes represent completely, partially, or not conserved residues. Residues targeted for mutations are indicted by arrowheads. The leader sequences are underlined and the initiation of matrix (MA) protein is indicated. (B) 293T cells were transfected with pCMV6-mSer5-FLAG (0.2μg) and pcDNA3.1-glycoMA-HA bearing G2A, Y36A, G2A/Y36A, P31A, L39A, and R63A mutations (2 μg for each mutant). Protein expression was detected by Western blotting using anti-HA, anti-FLAG, and anti-actin antibodies. The relative Ser5 expression was determined by quantifying their intensity on Western blots and is presented. Error bars indicate the SEM from three independent experiments. (C) pBJ5-iFLAG-mSer5 (1 μg) was expressed with pcDNA3.1-glycoMA-HA bearing indicated mutations (2 μg) in 293T cells, and the Ser5 expression on the cell surface was determined by flow cytometry using an anti-FLAG antibody. Error bars indicate the SEM from three independent experiments.
Four conserved residues P31, Y36, L39, and R63, and one nonconserved residue G2 were selected for site-directed mutagenesis by introducing G2A, P31A, Y36A, L39A, and R63A mutation, and a G2A/Y36A double mutation was also introduced. When Ser5 was expressed with these glycoMA mutants, the WT and the G2A mutant reduced the Ser5 expression at steady-state levels with similar efficiencies, whereas the P31A, Y36A, L39A, R63A, and G2A/Y36A mutants lost such activity (Fig. 2B). In addition, although the WT and the G2A mutant effectively downregulated Ser5 from the cell surface, the P31A, Y36A, L39A, and R63A mutants almost completely lost the activity (Fig. 2C). These results not only confirm the critical Y36XXL39 motif but also identify P31 and R63 as two critical residues for the glycoMA activity.
Detection of glycoMA and Ser5 interaction by BiFC.
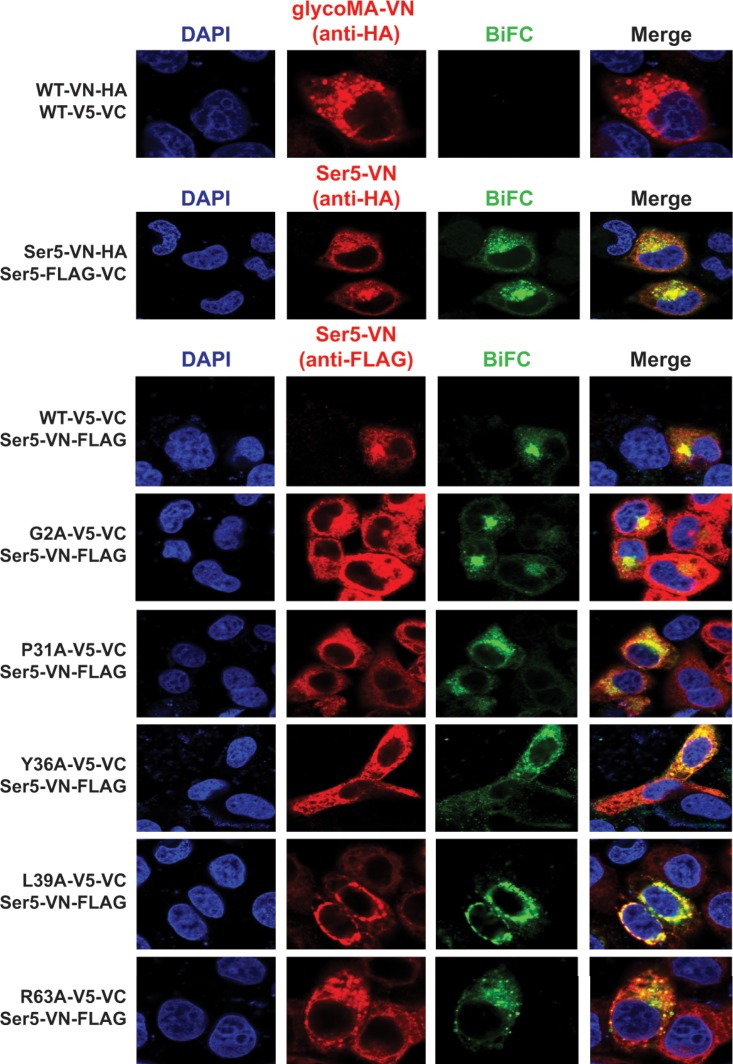
To understand how Ser5 is targeted by glycoGag, we used a bimolecular fluorescence complementation (BiFC) assay to detect the Ser5 and glycoMA interaction (25). Using this assay, we have demonstrated the Nef interaction with Ser5 (9). N-terminal residues 2 to 173 of Venus (VN) and its C-terminal residues 154 to 238 (VC) were fused to the C terminus of glycoMA or Ser5. After pairwise expression, their BiFC fluorescent signals were visualized by confocal microscopy. If glycoMA and Ser5 bind to each other, their interaction juxtaposes the two Venus fragments, resulting in structural complementation and a bright fluorescent signal. In addition, the assay also reports the subcellular localization of the interaction.
First, we tried to detect glycoMA and Ser5 oligomerization using this assay. The glycoMA-VN/glycoMA-VC or Ser5-VN/Ser5-VC pair was expressed, and their green BiFC fluorescent signals were detected by confocal microscopy. The glycoMA pair did not produce any BiFC signals, whereas strong BiFC signals were detected from the Ser5 pair (Fig. 3). The BiFC signals colocalized with the red fluorescent signals from specific detection of Ser5-VN protein, indicating that these BiFC signals were generated from the specific Ser5-Ser5 interactions. These results demonstrate that Ser5 oligomerizes, whereas glycoMA should not.
FIG 3.
Detection of glycoMA and Ser5 interaction by BiFC. HeLa cells were cotransfected with pcDNA3.1-glycoMA-VN-HA/pcDNA3.1-glycoMA-V5-VC (2 μg) or with pcDNA3.1-mSer5-VN-HA/pcDNA3.1-mSer5-FLAG-VC (1 μg). In addition, pcDNA3.1-mSer5-VN-HA (1 μg) was also cotransfected with pcDNA3.1-glycoMA-V5-VC (3 μg) or its mutants, as indicated. Their BiFC fluorescent signals were visualized by confocal microscopy.
Second, we tried to detect glycoMA-Ser5 interaction. When Ser5-VN was expressed with the WT glycoMA-VC, the specific BiFC fluorescent signals were detected in intracellular compartments (Fig. 3). This result demonstrates that glycoMA interacts with Ser5 and confirms that glycoMA internalizes Ser5, as shown in Fig. 1D. We also tested the G2A, P31A, Y36A, L39A, and R63A mutant interaction with Ser5. All of these mutants produced the BiFC signals with Ser5. Nonetheless, only the G2A mutant-Ser5 BiFC signals were found in intracellular compartments as the WT protein-Ser5 signals, whereas those P31A, Y36A, L39A, and R63A mutant-Ser5 BiFC signals were not. These results demonstrate that although P31, Y36, L39, and R63 are not involved in the glycoMA-Ser5 interaction, they play an important role in Ser5 intracellular trafficking and downregulation.
Ser5 is internalized by glycoMA via receptor-mediated endocytosis.
To understand how Ser5 is internalized by glycoGag, we studied the AP-2 pathway. The AP-2 complex is a heterotetramer consisting of two large subunits (α and β2), one medium subunit (μ), and one small subunit (σ) (26). We ectopically expressed the AP-2α and σ subunits and determined how Ser5 is downregulated by glycoMA. As a control, the AP-1μ subunit was tested. The decrease of Ser5 by glycoMA at steady-state levels was enhanced by AP-2σ but not by AP-1μ and AP-2α (Fig. 4A). To validate the AP-2σ activity, cells were treated with AP-2σ-specific shRNAs (Fig. 4B, lanes 1 and 2). The glycoMA-mediated Ser5 downregulation was completely disrupted by these AP-2σ shRNAs (Fig. 4B, lanes 3 to 6). In addition, we noted an increase in Ser5 molecular weights upon this shRNA treatment. These results demonstrate that AP-2 is involved in the Ser5 downregulation and confirm our previous observation that the endogenous AP-2σ expression is limited in this process (9).
FIG 4.
Ser5 is internalized by glycoMA via receptor-mediated endocytosis. (A) 293T cells were transfected with pCMV6-mSer5-FLAG (0.1μg) and pcDNA3.1-glycoMA-HA (3 μg) in the presence of AP-1μ, AP-2α, or AP-2σ expression vector (1 μg). The protein expression was analyzed by Western blotting. (B) 293T cells were transfected with pCMV6-mSer5-FLAG (0.1 μg) and pcDNA3.1-glycoMA-HA (3 μg) in the presence of AP-2σ shRNAs (4 μg). In addition, 293T cells were only transfected with AP-2σ shRNAs. The protein expression was analyzed by Western blotting. (C) 293T cells were transfected with pBJ5-iFLAG-mSer5 (1 μg) and pcDNA3.1-glycoMA-HA (2 μg). Cells were stained with an anti-FLAG antibody, and the antibody uptake was determined at 4 and 37°C by confocal microscopy. (D) The levels of Ser5 endocytosis in panel C were calculated by the detection frequency of cells where Ser5 was internalized from the plasma membrane. Error bars show the SEM from three independent experiments. (E) Experiments were conducted as in panel C, and the Ser5 expression on the cell surface was determined by flow cytometry. (F) The relative levels of Ser5 surface expression in panel E were calculated from three independent experiments, with error bars representing the SEM.
We then used an antibody uptake assay to monitor the AP-2 mediated Ser5 endocytosis. HeLa cells were transfected with Ser5 and glycoMA expression vector and then cultured for 24 h. The cells were stained with an anti-FLAG antibody to label Ser5 proteins on the cell surface and incubated at either 37°C to allow endocytosis, or at 4°C to block endocytosis. One hour later, Ser5 subcellular localization was determined by fluorescence microscopy. In the absence of glycoMA, cell surface Ser5 proteins were barely internalized even at 37°C (Fig. 4C, top panels, and Fig. 4D). However, when glycoMA was expressed, Ser5 was effectively internalized at 37°C but not at 4°C (Fig. 4C, bottom panels, and Fig. 4D). We also used a flow cytometry-based assay to demonstrate the Ser5 internalization by glycoMA. The cell surface Ser5 expression was effectively reduced by glycoMA at 37°C, but much less effectively at 4°C (Fig. 4E and F). Collectively, these results demonstrate that glycoGag internalizes Ser5 via receptor-mediated endocytosis.
Rab GTPases are involved in glycoMA-mediated Ser5 downregulation.
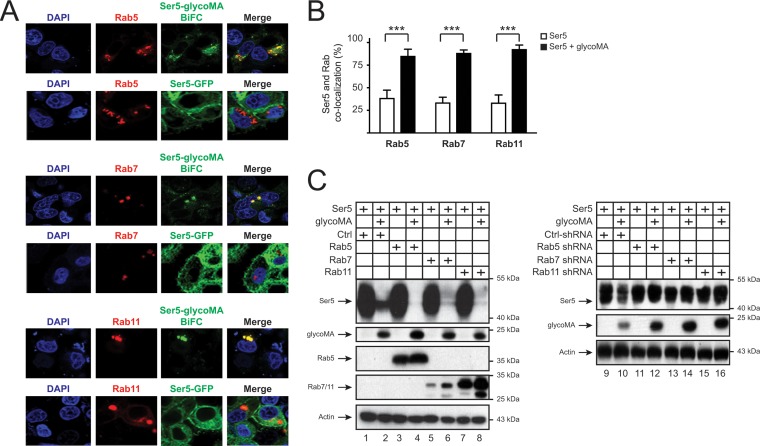
Our previous results imply that the Rab5+ early, Rab7+ late, and Rab11+ recycling endosomes are involved in the Ser5 internalization by glycoGag (Fig. 1D). We determined whether glycoMA-Ser5 complexes are associated with these endosomal compartments. When Ser5 was expressed alone, it was mainly distributed to the plasma membrane, and barely colocalized with Rab5, Rab7, or Rab11 (Fig. 5A and B). When Ser5 was expressed with glycoMA as a BiFC pair, Ser5 relocalized to Rab5+, Rab7+, and Rab11+ intracellular compartments (Fig. 5A and B). Thus, glycoGag promotes Ser5 trafficking into these endosomes.
FIG 5.
Rab GTPases are involved in glycoMA-mediated Ser5 downregulation. (A) pCMV-DsRed-HA-Rab5b (1 μg), pCMV-DsRed-2xHA-Rab7a (1 μg), or pCMV-DsRed-2xHA-Rab11a (1 μg) was expressed with the pcDNA3.1-mSer5-VN-HA (1 μg)/pcDNA3.1-glycoMA-V5-VC (2 μg) pair or with pEGFP-N1-mSer5-FLAG (1 μg) alone in HeLa cells. Fluorescence signals were detected by confocal microscopy. (B) The colocalization of Ser5 with Rab small GTPases in the presence or absence of glycoMA in panel A was statistically analyzed. Error bars indicate the SEM from three independent experiments. (C) pCMV6-mSer5-FLAG (0.1 μg) was expressed with pcDNA3.1-glycoMA-HA (3 μg) in the presence of either the Rab5, Rab7, and Rab11 expression vector (1 μg) or their shRNA expression vector (4 μg). The protein expression was determined by Western blotting.
Next, we investigated whether Rab5, Rab7, or Rab11 affects the Ser5 downregulation. Initially, we tested Ser5 downregulation by glycoMA in the presence of Rab5, Rab7, and Rab11 overexpression. The overexpression did not affect the Ser5 expression in the absence of glycoMA but further decreased the Ser5 expression at steady-state levels when glycoMA was expressed (Fig. 5C, lanes 1 to 8). Next, we repeated this experiment by decreasing Rab5, Rab7, and Rab11 expression via those shRNAs (Fig. 1D) (9). All three Rab-specific shRNAs effectively blocked the glycoMA-mediated Ser5 downregulation (Fig. 5C, lanes 9 to 16). Collectively, these results demonstrate that glycoGag-mediated Ser5 downregulation is dependent on these Rab5+ early, Rab7+ late, and Rab11+ recycling endosomes.
Ubiquitination plays an important role in glycoMA downregulation of Ser5.
To understand whether Ser5 is targeted to degradative pathways via these endosomes, we investigated how ubiquitination is involved in the Ser5 downregulation.
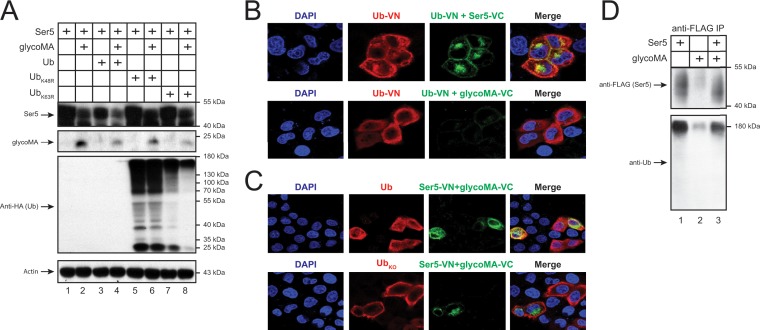
First, we determined whether ubiquitination via K48 and K63 plays a role in glycoMA downregulation of Ser5. Polyubiquitination through K48 and K63 occurs most frequently, which accounts for 52 or 38% of ubiquitination events (27). We expressed the WT and two ubiquitin mutants bearing a K48 or K63 substitution with an arginine residue (UbK48R and UbK63R) and then determined how the Ser5 downregulation by glycoMA was affected. The Ser5 downregulation was accelerated by the WT Ub but was compromised by UbK48R and UbK63R (Fig. 6A). In addition, the WT Ub expression was not detectable, although both UbK48R and UbK63R expression were detected, confirming the accelerated Ser5 downregulation by the WT Ub. These results demonstrate that polyubiquitination via K48 and K63 is involved in glycoMA-dependent Ser5 downregulation, which is consistent with our previous results (9).
FIG 6.
Ubiquitination plays an important role in glycoMA-mediated Ser5 downregulation. (A) pCMV-mSer5-FLAG (0.05μg) and pcDNA3.1-glycoMA-HA (2 μg) were expressed with a WT ubiquitin (Ub) or its mutant (UbK48R and UbK63R) expression vector (1 μg) in 293T cells. The protein expression was analyzed by Western blotting. (B) pcDNA3.1-mSer5-FLAG-VC (1 μg) or pcDNA3.1-glycoMA-V5-VC (2 μg) was transfected with pcDNA3.1-Ub-VN-HA (1 μg) into HeLa cells. Ectopic ubiquitin was detected by an anti-HA, followed by Alexa Fluor 647-conjugated secondary antibody and the fluorescence was observed by confocal microscopy. (C) pcDNA3.1-mSer5-VN-HA (1 μg) and pcDNA3.1-glycoMA-V5-VC (2 μg) were expressed with a WT or lysine-free ubiquitin (UbKO) expression vector in HeLa cells. The ectopic ubiquitin was detected similarly as in panel B, and the fluorescence was observed by confocal microscopy. (D) pCMV-mSer5-FLAG (0.2μg) and/or pcDNA3.1-glycoMA-HA (12 μg) was expressed in 293T cells. Proteins were immunoprecipitated with anti-FLAG beads and detected by anti-FLAG and anti-Ub antibodies.
Second, we detected Ser5 polyubiquitination. Initially, we tested Ser5 and glycoMA interaction with ubiquitin using BiFC. As reported previously, we could detect the specific BiFC signals from the Ub-VN/Ser5-VC pair expression in live cells (9), indicating that Ser5 interacts with ubiquitin in the absence of glycoMA (Fig. 6B). However, BiFC signals were not detected from the Ub-VN/glycoMA-VC pair, indicating that glycoMA should not interact with ubiquitin. In addition, the Ser5-glycoMA BiFC complex strongly colocalized with WT ubiquitin but not a lysine-free ubiquitin mutant (UbKO) (Fig. 6C). Next, Ser5 and/or glycoMA were expressed and, after immunoprecipitation, Ser5 protein expression was detected by Western blotting. The Ser5 expression was detectable by an anti-Ub antibody in the absence and presence of glycoMA (Fig. 6D, lanes 1 and 3), indicating that Ser5 is likely polyubiquitinated even in the absence of glycoMA. In addition, more polyubiquitinated Ser5 proteins were detected in the absence of glycoMA, which is consistent with glycoMA downregulation of Ser5. These results demonstrate that Ser5 is likely polyubiquitinated in the absence of glycoGag, which is required for the Ser5 downregulation.
Ser5 is targeted to lysosomes for degradation.
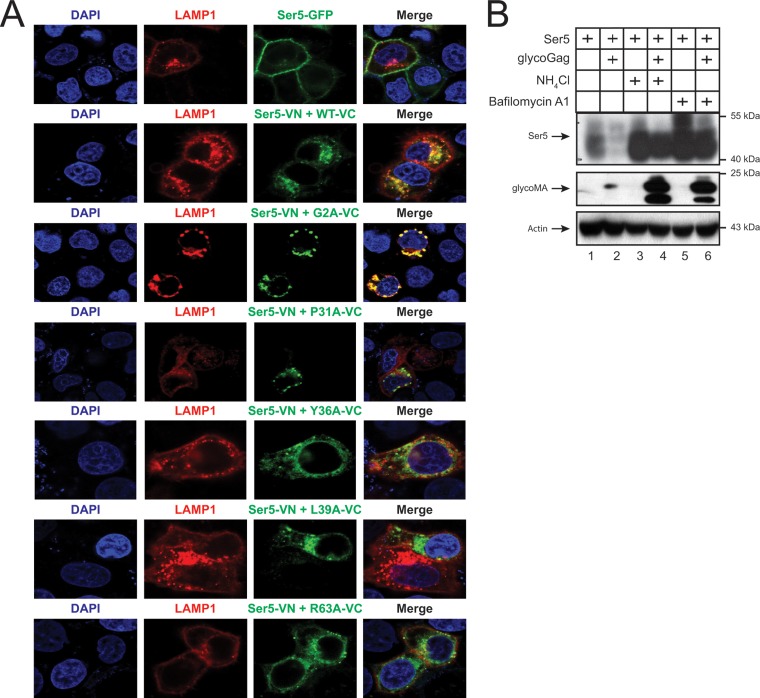
To determine whether Ser5 is targeted to lysosomes, lysosome-associated membrane protein 1 (LAMP1) was expressed with Ser5-GFP or the Ser5-VN/glycoMA-VC BiFC pair, and their subcellular colocalization was observed by confocal microscopy. Ser5-GFP showed a plasma membrane localization and did not colocalize with LAMP1 (Fig. 7A). In contrast, the Ser5-glycoMA WT and G2A mutant BiFC complexes were found in punctated perinuclear compartments, where it strongly colocalized with LAMP1. Nonetheless, the Ser5-glycoMA P31A, Y36A, L39A, and R63A mutant BiFC complexes were diffused in the cytoplasm and poorly colocalized with LAMP1. This result is consistent with their poor activity to downregulate Ser5.
FIG 7.
Ser5 is targeted to lysosomes for degradation. (A) pcDNA3.1-mSer5-VN-HA (1 μg) and pcDNA3.1-glycoMA-V5-VC (2 μg) were expressed with pCMV-DsRed-HA-LAMP1 (1 μg) in HeLa cells. As controls, glycoMA G2A, P31A, Y36A, L39A, and R63A mutants were used. The protein expression was analyzed by confocal microscopy. (B) pCMV-mSer5-FLAG (0.05μg) was expressed with pcDNA3.1-glycoMA-HA (3 μg) in 293T cells. After 24 h, the cells were treated with NH4Cl (20 μM) or bafilomycin A1 (100 nM). Protein expression was analyzed by Western blotting.
Next, Ser5 and glycoMA were expressed and treated with NH4Cl and bafilomycin A1 that block the lysosome function, and Ser5 and glycoMA expression at steady-state levels were determined by Western blotting. Although the strong glycoMA-mediated Ser5 downregulation was observed again, such downregulation was completely disrupted by these two inhibitors (Fig. 7B). Notably, the glycoMA expression was also strongly increased by these two inhibitors. These results demonstrate that glycoMA should target Ser5 into lysosomes for degradation.
Murine Ser1, Ser2, and Ser3 are downregulated by glycoMA.
To define the broadness of the glycoMA activity, we tested whether murine Ser1, Ser2, and Ser3 are also targeted by glycoMA. First, we determined whether these three proteins have any antiviral activities. WT and ΔN HIV-1 pseudoviruses were produced in the presence of murine Ser1, Ser2, Ser3, or Ser5, and viral infectivity was determined. Ser5 strongly reduced the ΔN, but not WT HIV-1 infectivity, whereas Ser1, Ser2, and Ser3 did not show any antiviral activity (Fig. 8A). Consistently, Nef reduced the Ser5 expression at steady-state levels (Fig. 8B). In addition, Nef also reduced the Ser1 and Ser3, but not the Ser2 expression. Second, we tested how glycoMA could affect Ser1, Ser2, and Ser3 expression. The Ser1, Ser2, and Ser3 expression were all decreased by glycoMA (Fig. 8C). Nonetheless, the decrease was effectively inhibited by NH4Cl, and the glycoMA expression was also strongly increased by NH4Cl. The NH4Cl treatment also decreased the molecular weights of these SERINC proteins. Thus, although murine Ser1, Ser2, and Ser3 do not have any antiviral activities, glycoMA still downregulates them, probably by a very similar mechanism.
FIG 8.
Ser1, Ser2, and Ser3 are downregulated by glycoMA. (A) WT and ΔN HIV-1 pseudoviruses were produced from 293T cells in the presence of pCMV6-mSer1, pCMV6-mSer2, pCMV6-mSer3, or a control (Ctrl) vector (0.05 μg). After being normalized by p24Gag ELISA, viral infectivity was determined in TZM-bI cells. The infectivity of WT viruses produced in the absence of Ser5 was set as 100%. Error bars indicate the SEM from three independent experiments. (B) pCMV6-mSer1-FLAG, pCMV6-mSer2-FLAG, pCMV6-mSer3-FLAG, and pCMV6-mSer5-FLAG (0.05 μg) were expressed with pNLenCAT or pNLenCAT-Xh (3 μg), and protein expression was analyzed by Western blotting. (C) pCMV6-mSer1-FLAG, pCMV6-mSer2-FLAG, and pCMV6-mSer3-FLAG (0.05 μg) were expressed with pcDNA3.1-glycoMA-HA (3 μg). After 24 h, cells were treated with NH4Cl (20 μM), and protein expression was analyzed by Western blotting.
DISCUSSION
Previous studies have shown that glycoGag inhibits Ser5 incorporation into HIV-1 virions and counteracts its antiviral activity (2, 3). These studies also find that glycoGag downregulates Ser5 from the cell surface and relocalizes Ser5 from the plasma membrane to perinuclear vesicles. We not only confirmed these observations but also demonstrate that glycoGag reduces the Ser5 expression at steady-state levels. Such a reduction was not observed in these earlier studies. We reported that Nef becomes unable to counteract Ser5 when Ser5 is expressed at relatively high levels (7). To observe the glycoGag effect, we decreased the Ser5 expression by using the pBJ5 vector that has a weak promoter or by using much less of the pCMV6 vector that has a strong promoter. Thus, a relatively higher glycoGag expression may be required for the decreased Ser5 expression. It was recently reported that human Ser5 is subjected to N-glycosylation at residue N294 (28). This N-glycosylation site is conserved in all human and murine SERINC family members. The major Ser5 species detected in our studies has a molecular weight slightly larger than 40 kDa, which should correspond to the low molecular weight form of the human Ser5 modified by high-mannose glycans. We also detected a change in SERINC protein molecular weights after treatments with AP-2σ shRNAs and NH4Cl, which may result from an influence on N-glycosylation.
To understand how Ser5 is targeted by glycoGag, we investigated their interactions via BiFC. Recently, we reported the detection of the Nef-Ser5 interactions using this assay and demonstrated that Nef membrane association and oligomerization are required for its interaction with Ser5 (9). Notably, although we could detect Ser5 oligomerization, the glycoMA oligomerization was not detectable via BiFC. Nonetheless, the glycoMA-Ser5 interaction was detected via BiFC, indicating that glycoGag oligomerization is not required for its interaction with Ser5. Thus, Nef and glycoMA have a differential dependence on their oligomerization for Ser5 interaction.
A previous study has mapped a crucial glycoGag region to residues 24 to 54 in the 88-amino-acid leader region, where the important Y36XXL39 motif is located (24). We identified an important P31 residue in this region and found another crucial R63 residue in the conserved membrane-proximal region. Inactivation of these four residues by introducing P31A, Y36A, L39A, and R63A mutation disabled glycoMA from Ser5 downregulation. Nonetheless, all of these mutants still interacted with Ser5 when measured by BiFC but did not relocalize Ser5 into lysosomes. These results suggest that P31, Y36, L39, and R63 should not determine glycoMA interaction with Ser5 and instead should play a role in glycoMA-Ser5 trafficking after the complex is formed.
Tyrosine-based sorting signals conforming to YXX∅ motifs (where “X” is any amino acid and “∅” is an amino acid with a bulky hydrophobic side chain), such as YXXL, are recognized by the μ subunits of heterotetrameric AP complexes such as AP-2μ (29, 30). AP-2 is responsible for internalization of transmembrane proteins on the plasma membrane via clathrin-mediated endocytosis (31). Both the glycoGag Y36XXL39 motif and the cellular AP-2 complex were found to be indispensable for the glycoGag activity to enhance viral replication (24). In support of these previous observations, we found that AP-2 is required for the Ser5 downregulation by glycoMA, as measured by Western blotting, and that glycoMA induces Ser5 endocytosis, as measured by an antibody uptake assay. glycoGag alone has been shown to exhibit a punctuated perinuclear distribution which is also stained with the late endosomal marker Rab7A (24). We reported that after triggering vigorous endocytosis, Nef relocalizes Ser5 into Rab5+ early, Rab7+ late, and Rab11+ recycling endosomes (9). We found that glycoMA also relocalizes Ser5 into these endosomes, and manipulation of Rab5, Rab7, and Rab11 expression positively regulates the Ser5 downregulation. Collectively, these results clearly demonstrate that endosomes are also required for glycoMA downregulation of Ser5.
To unveil where Ser5 is targeted from endosomes, we investigated the ubiquitination pathway. We reported that although Nef does not promote Ser5 ubiquitination, ubiquitination is required for Ser5 downregulation by Nef (9). Similarly, we found that the expression of ubiquitin mutants UbK48R and UbK63R compromises the Ser5 downregulation by glycoMA. In addition, although WT ubiquitin strongly colocalizes with the glycoMA-Ser5 complex, the lysine-free ubiquitin mutant (UbKO) does not. To understand which protein is targeted by ubiquitin, we tested ubiquitin interactions with glycoMA or Ser5 via BiFC. We found that ubiquitin selectively interacts with Ser5, but not glycoMA. Furthermore, WT glycoMA could target Ser5 into LAMP1+ compartments, whereas the P31A, Y36A, L39A, and R63A mutant could not. Finally, inhibition of the lysosome activity with NH4Cl and bafilomycin A1 resulted in a complete block of the decrease of Ser5 expression at steady-state levels by glycoMA. Collectively, these results demonstrate that glycoGag targets polyubiquitinated Ser5 via K48 and/or K63 linkage to lysosomes for destruction.
In summary, we have collected new insights into the poorly defined Ser5 antagonism by MLV glycoGag. GlycoGag promotes Ser5 endocytosis from plasma membrane via direct interactions and sorts polyubiquitinated Ser5 into endosomes, resulting in Ser5 degradation in lysosomes. Because glycoMA also decreases Ser1, Ser2, and Ser3 expression at steady-state levels via a similar mechanism, the Ser5 antagonism by glycoGag is conserved. Thus, further elucidating this glycoGag counteractive mechanism will provide a new understanding of the arms race between Ser5 and retroviruses.
MATERIALS AND METHODS
Cells.
Human embryonic kidney epithelium 293T cells, cervical cancer HeLa cells, and mouse embryonic fibroblast NIH 3T3 cells were obtained from American Type Culture Collection. The HIV-1 luciferase reporter TZM-bI cells were obtained from the NIH AIDS Reagent Program. All cells were cultured in Dulbecco modified Eagle medium (DMEM) with 10% fetal bovine serum (FBS; Sigma) and 100 mg/ml streptomycin and penicillin.
Plasmids.
The MLV provirus construct (MoMLV; accession no. J02255) pNCA was obtained from Stephen Goff (32). To generate glycoGag-defective MLV provirus plasmid (pNCAΔgG), the initiation codon CTG of glycoGag was mutated. The Env-deficient HIV-1 proviral vector pNLΔE (pNLenCAT), its Nef-deficient version pNLΔEΔN (pNLenCAT-Xh), and HIV-1 Env expression vector pNLnΔBS were provided by Kenzo Tokunaga (33). pLNCX2-Luc was reported previously (34).
pBJ5-glycoMA-HA was provided by Heinrich Gottlinger (24). pBJ5-mSer5-HA was generated by replacing glycoMA with mSer5 after XhoI/EcoRI digestion (m: Mouse). pBJ5-iFLAG-mSer5 that has a FLAG tag inserted between residues 291 and 292 of mSer5 was created by overlapping PCR amplification. pEGFP-N1-mSer5-FLAG was created by cloning the mSer5-FLAG gene into pEGFP-N1 vector after KpnI/AgeI digestion. pcDNA3.1-glycoMA-HA was created by cloning glycoMA-HA into pcDNA3.1-eGFP after HindIII/EcoRV digestion. An XhoI site was introduced before the ATG codon, and a BspEI site was introduced between glycoMA and the HA tag during this cloning. pcDNA3.1-SF2Nef-HA (Nef gene from HIV-1 SF2 strain) was created by cloning Nef-HA into pcDNA3.1-mSer5-VN-HA vector after XhoI/EcoRI digestion, and a BspEI site was introduced between Nef and the HA tag during this cloning.
pcDNA3.1-Ser5-FLAG-VC, pcDNA3.1-Ser5-VN-HA, pcDNA3.1-Ser5-VN-FLAG, pcDNA3.1-Nef-V5-VC, and pcDNA3.1-Ub-VN-HA were reported previously (9). To construct pcDNA3.1-mSer5-VN-HA, mSer5-VN-HA was amplified by overlapping PCR and cloned into pcDNA3.1-Ser5-VN-HA by XhoI/EcoRV digestion. An EcoRI site was created immediately before the EcoRV site during this cloning. pcDNA3.1-mSer5-FLAG-VC was created after replacing human Ser5 in pcDNA3.1-Ser5-FLAG-VC with mSer5 after HindIII/EcoRV digestion. pcDNA3.1-mSer5-VN-FLAG was created from pcDNA3.1-Ser5-VN-FLAG by homologous recombination after XhoI/BspEI digestion. pcDNA3.1-glycoMA-V5-VC was created by replacing Nef in pcDNA3.1-Nef-V5-VC via homologous recombination after Xho1/EcoR1 digestion. pcDNA3.1-glycoMA-VN-HA was created by replacing Ser5 in pcDNA3.1-Ser5-VN-HA via homologous recombination after XhoI/BspEI digestion. GlycoMA G2A, G2A/Y36A, P31A, Y36A, L39A, and R63A mutation in pcDNA3.1-glycoMA-HA and pcDNA3.1-glycoMA-V5-VC were created by site-directed mutagenesis, respectively. Ub K48R and K63R mutants were created in the pCMV-HA-Ub vector by site-directed mutagenesis. Primers and cloning methods are available upon request.
The following vectors were ordered from OriGene: pCMV6-AP-1μ1 (RC202366), pCMV-mSer1-FLAG (MR207184), pCMV-mSer2-FLAG (MR207238), pCMV-mSer3-FLAG (MR207539), pCMV-mSer5-FLAG (MR207354), and pGFP-C-shLenti vectors expressing shRNAs against AP-2σ (TL306655), Rab5a (TL309987), Rab7a (TL309983), and Rab11a (TL310015).
The following vectors were obtained via Addgene: pCMV-DsRed-2xHA-Rab7a (no. 12661) and pCMV-DsRed-2xHA-Rab11a (no. 12679) from Richard Pagano, pCMV-mRFP-Rab5a (catalog no. 14437) from Ari Helenius, pCMV-LAMP1-mGFP (no. 34831) from Esteban Dell’Angelica, pCMV-HA-Ub (no. 18712) from Edward Yeh, and pRK5-HA-Ubiquitin-KO (no. 17603) from Ted Dawson.
Effect of Ser5 on HIV-1 and MLV infectivity.
WT and ΔN HIV-1 pseudoviruses were produced from 293T cells. Cells were transfected with 1 μg of pBJ5-mSer5-HA that expresses mSer5, 0.2 μg of pNLnΔBS that expresses Env, and 1 μg of pNLenCAT that produces WT particles or with pNLenCAT-Xh that produces ΔN particles, using polyethylenimine from Polysciences as transfection reagent. The cells were cultured for 48 h after transfection. Viral production was quantified by p24Gag enzyme-linked immunosorbent assay (ELISA), and viral infectivity was detected via infecting the HIV-1 luciferase reporter cell line TZM-bI cells in a 96-well plate for 48 h. Intracellular luciferase activities were determined using the Bright-Glo luciferase assay system (Promega) and used to calculate viral infectivity.
WT and ΔgG MLV were produced from 293T cells after transfection of 1 μg of pNCA or pNCAΔgG with 1 μg of pLNCX2-Luc in the presence of 1 μg of pBJ5-mSer5-HA. Cells were cultured for 2 days before viruses were harvested. Virions were purified by ultracentrifugation and quantified by Gag by Western blotting. Infectivity was assayed by measuring the intracellular luciferase activities after 48 h infection of NIH 3T3 cells.
Effect of glycoMA on Ser5 endocytosis.
HeLa cells was transfected with pBJ5-iFLAG-mSer5 and pcDNA3.1-glycoMA-HA for 24 h. Cells were incubated with anti-FLAG monoclonal antibody (1:1,000) at 4°C for 30 m. After being washed three times with phosphate-buffered saline (PBS), the cells were cultured with DMEM at 4 or 37°C for 1 h. The cells were then fixed with 4% paraformaldehyde, followed by washing with PBS and permeabilization with 0.1% Triton X-100 for 5 min. The cells were incubated for 1 h at room temperature with Alexa Fluor 488-conjugated goat anti-mouse antibody (1:500) diluted in 5% FBS. After the cells were washed with PBS, DAPI (4′,6′-diamidino-2-phenylindole) was used to stain the nuclei. Ser5 internalization was detected by the laser scanning confocal microscope. The frequency of cells in which relocalization of Ser5 occurred was calculated to determine the level of endocytosis.
Western blotting.
In brief, 293T cells were seeded either in 6-well plates or in 10-cm dishes with an initial density of 5 × 105 cells per well or 2 × 106 cells per dish. At 24 or 48 h transfection, the cells were lysed with radioimmunoprecipitation assay buffer (Sigma), and the cytosolic cellular proteins were revolved by sodium dodecyl sulfate polyacrylamide gel electrophoresis. After transfer to Immun-Blot polyvinylidene difluoride membranes (Bio-Rad), the proteins were incubated with primary and secondary antibodies. The mouse anti-FLAG, mouse anti-HA, and mouse anti-actin monoclonal antibodies were purchased from Sigma; the mouse anti-V5 monoclonal antibodies were purchased from Invitrogen; the rabbit anti-Rab5 antibodies were purchased from Cell Signaling Technology; Horseradish peroxidase-conjugated anti-human, rabbit, or mouse immunoglobulin G secondary antibodies were purchased from Pierce. The enhanced chemiluminescence detection kit was purchased from Thermo Fisher.
Confocal microscopy.
Approximately 1.5 × 105 to 2.0 × 105 HeLa cells were seeded on poly-l-lysine-coated glass slides and transfected with indicated vectors using Lipofectamine 3000 as a transfection reagent (Thermo Fisher). After 24 h, the cells were fixed with 4% paraformaldehyde, permeabilized with 0.1% Triton X-100, and blocked with 5% bovine serum albumin (BSA). For immunofluorescence assay, the cells were incubated with primary antibodies against HA or FLAG tags at a 1:1,000 dilution in PBS with 5% BSA for 2 h. After five washes with PBS, the cells were stained using Alexa Fluor 488- or Alexa Fluor 647-conjugated secondary antibodies at a 1:500 dilution for 1 h. The cells were washed with PBS five times and then incubated with DAPI for 2 min for nuclear staining. After further washing with PBS, cells were observed under a confocal microscope (ZEISS LSM880). For imaging, a 63× oil objective was used. At least 100 random cells per slide were analyzed, and the most representative images from each slide were selected for presentation.
Flow cytometry.
293T cells were transfected with pBJ5-iFLAG-mSer5 and pcDNA3.1-glycoMA-HA for 24 h. After fixation with 4% paraformaldehyde, the cells were incubated with an anti-FLAG monoclonal antibody at 4°C overnight. After washing, the cells were incubated with Alexa Fluor 488-conjugated goat anti-mouse antibody for 1 h. Ser5 expression on the cell surface was determined by flow cytometry.
Statistical analysis.
Microsoft Excel was used for statistical tests. An unpaired two-tailed Student t test was used to evaluate the significance of differences between samples. In each group, the standard errors of the mean (SEM) were calculated to estimate the variance. All experiments were repeated independently at least three times; the results of a representative experiment are shown (*, P < 0.05; **, P < 0.01; ***, P < 0.001; NS, not significant [P > 0.05]).
ACKNOWLEDGMENTS
We thank Kenzo Tokunaga, Heinrich Gottlinger, Massimo Pizzato, Richard Pagano, Ari Helenius, Esteban Dell’Angelica, Edward Yeh, and Ted Dawson, as well as the NIH AIDS Reagent Program for providing various reagents.
S.L., Y.C., and B.W. are supported by grants 31700138, 31702270, and 31502089, respectively, from the National Natural Science Foundation of China. S.L. is also supported by grants from the Natural Science Foundation of Heilongjiang Province (QC2018037) and the China Postdoctoral Science Foundation (2017M620980). I.A. is supported by scholarships from the Chinese Academy of Agricultural Sciences and Beijing Government. Y.-H.Z. is supported by grants AI120189, AI122863, and AI138707 from the National Institutes of Health.
REFERENCES
- 1.Inuzuka M, Hayakawa M, Ingi T. 2005. SERINC, an activity-regulated protein family, incorporates serine into membrane lipid synthesis. J Biol Chem 280:35776–35783. doi: 10.1074/jbc.M505712200. [DOI] [PubMed] [Google Scholar]
- 2.Rosa A, Chande A, Ziglio S, De Sanctis V, Bertorelli R, Goh SL, McCauley SM, Nowosielska A, Antonarakis SE, Luban J, Santoni FA, Pizzato M. 2015. HIV-1 Nef promotes infection by excluding SERINC5 from virion incorporation. Nature 526:212–217. doi: 10.1038/nature15399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Usami Y, Wu Y, Gottlinger HG. 2015. SERINC3 and SERINC5 restrict HIV-1 infectivity and are counteracted by Nef. Nature 526:218–223. doi: 10.1038/nature15400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aiken C, Trono D. 1995. Nef stimulates human immunodeficiency virus type 1 proviral DNA synthesis. J Virol 69:5048–5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chowers MY, Spina CA, Kwoh TJ, Fitch NJ, Richman DD, Guatelli JC. 1994. Optimal infectivity in vitro of human immunodeficiency virus type 1 requires an intact nef gene. J Virol 68:2906–2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miller MD, Warmerdam MT, Gaston I, Greene WC, Feinberg MB. 1994. The human immunodeficiency virus-1 nef gene product: a positive factor for viral infection and replication in primary lymphocytes and macrophages. J Exp Med 179:101–113. doi: 10.1084/jem.179.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang X, Zhou T, Yang J, Lin Y, Shi J, Zhang X, Frabutt DA, Zeng X, Li S, Venta PJ, Zheng YH. 2017. Identification of SERINC5-001 as the predominant spliced isoform for HIV-1 restriction. J Virol 91:e00137-17. doi: 10.1128/JVI.00137-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sood C, Marin M, Chande A, Pizzato M, Melikyan GB. 2017. SERINC5 protein inhibits HIV-1 fusion pore formation by promoting functional inactivation of envelope glycoproteins. J Biol Chem 292:6014–6026. doi: 10.1074/jbc.M117.777714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi J, Xiong R, Zhou T, Su P, Zhang X, Qiu X, Li H, Li S, Yu C, Wang B, Ding C, Smithgall TE, Zheng YH. 2018. HIV-1 Nef antagonizes SERINC5 restriction by downregulation of SERINC5 via the endosome/lysosome system. J Virol 92:e00196-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heigele A, Kmiec D, Regensburger K, Langer S, Peiffer L, Sturzel CM, Sauter D, Peeters M, Pizzato M, Learn GH, Hahn BH, Kirchhoff F. 2016. The potency of Nef-mediated SERINC5 antagonism correlates with the prevalence of primate lentiviruses in the wild. Cell Host Microbe 20:381–391. doi: 10.1016/j.chom.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ahi YS, Zhang S, Thappeta Y, Denman A, Feizpour A, Gummuluru S, Reinhard B, Muriaux D, Fivash MJ, Rein A. 2016. Functional interplay between murine leukemia virus glycoGag, Serinc5, and surface glycoprotein governs virus entry, with opposite effects on gammaretroviral and ebolavirus glycoproteins. mBio 7:e01985-16. doi: 10.1128/mBio.01985-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chande A, Cuccurullo EC, Rosa A, Ziglio S, Carpenter S, Pizzato M. 2016. S2 from equine infectious anemia virus is an infectivity factor which counteracts the retroviral inhibitors SERINC5 and SERINC3. Proc Natl Acad Sci U S A 113:13197–13202. doi: 10.1073/pnas.1612044113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edwards SA, Fan H. 1979. gag-Related polyproteins of Moloney murine leukemia virus: evidence for independent synthesis of glycosylated and unglycosylated forms. J Virol 30:551–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Evans LH, Dresler S, Kabat D. 1977. Synthesis and glycosylation of polyprotein precursors to the internal core proteins of Friend murine leukemia virus. J Virol 24:865–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neil JC, Smart JE, Hayman MJ, Jarrett O. 1980. Polypeptides of feline leukemia virus: a glycosylated gag-related protein is released into culture fluids. Virology 105:250–253. doi: 10.1016/0042-6822(80)90173-7. [DOI] [PubMed] [Google Scholar]
- 16.Schultz AM, Rabin EH, Oroszlan S. 1979. Posttranslational modification of Rauscher leukemia virus precursor polyproteins encoded by the gag gene. J Virol 30:255–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prats AC, De Billy G, Wang P, Darlix JL. 1989. CUG initiation codon used for the synthesis of a cell surface antigen coded by the murine leukemia virus. J Mol Biol 205:363–372. doi: 10.1016/0022-2836(89)90347-1. [DOI] [PubMed] [Google Scholar]
- 18.Fujisawa R, McAtee FJ, Zirbel JH, Portis JL. 1997. Characterization of glycosylated Gag expressed by a neurovirulent murine leukemia virus: identification of differences in processing in vitro and in vivo. J Virol 71:5355–5360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pillemer EA, Kooistra DA, Witte ON, Weissman IL. 1986. Monoclonal antibody to the amino-terminal L sequence of murine leukemia virus glycosylated Gag polyproteins demonstrates their unusual orientation in the cell membrane. J Virol 57:413–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Renner TM, Belanger K, Lam C, Gerpe MCR, McBane JE, Langlois MA. 2018. Full-length glycosylated Gag of murine leukemia virus can associate with the viral envelope as a type I integral membrane protein. J Virol 92:e01530-17. doi: 10.1128/JVI.01530-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Corbin A, Prats AC, Darlix JL, Sitbon M. 1994. A nonstructural gag-encoded glycoprotein precursor is necessary for efficient spreading and pathogenesis of murine leukemia viruses. J Virol 68:3857–3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Portis JL, Fujisawa R, McAtee FJ. 1996. The glycosylated gag protein of MuLV is a determinant of neuroinvasiveness: analysis of second site revertants of a mutant MuLV lacking expression of this protein. Virology 226:384–392. doi: 10.1006/viro.1996.0666. [DOI] [PubMed] [Google Scholar]
- 23.Pizzato M. 2010. MLV glycosylated-Gag is an infectivity factor that rescues Nef-deficient HIV-1. Proc Natl Acad Sci U S A 107:9364–9369. doi: 10.1073/pnas.1001554107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Usami Y, Popov S, Gottlinger HG. 2014. The Nef-like effect of murine leukemia virus glycosylated gag on HIV-1 infectivity is mediated by its cytoplasmic domain and depends on the AP-2 adaptor complex. J Virol 88:3443–3454. doi: 10.1128/JVI.01933-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kerppola TK. 2008. Bimolecular fluorescence complementation (BiFC) analysis as a probe of protein interactions in living cells. Annu Rev Biophys 37:465–487. doi: 10.1146/annurev.biophys.37.032807.125842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park SY, Guo X. 2014. Adaptor protein complexes and intracellular transport. Biosci Rep 34 doi: 10.1042/BSR20140069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dammer EB, Na CH, Xu P, Seyfried NT, Duong DM, Cheng D, Gearing M, Rees H, Lah JJ, Levey AI, Rush J, Peng J. 2011. Polyubiquitin linkage profiles in three models of proteolytic stress suggest the etiology of Alzheimer disease. J Biol Chem 286:10457–10465. doi: 10.1074/jbc.M110.149633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharma S, Lewinski MK, Guatelli J. 2018. An N-glycosylated form of SERINC5 is specifically incorporated into HIV-1 virions. J Virol 92:e00753-18. doi: 10.1128/JVI.00753-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohno H, Stewart J, Fournier MC, Bosshart H, Rhee I, Miyatake S, Saito T, Gallusser A, Kirchhausen T, Bonifacino JS. 1995. Interaction of tyrosine-based sorting signals with clathrin-associated proteins. Science 269:1872–1875. doi: 10.1126/science.7569928. [DOI] [PubMed] [Google Scholar]
- 30.Traub LM. 2003. Sorting it out: AP-2 and alternate clathrin adaptors in endocytic cargo selection. J Cell Physiol 163:203–208. doi: 10.1083/jcb.200309175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pearse BM, Smith CJ, Owen DJ. 2000. Clathrin coat construction in endocytosis. Curr Opin Struct Biol 10:220–228. doi: 10.1016/S0959-440X(00)00071-3. [DOI] [PubMed] [Google Scholar]
- 32.Colicelli J, Goff SP. 1988. Sequence and spacing requirements of a retrovirus integration site. J Mol Biol 199:47–59. doi: 10.1016/0022-2836(88)90378-6. [DOI] [PubMed] [Google Scholar]
- 33.Tokunaga K, Kojima A, Kurata T, Ikuta K, Akari H, Koyama AH, Kawamura M, Inubushi R, Shimano R, Adachi A. 1998. Enhancement of human immunodeficiency virus type 1 infectivity by Nef is producer cell-dependent. J Gen Virol 79:2447–2453. doi: 10.1099/0022-1317-79-10-2447. [DOI] [PubMed] [Google Scholar]
- 34.Dang Y, Wang X, Esselman WJ, Zheng YH. 2006. Identification of APOBEC3DE as another antiretroviral factor from the human APOBEC family. J Virol 80:10522–10533. doi: 10.1128/JVI.01123-06. [DOI] [PMC free article] [PubMed] [Google Scholar]