Necroptosis is a programmed cell death that is inflammatory in nature owing to the release of danger-associated molecular patterns from the ruptured cell membrane. However, necroptosis also constitutes an important arm of host immune responses. Thus, a balanced inflammatory response determines the disease outcome. We report that the NS1 protein of IAV participates in necroptosis by interacting with MLKL, resulting in increased MLKL oligomerization and membrane translocation. These results reveal a novel function of the NS1 protein and the mechanism by which IAV induces necroptosis. Moreover, we show that this interaction enhances NLRP3 inflammasome activation and IL-1β processing and secretion. This information may contribute to a better understanding of the role of necroptosis in IAV-induced inflammation.
KEYWORDS: influenza A virus, MLKL, NS1 protein, inflammasome, necroptosis
ABSTRACT
Elimination of infected cells by programmed cell death is a well-recognized host defense mechanism to control the spread of infection. In addition to apoptosis, necroptosis is also one of the mechanisms of cell death that can be activated by viral infection. Activation of necroptosis leads to the phosphorylation of mixed-lineage kinase domain-like protein (MLKL) by receptor-interacting protein kinase 3 (RIPK3) and results in MLKL oligomerization and membrane translocation, leading to membrane disruption and a loss of cellular ion homeostasis. It has recently been reported that influenza A virus (IAV) infection induces necroptosis. However, the underlying mechanism of the IAV-mediated necroptosis process, particularly the roles of IAV proteins in necroptosis, remains unexplored. Here, we report that IAV infection induces necroptosis in macrophages and epithelial cells. We demonstrate that the NS1 protein of IAV interacts with MLKL. Coiled-coil domain 2 of MLKL has a predominant role in mediating the MLKL interaction with NS1. The interaction of NS1 with MLKL increases MLKL oligomerization and membrane translocation. Moreover, the MLKL-NS1 interaction enhances MLKL-mediated NLRP3 inflammasome activation, leading to increased interleukin-1β (IL-1β) processing and secretion.
IMPORTANCE Necroptosis is a programmed cell death that is inflammatory in nature owing to the release of danger-associated molecular patterns from the ruptured cell membrane. However, necroptosis also constitutes an important arm of host immune responses. Thus, a balanced inflammatory response determines the disease outcome. We report that the NS1 protein of IAV participates in necroptosis by interacting with MLKL, resulting in increased MLKL oligomerization and membrane translocation. These results reveal a novel function of the NS1 protein and the mechanism by which IAV induces necroptosis. Moreover, we show that this interaction enhances NLRP3 inflammasome activation and IL-1β processing and secretion. This information may contribute to a better understanding of the role of necroptosis in IAV-induced inflammation.
INTRODUCTION
Elimination of infected cells by programmed cell death is a well-recognized host defense mechanism to control the spread of infection. Recent studies involving programmed cell death have highlighted the fact that apart from apoptosis, other mechanisms of cell death may also contribute to host defense. Necroptosis is one such mechanism of cell death that can be activated by a number of factors, including bacterial and viral infections (1). Activation of the receptor-interacting protein (RIP) kinases (RIPKs), namely, RIPK1 and RIPK3, is central to necroptotic signaling. Both RIPK1 and RIPK3 possess a RIP homotypic interaction motif (RHIM). The interaction of RIPK1 and RIPK3 through the RHIM-RHIM interaction leads to the formation of a complex called the necrosome, resulting in the phosphorylation and activation of RIPK1 and RIPK3 (2, 3). The activation of RIPK3 leads to the phosphorylation of its substrate, mixed-lineage kinase domain-like protein (MLKL), which results in MLKL oligomerization and membrane translocation, leading to membrane disruption and a loss of cellular ion homeostasis (4–7). The oligomerization and membrane translocation of MLKL are mediated by the two coiled-coil domains of MLKL (4). Recent studies have shown that in addition to executing necroptosis, activated MLKL can also cause the activation of the NLRP3 inflammasome and the consequent processing and release of interleukin-1β (IL-1β) (8, 9).
A number of viruses have been reported to either induce or inhibit necroptosis (10). For example, murine cytomegalovirus (MCMV) employs its M45 protein (a RHIM domain-containing protein) to disrupt RHIM-RHIM interaction-dependent necroptosis signaling transduction and thus inhibit necroptosis in mouse cells (11). Similarly, herpes simplex virus 1 (HSV-1) and HSV-2 inhibit necroptosis in human cells by encoding ICP6 and ICP10 proteins, respectively, which inhibit the RHIM domain-dependent interaction of RIPK1 and RIPK3 (12). On the other hand, vaccinia virus has been reported to induce tumor necrosis factor alpha (TNF-α)-mediated necroptosis in both human and mouse cells (13, 14). Recent studies have also reported that influenza A virus (IAV) activates MLKL-dependent necroptosis (15–17). During IAV infection, the DNA-dependent activator of interferon (IFN)-regulatory factors (DAI) senses viral RNA and activates RIPK3, which leads to the activation of MLKL, resulting in necroptosis (15).
Although significant progress has been made in recent years in understanding the mechanism of IAV-mediated necroptosis in infected cells, the roles of various IAV proteins in necroptosis are still unclear. Here, we report that the NS1 protein of IAV participates in necroptosis by interacting with MLKL. NS1 is a multifunctional protein that is involved in the regulation of apoptotic cell death and host immune responses to ensure efficient virus replication and increased virulence during infection. It has been reported that NS1 interacts with the p85β subunit of phosphatidylinositol 3-kinase (PI3K) and activates the PI3K/AKT pathway. Phosphorylation of AKT leads to prolonged cell survival and thus increases virus replication (18–20). It has also been reported that as a result of its RNA binding activity, NS1 is able to inhibit double-stranded RNA-mediated protein kinase R (PKR) activation and the resultant IFN-β production (21). Similarly, the interaction of NS1 with the ubiquitin ligase TRIM25 has been implicated in the inhibition of RIG-I activation, which prevents IFN-β production (22). Moreover, we recently demonstrated that NS1 interacts with DDX3 and is able to counteract virus-induced stress granule formation and DDX3 localization in these stress granules (23). Here, we investigate the role of NS1 in mediating necroptosis and inflammation in immune cells. We demonstrate that the interaction of NS1 with MLKL increases MLKL oligomerization and membrane translocation. Moreover, the MLKL-NS1 interaction enhances NLRP3 inflammasome activation and increases IL-1β processing and secretion.
RESULTS
IAV infection induces necroptosis in macrophages and epithelial cells.
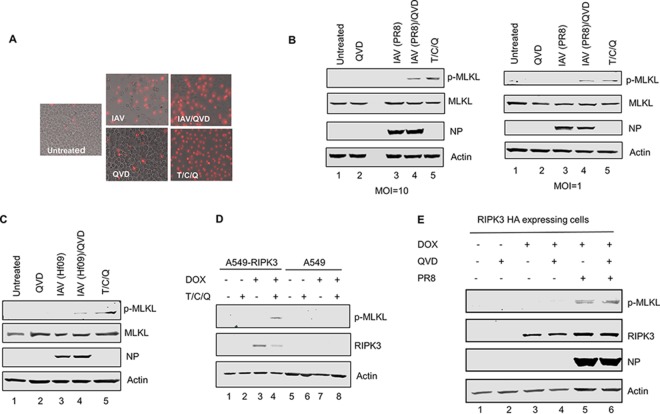
To examine if IAV infection can induce necroptosis in macrophages, we stained influenza A/Puerto Rico/8/34 (PR8) virus-infected (multiplicity of infection [MOI] of 1) differentiated THP1 cells with propidium iodide (PI) at 16 h postinfection (p.i.). Cells treated with a combination of TNF-α, cycloheximide, and the pancaspase inhibitor Q-VD-OPH (QVD) (T/C/Q) were used as a positive control. PI can penetrate necrotic but not apoptotic cells and hence is a good indicator of necroptotic cell death. As shown in Fig. 1A, PR8-infected and T/C/Q-treated cells showed increased PI uptake compared to uninfected cells, indicating an induction of necroptosis upon IAV infection. Moreover, the number of IAV-infected cells showing necroptosis increased when apoptosis was inhibited by the pancaspase inhibitor QVD.
FIG 1.
IAV infection induces necroptosis in THP1 cells and the A549 cell line inducibly expressing human RIPK3. (A) Representative images of differentiated THP1 cells that were either left untreated, infected with IAV (PR8) at an MOI of 1, treated with QVD, treated with QVD for 1 h and then infected with IAV, or treated with the combination of T/C/Q. Cell death was assessed by PI staining at 16 h p.i. (B) THP1 cells were infected with IAV (PR8) at the indicated MOIs or treated as described above for panel A, and at 8 h posttreatment, cells were harvested and examined for phosphorylated MLKL by Western blotting. (C) THP1 cells were infected with IAV (Hf09) at an MOI of 10 or treated as described above for panel A. At 8 h posttreatment, cells were harvested and examined for phosphorylated MLKL by Western blotting. (D) A549 cells and an A549 cell line inducibly expressing HA-tagged human RIPK3 were cultured in the absence or presence of 0.5 μg/ml of doxycycline (DOX) for 12 h and either left untreated or treated with the combination of T/C/Q (TNF-α, cycloheximide, and QVD). At 8 h posttreatment, cells were harvested and examined for RIPK3 and phosphorylated MLKL by Western blotting. (E) The A549 cell line inducibly expressing human RIPK3 was cultured in the absence or presence of 0.5 μg/ml of doxycycline for 12 h, and cells were either left untreated or treated with QVD for 1 h and then either mock infected or infected with IAV (PR8) at an MOI of 10. At 8 h p.i., cells were harvested and examined for phosphorylated MLKL by Western blotting.
Upon necroptosis induction, MLKL is phosphorylated at the threonine 357 (T357) and the serine 358 (S358) residues by RIPK3 (24). To further confirm that IAV infection induces necroptosis in THP1 cells, we examined the phosphorylation status of MLKL in IAV-infected THP1 cells by Western blotting. As seen in Fig. 1B (left), phosphorylation of MLKL can be observed in cells infected with IAV (PR8 at an MOI of 10) following pretreatment with QVD and in cells treated with T/C/Q (lanes 4 and 5) but not in mock-infected or QVD-treated cells (lanes 1 and 2). To further confirm that these results are not seen only with a high-MOI IAV infection, we infected THP1 cells with IAV at an MOI of 1 and examined the phosphorylation status of MLKL. As seen in Fig. 1B (right), phosphorylation of MLKL can be observed in cells infected with IAV following pretreatment with QVD and in cells treated with T/C/Q (lanes 4 and 5) but not in mock-infected or QVD-treated cells (lanes 1 and 2). To examine whether IAV-induced necroptosis in THP1 cells is not specific to PR8 virus, we infected THP1 cells with pandemic influenza A/Halifax/210/2009/H1N1 (Hf09) virus. Phosphorylation of MLKL can be observed in cells infected with IAV (Hf09) following pretreatment with QVD albeit at a lower level than with PR8 virus (Fig. 1C, lane 4). These results reinforce the observations that IAV infection can induce necroptosis in THP1 cells.
To extend our results to a physiologically relevant cell type, we decided to use A549 cells, a human lung epithelial cell line. However, necroptosis is repressed in A549 cells, as they are deficient in RIPK3 expression (25). We thus generated a stable A549 cell line, A549-RIPK3HA, that inducibly expresses human RIPK3. We first tested if supplementing RIPK3 in the A549 cell line would lead to the phosphorylation of MLKL. A549 and A549-RIPK3HA cells were cultured in the absence or the presence of 0.5 μg/ml of doxycycline for 12 h and either left untreated or treated with the combination of T/C/Q for 8 h. As seen in Fig. 1D, RIPK3 was detected in the A549-RIPK3HA cell line upon induction with doxycycline (lanes 3 and 4), whereas no RIPK3 expression could be detected from A549 cells (lanes 5 to 8) and A549-RIPK3HA cells that were not induced by doxycycline (lanes 1 and 2). Moreover, phosphorylation of MLKL can be observed following treatment with T/C/Q only in A549-RIPK3HA cells where RIPK3 expression was induced (lane 4). After confirming that the inducible A549-RIPK3HA cell line is competent in necroptosis pathway activation, we set out to see if IAV infection can activate necroptosis signaling in these cells. As seen in Fig. 1E, phosphorylation of MLKL can be observed in RIPK3-expressing cells that were infected with IAV with or without pretreatment with QVD (lanes 5 and 6). It cannot be detected in mock-infected cells regardless of the expression of RIPK3. These data thus confirm that IAV infection activates the necroptosis pathway not only in THP1 cells but also in A549 cells expressing RIPK3.
IAV-mediated cell death of THP1 cells involves both apoptosis and necroptosis.
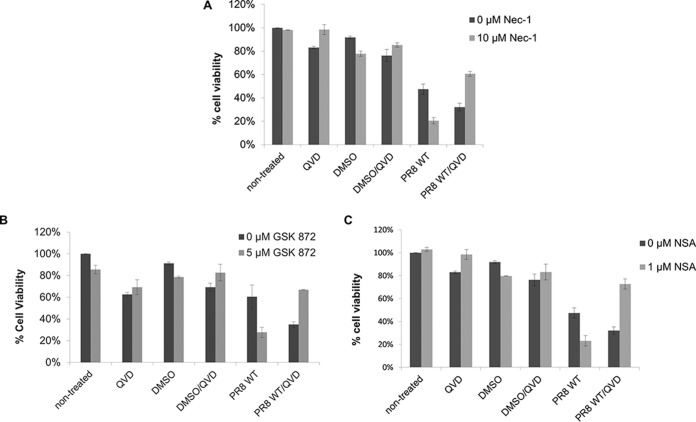
Since IAV infection induced necroptosis in THP1 cells, we hypothesized that the inhibition of necroptosis by the use of pharmaceutical inhibitors of key proteins involved in the necroptosis pathway would decrease IAV-induced cell death. As shown in Fig. 2, unexpectedly, inhibition of RIPK1 by Necrostatin-1 (Nec-1) (Fig. 2A), RIPK3 by GSK872 (Fig. 2B), or MLKL by necrosulfonamide (NSA) (Fig. 2C) in the absence of the pancaspase inhibitor QVD did not inhibit IAV-induced cell death; rather, it enhanced IAV-induced cell death. Also note that inhibition of caspase activity in the absence of a necroptosis inhibitor increased IAV-induced cell death, suggesting that inhibition of either cell death pathway may skew the cells toward an erroneous mode of cell death. In contrast, when both a caspase inhibitor and one of the necroptosis inhibitors were concomitantly used, IAV-induced cell death was significantly decreased. These results indicate that IAV-induced cell death of THP1 cells involves both apoptosis and necroptosis. A combined blockade of both pathways is therefore necessary for inhibiting IAV-mediated cell death. Furthermore, RIP1, RIPK3, and MLKL are all involved in IAV-induced necroptosis.
FIG 2.
IAV-induced cell death involves both apoptosis and necroptosis. Differentiated THP1 cells were either left untreated or treated for 1 h with the caspase inhibitor QVD (20 μM) and one of the following pharmacological inhibitors: Nec-1 (10 μM) for RIPK1 (A), GSK872 (5 μM) for RIPK3 (B), and NSA (1 μM) for MLKL (C). Following chemical treatment, the cells were either mock infected or infected with PR8 virus at an MOI of 5. At 18 to 20 h p.i., cell death was assessed by measuring the ATP concentration in cells using a Cell Titer-Glo luminescent viability assay.
The NS1 protein of IAV interacts with MLKL.
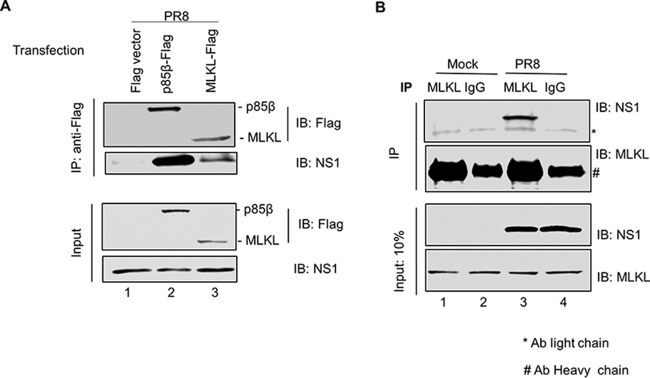
Viruses are known to modulate various cellular pathways by means of interactions of their proteins with cellular proteins. We hypothesized that one or more IAV proteins would interact with the key proteins involved in necroptosis. Since MLKL executes cell death during necroptosis, we decided to identify its interaction partners. To this end, we tested the interaction between Flag-tagged MLKL and viral proteins expressed during infection by coimmunoprecipitation (co-IP). HEK293T cells were transfected with either a Flag vector, Flag-tagged MLKL, or a Flag-tagged p85β plasmid (a known interactor of the viral NS1 protein) (19) followed by IAV infection at 24 h posttransfection. At 6 h p.i., the cell lysates were collected and immunoprecipitated with an antibody against the Flag tag. The precipitated proteins were then subjected to Western blotting with antibodies against the Flag tag and a panel of IAV proteins. Of these, we identified that the NS1 protein coprecipitated readily with Flag-MLKL (Fig. 3A, lane 3) and Flag-p85β (Fig. 3A, lane 2) but not with the Flag vector (Fig. 3A, lane 1). These data demonstrate that the viral protein NS1 interacts with MLKL during IAV infection.
FIG 3.
NS1 protein of IAV interacts with MLKL. (A) HEK293T cells were transfected with a Flag vector or the p85β-Flag or MLKL-Flag plasmid. At 24 h posttransfection, cells were infected with PR8 virus at an MOI of 10. Cell lysates were prepared at 6 h p.i. and subjected to IP with anti-Flag antibody. Precipitated proteins were subjected to Western blotting using antibodies against Flag and NS1 proteins. (B) Differentiated THP1 cells were either mock infected or infected with PR8 virus at an MOI of 10. Cell lysates were prepared at 8 h p.i. and subjected to IP with either normal rabbit IgG or anti-MLKL antibody (Ab). Precipitated proteins were subjected to Western blotting using antibodies against MLKL and NS1 proteins. IB, immunoblotting.
To further confirm that the interaction between NS1 and MLKL is not an artifact and is not because of the overexpression of MLKL, we determined the interaction of endogenous MLKL with NS1 by co-IP. THP1 cells were infected with IAV for 8 h, and the cell lysate was collected and immunoprecipitated with either an antibody against MLKL or normal rabbit IgG. The precipitated proteins were then subjected to Western blotting with antibodies against the MLKL and NS1 proteins. Consistent with the overexpression results, NS1 was coprecipitated readily by MLKL from the cell lysate of infected cells (Fig. 3B, lane 3). Moreover, NS1 could not be detected in pulldown samples when normal rabbit IgG was used for IP (Fig. 3B, lanes 2 and 4). These data thus confirm that NS1 indeed interacts with MLKL in THP1 cells during IAV infection.
Coiled-coil domain 2 of MLKL has a predominant role in mediating MLKL interaction with NS1.
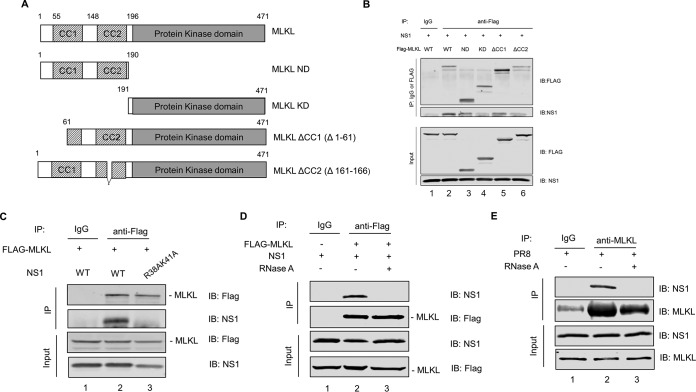
MLKL consists of two domains, including an N-terminal domain (ND) and a C-terminal pseudokinase domain (KD). The N-terminal domain is responsible for inducing necroptosis. Moreover, there are two coiled-coil motifs within the N-terminal domain. MLKL coiled-coil domain 1 (CC1) is responsible for MLKL membrane translocation, while coiled-coil domain 2 (CC2) is responsible for the oligomerization of MLKL (4, 6, 26). To delineate the region of MLKL involved in its interaction with NS1, we constructed Flag-tagged plasmids expressing different truncated MLKL mutants and examined their ability to interact with NS1. These include four truncation mutants expressing either the N-terminal domain (amino acids [aa] 1 to 190) (MLKL ND), the C-terminal domain (amino acids 191 to 471) (MLKL KD), MLKL with CC1 disrupted (amino acids 61 to 471) (MLKL ΔCC1), or MLKL with CC2 disrupted (amino acids 161 to 166 deleted) (MLKL ΔCC2) (Fig. 4A). To identify the domain of MLKL that was critical for the interaction with NS1, HEK293T cells were cotransfected with each of the Flag-tagged MLKL mutant plasmids and the plasmid expressing NS1. At 36 h posttransfection, the cell lysates were collected and subjected to co-IP. As seen in Fig. 4B, while the NS1 protein interacts with full-length MLKL and the N-terminal domain of MLKL (lanes 2 and 3), it does not interact with the C-terminal domain of MLKL (lane 4). Moreover, NS1 could still interact with MLKL when CC1 was disrupted (lane 5); however, the interaction was reduced when CC2 was disturbed in MLKL (lane 6). These data point to the fact that CC2 of MLKL has a predominant role in the interaction with NS1.
FIG 4.
CC2 of MLKL has a predominant role in mediating MLKL interaction with NS1. (A) Flag-tagged plasmids expressing different truncated MLKL proteins were constructed to identify the domains critical for the interaction of MLKL with the NS1 protein. A schematic representation of plasmids expressing mutant MLKL proteins is shown. (B) HEK293T cells were cotransfected with an NS1-expressing plasmid and either the Flag-tagged MLKL-expressing plasmid or one of the truncated MLKL-expressing plasmids. Cell lysates were prepared at 36 h posttransfection and subjected to IP with either normal rabbit IgG or anti-Flag antibody. Precipitated proteins were subjected to Western blotting using antibodies against the Flag tag or the NS1 protein. (C) HEK293T cells were cotransfected with the Flag-tagged MLKL-expressing plasmid and either the WT NS1- or NS1(R38A/K41A)-expressing plasmid. Cell lysates were prepared at 36 h posttransfection and subjected to IP with either normal rabbit IgG or anti-Flag antibody. Precipitated proteins were subjected to Western blotting using antibodies against the Flag tag and NS1 protein. (D) HEK293T cells were cotransfected with the Flag-tagged MLKL-expressing plasmid and NS1-expressing plasmid. At 36 h posttransfection, cell lysates were prepared, treated with RNase A at 10 μg/ml for 30 min at 4°C or left untreated, and subjected to IP with either normal rabbit IgG or anti-Flag antibody. Precipitated proteins were subjected to Western blotting using antibodies against the Flag tag and NS1 protein. (E) Differentiated THP1 cells were infected with PR8 virus at an MOI of 10. At 8 h p.i., cell lysates were prepared, treated with RNase A or left untreated, and subjected to IP with either normal rabbit IgG or anti-MLKL antibody. Precipitated proteins were subjected to Western blotting using antibodies against MLKL and NS1 proteins.
Additionally, we tested the interaction of MLKL with a mutant NS1 protein, NS1(R38A/K41A), which carries a replacement of the R38 and K41 amino acids by alanine. It is known that the replacement of the R38 and K41 amino acids by alanine in NS1 abolishes its RNA binding ability (27). We cotransfected plasmids expressing Flag-tagged MLKL and either the wild-type (WT) NS1 or the NS1(R38A/K41A) protein in HEK293T cells. At 36 h posttransfection, the cell lysates were collected and subjected to co-IP with the anti-Flag antibody. As seen in Fig. 4C, while WT NS1 coprecipitated readily with MLKL (lane 2), NS1(R38A/K41A) failed to interact with MLKL (lane 3). These data indicate that the R38/K41 site in NS1 is essential for the interaction with MLKL. To further investigate if the interaction between NS1 and MLKL is mediated by RNA, we cotransfected plasmids expressing NS1 and Flag-MLKL in HEK293T cells. The cell lysates were pretreated with RNase A before being subjected to IP with either Flag antibody or normal rabbit IgG. As seen in Fig. 4D, while NS1 interacted with MLKL when the cell lysate was not treated with RNase A (lane 2), the NS1-MLKL interaction was abrogated by RNase A treatment (lane 3), indicating that the NS1-MLKL interaction is RNA dependent. Furthermore, we infected THP1 cells with IAV for 8 h, and the cell lysates with or without RNase A treatment were subjected to IP with MLKL antibody. As seen in Fig. 4E, while NS1 interacted with MLKL when the cell lysate was not treated with RNase A (lane 2), the NS1-MLKL interaction was abrogated by RNase A treatment (lane 3), confirming that the NS1-MLKL interaction is indeed RNA dependent.
NS1 enhances MLKL oligomerization and membrane translocation.
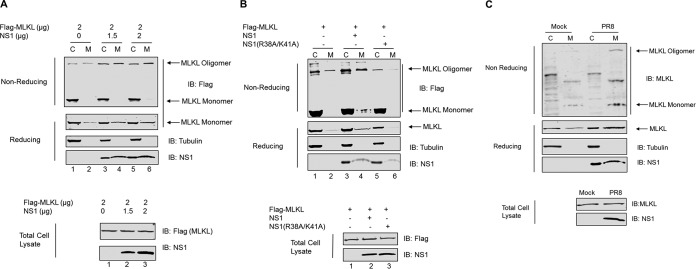
During necroptosis, MLKL is phosphorylated by RIPK3, which leads to its oligomerization and membrane translocation. The oligomerization and membrane translocation of MLKL are mediated by the two coiled-coil domains of MLKL (4). Since coiled-coil domain 2 of MLKL has a predominant role in mediating the MLKL interaction with NS1, we sought to determine if NS1 affects MLKL oligomerization and membrane translocation. For this, we used overexpression of MLKL in HEK293T cells as a model. We overexpressed MLKL in either the absence or the presence of increasing amounts of NS1 in HEK293T cells and separated the crude membrane and cytosolic fractions. MLKL protein expression was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) under reducing and nonreducing conditions. As seen in Fig. 5A, under nonreducing conditions, most of the MLKL protein detected in the crude membrane fraction was in the oligomer form, whereas most of the MLKL protein in the cytosolic fraction is in the monomer form. Moreover, the amount of MLKL present in the oligomer form increased when NS1 was concomitantly expressed (lane 4). The amount of MLKL in the oligomer form further slightly increased when NS1 was expressed in increasing amounts (lane 6). Similar results were observed when MLKL expression was analyzed by SDS-PAGE under reducing conditions. The amount of MLKL in the membrane fraction increased when NS1 was concomitantly expressed (lane 4, reducing gel). Interestingly, the amount of MLKL in the membrane fraction was also slightly increased when NS1 was expressed in increasing amounts (lane 6, reducing gel). The fractions were also probed with the antitubulin antibody to confirm the purity of the fractions; no tubulin could be detected in the membrane fractions. We also analyzed the whole-cell lysate and found that the total expression levels of MLKL were similar regardless of the presence of NS1 (Fig. 5A, bottom).
FIG 5.
NS1 enhances MLKL oligomerization and membrane translocation. (A) HEK293T cells were cotransfected with the Flag-tagged MLKL-expressing plasmid and increasing amounts of the NS1-expressing plasmid. At 36 h posttransfection, cell lysates were separated into the total, cytoplasmic (C), or membrane (M) fraction. The cell lysates were resolved on SDS-PAGE gels with (reducing) or without (nonreducing) β-mercaptoethanol and analyzed by Western blotting using the indicated antibodies. (B) HEK293T cells were cotransfected with the Flag-tagged MLKL-expressing plasmid and either the WT NS1- or NS1(R38A/K41A)-expressing plasmid. At 36 h posttransfection, cell lysates were harvested and subjected to assays as described above for panel A. (C) Differentiated THP1 cells were either mock infected or infected with PR8 virus at an MOI of 10. At 8 h p.i., cell lysates were harvested and subjected to assays as described above for panel A.
To further confirm that the observed increases in the oligomerization and membrane translocation of MLKL in the presence of NS1 resulted from the interaction between MLKL and NS1, we analyzed the oligomerization state and membrane translocation of MLKL in the presence of NS1(R38A/K41A), which did not interact with MLKL. We again overexpressed MLKL alone or together with either WT NS1 or NS1(R38A/K41A) in HEK293T cells and separated the crude membrane and cytosolic fractions. MLKL protein expression was analyzed by SDS-PAGE under both reducing and nonreducing conditions (Fig. 5B). We consistently observed that under nonreducing conditions, the amount of MLKL present in the oligomer form increased when WT NS1 was supplemented (lane 4). However, when NS1(R38A/K41A) was expressed together with MLKL, there was no increase in the amount of oligomerized MLKL (lane 6). Similarly, under reducing conditions, the amount of MLKL present in the membrane fraction increased when WT NS1 was provided (lane 4 of the reducing gel). In contrast, NS1(R38A/K41A) did not give rise to an increased amount of MLKL in the membrane fraction (lane 6).
To investigate if MLKL oligomerization and membrane translocation are altered upon IAV infection, THP1 cells were infected with IAV, and MLKL protein expression was analyzed by SDS-PAGE under reducing and nonreducing conditions (Fig. 5C). We observed that under nonreducing conditions, more MLKL was present in the oligomer form in the membrane fraction of infected cells than in mock-infected cells (lanes 2 and 4). Similarly, under reducing conditions, more MLKL could be detected in the membrane fraction of infected cells than in mock-infected THP1 cells. Taken together, these results indicate that the interaction of NS1 with MLKL leads to increased oligomerization and membrane translocation of MLKL during IAV infection.
Interaction of NS1 with MLKL enhances MLKL-mediated NLRP3 inflammasome activation and IL-1β processing and secretion.
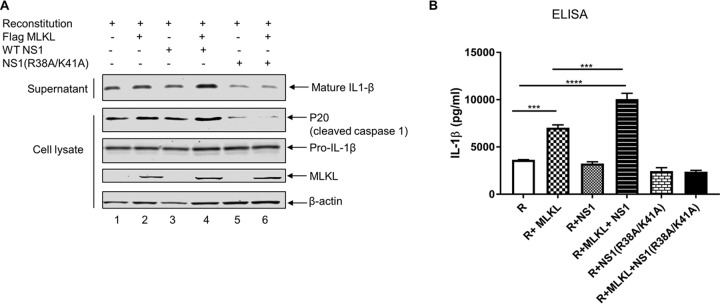
In addition to necroptosis, activated MLKL also causes the activation of the NLRP3 inflammasome and the consequent processing and release of IL-1β (8, 9). Since we observed that NS1 increases MLKL oligomerization and membrane translocation, we hypothesized that NS1 would increase MLKL-mediated NLRP3 inflammasome activation and IL-1β processing and secretion. To test this, we took advantage of the NLRP3 reconstitution system in HEK293T cells, as we described previously (28). As seen in Fig. 6A, cotransfection of all four plasmids expressing constituents of the NLRP3 inflammasome led to the secretion of IL-1β in the supernatant (lane 1). While the pro-IL-1β level remained the same in all samples, the secretion of IL-1β was slightly increased in the supernatant when MLKL was coexpressed together with constituents of the NLRP3 inflammasome (lane 2). Moreover, the secretion of IL-1β was significantly increased when NS1 was coexpressed together with MLKL and the constituents of the NLRP3 inflammasome (lane 4). No such increase in IL-1β secretion was observed when WT NS1 or NS1(R38A/K41A) was coexpressed alone with the constituents of the NLRP3 inflammasome (lanes 3 and 5, respectively). Similarly, no increase in IL-1β secretion was observed when NS1(R38A/K41A) was coexpressed together with MLKL and constituents of the NLRP3 inflammasome (lane 6). To further confirm our results, we measured IL-1β protein levels in the supernatants of transfected cells by an enzyme-linked immunosorbent assay (ELISA). In agreement with the Western blotting results, the secretion of IL-1β was increased when MLKL was coexpressed with constituents of the NLRP3 inflammasome. The coexpression of NS1 together with MLKL and the constituents of the NLRP3 inflammasome further increased the secretion of pro-IL-1β (Fig. 6B).
FIG 6.
Interaction of NS1 with MLKL enhances MLKL-mediated NLRP3 inflammasome activation and IL-1β processing and secretion. (A) HEK293T cells were transfected with plasmids expressing NLRP3 inflammasome components and pro-IL-1β along with the indicated plasmids expressing null (empty vector), MLKL, NS1, or NS1(R38A/K41A). The supernatant and cell lysates were collected at 20 h posttransfection and analyzed by Western blotting with the indicated antibodies. (B) HEK293T cells were transfected as described above for panel A, and IL-1β levels in the supernatants of the above-described samples were measured by an ELISA. R, reconstitution (pcDNA-NLRP3 plus pcDNA-ASC plus pCMV-Flag-pro-caspase 1 plus pcDNA-pro-IL-1β). ***, P < 0.001; ****, P < 0.0001. Results are representative of data from three independent experiments performed in duplicates.
DISCUSSION
IAV has the ability to activate parallel pathways of apoptotic and necroptotic cell death in fibroblasts and respiratory epithelial cells (16). In this study, we demonstrate that IAV infection induces necroptosis in THP1 cells and RIPK3-expressing A549 cells. To further understand the cell death pathways activated by IAV in THP1 cells, we used pharmaceutical inhibitors to inhibit necroptosis and apoptosis. A dramatic decrease in IAV-mediated cell death could be observed only when the pancaspase inhibitor QVD was used along with either the RIPK1 inhibitor, the RIPK3 inhibitor, or the MLKL inhibitor, thus indicating that IAV activates both apoptotic and necroptotic cell death pathways in THP1 cells.
Apoptotic signaling during IAV infection has been extensively studied, and the roles of various host and IAV proteins in manipulation of the apoptotic pathway are well understood (29). Similarly, a number of recent studies have demonstrated that IAV induces necroptosis (15–17), and significant progress has been made in understanding the mechanism of IAV-mediated necroptotic cell death and the roles played by the various cellular proteins involved. However, there is still a gap in the understanding of the roles played by various IAV proteins during necroptosis. In our quest for IAV proteins that participate in necroptosis, we identified that the NS1 protein of IAV directly interacts with MLKL. MLKL, the effector of the necroptotic pathway, is a pseudokinase that contains an N-terminal four-helix bundle domain and a C-terminal pseudokinase domain. The killer function of MLKL has been attributed to the N-terminal four-helix bundle domain (7, 30). RIPK3-mediated phosphorylation of MLKL leads to a conformational change in MLKL resulting in the unleashing of the N-terminal effector domain (7). Activated MLKL then oligomerizes and translocates to the plasma membrane, where it causes pore formation, cellular swelling, and, eventually, cell death (4, 5). N-terminal CC1 and CC2 of MLKL have been implicated in membrane translocation and oligomerization of MLKL, respectively (4). MLKL oligomerization and membrane translocation are hallmarks of the necroptosis pathway; however, the underlying mechanisms regulating these events are not fully understood. Several cellular factors have been hypothesized to regulate MLKL oligomerization and membrane trafficking. Heat shock protein 90 (HSP90) has been shown to stabilize MLKL and prevent MLKL degradation through the proteasomal pathway. Moreover, HSP90, when overexpressed in HEK293T cells, increases MLKL oligomerization (31, 32). Similarly, it has been hypothesized that phosphatidylinositol transfer protein alpha (PITPα) recruits MLKL to the plasma membrane through its interaction with MLKL (33). In our study, by using a co-IP assay in IAV-infected as well as NS1-transfected cells, we identified that NS1 interacts with MLKL in an RNA-dependent manner and that the CC2 region of MLKL is responsible for interacting with NS1. Moreover, we demonstrated that the interaction of NS1 with MLKL increases MLKL oligomerization and membrane translocation. While the mechanism by which NS1 increases MLKL oligomerization and membrane translocation requires further investigation, it is tempting to speculate that NS1 provides a platform where several MLKL molecules can interact to form oligomers. Another possibility is that the interaction of MLKL with NS1 induces a conformational change in MLKL predisposing it to oligomerization. NS1 may also induce a conformational change in MLKL that enhances or impedes its interaction with other regulatory proteins. The phosphorylation of MLKL by RIPK3 triggers a conformational change in MLKL leading to its transition from an inactive to an activated state. The interaction of NS1 with MLKL might increase the susceptibility of MLKL to RIPK3-mediated phosphorylation.
Activated MLKL also causes an activation of the NLRP3 inflammasome and the consequent processing and release of IL-1β, which correlate with MLKL oligomerization and translocation (8, 9). Since the NS1-MLKL interaction led to increased MLKL oligomerization and translocation to the plasma membrane, we further examined if the MLKL-mediated activation of the NLRP3 inflammasome and IL-1β production was modulated by NS1. As expected, our results indicate that the interaction of NS1 with MLKL enhances MLKL-mediated NLRP3 inflammasome activation and IL-1β processing. Various RNA viruses, including IAV, vesicular stomatitis virus (VSV), respiratory syncytial virus (RSV), Zika virus, and dengue virus, have been reported to activate the NLRP3 inflammasome (28, 34, 35). The activated NLRP3 inflammasome can be beneficial to the host, as it can lead to an enhancement of antiviral defense resulting in virus clearance and resolution of tissue damage. For example, Nogusa et al. reported that IAV infection triggers RIPK3, which then activates both MLKL-driven necroptosis and Fas-associated death domain protein (FADD)-mediated apoptosis to protect the host against IAV infection (16). On the other hand, inflammasome activation can also be beneficial to the virus, as it can increase viral pathogenesis and tissue damage and help in the dissemination of the virus. Whether NLRP3 inflammasome activation will be beneficial to the host or virus is dependent upon the magnitude of NLRP3 activation and the stage of viral infection when the NLRP3 inflammasome is activated (34–36). A recent study reassessed the role of the NLRP3 inflammasome in IAV pathogenesis by using the NLRP3-specific inhibitor MCC950 at different stages of infection (37). It was reported that inhibition of NLRP3 activation during the early phase of infection (1 day postchallenge) resulted in increased mortality in mice. In contrast, inhibition of NLRP3 at a later stage (day 3 or 7 postchallenge) protected mice from IAV-induced death owing to decreased levels of proinflammatory cytokines and a reduction in the recruitment of inflammatory cells in the airways. Thus, these finding suggested that NLRP3 plays a detrimental role later in IAV infection. The contribution of the inflammasome to virus pathogenesis was also reported for other viruses. For example, one recent study using mice reported that the NLRP3 inflammasome is activated following Zika virus infection and that IL-1β secretion and a strong inflammatory response following the activation of the NLRP3 inflammasome are major factors contributing to Zika virus pathogenicity (38). Similarly, one of the major causes of increased vascular permeability and hemoconcentration in dengue fever patients is the shedding of IL-1β-containing microparticles, which are generated as a result of NLRP3 inflammasome activation in platelets by dengue virus (39). Furthermore, NLRP3 inflammasome activation following severe acute respiratory syndrome coronavirus (SARS-CoV) infection leads to excessive IL-1β production, resulting in pulmonary damage, edema, and death (40). A similar association between the activation of NLRP3 and increased pathogenesis has been reported for HIV-1 (41), hepatitis C virus (HCV) (42, 43), and Sindbis virus (44). Our finding that NS1 of IAV interacts with MLKL, resulting in inflammasome activation and increased IL-1β processing and release, may fit into the finding that NS1 activates necroptosis and inflammasome pathways to benefit IAV pathogenesis through excessive IL-1β production and a strong inflammatory response.
In conclusion, our study demonstrates that the IAV NS1 protein participates in the necroptosis process by interacting with MLKL. This interaction results in increased MLKL oligomerization and membrane translocation, which consequently increases MLKL-mediated NLRP3 inflammasome activation and IL-1β production. Collectively, these data provide insights into the critical role of NS1 in regulating pathogenesis and immunopathology during IAV infection by regulation of cell death and inflammatory responses.
MATERIALS AND METHODS
Cells and viruses.
Human embryonic kidney HEK293T and A549 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Sigma) supplemented with 10% fetal bovine serum (FBS; Sigma). THP1 cells were cultured in RPMI 1640 medium (Gibco) supplemented with 10% FBS and 0.05 mM β-mercaptoethanol (Gibco). Phorbol 12-myristate 13-acetate (PMA; Sigma-Aldrich) was added to the THP1 culture (100 ng/ml) at least overnight to differentiate THP1 cells into macrophage-like cells. The A549 cell line inducibly expressing the hemagglutinin (HA)-tagged human RIPK3 protein was cultured in DMEM supplemented with 10% tetracycline-free FBS (Clontech) and 2 μg/ml puromycin (Invivogen).
Influenza A/Puerto Rico/8/34 (PR8) virus was propagated in 11-day-old embryonated chicken eggs as described previously (18). Influenza A/Halifax/210/2009/H1N1 (Hf09) virus was grown in MDCK cells as described previously (28).
Antibodies and reagents.
Rabbit polyclonal NS1 and NP antibodies were generated in our laboratory as previously described (18). The following antibodies were purchased from different commercial sources: rabbit anti-MLKL (58-70) (catalog number M6697; Sigma), rabbit monoclonal (EPR17514) anti-MLKL (catalog number ab184718; Abcam), rabbit monoclonal anti-MLKL (phospho-S358) antibody (EPR9514) (catalog number ab187091; Abcam), rabbit monoclonal anti-RIP3 (catalog number 13526; Cell Signaling), mouse monoclonal anti-RIP1 (catalog number sc-133102; Santa Cruz), mouse monoclonal anti-Flag (catalog number F1804; Sigma), rabbit anti-Flag DYKDDDDK tag (catalog number 2368S; Cell Signaling), mouse monoclonal antibody to β-actin (Cell Signaling), goat polyclonal anti-IL-1β antibody (catalog number BAF681; R&D Systems), rabbit polyclonal anti-caspase 1 (p20) antibody (catalog number PAB592Po01; Cloud-Clone Corp.), mouse monoclonal anti-porcine IL-1β antibody (catalog number MAB6811; R&D Systems), and goat anti-porcine IL-1β biotinylated antibody (catalog number BAF681; R&D Systems). The following secondary antibodies were purchased from commercial sources: IRDye 680RD anti-rabbit antibody (Li-Cor), IRDye 800CW anti-mouse antibody (Li-Cor), and goat anti-rabbit IgG secondary antibody.
The reagents used in this study include recombinant human TNF-α (catalog number 210-TA-020; R&D Systems), porcine IL-1β protein (catalog number 681-PI-010; R&D Systems), alkaline phosphatase-streptavidin (catalog number 016-050-084; Jackson ImmunoResearch), propidium iodide (catalog number 638; ImmunoChemistry), cycloheximide (catalog number C4859; Sigma), Halt protease and phosphatase inhibitor cocktail (catalog number 1861281; Thermo Scientific), the pancaspase inhibitor Q-VD-OPH (Sigma), the RIPK1 inhibitor Necrostatin-1 (Nec-1; Enzo Life Science), the MLKL inhibitor necrosulfonamide (NSA; EMD Millipore), and the RIPK3 inhibitor GSK872 (Biovision).
Plasmid construction.
Plasmid expressing wild-type PR8 NS1 (pcDNA-PR8-NS1) and mutant NS1(R38A/K41A) were previously described (23). Similarly, plasmids pcDNA-NLRP3, pCMV-Flag-pro-caspase1, pcDNA-pro-IL-1β, and pcDNA-ASC were previously described (28). The complete MLKL open reading frame (ORF) was amplified by reverse transcription-PCR (RT-PCR) using the total mRNA from THP1 cells as the template and was cloned into plasmid pCMV-3×Flag. The resulting plasmid was designated pCMV-3×Flag-hMLKL. Truncated MLKL with amino acids (aa) 1 to 190, 191 to 471, or 61 to 471 was cloned into the pCMV-3×Flag vector, generating pCMV-3×Flag-MLKL-ND (aa 1 to 190), pCMV-3×Flag-MLKL-KD (aa 191 to 471), and pCMV-3×Flag-MLKL ΔCC1 (aa 61 to 471), respectively. Similarly, plasmid pCMV-3×Flag-MLKL ΔCC2 was generated by deleting aa 161 to 166 from full-length MLKL and then cloned into pCMV-3×Flag. The hRIP3-GFP (human RIP3 fused with GFP) plasmid was obtained from Addgene (plasmid 41387) and was used as the template to amplify and clone the complete human RIPK3 ORF into plasmid pcDNA-3×HA. The resulting plasmid was designated pcDNA-hRIPK3-3×HA and was used to amplify and clone the complete human RIPK3 ORF along with the HA tag in the Tet-on-inducible vector pTripZ to generate plasmid pTripZ-hRIPK3HA.
Generation of A549 cells inducibly expressing HA-tagged human RIPK3.
For the generation of lentivirus, HEK293T cells were cotransfected with plasmids pTripZ‐hRIPK3HA, psPAX2, and pMD2.G. After 48 h posttransfection, the lentivirus-containing medium was collected and passed through a 0.45-μm filter. The A549 cells were treated with 8 μg/ml of Polybrene (Sigma) for 15 min and transduced with the lentivirus. After 24 h posttransduction, the cells were subjected to puromycin selection by using selection medium containing puromycin (Invivogen) at a concentration of 2 μg/ml. The selection medium was changed every 3 days until the appearance of puromycin-resistant clones. The stable cell lines were characterized by examining the expression of HA-tagged RIPK3 protein by Western blotting after culturing them in the presence of 0.5 μg/ml of doxycycline (Sigma) for 24 h.
Transfection and immunoprecipitation.
Cells seeded into six-well plates were transfected with 1 μg of each plasmid expressing the protein of interest using the TransIT-LT1 transfection reagent (catalog number MIR2300; Mirus Bio) according to the manufacturer’s instructions. At the indicated time points posttransfection, the cell lysates were prepared using a co-IP lysis buffer. For co-IP, 1.5 μg of the indicated antibody was added to the cell lysate and incubated overnight with gentle rocking at 4°C. On the next day, 40 μl of Dynabeads-protein G (Life technologies) was added to the lysate and incubated for 2 h with gentle rocking at 4°C. The beads were subsequently washed 5 times with the co-IP lysis buffer, and the immunoprecipitated proteins were subjected to SDS-PAGE and Western blotting with the indicated antibodies.
Western blotting.
Cell lysates or co-IP samples were resolved by SDS-PAGE and transferred onto nitrocellulose membranes. The membranes were blocked with 5% skim milk in Tris-buffered saline (TBS) with 0.1% Tween 20 (TBST) for 1 h and then incubated with primary antibodies at 4°C overnight. The membranes were washed with TBST and incubated with the secondary antibodies at room temperature for 1 h. The membranes were visualized in an Odyssey infrared imager (Li-Cor Biosciences). All the experiments were repeated at least three times.
Propidium iodide staining.
THP1 cells (1 × 105) were differentiated in 8-well chamber slides (Nunc Lab-Tek II CC2 chamber slide system) with 100 ng/ml PMA at least overnight. Cells were either left untreated or treated with 20 μM QVD for 1 h and then infected with PR8 virus at a multiplicity of infection (MOI) of 1. Cells treated with a combination of TNF-α (100 ng/ml), QVD (20 μM), and cycloheximide (10 μg/ml) were used as a positive control. Sixteen hours after treatment or infection, the treated or infected THP1 cells were stained with PI in sample medium at 0.5% (vol/vol) for 10 min at room temperature. The stained cells were visualized using a fluorescence microscope with excitation at 488 nm and emission at 635 nm (Leica CTR6000, N3 filter). Data shown are representative of results from three independent experiments with triplicates.
Analysis of cell viability.
THP1 cells (3 × 104) were differentiated in 96-well plates (catalog number 6005580; PerkinElmer) with 100 ng/ml PMA overnight. The cells were either left untreated or treated with the indicated chemicals for 1 h. Following chemical treatment, the cells were either mock infected or infected with the PR8 virus. At 18 to 20 h postinfection (p.i.), the ATP concentration in cells was measured using a Cell Titer-Glo luminescent cell viability assay kit according to the manufacturer’s instructions (Promega, Madison, WI). Briefly, 100 μl of Cell Titer-Glo reagent was added to each well, and the plate was shaken orbitally for 2 min at room temperature. Afterwards, the plate was placed in a microplate reader for 10 min to stabilize the luminescence signal. The signal was read in a Victor3 V 1420 multilabel counter (PerkinElmer). Control wells with only the medium and untreated cells were used to obtain the background and maximal luminescence. Data are shown as means ± standard deviations (SD) from three independent experiments performed in triplicate.
NLRP3 inflammasome reconstitution assay.
A porcine NLRP3 inflammasome reconstitution assay was conducted as described previously (28). Briefly, 1.8 × 105 HEK293T cells were seeded per well of a 24-well plate and cotransfected with pcDNA-NLRP3 (30 ng), pcDNA-ASC (20 ng), pCMV-Flag-pro-caspase-1 (20 ng), and pcDNA-pro-IL-1β (100 ng) along with a plasmid expressing either human MLKL (300 ng), WT NS1 (300 ng), or NS1(R38A/K41A) (300 ng), alone or in combination, as indicated. At 20 h posttransfection, the cell lysates were prepared and analyzed by Western blotting, while the supernatants were harvested and analyzed by an IL-1β ELISA and Western blotting.
IL-1β ELISA.
The IL-1β ELISA for supernatants from HEK293T cells (reconstitution system) was performed as previously described (28). Briefly, the mouse monoclonal anti-porcine IL-1β antibody in phosphate-buffered saline (PBS) was used to coat Immulon 2 HB U plates (Thermo Fisher) overnight at room temperature. The plates were washed four times with TBST between each step. Blocking was conducted for 1 h at room temperature using 1% bovine serum albumin (BSA). Twofold serial dilutions of recombinant porcine IL-1β protein in diluent were used as the standard. The plates were incubated with either samples or the standard for 2 h. Next, the plates were incubated with a goat anti-porcine IL-1β biotinylated antibody for 1 h, followed by incubation with alkaline phosphatase-streptavidin for another 1 h. p-Nitrophenyl phosphate in diethanolamine buffer was then added for color development, and the optical densities were measured at 405 nm with reference at 490 nm in an xMark microplate absorbance spectrophotometer (Bio-Rad). The experiments were conducted at least three times in duplicates for each sample.
Fractionation and MLKL oligomerization.
Cytosolic and membrane fractionation for analysis of MLKL oligomerization was performed as previously described (7). Briefly, transfected HEK293T cells or mock-infected or infected THP1 cells were harvested and permeabilized in a permeabilization buffer containing 0.025% digitonin and protease inhibitors. The samples were then centrifuged at 11,000 × g for 5 min to separate the cytosolic and crude membrane fractions. The crude membrane fraction was further solubilized in permeabilization buffer with 1% digitonin and clarified by centrifugation. The cytosolic and membrane fractions were then resolved on SDS-PAGE gels under either reducing (with β-mercaptoethanol) or nonreducing (without β-mercaptoethanol) conditions.
Statistical analysis.
The data were analyzed using GraphPad Prism 7 by one-way analysis of variance (ANOVA) with Tukey’s multiple-comparison test. The bars indicate the means ± SD. The data shown are representative of results from three independent experiments performed in duplicates unless otherwise indicated. A P value of less than 0.05 was considered to be statistically significant.
ACKNOWLEDGMENT
This work was supported by a grant from the Natural Sciences and Engineering Research Council of Canada (NSERC) to Y.Z.
REFERENCES
- 1.Sridharan H, Upton JW. 2014. Programmed necrosis in microbial pathogenesis. Trends Microbiol 22:199–207. doi: 10.1016/j.tim.2014.01.005. [DOI] [PubMed] [Google Scholar]
- 2.Galluzzi L, Vanden Berghe T, Vanlangenakker N, Buettner S, Eisenberg T, Vandenabeele P, Madeo F, Kroemer G. 2011. Programmed necrosis from molecules to health and disease. Int Rev Cell Mol Biol 289:1–35. doi: 10.1016/B978-0-12-386039-2.00001-8. [DOI] [PubMed] [Google Scholar]
- 3.Li J, McQuade T, Siemer AB, Napetschnig J, Moriwaki K, Hsiao YS, Damko E, Moquin D, Walz T, McDermott A, Chan FK, Wu H. 2012. The RIP1/RIP3 necrosome forms a functional amyloid signaling complex required for programmed necrosis. Cell 150:339–350. doi: 10.1016/j.cell.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cai Z, Jitkaew S, Zhao J, Chiang HC, Choksi S, Liu J, Ward Y, Wu LG, Liu ZG. 2014. Plasma membrane translocation of trimerized MLKL protein is required for TNF-induced necroptosis. Nat Cell Biol 16:55–65. doi: 10.1038/ncb2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang H, Sun L, Su L, Rizo J, Liu L, Wang LF, Wang FS, Wang X. 2014. Mixed lineage kinase domain-like protein MLKL causes necrotic membrane disruption upon phosphorylation by RIP3. Mol Cell 54:133–146. doi: 10.1016/j.molcel.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 6.Dondelinger Y, Declercq W, Montessuit S, Roelandt R, Goncalves A, Bruggeman I, Hulpiau P, Weber K, Sehon CA, Marquis RW, Bertin J, Gough PJ, Savvides S, Martinou JC, Bertrand MJ, Vandenabeele P. 2014. MLKL compromises plasma membrane integrity by binding to phosphatidylinositol phosphates. Cell Rep 7:971–981. doi: 10.1016/j.celrep.2014.04.026. [DOI] [PubMed] [Google Scholar]
- 7.Hildebrand JM, Tanzer MC, Lucet IS, Young SN, Spall SK, Sharma P, Pierotti C, Garnier JM, Dobson RC, Webb AI, Tripaydonis A, Babon JJ, Mulcair MD, Scanlon MJ, Alexander WS, Wilks AF, Czabotar PE, Lessene G, Murphy JM, Silke J. 2014. Activation of the pseudokinase MLKL unleashes the four-helix bundle domain to induce membrane localization and necroptotic cell death. Proc Natl Acad Sci U S A 111:15072–15077. doi: 10.1073/pnas.1408987111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conos SA, Chen KW, De Nardo D, Hara H, Whitehead L, Nunez G, Masters SL, Murphy JM, Schroder K, Vaux DL, Lawlor KE, Lindqvist LM, Vince JE. 2017. Active MLKL triggers the NLRP3 inflammasome in a cell-intrinsic manner. Proc Natl Acad Sci U S A 114:E961–E969. doi: 10.1073/pnas.1613305114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gutierrez KD, Davis MA, Daniels BP, Olsen TM, Ralli-Jain P, Tait SW, Gale M Jr, Oberst A. 2017. MLKL activation triggers NLRP3-mediated processing and release of IL-1beta independently of Gasdermin-D. J Immunol 198:2156–2164. doi: 10.4049/jimmunol.1601757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Upton JW, Shubina M, Balachandran S. 2017. RIPK3-driven cell death during virus infections. Immunol Rev 277:90–101. doi: 10.1111/imr.12539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Upton JW, Kaiser WJ, Mocarski ES. 2010. Virus inhibition of RIP3-dependent necrosis. Cell Host Microbe 7:302–313. doi: 10.1016/j.chom.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo H, Omoto S, Harris PA, Finger JN, Bertin J, Gough PJ, Kaiser WJ, Mocarski ES. 2015. Herpes simplex virus suppresses necroptosis in human cells. Cell Host Microbe 17:243–251. doi: 10.1016/j.chom.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cho YS, Challa S, Moquin D, Genga R, Ray TD, Guildford M, Chan FK. 2009. Phosphorylation-driven assembly of the RIP1-RIP3 complex regulates programmed necrosis and virus-induced inflammation. Cell 137:1112–1123. doi: 10.1016/j.cell.2009.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan FK, Shisler J, Bixby JG, Felices M, Zheng L, Appel M, Orenstein J, Moss B, Lenardo MJ. 2003. A role for tumor necrosis factor receptor-2 and receptor-interacting protein in programmed necrosis and antiviral responses. J Biol Chem 278:51613–51621. doi: 10.1074/jbc.M305633200. [DOI] [PubMed] [Google Scholar]
- 15.Thapa RJ, Ingram JP, Ragan KB, Nogusa S, Boyd DF, Benitez AA, Sridharan H, Kosoff R, Shubina M, Landsteiner VJ, Andrake M, Vogel P, Sigal LJ, tenOever BR, Thomas PG, Upton JW, Balachandran S. 2016. DAI senses influenza A virus genomic RNA and activates RIPK3-dependent cell death. Cell Host Microbe 20:674–681. doi: 10.1016/j.chom.2016.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nogusa S, Thapa RJ, Dillon CP, Liedmann S, Oguin TH III, Ingram JP, Rodriguez DA, Kosoff R, Sharma S, Sturm O, Verbist K, Gough PJ, Bertin J, Hartmann BM, Sealfon SC, Kaiser WJ, Mocarski ES, Lopez CB, Thomas PG, Oberst A, Green DR, Balachandran S. 2016. RIPK3 activates parallel pathways of MLKL-driven necroptosis and FADD-mediated apoptosis to protect against influenza A virus. Cell Host Microbe 20:13–24. doi: 10.1016/j.chom.2016.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuriakose T, Man SM, Subbarao Malireddi RK, Karki R, Kesavardhana S, Place DE, Neale G, Vogel P, Kanneganti TD. 2016. ZBP1/DAI is an innate sensor of influenza virus triggering the NLRP3 inflammasome and programmed cell death pathways. Sci Immunol 1:aag2045. doi: 10.1126/sciimmunol.aag2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shin YK, Liu Q, Tikoo SK, Babiuk LA, Zhou Y. 2007. Influenza A virus NS1 protein activates the phosphatidylinositol 3-kinase (PI3K)/Akt pathway by direct interaction with the p85 subunit of PI3K. J Gen Virol 88:13–18. doi: 10.1099/vir.0.82419-0. [DOI] [PubMed] [Google Scholar]
- 19.Li Y, Anderson DH, Liu Q, Zhou Y. 2008. Mechanism of influenza A virus NS1 protein interaction with the p85beta, but not the p85alpha, subunit of phosphatidylinositol 3-kinase (PI3K) and up-regulation of PI3K activity. J Biol Chem 283:23397–23409. doi: 10.1074/jbc.M802737200. [DOI] [PubMed] [Google Scholar]
- 20.Hale BG, Jackson D, Chen YH, Lamb RA, Randall RE. 2006. Influenza A virus NS1 protein binds p85beta and activates phosphatidylinositol-3-kinase signaling. Proc Natl Acad Sci U S A 103:14194–14199. doi: 10.1073/pnas.0606109103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Donelan NR, Basler CF, Garcia-Sastre A. 2003. A recombinant influenza A virus expressing an RNA-binding-defective NS1 protein induces high levels of beta interferon and is attenuated in mice. J Virol 77:13257–13266. doi: 10.1128/JVI.77.24.13257-13266.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gack MU, Albrecht RA, Urano T, Inn KS, Huang IC, Carnero E, Farzan M, Inoue S, Jung JU, Garcia-Sastre A. 2009. Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I. Cell Host Microbe 5:439–449. doi: 10.1016/j.chom.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thulasi Raman SN, Liu G, Pyo HM, Cui YC, Xu F, Ayalew LE, Tikoo SK, Zhou Y. 2016. DDX3 interacts with influenza A virus NS1 and NP proteins and exerts antiviral function through regulation of stress granule formation. J Virol 90:3661–3675. doi: 10.1128/JVI.03010-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun L, Wang H, Wang Z, He S, Chen S, Liao D, Wang L, Yan J, Liu W, Lei X, Wang X. 2012. Mixed lineage kinase domain-like protein mediates necrosis signaling downstream of RIP3 kinase. Cell 148:213–227. doi: 10.1016/j.cell.2011.11.031. [DOI] [PubMed] [Google Scholar]
- 25.Koo GB, Morgan MJ, Lee DG, Kim WJ, Yoon JH, Koo JS, Kim SI, Kim SJ, Son MK, Hong SS, Levy JM, Pollyea DA, Jordan CT, Yan P, Frankhouser D, Nicolet D, Maharry K, Marcucci G, Choi KS, Cho H, Thorburn A, Kim YS. 2015. Methylation-dependent loss of RIP3 expression in cancer represses programmed necrosis in response to chemotherapeutics. Cell Res 25:707–725. doi: 10.1038/cr.2015.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murphy JM, Czabotar PE, Hildebrand JM, Lucet IS, Zhang JG, Alvarez-Diaz S, Lewis R, Lalaoui N, Metcalf D, Webb AI, Young SN, Varghese LN, Tannahill GM, Hatchell EC, Majewski IJ, Okamoto T, Dobson RC, Hilton DJ, Babon JJ, Nicola NA, Strasser A, Silke J, Alexander WS. 2013. The pseudokinase MLKL mediates necroptosis via a molecular switch mechanism. Immunity 39:443–453. doi: 10.1016/j.immuni.2013.06.018. [DOI] [PubMed] [Google Scholar]
- 27.Wang W, Riedel K, Lynch P, Chien CY, Montelione GT, Krug RM. 1999. RNA binding by the novel helical domain of the influenza virus NS1 protein requires its dimer structure and a small number of specific basic amino acids. RNA 5:195–205. doi: 10.1017/S1355838299981621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park HS, Liu G, Raman SNT, Landreth SL, Liu Q, Zhou Y. 2018. NS1 protein of 2009 pandemic influenza A virus inhibits porcine NLRP3 inflammasome-mediated interleukin-1 beta production by suppressing ASC ubiquitination. J Virol 92:e00022-18. doi: 10.1128/JVI.00022-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herold S, Ludwig S, Pleschka S, Wolff T. 2012. Apoptosis signaling in influenza virus propagation, innate host defense, and lung injury. J Leukoc Biol 92:75–82. doi: 10.1189/jlb.1011530. [DOI] [PubMed] [Google Scholar]
- 30.Murphy JM, Zhang Q, Young SN, Reese ML, Bailey FP, Eyers PA, Ungureanu D, Hammaren H, Silvennoinen O, Varghese LN, Chen K, Tripaydonis A, Jura N, Fukuda K, Qin J, Nimchuk Z, Mudgett MB, Elowe S, Gee CL, Liu L, Daly RJ, Manning G, Babon JJ, Lucet IS. 2014. A robust methodology to subclassify pseudokinases based on their nucleotide-binding properties. Biochem J 457:323–334. doi: 10.1042/BJ20131174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jacobsen AV, Lowes KN, Tanzer MC, Lucet IS, Hildebrand JM, Petrie EJ, van Delft MF, Liu Z, Conos SA, Zhang JG, Huang DC, Silke J, Lessene G, Murphy JM. 2016. HSP90 activity is required for MLKL oligomerisation and membrane translocation and the induction of necroptotic cell death. Cell Death Dis 7:e2051. doi: 10.1038/cddis.2015.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao XM, Chen Z, Zhao JB, Zhang PP, Pu YF, Jiang SH, Hou JJ, Cui YM, Jia XL, Zhang SQ. 2016. Hsp90 modulates the stability of MLKL and is required for TNF-induced necroptosis. Cell Death Dis 7:e2089. doi: 10.1038/cddis.2015.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jing L, Song F, Liu Z, Li J, Wu B, Fu Z, Jiang J, Chen Z. 2018. MLKL-PITPalpha signaling-mediated necroptosis contributes to cisplatin-triggered cell death in lung cancer A549 cells. Cancer Lett 414:136–146. doi: 10.1016/j.canlet.2017.10.047. [DOI] [PubMed] [Google Scholar]
- 34.Yu J, Wu Y, Wang J. 2017. Activation and role of NACHT, LRR, and PYD domains-containing protein 3 inflammasome in RNA viral infection. Front Immunol 8:1420. doi: 10.3389/fimmu.2017.01420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schroder K, Tschopp J. 2010. The inflammasomes. Cell 140:821–832. doi: 10.1016/j.cell.2010.01.040. [DOI] [PubMed] [Google Scholar]
- 36.Lupfer CR, Kanneganti TD. 2012. The role of inflammasome modulation in virulence. Virulence 3:262–270. doi: 10.4161/viru.20266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tate MD, Ong JD, Dowling JK, McAuley JL, Robertson AB, Latz E, Drummond GR, Cooper MA, Hertzog PJ, Mansell A. 2016. Reassessing the role of the NLRP3 inflammasome during pathogenic influenza A virus infection via temporal inhibition. Sci Rep 6:27912. doi: 10.1038/srep27912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang W, Li G, De W, Luo Z, Pan P, Tian M, Wang Y, Xiao F, Li A, Wu K, Liu X, Rao L, Liu F, Liu Y, Wu J. 2018. Zika virus infection induces host inflammatory responses by facilitating NLRP3 inflammasome assembly and interleukin-1beta secretion. Nat Commun 9:106. doi: 10.1038/s41467-017-02645-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hottz ED, Lopes JF, Freitas C, Valls-de-Souza R, Oliveira MF, Bozza MT, Da Poian AT, Weyrich AS, Zimmerman GA, Bozza FA, Bozza PT. 2013. Platelets mediate increased endothelium permeability in dengue through NLRP3-inflammasome activation. Blood 122:3405–3414. doi: 10.1182/blood-2013-05-504449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nieto-Torres JL, Verdiá-Báguena C, Jimenez-Guardeño JM, Regla-Nava JA, Castaño-Rodriguez C, Fernandez-Delgado R, Torres J, Aguilella VM, Enjuanes L. 2015. Severe acute respiratory syndrome coronavirus E protein transports calcium ions and activates the NLRP3 inflammasome. Virology 485:330–339. doi: 10.1016/j.virol.2015.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mamik MK, Hui E, Branton WG, McKenzie BA, Chisholm J, Cohen EA, Power C. 2017. HIV-1 viral protein R activates NLRP3 inflammasome in microglia: implications for HIV-1 associated neuroinflammation. J Neuroimmune Pharmacol 12:233–248. doi: 10.1007/s11481-016-9708-3. [DOI] [PubMed] [Google Scholar]
- 42.Farag NS, Breitinger U, El-Azizi M, Breitinger HG. 2017. The p7 viroporin of the hepatitis C virus contributes to liver inflammation by stimulating production of interleukin. Biochim Biophys Acta 1863:712–720. doi: 10.1016/j.bbadis.2016.12.006. [DOI] [PubMed] [Google Scholar]
- 43.Negash AA, Ramos HJ, Crochet N, Lau DT, Doehle B, Papic N, Delker DA, Jo J, Bertoletti A, Hagedorn CH, Gale M Jr. 2013. IL-1beta production through the NLRP3 inflammasome by hepatic macrophages links hepatitis C virus infection with liver inflammation and disease. PLoS Pathog 9:e1003330. doi: 10.1371/journal.ppat.1003330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prow NA, Irani DN. 2008. The inflammatory cytokine, interleukin-1 beta, mediates loss of astroglial glutamate transport and drives excitotoxic motor neuron injury in the spinal cord during acute viral encephalomyelitis. J Neurochem 105:1276–1286. doi: 10.1111/j.1471-4159.2008.05230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]