Oncolytic herpes simplex virus 1 is a promising agent for cancer immunotherapy. Due to a complex virus-host interaction, less is clear about what viral signature(s) constitutes a potent oncolytic backbone. Through molecular or genetic dissection, we showed that selective editing of the γ134.5 gene enables viral replication in malignant cells, activation of transcription factor IRF3, and subsequent induction of type I IFN. This translates into profoundly reduced primary tumor growth and metastasis burden in an aggressive breast carcinoma model in vivo. Our work reveals a distinct oncolytic platform that is amendable for further development.
KEYWORDS: herpes simplex virus, oncolytic viruses, viral replication, virus-host interactions
ABSTRACT
Oncolytic herpes simplex virus 1 (HSV-1), devoid of the γ134.5 gene, exerts antitumor activities. However, the oncolytic effects differ, ranging from pronounced to little responses. Although viral and host factors are involved, much remains to be deciphered. Here we report that engineered HSV-1 ΔN146, bearing amino acids 147 to 263 of γ134.5, replicates competently in and lyses malignant cells refractory to the γ134.5 null mutant. Upon infection, ΔN146 precludes phosphorylation of translation initiation factor eIF2α (α subunit of eukaryotic initiation factor 2), ensuring viral protein synthesis. On the other hand, ΔN146 activates interferon (IFN) regulatory factor 3 (IRF3) and IFN expression, known to prime immunity against virus and tumor. Nevertheless, ΔN146 exhibits sustained replication even exposed to exogenous IFN-α. In a 4T1 tumor model, ΔN146 markedly reduces tumor growth and metastasis formation. This coincides with viral replication or T cell infiltration in primary tumors. ΔN146 is undetectable in normal tissues in vivo. Targeted HSV-1 editing results in a unique antineoplastic agent that enables inflammation without major interference of viral growth within tumor cells.
IMPORTANCE Oncolytic herpes simplex virus 1 is a promising agent for cancer immunotherapy. Due to a complex virus-host interaction, less is clear about what viral signature(s) constitutes a potent oncolytic backbone. Through molecular or genetic dissection, we showed that selective editing of the γ134.5 gene enables viral replication in malignant cells, activation of transcription factor IRF3, and subsequent induction of type I IFN. This translates into profoundly reduced primary tumor growth and metastasis burden in an aggressive breast carcinoma model in vivo. Our work reveals a distinct oncolytic platform that is amendable for further development.
INTRODUCTION
Oncolytic herpes simplex virus 1 (HSV-1) is an attractive agent for cancer immunotherapy (1). Upon infection, HSV-1 undergoes sequential gene expression, DNA replication, assembly, and egress, resulting in tumor cell destruction. This is accompanied by release of danger signals and neoantigens that activate adaptive antitumor immunity. A range of oncolytic HSVs is under various stages of development (1). The most clinically advanced agent is talimogene laherparepvec (T-VEC), recently approved by FDA for treating advanced melanoma (2). Additional examples of oncolytic HSVs are G207, HSV1716, and ΔG47, which have undergone or are in clinical trials (3–7). Although differing in the backbone design, these oncolytic HSVs have originally deleted the γ134.5 gene, which codes for a virulence factor (8, 9).
HSV γ134.5 contains a large amino-terminal domain (amino acids [aa] 1 to 146), linker region, and carboxyl-terminal domain (aa 147 to 263) (10). In infected cells, HSV-1 activates double-stranded RNA-dependent kinase (PKR), which shuts off protein synthesis by phosphorylation of translation initiation factor eIF2α (α subunit of eukaryotic initiation factor 2) (11). Consequently, γ134.5 redirects protein phosphatase 1 (PP1) to dephosphorylate eIF2α (12). Notably, site-specific disruption of the γ134.5-PP1 interaction abrogates viral virulence (13, 14). HSV γ134.5 is also reported to affect glycoprotein processing and viral spread (15, 16). Furthermore, evidence suggests that the γ134.5 protein displays additional activities. These include inhibition of autophagy, interferon (IFN) induction by TANK binding kinase 1, and dendritic cell maturation by Toll-like receptors and acceleration of nuclear egress (17–21). Although the γ134.5 protein shuttles between the nucleus and cytoplasm (22, 23), its precise interplay with the host, in particular malignant cells, remains obscure.
There is abundant evidence that HSV-1 mutants deficient in the γ134.5 gene have oncolytic activities. This has been shown for tumors, including in the brain, colon, ovarian, breast, liver, and skin, in immunodeficient as well as in immunocompetent preclinical models (24–31). However, the antitumor outcomes vary widely. For example, HSV1716 is highly potent against hepatocellular carcinoma (30). With respect to neuroblastomas, HSV1716 exhibits activities from a complete response in the CHP-134 model to a mild effect in the SK-N-AS model (31). The underlying events are complex, but the nature of virus-host interactions seems a determinant. It has been suggested that the activation of mitogen-activated protein kinase or RAS oncogene in tumor cells inhibits PKR and thereby permits viral replication (32, 33). On the other hand, genetic or epigenetic suppression of stimulator of interferon gene (STING), a mediator of IFN induction, is reported to license the γ134.5 null mutant for tumor destruction (34, 35).
Type I IFNs are a family of cytokines that upregulate a spectrum of molecules with various functions (36). While antiviral in nature, type I IFN also critically primes antitumor immunity (37, 38). In light of these observations, we hypothesize that a desirable oncolytic HSV backbone would instigate IFN production while retaining robust replication within tumor cells. Here we report that targeted editing of the γ134.5 gene enables viral replication and IFN induction. Such a unique agent markedly reduces tumor growth and metastasis in vivo. Our work suggests that a selective alteration of virus-cell interactions favors tumor destruction.
RESULTS
The γ134.5 mutant bearing amino acids 147 to 263 replicates efficiently in tumor cells.
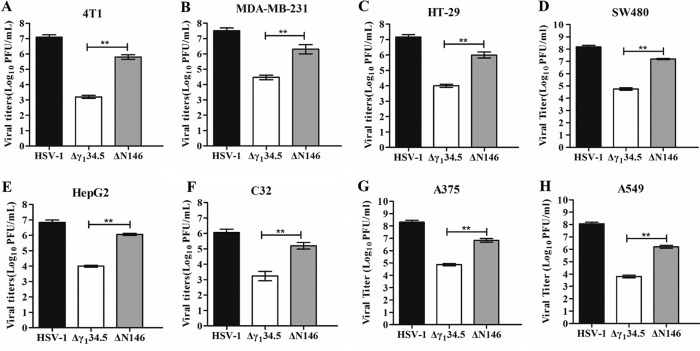
We recently reported that an HSV γ134.5 mutant (ΔN146), with only amino acids 147 to 263, is substantially impaired for viral growth in normal cells or tissues (18, 39). To explore its property in malignant cells, we determined viral replication. As illustrated in Fig. 1A, in 4T1 (murine breast carcinoma) cells, wild-type HSV-1 replicated to 1 × 107 PFU/ml, whereas the γ134.5 null mutant (Δγ134.5) reached only 1 × 103 PFU/ml. However, ΔN146 grew to 1 × 106 PFU/ml, indicative of robust replication. A similar trend was observed in MDA-MB-231 (human breast adenocarcinoma) cells, in which ΔN146 replicated 100-fold better than Δγ134.5 (Fig. 1B). Moreover, these phenotypes were recapitulated in a range of other tumor cells, including human HT29 (colon), SW480 (colon), HepG2 (liver), C32 (melanoma), A375 (melanoma), and A549 (lung) cells (Fig. 1C to H).
FIG 1.
Comparison of viral replication in tumor cell lines. 4T1 (A), MDA-MB-231 (B), HT-29 (C), SW480 (D), HepG2 (E), C32 (F), A375 (G), and A549 (H) cells were infected with HSV-1, Δγ134.5, or ΔN146 (0.01 PFU/cell). At 48 h postinfection, viral yields were determined on Vero cells. The data are representative of those from three experiments with triplicate samples. Differences between the selected groups were statistically assessed by a two-tailed Student t test. **, P < 0.01.
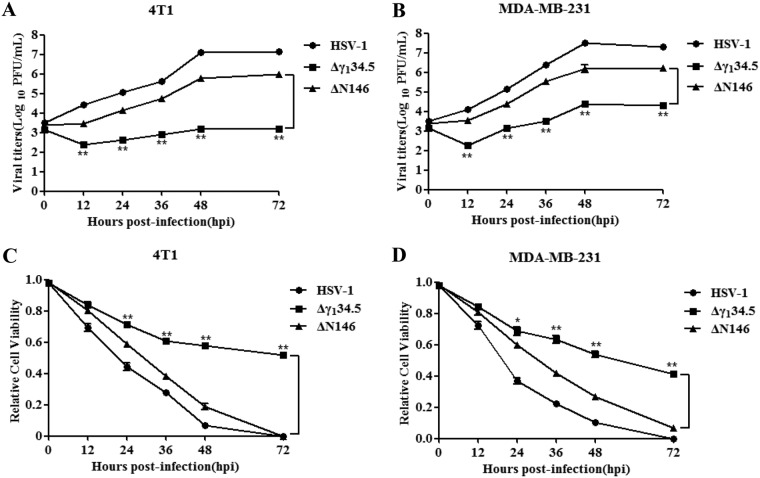
Next, we examined the kinetics of viral growth. As presented in Fig. 2A, wild-type HSV-1 grew steadily in 4T1 cells as infection progressed, with a titer increased to 1 × 107 PFU/ml by 72 h postinfection. ΔN146 replicated to 1 × 106 PFU at a slightly lower level. And Δγ134.5 barely replicated, with a titer of 1 × 103 PFU/ml throughout infection. A similar trend was observed in MDA-MB-231 cells, in which ΔN146 replicated 100-fold better than Δγ134.5 (Fig. 2B). To assess viral cytolytic activity, we measured cell viability. Figure 2C shows that similar to wild-type virus, ΔN146 lysed almost 95% of 4T1 cells by 72 h, with slightly delayed kinetics. Δγ134.5 destroyed approximately 40% of cells. Such effects were also mirrored in MDA-MB-231 cells (Fig. 2D). Together, these results suggest that ΔN146 replicates in and lyses tumor cells more effectively than the γ134.5 null mutant.
FIG 2.
(A and B) Kinetics of viral growth in 4T1 (A) and MDA-MB-231 (B) cells. Cells were infected with indicated viruses (0.01 PFU/cell). Viral yields were measured at different time points postinfection. (C and D) Viral cytolytic effects on 4T1 (C) and MDA-MB-231 (D) cells. Cells were infected with viruses (0.1 PFU/cell) for 2 h, and then cell viability was determined by CellTiter-Glo luminescent cell viability assay at the indicated times. The relative cell viability is normalized to that of the mock control. All the data are representative of those from three experiments with triplicate samples. Differences between the selected groups were statistically assessed by a two-tailed Student t test. *, P < 0.05; **, P < 0.01.
ΔN146 promotes viral protein production through a block of eIF2α phosphorylation in tumor cells.
HSV infection proceeds in a temporal manner, with sequential expression of α, β, and γ genes. Onset of viral DNA replication invokes the cessation of protein synthesis in the absence of γ134.5 (11). To investigate ΔN146 in breast cancer cells, we analyzed ICP27 (α protein) and gC (γ protein), whose expression relies on viral DNA replication. Cells were mock infected or infected with viruses. At 12 h postinfection, samples were subjected to Western blot analysis. As shown in Fig. 3A and B, wild-type virus expressed both ICP27 and gC in 4T1 and MDA-MB-231 cells. However, Δγ134.5 expressed little gC. Under this condition, ΔN146 produced comparable levels of ICP27 and gC. In this respect, ΔN146 resembles wild-type virus in blocking translational arrest initiated by viral DNA replication.
FIG 3.
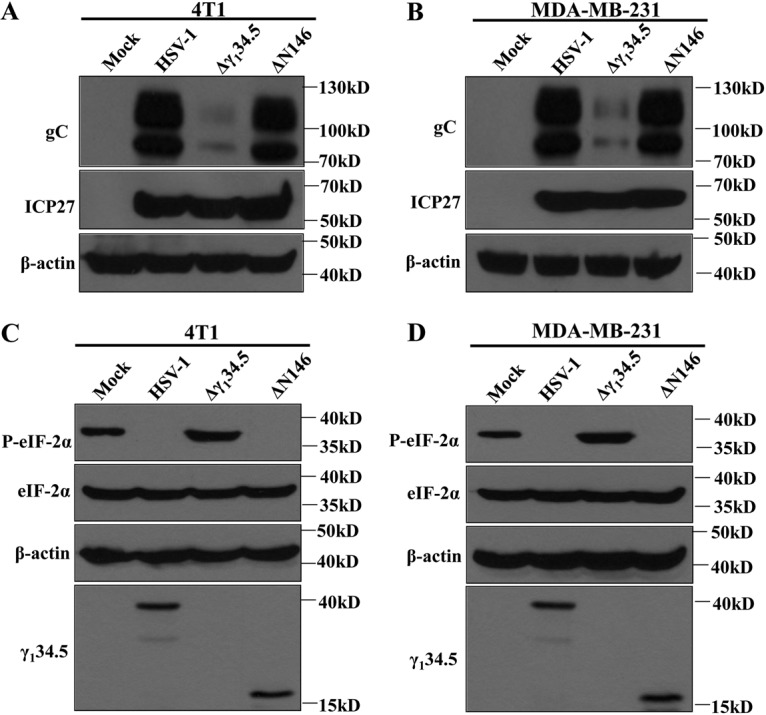
(A and B) Production of gC and ICP27 in virus-infected cells. 4T1 (A) and MDA-MB-231 (B) cells were mock infected or infected with the indicated viruses at 5 PFU per cell. At 12 h postinfection, cells were harvested and subjected to Western blot analysis with antibodies against gC, ICP27, and β-actin. (C and D) Effects of viral infection on eIF2α phosphorylation. 4T1 (C) and MDA-MB-231 (D) cells were mock infected or infected with viruses. At 12 h postinfection, lysates of cells were subjected to immunoblotting analysis with antibodies against eIF2α, phosphorylated eIF2α (Ser51), γ134.5, and β-actin. The data are representative of those from three independent experiments.
As eIF2α is coupled to protein synthesis, we monitored eIF2α, which, upon phosphorylation by stress kinases PKR, PERK, and GCN2 (40), arrests translation. As shown in Fig. 3C and D, levels of expression of eIF2α were comparable in mock- and virus-infected cells. Interestingly, phosphorylated eIF2α was presented in mock-infected cells, which is presumably due to oncogenic stress (41, 42). While wild-type virus inhibited eIF2α phosphorylation, Δγ134.5 aggravated its phosphorylation. Notably, ΔN146 completely abrogated eIF2α phosphorylation. It appears that in tumor cells, the coding region spanning amino acids 147 to 263 from γ134.5 effectively inhibits eIF2α phosphorylation.
ΔN146 stimulates the inflammatory response in tumor cells.
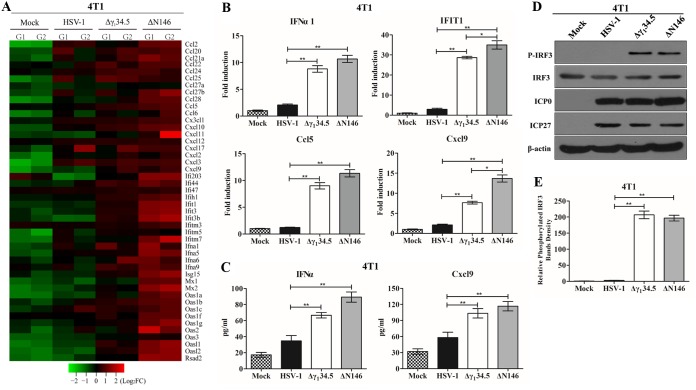
To further investigate the footprint of ΔN146, we performed transcriptome analysis. We found that numerous genes in diverse cellular pathways were expressed differentially in 4T1 cells mock infected and infected with viruses. Of note, many genes in the innate immune pathways were evidently upregulated in response to ΔN146. Among 46 genes listed in Fig. 4A, most remained unchanged or marginally expressed in cells mock infected or infected with wild-type virus. However, they were upregulated in cells infected with Δγ134.5, albeit to a different extent. Notably, gene induction was more pronounced in cells infected with ΔN146, suggesting that ΔN146 has a propensity to stimulate the inflammatory response.
FIG 4.
(A) Transcriptome analysis of 4T1 cells. Cells were mock infected or infected with the indicated viruses (5 PFU/cell). At 6 h postinfection, samples were processed for microarray analysis. The heat map includes 46 chemokines or interferon-related genes (IRGs). G1 and G2 represent distinct experimental replicates. The data represents log2 fold changes. (B) Expression of IFN-α1, IFIT1, Ccl5, and Cxcl9. The RNA samples were analyzed by quantitative PCR. Results are expressed as fold activation with SDs among triplicate samples. Differences between the selected groups were statistically assessed by a two-tailed Student t test. (C) 4T1 cells were mock infected or infected with HSV-1(F), Δγ134.5, or ΔN146 (5 PFU/cell) for 16 h. Cell supernatants were collected to determine IFN-α and Cxcl9 levels using a commercial ELISA kit. The data from triplicate samples were statistically assessed by a two-tailed Student test. (D) IRF3 phosphorylation detection after HSV-1(F), Δγ134.5, or ΔN146 infection. 4T1 cells were infected with the indicated viruses at 5 PFU/cell, and cell lysates were subjected to immunoblotting analysis with antibodies against IRF3, phosphorylated IRF3 (Ser396), ICP27, ICP0, and β-actin at 6 h postinfection. (E) Quantification of IRF3 phosphorylation. The protein bands shown in panel D were quantified using NIH ImageJ software. The data are presented as the relative amount of phosphorylated IRF3 normalized to the total level of IRF3 in each sample, with mock infection arbitrarily set at 1.0. The data are averages from three independent experiments and were statistically assessed by a two-tailed Student t test. *, P < 0.05; **, P < 0.01.
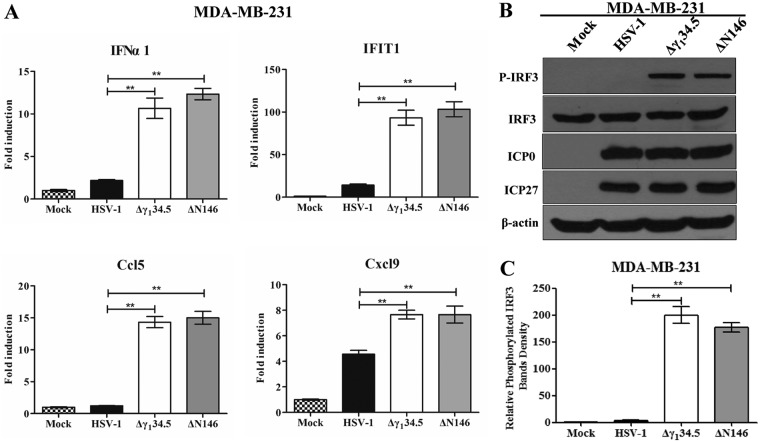
To validate these results, we determined the expression of selected cytokines and interferon-stimulated genes by real-time PCR (Fig. 4B). As expected, wild-type virus triggered little expression of IFN-α1, IFIT1, Ccl5, and Cxcl9, whereas Δγ134.5 or ΔN146 sharply induced these genes. This was corroborated by the levels of cytokine production in enzyme-linked immunosorbent assay (ELISA) (Fig. 4C). To dissect the molecular basis, we analyzed interferon regulatory factor 3 (IRF3), which activates immune responses. As illustrated in Fig. 4D, IRF3 was unphosphorylated in 4T1 cells mock infected or infected with wild-type HSV-1. In contrast, it became phosphorylated in cells infected with Δγ134.5 or ΔN146. This was not due to differences in viral infectivity, as indicated by the normal expression of ICP0 and ICP27. And these results were confirmed in multiple experiments (Fig. 4E). Moreover, these phenotypes were seen in human MDA-MB-231 cells (Fig. 5). We conclude that like Δγ134.5, ΔN146 is immunostimulatory upon infection of malignant cells.
FIG 5.
(A) Cytokine expression in MDA-MB-231 cells. Cells were either mock infected or infected with HSV-1(F), Δγ134.5, or ΔN146 (5 PFU/cell). At 6 h after infection, total RNA extracted from cells was subjected to quantitative real-time PCR amplification for the expression of IFN-α1, IFIT1, Ccl5, and Cxcl9 on MDA-MB-231 cells. The results are representative of those from three experiments with triplicate samples and were statistically assessed by a two-tailed Student t test. (B) IRF3 phosphorylation viral infection. MDA-MB-231 cells were infected with the indicated viruses at 5 PFU/cell, and cell lysates were subjected to immunoblotting analysis with antibodies against IRF3, phosphorylated IRF3 (Ser396), ICP27, ICP0, and β-actin at 6 h postinfection. (C) Quantification of IRF3 phosphorylation. The protein bands shown in panel B were quantified using NIH ImageJ software. The data are averages from three independent experiments and were statistically assessed by a two-tailed Student t test. **, P < 0.01.
ΔN146 replicates competently in tumor cells treated with IFN-α.
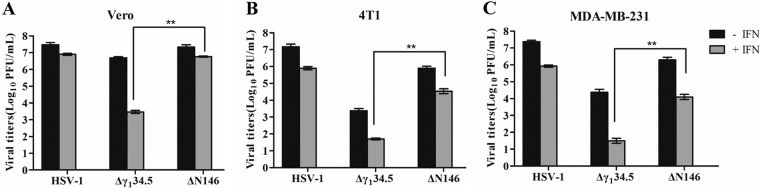
Type I IFN is necessary to prime immunity against tumors (37, 38). On the other hand, it mediates antiviral responses (36). To determine whether ΔN146 is refractory to clearance by IFN, we examined viral growth. As proof of concept, we first determined the viral response to IFN in Vero cells, which are devoid of IFN-α/β genes (Fig. 6A). Treatment with IFN-α had little effect on replication of HSV-1(F) but drastically reduced replication of Δγ134.5, approximately 1,000-fold. However, IFN-α only modestly decreased replication of ΔN146. Furthermore, when tested in 4T1 and MDA-MB-231 cells, a similar trend was observed (Fig. 6B and 6C). While IFN-α reduced viral replication in general, the effect was smaller on wild-type HSV-1 or ΔN146. Indeed, ΔN146 consistently replicated 500- to 1,000-fold higher than Δγ134.5 in the presence of exogenous IFN-α. Thus, amino acids 147 to 263 from γ134.5 are sufficient to confer viral resistance to IFN.
FIG 6.
Viral response to type I interferon. (A)V ero cells were untreated or pretreated with human IFN-α (Sigma) at 500 U/ml for 20 h. Cells were then infected with indicated viruses at 0.01 PFU per cell. Viral yields were determined at 48 h postinfection. (B) 4T1 cells were left untreated or pretreated with mouse IFN-α (Sigma) at 250 U/ml for 20 h. Cells were then infected with the indicated viruses at 0.01 PFU per cell. Virus yields were determined at 48 h postinfection. (C) MDA-MB-231 cells were treated as for panel A, and virus yields were determined at 48 h postinfection. The data are representative of those from three independent experiments. Differences between the selected groups were statistically assessed by a two-tailed Student t test. **, P < 0.01.
ΔN146 reduces primary tumor growth and metastasis in vivo.
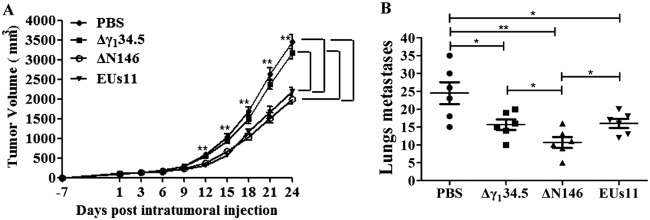
Based on the above-described analyses, we hypothesize that the capacity of ΔN146 to replicate and activate inflammation may facilitate tumor destruction in vivo. To test this, we chose an aggressive 4T1 mammary carcinoma that spontaneously metastasizes, a process analogous to that in human mammary tumors (43). For comparison, we used Δγ134.5, which resembles HSV1716 (4, 6). In addition, we included recombinant HSV EUs11 (44), which is structurally equivalent to the oncolytic backbone for talimogene laherparepvec (45). Tumors formed subcutaneously in the flanks of mice were injected with phosphate-buffered saline (PBS), Δγ134.5, ΔN146, or EUs11 (1 × 107 PFU) on days 1, 3, and 6. Tumor size was then monitored. As illustrated in Fig. 7A, control tumors treated with PBS grew at a higher rate over time. Treatment with the γ134.5 null virus marginally reduced local tumor growth. However, intratumor inoculation with ΔN146 or EUs11 slowed tumor growth. And reduction in tumor size became more apparent as the treatment progressed. On day 24, ΔN146 as well as EUs11 reduced the tumor size by nearly 45% compared to that with the mock control or Δγ134.5. Hence, while comparable to EUs11, ΔN146 is superior against primary tumors compared with Δγ134.5.
FIG 7.
(A) ΔN146 reduces local tumor growth. 4T1 cells were implanted subcutaneously into mice (day −7). Tumors formed were injected with PBS, Δγ134.5, ΔN146, or EUs11 suspended in PBS on days 1, 3, and 6 as described in Materials and Methods. Tumor sizes were measured periodically (x axis) until day 24 (n = 6 each group). Average tumor volumes over time are shown on the y axis. Asterisks indicate statistical significance by nonparametric analysis. (B) Mice were sacrificed on day 24 after the initiation of treatment. The lungs were collected and fixed in formalin. The number of lung metastases was quantified by counting under a light microscope. The results shown are from one of three independent experiments. Differences between the selected groups were statistically assessed by a two-tailed Student t test. *, P < 0.05; **, P < 0.01.
To assess the viral impact on metastasis, we analyzed lung tumor formation on day 24. Figure 7B shows that pulmonary metastasis was readily detectable in control mice, with an average of 25 nodules per animal as measured by microscopic analysis. Treatment with Δγ134.5 or EUs11 reduced metastasis incidence, with an average of 15 nodules per animal. Notably, ΔN146 further reduced metastasis burden, with an average of 10 nodules. Therefore, although all virus constructs reduce pulmonary metastasis, ΔN146 exerts the most notable effect.
ΔN146 replicates in primary tumor but not normal tissues.
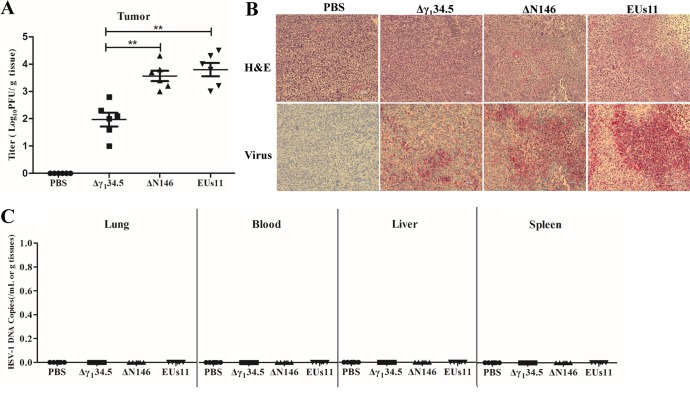
To assess the extent of viral replication, we first measured viral yields in primary tumors collected on day 9. As illustrated in Fig. 8A, Δγ134.5 maintained at an average titer of 1 × 102 PFU/g of tumor tissue as measured by plaque assay. On the other hand, EUs11 grew at an average titer of 7 × 103 PFU/g of tumor tissue. Similarly, ΔN146 grew at an average titer of 5 × 103 PFU/g of tumor tissue. Apparently, like EUs11, ΔN146 replicated approximately 50-fold better than Δγ134.5. In line with this, viral antigens were detected in thin sections of the tumor beds (Fig. 8B), where ΔN146 and EUs11 spread more extensively than Δγ134.5. This correlated with the degree of necrosis of the tumor tissues.
FIG 8.
(A) Viral growth in 4T1 tumors. Tumors treated with PBS, Δγ134.5, ΔN146, and EUs11 suspended in PBS were collected on day 9, and infectious viruses present in tumors were quantified by plaque assay (n = 6). (B) Hematoxylin and eosin (H&E) and immunostaining showing the HSV-1 antigens in the tumors. (C) Quantification PCR analysis of HSV-1 DNA in blood, liver, and spleen (n = 6). All the data are representative of those from three experiments. Differences between the selected groups were statistically assessed by a two-tailed Student t test. **, P < 0.01.
To gauge whether viruses spread to the normal tissues, we analyzed for Δγ134.5, ΔN146, and EUs11 in the lung, blood, liver, and spleen by qPCR assay. As shown in Fig. 8C, no viruses were detectable in these tissues on day 9, although they were readily found in the tumors (data not shown). These results suggest that like that of Δγ134.5 or EUs11, replication of ΔN146 is limited to the tumor tissues in vivo.
To verify that viral replication indeed occurs actively in the tumors, we performed triple therapy of 4T1 primary tumors and measured viral yields on days 7, 9, and 15. As indicated in Fig. 9, viruses were detectable at about 2 × 102 PFU/g of tumor tissue on day 7 by plaque assay. As treatment progressed, the quantity of Δγ134.5 remained unchanged initially and then was reduced to 1 × 10 PFU/g of tumor tissue by day 15. However, under this condition, the level of ΔN146 increased to 1 × 104 PFU/g of tumor tissue on day 9, which subsequently decreased to 1 × 103 PFU/g of tumor tissue by day 15. EUs11 displayed a similar growth pattern. Therefore, unlike Δγ134.5, ΔN146 and EUs11 are able to replicate within tumor in vivo.
FIG 9.
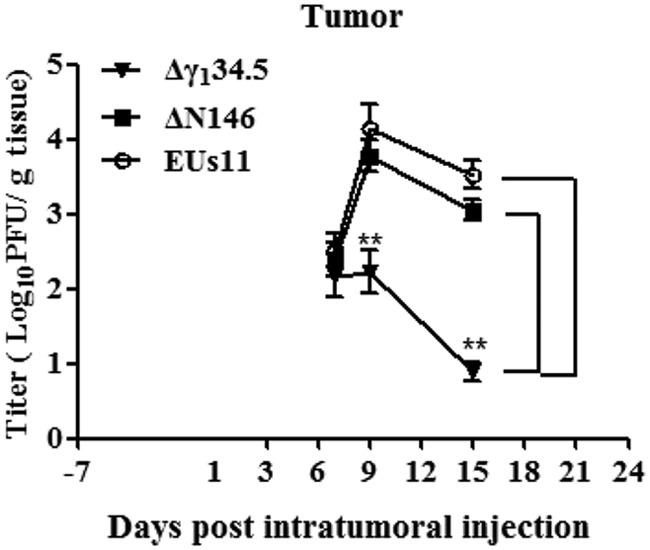
Kinetics of viral replication in tumors. Mice with established 4T1 tumors were given triple therapy on day 1, 3, and 6 as described in Materials and Methods. Tumor tissues were harvested on days 7, 9, and 15 (n = 4 per time point) and subsequently examined for viral growth by plaque assay. The data from a representative experiment were statistically assessed by a two-tailed Student t test. **, P < 0.01.
ΔN146 enhances infiltration of CD4+ and CD8+ T cells into primary tumors.
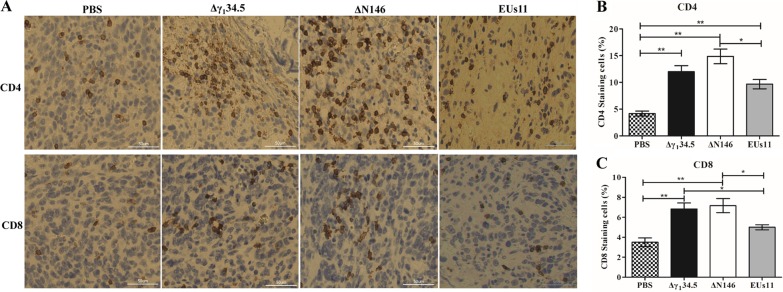
Previous work suggested that oncolytic HSV with deletion of γ134.5 activates systemic antitumor immunity (28, 46). As intratumor virus injection reduced both local tumor growth and metastasis formation, we asked whether there is induction of adaptive immunity. To investigate this, we probed for CD4+ and CD8+ T cells by immunohistochemistry analysis. Primary tumors collected on day 24 were processed and stained for the presence of CD4+ and CD8+ T cells. As illustrated in Fig. 10, in mock-infected tumors, a few CD4+ or CD8+ T cells (<4%) were detectable. However, in tumors treated with Δγ134.5, CD4+ T cells rose to 12% and CD8+ T cells to 7%. Similarly, ΔN146 accounted for 15% of CD4+ and 8% CD8+ T cells, respectively. Although EUs11 triggered immune cell infiltration, the observed effect was reduced for both CD4+ (10%) and CD8+ (5%) T cells. These results suggest that similar to Δγ134.5, ΔN146 induces T cell infiltration, whereas EUs11 appears to dampen this process.
FIG 10.
(A) Infiltration of CD4+ and CD8+ cells into 4T1 tumors. Tumor tissues were harvested from mice with triple therapy on day 24. Thin sections were processed and stained with antibodies against CD4+ and CD8+ T cells. CD4+ and CD8+ T cells are brown. (B) CD4+ T cells were quantified using NIH ImageJ software in tumor tissues (n = 6). (C) CD8+ T cells were quantified in tumor tissues (n = 6). Differences between the selected groups were statistically assessed by a two-tailed Student t test. *, P < 0.05; **, P < 0.01.
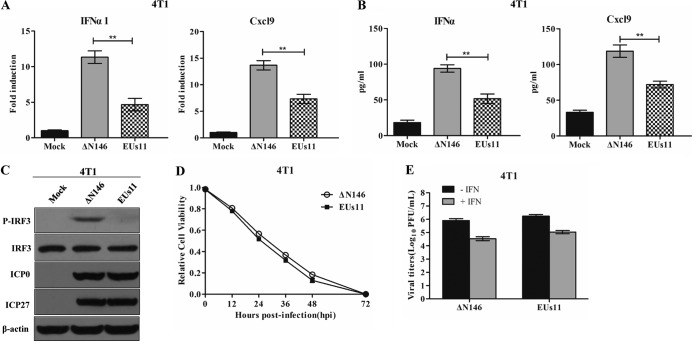
ΔN146 and EUs11 interact with tumor cells differently.
To determine whether ΔN146 and EUs11 interact with tumor cells differently, we carried out in vitro analyses. As shown in Fig. 11A, ΔN146 infection stimulated transcription of the IFN-α1 and Cxcl9 genes. In contrast, EUs11 suppressed gene expression. This paralleled the levels of cytokine production as measured by ELISA (Fig. 11B). Consistently, ΔN146 stimulated phosphorylation of IRF3 and EUs11 failed to do so (Fig. 11C), suggesting that EUs11 mediates immunosuppression upon virus infection.
FIG 11.
Comparative analysis of ΔN146 and EUs11 in vitro. (A) Viral effects on the expression of IFN-α1 and Cxcl9. 4T1 cells were mock infected or infected with ΔN146 or EUs11 (5 PFU/cell). At 6 h postinfection, RNA samples were analyzed by quantitative PCR. (B) Virus effects on cytokine production. 4T1 cells were infected as for panel A. At 16 h, cell supernatants were collected to determine the levels of IFN-α and Cxcl9 by ELISA. All the data are representative of those from three experiments with triplicate samples. Differences between the selected groups were statistically assessed by a two-tailed Student t test. **, P < 0.01. (C) Viral effects on IRF3 phosphorylation. 4T1 cells were mock infected or infected with the indicated viruses (5 PFU/cell). At 6 h postinfection, lysates of cells were subjected to immunoblotting analysis with antibodies against IRF3, phosphorylated IRF3 (Ser396), ICP27, ICP0, and β-actin. (D) Viral cytolytic effects on 4T1 cells. Cells were infected with viruses (0.1 PFU/cell), and cell viability was determined by CellTiter-Glo luminescent cell viability assay at the indicated time points. The relative cell viability is normalized to that of the mock control. (E) Viral response to interferon. 4T1 cells were left untreated or pretreated with IFN-α (Sigma) at 250 U/ml for 20 h. Cells were then infected with the indicated viruses (0.01 PFU/cell), and virus yields were determined at 48 h postinfection. All data are representative of those from three experiments among triplicate samples with SDs.
To assess the viral capacity to destroy tumor cells, we measured cell viability. Figure 11D shows that like EUs11, ΔN146 lysed almost 95% of 4T1 cells by 72 h. Thus, both ΔN146 and EUs11 lysed tumor cells efficiently. We further determined viral replication in 4T1 cells with or without IFN treatment. Figure 11E shows that in the absence of IFN-α, both ΔN146 and EUs11 replicated efficiently, with a titer reaching about 1 × 106 PFU/ml. Addition of exogenous IFN-α modestly reduced viral replication for ΔN146 and EUs11, with a titer of 5 × 104 PFU/ml, suggesting that they are equally resistant to type I IFN.
DISCUSSION
Several oncolytic HSV agents, deficient in the γ134.5 gene, have moved into or completed clinical trials (3–7). While these represent a milestone, much remains to be deciphered. This is particularly evident with respect to virus-tumor cell interactions. In the present study, we showed that selective editing of the γ134.5 gene results in a distinct oncolytic HSV backbone which propagates robustly in tumor cells, instigates inflammatory cytokine expression, and mediates antitumor effects in vivo.
Our work indicates that the γ134.5 domain spanning amino acids 147 to 263 is sufficient to promote oncolytic activity. We observed that ΔN146 replicated competently in and lysed the tumor cells refractory to Δγ134.5. We suspect that only a subset of γ134.5 functions is required for efficient productive infection of the tumor cells and cytotoxicity, which might occur through multiple pathways. As ΔN146 prevents translation arrest of glycoprotein C and eIF2α phosphorylation, a notable oncolytic mechanism would be that the phosphatase 1 regulatory motif conserved in γ134.5 and cellular Myd116 precludes eIF2α phosphorylation by PKR (12, 47), which partly accounts for viral resistance to IFN (48). This is functionally analogous to Us11, which directly inhibits PKR when expressed by the EUs11 virus (44, 49, 50). Another possibility is that ΔN146 may abrogate eIF2α phosphorylation mediated by PERK or GCN2, both of which are often activated under oncogenic stress in tumor cells (41, 42, 51, 52). An additional possibility is to block the antiviral action of interferon-stimulated genes, such as IFIT1, IFIT3, Rsad2 (viperin), and Mx2. Two lines of evidence are consistent with this model. First, upregulation of these antiviral effectors barely affected ΔN146 replication. Second, like EUs11, ΔN146 was more resistant to IFN-α treatment than was Δγ134.5. Additional work is required to test these hypotheses.
ΔN146 stimulates the expression of inflammatory cytokines without major interference of viral growth in tumor cells. Consistently, ΔN146 induced phosphorylation of IRF3 upon infection of 4T1 and MDA-MB-231 cells. This is attributable to the deletion in the N-terminal domain from γ134.5 necessary to inhibit STING or TBK1 (18, 53). The immunostimulatory activity, coupled with its robust replication in tumor cells, suggests that ΔN146 is a unique oncolytic platform. This is in contrast to the EUs11 virus, which mediates an immunosuppressive effect. This may operate through early expression of the Us11 protein to disrupt the HSP90-TBK1 complex (44). Considering a costimulatory profile of IFN in cancer immunotherapy (38), it is attractive to speculate that ΔN146 may prime antitumor responses in an IFN-dependent manner in vivo. However, the precise mechanism(s) of ΔN146 action is to be established. Several DNA sensors, including cGAMP synthase, interferon-inducible protein 16, and DEAD box helicase 41, recognize HSV-1 (54–56). It is possible that ΔN146 may trigger one or more these receptors, leading to inflammatory gene expression. Alternatively, ΔN146 may engage Toll-like receptor 3, retinoid acid-inducible gene I, or melanoma differentiation-associated gene 5, which mediates cytokine production in the tumor microenvironment (57).
The 4T1 breast carcinoma model closely resembles stage IV human breast cancer (43). Its gene signature is suggested to predict poor survival in human malignancies (58). Although the tumor grew aggressively and spread spontaneously, ΔN146 markedly decreased primary tumor growth, like EUs11. This contrasts to a marginal effect with Δγ134.5, which lacks the γ134.5 gene. These phenotypes correlated with the extent of viral replication in primary tumor, suggesting that expression of only amino acids 147 to 263 from γ134.5 is adequate to enhance oncolytic potency. Importantly, intratumor virus administration also reduced lung metastasis. In this regard, ΔN146 mediated the most pronounced effect, which cannot be explained solely by the difference in viral replication. In line with these results, treatment with ΔN146, EUs11, or Δγ134.5 induced infiltrations of CD4+ and CD8+ T cells with a different magnitude. This is consistent with the early observations that the γ134.5 null mutants activated systemic adaptive immunity against tumors (28, 46). We speculate that besides cytolytic destruction, ΔN146 may favorably activate antitumor immunity to prevent metastasis into distal organs.
Deletion in the N-terminal domain from γ134.5 limits viral replication to the malignant tissues. Infectious viruses were recovered from the tumor beds after intratumor inoculation. Remarkably, ΔN146 was present at a much higher level than Δγ134.5, which might reflect active virus replication or a delay in clearance. This resembled the EUs11 virus, in which expression of Us11 recused viral replication. However, no viruses were detectable in the lung, kidney, liver, or blood. Given that ΔN146 is avirulent, as demonstrated in our previous work (18), these results support the model that an intact γ134.5 protein is required for viral replication in normal tissues. Because the N-terminal domain is associated with multiple functions, including virus egress and the inhibition of IFN expression, dendritic cell maturation, and autophagy (17–21), its removal may render the virus unable to overcome host restrictions, possibly at multiple levels. Work is in progress to investigate this possibility.
MATERIALS AND METHODS
Cells and viruses.
Vero, HT-29, SW480, C32, A375, MDA-MB-231, 4T1, HepG2, and A549 cells were obtained from the American Type Culture Collection. Vero, SW480, C32, A375, MDA-MB-231, and A549 cells were propagated in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum. HT-29, 4T1, and HepG2 cells were propagated in RPMI 1640 supplemented with 10% fetal bovine serum. HSV-1(F) is a prototype HSV-1 strain used in this study (59). In recombinant virus Δγ134.5, a 1-kb fragment from the coding region of the γ134.5 gene was deleted (8). In ΔN146, the sequences of the γ134.5 gene encoding amino acids 1 to 146 were deleted (18). In EUs11, the γ134.5 gene was deleted but with the Us11 gene driven by the α-47 promoter (44). Preparation of viral stock and titration of infectivity were carried out as described previously (18, 60).
Virus infection.
Virus infections were carried out at the desired multiplicities of infection (18). Cells were then harvested and processed for immunoblot, real-time PCR, or viral growth analysis (18, 21). The cell viability was determined by CellTiter-Glo luminescent cell viability assay (Promega) according to the manufacturer protocols. For the interferon assay, Vero and MDA-MB-231 cells were left untreated or treated with human IFN-α (SRP4596; Sigma), and 4T1 cells were treated with mouse IFN-α (I8782-1VL; Sigma) for 20 h. Cells were then infected with viruses, and virus yields were determined at 48 h postinfection.
Immunoblot analysis and ELISA.
Cells were harvested, washed with phosphate-buffered saline (PBS), and lysed as described previously (18). Samples were then subjected to electrophoresis on denaturing polyacrylamide gels, transferred to nitrocellulose membranes, and reacted with antibodies against gC (61), γ134.5 (22), ICP27 (P1113; Virusys Inc.), ICP0 (sc-53070; Santa Cruz), eIF2α (5324; Cell Signaling Technology, Inc.), phosphorylated eIF2α (3398; Cell Signaling Technology, Inc.), IRF3 (4302; Cell Signaling Technology, Inc.), phosphorylated IRF3 (4947; Cell Signaling Technology, Inc.), and β-actin (A5316; Sigma). The membranes were rinsed in PBS, reacted with either donkey anti-rabbit or anti-mouse immunoglobulin conjugated to horseradish peroxidase, and developed with an enhanced chemiluminescence Western blot detection system kit (Amersham Pharmacia Biotechnology, Inc.). To perform enzyme-linked immunosorbent assays (ELISA), supernatants of cell culture were collected to analyze IFN-α (42120-1) and Cxcl9 (DY492) according to the manufacturer’s instructions (R&D Systems).
Transcriptome analysis.
4T1 cells were mock infected or infected with viruses (5 PFU/cell). At 6 h postinfection, RNA was extracted from the cells using an RNase Plus minikit (Qiagen) and treated with DNase I digestion (New England BioLabs). Duplicate RNA samples were processed using Clariom S Affymetrix array at the Center for Genomic Research at the University of Illinois. Raw data generated from the Clariom S mouse array were processed in R using package Oligo (62). Feature intensity values from each CEL file was converted into normalized expression values using Robust Multiarray Average (RMA) with default settings. All the positive- and negative-control probes, along with Affymetrics report genes (RPTR), were removed before performing the downstream analysis. Principal-component analysis (PCA) plots were generated to check for any batch effect. Differential gene expression analysis was performed using the limma package (63). Significantly expressed genes were filtered for an adjusted P value of <0.05. Heat maps were produced from the primary data (the normalized expression value) using the R package pheatmap v1.0.8.
Quantitative real-time PCR assay.
Cells were mock infected or infected with viruses. At 6 h after infection, total RNA was harvested and analyzed by real-time PCR (18). Gene expression levels were normalized to endogenous control 18S rRNA. Relative gene expression was determined by the threshold cycle (2−ΔΔCT) method (64). Primers for each gene were chosen according to the recommendation of the qPrimerDepot database (12). Primer sequences were as follows: mouse IFN-α1, GCC TTG ACA CTC CTG GTA CAA ATG AG and CAG CAC ATT GGC AGA GGA AGA CAG; mouse IFIT1, CAA GGC AGG TTT CTG AGG AG and AAG CAG ATT CTC CAT GAC CTG; mouse Ccl5, CTG CTG CTT TGC CTA CCT CT and CAC TTC TTC TCT GGG TTG GC; mouse Cxcl9, TCC TTC CTT CCT TCC TTC CTT CC and AGG CTC TTT TTC ACC CTG TCT GG; human IFN-α1, GGC CTT GAC CTT TGC TTT ACT G and CAC AGA GCA GCT TGA CTT GCA; human IFIT1, CCT CCT TGG GTT CGT CTA CA and AGT GGC TGA TAT CTG GGT GC; human Ccl5, CCT GCT GCT TTG CCT ACA TT and ACA CAC TTG GCG GTT CTT TC; human Cxcl9, CCC TGT TTC TTC CAC AGT GCC TA and GAG ACA ATG GTC TGG TTG CCA TC; 18S rRNA, CCT GCG GCT TAA TTT GAC TC and AAC CAG ACA AAT CGC TCC AC.
Mouse studies.
Five-week-old BALB/c mice were purchased from Harlan Sprague Dawley Inc. and housed under specific-pathogen-free conditions and biosafety level 2 containment. All experimental procedures involving animals were approved by the institutional animal care and use committee of University of Illinois at Chicago. At 6 weeks of age, 1 × 105 viable 4T1 cells suspended in PBS were inoculated subcutaneously into the right flanks of the mice (day −7). When the tumor reached a volume of approximately 100 mm3 8 days after, mice were randomly assigned into three groups. Mice then received three intratumor injections of Δγ134.5, ΔN146, or PBS on days 1, 3, and 6. Each tumor was injected slowly with total of 1 × 107 PFU of virus or PBS in a volume of 0.1 ml. The tumor growth was monitored by measuring two perpendicular tumor diameters the height and with a digital caliper. Tumor volumes were calculated using the following formula: volume = (length × width × height)/2. On day 24 after tumor inoculation, mice were euthanized by CO2 inhalation.
Tissue analysis.
On selected days after the last intratumor injection, six mice from each treatment group were sacrificed to collect the tumor, lungs, liver, spleen, and blood. To measure viral growth, the samples were minced, homogenized, subjected to bead beating, freeze-thawed three times, and sonicated in DMEM. After centrifugation, the tumor supernatants were used for plaque assay. The supernatants from the lungs, liver, spleen, and blood were used for quantitative real-time PCR assay. Briefly, the supernatants were suspended in buffer containing 1% SDS, 50 mM Tris (pH 7.5), and 10 mM EDTA. After incubation with proteinase K (50 μg/ml) at 37°C, viral DNA was extracted and quantified by real-time PCR using HSV-1 gD-specific primers (TAC AAC CTG ACC ATC GCT TG and GCC CCC AGA GAC TTG TTG TA). For metastatic formation assay, the lungs from mice were excised and fixed in formalin. The number of lung metastases was quantified by counting under a light microscope (65).
Immunohistochemistry analysis.
Tissue sections were processed and HSV-1 antigens were detected with antibody against HSV-1 (Dako) as described previously (18). CD4 (25229; Cell Signaling Technology, Inc.) and CD8 (98941; Cell Signaling Technology, Inc.) were straining according to the manufacturer protocol. Samples were incubated with primary antibody prior to the addition of biotinylated anti-rabbit immunoglobulin secondary antibody, avidin-horseradish peroxidase, and 3,3-diaminobenzidine tetrahydrochloride (0.04%) in 0.05 M Tris-HCl (pH 7.4) and 0.025% H2O2 as a chromogen (Ventana Medical Systems, Tucson, AZ).
ACKNOWLEDGMENTS
We thank Gary Cohen and Roselyn Eisenberg for anti-gC antibody.
This work was supported in part by grant from the National Institute of Allergy and Infectious Diseases (AI112755 to B.H.) and department funds.
REFERENCES
- 1.Peters C, Rabkin SD. 2015. Designing herpes viruses as oncolytics. Mol Ther Oncolytics 2:15010. doi: 10.1038/mto.2015.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andtbacka RH, Kaufman HL, Collichio F, Amatruda T, Senzer N, Chesney J, Delman KA, Spitler LE, Puzanov I, Agarwala SS, Milhem M, Cranmer L, Curti B, Lewis K, Ross M, Guthrie T, Linette GP, Daniels GA, Harrington K, Middleton MR, Miller WH Jr, Zager JS, Ye Y, Yao B, Li A, Doleman S, VanderWalde A, Gansert J, Coffin RS. 2015. Talimogene laherparepvec improves durable response rate in patients with advanced melanoma. J Clin Oncol 33:2780–2788. doi: 10.1200/JCO.2014.58.3377. [DOI] [PubMed] [Google Scholar]
- 3.Markert JM, Medlock MD, Rabkin SD, Gillespie GY, Todo T, Hunter WD, Palmer CA, Feigenbaum F, Tornatore C, Tufaro F, Martuza RL. 2000. Conditionally replicating herpes simplex virus mutant, G207 for the treatment of malignant glioma: results of a phase I trial. Gene Ther 7:867–874. doi: 10.1038/sj.gt.3301205. [DOI] [PubMed] [Google Scholar]
- 4.Rampling R, Cruickshank G, Papanastassiou V, Nicoll J, Hadley D, Brennan D, Petty R, MacLean A, Harland J, McKie E, Mabbs R, Brown M. 2000. Toxicity evaluation of replication-competent herpes simplex virus (ICP 34.5 null mutant 1716) in patients with recurrent malignant glioma. Gene Ther 7:859–866. doi: 10.1038/sj.gt.3301184. [DOI] [PubMed] [Google Scholar]
- 5.MacKie RM, Stewart B, Brown SM. 2001. Intralesional injection of herpes simplex virus 1716 in metastatic melanoma. Lancet 357:525–526. doi: 10.1016/S0140-6736(00)04048-4. [DOI] [PubMed] [Google Scholar]
- 6.Streby KA, Geller JI, Currier MA, Warren PS, Racadio JM, Towbin AJ, Vaughan MR, Triplet M, Ott-Napier K, Dishman DJ, Backus LR, Stockman B, Brunner M, Simpson K, Spavin R, Conner J, Cripe TP. 2017. Intratumoral injection of HSV1716, an oncolytic herpes virus, is safe and shows evidence of immune response and viral replication in young cancer patients. Clin Cancer Res 23:3566–3574. doi: 10.1158/1078-0432.CCR-16-2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fukuhara H, Ino Y, Todo T. 2016. Oncolytic virus therapy: a new era of cancer treatment at dawn. Cancer Sci 107:1373–1379. doi: 10.1111/cas.13027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chou J, Kern ER, Whitley RJ, Roizman B. 1990. Mapping of herpes simplex virus-1 neurovirulence to g134.5, a gene nonessential for growth in culture. Science 250:1262–1266. doi: 10.1126/science.2173860. [DOI] [PubMed] [Google Scholar]
- 9.MacLean AR, Ul-Fareed M, Robertson L, Harland J, Brown SM. 1991. Herpes simplex virus type 1 deletion variants 1714 and 1716 pinpoint neurovirulence-related sequences in Glasgow strain 17+ between immediate early gene 1 and the “a” sequence. J Gen Virol 72:631–639. doi: 10.1099/0022-1317-72-3-631. [DOI] [PubMed] [Google Scholar]
- 10.Chou J, Roizman B. 1990. The herpes simplex virus 1 gene for ICP34.5, which maps in inverted repeats, is conserved in several limited-passage isolates but not in strain 17syn+. J Virol 64:1014–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chou J, Roizman B. 1992. The γ134.5 gene of herpes simplex virus 1 precludes neuroblastoma cells from triggering total shutoff of protein synthesis characteristic of programed cell death in neuronal cells. Proc Natl Acad Sci U S A 89:3266–3270. doi: 10.1073/pnas.89.8.3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He B, Gross M, Roizman B. 1997. The γ134.5 protein of herpes simplex virus 1 complexes with protein phosphatase 1alpha to dephosphorylate the alpha subunit of the eukaryotic translation initiation factor 2 and preclude the shutoff of protein synthesis by double-stranded RNA-activated protein kinase. Proc Natl Acad Sci U S A 94:843–848. doi: 10.1073/pnas.94.3.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Verpooten D, Feng Z, Valyi-Nagy T, Ma Y, Jin H, Yan Z, Zhang C, Cao Y, He B. 2009. Dephosphorylation of eIF2α mediated by the γ134.5 protein of herpes simplex virus 1 facilitates viral neuroinvasion. J Virol 83:12626–12630. doi: 10.1128/JVI.01431-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilcox DR, Muller WJ, Longnecker R. 2015. HSV targeting of the host phosphatase PP1alpha is required for disseminated disease in the neonate and contributes to pathogenesis in the brain. Proc Natl Acad Sci U S A 112:E6937–E6944. doi: 10.1073/pnas.1513045112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bower JR, Mao H, Durishin C, Rozenbom E, Detwiler M, Rempinski D, Karban TL, Rosenthal KS. 1999. Intrastrain variants of herpes simplex virus type 1 isolated from a neonate with fatal disseminated infection differ in the ICP34.5 gene, glycoprotein processing, and neuroinvasiveness. J Virol 73:3843–3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mao H, Rosenthal KS. 2003. Strain-dependent structural variants of herpes simplex virus type 1 ICP34.5 determine viral plaque size, efficiency of glycoprotein processing, and viral release and neuroinvasive disease potential. J Virol 77:3409–3417. doi: 10.1128/JVI.77.6.3409-3417.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Orvedahl A, Alexander D, Talloczy Z, Sun Q, Wei Y, Zhang W, Burns D, Leib DA, Levine B. 2007. HSV-1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein. Cell Host Microbe 1:23–35. doi: 10.1016/j.chom.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 18.Ma Y, Jin H, Valyi-Nagy T, Cao Y, Yan Z, He B. 2012. Inhibition of TANK binding kinase 1 by herpes simplex virus 1 facilitates productive infection. J Virol 86:2188–2196. doi: 10.1128/JVI.05376-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin H, Yan Z, Ma Y, Cao Y, He B. 2011. A herpesvirus virulence factor inhibits dendritic cell maturation through protein phosphatase 1 and Ikappa B kinase. J Virol 85:3397–3407. doi: 10.1128/JVI.02373-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brown SM, MacLean AR, Aitken JD, Harland J. 1994. ICP34.5 influences herpes simplex virus type 1 maturation and egress from infected cells in vitro. J Gen Virol 75:3679–3686. doi: 10.1099/0022-1317-75-12-3679. [DOI] [PubMed] [Google Scholar]
- 21.Wu S, Pan S, Zhang L, Baines J, Roller R, Ames J, Yang M, Wang J, Chen D, Liu Y, Zhang C, Cao Y, He B. 2016. Herpes simplex virus 1 induces phosphorylation and reorganization of lamin A/C through the γ134.5 protein that facilitates nuclear egress. J Virol 90:10414–10422. doi: 10.1128/JVI.01392-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheng G, Brett ME, He B. 2002. Signals that dictate nuclear, nucleolar, and cytoplasmic shuttling of the γ134.5 protein of herpes simplex virus type 1. J Virol 76:9434–9445. doi: 10.1128/JVI.76.18.9434-9445.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mao H, Rosenthal KS. 2002. An N-terminal arginine rich cluster and a proline-alanine-threonine repeat region determines the cellular localization of the herpes simplex virus type-1 ICP34.5 protein and its ligand, protein phosphatase 1. J Biol Chem 11:11. [DOI] [PubMed] [Google Scholar]
- 24.Mineta T, Rabkin SD, Yazaki T, Hunter WD, Martuza RL. 1995. Attenuated multi-mutated herpes simplex virus-1 for the treatment of malignant gliomas. Nat Med 1:938–943. doi: 10.1038/nm0995-938. [DOI] [PubMed] [Google Scholar]
- 25.Toda M, Rabkin SD, Martuza RL. 1998. Treatment of human breast cancer in a brain metastatic model by G207, a replication-competent multimutated herpes simplex virus 1. Hum Gene Ther 9:2177–2185. doi: 10.1089/hum.1998.9.15-2177. [DOI] [PubMed] [Google Scholar]
- 26.Chambers R, Gillespie GY, Soroceanu L, Andreansky S, Chatterjee S, Chou J, Roizman B, Whitley RJ. 1995. Comparison of genetically engineered herpes simplex viruses for the treatment of brain tumors in a scid mouse model of human malignant glioma. Proc Natl Acad Sci U S A 92:1411–1415. doi: 10.1073/pnas.92.5.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Randazzo BP, Kesari S, Gesser RM, Alsop D, Ford JC, Brown SM, Maclean A, Fraser NW. 1995. Treatment of experimental intracranial murine melanoma with a neuroattenuated herpes simplex virus 1 mutant. Virology 211:94–101. doi: 10.1006/viro.1995.1382. [DOI] [PubMed] [Google Scholar]
- 28.Thomas DL, Fraser NW. 2003. HSV-1 therapy of primary tumors reduces the number of metastases in an immune-competent model of metastatic breast cancer. Mol Ther 8:543–551. doi: 10.1016/S1525-0016(03)00236-3. [DOI] [PubMed] [Google Scholar]
- 29.Coukos G, Makrigiannakis A, Kang EH, Rubin SC, Albelda SM, Molnar-Kimber KL. 2000. Oncolytic herpes simplex virus-1 lacking ICP34.5 induces p53-independent death and is efficacious against chemotherapy-resistant ovarian cancer. Clin Cancer Res 6:3342–3353. [PubMed] [Google Scholar]
- 30.Braidwood L, Learmonth K, Graham A, Conner J. 2014. Potent efficacy signals from systemically administered oncolytic herpes simplex virus (HSV1716) in hepatocellular carcinoma xenograft models. J Hepatocell Carcinoma 1:149–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang PY, Swain HM, Kunkler AL, Chen CY, Hutzen BJ, Arnold MA, Streby KA, Collins MH, Dipasquale B, Stanek JR, Conner J, van Kuppevelt TH, Glorioso JC, Grandi P, Cripe TP. 2016. Neuroblastomas vary widely in their sensitivities to herpes simplex virotherapy unrelated to virus receptors and susceptibility. Gene Ther 23:135–143. doi: 10.1038/gt.2015.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Farassati F, Yang AD, Lee PW. 2001. Oncogenes in Ras signalling pathway dictate host-cell permissiveness to herpes simplex virus 1. Nat Cell Biol 3:745–750. doi: 10.1038/35087061. [DOI] [PubMed] [Google Scholar]
- 33.Smith KD, Mezhir JJ, Bickenbach K, Veerapong J, Charron J, Posner MC, Roizman B, Weichselbaum RR. 2006. Activated MEK suppresses activation of PKR and enables efficient replication and in vivo oncolysis by Deltagamma(1)34.5 mutants of herpes simplex virus 1. J Virol 80:1110–1120. doi: 10.1128/JVI.80.3.1110-1120.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xia T, Konno H, Ahn J, Barber GN. 2016. Deregulation of STING signaling in colorectal carcinoma constrains DNA damage responses and correlates with tumorigenesis. Cell Rep 14:282–297. doi: 10.1016/j.celrep.2015.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xia T, Konno H, Barber GN. 2016. Recurrent loss of STING signaling in melanoma correlates with susceptibility to viral oncolysis. Cancer Res 76:6747–6759. doi: 10.1158/0008-5472.CAN-16-1404. [DOI] [PubMed] [Google Scholar]
- 36.Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM. 2011. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Woo SR, Fuertes MB, Corrales L, Spranger S, Furdyna MJ, Leung MY, Duggan R, Wang Y, Barber GN, Fitzgerald KA, Alegre ML, Gajewski TF. 2014. STING-dependent cytosolic DNA sensing mediates innate immune recognition of immunogenic tumors. Immunity 41:830–842. doi: 10.1016/j.immuni.2014.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parker BS, Rautela J, Hertzog PJ. 2016. Antitumour actions of interferons: implications for cancer therapy. Nat Rev Cancer 16:131–144. doi: 10.1038/nrc.2016.14. [DOI] [PubMed] [Google Scholar]
- 39.Ma Y, Chen M, Jin H, Prabhakar BS, Valyi-Nagy T, He B. 2017. An engineered herpesvirus activates dendritic cells and induces protective immunity. Sci Rep 7:41461. doi: 10.1038/srep41461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Donnelly N, Gorman AM, Gupta S, Samali A. 2013. The eIF2alpha kinases: their structures and functions. Cell Mol Life Sci 70:3493–3511. doi: 10.1007/s00018-012-1252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feng YX, Sokol ES, Del Vecchio CA, Sanduja S, Claessen JH, Proia TA, Jin DX, Reinhardt F, Ploegh HL, Wang Q, Gupta PB. 2014. Epithelial-to-mesenchymal transition activates PERK-eIF2alpha and sensitizes cells to endoplasmic reticulum stress. Cancer Discov 4:702–715. doi: 10.1158/2159-8290.CD-13-0945. [DOI] [PubMed] [Google Scholar]
- 42.Feng YX, Jin DX, Sokol ES, Reinhardt F, Miller DH, Gupta PB. 2017. Cancer-specific PERK signaling drives invasion and metastasis through CREB3L1. Nat Commun 8:1079. doi: 10.1038/s41467-017-01052-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pulaski BA, Ostrand-Rosenberg S. 1998. Reduction of established spontaneous mammary carcinoma metastases following immunotherapy with major histocompatibility complex class II and B7.1 cell-based tumor vaccines. Cancer Res 58:1486–1493. [PubMed] [Google Scholar]
- 44.Liu X, Main D, Ma Y, He B. 2018. Herpes simplex virus 1 inhibits TANK-binding kinase 1 through formation of the Us11-Hsp90 complex. J Virol 92:e00402-18. doi: 10.1128/JVI.00402-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu BL, Robinson M, Han ZQ, Branston RH, English C, Reay P, McGrath Y, Thomas SK, Thornton M, Bullock P, Love CA, Coffin RS. 2003. ICP34.5 deleted herpes simplex virus with enhanced oncolytic, immune stimulating, and anti-tumour properties. Gene Ther 10:292–303. doi: 10.1038/sj.gt.3301885. [DOI] [PubMed] [Google Scholar]
- 46.Toda M, Rabkin SD, Kojima H, Martuza RL. 1999. Herpes simplex virus as an in situ cancer vaccine for the induction of specific anti-tumor immunity. Hum Gene Ther 10:385–393. doi: 10.1089/10430349950018832. [DOI] [PubMed] [Google Scholar]
- 47.Andreansky SS, He B, Gillespie GY, Soroceanu L, Markert J, Chou J, Roizman B, Whitley RJ. 1996. The application of genetically engineered herpes simplex viruses to the treatment of experimental brain tumors. Proc Natl Acad Sci U S A 93:11313–11318. doi: 10.1073/pnas.93.21.11313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cheng G, Brett M-E, He B. 2001. Val193 and Phe195 of the γ134.5 protein of herpes simplex virus 1 are required for viral resistance to interferon-α/β. Virology 290:115–120. doi: 10.1006/viro.2001.1148. [DOI] [PubMed] [Google Scholar]
- 49.Cassady KA, Gross M, Roizman B. 1998. The herpes simplex virus US11 protein effectively compensates for the gamma1(34.5) gene if present before activation of protein kinase R by precluding its phosphorylation and that of the alpha subunit of eukaryotic translation initiation factor 2. J Virol 72:8620–8626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Poppers J, Mulvey M, Khoo D, Mohr I. 2000. Inhibition of PKR activation by the proline-rich RNA binding domain of the herpes simplex virus type 1 Us11 protein. J Virol 74:11215–11221. doi: 10.1128/JVI.74.23.11215-11221.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ye J, Kumanova M, Hart LS, Sloane K, Zhang H, De Panis DN, Bobrovnikova-Marjon E, Diehl JA, Ron D, Koumenis C. 2010. The GCN2-ATF4 pathway is critical for tumour cell survival and proliferation in response to nutrient deprivation. EMBO J 29:2082–2096. doi: 10.1038/emboj.2010.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nguyen HG, Conn CS, Kye Y, Xue L, Forester CM, Cowan JE, Hsieh AC, Cunningham JT, Truillet C, Tameire F, Evans MJ, Evans CP, Yang JC, Hann B, Koumenis C, Walter P, Carroll PR, Ruggero D. 2018. Development of a stress response therapy targeting aggressive prostate cancer. Sci Transl Med 10:eaar2036. doi: 10.1126/scitranslmed.aar2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pan S, Liu X, Ma Y, Cao Y, He B. 2018. Herpes simplex virus 1 γ134.5 protein inhibits STING activation that restricts viral replication. J Virol 92:e01015-18. doi: 10.1128/JVI.01015-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sun L, Wu J, Du F, Chen X, Chen ZJ. 2013. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS, Fitzgerald KA, Paludan SR, Bowie AG. 2010. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol 11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang Z, Yuan B, Bao M, Lu N, Kim T, Liu YJ. 2011. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat Immunol 12:959–965. doi: 10.1038/ni.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ma Y, He B. 2014. Recognition of herpes simplex viruses: Toll-like receptors and beyond. J Mol Biol 426:1133–1147. doi: 10.1016/j.jmb.2013.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ouzounova M, Lee E, Piranlioglu R, El Andaloussi A, Kolhe R, Demirci MF, Marasco D, Asm I, Chadli A, Hassan KA, Thangaraju M, Zhou G, Arbab AS, Cowell JK, Korkaya H. 2017. Monocytic and granulocytic myeloid derived suppressor cells differentially regulate spatiotemporal tumour plasticity during metastatic cascade. Nat Commun 8:14979. doi: 10.1038/ncomms14979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ejercito PM, Kieff ED, Roizman B. 1968. Characterization of herpes simplex virus strains differing in their effects on social behaviour of infected cells. J Gen Virol 2:357–364. doi: 10.1099/0022-1317-2-3-357. [DOI] [PubMed] [Google Scholar]
- 60.Goins WF, Krisky DM, Wechuck JB, Wolfe D, Huang S, Glorioso JC. 2011. Generation of replication-competent and -defective HSV vectors. Cold Spring Harb Protoc 2011:pdb.prot5615. doi: 10.1101/pdb.prot5615. [DOI] [PubMed] [Google Scholar]
- 61.Jing X, Cerveny M, Yang K, He B. 2004. Replication of herpes simplex virus 1 depends on the γ134.5 functions that facilitate virus response to interferon and egress in the different stages of productive infection. J Virol 78:7653–7666. doi: 10.1128/JVI.78.14.7653-7666.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Carvalho BS, Irizarry RA. 2010. A framework for oligonucleotide microarray preprocessing. Bioinformatics 26:2363–2367. doi: 10.1093/bioinformatics/btq431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. 2015. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schmittgen TD, Livak KJ. 2008. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 65.Pourchet A, Fuhrmann SR, Pilones KA, Demaria S, Frey AB, Mulvey M, Mohr I. 2016. CD8(+) T-cell immune evasion enables oncolytic virus immunotherapy. EBioMedicine 5:59–67. doi: 10.1016/j.ebiom.2016.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]