The human MxA protein efficiently blocks the replication of IAV from nonhuman species. In rare cases, however, these IAV overcome the species barrier and become pandemic. All known pandemic viruses have acquired and maintained MxA escape mutations in the viral NP and thus are not efficiently controlled by MxA. Intriguingly, partial MxA resistance can also be acquired in other hosts that express antivirally active Mx proteins, such as swine. To perform a risk assessment of IAV circulating in the European swine population, we analyzed the degree of MxA resistance of Eurasian avian-like swine IAV. Our data demonstrate that these viruses carry formerly undescribed Mx resistance mutations in the NP that mediate efficient escape from human MxA. We conclude that Eurasian avian-like swine IAV possess substantial zoonotic potential.
KEYWORDS: Eurasian avian-like swine IAV, influenza A virus, MxA, host barrier, innate immunity, pH1N1, pandemic
ABSTRACT
To cross the human species barrier, influenza A viruses (IAV) of avian origin have to overcome the interferon-induced host restriction factor MxA by acquiring distinct mutations in their nucleoprotein (NP). We recently demonstrated that North American classical swine IAV are able to partially escape MxA restriction. Here we investigated whether the Eurasian avian-like swine IAV lineage currently circulating in European swine would likewise evade restriction by human MxA. We found that the NP of the influenza virus isolate A/Swine/Belzig/2/2001 (Belzig-NP) exhibits increased MxA escape, similar in extent to that with human IAV NPs. Mutational analysis revealed that the MxA escape mutations in Belzig-NP differ from the known MxA resistance cluster of the North American classical swine lineage and human-derived IAV NPs. A mouse-adapted avian IAV of the H7N7 subtype encoding Belzig-NP showed significantly greater viral growth in both MxA-expressing cells and MxA-transgenic mice than control viruses lacking the MxA escape mutations. Similarly, the growth of the recombinant Belzig virus was only marginally affected in MxA-expressing cells and MxA-transgenic mice, in contrast to that of Belzig mutant viruses lacking MxA escape mutations in the NP. Phylogenetic analysis of the Eurasian avian-like swine IAV revealed that the NP amino acids required for MxA escape were acquired successively and were maintained after their introduction. Our results suggest that the circulation of IAV in the swine population can result in the selection of NP variants with a high degree of MxA resistance, thereby increasing the zoonotic potential of these viruses.
IMPORTANCE The human MxA protein efficiently blocks the replication of IAV from nonhuman species. In rare cases, however, these IAV overcome the species barrier and become pandemic. All known pandemic viruses have acquired and maintained MxA escape mutations in the viral NP and thus are not efficiently controlled by MxA. Intriguingly, partial MxA resistance can also be acquired in other hosts that express antivirally active Mx proteins, such as swine. To perform a risk assessment of IAV circulating in the European swine population, we analyzed the degree of MxA resistance of Eurasian avian-like swine IAV. Our data demonstrate that these viruses carry formerly undescribed Mx resistance mutations in the NP that mediate efficient escape from human MxA. We conclude that Eurasian avian-like swine IAV possess substantial zoonotic potential.
INTRODUCTION
Influenza A viruses (IAV) are significant human pathogens, all originating from their natural reservoir, wild aquatic waterfowl (1, 2). Human infection by avian IAV occurs sporadically but is usually restricted to individuals. In rare cases, however, avian IAV obtain sufficient adaptive mutations to facilitate sustained human-infection chains, incidents that can result in devastating pandemics (1).
Swine seem to play a particularly important role in the emergence of pandemic viruses. This is in part due to their high susceptibility to avian as well as human IAV, which can result in coinfection with IAV from both origins and exchange of whole gene segments, termed gene “reassortment” (3). Reassortment in swine has occurred at a highly elevated frequency since the late 1990s (4, 5), and the 2009 pandemic “swine flu” virus (pH1N1) represents a complex reassortant virus, comprising gene segments from human seasonal IAV as well as genes from the North American avian IAV, the North American classical swine IAV, and the Eurasian avian-like swine IAV lineage.
The interferon-induced and antivirally active MxA GTPase found in humans is believed to represent a major barrier to the establishment of a new lineage in the human population by avian IAV (6–9). As a consequence, all human-adapted IAV, including the pandemic 1918 and pH1N1 viruses, as well as all their descendants, encode adaptive mutations in their nucleoproteins (NPs) that enable them to escape MxA restriction, whereas avian IAV lack such amino acids (8). The acquisition of MxA resistance mutations seems, however, to be frequently associated with a loss of viral fitness and thus to be a difficult process (8, 10). This might explain why a novel NP with sufficient MxA escape amino acids has been introduced into the human population only twice in the last 100 years, in 1918 and in 2009. In the case of the pH1N1 virus, we found strong evidence that the acquisition of the required escape mutations in the NP was driven by porcine Mx1 (poMx1) during the circulation of the pH1N1 precursor in pigs (8). This virus of the classical North American swine IAV lineage gradually obtained sufficient NP mutations to counteract poMx1 and concomitantly to partially resist the activity of human MxA. The pH1N1 virus acquired this preadapted NP through reassortment and gained additional mutations enabling complete escape from the more potent human MxA (8).
In Europe, the IAV most frequently isolated from pigs belong to a lineage different from the North American classical swine IAV lineage, namely, the Eurasian avian-like swine IAV lineage (4, 11, 12). Here, using polymerase reconstitution assays, we found that specific amino acids in the NP of the Eurasian avian-like influenza virus strain A/swine/Belzig/2/2001 (Belzig) allow escape from human MxA comparable in degree to that with the NP of the pH1N1 virus. This pronounced resistance to MxA was validated in MxA-overexpressing cells both by using a recombinant Belzig virus and by replacing the NP of an otherwise MxA-sensitive virus with Belzig-NP. In addition, we could demonstrate, by use of a transgenic MxA mouse model, that Belzig-NP is able to render an avian IAV resistant to MxA. Finally, phylogenetic analysis revealed that the critical MxA escape amino acids have been introduced successively over time and were maintained in the Eurasian avian-like swine IAV, suggesting positive selection by poMx1.
RESULTS
Identification of residues in Eurasian avian-like swine NP that are responsible for MxA escape.
We have demonstrated previously that the MxA-sensitive influenza virus A/Thailand/1(KAN-1)/2004 (H5N1)-based polymerase reconstitution assay can be readily utilized to identify MxA escape mutations within the nucleoproteins (NPs) of human-adapted influenza A viruses (IAV) such as the 1918 and 2009 (pH1N1) pandemic strains (8). Therefore, we employed a similar approach to determine the level of MxA sensitivity conferred by the NP of the Eurasian avian-like swine isolate A/Swine/Belzig/2/2001 (Belzig), which differs from H5N1-NP at 18 amino acid positions (Fig. 1A; see also Fig. S1A in the supplemental material). In our assay, H5N1 polymerase activity is measured not only in the presence of MxA but also in the presence of the antivirally inactive variant with a T-to-A change at position 103 (MxA-T103A) in order to account for differences in general polymerase activity in the context of different NPs. The relative activity (ratio of MxA to MxA-T103A) (see also Materials and Methods) indicates whether an NP is able to resist the antiviral effect of MxA. As expected, the polymerase activity of H5N1-NP was strongly inhibited by MxA, while the pH1N1-NP from influenza virus A/Hamburg/4/2009 largely resisted MxA inhibition (Fig. 1B). Intriguingly, however, Belzig-NP not only partially escaped restriction by MxA but conferred a level of MxA resistance comparable to that of pH1N1-NP, suggesting that Belzig-NP harbors MxA escape mutations of high potency.
FIG 1.
The nucleoprotein of Eurasian avian-like swine influenza viruses confers MxA resistance in polymerase reconstitution assays. (A) Amino acid differences between the NPs of H5N1, H7N7 (dark gray), pH1N1 (red), and Belzig (green) strains (for a complete alignment, see Fig. S1A in the supplemental material). Underlined positions have been shown previously to influence the MxA sensitivity phenotype. (B) (Top) The ratio of the activity of MxA to the activity of antivirally inactive MxA-T103A was normalized to that for pH1N1-NP (set at 1) and is shown as relative activity (shaded bars). (Center) Viral polymerase activity in the presence of MxA (filled bars) or MxA-T103A (open bars). HEK293T cells were transfected with expression plasmids encoding H5N1-PB2, -PB1, and -PA (10 ng), the indicated NP (100 ng), MxA (50 ng), and an artificial minigenome encoding firefly luciferase under the control of the polI promoter (100 ng). Additionally, a plasmid encoding Renilla luciferase (30 ng) was transfected as a control for transfection efficiency. Error bars indicate the standard errors of the means from at least four independent experiments. Student’s t test was performed to determine the P value for the difference from the respective wild-type NP. *, P < 0.05; **, P < 0.01; ***, P < 0.001. (Bottom) Expression levels of MxA and NP were detected via Western blotting. Actin was used as a loading control. (C) MxA escape mutations previously identified in pH1N1-NP and the newly identified mutations in Belzig-NP are shown in red and green, respectively, in the structural model of influenza virus A/HK/483/97 (H5N1) NP (PDB code 2Q06).
Aiming to identify the Belzig-NP amino acids responsible for increased MxA resistance, we aligned the amino acid sequence of Belzig-NP to that of the MxA-sensitive H5N1-NP as well as to the H7N7-NP from the MxA-sensitive (9, 10) strain A/Seal/Massachusetts/1/1980 (H7N7) and to the MxA-resistant NP of the pH1N1 virus (Fig. 1A; also Fig. S1A in the supplemental material). Carboxy-terminal amino acids beyond amino acid 357 were excluded, since they had been shown previously not to contribute to MxA resistance (8). As expected, the amino acid sequence of H7N7-NP shows high similarity to that of H5N1-NP, while Belzig-NP and, even more, pH1N1-NP show increased numbers of differences. Surprisingly, however, none of the major pH1N1-NP-specific MxA escape mutations are found in Belzig-NP. Therefore, by utilizing the alignment as well as the published IAV NP crystal structure (13) (PDB code 2Q06), surface-exposed Belzig-NP-specific amino acids in close proximity to the known pH1N1-NP MxA escape cluster were selected for further analysis (Fig. 1A; also Fig. S1B).
Using site-directed mutagenesis, we constructed expression vectors encoding Belzig-NP mutant proteins with H5N1-specific residues at the selected amino acid positions (Fig. 1B). The H5N1 polymerase reconstitution assay revealed that MxA escape by Belzig-NP relies particularly on amino acids 48Q and 99K, since mutation to the H5N1-specific residue 48K or 99R led to significantly decreased MxA resistance (Fig. 1B). Since Belzig-NP amino acid 98K is found directly adjacent to 48Q and 99K in the NP crystal structure (Fig. 1C; also Fig. S1B in the supplemental material), we combined the replacements of H5N1-NP-specific amino acids for these positions. While the combination of two mutations hardly affected MxA resistance relative to that of the single mutants Belzig-NP-Q48K and Belzig-NP-K99R, the combination of all three mutations (Q48K, K98R, and K99R) in Belzig-NP reduced MxA resistance to a level comparable to that of H5N1-NP. Vice versa, mutation of all three H5N1-NP amino acids at positions 48, 98, and 99 to Belzig-NP-specific residues (H5N1-NP-K48Q, R98K, R99K) was required to render H5N1-NP as MxA resistant as Belzig-NP (Fig. 1B). Finally, we investigated whether the same three Belzig-NP amino acids would be sufficient to escape restriction by porcine Mx1 (poMx1). The inhibitory activity of poMx1 had been demonstrated to be less pronounced than that of MxA (8). Therefore, the polymerase activity of H5N1-NP was inhibited only moderately in the presence of poMx1 (Fig. S1C). In line with our results obtained using MxA, H5N1-NP carrying all three Belzig-NP MxA escape mutations (H5N1-NP-K48Q, R98K, R99K) displayed increased poMx1 resistance, comparable to that of wild type Belzig-NP. All three identified amino acids of Belzig-NP localize to an area that is adjacent to the MxA escape amino acids of pH1N1-NP (Fig. 1C).
Belzig-NP confers MxA resistance comparable to that of human-adapted viruses via amino acids 48Q, 98K, and 99K.
To evaluate the MxA resistance conferred by Belzig-NP in a viral context, we generated recombinant viruses. For this purpose, we used the established MxA-sensitive influenza virus strain A/Seal/Massachusetts/1/1980 (H7N7), which had been used successfully previously to study MxA resistance in cell culture as well as in MxA-transgenic mice (9, 10). As shown before, MxA resistance-conferring mutations can reduce viral fitness. Therefore, we first investigated viral growth in MDCKII cells, which do not express antivirally active Mx proteins (8, 14), by infecting them at a multiplicity of infection (MOI) of 0.001 and determining viral titers by use of plaque assays after 12, 24, 36, and 48 h.
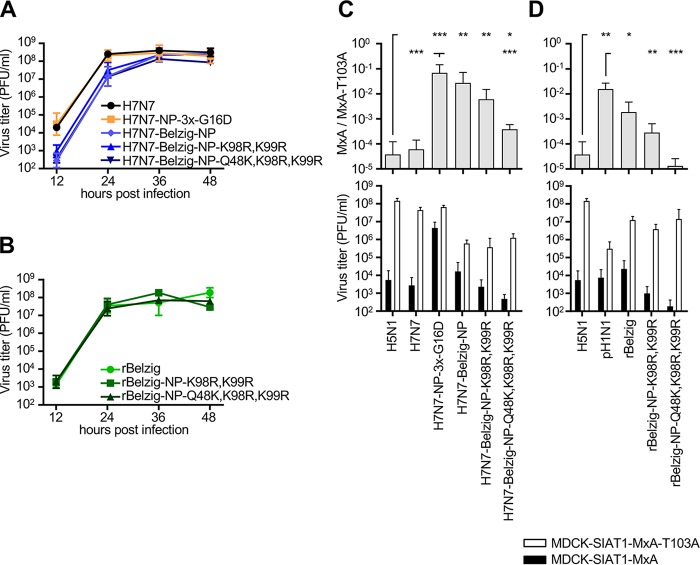
As demonstrated before (10), H7N7-NP-3x-G16D, harboring all three MxA escape mutations (R100V, L283P, and F313Y) of the pandemic 1918 strain as well as the stabilizing mutation G16D, grew to titers similar to those of the wild-type H7N7 virus (Fig. 2A). The introduction of Belzig-NP into the H7N7 virus (H7N7-Belzig-NP), however, resulted in a >1.5-log10 reduction in viral titers at early time points. This loss in viral fitness could not be overcome by the introduction of the MxA resistance-decreasing amino acids K98R and K99R (H7N7-Belzig-NP-K98R, K99R) or K98R, K99R, and Q48K (H7N7-Belzig-NP-Q48K, K98R, K99R). Therefore, the slight attenuation observed in the context of the H7N7 virus seems to result from factors other than MxA resistance. We also generated a recombinant wild-type Belzig virus (rBelzig) and recombinant Belzig viruses with a combination of the mutations K98R and K99R (rBelzig-NP-K98R, K99R) or K98R, K99R, and Q48K (rBelzig-NP-Q48K, K98R, K99R). Both the wild-type and mutant viruses grew efficiently and to similar viral titers (Fig. 2B) except at the last time point, 48 h, when the mutants were delayed by 0.5 to 1 log10 PFU/ml. Additionally, we investigated growth in swine NPTr cells. Here, the two mutant Belzig viruses lacking MxA escape mutations showed impaired replication relative to that of the wild-type Belzig virus (Fig. S1D in the supplemental material).
FIG 2.
Influenza A viruses encoding Belzig-NP escape MxA restriction in MxA-overexpressing cells. (A and B) MDCKII cells were infected at an MOI of 0.001 with wild-type or mutant H7N7 (A) or rBelzig (B) viruses and were incubated at 37°C. Viral titers were determined at 12, 24, 36, and 48 h postinfection via plaque assay. Error bars indicate the standard errors of the means from at least three independent experiments. (C and D) MDCK-SIAT1 cells stably expressing either MxA (filled bars) or the antivirally inactive MxA-T103A mutant (open bars) were infected at an MOI of 0.001 with wild-type or mutant H7N7 (C) or rBelzig (D) viruses and were incubated at 37°C. The H5N1 and pH1N1 viruses were used as controls for MxA-sensitive and -resistant viruses, respectively. Viral titers were determined 24 h postinfection via plaque assay. Error bars indicate the standard errors of the means from at least three independent experiments. Student’s t test was performed to determine the P value. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
To evaluate the MxA sensitivity of the viruses generated, we made use of the established cell line MDCK-SIAT1-MxA and the control cell line MDCK-SIAT1-MxA-T103A, stably overexpressing MxA or antivirally inactive MxA-T103A, respectively (15). Cells were infected at an MOI of 0.001, and viral titers were determined 24 h postinfection. The ratios of the viral titers in MDCK-SIAT1-MxA cells to those in MDCK-SIAT1-MxA-T103A cells were calculated in order to more easily assess the MxA sensitivity phenotype. All viruses grew to reasonable titers in the absence of antivirally active MxA (Fig. 2C, bottom, open bars). As expected, the growth of the wild-type H5N1 and H7N7 viruses was strongly inhibited by MxA, with >4-log10 reductions in titers in both cases, while the positive control, H7N7-NP-3x-G16D, was only minimally affected by the activity of MxA (∼1-log10 reduction). A similar phenotype of highly increased MxA resistance was observed with H7N7-Belzig-NP (<1.5-log10 reduction). In accord with our results presented above (Fig. 1B), the introduction of the two avian influenza virus amino acids 98R and 99R into H7N7 virus-encoded Belzig-NP resulted in decreased MxA resistance, reducing viral titers by >2 log10 PFU/ml in the presence of MxA. Finally, the replacement of all three MxA escape mutations in Belzig-NP with amino acids of avian influenza virus origin (Q48K, K98R, and K99R) led to a phenotype comparable to those of the MxA-sensitive H5N1 and H7N7 viruses: the difference in viral titers between active and inactive MxA reached >3 log10 units (Fig. 2C).
Next, we compared the viral growth of the rBelzig and 2009 pandemic pH1N1 influenza virus strains in MDCK-SIAT1-MxA and MDCK-SIAT1-MxA-T103A cells. As shown in Fig. 2D, rBelzig showed a level of MxA resistance comparable to that of the pH1N1 virus, with titers reduced by <2.5 log10 units in the presence of MxA. In contrast, viral replication of rBelzig-NP-K98R, K99R and, in particular, rBelzig-NP-Q48K, K98R, K99R was strongly reduced in the MxA-overexpressing cell line, reaching a >4.5-log10 reduction in the case of the triple mutant and thus matching the phenotype of the MxA-sensitive H5N1 virus. In summary, these data show that Belzig-NP confers a level of MxA resistance comparable to that of human-adapted pandemic viruses and that this resistance is due to the three mutations K48Q, R98K, and R99K.
Belzig-NP mediates increased MxA resistance in MxA-transgenic mice.
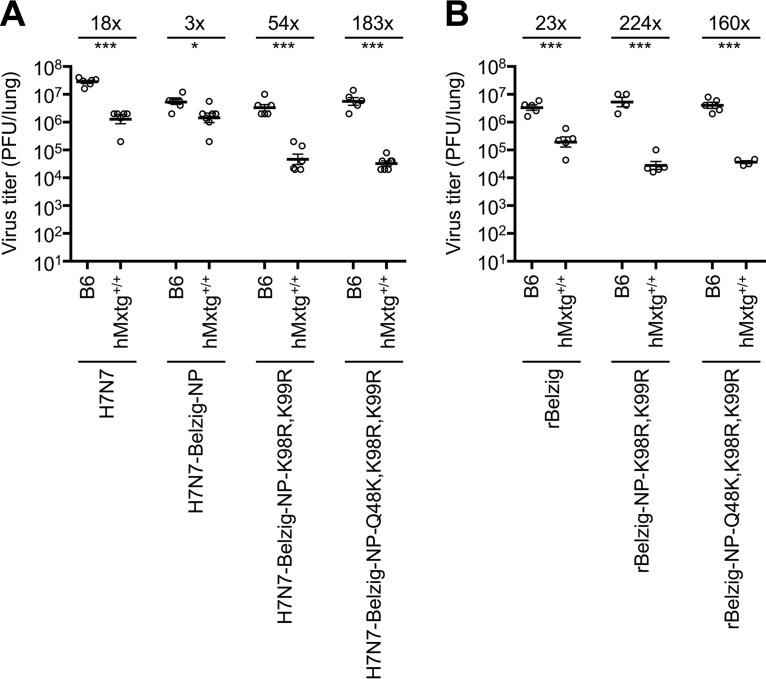
MxA-transgenic mice have recently been shown to be a bona fide animal model for distinguishing IAV strains susceptible to inhibition by MxA from MxA-resistant strains (9). We aimed to use this model to investigate whether Belzig-NP also confers MxA resistance in vivo. For this purpose, C57BL/6N (B6) control mice or homozygous MxA-transgenic (hMxtg+/+) mice were infected intranasally using an inoculum of 104 PFU/40 μl. Three days postinfection (dpi), lungs were harvested, and viral titers were determined using plaque assays. The wild-type H7N7 virus grew to high titers of ∼5 × 107 PFU/lung in B6 mice (Fig. 3A). As expected from the earlier cell culture experiments (Fig. 2A and C), titers of H7N7-Belzig-NP and the mutant viruses (H7N7-Belzig-NP-K98R, K99R and H7N7-Belzig-NP-Q48K, K98R, K99R) were reduced by about 1 log10 unit (Fig. 3A). In hMxtg+/+ mice, on the other hand, the titer of the H7N7 virus was clearly about 18-fold lower than the titer observed in B6 mice. In contrast, H7N7-Belzig-NP largely resisted MxA activity in hMxtg+/+ mice, and titers were hardly affected (Fig. 3A). As expected, the introduction of 98R and 99R or of all three MxA-sensitive amino acids, 98R, 99R, and 48K (H7N7-Belzig-NP-K98R, K99R or H7N7-Belzig-NP-Q48K, K98R, K99R), resulted in a pronounced >2-log10 reduction in viral titers in hMxtg+/+ mice. Recombinant wild-type Belzig and mutant viruses grew to comparable titers of around 6 × 106 PFU/ml in B6 mice, indicating that amino acids 48Q, 98K, and 99K do not impair viral replication (Fig. 3B). However, the titers of rBelzig-NP-K98R, K99R and rBelzig-NP-Q48K, K98R, K99R were strongly decreased in hMxtg+/+ mice, by 224- and 160-fold, respectively, whereas the titer of wild-type Belzig virus was reduced by only 23-fold. These data demonstrate that Belzig-NP confers MxA resistance, which relies mainly on amino acids 48Q, 98K, and 99K, not only in cell culture but also in vivo.
FIG 3.
Influenza A viruses encoding Belzig-NP escape restriction in MxA-transgenic mice. (A and B) C57BL/6N (B6) mice lacking functional Mx proteins and B6 mice homozygous for the human MxA transgene (hMxtg+/+) were challenged intranasally with 104 PFU of the indicated viruses with the H7N7 (A) or rBelzig (B) backbone. Viral titers in the lungs at 3 days postinfection were determined via plaque assay. Error bars indicate the standard errors of the means for at least four mice. The fold decrease in the titer in hMxtg+/+ mice from that in B6 mice is shown at the top. Student’s t test was performed to determine the P value for the difference between B6 and hMxtg+/+ mice. *, P < 0.05; ***, P < 0.001.
Positive selection of Mx resistance-increasing mutations in swine.
We recently showed that the acquisition of Mx resistance-conferring mutations in NP is evolutionarily driven by positive selection through antivirally active Mx proteins (8). Based on these data, it is therefore conceivable that the three Belzig-NP amino acids 48Q, 98K, and 99K were preferentially selected by porcine Mx1 during the circulation of the Eurasian avian-like swine IAV lineage in swine. To test this hypothesis, we generated a phylogenetic tree comprising 267 Eurasian avian-like swine lineage isolates collected at different time points between 1979 and 2013. Additionally, a number of characteristic isolates of several other lineages, including human-adapted seasonal descendants of pandemic IAV, were added to provide an elaborate overview through IAV NP evolution (Fig. 4 shows a condensed phylogenetic tree, whereas Fig. S2 in the supplemental material demonstrates the complete phylogenetic tree). As expected, the amino acids 48Q, 98K, and 99K appear in the NPs of the Eurasian avian-like swine IAV lineage in a consecutive manner, separated by several years. The mutation with the most pronounced MxA escape effect, R99K, was observed first, in an isolate from 1980, followed by the K48Q mutation in 1989. R98K was found in isolates last, beginning in 1992. The phylogenetic tree also demonstrates that once they emerged, the amino acid changes were maintained. Intriguingly, viruses of the Eurasian avian-like swine IAV lineage were isolated only from humans (zoonotic events highlighted with asterisks in Fig. 4) since the time when all three mutations were acquired.
FIG 4.
Phylogenetic analysis of representative NP sequences and the presence of MxA resistance-enhancing mutations. Shown is a maximum-likelihood tree of 140 aligned representative NP sequences. Nucleotide sequences were retrieved from GenBank, manually aligned, and used for Bayesian tree inference with MrBayes, v3.2. The GTR+G+I substitution model was used. Convergence was reached after 3 million generations. The bar indicates substitutions per site. Avian sequences are printed in gray, human seasonal sequences (H1N1, H3N2) in blue, Eurasian porcine sequences in green, and the classical swine-derived H1N1 sequences in red. Amino acid substitutions resulting in MxA resistance are indicated. The complete phylogenetic tree is shown in Fig. S2 in the supplemental material. Zoonotic events are highlighted with asterisks.
To complement these data, we furthermore analyzed the frequencies of amino acids 48Q, 98K, and 99K in avian, swine, and human IAV (data obtained from the NCBI Influenza Virus Sequence Database [16]). As summarized in Table 1, all three mutations are highly conserved among 737 viruses of the Eurasian avian-like swine IAV lineage isolated from swine, with frequencies of at least 96.7%, and are completely conserved in the 6 Eurasian avian-like swine IAV isolated from humans to date. This high level of conservation of MxA resistance-conferring amino acids is similar to the frequencies previously determined for human-adapted IAV (8) (see Table S1 in the supplemental material). It is noteworthy that the amino acid 98K is also rather frequently found in the descendants of the 1918 virus, with 91.5% frequency for seasonal viruses of the H1N1 subtype and 11.6% frequency for viruses of the H1N2, H2N2, and H3N2 subtypes. Furthermore, 98K is even present in a small number of avian virus isolates (2.4%).
TABLE 1.
Conservation of amino acids in NP that confer MxA resistancea
| Host | Subtype(s) or lineage | Frequency (%) of the following conserved residue: |
No. of strains analyzed | ||
|---|---|---|---|---|---|
| 48Q | 98K | 99K | |||
| Avian | All | 0.1 | 2.4 | 0.0 | 15,871 |
| Human | H1N1 seasonal | 0.0 | 91.5 | 0.1 | 1,609 |
| Human | H1N2/H2N2/H3N2 seasonal | 0.0 | 11.6 | 0.0 | 12,911 |
| Swine | Classical North American swine | 0.0 | 0.5 | 0.2 | 2,842 |
| Human | pH1N1 | 0.0 | 0.1 | 0.0 | 8,998 |
| Swine | pH1N1 | 0.0 | 0.3 | 0.1 | 1,664 |
| Human | Eurasian avian-like swine | 100.0 | 100.0 | 100.0 | 6 |
| Swine | Eurasian avian-like swine | 97.0 | 96.7 | 97.6 | 737 |
Full-length NP protein sequences of the indicated subtype and host were downloaded on 23 August 2018 from the NCBI influenza virus resource (16).
DISCUSSION
In this study, we challenged the hypothesis that IAV nucleoproteins (NPs) are under positive selection by poMx1 in swine by studying the Eurasian avian-like swine IAV lineage. To our surprise, we found that MxA resistance conferred by the NP of the Eurasian avian-like swine IAV A/Swine/Belzig/2/2001 enables the virus to escape the activity of MxA in a manner comparable to that of the human-adapted pH1N1-NP. Using polymerase reconstitution assays, we could show that amino acids 48Q, 98K, and 99K in Belzig-NP are sufficient to provide resistance to human MxA, since replacement of these MxA escape mutations with avian virus amino acids completely abrogated MxA resistance. In agreement with those results, the introduction of 48Q, 98K, and 99K into the MxA-sensitive H5N1-NP conferred MxA resistance to the same extent as that of Belzig-NP and pH1N1-NP. These data could be validated in MxA-overexpressing MDCK-SIAT1 cells and in an MxA-transgenic mouse model by use of an avian IAV encoding Belzig-NP or a recombinant wild-type Belzig virus. Finally, phylogenetically, the three MxA escape mutations were successively introduced into the NP of the relatively young Eurasian avian-like swine IAV lineage in a short time frame of about 1 decade and were maintained as would have been predicted assuming positive selection under constant pressure of the restriction factor poMx1.
While 48Q, 98K, and 99K are novel MxA escape mutations, which have not yet been reported for any other IAV in this context, they localize in close proximity to the known MxA escape clusters determined for pH1N1-NP and 1918-NP (8). The K48Q and R99K mutations are unique to the Eurasian avian-like swine influenza virus lineage and hardly ever appear in other IAV isolates. Intriguingly, however, the R99K substitution could, among other substitutions, be observed after serial passaging of an avian H5N1 virus in ferrets, which rendered the virus transmissible by respiratory droplets in this animal model (17). The amino acid 98K, on the other hand, is not unique and can frequently be found in the NPs of descendants of the 1918 virus and even in a small number of avian isolates. However, the almost complete conservation of 98K in the Eurasian avian-like swine IAV lineage and its localization adjacent to the critical position 100 in the MxA resistance patch of human IAV suggest that it plays a crucial role in the evolution of recent Eurasian avian-like swine lineage IAV.
We demonstrated previously that the most potent MxA escape mutations cause a loss of viral fitness and are therefore not selected in the absence of Mx proteins similar in potency to human MxA (8, 10). Consistently, the most potent pH1N1-NP MxA escape mutation (E53D), which emerged during the circulation of classical North American swine IAV in swine, is frequently lost after the reintroduction of the pH1N1 virus into swine, most likely due to a lower selective pressure exerted by the less potent porcine Mx1 (8). It is therefore surprising that swine-derived Belzig-NP reached a degree of MxA resistance comparable to that of human pH1N1-NP. It is noteworthy, however, that the three Belzig-NP-specific mutations K48Q, R98K, and R99K seem to reduce viral fitness only minimally if at all, since in the absence of MxA, their replacement in Belzig-NP with avian IAV amino acids fails to show an increase in viral titers of the avian H7N7 or the Belzig wild-type virus. We therefore speculate that the acquisition of MxA escape mutations in Belzig-NP generated no obvious growth deficit, and as a result, no counterselection of these amino acids occurred. We cannot, however, completely exclude a slight impact on viral fitness unless the NP mutations are tested individually in avian viruses, such as H7N7 or H5N1 viruses.
We assumed previously that due to the fitness loss associated with MxA escape, only two NPs with mutations sufficient to overcome MxA restriction have emerged during the last 100 years, namely, 1918-NP and pH1N1-NP (8). These two NPs were used by all IAV causing pandemics during this time frame, and therefore, efficient MxA escape clusters appear to be a prerequisite for overcoming the human species barrier (Fig. 5). Consistently, the mutations were maintained in all viruses able to circulate stably in the human population. Unexpectedly, we now identified a third MxA escape cluster with an independent evolutionary history but comparable properties, indicating a zoonotic or even pandemic potential of Eurasian avian-like swine IAV. Accordingly, human Eurasian avian-like isolates have only started to emerge since all three MxA escape mutations were acquired, but symptomatic infections have been sparse so far (18). Nevertheless, efficient airborne transmission of Eurasian avian-like swine viruses could be demonstrated in ferrets, indicating an already advanced adaptation to the mammalian, and therefore also to the human, IAV transmission route (19). Additionally, since the late 1990s, the rate of reassortment in pigs has increased dramatically (4), especially in Asia, where most IAV lineages constantly cocirculate (5). Furthermore, it has been shown that reassortment of IAV with viruses of the Eurasian avian-like swine lineage is possible, since two of the eight gene segments of the pandemic pH1N1 virus originate from this lineage (1). Therefore, even if the Eurasian avian-like swine IAV fail to establish sustained infection chains in humans, the fact that a novel MxA-resistant nucleoprotein circulates in pigs is alarming, since we could show that H7N7 viruses incorporating the Belzig-NP could withstand MxA restriction in vivo in the MxA transgenic mouse model. This indicates that the Belzig-NP gene might be capable of being combined with genes from many other IAV, including highly pathogenic avian IAV, during reassortment.
FIG 5.
Temporal appearance of MxA resistance-enhancing amino acids in NPs of Eurasian avian-like swine IAV. Shown is the consecutive acquisition of MxA resistance-enhancing NP mutations in the 1918, “classical” North American swine, and Eurasian avian-like swine lineages. For more-detailed information, see reference 8, Fig. 4, and Fig. S2 in the supplemental material.
In summary, we identified a novel MxA-resistant nucleoprotein that is encoded by the Eurasian avian-like swine IAV lineage currently circulating in swine populations in Eurasia. Since escape from MxA is mandatory for overcoming the human species barrier, this MxA-resistant NP presumably increases the pandemic risk of Eurasian avian-like swine IAV. Furthermore, acquisition of the NP-encoding segment of IAV from this lineage during coinfection events can result in reassortant viruses with high pandemic potential, as exemplified best by the quadruple reassortant pandemic pH1N1 virus of 2009, which gained an MxA-preadapted NP from the classical swine IAV lineage (1, 8).
MATERIALS AND METHODS
Ethics statement.
All animal experiments were performed in compliance with the German animal protection law (TierSchG). The mice were housed and handled in accordance with good animal practices as defined by FELASA and the national animal welfare body GV-SOLAS. The animal welfare committees of the University of Freiburg, as well as the local authorities (Regierungspräsidium Freiburg), approved all animal experiments.
Cells.
Canine MDCKII, MDCK-SIAT1-MxA, and MDCK-SIAT1-MxA-T103A cells (15) (kindly provided by Jesse D. Bloom, Fred Hutchinson Cancer Research Center, USA) and human HEK293T cells were maintained with Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum, 2 mM l-glutamine, and 1% penicillin-streptomycin. The cells were kept at 37°C under a 5% CO2 atmosphere in a tissue culture incubator.
Plasmid construction.
pHW2000 rescue plasmids (20) and pCAGGS (21) expression plasmids coding for NP were used for site-directed mutagenesis via QuikChange PCR or two-step assembly PCR, respectively.
Generation of recombinant influenza A viruses.
The recombinant viruses A/Swine/Belzig/2/2001 (H1N1), A/Hamburg/4/2009 (pH1N1), A/Thailand/1(KAN-1)/2004 (H5N1), and A/Seal/Massachusetts/1/1980 (H7N7), as well as the NP mutant viruses, were generated by the eight-plasmid reverse-genetics system as described previously (22). All recombinant viruses were plaque purified on MDCKII cells. Virus stocks were prepared on MDCKII cells, and titers were determined by plaque assay. The presence of the NP mutations introduced was confirmed via cDNA synthesis of isolated NP viral RNA using the One-Step RT-PCR kit (Qiagen) and NP-specific primers. The PCR products were then analyzed via Sanger sequencing.
Polymerase reconstitution assays.
Polymerase activity was measured by transfecting HEK293T cells with 10 ng of pCAGGS plasmids encoding PB2, PB1, and PA, 100 ng of pCAGGS plasmids encoding the specific NP, and 100 ng of the artificial viral minigenome construct pPolI-FFLuc-RT. Transfection efficiency was measured by cotransfecting 30 ng of pRL-SV40 expressing Renilla luciferase under the control of the simian virus 40 (SV40) promoter. Additionally, 50 ng of the pCAGGS plasmid expressing either MxA or the antivirally inactive MxA-T103A mutant was transfected. Alternatively, 200 ng of the pCAGGS plasmid expressing either poMx1 or antivirally inactive MxA-T103A was transfected. The cells were lysed 24 h posttransfection, and firefly and Renilla luciferase activities were measured using the dual-reporter assay (Promega) according to the manufacturer’s protocol.
Virus infections in cell culture.
NPTr, MDCKII, MDCK-SIAT1-MxA, and MDCK-SIAT1-MxA-T103A cells were seeded in 6-well plates and were infected with virus at a multiplicity of infection (MOI) of 0.001 in 2 ml infection medium (DMEM supplemented with 0.2% bovine serum albumin [BSA] and 1% penicillin-streptomycin). For pH1N1 and the rBelzig viruses, 1 μg/ml tosylsulfonyl phenylalanyl chloromethyl ketone (TCPK)-treated trypsin was added to the infection medium. Virus titers in cell culture supernatants were determined by plaque assay.
Animal experiments.
C67BL/6N mice were obtained from Janvier (Le Genest-Saint-Isle, France), and C67BL/6N mice transgenic for human MxA (hMxtg+/+) (9) were bred locally. Eight- to 10-week-old mice were anesthetized with a mixture of ketamine (100 μg per g of body weight) and xylazine (5 μg per g of body weight) administered intraperitoneally (i.p.) and were inoculated with the indicated doses of viruses in 40 μl Opti-MEM supplemented with 0.3% BSA. The lungs of the infected animals were harvested 3 dpi and were homogenized in 800 μl phosphate-buffered saline (PBS). Virus titers in the organ homogenates were determined via plaque assays.
Molecular modeling.
The PyMOL program (www.pymol.org) was used to assign the indicated positions in the structural model of the NP of influenza virus A/Hong Kong/483/97 (H5N1) (PDB code 2Q06).
Alignments and phylogenetic analyses.
NP sequences were retrieved from GenBank and were aligned with MEGA5 (23). For maximum-likelihood (ML) tree inference, Metropolis-coupled Markov chains were computed with MrBayes, v3.2 (24). The general time-reversible (GTR) substitution model assuming gamma distribution (+G) (four gamma categories) and invariant sites (+I) was selected on the basis of the Bayesian information criterion (BIC) and the corrected Akaike information criterion (AICc) using a model test implemented in MEGA5. Two runs were conducted until convergence was reached (3,000,000 and 5,000,000 generations, respectively).
Supplementary Material
ACKNOWLEDGMENTS
We thank Jürgen Stech for providing rescue plasmids for the Eurasian avian-like swine influenza virus isolate A/Swine/Belzig/2/2001 (H1N1), Jesse D. Bloom for providing MDCK-SIAT1 cells stably expressing MxA or MxA-T103A, and Georg Kochs for providing the MxA-specific antibody.
This study was supported in part by funds from the German Research Foundation (DFG; SFB 1160, project 13) to M.S. and the Excellence Initiative of the German Research Foundation (GSC-4, Spemann Graduate School) to E.H. The funders had no role in study design, data collection and analysis, decision to publish, or the preparation of the manuscript.
D.D. and M.S. conceived and designed the experiments. P.P.P., D.D., E.H., and R.Z. performed the experiments. D.D., M.S., P.P.P., and R.Z. analyzed the data. D.D. and M.S. wrote the paper.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.00997-18.
REFERENCES
- 1.Taubenberger JK, Kash JC. 2010. Influenza virus evolution, host adaptation, and pandemic formation. Cell Host Microbe 7:440–451. doi: 10.1016/j.chom.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Horimoto T, Kawaoka Y. 2001. Pandemic threat posed by avian influenza A viruses. Clin Microbiol Rev 14:129–149. doi: 10.1128/CMR.14.1.129-149.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ito T, Couceiro JN, Kelm S, Baum LG, Krauss S, Castrucci MR, Donatelli I, Kida H, Paulson JC, Webster RG, Kawaoka Y. 1998. Molecular basis for the generation in pigs of influenza A viruses with pandemic potential. J Virol 72:7367–7373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vincent A, Awada L, Brown I, Chen H, Claes F, Dauphin G, Donis R, Culhane M, Hamilton K, Lewis N, Mumford E, Nguyen T, Parchariyanon S, Pasick J, Pavade G, Pereda A, Peiris M, Saito T, Swenson S, Van Reeth K, Webby R, Wong F, Ciacci-Zanella J. 2014. Review of influenza A virus in swine worldwide: a call for increased surveillance and research. Zoonoses Public Health 61:4–17. doi: 10.1111/zph.12049. [DOI] [PubMed] [Google Scholar]
- 5.Zhu H, Webby R, Lam TT, Smith DK, Peiris JS, Guan Y. 2013. History of swine influenza viruses in Asia. Curr Top Microbiol Immunol 370:57–68. doi: 10.1007/82_2011_179. [DOI] [PubMed] [Google Scholar]
- 6.Haller O, Staeheli P, Schwemmle M, Kochs G. 2015. Mx GTPases: dynamin-like antiviral machines of innate immunity. Trends Microbiol 23:154–163. doi: 10.1016/j.tim.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 7.Verhelst J, Hulpiau P, Saelens X. 2013. Mx proteins: antiviral gatekeepers that restrain the uninvited. Microbiol Mol Biol Rev 77:551–566. doi: 10.1128/MMBR.00024-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Manz B, Dornfeld D, Gotz V, Zell R, Zimmermann P, Haller O, Kochs G, Schwemmle M. 2013. Pandemic influenza A viruses escape from restriction by human MxA through adaptive mutations in the nucleoprotein. PLoS Pathog 9:e1003279. doi: 10.1371/journal.ppat.1003279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deeg CM, Hassan E, Mutz P, Rheinemann L, Gotz V, Magar L, Schilling M, Kallfass C, Nurnberger C, Soubies S, Kochs G, Haller O, Schwemmle M, Staeheli P. 2017. In vivo evasion of MxA by avian influenza viruses requires human signature in the viral nucleoprotein. J Exp Med 214:1239–1248. doi: 10.1084/jem.20161033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gotz V, Magar L, Dornfeld D, Giese S, Pohlmann A, Hoper D, Kong BW, Jans DA, Beer M, Haller O, Schwemmle M. 2016. Influenza A viruses escape from MxA restriction at the expense of efficient nuclear vRNP import. Sci Rep 6:23138. doi: 10.1038/srep23138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watson SJ, Langat P, Reid SM, Lam TT, Cotten M, Kelly M, Van Reeth K, Qiu Y, Simon G, Bonin E, Foni E, Chiapponi C, Larsen L, Hjulsager C, Markowska-Daniel I, Urbaniak K, Durrwald R, Schlegel M, Huovilainen A, Davidson I, Dan A, Loeffen W, Edwards S, Bublot M, Vila T, Maldonado J, Valls L, Brown IH, Pybus OG, Kellam P. 2015. Molecular epidemiology and evolution of influenza viruses circulating within European swine between 2009 and 2013. J Virol 89:9920–9931. doi: 10.1128/JVI.00840-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zell R, Scholtissek C, Ludwig S. 2013. Genetics, evolution, and the zoonotic capacity of European swine influenza viruses. Curr Top Microbiol Immunol 370:29–55. doi: 10.1007/82_2012_267. [DOI] [PubMed] [Google Scholar]
- 13.Ng AK, Zhang H, Tan K, Li Z, Liu JH, Chan PK, Li SM, Chan WY, Au SW, Joachimiak A, Walz T, Wang JH, Shaw PC. 2008. Structure of the influenza virus A H5N1 nucleoprotein: implications for RNA binding, oligomerization, and vaccine design. FASEB J 22:3638–3647. doi: 10.1096/fj.08-112110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seitz C, Frensing T, Hoper D, Kochs G, Reichl U. 2010. High yields of influenza A virus in Madin-Darby canine kidney cells are promoted by an insufficient interferon-induced antiviral state. J Gen Virol 91:1754–1763. doi: 10.1099/vir.0.020370-0. [DOI] [PubMed] [Google Scholar]
- 15.Ashenberg O, Padmakumar J, Doud MB, Bloom JD. 2017. Deep mutational scanning identifies sites in influenza nucleoprotein that affect viral inhibition by MxA. PLoS Pathog 13:e1006288. doi: 10.1371/journal.ppat.1006288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bao Y, Bolotov P, Dernovoy D, Kiryutin B, Zaslavsky L, Tatusova T, Ostell J, Lipman D. 2008. The influenza virus resource at the National Center for Biotechnology Information. J Virol 82:596–601. doi: 10.1128/JVI.02005-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herfst S, Schrauwen EJ, Linster M, Chutinimitkul S, de Wit E, Munster VJ, Sorrell EM, Bestebroer TM, Burke DF, Smith DJ, Rimmelzwaan GF, Osterhaus AD, Fouchier RA. 2012. Airborne transmission of influenza A/H5N1 virus between ferrets. Science 336:1534–1541. doi: 10.1126/science.1213362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krueger WS, Gray GC. 2013. Swine influenza virus infections in man. Curr Top Microbiol Immunol 370:201–225. doi: 10.1007/82_2012_268. [DOI] [PubMed] [Google Scholar]
- 19.Yang H, Chen Y, Qiao C, He X, Zhou H, Sun Y, Yin H, Meng S, Liu L, Zhang Q, Kong H, Gu C, Li C, Bu Z, Kawaoka Y, Chen H. 2016. Prevalence, genetics, and transmissibility in ferrets of Eurasian avian-like H1N1 swine influenza viruses. Proc Natl Acad Sci U S A 113:392–397. doi: 10.1073/pnas.1522643113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stech J, Stech O, Herwig A, Altmeppen H, Hundt J, Gohrbandt S, Kreibich A, Weber S, Klenk HD, Mettenleiter TC. 2008. Rapid and reliable universal cloning of influenza A virus genes by target-primed plasmid amplification. Nucleic Acids Res 36:e139. doi: 10.1093/nar/gkn646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reuther P, Manz B, Brunotte L, Schwemmle M, Wunderlich K. 2011. Targeting of the influenza A virus polymerase PB1-PB2 interface indicates strain-specific assembly differences. J Virol 85:13298–13309. doi: 10.1128/JVI.00868-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zimmermann P, Manz B, Haller O, Schwemmle M, Kochs G. 2011. The viral nucleoprotein determines Mx sensitivity of influenza A viruses. J Virol 85:8133–8140. doi: 10.1128/JVI.00712-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.