Abstract
The purpose of our study was to better understand the effects of mitochondrial-division inhibitor 1 (Mdivi-1) on mitochondrial fission, mitochondrial biogenesis, electron transport activities and cellular protection. In recent years, researchers have found excessive mitochondrial fragmentation and reduced fusion in a large number of diseases with mitochondrial dysfunction. Therefore, several groups have developed mitochondrial division inhibitors. Among these, Mdivi-1 was extensively studied and was found to reduce dynamin-related protein 1 (Drp1) levels and excessive mitochondrial fission, enhance mitochondrial fusion activity and protect cells. However, a recent study by Bordt et al. (1) questioned earlier findings of the beneficial, inhibiting effects of Mdivi-1. In the current study, we studied the protective effects of Mdivi-1 by studying the following: mRNA and protein levels of electron transport chain (ETC) genes; mitochondrial dynamics and biogenesis genes; enzymatic activities of ETC complexes I, II, III and IV; the mitochondrial network; mitochondrial size & number; Drp1 GTPase enzymatic activity and mitochondrial respiration (1) in N2a cells treated with Mdivi-1, (2) overexpressed with full-length Drp1 + Mdivi-1-treated N2a cells and (3) Drp1 RNA silenced+Mdivi-1-treated N2a cells. We found reduced levels of the fission genes Drp1 and Fis1 levels; increased levels of the fusion genes Mfn1, Mfn2 and Opa1; and the biogenesis genes PGC1α, nuclear respiration factor 1, nuclear respiratory factor 2 and transcription factor A, mitochondrial. Increased levels mRNA and protein levels were found in ETC genes of complexes I, II and IV genes. Immunoblotting data agreed with mRNA changes. Transmission electron microscopy analysis revealed reduced numbers of mitochondria and increased length of mitochondria (1) in N2a cells treated with Mdivi-1, (2) cells overexpressed with full-length Drp1 + Mdivi-1-treated N2a cells and (3) Drp1 RNA silenced+Mdivi-1-treated N2a cells. Immunofluorescence analysis revealed that mitochondrial network was increased. Increased levels of complex I, II and IV enzymatic activities were found in all three groups of cells treated with low concentration of Mdivi-1. Mitochondrial function was increased and GTPase Drp1 activity was decreased in all three groups of N2a cells. These observations strongly suggest that Mdivi-1 is a Drp1 inhibitor and directly reduces mitochondrial fragmentation and further, Mdivi-1 is a promising molecule to treat human diseases with ETC complexes, I, II and IV.
Introduction
Mitochondria are critical for cell survival and death. Mitochondria are dynamic cytoplasmic organelles that arise from a symbiotic association between glycolytic proto-eukaryotic cells and oxidative bacteria (2–5). Several features of mitochondria that reflect their endosymbiotic origin are their double-membrane structure and their circular genome with mitochondria-specific transcription, translation and protein-assembly systems. Structurally, mitochondria are compartmentalized into two lipid membranes: an outer and an inner membrane. The outer membrane is highly porous and allows the flow of small molecules and metabolites into its inner membrane. The inner membrane covers the matrix, which contains beta oxidation and tricarboxylic acid. The inner membrane harbors the electron transport chain (ETC), is highly non-porous and restricts ionic flow into the mitochondrial matrix. The ETC participates in oxidative phosphorylation (OXPHOS) and produces adenosine triphosphate (ATP) (3). The reactive oxygen species (ROS) that is produced in mitochondria is a physiological by-product of the ETC. Mitochondria perform several important cellular functions, including producing ATP, regulating intracellular calcium (Ca2+) levels, regulating sites of production of free radicals, scavenging free radicals, releasing proteins that activate the caspase family of proteases and altering the reduction–oxidation potential of cells. In a diseased state, such as in Alzheimer’s disease (AD), mitochondrial function is defective because of mutant AD proteins associated with mitochondria (5).
Mammalian mitochondrial DNA (mtDNA) consists of a 16.5 kb, double-stranded circular DNA molecule. mtDNA contains 13 polypeptide genes, all of which encode essential components of the ETC. All 13 polypeptide genes in the mtDNA are involved in producing components of mitochondrial complexes. mtDNA encodes 7 subunits (ND1, 2, 3, 4, 4L, 5 and 6) of the 43 subunits of complex I, 1 (cytochrome b) of the 11 subunits of complex III, 3 (COX1, COX2 and COX3) of the 13 subunits of complex IV and 2 (ATPase 6 and ATPase 8) of the 17 subunits of complex V. Mitochondrial toxins, mutant proteins and age-related injuries are reported to alter ETC activities and mitochondrial ATP levels (4). Protective compounds and mitochondria-targeted molecules are reported to maintain ETC activities (6).
An accumulation of mtDNA mutations could damage mitochondrial structures and functions, thus altering enzymatic activities, disrupting mitochondrial pore gating, altering mitochondrial calcium levels and lowering ATP—ultimately leading to cellular senescence (4).
Mitochondrial biogenesis is another important activity that is affected by toxins and an accumulation of mtDNA mutations. Mitochondrial biogenesis is the process by which new mitochondria are synthesized in the cell and is activated by numerous different signals during times of cellular stress. There are four genes that are involved in mitochondrial biogenesis: PGC1α (PPAR (peroxisome proliferator-activated receptor)-γ coactivator-1α), NRF1 (nuclear respiratory factor 1), NRF2 (nuclear respiratory factor 2) and TFAM (transcription factor A, mitochondrial). In a diseased state, such as AD, mitochondrial biogenesis is reduced or defective, mainly because of the association of mutant proteins with mitochondria and increased free radical production (5,7–9). Therefore, it may be productive to study mitochondrial biogenesis to further elucidate mitochondrial function in a diseased state and to determine whether a protective agent, such as mitochondrial-division inhibitor 1 (Mdivi-1), and/or toxic compounds, such as rotenone or oligomycin, affect the mitochondrial biogenesis process.
Mitochondrial division is regulated via a highly evolutionarily conserved GTPase gene, dynamin-related protein 1 (Drp1). Drp1 is critical for the division, size and shape of mitochondria and for the distribution of mitochondria throughout the neuron, from cell body to axons, dendrites, nerve terminals and synapses where energy is in high demand (5–6,10).
Drp1 phosphorylation at Ser 656 is essential for maintaining the shape of mitochondria and regulating mitochondrial apoptosis (11). Phosphorylated Drp1 may alter Drp1 function and mitochondrial morphology, which in turn is known to regulate mitochondrial division (12). Upon the induction of apoptosis, Drp1 translocates from the cytosol to the mitochondrial outer membrane, where it induces mitochondrial division. Inhibition of Drp1 by the overexpression of a dominant-negative mutant counteracts the conversion to a puncti form of the mitochondrial phenotype, which acts to prevent the loss of the mitochondrial membrane potential and the release of cytochrome c and to reveal a reproducible swelling of mitochondria. In addition, the inhibition of Drp1 has been found to block cell death by implicating mitochondrial fission during apoptosis (13). These findings suggest that Drp1 activation may be crucial for mitochondrial fragmentation, which in turn may initiate apoptotic cell death.
Mitochondrial fragmentation occurs in several ways: (1) excessive ROS production, which activates fission proteins and increases GTPase Drp1 enzymatic activity, ultimately affecting the structural integrity of mitochondria and increasing mitochondrial fission; (2) interaction of a mutant protein(s), such as mutant Htt, Aβ or DJ1/LRRK2 with Drp1 and a subsequent increase in GTPase Drp1 enzymatic activity, which, in turn, increases mitochondrial fission and creates an imbalance in mitochondrial dynamics; (3) S-nitrosylation of Drp1, which enhances GTPase Drp1 activity, causing excessive mitochondrial fission and (4) phosphorylated Drp1 at Ser 616, Ser 585 and Ser 637 sites, which alters GTPase activity, causing defective mitochondrial fission (14).
Several studies suggest that Drp1 is involved in increased mitochondrial division and decreased fusion, and a loss of Drp1 function is involved in increased mitochondrial fusion and mitochondrial connectivity (15). Knockdown of wild-type Drp1 in primary neurons was found to cause impaired mitochondrial distribution (16–17). In contrast, an overexpressed dominant-negative mutation of Drp1 has been found to lead to increased mitochondrial fusion. Thus, the movement or distribution of mitochondria into dendrites appears essential to support synapses, and synaptic activity appears to modulate mitochondrial motility and the fusion–fission balance (16–17).
Interestingly, several groups have found that increased levels of Drp1 in postmortem AD brains (18), in brain tissues from AD mouse and cell models (19–23) and in AD cybrids (24) enhance Drp1 GTPase activities, ultimately leading to excessive fragmentation of mitochondria, reduced mitochondria fusion, increased free radical production and defective mitochondrial function (18–20,24). Since mitochondrial fission has been found to be increased in affected neurons of neurodegenerative diseases, inhibitors of mitochondrial fission may hold promise as therapeutic targets to treat patients diagnosed with such neurodegenerative diseases as AD and Huntington’s disease (HD). In the past 10 years, there has been some progress in identifying and developing inhibitors of mitochondrial fission, including the molecules Mdivi 1 (15), P110 (25), Dynasore (26) and mitochondrial division dynamin (27).
Following the discovery of Mdivi-1 reported by Cassidy-Stone and colleagues in 2008 (15), over 194 papers (Pubmed search, September 13, 2018) have been published on Mdivi-1, noting that Mdivi-1 inhibits excessive mitochondrial fission and enhances mitochondrial fusion activity, leading to elongated mitochondria and the protection of cells from toxic insults. Mechanistically, researchers found that excessive mitochondrial fragmentation can be reduced by directly decreasing GTPase Drp1 enzymatic activity, leading to the conclusion that Mdivi-1 reduces fission activity.
However, Bordt and colleagues (1) questioned whether Mdivi-1 has any effect on mitochondrial fission, GTPase Drp1 activity or mitochondrial elongation. They argue that Mdivi-1 reversibly inhibits respiration at complex I and that the effects of Mdivi-1 on respiration and ROS are independent of Drp1. To clarify this apparent controversy about whether Mdivi-1 reduces Drp1 levels and reduces Drp1-GTPase activity, we used (1) healthy N2a cells, (2) N2a cells transfected with human full-length Drp1 cDNA and (3) Drp1 RNA silenced in N2a cells in order to quantify (1) mRNA and protein levels of mitochondrial dynamics, mitochondrial biogenesis and ETC genes in treated and untreated N2a cells with Mdivi-1 (25 and 75 μm); (2) enzymatic activities of ETC complexes I, II, III and IV; (3) the mitochondrial network; (4) mitochondrial morphology, including size and number; (5) the extent of GTPase Drp1 enzymatic activity and (6) the degree of mitochondrial respiration, using a Seahorse XFe96 Extracellular Flux Analyzer (Seahorse Bioscience, North Billerica, MA, USA).
Results
mRNA levels in N2a cells treated with Mdivi-1
To better understand the effects of Mdivi-1 on mitochondrial dynamics, mitochondrial biogenesis and the ETC, we performed real-time quantitative reverse transcription PCR (qRT-PCR) and assessed mRNA levels of mitochondrial dynamics, mitochondrial biogenesis and ETC genes in untreated mouse neuroblastoma (N2a) cells and in N2a cells treated with Mdivi-1.
Mitochondrial dynamics genes
We found significantly reduced levels of mRNA expressions of fission genes Drp1 (by 1.3-fold in Mdivi-1-treatments of 25 μm and 1.6-fold in Mdivi-1-treatments of 75 μm) and Fis1 (by 2.1-fold in 25 μm and 2.4-fold in 75 μm) in Mdivi-1-treated N2a cells relative to untreated cells (Table 1). In contrast, increased levels of mRNA expression of the mitochondrial fusion genes Mfn1 (by 1.3-fold in 25 μm and 1.8-fold in 75 μm), Mfn2 (by 1.3-fold in 25 μm and 1.6-fold in 75 μm) and Opa1 (by 1.6-fold in 25 μm and 1.8-fold in 75 μm) were found in Mdivi-1-treated N2a cells relative to the untreated N2a cells. These findings indicate that Mdivi-1 reduces fission activity and increases fusion activity in N2a cells.
Table 1.
Fold changes of mRNA expression in mitochondrial dynamics, biogenesis and OXOPHOS genes in Mdivi-l-treated N2a cells compared with untreated N2a cells
| Genes | mRNA fold changes | ||
|---|---|---|---|
| N2a + Mdivi-1 25 μm | N2a + Mdivi-1 75 μm | ||
| Mitochondrial dynamics genes | Drp1 | −1.3 | −1.6* |
| Fis1 | −2.1** | −2.4** | |
| Mfn1 | 1.3 | 1.8* | |
| Mfn2 | 1.3 | 1.6* | |
| OPA1 | 1.6* | 1.8* | |
| Biogenesis genes | PGC1α | 1.8* | 2.2** |
| Nrf1 | 1.8* | 1.6* | |
| Nrf2 | 1.4 | 1.8* | |
| TFAM | 1.2 | 1.8* | |
| ETC-complex I genes | ETC-CI sub1 | 1.3 | −1.4 |
| ETC-CI sub2 | 1.3 | −1.3 | |
| ETC-CI sub3 | 1.4 | −1.5* | |
| ETC-CI sub4 | 1.4 | −1.2 | |
| ETC-CI sub5 | 1.6* | −1.3 | |
| ETC-CI sub6 | 1.6* | −1.4 | |
| ETC-complex III genes | Cyt B | 1.0 | 1.0 |
| ETC-complex IV genes | COX1 | 1.4 | −1.5* |
| COX2 | 1.6* | −1.2 | |
| COX3 | 1.5* | −1.3 | |
| ETC-complex V genes | ATP6 | 1.0 | 1.1 |
| ATP8 | 1.0 | 1.0 | |
P-values, *P < 0.05; **P < 0.005.
Mitochondrial biogenesis genes
Increased levels of biogenesis genes, PGC1α (by 1.8-fold in 25 μm and 2.2-fold in 75 μm), NRF1 (by 1.8-fold in 25 μm and 1.6-fold in 75 μm), NRF2 (by 1.4-fold in 25 μm and 1.8-fold in 75 μm) and TFAM (by 1.2-fold in 25 μm and 1.8-fold by 75 μm) in Mdivi-1-treated N2a cells relative to untreated N2a cells. These findings suggest that Mdivi-1 enhances mitochondrial biogenesis activity in N2a cells.
Mitochondrial ETC genes
Increased mRNA levels were found in the ETC subunit 1 (by 1.3-fold), subunit 2 (by 1.3-fold), subunit 3 (by 1.4-fold), subunit 4 (by 1.4-fold), subunit 5 (by 1.6-fold) and subunit 6 (by 1.6-fold) in N2a cells treated with 25 μm Mdivi-1 relative to untreated N2a cells. Statistical significance was found only in subunits 5 and 6. mRNA levels were unchanged for complex III, CytB, complex V, genes ATP6 and ATP8 genes. mRNA levels were increased for complex IV, the genes COX1 (by 1.4-fold), COX2 (by 1.6-fold) and COX3 (by 1.5-fold). In contrast, mRNA levels were reduced for all subunits in N2a cells treated with 75 μm Mdivi-1 (Table 1). These findings indicate that, at a higher concentration of Mdivi-1, Mdivi-1 can reduce ETC activity in N2a cells.
mRNA levels in N2a cells transfected with full-length Drp1 and treated with Mdivi-1
To elucidate the effects of full-length Drp1 in mitochondrial dynamics, biogenesis and ETC genes, we transfected N2a cells with human, full-length Drp1 cDNA and then assessed the activities of mitochondrial dynamics, biogenesis and the ETC genes. We also treated Drp1-transfected N2a cells with Mdivi-1 in order to better understand whether Mdivi-1 treatments at 25 and 75 μm alter fission, fusion and ETC activities in Drp1 overexpressed N2a cells. We also performed real-time qRT-PCR and assessed mRNA levels of mitochondrial dynamics, mitochondrial biogenesis and ETC genes. Results from these Mdivi-1 experiments are given below.
Mitochondrial dynamics genes
We found significantly reduced levels of mRNA expressions of fission genes Drp1 (by 3.2-fold in Mdivi-1 treatments of 25 μm and 3.7-fold in Mdivi-1 treatments of 75 μm) and Fis1 (by 2.6-fold in 25 μm and 3.2-fold in 75 μm) in FL-Drp1 + Mdivi-1-treated N2a cells relative to FL-Drp1 N2a cells and untreated N2a cells (Table 2). In contrast, increased levels of mRNA expression of the mitochondrial fusion genes Mfn1 (by 1.9-fold in 25 μm and 1.1-fold in 75 μm), Mfn2 (by 1.4-fold in 25 μm and 2.0-fold in 75 μm) and Opa1 (by 2.0-fold in 25 μm and 2.8-fold in 75 μm) were found in FL-Drp1 + Mdivi-1-treated N2a cells relative to FL-Drp1 N2a cells and untreated N2a cells (Table 2). These findings indicate that Mdivi-1 reduces fission activity and increases fusion activity in Drp1-overexpressed N2a cells.
Table 2.
Fold changes of mRNA expression in mitochondrial dynamics, biogenesis and ETC genes in FL-Drp1 overexpressed N2a cells + Mdivi-1 compared with untreated FL-Drp1 overexpressed N2a cells
| Genes | mRNA fold changes | ||
|---|---|---|---|
| FL-Drp1 + Mdivi-1 25 μm | FL-Drp1 + Mdivi-1 75 μm | ||
| Mitochondrial dynamics genes | Drp1 | −3.2** | −3.7** |
| Fis1 | −2.6** | −3.2** | |
| Mfn1 | 1.9* | 1.1 | |
| Mfn2 | 1.4* | 2.0** | |
| OPA1 | 2.0** | 2.8** | |
| Biogenesis genes | PGC1α | 1.8* | 2.6** |
| Nrf1 | 2.8** | 2.8** | |
| Nrf2 | 1.4 | 1.8* | |
| TFAM | 1.8* | 2.2** | |
| ETC-complex I genes | ETC-CI sub1 | −1.1 | −1.7* |
| ETC-C1 sub2 | −1.2 | −1.8* | |
| ETC-C1 sub3 | −1.3 | −2.1** | |
| ETC-C1 sub4 | −1.2 | −2.0** | |
| ETC-C1 sub5 | −1.2 | −1.4 | |
| ETC-C1 sub6 | −1.2 | −1.6* | |
| ETC-complex III genes | Cyt B | 1.0 | 1.0 |
| ETC-complex IV genes | COX1 | 1.0 | −1.8* |
| COX2 | −1.3 | −1.7* | |
| COX3 | 1 | −1.3 | |
| ETC-complex V genes | ATP6 | −1.1 | 1.0 |
| ATP8 | 1.0 | 1.2 | |
P-values, *P < 0.05; **P < 0.005.
Mitochondrial biogenesis genes
Increased levels of the biogenesis genes PGC1α (by 1.8-fold in Mdivi-1 treatments of 25 μm and 2.6-fold in Mdivi-1 treatments of 75 μm), NRF1 (by 2.8-fold in 25 μm and 2.8-fold in 75 μm), NRF2 (by 1.4-fold in 25 μm and 1.8-fold in 75 μm) and TFAM (by 1.8-fold in 25 μm and 2.2-fold by 75 μm) in FL-Drp1 + Mdivi-1-treated N2a cells relative to FL-Drp1 N2a cells and untreated N2a cells. These results suggest that Mdivi-1 enhances mitochondrial biogenesis activity in FL-Drp1-overexpressed N2a cells.
Mitochondrial ETC genes
Reduced mRNA levels were found in ETC genes in FL-Drp1 + Mdivi-1-treated N2a cells relative to FL-Drp1 N2a cells and untreated N2a cells. These findings indicate that Mdivi-1 reduces ETC activity in FL-Drp1 + Mdivi-1-treated N2a cells (Table 2).
RNA silencing of Drp1 in N2a cells treated with Mdivi-1
Mitochondrial dynamics genes
Significantly reduced levels of mRNA expressions of fission genes Drp1 (by 1.5-fold in Mdivi-1-treatments of 25 μm and 1.6-fold in Mdivi-1-treatments of 75 μm) and Fis1 (by 1.4-fold in 25 μm and 1.8-fold in 75 μm) in Drp1 RNA silenced Mdivi-1-treated N2a cells relative to Drp1 RNA silenced cells (Table 3). In contrast, increased levels of mRNA expression of the mitochondrial fusion genes Mfn1 (by 1.4-fold in 25 μm and 1.4-fold in 75 μm), Mfn2 (by 1.8-fold in 25 μm and 1.8-fold in 75 μm) and Opa1 (by 1.6-fold in 25 μm and 1.8-fold in 75 μm) were found in Mdivi-1-treated Drp1 RNA silenced N2a cells relative to the untreated Drp1 RNA silenced N2a cells. These findings indicate that Mdivi-1 reduces fission activity and increases fusion activity in Drp1 RNA silenced N2a cells.
Table 3.
Fold changes of mRNA expression of mitochondrial dynamics, biogenesis and OXOPHOS genes in siRNA-Drp1 N2a cells after Mdivi1 treatment relative to untreated siRNA-Drp1 N2a cells
| Genes | mRNA fold changes | ||
|---|---|---|---|
| siRNA-Drp1 N2a + Mdivi1 25 μm | siRNA-Drp1 N2a + Mdivi1 75 μm | ||
| Mitochondrial dynamics genes | Drp1 | −1.5* | −1.6* |
| Fis1 | −1.4 | −1.8* | |
| Mfn1 | 1.4 | 1.4 | |
| Mfn2 | 1.8* | 1.8* | |
| OPA1 | 1.6* | 1.8* | |
| Biogenesis genes | PGC1α | 1.3 | 1.4 |
| Nrf1 | 1.8* | 1.6* | |
| Nrf2 | 1.8* | 2.0* | |
| TFAM | 1.4 | 1.4 | |
| ETC-complex I genes | ETC-CI sub1 | 1.3 | −1.4 |
| ETC-CI sub2 | 1.3 | −1.3 | |
| ETC-CI sub3 | 1.4 | −1.5* | |
| ETC-CI sub4 | 1.4 | −1.2 | |
| ETC-CI sub5 | 1.3 | −1.3 | |
| ETC-CI sub6 | 1.6* | −1.4 | |
| ETC-complex III genes | Cyt B | 1.2 | 1.3 |
| ETC-complex IV genes | COX1 | 1.4 | −1.2 |
| COX2 | 1.3 | −1.2 | |
| COX3 | 1.5* | −1.3 | |
| ETC-complex V genes | ATP6 | 1.2 | 1.3 |
| ATP8 | 1.4 | 1.3 | |
P-values, *P < 0.05; **P < 0.005.
Mitochondrial biogenesis genes
Increased levels of biogenesis genes, PGC1α (by 1.3-fold in 25 μm and 1.4-fold in 75 μm), NRF1 (by 1.8-fold in 25 μm and 1.6-fold in 75 μm), NRF2 (by 1.8-fold in 25 μm and 2.0-fold in 75 μm) and TFAM (by 1.4-fold in 25 μm and 1.4-fold by 75 μm) in Mdivi-1-treated Drp1 RNA silenced N2a cells relative to Drp1 RNA silenced N2a cells. These findings suggest that Mdivi-1 enhances mitochondrial biogenesis activity in Drp1 RNA silenced N2a cells.
Mitochondrial ETC genes
Increased mRNA levels were found in the ETC subunit 1 (by 1.3-fold), subunit 2 (by 1.3-fold), subunit 3 (by 1.4-fold), subunit 4 (by 1.4-fold), subunit 5 (by 1.3-fold) and subunit 6 (by 1.6-fold) in Drp1 RNA silenced N2a cells treated with 25 μm Mdivi-1 relative to Drp1 RNA silenced untreated N2a cells. Statistical significance was found only in subunit 6. mRNA levels were increased for complex III, CytB (by 1.2-fold), complex IV (COX1 by 1.4-fold, COX2 1.3-fold and COX3 1.5-fold) and complex V ATP6 (by 1.2-fold) and ATP8 (by 1.4-fold) genes. In contrast, mRNA levels were reduced for all subunits in N2a cells treated with 75 μm Mdivi-1 (Table 3). These findings indicate that, at a higher concentration of Mdivi-1, Mdivi-1 can reduce ETC activity in N2a cells. These observations strongly suggest that at low concentration, Mdivi-1 is protective and at high concentration, Mdivi-1 is toxic to cells.
Immunoblotting analysis in N2a cells treated with Mdivi-1
To determine the effects of Mdivi-1 on mitochondrial fission and fusion proteins, we performed immunoblotting analysis of protein lysates prepared from Mdivi-1-treated and untreated N2a cells.
Mitochondrial dynamics proteins
As shown in Figure 1A–D, significantly reduced levels of the fission proteins Drp1 (P = 0.001 in Mdivi-1 treatments of 25 μm and P = 0.004 in Mdivi-1 treatments of 75 μm) and Fis1 (P = 0.02 in 25 μm and P = 0.001 in 75 μm) were found in Mdivi-1-treated N2a cells relative to untreated N2a cells. In contrast, mitochondrial fusion proteins Mfn1 (P = 0.04 in 25 μm and P = 0.02 in 75 μm), Mfn2 (P = 0.02 in 25 μm and P = 0.001 in 75 μm) and Opa1 (P = 0.003 in 25 μm and P = 0.001 in 75 μm) were significantly increased in Mdivi-1-treated N2a cells relative to the untreated cells (Fig. 1A–E).
Figure 1.
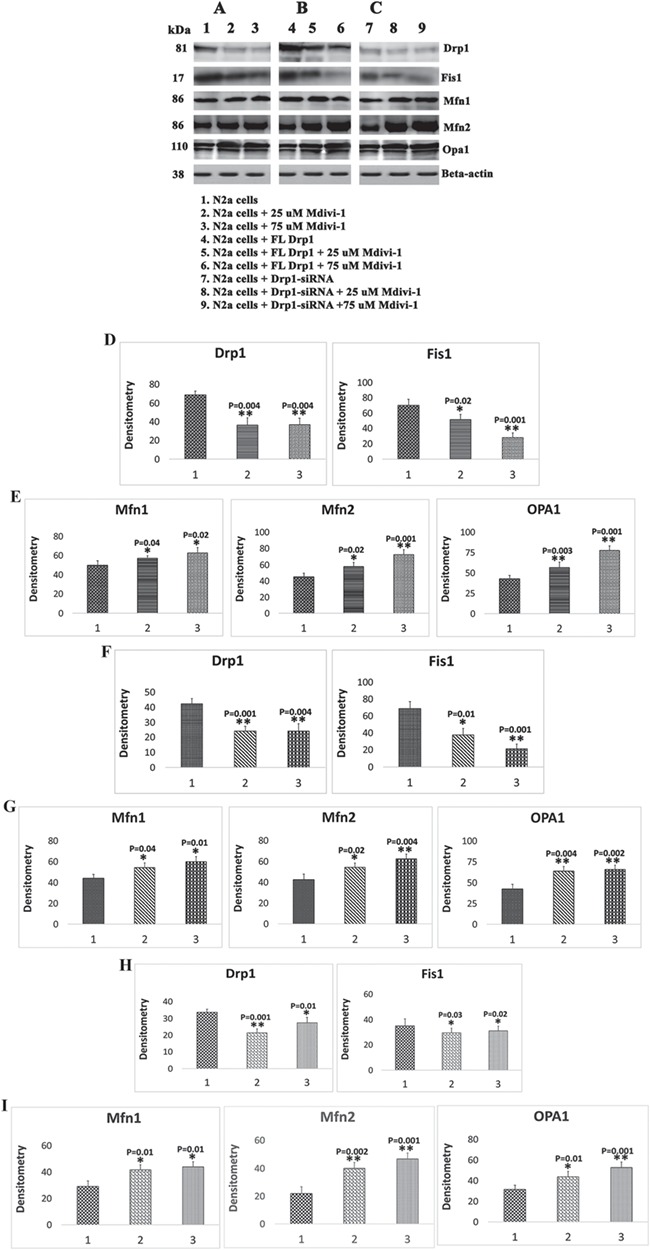
Immunoblotting analysis of mitochondrial dynamics proteins in Mdivi-1-treated N2a cells. (A) Representative immunoblotting analysis of Mdivi-1-treated N2a cells. (B) Representative immunoblotting analysis of N2a cells transfected with human full-length Drp1 + Mdivi-1-treated. (C) Representative immunoblotting analysis RNA silenced Drp1 in N2a cells+Mdivi-1-treated. (D) Quantitative densitometry analysis of mitochondrial dynamics for fission proteins (Drp1, Fis1) and (E) fusion proteins (Mfn1, Mfn2 and Opa1) of Mdivi-1-treated N2a cells. (F) Quantitative densitometry analysis of mitochondrial fission proteins and (G) fusion proteins of full-length Drp1 transfected+Mdivi-1-treated N2a cells. (H) Quantitative densitometry analysis of mitochondrial dynamics for fission proteins of Drp1 RNA silenced in N2a cells+Mdivi-1-treated. (I) Fusion proteins of Drp1 RNA silenced in N2a cells+Mdivi-1-treated.
Mitochondrial biogenesis
As shown in Figure 2A and D, significantly increased levels of the biogenesis proteins PGC1α (P = 0.03 in Mdivi-1 treatments of 25 μm and P = 0.004 in Mdivi-1 treatments of 75 μm), NRF1 (P = 0.01 in 25 μm and P = 0.01 in 75 μm), NRF2 (P = 0.003 in 25 μm and P = 0.002 in 75 μm) and TFAM (P = 0.02 in 25 μm and P = 0.04 in 75 μm) were found in Mdivi-1-treated N2a cells relative to Mdivi-1-untreated N2a cells (Fig. 2C and D), indicating that mitochondrial biogenesis increasein the Mdivi-1-treated cells.
Figure 2.
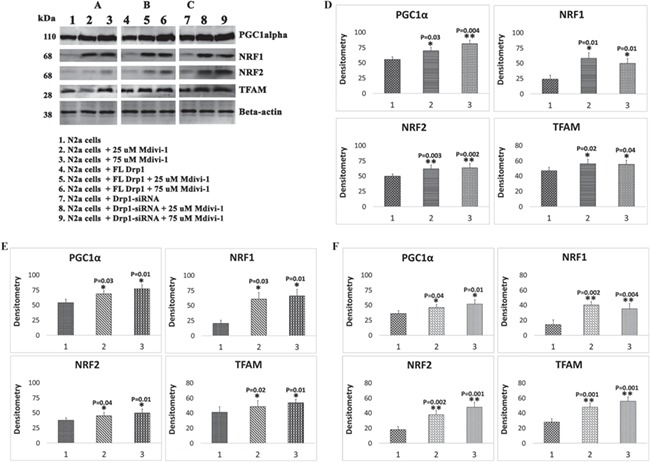
Immunoblotting analysis of mitochondrial biogenesis proteins in Mdivi-1-treated N2a cells. (A) Representative immunoblotting analysis of Mdivi-1-treated N2a cells. (B) Representative immunoblotting analysis of human, full-length Drp1 + Mdivi-1-treated N2a cells. (C) Representative immunoblotting analysis Drp1 RNA silenced in N2a cells+Mdivi-1-treated. (D) Quantitative densitometry analysis of biogenesis proteins (PGC1α, NRF1, NRF2 and TFAM) for Mdivi-1-treated N2a cells. (E) Quantitative densitometry analysis of these same biogenesis proteins for full-length, Drp1 transfected+Mdivi-1-treated N2a cells. (F) Quantitative densitometry analysis of these same biogenesis proteins for Drp1 RNA silenced in N2a cells+Mdivi-1-treated.
Mitochondrial ETC proteins
Significantly increased levels of complex I (P = 0.04), complex II (P = 0.04) and complex IV (P = 0.03) proteins were found in N2a cells treated with 25 μm Mdivi-1 (Fig. 3A and D). Interestingly, significantly reduced levels of complex I (P = 0.001), complex II (P = 0.01) and complex IV (P = 0.001) proteins were found in the N2a cells treated with 75 μm Mdivi-1 (Fig. 3A and D). Protein levels in complexes III and V were unchanged. These findings suggest that mitochondrial complexes I, II and IV are affected by Mdivi-1 treatments.
Figure 3.
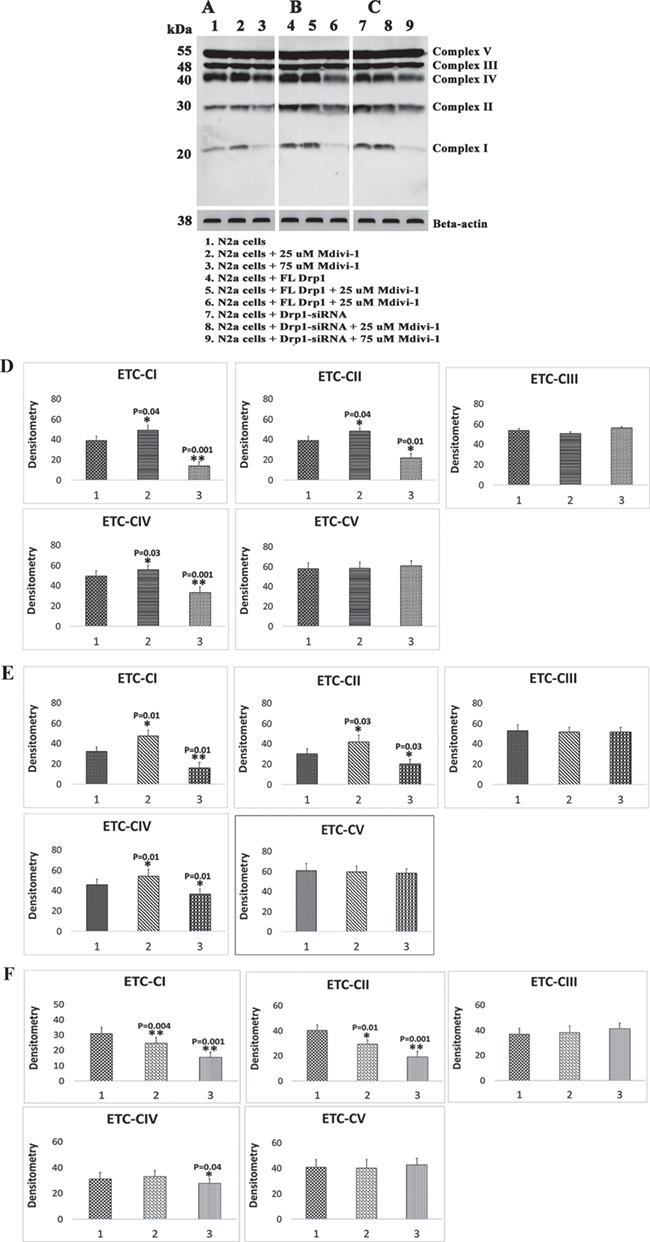
Immunoblotting analysis of ETC proteins in Mdivi-1-treated N2a cells. (A) Representative immunoblotting analysis of Mdivi-1-treated N2a cells. (B) Representative immunoblotting analysis of human, full-length Drp1 + Mdivi-1-treated N2a cells. (C) Representative immunoblotting analysis of RNA silenced Drp1 in N2a cells+Mdivi-1-treated. (D) Quantitative densitometry analysis of ETC complexes I–V proteins of Mdivi-1-treated N2a cells. (E) Quantitative densitometry analysis of ETC complexes I–V proteins of full-length, Drp1 transfected+Mdivi-1-treated N2a cells. (F) Quantitative densitometry analysis of ETC complexes I–V proteins of Drp1 RNA silenced in N2a cells.
Immunoblotting analysis in N2a cells transfected with full-length Drp1 and treated with Mdivi-1
To determine the effects of full-length Drp1 on mitochondrial fission and fusion proteins, we performed immunoblotting analysis of protein lysates prepared from N2a cells transfected with human, full-length Drp1. We also assessed the effects of Midivi-1 in FL-Drp1 transfected cells relative to FL-Drp1 transfected N2a cells.
Mitochondrial dynamics proteins
As shown in Figure 1B and F, significantly reduced levels of the fission proteins Drp1 (P = 0.001 in Mdivi-1 treatments of 25 μm and P = 0.004 in Mdivi-1 treatments of 75 μm) and Fis1 (P = 0.01 in 25 μm and P = 0.001 in 75 μm) were found in FL-Drp1 transfected+Mdivi-1-treated N2a cells relative to FL-Drp1-transfected and Mdivi-1-untreated N2a cells. In contrast, the mitochondrial fusion proteins Mfn1 (P = 0.04 in Mdivi-1 treatments of 25 μm and P = 0.01 in Mdivi-1 treatments of 75 μm), Mfn2 (P = 0.02 in 25 μm and P = 0.004 in 75 μm) and Opa1 (P = 0.004 in 25 μm and P = 0.002 in 75 μm) were significantly increased in FL-Drp1 transfected+Mdivi-1-treated N2a cells relative to FL-Drp1-transfected and Mdivi-1-untreated cells (Fig. 1B and G).
Mitochondrial biogenesis
As shown in Figure 2B and E, significantly increased levels of the biogenesis proteins PGC1α (P = 0.03 in Mdivi-1 treatments of 25 μm and P = 0.01 in Mdivi-1 treatments of 75 μm), NRF1 (P = 0.03 in 25 μm and P = 0.01 in 75 μm), NRF2 (P = 0.04 in 25 μm and P = 0.01 in 75 μm) and TFAM (P = 0.02 in 25 μm and P = 0.01 in 75 μm) were found in FL-Drp1-transfected+Mdivi-1-treated N2a cells relative to FL-Drp1-transfected and untreated cells, indicating that mitochondrial biogenesis increased in FL-Drp1-transfected+Mdivi-1-treated N2a cells.
Mitochondrial ETC proteins
Significantly increased levels of complex I (P = 0.01), complex II (P = 0.03) and complex IV (P = 0.01) proteins were found in FL-Drp1-transfected N2a cells treated with 25 μm Mdivi-1 (Fig. 3B and E). In contrast, significantly reduced levels of complex I (P = 0.01), complex II (P = 0.03) and complex IV (P = 0.01) proteins were found in FL-Drp1-transfected N2a cells treated with 75 μm Mdivi-1. Protein levels of complexes III and V were unchanged when treated with Mdivi-1 of each dosage. These findings suggest that mitochondrial complexes I, II and IV are affected and increased at the lower concentration of Mdivi-1 (25 μm) and reduced at the higher concentration (75 μm) of Mdivi-1.
Immunoblotting analysis of Drp1 RNA silenced in N2a cells treated with Mdivi-1
Mitochondrial dynamics proteins
As shown in Figure 1C–H, significantly reduced levels of the fission proteins Drp1 (P = 0.001 in Mdivi-1 treatments of 25 μm and P = 0.001 in Mdivi-1 treatments of 75 μm) and Fis1 (P = 0.03 in 25 μm and P = 0.03 in 75 μm) were found in Drp1 RNA silenced and Mdivi-1-treated N2a cells relative to Drp1 RNA silenced untreated N2a cells. In contrast, mitochondrial fusion proteins Mfn1 (P = 0.01 in 25 μm and P = 0.03 in 75 μm), Mfn2 (P = 0.002 in 25 μm and P = 0.002 in 25 μm) and Opa1 (P = 0.01 in 25 μm and P = 0.001 in 75 μm) were significantly increased in Drp1 RNA silenced Mdivi-1-treated N2a cells relative to the Drp1 RNA silenced untreated cells (Fig. 1C–I).
Mitochondrial biogenesis
As shown in Figure 2C and F, significantly increased levels of the biogenesis proteins PGC1α (P = 0.04 in Mdivi-1 treatments of 25 μm and P = 0.01 in Mdivi-1 treatments of 75 μm), NRF1 (P = 0.002 in 25 μm and P = 0.004 in 75 μm), NRF2 (P = 0.002 in 25 μm and P = 0.001 in 75 μm) and TFAM (P = 0.001 in 25 μm and P = 0.001 in 75 μm) were found in RNA silenced and Mdivi-1-treated N2a cells relative to RNA silenced Mdivi-1-untreated N2a cells (Fig. 2C and F), indicating that mitochondrial biogenesis increased in the Mdivi-1-treated cells.
Mitochondrial ETC proteins
Significantly increased levels of complex I (P = 0.004), complex II (P = 0.01) and complex IV (P = 0.04) proteins were found in Drp1 RNA silenced and Mdivi-1-treated N2a cells with 25 μm Mdivi-1 (Fig. 3C and F). Protein levels in complexes III and V were unchanged. These findings suggest that mitochondrial complexes I, II and IV are affected by Mdivi-1 treatments in Drp1 RNA silenced cells.
Immunoblotting analysis of phosphorylated Drp1 in N2a cells treated with Mdivi-1
To determine the effect of Mdivi-1 on Drp1 and phosphorylated Drp1, 616 and 637, we treated N2a cells for 24 h with five different concentrations of Mdivi-1: 12.5, 25, 50, 75 and 100 μm. We then performed immunoblotting using protein lysates from Mdivi-1-treated and untreated N2a cells. As shown in Figure 4A–D, we found significantly reduced Drp1 (P = 0.04 at Mdivi-1 treatments of 12.5 μm, P = 0.02 at 25 μm, P = 0.01 at 50 μm, P = 0.02 at 75 μm and P = 0.003 at 100 μm), phosphorylated Drp1 637 (P = 0.02 at 25 μm, P = 0.01 at 50 μm, P = 0.004 at 75 μm and P = 0.001 at 100 μm) and phosphorylated Drp1 616 (P = 0.01 at 12.5 μm, P = 0.01 at 25 μm, P = 0.004 at 50 μm, P = 0.002 at 75 μm and P = 0.001 at 100 μm) in Mdivi-1-treated N2a cells relative to untreated cells. These results indicate that Mdivi-1 reduces both full-length Drp1 and phosphorylated Drp1.
Figure 4.
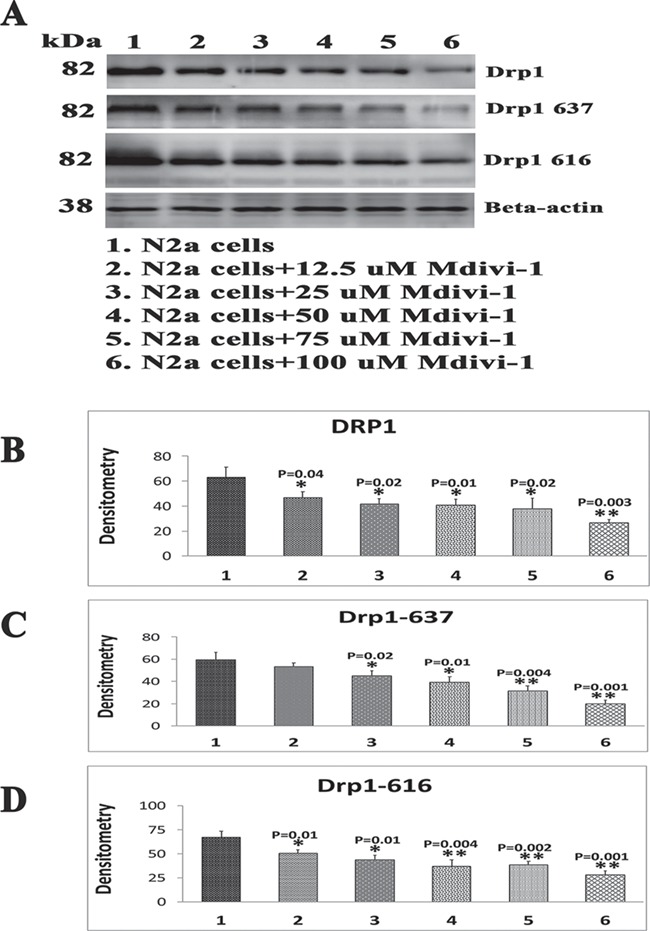
Immunoblotting analysis of phosphorylated Drp1 proteins in Mdivi-1-treated N2a cells. (A) Representative immunoblotting analysis of five different dilutions of Mdivi-1-treated N2a cells. (B–D) Quantitative densitometry analysis of five different dilutions of Mdivi-1-treated N2a cells.
GTPase Drp1 activity in N2a cells treated with Mdivi-1
To determine the effects of Mdivi-1 on GTPase Drp1 activity in N2a cells, using purified Drp1, we performed immunoprecipitation with an anti-Drp1 antibody on N2a cells treated with Mdivi-1 at 25 and 75 μm concentrations and untreated cells and untransfected cells. As shown in Figure 5A, we found significantly reduced GTPase Drp1 activity in Mdivi-1-treated N2a cells at 25 μm (P = 0.01) and 75 μm (P = 0.005) concentrations, indicating that Mdivi-1 reduces GTPase Drp1 activity in N2a cells.
Figure 5.
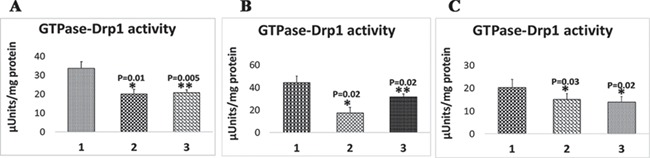
GTPase Drp1 enzymatic activity in Mdivi-treated N2a cells. (A) Representative GTPase Drp1 enzymatic activity in purified Drp1 of Mdivi-1-treated N2a cells. (B) GTPase Drp1 enzymatic activity in purified Drp1 of full-length Drp1 transfected+Mdivi-1-treated N2a cells. (C) GTPase Drp1 enzymatic activity in purified Drp1 of RNA silenced Drp1 + Mdivi-1-treated N2a cells.
GTPase Drp1 activity in N2a cells transfected with full-length Drp1 and treated with Mdivi-1
As shown in Figure 5B, significantly reduced GTPase Drp1 activity was found in FL-Drp1 transfected+Mdivi-1 cells at 25 μm (P = 0.02) and 75 μm (P = 0.02) concentrations relative to FL-Drp1-transfected and untreated N2a cells, indicating that Mdivi-1 reduces GTPase Drp1 activity in FL-Drp1 transfected N2a cells.
GTPase Drp1 activity in Drp1 RNA silenced N2a cells and treated with Mdivi-1
As shown in Figure 5C, significantly reduced GTPase Drp1 activity was found in Drp1 RNA silenced+Mdivi-1-treated cells at 25 μm (P = 0.03) and 75 μm (P = 0.02) concentrations relative to Drp1 RNA silenced and Mdivi-1 untreated N2a cells, indicating that Mdivi-1 reduces GTPase Drp1 activity in Drp1 RNA silenced N2a cells.
Mitochondrial function in N2a cells treated with Mdivi-1
Mitochondrial function was assessed in Mdivi-1-treated and untreated N2a cells by measuring hydrogen peroxide (H2O2), lipid peroxidation and mitochondrial ATP.
H2O2 production
As shown in Figure 6A, significantly reduced levels of H2O2 were found in N2a cells (P = 0.03) treated at concentrations of 25 μm relative to untreated N2a cells. In contrast, H2O2 was found to be increased in Mdivi-1-treated N2a cells at concentrations of 75 μm (P = 0.04). These findings indicate that, at lower concentrations (e.g. 25 μm) of Mdivi-1, Mdivi-1 is protective of N2a cells, and at high concentrations (e.g. 75 μm), Mdivi-1 is toxic to cells.
Figure 6.
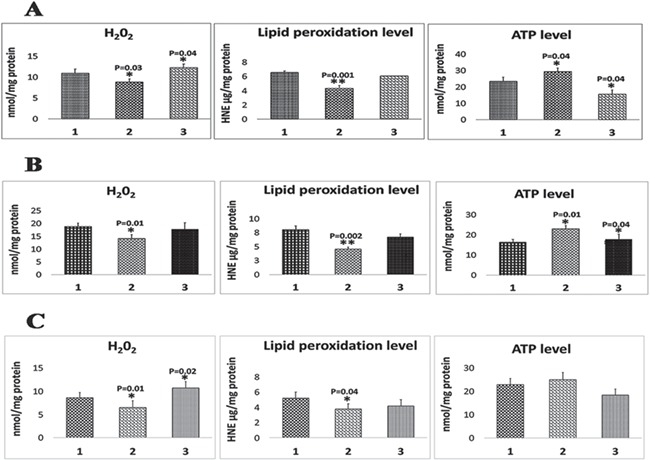
Mitochondrial functional parameters in Mdivi-1-treated cells. (A) H2O2 production, lipid peroxidation and ATP levels in Mdivi-1-treated cells. (B) H2O2 production, lipid peroxidation and ATP levels in full-length, Drp1 transfected+Mdivi-1-treated cells. (C) H2O2 production, lipid peroxidation and ATP levels in Drp1 RNA silenced+Mdivi-1-treated N2a cells.
Lipid peroxidation
Similar to H2O2, levels of 4-hydroxy-2-nonenol (HNE), an indicator of lipid peroxidation, were significantly reduced in N2a cells treated with Mdivi-1 at 25 μm (P = 0.001) relative to untreated N2a cells (Fig. 6A). In N2a cells treated with Mdivi-1 at 75 μm, lipid peroxidation levels were unchanged.
ATP production
As shown in Figure 6A, significantly increased levels of mitochondrial ATP were found in Mdivi-1-treated N2a cells (P = 0.04) relative to untreated N2a cells, indicating that Mdivi-1 is protective to cells at 25 μm concentration treatment. On the other hand, significantly reduced levels of mitochondrial ATP were found in 75 μm Mdivi-1-treated N2a cells (P = 0.04), relative to Mdivi-1-untreated cells. These observations indicate that at high concentration (75 μm), Mdivi-1 is toxic to cells.
Mitochondrial function in N2a cells transfected with full-length Drp1 and treated with Mdivi-1
Mitochondrial function was assessed in N2a cells transfected with full-length Drp1 + Mdivi-1-treated N2a cells by measuring H2O2, lipid peroxidation and mitochondrial ATP.
H2O2 production
As shown in Figure 6B, significantly reduced levels of H2O2 were found in FL-Drp1 transfected+25 μm Mdivi-1-treated N2a cells (P = 0.01) relative to FL-Drp1 transfected N2a cells. In contrast, H2O2 levels were unchanged in FL-Drp1 transfected+75 μm Mdivi-1-treated N2a cells. These results indicate that, at lower concentrations (e.g. 25 μm) of Mdivi-1, Mdivi-1 is protective of N2a cells.
Lipid peroxidation
Similar to H2O2, levels of HNE were significantly reduced in FL-Drp1 transfected+Mdivi-1-treated N2a cells (P = 0.002) relative to FL-Drp1 transfected N2a cells (Fig. 6B). In N2a cells transfected with FL-Drp1 + treated with 75 μm concentration of Mdivi-1, lipid peroxidation levels were unchanged.
ATP production
As shown in Figure 6B, significantly increased levels of mitochondrial ATP were found in FL-Drp1 transfected+Mdivi-1-treated N2a cells (P = 0.01 at 25 μm and P = 0.04 at 75 μm) relative to FL-Drp1-transfected N2a cells, indicating that Mdivi-1 is protective of cells at the low concentration of 25 μm.
Mitochondrial function in Drp1 RNA silenced N2a cells treated with Mdivi-1
H2O2 production
As shown in Figure 6C, significantly reduced levels of H2O2 were found in Drp1 RNA silenced+Mdivi1-treated N2a cells (P = 0.01) treated at concentrations of 25 μm relative to Drp1 RNA silenced and Mdivi-1-untreated N2a cells. In contrast, H2O2 was found to be increased in Mdivi-1-treated N2a cells at concentrations of 75 μm (P = 0.02). These findings indicate that, at lower concentrations (e.g. 25 μm) of Mdivi-1, Mdivi-1 is protective of Drp1 RNA silenced N2a cells, and at high concentrations (e.g. 75 μm), Mdivi-1 is toxic to cells.
Lipid peroxidation
Similar to H2O2, levels of HNE, an indicator of lipid peroxidation, were significantly reduced in Drp1 RNA silenced in N2a cells treated with Mdivi-1 at 25 μm (P = 0.04) relative to Drp1 RNA silenced+Mdivi-1-untreated N2a cells (Fig. 6C).
ATP production
As shown in Figure 6C, increased levels of mitochondrial ATP were found in RNA silenced Drp1 + Mdivi-1-treated N2a cells relative to RNA silenced Drp1 + Mdivi-1-untreated N2a cells, but not significant, indicating that Mdivi-1 is protective to cells at 25 μm concentration treatment. On the other hand, reduced levels of mitochondrial ATP were found in 75 μm Mdivi-1-treated N2a cells, relative to Mdivi-1-untreated cells.
ETC Enzymatic activities in N2a cells treated with Mdivi-1
To determine the effects of Mdivi-1 on enzymatic activities of the mitochondrial ETC, we treated N2a cells with 25 and 75 μm concentrations of Mdivi-1 and assessed enzymatic activities of complexes I, III and IV.
Complex I
As shown in Figure 7A, we found significantly increased levels of complex I in 25 μm Mdivi-1-treated N2a cells (P = 0.03) relative to untreated N2a cells. In contrast, in N2a cells treated with 75 μm Mdivi-1, we found significantly reduced levels of complex I (P = 0.01), indicating that 75 μm Mdivi-1 is toxic to N2a cells.
Figure 7.
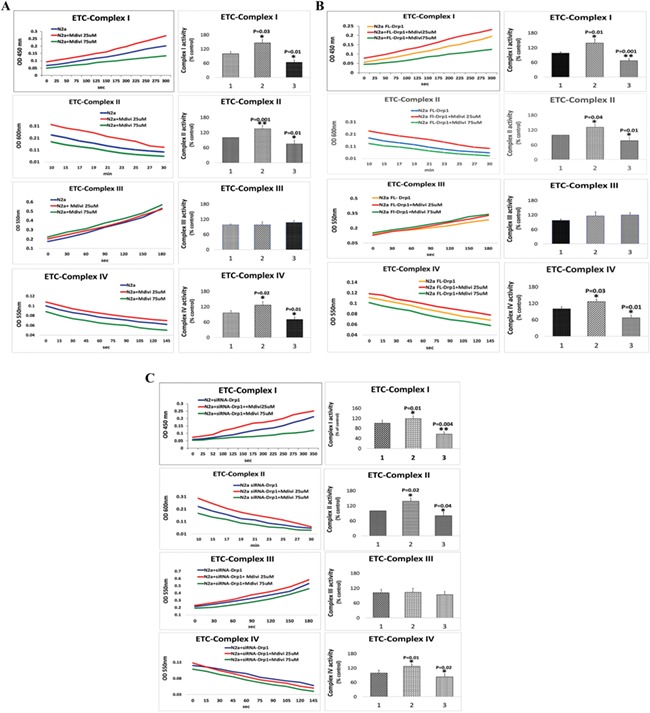
ETC enzymatic activities in Mdivi-1-treated N2a cells. (A) Enzymatic activities of complexes I, II, III and IV in Mdivi-1-treated N2a cells. (B) Enzymatic activities of complexes I, II, III and IV of full-length Drp1 overexpressed+Mdivi-1-treated N2a cells and (C) Enzymatic activities of complexes I, II, III and IV of Drp1 RNA silenced+Mdivi-1-treated N2a cells.
Complex II
Significantly increased levels of complex II enzymatic activity were found in 25 μm Mdivi-1-treated N2a cells (P = 0.001) relative to untreated N2a cells (Fig. 7A). On the other hand, significantly reduced levels of complex II were found in 75 μm Mdivi-1-treated N2a cells (P = 0.01) relative to untreated N2a cells.
Complex III
In N2a cells treated with 25 and 75 μm concentrations of Mdivi-1, we found no changes in complex III enzymatic activity (Fig. 7A).
Complex IV
Significantly increased levels of complex IV were found in 25 μm Mdivi-1-treated N2a cells (P = 0.02) relative to untreated N2a cells (Fig. 7A). In contrast, in N2a cells treated with 75 μm Mdivi-1, we found significantly reduced levels of complex IV (P = 0.01), indicating that 75 μm Mdivi-1 is toxic to N2a cells.
ETC enzymatic activities in full-length Drp1 transfected N2a cells and treated with Mdivi-1.
To determine the effect of full-length Drp1 on ETC enzymatic activities, we transfected human, full-length Drp1 into N2a cells and assessed enzymatic activities. We also assessed the effects of Mdivi-1 at concentrations of 25 and 75 μm on enzymatic activities in overexpressed, full-length Drp1 in N2a cells.
Complex I
We found significantly increased levels of complex I in full-length Drp1 + Mdivi-1-treated N2a cells (P = 0.01) at a concentration of 25 μm Mdivi-1 relative to full-length, Drp1-transfected N2a cells (Fig. 7B). In contrast, in transfected full-length Drp1 + Mdivi-1-treated N2a cells at a concentration of 75 μm Mdivi-1, we found significantly reduced levels of complex I activity (P = 0.001), indicating that a 75 μm concentration of Mdivi-1 is toxic to N2a cells (Fig. 7B).
Complex II
As shown in Figure 7B, significantly increased levels of complex II enzymatic activity were found in full-length Drp1 + Mdivi-1-treated N2a cells (Mdivi-1 concentration at 25 μm) (P = 0.04) relative to full-length Drp1-transfected N2a cells (Fig. 7B). In contrast, significantly reduced complex II enzymatic activity levels were found in full-length Drp1 + Mdivi-1-treated N2a cells (Mdivi-1 concentration at 75 μm) (P = 0.01) relative to full-length Drp1-transfected N2a cells (Fig. 7B).
Complex III
N2a cells transfected with full-length Drp1 + Mdivi-1, at 25 and 75 μm concentrations of Mdivi-1), did not show significantly altered complex III enzymatic activity (Fig. 7B).
Complex IV
As shown in Figure 7B, we found significantly increased levels of complex I in full-length Drp1 + Mdivi-1-treated N2a cells (Mdivi-1 concentration at 25 μm) (P = 0.03) relative to full-length Drp1-transfected N2a cells (Fig. 7B). In contrast, in transfected full-length Drp1 + Mdivi-1-treated N2a cells (Mdivi-1 concentration at 75 μm), we found significantly reduced levels of complex I activity (P = 0.01), indicating that 75 μm concentration of Mdivi-1 is toxic to cells.
ETC enzymatic activities in RNA silenced Drp1 N2a cells and treated with Mdivi-1
Complex I
As shown in Figure 7C, significantly increased levels of complex I enzymatic activity were found in RNA silenced Drp1 + Mdivi-1-treated N2a cells (P = 0.01) at a concentration of 25 μm Mdivi-1 relative to Drp1 RNA silenced and Mdivi-1-untreated N2a cells (Fig. 7C). In contrast, in Drp1 RNA silenced+Mdivi-1-treated N2a cells at a concentration of 75 μm Mdivi-1, we found significantly reduced levels of complex I activity (P = 0.004), indicating that a 75 μm concentration of Mdivi-1 is toxic (Fig. 7C).
Complex II
Significantly increased levels of complex II enzymatic activity were found in RNA silence Drp1 + Mdivi-1-treated N2a cells (Mdivi-1 concentration at 25 μm) (P = 0.02) relative to Drp1 RNA silenced + Mdivi-1-untreated N2a cells (Fig. 7C). On the contrary, significantly reduced complex II enzymatic activity was found in Drp1 RNA silenced+Mdivi-1-treated N2a cells (Mdivi-1 concentration at 75 μm) (P = 0.04) relative to Drp1 RNA silenced + Mdivi-1-untreated N2a cells.
Complex III
Drp1 RNA silenced N2a cells+Mdivi-1, at 25 and 75 μm concentrations of Mdivi-1, did not show significantly altered complex III enzymatic activity (Fig. 7C) relative to Drp1 RNA silenced +Mdivi-1-untreated N2a cells.
Complex IV
As shown in Figure 7C, we found significantly increased levels of complex I in RNA silenced Drp1 + Mdivi-1-treated N2a cells (Mdivi-1 concentration at 25 μm) (P = 0.01) relative to RNA silenced Drp1 and Mdivi-1-untreated N2a cells (Fig. 7C). In contrast, in Drp1 RNA silenced+ Mdivi-1-treated N2a cells (Mdivi-1 concentration at 75 μm), we found significantly reduced levels of complex I activity (P = 0.02), indicating that 75 μm concentration of Mdivi-1 is toxic to cells.
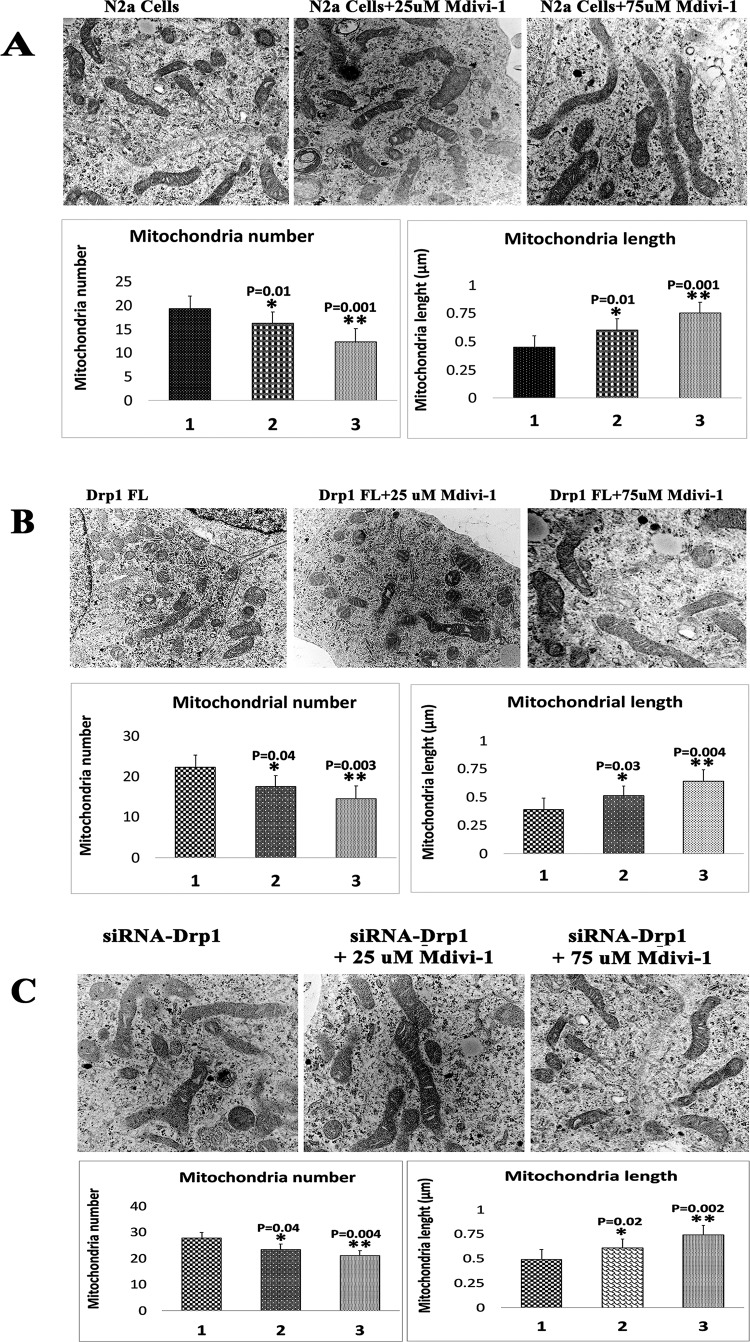
Transmission electron microscopy in N2a cells treated with Mdivi-1
To determine the effects of Midivi-1 on the number and length of mitochondria, we used transmission electron microscopy on Mdivi-1-treated and untreated N2a cells.
Mitochondrial number in Mdivi-1-treated N2a cells
We found a significantly reduced number of mitochondria in N2a cells treated with Mdivi-1 at concentrations of 25 (P = 0.01) and 75 μm (P = 0.001) relative to untreated N2a cells (Fig. 8A), suggesting that Mdivi-1 reduces mitochondrial fission.
Figure 8.
Number and length of mitochondria in Mdivi-1-treated N2a cells. (A) Representative transmission electron microscopy images of mitochondria in Mdivi-1-treated cells. (B) Representative transmission electron microscopy images of mitochondria in full-length, Drp1 transfected+Mdivi-1-treated cells. (C) Representative transmission electron microscopy images of mitochondria in Drp1 RNA silenced+Mdivi-1-treated cells.
Mitochondrial length in Mdivi-1-treated N2a cells
We found significantly increased mitochondrial length in N2a cells treated with Mdivi-1 at concentrations of 25 (P = 0.01) and 75 μm (P = 0.001) relative to Mdivi-1-untreated cells (Fig. 8A).
The significantly reduced number and increased length of mitochondria in Mdivi-1-treated N2a cells strongly suggest that Mdivi-1 reduces fission machinery and enhances fusion activity.
Transmission electron microscopy in N2a cells transfected with full-length Drp1 and treated with Mdivi-1
We determined the effects of full-length Drp1 on mitochondrial number and length in the presence and absence of Mdivi-1 in FL-Drp1 + Mdivi-1-treated N2a cells.
Mitochondrial number
We found significantly reduced numbers of mitochondria in FL-Drp1 transfected+Midivi-1-treated N2a cells at Mdivi-1 concentrations of 25 (P = 0.04) and 75 μm (P = 0.003) relative to FL-Drp1transfected and Mdivi-1 untreated cells (Fig. 8B), suggesting that Mdivi-1 reduces mitochondrial fission in overexpressed Drp1 cells.
Mitochondrial length
We found significantly reduced length of FL-Drp1 transfected+Midivi-1-treated N2a cells at Mdivi-1 concentrations of 25 (P = 0.03) and 75 μm (P = 0.004) relative to FL-Drp1transfected and Mdivi-1 untreated N2a cells (Fig. 8B), suggesting that Mdivi-1 increases mitochondrial length in overexpressed Drp1 cells.
Transmission electron microscopy in Drp1 RNA silence N2a cells treated with Mdivi-1
Mitochondrial number in Drp1 RNA silenced N2a cells treated with Mdivi-1
As shown in Figure 8C, significantly reduced number of mitochondria were found in Drp1 RNA silenced N2a cells treated with Mdivi-1 at concentrations of 25 (P = 0.04) and 75 μm (P = 0.004) relative to Drp1 RNA silenced+Mdivi-1-untreated N2a cells (Fig. 8C), suggesting that Mdivi-1 reduces mitochondrial fission.
Mitochondrial length in Drp1 RNA silenced N2a cells treated with Mdivi-1
Significantly increased mitochondrial length in Drp1 RNA silenced N2a cells treated with Mdivi-1 at concentrations of 25 (P = 0.02) and 75 μm (P = 0.002) relative to Drp1 RNA silenced+Mdivi-1-untreated cells (Fig. 8C).
The significantly reduced number and increased length of mitochondria in Mdivi-1-treated N2a cells strongly suggest that Mdivi-1 reduces fission machinery and enhances fusion activity.
Mitochondrial network in N2a cells treated with Mdivi-1
To determine the effect of Mdivi-1 on the mitochondrial network, we treated N2a cells with 25 and 75 μm concentrations of Mdivi-1 and performed single- and double-immunofluorescence analyses using Drp1 and TOM20 antibodies.
As shown in Figure 9A and B, we found significantly reduced Drp1 immunoreactivity in N2a cells treated with 25 μm Mdivi-1 relative to untreated N2a cells (P = 0.01). Statistical significance was not observed in N2a cells treated with 75 μm Mdivi-1. In N2a cells treated with 25 μm Mdivi-1, TOM20, a marker for the mitochondrial network, showed significantly increased immunoreactivity (P = 0.03). Our double analysis of Drp1 and TOM20 revealed the colocalization of Drp1 and TOM20, indicating reduced fission and an increased mitochondrial network in N2a cells treated with 25 μm Mdivi-1.
Figure 9.
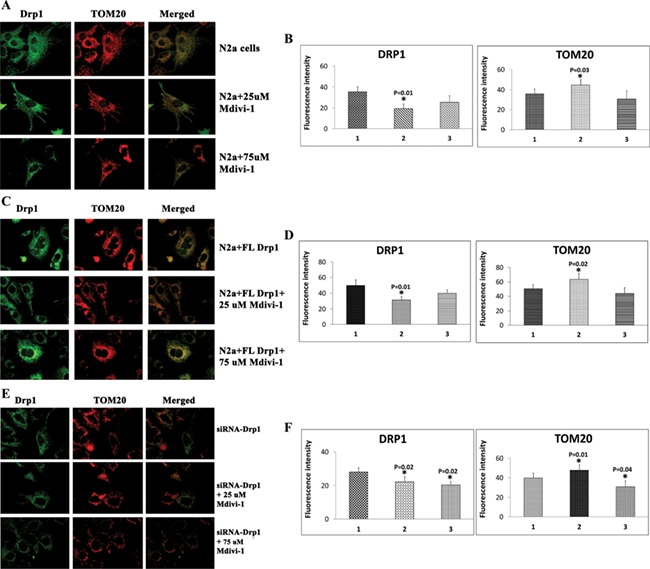
Immunofluorescence analysis of Drp1 and TOM20 in Mdivi-1-treated N2a cells. (A) Representative images of immunofluorescence analysis of Drp1 (green), TOM20 (red) and double-labeling (merged) in Mdivi-1-treated cells. (B) Quantitative analysis of Drp1 and TOM20. (C) Representative images of immunofluorescence analysis of Drp1 (green), TOM20 (red) and double-labeling (merged) in full-length, Drp1 transfected+Mdivi-1-treated cells. (D) Quantitative analysis of Drp1 and TOM20. (E) Representative images of immunofluorescence analysis of Drp1 (green), TOM20 (red) and double-labeling (merged) in Drp1 RNA silenced+Mdivi-1. (F) Quantitative analysis of Drp1 and TOM20.
Mitochondrial network in full-length Drp1-transfected N2a cells and N2a cells treated with Mdivi-1
We also assessed the mitochondrial network in N2a cells treated with 25 and 75 μm Mdivi-1 and performed single- and double-immunofluorescence analyses using Drp1 and TOM20 antibodies. We found significantly decreased Drp1 immunoreactivity (P = 0.01) and increased TOM20 immunoreactivity (P = 0.02) in FL-Drp1 transfected+25 μm Mdivi-1-treated N2a cells relative to FL-Drp1 transfected and untreated N2a cells (Fig. 9C and D). These observations indicate that Mdivi-1 reduces fission and enhances the mitochondrial network.
Mitochondrial network in Drp1 RNA silenced+Mdivi-1 N2a cells
Significantly decreased Drp1 immunoreactivity were found in Drp1 RNA silenced+Mdivi-1-treated N2a cells (P = 0.02 in 25 μm concentration and P = 0.02 at 75 μm concentration of Mdivi-1) and significantly increased TOM20 immunoreactivity in RNA silenced+Mdivi-1 (P = 0.01 in 25 μm concentration of Mdivi-1) relative to Drp1 RNA silenced N2a cells alone (Fig. 9E and F). These observations indicate that Mdivi-1 reduces fission and enhances the mitochondrial network.
Mitochondrial respiration in Mdivi-1-treated cells
To determine the effects of Mdivi-1 on mitochondrial respiration, using an XFe96-well Extracellular Flux Analyzer (Seahorse Bioscience), we assessed the maximal oxygen consumption rate (OCR) in N2a cells treated with Mdivi-1 at concentrations of 25 and 75 μm and in untreated N2a cells. As shown in Figure 10A, significantly increased maximal OCR in N2a cells treated with Mdivi-1 at 25 μm concentration (P = 0.04) relative to untreated N2a cells, indicating that Mdivi-1 enhances maximal OCR. On the contrary, significantly reduced maximal OCR was found in N2a cells treated with Mdivi-1 at 75 μm concentration (P = 0.01) relative to untreated N2a cells, indicating that Mdivi-1 is toxic at higher concentration.
Figure 10.
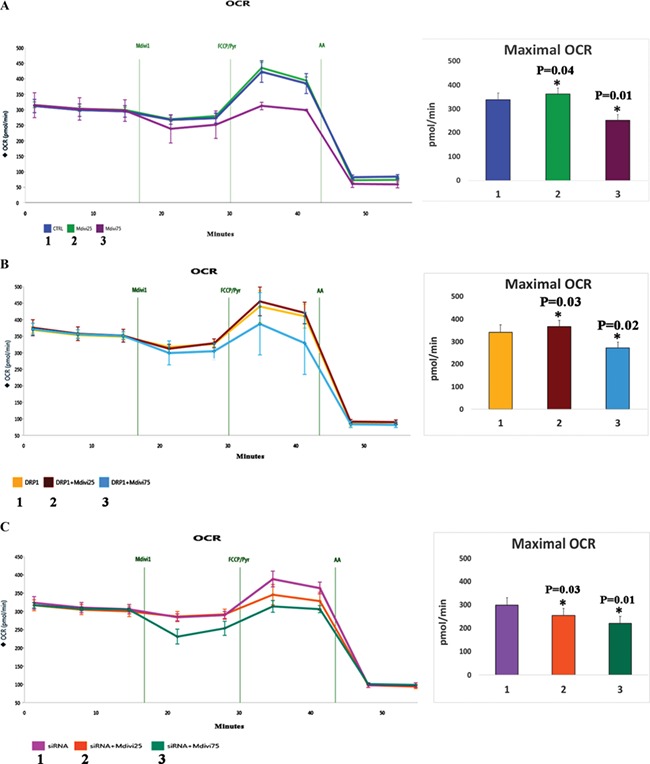
Measurement of mitochondrial respiration using Seahorse Bioanalyzer in Mdivi-1-treated N2a cells. (A) Maximal OCR in Mdivi-1-treated N2a cells. (B) Maximal OCR in Drp1 transfected+Mdivi-1 in N2a cells. (C) Maximal OCR in Drp1 RNA silenced+Mdivi-1-treated N2a cells.
Mitochondrial respiration in full-length Drp1 overexpressed and Mdivi-1-treated N2a cells
As shown in Figure 10B, increased maximal OCR in FL overexpressed Drp1+ 25 μm concentration of Mdivi-1-treated N2a cells (P = 0.03) relative to FL overexpressed Drp1 alone in N2a cells. On the contrary, significantly reduced maximal OCR was found in FL overexpressed Drp1 + 75 μm concentration of Mdivi-1-treated N2a cells (P = 0.02) relative to FL overexpressed Drp1 alone in N2a cells.
Mitochondrial respiration in RNA silenced Drp1 and Mdivi-1-treated N2a cells
As shown in Figure 10C, significantly reduced maximal OCR was found in Drp1 RNA silenced N2a cells+Mdivi-1 at 25 (P = 0.03) and 75 μm treated N2a cells (P = 0.01) relative to Drp1 RNA silenced cells alone.
Discussion
The objective of our study was to resolve the controversy regarding whether Mdivi-1 reduces Drp1 levels and Drp1-GTPase activity. To this end, we measured mRNA and protein levels of genes of mitochondrial dynamics, biogenesis and ETC complexes I, III, IV and V of untreated N2a cells and N2a cells treated with Mdivi-1 at two concentrations (25 and 75 μm) in order to verify the effects of Mdivi-1 on mitochondrial fission–fusion activities, mitochondrial biogenesis and ETC activities. Using biochemical assays, we measured mitochondrial function and GTPase Drp1 enzymatic activity and ETC enzymatic activities of complexes I, II, III and IV. Using transmission electron microscopy and immunofluorescence analyses, we measured mitochondrial network and mitochondrial morphology. Further, using Seahorse XFe96 Extracellular Flux Analyzer, we measured mitochondrial respiration. We also overexpressed the full-length Drp1 in N2a cells and further RNA silenced mouse Drp1 in N2a cells and treated Mdivi-1 in both Drp1 overexpressed and Drp1 RNA silenced N2a cells and studied all of the above parameters.
At the low concentration of Mdivi-1 (25 μm), we found that the treated N2a cells exhibited reduced levels of fission genes (Drp1 and Fis1), increased levels of fusion genes (Mfn1, Mfn2 and Opa1) and increased levels of biogenesis genes (PGC1α, Nrf1, Nrf2 and TFAM) genes. Increased levels of complex I and IV genes were also found in Mdivi-1-treated N2a cells. Immunoblotting data agreed with mRNA changes for all genes studied. In full-length Drp1 + Mdivi-1-treated N2a cells and Drp1 RNA silenced cells, we found reduced levels of fission genes, increased levels of fusion and biogenesis genes. Transmission electron microscopy analysis revealed reduced numbers of mitochondria and increased mitochondrial length in N2a cells treated with 25 μm concentration of Mdivi-1 and also overexpressed with full-length Drp1 and Drp1 RNA silenced in N2a cells and treated with 25 μm Mdivi-1. The mitochondrial network was increased in cells treated with 25 μm Mdivi-1. Increased levels of CI, CII and CIV enzymatic activities were found in N2a cells treated with 25 μm Mdivi-1 and also in N2a cells transfected with full-length Drp1 + Mdivi-1 at 25 μm. Mitochondrial function is increased in cells treated with 25 μm Mdivi-1 and also in N2a cells transfected with full-length Drp1 + treated at 25 μm Mdivi-1. GTPase Drp1 activity decreased in cells treated with 25 μm and also cells transfected with full-length Drp1 + treated with 25 μm Mdivi-1 and also in Drp1 RNA silenced cells at both 25 and 75 μm concentrations. Our results strongly suggest that Mdivi-1 reduces Drp1 levels and mitochondrial fragmentation in N2a cells, in full-length Drp1 overexpressed and Drp1 RNA silenced in N2a cells.
Mdivi-1 and reduced Drp1 levels
At the low concentration of Mdivi-1 treatment (25 μm), both mRNA and protein levels of Drp1 were significantly reduced as Fis1, strongly suggesting that Mdivi-1 is capable of reducing Drp1 levels and fission activity. Further, interestingly, Mdivi-1 enhances fusion proteins at both low and high concentrations of Mdiv-1 treatments. Our current study findings were supported by a large number of studies of mitochondrial fission in AD (28–30) and HD (31). The effect of Mdivi 1 on mitochondrial fission has also been extensively studied, using experimental rodent models of epilepsy and epileptic seizures (32–33), ischemia/reperfusion injury (34–36) and oxygen–glucose deprivation (37). In all of these diseased states and conditions, Mdivi-1 was found to reduce excessive mitochondrial fission, to enhance mitochondrial fusion and to maintain the normal functioning of cells. Our current study, together with these earlier studies, strongly suggests that Mdivi-1 is Drp1 inhibitor.
Mdivi-1 and levels of ETC and biogenesis proteins
To determine the effect of Mdivi-1 on mRNA and protein levels of ETC subunits, we assessed mRNA and protein levels. Interestingly, significantly increased mRNA and protein levels of complexes I and IV subunits were observed in N2a cells receiving 25 μm Mdivi-1. Similar findings were observed in Drp1 overexpressed + Mdivi-1 (25 μm) treated cells. On the contrary, reduced mRNA and protein levels were observed for complexes I and IV in cells treated with 75 μm concentration of Mdivi-1. These observations indicate Mdivi-1 has protective effect at low concentration but not at high concentrations.
Increased levels of mRNA and protein levels of mitochondrial biogenesis levels were observed cells treated with Mdivi-1 at 25 and 75 μm concentrations and also in Drp1 overexpressed plus Mdivi-1 (25 and 75 μm concentrations) treated cells. These observations suggest that Mdivi-1 enhances mitochondrial biogenesis activity.
Mdivi-1 and reduced GTPase Drp1 activity
It has been well established that increased Drp1 activity produces excessively fragmented mitochondria in AD neurons (18,23,38–40). Excessive mitochondrial fragmentation is a major feature in neurodegenerative diseases (16–18,20,23,40–44). Reduction in GTPase Drp1 activity is considered as a therapeutic strategy in diseases that involve increased mitochondrial fragmentation and mitochondrial dysfunction (18,20,31). The levels of GTPase Drp1 activity are directly correlated with Drp1 levels. Therefore, in the current study, we assessed GTPase Drp1 activity (1) in Mdivi-1-treated N2a cells, (2) in Drp1 overexpressed plus Mdivi-1-treated cells and (3) in Drp1 RNA silenced plus Mdivi-1-treated N2a cells at low and high concentrations.
Our finding, that reduced levels of GTPase Drp1 activity occur in Mdivi-1-treated cells at 25 and 75 μm concentrations in all three conditions, supports the hypothesis that GTPase Drp1 activity is essential for mitochondrial division and increased GTPase Drp1 activity levels enhance mitochondrial fragmentation and reduced Drp1 levels decrease mitochondrial fragmentation. These findings are also supported by our earlier research of genetic crossings of heterozygote knockout mice for Drp1 with APP and Tau mice, in which we found reduced Drp1 levels decreases mitochondrial fragmentation and enhances mitochondrial fusion and network (38,39).
Mdivi-1 and mitochondrial number, length and network
Our findings, that Mdivi-1-treated N2a cells significantly reduced mitochondrial number, indicate that Mdivi-1 reduces fragmentation of mitochondria. This reduction is mainly because of reduced Drp1 levels and GTPase Drp1 enzymatic activity. Our observations are strongly supported by earlier cell culture studies in AD (28,40) and HD (31). Another supporting evidence based on transmission electron microscopy (1) in N2a cells treated with Mdivi-1, (2) Drp1 overexpressed plus Mdivi-1 and (3) Drp1 RNA silenced plus Mdivi-1-treated is that significantly increased length of mitochondria and reduced mitochondrial number are likely because of reduced GTPase Drp1 enzymatic activity.
Mdivi-1 and ETC enzymatic activities
Based on our findings (Fig. 7A–C), enzymatic activities of complexes I, II and IV are increased by Mdivi-1 at concentrations of 25 μm in Mdivi-treated N2a and Drp1 overexpressed+Mdivi-1-treated N2a cells, strongly suggesting that Mdivi-1 affects complexes I, II and IV. Immunoblotting data using ETC cocktail antibody further confirm our observations of RNA and ETC enzymatic activities.
Summary and conclusions
Overall, findings from our study strongly suggest that it is important to consider the levels of RNA and protein, GTPase Drp1 activity, ETC enzymatic activities, mitochondrial morphology, including number & length and network and mitochondrial respiration in future studies of mitochondrial therapeutics.
In summary, we have found that (1) at the low concentration of 25 μm, Mdivi-1 reduces Drp1 levels, mitochondrial fragmentation and enhances fusion; (2) Mdivi-1 reduced GTPase Drp1 enzymatic activity at both low and high concentrations; (3) at low and high concentrations, Mdivi-1 reduces the mitochondrial number and increases length of mitochondria; (4) Mdivi-1 enhances mitochondrial network; and (5) Mdivi-1 increases enzymatic activities of complexes I, II and IV. These observations strongly suggest that Mdivi-1 is a Drp1 inhibitor and directly reduces mitochondrial fragmentation and further, Mdivi-1 is a promising molecule to treat human diseases with ETC complexes, I, II and IV.
Materials and Methods
Tissue culture work
N2a cells were grown in DMEM/F12 medium plus 10% fetal bovine serum (FBS) and 1X penicillin and streptomycin (Invitrogen, Carlsbad, CA, USA) until the cells were 75–90% confluent. Four independent cell cultures and transfections were performed for all cell culture experiments. For overexpression of Drp1, we transfected human full-length Drp1 cDNA construct into N2a cells using standard conditions. We received human Drp1 cDNA constructs from Dr van der Bliek, University of California, Los Angeles.
We also performed RNA silencing of Drp1 in N2a cells using standard conditions, following manufacturer recommendations. Mouse Drp1 siRNA (Cat # SC-45953) was purchased from Santa Cruz Biotechnology, Inc. (CA, USA). Using mouse Drp1 siRNA reagents, we performed Drp1 RNA silencing experiments in N2a cells following manufacturer guidelines (Santa Cruz Biotechnology, Inc). Cell pellets were collected and used to assess mRNA, protein levels, mitochondrial ETC enzymatic activities and transmission electron microscopy studies.
Mitochondrial respiration using Seahorse XFe96 Extracellular Flux Analyzer
Mouse brain neuroblastoma N2a cells were seeded at 17 500/well overnight adhere on a poly-L-Lysis-coated XF 96 well tissue culture microplates; next day N2a cells were transfected with human full-length Drp1 for 24 h. For Drp1 RNA silencing experiment, we silenced mouse Drp1 in N2a cells (Santa Cruz Biotechnology, Inc., Cat #sc-45953) for 24 h. Sensor Cartridge with 200 μL/well of XF Calibrant was kept into a non-CO2 incubator at 37°C overnight. OCR was determined using the Seahorse XFe96 Extracellular Flux Analyzer (Seahorse Bioscience). The following day cells were washed three times in assay media and incubated in 175 μL of the same assay media kept into a non-CO2 incubator at 37°C for 1 h. The assay medium was used as described by Bordt et al. (1), which consisted of 120 mm NaCl, 3.5 mm KCl, 1.3 mm CaCl2, 0.4 mm KH2PO4, 1 mm MgCl2, 5 mm HEPES, 15 mm glucose, 4 mg/ml fatty acid free BSA and supplemented with complex I substrates 5 mm sodium pyruvate add with 5 mm malate, pH = 7.4; the complex I substrates were freshly prepared. After Sensor Cartridge calibrated, the cells were loaded into the XFe96 extracellular flux analyzer, three time cycles of 3 min mix and 3 min measurement were measured as baseline after equilibrated, then cells were sequentially injected with DMSO (as control) or Mdivi-1 (25 and 75 μm); 0.25 μm FCCP plus 10 mm sodium pyruvate and final with l μm antimycin A. The 3 min mix and 3 min measurement cycles were repeated two times after each compound injection, all measurement cycles were performed at 37°C. Data shown are mean ± standard error of the mean from five to eight wells.
Mitochondrial functional assays involved measuring H2O2, lipid peroxidation and ATP levels. Also assessed were ETC enzymatic activities and GTPase Drp1 enzymatic activities. Mitochondrial ATP was measured from isolated mitochondria in the N2a cells. All cell culture experiments were performed four times (n = 4) in the current study.
Quantification of mRNA expression of mitochondrial dynamics, mitochondrial biogenesis and synaptic genes using real-time RT-PCR
Using the reagent TriZol (Invitrogen), we isolated total RNA from transfected, untransfected and Mdivi-1-treated N2a cells. Using primer express Software (Applied Biosystems, Foster City, CA, USA), we designed the oligonucleotide primers for the housekeeping genes and for mitochondrial dynamics, mitochondrial biogenesis and ETC genes. The primer sequences and amplicon sizes are listed in Table 4. With SYBR-Green chemistry-based quantitative real-time RT-PCR, mRNA expression of the genes mentioned in the previous sentence was measured, as described by Manczak et al. (18). Briefly, 2 μg of DNAse-treated total RNA was used as starting material, to which we added 1 μl of oligo (dT), 1 μl of 10 mm dNTPs, 4 μl of 5× first strand buffer, 2 μl of 0.1 M DTT and 1 μl RNAseOUT and dT to oligo (dT). The reagents RNA, dT and dNTPs were mixed then heated at 65°C for 5 min and subsequently chilled on ice until the remaining components were added. The samples were incubated at 42°C for 2 min and then 1 μl of Superscript III (40 U/μl) was added. The samples were then incubated at 42°C for 50 min, at which time the reaction was inactivated by heating at 70°C for 15 min.
Table 4.
Summary of qRT-PCR oligonucleotide primers used in measuring mRNA expression in mitochondrial dynamics and mitochondrial biogenesis genes and mitochondrial ETC genes in Mdivi-l-treated N2a cells and FL-Drp1 N2a cells treated with Mdivi-1
| Gene | DNA sequence (5′–3′) | PCR product size |
|---|---|---|
| Mitochondrial dynamics genes | ||
| Drp1 | Forward Primer ATGCCAGCAAGTCCACAGAA | 86 |
| Reverse Primer TGTTCTCGGGCAGACAGTTT | ||
| Fis1 | Forward Primer CAAAGAGGAACAGCGGGACT | 95 |
| Reverse Primer ACAGCCCTCGCACATACTTT | ||
| Mfn1 | Forward Primer GCAGACAGCACATGGAGAGA | 83 |
| Reverse Primer GATCCGATTCCGAGCTTCCG | ||
| Mfn2 | Forward Primer TGCACCGCCATATAGAGGAAG | 78 |
| Reverse Primer TCTGCAGTGAACTGGCAATG | ||
| Opa1 | Forward Primer ACCTTGCCAGTTTAGCTCCC | 82 |
| Reverse Primer TTGGGACCTGCAGTGAAGAA | ||
| Mitochondrial biogenesis genes | ||
| PGC1α | Forward Primer GCAGTCGCAACATGCTCAAG | 83 |
| Reverse Primer GGGAACCCTTGGGGTCATTT | ||
| Nrf1 | Forward Primer AGAAACGGAAACGGCCTCAT | 96 |
| Reverse Primer CATCCAACGTGGCTCTGAGT | ||
| Nrf2 | Forward Primer ATGGAGCAAGTTTGGCAGGA | 96 |
| Reverse Primer GCTGGGAACAGCGGTAGTAT | ||
| TFAM | Forward Primer TCCACAGAACAGCTACCCAA | 84 |
| Reverse Primer CCACAGGGCTGCAATTTTCC | ||
| Reverse Primer AGACGGTTGTTGATTAGGCGT | ||
| Mitochondrial ETC genes | ||
| ETC-CI sub1 | Forward Primer ATTACTTCTGCCAGCCTGACC | 70 |
| Reverse Primer GGCCCGGTTTGTTTCTGCTA | ||
| ETC-CI sub2 | Forward Primer ATAAAACTAGGCCTCGCCCC | 74 |
| Reverse Primer AGTCCTATGTGCAGTGGGAT | ||
| ETC-CI sub3 | Forward Primer TTGCATTCTGACTCCCCCAAAT | 77 |
| Reverse Primer GCTTGTAGGGTCGAATCCGC | ||
| ETC-CI sub4 | Forward Primer TAATCGCACATGGCCTCACA | 71 |
| Reverse Primer GCTGTGGATCCGTTCGTAGT | ||
| ETC-CI sub5 | Forward Primer ATGGTACGGACGAACAGACG | 71 |
| Reverse Primer CGATGTCTCCGATGCGGTTA | ||
| ETC-CI sub6 | Forward Primer CCGCAAACAAAGATCACCCAG | 79 |
| Reverse Primer GAAGGAGGGATTGGGGTAGC | ||
| ETC-CIII CytB | Forward Primer GGCTACGTCCTTCCATGAGG | 75 |
| Reverse Primer TGGGATGGCTGATAGGAGGT | ||
| ETC-CIV COX1 | Forward Primer ATCACTACCAGTGCTAGCCG | 84 |
| Reverse Primer CCTCCAGCGGGATCAAAGAA | ||
| ETC-CIV COX2 | Forward Primer CCGAGTCGTTCTGCCAATAGA | 75 |
| Reverse Primer ACTGCTCATGAGTGGAGGAC | ||
| ETC-CIV COX3 | Forward Primer TGCAGGATTCTTCTGAGCGTT | 70 |
| Reverse Primer AGGTCAGCAGCCTCCTAGAT | ||
| ETC-CV ATP6 | Forward Primer TCCCAATCGTTGTAGCCATCA | 76 |
| Reverse Primer AGACGGTTGTTGATTAGGCGT | ||
| ETC-CV ATP8 | Forward Primer AACATTCCCACTGGCACCTT | 77 |
| Reverse Primer TCGTTCATTTTAATTCTCAAGGGGT | ||
| Housekeeping genes | ||
| B-actin | Forward Primer AGAAGCTGTGCTATGTTGCTCTA | 91 |
| Reverse Primer TCAGGCAGCTCATAGCTCTTC | ||
| GAPDH | Forward Primer TTCCCGTTCAGCTCTGGG | 59 |
| Reverse Primer CCCTGCATCCACTGGTGC | ||
Quantitative real-time PCR amplification reactions were performed in an ABI Prism 7900 sequence detection system (Applied Biosystems) in a 25 μl volume of total reaction mixture, as described in Manczak et al. (18). All RT-PCR reactions were carried out in triplicate, with no template control. The PCR conditions were 50°C for 2 min and 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. The fluorescent spectra were recorded during the elongation phase of each PCR cycle. To distinguish specific amplicons from non-specific amplifications, a dissociation curve was generated. The cycle threshold (CT)-values were calculated with sequence detection system software V1.7 (Applied Biosystems) and an automatic setting of base line, which was the average value of PCR, cycles 3–15, plus CT generated 10 times its standard deviation. The amplification plots and CT values were exported from the exponential phase of PCR directly into a Microsoft Excel worksheet for further analysis.
The mRNA transcript level was normalized against β-actin and the GAPDH at each dilution. The standard curve was the normalized mRNA transcript level, plotted against the log value of the input cDNA concentration at each dilution. The ΔCT-value was obtained by subtracting the average β-actin CT value from the average CT value of for the genes of interest. The ΔCT of N2a cells was used as the calibrator. The fold change was calculated according to the formula 2 − (ΔΔCT), where ΔΔCT is the difference between ΔCT and the ΔCT calibrator value. Statistical significance was calculated between mRNA expression in N2a cells treated with Mdivi-1 cells and untreated cells, using the CT value difference and also N2a cells transfected cells with full-length Drp1 + Mdivi-1-treated cells and also Drp1 RNA silenced+Mdivi-1-treated cells.
Immunoblotting analysis
Immunoblotting analysis was performed using protein lysates prepared for untreated N2a cells, N2a cells treated with Mdivi-1 and N2a cells transfected with human Drp1 cDNA and Drp1 RNA silenced+Mdivi-1-treated N2a cells, as described in Manczak et al. (20). We also performed immunoblotting analysis for mitochondrial dynamics, biogenesis and ETC genes. Details of antibody dilutions are given in Table 4. Forty μg protein lysates were resolved on a 4–12% Nu-PAGE gel (Invitrogen). The resolved proteins were transferred to nylon membranes (Novax Inc., San Diego, CA, USA) and were then incubated for 1 h at room temperature (RT) with a blocking buffer (5% dry milk dissolved in a tris buffered saline with tween® 20 (TBST) buffer). The nylon membranes were incubated overnight with the primary antibodies. The membranes were washed with a TBST buffer three times at 10 min intervals and were then incubated for 2 h with appropriate secondary antibody Sheep anti-mouse HRP 1:10 000, followed by three additional washes at 10 min intervals. Proteins were detected with chemilumniscence reagents (Pierce Biotechnology, Rockford, IL, USA), and the bands from immunoblots were visualized.
Immunofluorescence analysis and quantification
To determine immunoreactivities of single- and double-labeling Drp1 and TOM20 proteins, immunofluorescence analysis was performed, using untreated N2a cells, N2a cells treated with Mdivi-1, Drp1 RNA silenced in N2a cells, N2a cells transfected with full-length Drp1 and treated with Mdivi-1 and N2a cells transfected with full-length Drp1, as described in Manczak et al. (20). Details of antibody dilutions are given in Table 5. The cells were washed with warm phosphate-buffered saline (PBS), fixed in freshly prepared 4% paraformaldehyde in PBS for 10 min and then washed with PBS and permeabilized with 0.1% Triton X-100 in PBS. They were blocked with a 1% blocking solution (Invitrogen) for 1 h at RT. All sections were incubated overnight with antibodies and dilutions, described in Table 3. After incubation, the cells were washed three times with PBS for 10 min each and were then incubated with a secondary antibody conjugated with Fluor 488 and Fluor 594 (Invitrogen) for 1 h at RT. The cells were washed three times with PBS and mounted on slides. Photographs were taken with a multiphoton laser scanning microscope system (ZeissMeta LSM510, 1680 Campus Delivery, Fort Collins, Colorado, USA). To quantify the immunoreactivities of antibodies, 10–15 photographs were taken at ×40 magnification, and statistical significance was assessed.
Table 5.
Summary of antibody dilutions used in the immunoblotting analysis of mitochondrial dynamics, mitochondrial biogenesis proteins and mitochondrial ETC proteins in Mdivi-l-treated N2a cells and FL-Drp1 N2a cells treated with Mdivi-1
| Marker | Primary antibody—species and dilution | Purchased from company, city & state | Secondary antibody, dilution | Purchased from company, city & state |
|---|---|---|---|---|
| Drp1 | Rabbit Polyclonal 1:500 | Novus Biological, Littleton, CO | Donkey anti-rabbit HRP 1:10 000 | GE Healthcare Amersham, Piscataway, NJ |
| Fis1 | Rabbit Polyclonal 1:500 | Protein Tech Group, Inc Chicago, IL | Donkey anti-rabbit HRP 1:10 000 | GE Healthcare Amersham, Piscataway, NJ |
| Mfn1 | Rabbit Polyclonal 1:400 | Abcam, Cambridge, MA | Donkey anti-rabbit HRP 1:10 000 | GE Healthcare Amersham, Piscataway, NJ |
| Mfn2 | Rabbit Polyclonal 1:400 | Abcam, Cambridge, MA | Donkey anti-rabbit HRP 1:10 000 | GE Healthcare Amersham, Piscataway, NJ |
| OPA1 | Rabbit Polyclonal 1:500 | Novus Biological, Littleton, CO | Donkey anti-rabbit HRP 1:10 000 | GE Healthcare Amersham, Piscataway, NJ |
| PGC1α | Rabbit Polyclonal 1:500 | Novus Biological, Littleton, CO | Donkey anti-rabbit HRP 1:10 000 | GE Healthcare Amersham, Piscataway, NJ |
| NRF2 | Rabbit Polyclonal 1:300 | Novus Biological, Littleton, CO | Donkey anti-rabbit HRP 1:10 000 | GE Healthcare Amersham, Piscataway, NJ |
| TFAM | Rabbit Polyclonal 1:300 | Novus Biological, Littleton, CO | Donkey anti-rabbit HRP 1:10 000 | GE Healthcare Amersham, Piscataway, NJ |
| Total OXPHOS cocktail | Mouse Monoclonal 1:250 | Abcam, Cambridge, MA | Sheep anti-mouse HRP 1:10 000 | GE Healthcare Amersham, Piscataway, NJ |
| B-actin | Mouse Monoclonal 1:500 | Sigma-Aldrich, St Luis, MO | Sheep anti-mouse HRP 1:10 000 | GE Healthcare Amersham, Piscataway, NJ |
Table 6.
Summary of antibody dilutions used in immunohisto-immunofluorescence analysis of mitochondrial dynamics, mitochondrial biogenesis and mitochondrial ETC proteins in Mdivi-l-treated N2a cells and FL-Drp1 N2a cells treated with Mdivi-1
| Marker | Primary antibody—species and dilution | Purchased from company, city & state | Secondary antibody, dilution, Alexa Fluor dye | Purchased from company, city & state |
|---|---|---|---|---|
| Drp1 | Rabbit Polyclonal 1:200 | Novus Biological, Littleton, CO | Donkey anti-rabbit IgG Alexa Fluor 488 | Thermo Fisher Scientific, Waltham, MA |
| TOM20 | Mouse Monoclonal 1:100 | Abcam, Cambridge, MA | Donkey anti-mouse IgG Alexa Fluor 594 | Thermo Fisher Scientific, Waltham, MA |
Mitochondrial functional assays
Mitochondrial function was assessed by measuring H202, lipid peroxidation and ATP in untreated N2a cells, N2a cells treated with Mdivi-1, Drp1 RNA silenced in N2a cells, N2a cells transfected with full-length Drp1 and treated with Mdivi-1 and N2a cells transfected with full-length Drp1. All experiments were conducted four times and assessed for statistical significance.
H2O2 production
Using an Amplex® Red H2O2 Assay Kit (Molecular Probes, Eugene, OR, USA), the production of H2O2 was measured using cell pellets from all groups of N2a cells, as described in Reddy et al. (38). Briefly, H2O2 production was measured in N2a cells and in untreated N2a cells and in N2a cells transfected with human, Drp1 cDNA and treated with Mdivi-1 and Drp1 RNA silenced+Mdivi-1-treated cells. A BCA Protein Assay Kit (Pierce Biotechnology) was used to estimate protein concentration. The reaction mixture contained mitochondrial proteins (μg/μl), Amplex red reagents (50 μm), horseradish peroxidase (0.1 U/ml) and a reaction buffer (1X). The mixture was incubated at RT for 30 min, followed by spectrophotometer readings of fluorescence (570 nm). Finally, H2O2 production was determined, using a standard curve equation expressed in nmol/μg mitochondrial protein.
Lipid peroxidation assay
Lipid peroxidates are unstable indicators of oxidative stress in the brain. The final product of lipid peroxidation is HNE, which was measured in all groups of N2a cells. An HNE-His ELISA Kit (Cell BioLabs, Inc., San Diego, CA, USA) was used, as described in Manczak and Reddy (45). Briefly, freshly prepared proteins were added to the 96-well plate first coated with HNE conjugate and incubated overnight at 4°C. It was then washed three times with a buffer. After the last wash, the anti-HNE-His antibody was added to the protein in the wells, which was then incubated for 2 h at RT and was washed again three times. Next, the samples were incubated with a secondary antibody conjugated with HRP for 2 h at RT, followed by incubation with an enzyme substrate. Optical density was measured (at 450 nm) to quantify HNE levels.
ATP levels
ATP levels were measured in isolated mitochondria from all groups of N2a cells using an ATP determination kit (Molecular Probes). The reaction of ATP with recombinant firefly luciferase and its substract luciferin form the basis of the bioluminescence assay. Luciferase catalyzes the formation of light from ATP and luciferin. It is the emitted light that is linearly related to ATP concentration, which is measured with a luminometer. ATP levels were measured from mitochondrial pellets using a standard curve method.
GTPase Drp1 enzymatic activity
Using a GTPase Assay Kit (Cat # 602–0120 Innova Bioscience, Waltham, MA), GTPase Drp1 enzymatic activity was measured in all groups of cells (untreated N2a, N2a cells treated with Mdivi-1, Drp1 RNA silenced in N2a cells, N2a cells transfected with full-length Drp1 and treated with Mdivi-1 and N2a cells transfected with full-length Drp1), using purified Drp1 after immunoprecipitation with anti-Drp1 antibody. This assay is based on GTP hydrolyzing to GDP and to inorganic Pi. GTPase activity was measured, based on the amount of Pi that the GTP produces. By adding the ColorLock Gold (orange) substrate to the Pi generated from GTP, GTP activity was assessed based on the inorganic complex solution (green). Colorimetric measurements (green) were read in the wavelength range of 650 nm.
Transmission electron microscopy
Using transmission electron miscopy, we assessed mitochondrial number and length in all control and experimental groups of cells (untreated N2a, N2a cells treated with Mdivi-1, Drp1 RNA silenced in N2a cells, N2a cells transfected with full-length Drp1 and treated with Mdivi-1 and N2a cells transfected with full-length Drp1).
Cells were fixed in 100 μm sodium cacodylate (pH 7.2), 2.5% glutaraldehyde, 1.6% paraformaldehyde, 0.064% picric acid and 0.1% ruthenium red. They were gently washed and post-fixed for 1 h in 1% osmium tetroxide plus 08% potassium ferricyanide, in 100 mm sodium cacodylate, pH 7.2. After a thorough rinsing in water, the N2a neuron were dehydrated, infiltrated overnight in 1:1 acetone:Epon 812 and infiltrated for 1 h with 100% Epon 812 resin. They were then embedded in the resin. After polymerization, 60–80 nm thin sections were cut on a Reichert ultramicrotome (1680 Campus Delivery, Fort Collins, Colorado, USA) and stained for 5 min in lead citrate. They were rinsed and post-stained for 30 min in uranyl acetate and then were rinsed again and dried. Electron microscopy was performed at 60 kV on a Morgagni TEM Philips (1680 Campus Delivery, Fort Collins, Colorado, USA) equipped with a CCD, and images were collected at magnifications of ×1000–37 000. The numbers of mitochondria and mitochondrial length were counted in all groups of cells and statistical significance was determined, using one-way analysis of variance.
ETC enzymatic activities
Enzymatic activities of complexes I, II, III and IV were measured in mitochondria from all groups of cells (untreated N2a, N2a cells treated with Mdivi-1, Drp1 RNA silenced in N2a cells, N2a cells transfected with full-length Drp1 and treated with Mdivi-1 and N2a cells transfected with full-length Drp1). We used Mitochondria Isolation Kit for Culture Cells (ThermoFisher Scientific Cat# 89874, 1680 Campus Delivery, Fort Collins, Colorado, USA) to isolate intact mitochondria.
Complex I
activity was determined using Complex I Enzyme Activity Microplate Assay Kit (abcam Cat # ab109721). The assay measures oxidation of NADH to NAD+ which leads to increased absorbance at optical density (OD) 450 nm. Briefly isolated mitochondria were added to the pre-coated with anti-complex I antibody microplate, incubate for 3 h at RT next the assay solution contains NADH and dye was added and absorbance was measured at 450 nm in kinetic mode for 5 min.
Complex II
activity was determined using Complex II Enzyme Activity Microplate Assay Kit (abcam Cat # ab109908). The assay measures the production of ubiquinol by the enzyme is coupled to the reduction of the dye DCPIP and decreases in its absorbance at OD 600 nm oxidation. Briefly isolated mitochondria were added to the pre-coated with anti-complex II antibody microplate, incubate for 2 h at RT next the assay solution contains succinate, ubiquinone 2 and DCIP dye was added and absorbance was measured at 600 nm in kinetic mode for 40 min with 3 min interval.
Complex III
activity was determined using MitoTox™ Complex II+ III OXPHOS Activity Assay Kit (abcam Cat # ab109905). The assay monitoring the conversion of oxidized cytochrome c into reduced form, which can be observed as increase in absorbance at OD 550 nm. We used rotenone (Complex I inhibitor) and addition of KCN (Complex IV inhibitor) ensures that there is no re-oxidation of cytochrome c. Mitochondria were incubated with oxidized cytochrome c on pre-coated 96-well microplate next absorbance was measured at 550 nm in kinetic mode for 5 min in RT.
Complex IV
activity was measured using Complex IV Rodent Enzyme Activity Microplate Assay Kit (abcam Cat # ab109911). The assay monitoring the conversion of reduced cytochrome c into oxidized form which leads to decrease in absorbance at 550 nm. Mitochondria were incubated for 3 h at RT in microplate previously pre-coated with anti-Complex IV antibody after adding reduced cytochrome c absorbance was measured at 550 nm in kinetic mode for 5 min in RT.
Statistical considerations
Statistical analyses were conducted for untreated N2a cells and N2a cells treated with Mdivi-1 and overexpressed full-length Drp1+ Mdivi-1 in N2a cells and overexpressed full-length Drp1 in N2a cells and Drp1 RNA silenced + Mdivi-1-treated cell using the Student t-test. The parameters included mRNA and protein levels, H2O2, enzymatic activities of ETC, complexes I, II, III and IV, lipid peroxidation, ATP production, GTPase Drp1 activity and mitochondrial length, number and network.
Conflict of Interest statement. None declared.
Funding
National Institutes of Health (AG042178, AG047812 and NS105473); Garrison Family Foundation; The CH Foundation; and Alzheimer’s Association SAGA grant (to P.H.R.).
References
- 1. Bordt E.A., Clerc P., Roelofs B.A., Saladino A.J., Tretter L., Adam-Vizi V., Cherok E., Khalil A., Yadava N., Ge S.X. et al. (2017) The putative drp1 inhibitor mdivi-1 is a reversible mitochondrial complex I inhibitor that modulates reactive oxygen species. Dev. Cell, 40, 583–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Reddy P.H. and Beal M.F. (2005) Are mitochondria critical in the pathogenesis of Alzheimer's disease? Brain Res. Brain Res. Rev., 49, 618–632. [DOI] [PubMed] [Google Scholar]
- 3. Wallace D.C. (1999) Mitochondrial diseases in man and mouse. Science, 283, 51482–55148. [DOI] [PubMed] [Google Scholar]
- 4. Wallace D.C. (2005) A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu. Rev. Genet., 39, 359–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Reddy P.H., Reddy T.P., Manczak M., Calkins M.J., Shirendeb U. and Mao P. (2011) Dynamin-related protein 1 and mitochondrial fragmentation in neurodegenerative diseases. Brain Res. Rev., 67, 103–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Reddy P.H., Tripathi R., Troung Q., Tirumala K., Reddy T.P., Anekonda V., Shirendeb U.P., Calkins M.J., Reddy A.P., Mao P. et al. (2012) Abnormal mitochondrial dynamics and synaptic degeneration as early events in Alzheimer's disease: implications to mitochondria-targeted antioxidant therapeutics. Biochim. Biophys. Acta, 1822, 639–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lin M.T. and Beal M.F. (2006) Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature, 443, 787–795. [DOI] [PubMed] [Google Scholar]
- 8. Selfridge J.E., E L., Lu J. and Swerdlow R.H. (2013) Role of mitochondrial homeostasis and dynamics in Alzheimer's disease. Neurobiol. Dis., 51, 3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Valero T. (2014) Mitochondrial biogenesis: pharmacological approaches. Curr. Pharm. Des., 35, 5507–5509. [DOI] [PubMed] [Google Scholar]
- 10. Kandimalla R. and Reddy P.H. (2016) Multiple faces of dynamin-related protein 1 and its role in Alzheimer's disease pathogenesis. Biochim. Biophys. Acta, 1862, 814–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cribbs J.T. and Strack S. (2007) Reversible phosphorylation of drp1 by cyclic AMP-dependent protein kinase and calcineurin regulates mitochondrial fission and cell death. EMBO Rep., 8, 939–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chang C.R. and Blackstone C. (2007) Cyclic AMP-dependent protein kinase phosphorylation of drp1 regulates its GTPase activity and mitochondrial morphology. J. Biol. Chem., 282, 21583–21587. [DOI] [PubMed] [Google Scholar]
- 13. Frank S., Gaume B., Bergmann-Leitner E.S., Leitner W.W., Robert E.G., Catez F., Smith C.L. and Youle R.J. (2001) The role of dynamin-related protein 1, a mediator of mitochondrial fission, in apoptosis. Dev. Cell, 1, 515–525. [DOI] [PubMed] [Google Scholar]
- 14. Reddy P.H. (2014) Inhibitors of mitochondrial fission as a therapeutic strategy for diseases with oxidative stress and mitochondrial dysfunction. J. Alzheimers Dis., 40, 245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cassidy-Stone A., Chipuk J.E., Ingerman E., Song C., Yoo C., Kuwana T., Kurth M.J., Shaw J.T., Hinshaw J.E., Green D.R. and Nunnari J. (2008) Chemical inhibition of the mitochondrial division dynamin reveals its role in Bax/Bak-dependent mitochondrial outer membrane permeabilization. Dev. Cell, 14, 193–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang X., Su B., Siedlak S.L., Moreira P.I., Fujioka H., Wang Y., Casadesus G. and Zhu X. (2008) Amyloid-beta overproduction causes abnormal mitochondrial dynamics via differential modulation of mitochondrial fission/fusion proteins. Proc. Natl. Acad. Sci. USA, 105, 19318–18323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang X., Su B., Lee H.G., Li X., Perry G., Smith M.A. and Zhu X. (2009) Impaired balance of mitochondrial fission and fusion in Alzheimer's disease. J. Neurosci., 29, 9090–9103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Manczak M., Calkins M.J. and Reddy P.H. (2011) Impaired mitochondrial dynamics and abnormal interaction of amyloid beta with mitochondrial protein drp1 in neurons from patients with Alzheimer's disease: implications for neuronal damage. Hum. Mol. Genet., 20, 2495–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Calkins M.J., Manczak M., Mao P., Shirendeb U. and Reddy P.H. (2010) Impaired mitochondrial biogenesis, defective axonal transport of mitochondria, abnormal mitochondrial dynamics and synaptic degeneration in a mouse model of Alzheimer's disease. Hum. Mol. Genet., 20, 4515–4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Manczak M., Mao P., Calkins M.J., Cornea A., Reddy A.P., Murphy M.P., Szeto H.H., Park B. and Reddy P.H. (2010) Mitochondria-targeted antioxidants protect against amyloid-beta toxicity in Alzheimer's disease neurons. J. Alzheimers Dis., 20, S609–S631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Manczak M., Kandimalla R., Yin X. and Reddy P.H. (2018) Hippocampal mutant APP and amyloid beta-induced cognitive decline, dendritic spine loss, defective autophagy, mitophagy and mitochondrial abnormalities in a mouse model of Alzheimer's disease. Hum. Mol. Genet., 27, 1332–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kandimalla R., Manczak M., Yin X., Wang R. and Reddy P.H. (2018) Hippocampal phosphorylated tau induced cognitive decline, dendritic spine loss and mitochondrial abnormalities in a mouse model of Alzheimer's disease. Hum. Mol. Genet., 27, 30–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Reddy P.H., Yin X., Manczak M., Kumar S., Pradeepkiran J.A., Vijayan M. and Reddy A.P. (2018) Mutant APP and amyloid beta-induced defective autophagy, mitophagy, mitochondrial structural and functional changes and synaptic damage in hippocampal neurons from Alzheimer's disease. Hum. Mol. Genet., 27, 2502–2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Silva D.F., Selfridge J.E., Lu J., E L., Roy N., Hutfles L., Burns J.M., Michaelis E.K., Yan S., Cardoso S.M. and Swerdlow R.H. (2013) Bioenergetic flux, mitochondrial mass and mitochondrial morphology dynamics in AD and MCI cybrid cell lines. Hum. Mol. Genet., 22, 3931–3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Qi X., Qvit N., Su Y.C. and Mochly-Rosen D. (2013) A novel drp1 inhibitor diminishes aberrant mitochondrial fission and neurotoxicity. J. Cell Sci., 126, 789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Macia E., Ehrlich M., Massol R., Boucrot E., Brunner C. and Kirchhausen T. (2006) Dynasore, a cell permeable inhibitor of dynamin. Dev. Cell, 10, 839–850. [DOI] [PubMed] [Google Scholar]
- 27. Mallat A., Uchiyama L.F., Lewis S.C., Fredenburg R.A., Terada Y., Ji N., Nunnari J. and Tseng C.C. (2018) Discovery and characterization of selective small molecule inhibitors of the mammalian mitochondrial division dynamin, DRP1. Biochem. Biophys. Res. Commun., 499, 556–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Reddy P.H., Manczak M. and Yin X. (2017) Mitochondria-division inhibitor 1 protects against amyloid-β induced mitochondrial fragmentation and synaptic damage in Alzheimer's disease. J. Alzheimers Dis., 58, 147–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Baek S.H., Park S.J., Jeong J.I., Kim S.H., Han J., Kyung J.W., Baik S.H., Choi Y., Choi B.Y., Park J.S. et al. (2017) Inhibition of drp1 ameliorates synaptic depression, Aβ deposition, and cognitive impairment in an Alzheimer's disease model. J. Neurosci., 37, 5099–5110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang W., Yin J., Ma X., Zhao F., Siedlak S.L., Wang Z., Torres S., Fujioka H., Xu Y., Perry G. and Zhu X. (2017) Inhibition of mitochondrial fragmentation protects against Alzheimer's disease in rodent model. Hum. Mol. Genet., 26, 4118–4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Manczak M. and Reddy P.H. (2015) Mitochondrial division inhibitor 1 protects against mutant huntingtin-induced abnormal mitochondrial dynamics and neuronal damage in Huntington's disease. Hum. Mol. Genet., 24, 7308–7325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Qiu X., Cao L., Yang X., Zhao X., Liu X., Han Y., Xue Y., Jiang H. and Chi Z. (2013) Role of mitochondrial fission in neuronal injury in pilocarpine-induced epileptic rats. Neuroscience, 245, 157–165. [DOI] [PubMed] [Google Scholar]
- 33. Xie N., Wang C., Lian Y., Zhang H., Wu C. and Zhang Q. (2013) A selective inhibitor of drp1, mdivi-1, protects against cell death of hippocampal neurons in pilocarpine-induced seizures in rats. Neurosci. Lett., 545, 64–68. [DOI] [PubMed] [Google Scholar]
- 34. Ong S.B., Subrayan S., Lim S.Y., Yellon D.M., Davidson S.M. and Hausenloy D.J. (2010) Inhibiting mitochondrial fission protects the heart against ischemia/reperfusion injury. Circulation, 121, 2012–2022. [DOI] [PubMed] [Google Scholar]
- 35. Zhang X., Yan H., Yuan Y., Gao J., Shen Z., Cheng Y., Shen Y., Wang R.R., Wang X., Hu W.W. et al. (2013) Cerebral ischemia-reperfusion-induced autophagy protects against neuronal injury by mitochondrial clearance. Autophagy, 9, 1321–1333. [DOI] [PubMed] [Google Scholar]
- 36. Park S.W., Kim K.Y., Lindsey J.D., Dai Y., Heo H., Nguyen D.H., Ellisman M.H., Weinreb R.N. and Ju W.K. (2011) A selective inhibitor of drp1, mdivi-1, increases retinal ganglion cell survival in acute ischemic mouse retina. Invest. Ophthalmol. Vis. Sci., 52, 2837–2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wappler E.A., Institoris A., Dutta S., Katakam P.V. and Busija D.W. (2013) Mitochondrial dynamics associated with oxygen-glucose deprivation in rat primary neuronal cultures. PLoS One, 8, e63206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Manczak M., Kandimalla R., Fry D., Sesaki H. and Reddy P.H. (2016) Protective effects of reduced dynamin-related protein 1 against amyloid beta-induced mitochondrial dysfunction and synaptic damage in Alzheimer's disease. Hum. Mol. Genet., 25, 5148–5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kandimalla R., Manczak M., Fry D., Suneetha Y., Sesaki H. and Reddy P.H. (2016) Reduced dynamin-related protein 1 protects against phosphorylated tau-induced mitochondrial dysfunction and synaptic damage in Alzheimer's disease. Hum. Mol. Genet., 25, 4881–4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Reddy P.H., Manczak M., Yin X. and Reddy A.P. (2018) Synergistic protective effects of mitochondrial division inhibitor 1 and mitochondria-targeted small peptide SS31 in Alzheimer's disease. J. Alzheimers Dis., 62, 1549–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shirendeb U., Reddy A.P., Manczak M., Calkins M.J., Mao P., Tagle D.A. and Reddy P.H. (2011) Abnormal mitochondrial dynamics, mitochondrial loss and mutant huntingtin oligomers in Huntington's disease: implications for selective neuronal damage. Hum. Mol. Genet., 20, 1438–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shirendeb U.P., Calkins M.J., Manczak M., Anekonda V., Dufour B., McBride J.L., Mao P. and Reddy P.H. (2012) Mutant huntingtin's interaction with mitochondrial protein drp1 impairs mitochondrial biogenesis and causes defective axonal transport and synaptic degeneration in Huntington's disease. Hum. Mol. Genet., 21, 406–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Song W., Chen J., Petrilli A., Liot G., Klinglmayr E., Zhou Y., Poquiz P., Tjong J., Pouladi M.A., Hayden M.R. et al. (2011) Mutant huntingtin binds the mitochondrial fission GTPase dynamin-related protein-1 and increases its enzymatic activity. Nat. Med., 17, 377–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Trushina E., Nemutlu E., Zhang S., Christensen T., Camp J., Mesa J., Siddiqui A., Tamura Y., Sesaki H., Wengenack T.M., Dzeja P.P. and Poduslo J.F. (2012) Defects in mitochondrial dynamics and metabolomic signatures of evolving energetic stress in mouse models of familial Alzheimer's disease. PLoS One, 7, e32737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Manczak M. and Reddy P.H. (2012) Abnormal interaction between the mitochondrial fission protein drp1 and hyperphosphorylated tau in Alzheimer's disease neurons: implications for mitochondrial dysfunction and neuronal damage. Hum. Mol. Genet., 21, 2538–2547. [DOI] [PMC free article] [PubMed] [Google Scholar]