Abstract
Orthotopic liver transplantation remains the only curative therapy for inborn errors of metabolism. Given the tremendous success for primary immunodeficiencies using ex-vivo gene therapy with lentiviral vectors, there is great interest in developing similar curative therapies for metabolic liver diseases. We have previously generated a pig model of hereditary tyrosinemia type 1 (HT1), an autosomal recessive disorder caused by deficiency of fumarylacetoacetate hydrolase (FAH). Using this model, we have demonstrated curative ex-vivo gene and cell therapy using a lentiviral vector to express FAH in autologous hepatocytes. To further evaluate the long-term clinical outcomes of this therapeutic approach, we continued to monitor one of these pigs over the course of three years. The animal continued to thrive off the protective drug NTBC, gaining weight appropriately, and maintaining sexual fecundity for the course of his life. The animal was euthanized 31 months after transplantation to perform a thorough biochemical and histological analysis. Biochemically, liver enzymes and alpha-fetoprotein levels remained normal and abhorrent metabolites specific to HT1 remained corrected. Liver histology showed no evidence of tumorigenicity and Masson’s trichrome staining revealed minimal fibrosis and no evidence of cirrhosis. FAH-immunohistochemistry revealed complete repopulation of the liver by transplanted FAH-positive cells. A complete histopathological report on other organs, including kidney, revealed no abnormalities. This study is the first to demonstrate long-term safety and efficacy of hepatocyte-directed gene therapy in a large animal model. We conclude that hepatocyte-directed ex-vivo gene therapy is a rational choice for further exploration as an alternative therapeutic approach to whole organ transplantation for metabolic liver disease, including HT1.
Keywords: ex-vivo gene therapy, hepatocyte transplantation, hereditary tyrosinemia type 1, porcine model, fumarylacetoacetate hydrolase
Introduction
Hereditary tyrosinemia type I (HT1) is an autosomal recessive metabolic liver disease caused by a deficiency in fumarylacetoacetate hydrolase (FAH), the final enzyme in the catabolism of tyrosine1,2. Liver transplant remains the only curative treatment but is limited by a shortage of donor organs, the need for lifelong immunosuppression and the relatively high graft-loss rates in pediatric populations3,4. We previously reported the first demonstration of curative ex-vivo gene therapy in a pig model of HT15–7 using an integrating lentiviral vector expressing the porcine Fah cDNA under the control of a hepatocyte-specific promoter8. In this previous study, autologous hepatocytes from four pigs were isolated after partial hepatectomy and re-transplanted to the same pigs after gene therapy9. The short-term (12 months) results from that study demonstrated extensive repopulation of the recipient livers by FAH-positive cells and amelioration of biochemical markers indicative of HT1. However, the long-term consequences of this therapeutic approach were unknown, and no reports of long-term safety and efficacy have been reported in a clinically-relevant model. Therefore, the goal of the present study was to assess the long-term safety and efficacy of hepatocyte-directed ex-vivo gene therapy using an integrating lentiviral vector in a single pig. Herein, we demonstrate that no adverse events were noted and that, histologically and biochemically, the animal appeared normal, with stable amelioration of markers indicative of HT1 after 31 months. We anticipate that these data will lay the foundation for further studies to assess the clinical relevance of this novel therapeutic approach for HT1 and other metabolic liver diseases.
Materials and Methods
Animals and Animal Care
All animals received humane care in compliance with the regulations of the institutional animal care and use committee at Mayo Clinic, Rochester, MN. Daily observations were performed by animal care/laboratory staff and any clinical concerns were addressed by on-site veterinarians. Animals were weighed weekly until NTBC independence was confirmed and then continued to be weighed at various time points throughout the study. Since growing pigs are prone to gastric ulceration and NTBC cycling can cause inappetence, animals were given prophylactic omeprazole orally at a dose of 1 mg/kg with a maximum dose of 80 mg/day for the duration of the study. At the onset of sexual maturity, semen collection was carried out weekly with average yield and concentration evaluated on a SpermaCue photometer (MiniTube of America, Delavan, WI, USA).
Biochemical Analysis
For biochemical analysis of alkaline phosphatase (ALP), alanine aminotransferase (ALT), aspartate aminotransferase (AST), albumin and total bilirubin (TBIL), plasma or serum was analyzed with the VetScan VS2 benchtop analyzer (Mammalian Liver Profile, Abaxis, Union City, CA, USA) according to the manufacturer’s instructions. Alpha-fetoprotein (AFP) was analyzed in serum with the Beckman Coulter Access AFP immunoenzymatic assay on the Beckman Coulter UniCel DXI 800 (Beckman Coulter Inc., Fullerton, CA, USA). Tyrosine values were determined using tandem mass spectrometry and chromatography via Mayo Clinic’s internal biochemical PKU test.
Histology Analysis
For histological analysis, tissue samples were fixed in 10% neutral buffered formalin (Protocol, Fisher-Scientific, Pittsburgh, PA, USA) and processed for paraffin embedding and sectioning. For hematoxylin and eosin staining, slides were prepared with standard protocols. FAH or Ki-67 immunohistochemistry using a polyclonal rabbit anti-FAH primary antibody10 or a monoclonal anti-Ki67 primary antibody (MIB-1; Dako/Agilent, Santa Clara, CA, USA) was performed with a Bond III automatic stainer (Leica, Buffalo Grove, IL, USA) with a 20 min antigen-retrieval step using Bond Epitope Retrieval Solution 2 (Leica, Buffalo Grove, IL, USA), and stained with diaminobenzidine (Leica, Buffalo Grove, IL, USA). Slides used to quantify fibrosis were stained with Masson’s trichrome stain using standard protocols. Fifteen rectangular areas totaling 2.39 × 107 µm2 per slide were randomly selected and analyzed using an Aperio ImageScope algorithm that quantifies pixel hue. Blue pixels were counted as being positive for collagen. Reported results are total percentage of positive pixels among those analyzed. Similarly, Ki-67 quantification was performed by selecting 15 random rectangular areas totaling 2.39 × 107 µm2 per slide. Areas were analyzed and quantified using an Aperio ImageScope algorithm that quantifies nuclear staining. Results are reported as percentage of nuclear positivity among cells analyzed.
Integration Site Analysis
Genomic DNA was isolated from snap-frozen tissue fragments of liver collected at necropsy using a Gentra Puregene Tissue Kit (Qiagen, Hilden, Germany). Ligation-mediated PCR (LM-PCR) was used for amplification of integration sites. MseI restriction digestions of genomic DNA samples was performed; the digested DNA samples were then ligated to linkers and treated with ApoI to limit amplification of the internal vector fragment downstream of the 5′ LTR. Samples were amplified by nested PCR and sequenced on the Illumina HiSeq 2500 Next-Generation Sequencing System (San Diego, CA, USA). Integration sites were judged to be authentic if the sequences began within three base pairs of the lentiviral 3′ LTR end, showed a > 98% sequence match, and yielded a unique best match when aligned to the pig genome using the Burrows-Wheeler Alignment algorithm (BWA-MEM). Quality control of the sequencing reads was performed with FastQC. The reads were then trimmed by: (a) viral sequence trimming – the viral sequence was trimmed using Cutadapt with a mismatch rate (e = 0.3) from the 5′ end of the first read (R1) and the 3′ end of the second read (R2) of sequenced pairs; (b) linker trimming – the linker sequence was trimmed using Cutadapt from the 3′ end of R1 and the 5′ end of R2. After each trimming step, reads that became too short (< 15 bp) were filtered out due to low fidelity of matches. After trimming, the reads were mapped to the susScr11 build of the pig reference genome using BWA-MEM in single-end mode using default parameters. The single-end mode was used instead of paired-end for mapping as the insert size of read pairs was determined to be too short and resulted in overlapping mates. Reads were also checked for contamination from potential bacterial, viral and model organism sources other than lentivirus by randomly selecting one million unmapped reads after alignment and using BLAST to querying these reads against the GenBank database (NCBI).
The aligned reads were subjected to multiple filtering criteria to be selected for viral integration point identification. A site of integration is defined by the position on the pig genome of the first base immediately following the viral sequence that was trimmed by Cutadapt. Positive reads were considered to: (a) have been successfully trimmed with the viral sequence, yielding a read length shorter than the original read length (< 150 bp); and (b) map uniquely to the genome with a BWA-MEM mapping quality score of greater than zero. Since the first read (R1) of a sequenced read pair predominantly contained the trimmed viral sequence, and the second read (R2) largely overlapped the first read, the second read was excluded from the analysis as it did not provide any additional value for detecting integration points. Using this approach, unique integration points were identified across the genome without any constraints on coverage. However, for downstream annotation and analysis, only those integration points which had a total read coverage of at least 5X were used to minimize false positives.
The integration points were then extensively annotated using in-house Python scripts with gene and feature information using the susScr11 Refflat file of the pig genome obtained from the UCSC Table Browser. Annotations of integration points were characterized by genomic features such as exons, introns and intergenic regions. To avoid conflict of feature categorization arising from multiple overlapping transcript definitions of the same gene, only the longest transcript of each gene was used for the annotation process. Additionally, the distance of each integration point from the transcription start site (TSS) of the nearest gene was calculated. The integration points were further annotated to assess their presence or absence in CpG-rich regions of the genome (‘CpG islands’) using the susScr11 CpG island definitions obtained from the UCSC Table Browser. Additionally, an in-house Mayo Large NGS Tumor Panel was evaluated from the resulting reads, which contained targeted regions from approximately 745 human genes to assess the distribution and density of the integration points in the pigs within these tumor-associated genes. This approach was based on the assumption that the tumor-associated genes are generally functionally homologous between these species and any viral integration in these tumor-associated genes in the pig would therefore result in a clinically translatable gene expression profile.
Statistical Analysis
Data were analyzed using GraphPad Prism software version 7. Experimental analyses with only one comparison were compared using a two-tailed Welch’s t-test. Differences between multiple groups were compared using a one-way ANOVA followed by Tukey’s multiple comparisons test; P < 0.05 was considered statistically significant.
Results
Pig Y842 Remained Phenotypically Stable for the Duration of the Study
Y842 was a male homozygous Fah-/- pig with a 50% Large White and 50% Landrace pedigree. At 39 days of age, weighing 11 kg, this pig underwent a partial hepatectomy to remove approximately 10% of its liver. Hepatocytes were isolated and transduced with a lentiviral vector (LV) expressing the porcine Fah cDNA under the control of a hepatocyte-specific promoter8. LV-transduced autologous hepatocytes were transplanted back to the same pig by percutaneous portal vein injection. Y842 was cycled on/off the protective drug NTBC for four cycles and became NTBC-independent 95 days after transplantation. At 12 months of age, liver biopsies and biochemical analyses revealed complete amelioration of symptoms characteristic of HT1.
In this study, we continued to monitor the phenotype of Y842 over the course of 949 days after transplantation. At 532 days of age, the pig weighed 265 kg, which was at the functional limit of the available scale (Fig. 1A). The animal continued to thrive and remained NTBC independent for the course of this study. As a measure of health, the animal maintained sexual fecundity, siring a total of 57 piglets from five breedings; the last litter of piglets was sired 860 days after transplantation. Semen collection was carried out weekly with an average yield of 275 ml at a concentration of 265×106 sperm/ml. At 988 days after birth, Y842 was euthanized at the request of the institute’s veterinarians, owing to the inability to humanely house such a large animal. No clinical manifestations of liver disease were noted at the time of euthanasia.
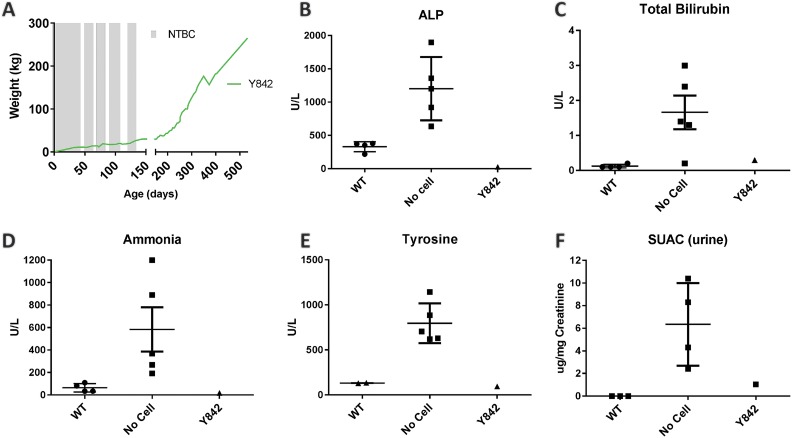
Fig. 1.
Biochemical analyses confirmed amelioration of metabolic disease. (A) Long-term weight data on Y842 from birth (day 0) though 532 days of age. Time on NTBC is represented by shaded bars. (B–E) Plasma or serum from the time of sacrifice was compared to historical wildtype (WT; n = 4) and Fah-/- controls that received no cell transplant (No Cell; n = 5). (F) Urine succinylacetone (SUAC) from the time of sacrifice was compared to historical wildtype (n = 3) and Fah-/- untreated controls (n = 4).
Biochemical Parameters of Liver Function Were Normal
At the time of euthanasia, a complete biochemical and histological analysis was performed. Due to the lack of appropriate age-matched controls (large pigs are not routinely kept at institutes for this prolonged period of time), we compared biochemical values from Y842 to younger historical Fah+/+ (wildtype) and Fah-/- (No Cell) controls (Fig. 1B–F). Biochemical liver function markers, including alkaline phosphatase (ALP), total bilirubin and ammonia were within normal limits, indicating normal liver function. Next, we looked at metabolites specific to HT1. Tyrosine and succinylacetoacetate levels were also normalized, indicating continued amelioration of tyrosine catabolism. Finally, to investigate whether any biochemical indication of hepatocellular carcinoma (HCC) could be detected, we determined the concentration of serum AFP by ELISA to be 0.8 ng/ml, which was within normal limits11.
Histological Assessment of the Liver Revealed Normal Tissue Architecture
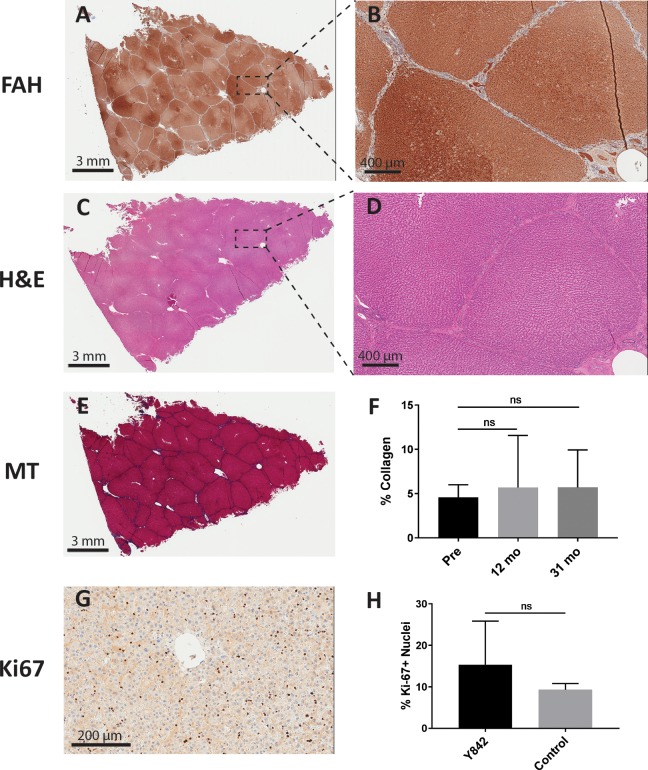
We next assessed by histology and immunohistochemistry whether any abnormal pathology could be detected in the liver. Macroscopically, the liver had no gross lesions or tumors and there was no evidence of surface nodularity or extrahepatic collateral vein development, suggesting the absence of cirrhosis (Fig. 2). FAH immunohistochemistry of the liver revealed complete repopulation of the Fah-/- liver by the LV-Fah-transduced hepatocytes (Fig. 3A–B). Dividing the liver into 1–2 cm sections and performing H&E (Fig. 3C–D) and Masson’s Trichrome staining (Fig. 3E–F) throughout the liver revealed no significant pathology, including no evidence of HCC – supporting the negative AFP findings in the serum. Only a mild increase in periportal fibrosis was noted, which was in contrast to mild and moderate fibrosis detected at the 12-month time point – providing provocative data that moderate fibrosis in HT1 may be reversible after FAH+ hepatocyte repopulation. Quantification of collagen staining revealed no significant changes in fibrosis over the course of 31 months, a striking contrast to Fah-/- pigs that succumb to severe fibrosis within six months in the absence of intervention7. Finally, we quantified Ki-67-positive cells in the liver to determine if abnormal proliferation was occurring.
Fig. 2.
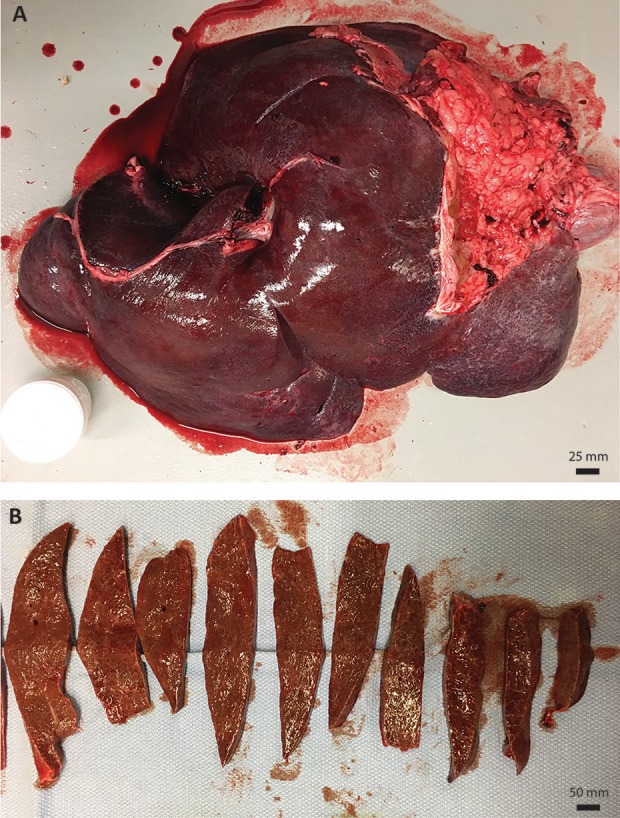
(A) Complete liver of Y842 at time of euthanasia. (B) Representative cross-sections of liver showing no significant pathology.
Fig. 3.
Histological analyses revealed no abnormal liver pathology. (A–B) Representative low (left) and high (right) magnification images from Y842 at time of euthanasia after FAH IHC. (C–D) Representative low (left) and high (right) magnification images from Y842 at time of euthanasia after H&E staining. (E) Masson’s Trichrome (MT) staining in a representative liver section at time of euthanasia. (F) Quantification of collagen staining was compared in the same animal from time of transplant (Pre), 1-year post-transplantation biopsy (12 mo), and at time of euthanasia (31 mo). (G) Ki-67 staining in a representative liver section at time of euthanasia in Y842. (H) Quantification of Ki-67-positive nuclei staining was compared in Y842 and a 42-month-old control pig.
No significant difference in Ki67-positive cells was noted between multiple segments of the liver from Y842 and a 42-month-old control boar (Fig. 3G–H).
Histological Assessment of the Kidney Revealed Normal Tissue Architecture
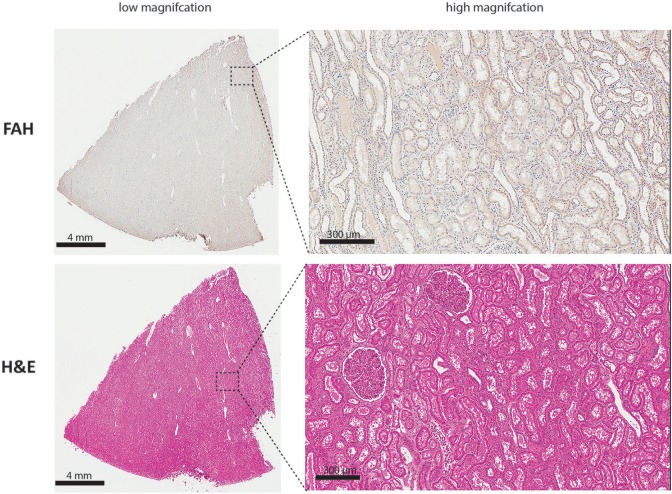
As histopathological abnormities of the kidney are commonly associated with HT1 clinically, we went on to analyze the kidneys of Y842 (Figs 4 & 5). As expected, no FAH+ renal tubular cells were detected by IHC, and H&E staining revealed no significant pathologies except for a mild increase in tubular degeneration with regeneration. Additionally, primarily medullary, inner and outer stripe and some medullary rays were noted with an increase in medullary interstitial stroma. In addition to the liver and kidneys, no significant pathology was detected in the rest of the animal that would indicate an adverse event resultant from the ex vivo gene therapy (data not shown).
Fig. 4.
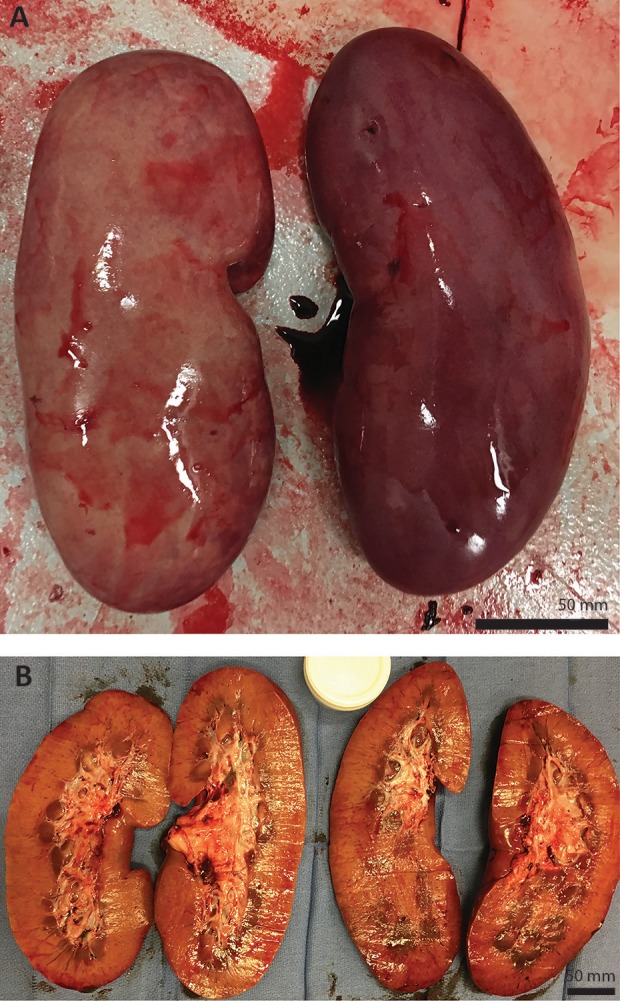
(A) Whole kidneys of Y842 at time of euthanasia. (B) Cross-sections of both kidneys showing no significant pathology.
Fig. 5.
Representative low (left) and high (right) magnification images of the kidney of Y842 at time of euthanasia after FAH IHC. (Top) Representative low (left) and high (right) magnification images from kidney of Y842 at time of euthanasia after H&E staining (Bottom).
Bioinformatics Showed a Benign Profile of Lentiviral Integration into the Pig Genome
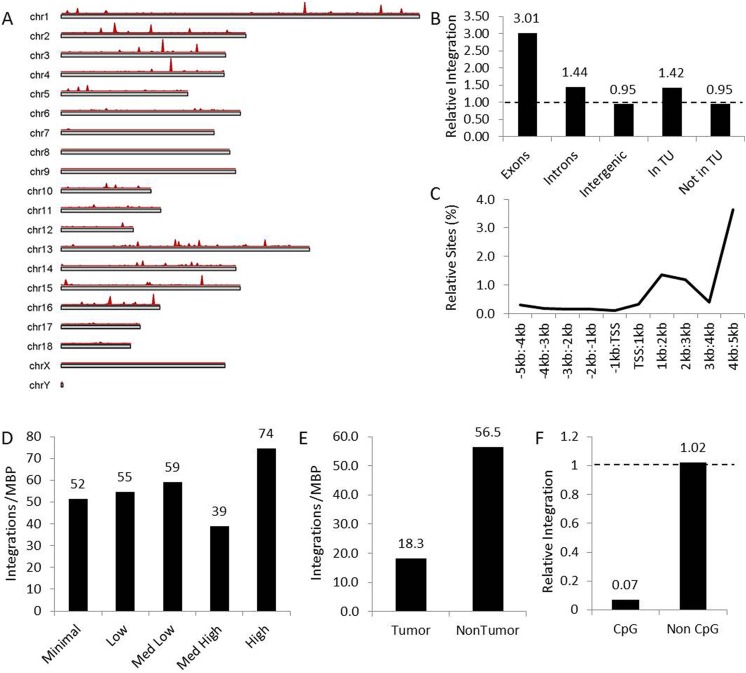
Although no tumorigenicity was present in this animal, the pattern of lentiviral vector insertion into the pig genome was evaluated to further determine safety of genomic integration. The top 15 sites for lentiviral integration are indicated in Table 1. Chromosomal mapping showed integration throughout the pig genome, with a few hot spots on multiple chromosomes (Fig. 6A, red peaks). Relative integration site profiling showed that the vector preferred integrating into transcriptional units, specifically exons (Fig. 6B), while the majority of integration sites occurred downstream of the transcriptional start site (Fig. 6C) regardless of coding strand. There was a trend for integration into more actively transcribed genes across 5 expression categories (Fig. 6D) and an approximately three-fold higher preference for integration into non-tumor coding genes (Fig. 6E). Lastly, the lenti vector seldom integrated into CpG-rich regions of the genome (Fig. 6F), suggesting minimal opportunity to activate proximal genes. Collectively, this pattern of integration suggests minimal potential for tumorigenicity due to lack of integration into promotor regions or tumor-associated genes, preferring coding regions of highly expressed genes as sites of integration.
Table 1.
Top 15 genes for lentiviral integration as ranked by the percent of total reads. Unique integration sites within each gene is also presented, indicating the number of possible disruptions of each gene in different clones of transduced hepatocytes.
| Rank | Gene | Unique sites | Percent of reads |
|---|---|---|---|
| 1 | GHITM | 13 | 9.00 |
| 2 | CS | 29 | 8.66 |
| 3 | ACVR2A | 48 | 8.59 |
| 4 | SLC33A1 | 24 | 6.08 |
| 5 | ST8SIA4 | 29 | 5.51 |
| 6 | CLDN16 | 8 | 5.29 |
| 7 | RSPO3 | 25 | 4.81 |
| 8 | SPATA16 | 33 | 4.26 |
| 9 | PROK1 | 7 | 4.08 |
| 10 | PHYHIPL | 16 | 3.02 |
| 11 | MIR216-1 | 67 | 2.84 |
| 12 | CHMP2B | 31 | 2.67 |
| 13 | REG3G | 27 | 2.33 |
| 14 | IFl44 | 20 | 1.87 |
| 15 | FHL5 | 16 | 1.59 |
Fig. 6.
Bioinformatics demonstrated a benign integration profile for lentiviral vector into treated hepatocytes. (A) Chromosomal map of the pig genome showing regions of integration (red peaks) for each chromosome based on frequency. (B) Relative integration of lentiviral vector in genomic features, including exons, introns, intergenic regions and regions in and out of transcription units (TU), note 1 = random distribution (dashed line). (C) Integration of lentivirus relative to the transcription start sites (TSS) showing preference for downstream integration. (D) Lentiviral vector integrations into genes categorized by increasing transcriptional activity normalized by million base pairs (MBP) of each. (E) Lentiviral vector integrations into tumor and non-tumor associated genes normalized by million base pairs (MBP) of each. (F) Relative integration of lentiviral vector in CpG islands normalized to the percent of the pig genome represented by each, note 1 = random distribution (dashed line).
Discussion
A significant advance in the treatment of HT1 occurred with the development of NTBC12. The early acute liver failure seen could easily be ameliorated, and the fibrosis, cirrhosis, and eventual HCC that occurred in the early lives of these patients improved significantly13. However, with current NTBC protocols and therapies in place for more than 20 years we have learned about the potential shortcomings of prolonged NTBC therapy. Lifelong elevated tyrosine levels appear to be detrimental, causing progressive neurocognitive decline and ocular degeneration14. Moreover, either through issues of poor compliance or ongoing fibrosis and injury, HCC has been discovered in many HT1 patients on NTBC therapy in their second and third decades of life15–17. The intense regimen related to daily NTBC therapy, frequent blood and imaging screening, along with poor compliance has made this therapy insufficient and difficult to adhere to for patients. Finally, there appears to be a percentage of patients who are refractory to NTBC and require a liver transplant to treat their disease18. Overall then, there still exists a clinical need for the development of better therapies for the long-term treatment of HT1.
Ex-vivo hepatocyte-directed gene therapy is a potential curative therapy for the multiple inborn errors of metabolism that are currently only curable by orthotopic liver transplantation. Early experiences using ex-vivo transduction of hepatocytes with gamma-retroviral vectors demonstrated efficacy in a number of animal models19–21. These successes led to the first – and to date only – clinical trial of ex-vivo gene therapy in five patients with familial hypercholesterolemia22. While these clinical results demonstrated proof-of-concept for this therapeutic approach, the results were modest and not curative. The underwhelming results were most likely due to poor transduction efficiencies of gammaretroviral vectors into hepatocytes, the need to culture hepatocytes ex vivo for three days and the absence of a strong selective pressure for corrected cells to proliferate once engrafted. The advent of lentiviral vectors heralded a new day for gene therapy as these vectors demonstrated improved transduction efficiency and safety in a number of cell types and species. To date, a number of seminal clinical studies have demonstrated ex-vivo transduction of hematopoietic stem cells to be curative for a number of primary immunodeficiencies, including Wiskott Aldrich Syndrome, Metachromatic Leukodystrophy and Cerebral Adrenoleukodystrophy23–25.
Given the tremendous success in the treatment of these hematopoietic-related diseases using ex-vivo transduction of hematopoietic stem cells with lentiviral vectors, there is great interest in developing clinically-relevant ex-vivo gene therapy approaches for metabolic liver diseases. In addition to successful cures of small animal models of inborn errors of metabolism by hepatocyte transplantation after ex-vivo gene therapy, a number of preclinical studies have demonstrated safety and reproducibility of this approach in large animal models, including non-human primates26. Encouraged by these results, we pursued this approach in a clinically-relevant pig model of HT1 (Fah-/- pigs). HT1 is an interesting therapeutic indication for several reasons: (a) it is the result of a single gene mutation; (b) it is phenotypically evident in hepatocytes, which are an accessible cell type for gene therapy; and (c) corrected hepatocytes have a strong selective advantage over diseased cells once present in the liver. This selective advantage overcomes a major hurdle in gene therapy, which is categorically limited by the necessity to affect a phenotypically relevant number of cells.
We previously reported curative ex-vivo gene therapy and autologous hepatocyte transplantation in a cohort of HT1 pigs using a VSV-G pseudotyped, self-inactivating lentiviral vector expressing the porcine Fah cDNA under the control of a hepatocyte specific promoter. This study demonstrated robust engraftment and proliferation of corrected cells in the Fah-/- liver which completely cured the disease by preventing the onset of severe fibrosis and liver failure, which is characteristic of untreated Fah-/- pigs. With this chronic follow up presented herein, we demonstrate that this phenotypic correction was durable and, in a single pig, did not result in cirrhosis or HCC, which is an important observation when considering the clinical potential of gene therapy for HT1. Indeed, the data described in this paper demonstrate conclusively that ex-vivo gene therapy for HT1 is feasible, efficacious and safe, providing a durable cure in a clinically relevant model of this disease.
Lastly, our lentiviral vector demonstrated a benign integration profile consistent with other evaluations27, showing preference to integrate downstream of the transcriptional start site, preferably in the exons of non-tumor associated genes. This genetic integration profile is unique in the literature because it is the product of years of potential selection/expansion in a large animal model, which is arguably a worst-case scenario to allow for the development of a dominant clone and subsequent tumor formation. Indeed, the current data support that there was no dominant clone as no individual gene represented 10% or more of the total integration sites read, and each of the top three genes had at least 13 unique integration sites present. Collectively with the phenotypic absence of tumors in any tissue, especially the liver, these integration data make a compelling argument for the safety of lentiviral vectors as gene delivery tools, here in the specific context of HT1, assuaging concerns of tumorigenicity from genetic integration in this and other disease indications.
In conclusion, a human trial aimed at treating HT1 is now a reasonable option. After the long-term safety demonstrated in this HT1 pig model and the well-documented complications related to the current therapies for HT1, there is now equipoise in moving on to human trials. Future work in this area should be focused on studies addressing any remaining safety concerns prior to beginning work in humans, such as defining an appropriate first-in-human dose level.
Supplemental Material
Supplemental Material, Supp_Figure_1JPG for Autologous Gene and Cell Therapy Provides Safe and Long-Term Curative Therapy in A Large Pig Model of Hereditary Tyrosinemia Type 1 by Raymond D. Hickey, Clara T. Nicolas, Kari Allen, Shennen Mao, Faysal Elgilani, Jaime Glorioso, Bruce Amiot, Caitlin VanLith, Rebekah Guthman, Zeji Du, Harvey Chen, Cary O. Harding, Robert A. Kaiser, Scott L. Nyberg, and Joseph B. Lillegard in Cell Transplantation
Supplemental Material
Supplemental Material, Supp_Figure_2JPG for Autologous Gene and Cell Therapy Provides Safe and Long-Term Curative Therapy in A Large Pig Model of Hereditary Tyrosinemia Type 1 by Raymond D. Hickey, Clara T. Nicolas, Kari Allen, Shennen Mao, Faysal Elgilani, Jaime Glorioso, Bruce Amiot, Caitlin VanLith, Rebekah Guthman, Zeji Du, Harvey Chen, Cary O. Harding, Robert A. Kaiser, Scott L. Nyberg, and Joseph B. Lillegard in Cell Transplantation
Supplemental Material
Supplemental Material, Supp_Figure_3JPG for Autologous Gene and Cell Therapy Provides Safe and Long-Term Curative Therapy in A Large Pig Model of Hereditary Tyrosinemia Type 1 by Raymond D. Hickey, Clara T. Nicolas, Kari Allen, Shennen Mao, Faysal Elgilani, Jaime Glorioso, Bruce Amiot, Caitlin VanLith, Rebekah Guthman, Zeji Du, Harvey Chen, Cary O. Harding, Robert A. Kaiser, Scott L. Nyberg, and Joseph B. Lillegard in Cell Transplantation
Acknowledgments
We thank LouAnn Gross and Tony Blahnik for histology and immunohistochemistry support; Aditya Bhagwate, Daniel O'Brien and Jean-Pierre Kocher, PhD for bioinformatics analysis.
Footnotes
Ethical Approval: This study was approved by the institutional animal care and use committee at Mayo Clinic, Rochester, MN.
Statement of Human and Animal Rights: All experimental procedures involving animals were carried out in compliance with the regulations of the institutional animal care and use committee at Mayo Clinic, Rochester, MN.
Statement of Informed Consent: There are no human subjects in this study and informed consent is not applicable.
Data Availability: All data generated for this study are available from the corresponding author on request.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: R.D.H. was funded through an NIH K01 DK106056 award and a Mayo Clinic Center for Regenerative Medicine Career Development Award. S.L.N. was funded through an NIH R41 DK092105 award, the Wallace H. Coulter Foundation, Marriot Foundation, Darwin Deason Family Foundation, and Mayo Foundation. J.B.L. was supported by the Children’s Hospital of Minnesota Foundation. Additional research support was provided by Regenerative Medicine Minnesota.
Supplemental Material: Supplemental material for this article is available online
References
- 1. Grompe M. The pathophysiology and treatment of hereditary tyrosinemia type 1. Semin Liver Dis. 2001;21(4):563–571. [DOI] [PubMed] [Google Scholar]
- 2. Simoncelli M, Samson J, Bussières JF, Lacroix J, Dorais M, Battista R, Perreault S. Cost-consequence analysis of nitisinone for treatment of tyrosinemia type I. Can J Hosp Pharm. 2015;68(3):210–217. [PMC free article] [PubMed] [Google Scholar]
- 3. Malik S, Kassai B, Cochat P. Overview of pediatric organ transplantation: current opinion and future perspectives on immunosuppression. Curr Opin Organ Transplant. 2015;20(5):527–535. [DOI] [PubMed] [Google Scholar]
- 4. Perito ER, Rhee S, Roberts JP, Rosenthal P. Pediatric liver transplantation for urea cycle disorders and organic acidemias: united network for organ sharing data for 2002–2012. Liver Transpl. 2014;20(1):89–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hickey RD, Lillegard JB, Fisher JE, McKenzie TJ, Hofherr SE, Finegold MJ, Nyberg SL, Grompe M. Efficient production of Fah-null heterozygote pigs by chimeric adeno-associated virus-mediated gene knockout and somatic cell nuclear transfer. Hepatology. 2011;54(4):1351–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hickey RD, Mao SA, Glorioso J, Lillegard JB, Fisher JE, Amiot B, Rinaldo P, Harding CO, Marler R, Finegold MJ, Grompe M, Nyberg SL. Fumarylacetoacetate hydrolase deficient pigs are a novel large animal model of metabolic liver disease. Stem Cell Res. 2014;13(1):144–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Elgilani F, Mao SA, Glorioso JM, Yin M, Iankov ID, Singh A, Amiot B, Rinaldo P, Marler RJ, Ehman RL, Grompe M, Lillegard JB, Hickey RD, Nyberg SL. Chronic phenotype characterization of a large animal model of hereditary tyrosinemia type 1. Am J Pathol. 2017;187(1):33–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hickey RD, Mao SA, Glorioso J, Elgilani F, Amiot B, Chen H, Rinaldo P, Marler R, Jiang H, DeGrado TR, Suksanpaisan L, O’Connor MK, Freeman BL, Ibrahim SH, Peng KW, Harding CO, Ho CS, Grompe M, Ikeda Y, Lillegard JB, Russell SJ, Nyberg SL. Curative ex vivo liver-directed gene therapy in a pig model of hereditary tyrosinemia type 1. Sci Transl Med. 2016;8(349):349ra399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kaiser RA, Mao SA, Glorioso J, Amiot B, Nicolas CT, Allen KL, Du Z, VanLith CJ, Hickey RD, Nyberg SL, Lillegard JB. Lentiviral Vector-mediated Gene Therapy of Hepatocytes Ex Vivo for Autologous Transplantation in Swine. J Vis Exp. (141):e58399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang X, Montini E, Al-Dhalimy M, Lagasse E, Finegold M, Grompe M. Kinetics of liver repopulation after bone marrow transplantation. Am J Pathol. 2002;161(2):565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ingvarsson BI, Carlsson RN, Karlsson BW. Synthesis of alpha-fetoprotein, albumin and total serum protein in neonatal pigs. Biol Neonate. 1978;34(5–6):259–268. [DOI] [PubMed] [Google Scholar]
- 12. Lindstedt S, Holme E, Lock EA, Hjalmarson O, Strandvik B. Treatment of hereditary tyrosinaemia type I by inhibition of 4-hydroxyphenylpyruvate dioxygenase. Lancet. 1992;340(8823):813–817. [DOI] [PubMed] [Google Scholar]
- 13. Larochelle J, Alvarez F, Bussieres JF, Chevalier I, Dallaire L, Dubois J, Faucher F, Fenyves D, Goodyer P, Grenier A, Holme E, Laframboise R, Lambert M, Lindstedt S, Maranda B, Melancon S, Merouani A, Mitchell J, Parizeault G, Pelletier L, Phan V, Rinaldo P, Scott CR, Scriver C, Mitchell GA. Effect of nitisinone (NTBC) treatment on the clinical course of hepatorenal tyrosinemia in Quebec. Mol Genet Metab. 2012;107(1–2):49–54. [DOI] [PubMed] [Google Scholar]
- 14. Thimm E, Richter-Werkle R, Kamp G, Molke B, Herebian D, Klee D, Mayatepek E, Spiekerkoetter U. Neurocognitive outcome in patients with hypertyrosinemia type I after long-term treatment with NTBC. J Inherit Metab Dis. 2012;35(2):263–268. [DOI] [PubMed] [Google Scholar]
- 15. van Spronsen FJ, Bijleveld CM, van Maldegem BT, Wijburg FA. Hepatocellular carcinoma in hereditary tyrosinemia type I despite 2-(2 nitro-4-3 trifluoro- methylbenzoyl)-1, 3-cyclohexanedione treatment. J Pediatr Gastroenterol Nutr. 2005;40(1):90–93. [DOI] [PubMed] [Google Scholar]
- 16. Masurel-Paulet A, Poggi-Bach J, Rolland MO, Bernard O, Guffon N, Dobbelaere D, Sarles J, de Baulny HO, Touati GNTBC. treatment in tyrosinaemia type I: long-term outcome in French patients. J Inherit Metab Dis. 2008;31(1):81–87. [DOI] [PubMed] [Google Scholar]
- 17. Mayorandan S, Meyer U, Gokcay G, Segarra NG, de Baulny HO, van Spronsen F, Zeman J, de Laet C, Spiekerkoetter U, Thimm E, Maiorana A, Dionisi-Vici C, Moeslinger D, Brunner-Krainz M, Lotz-Havla AS, Cocho de Juan JA, Couce Pico ML, Santer R, Scholl-Burgi S, Mandel H, Bliksrud YT, Freisinger P, Aldamiz-Echevarria LJ, Hochuli M, Gautschi M, Endig J, Jordan J, McKiernan P, Ernst S, Morlot S, Vogel A, Sander J, Das AM. Cross-sectional study of 168 patients with hepatorenal tyrosinaemia and implications for clinical practice. Orphanet J Rare Dis. 2014;9:107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Holme E, Lindstedt S. Nontransplant treatment of tyrosinemia. Clin Liver Dis. 2000;4(4):805–814. [DOI] [PubMed] [Google Scholar]
- 19. Grompe M, Jones SN, Loulseged H, Caskey CT. Retroviral-mediated gene transfer of human ornithine transcarbamylase into primary hepatocytes of spf and spf-ash mice. Hum Gene Ther. 1992;3(1):35–44. [DOI] [PubMed] [Google Scholar]
- 20. Overturf K, Al-Dhalimy M, Tanguay R, Brantly M, Ou CN, Finegold M, Grompe M. Hepatocytes corrected by gene therapy are selected in vivo in a murine model of hereditary tyrosinaemia type I. Nature genetics. 1996;12(3):266–273. [DOI] [PubMed] [Google Scholar]
- 21. Chowdhury JR, Grossman M, Gupta S, Chowdhury NR, Baker JR, Jr, Wilson JM. Long-term improvement of hypercholesterolemia after ex vivo gene therapy in LDLR-deficient rabbits. Science. 1991;254(5039):1802–1805. [DOI] [PubMed] [Google Scholar]
- 22. Grossman M, Rader DJ, Muller DW, Kolansky DM, Kozarsky K, Clark BJ, 3rd, Stein EA, Lupien PJ, Brewer HB, Jr, Raper SE, Wilson JM, et al. A pilot study of ex vivo gene therapy for homozygous familial hypercholesterolaemia. Nat Med. 1995;1(11):1148–1154. [DOI] [PubMed] [Google Scholar]
- 23. Aiuti A, Biasco L, Scaramuzza S, Ferrua F, Cicalese MP, Baricordi C, Dionisio F, Calabria A, Giannelli S, Castiello MC, Bosticardo M, Evangelio C, Assanelli A, Casiraghi M, Di Nunzio S, Callegaro L, Benati C, Rizzardi P, Pellin D, Di Serio C, Schmidt M, Von Kalle C, Gardner J, Mehta N, Neduva V, Dow DJ, Galy A, Miniero R, Finocchi A, Metin A, Banerjee PP, Orange JS, Galimberti S, Valsecchi MG, Biffi A, Montini E, Villa A, Ciceri F, Roncarolo MG, Naldini L. Lentiviral hematopoietic stem cell gene therapy in patients with Wiskott-Aldrich syndrome. Science. 2013;341(6148):1233151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Biffi A, Montini E, Lorioli L, Cesani M, Fumagalli F, Plati T, Baldoli C, Martino S, Calabria A, Canale S, Benedicenti F, Vallanti G, Biasco L, Leo S, Kabbara N, Zanetti G, Rizzo WB, Mehta NA, Cicalese MP, Casiraghi M, Boelens JJ, Del Carro U, Dow DJ, Schmidt M, Assanelli A, Neduva V, Di Serio C, Stupka E, Gardner J, von Kalle C, Bordignon C, Ciceri F, Rovelli A, Roncarolo MG, Aiuti A, Sessa M, Naldini L. Lentiviral hematopoietic stem cell gene therapy benefits metachromatic leukodystrophy. Science. 2013;341(6148):1233158. [DOI] [PubMed] [Google Scholar]
- 25. Eichler F, Duncan C, Musolino PL, Orchard PJ, De Oliveira S, Thrasher AJ, Armant M, Dansereau C, Lund TC, Miller WP, Raymond GV, Sankar R, Shah AJ, Sevin C, Gaspar HB, Gissen P, Amartino H, Bratkovic D, Smith NJC, Paker AM, Shamir E, O’Meara T, Davidson D, Aubourg P, Williams DA. Hematopoietic stem-cell gene therapy for cerebral adrenoleukodystrophy. New Engl J Med. 2017;377(17):1630–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Menzel O, Birraux J, Wildhaber BE, Jond C, Lasne F, Habre W, Trono D, Nguyen TH, Chardot C. Biosafety in ex vivo gene therapy and conditional ablation of lentivirally transduced hepatocytes in nonhuman primates. Mol Ther. 2009;17(10):1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Biffi A, Bartolomae CC, Cesana D, Cartier N, Aubourg P, Ranzani M, Cesani M, Benedicenti F, Plati T, Rubagotti E, Merella S, Capotondo A, Sgualdino J, Zanetti G, von Kalle C, Schmidt M, Naldini L, Montini E. Lentiviral vector common integration sites in preclinical models and a clinical trial reflect a benign integration bias and not oncogenic selection. Blood. 2011;117(20):5332–5339. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material, Supp_Figure_1JPG for Autologous Gene and Cell Therapy Provides Safe and Long-Term Curative Therapy in A Large Pig Model of Hereditary Tyrosinemia Type 1 by Raymond D. Hickey, Clara T. Nicolas, Kari Allen, Shennen Mao, Faysal Elgilani, Jaime Glorioso, Bruce Amiot, Caitlin VanLith, Rebekah Guthman, Zeji Du, Harvey Chen, Cary O. Harding, Robert A. Kaiser, Scott L. Nyberg, and Joseph B. Lillegard in Cell Transplantation
Supplemental Material, Supp_Figure_2JPG for Autologous Gene and Cell Therapy Provides Safe and Long-Term Curative Therapy in A Large Pig Model of Hereditary Tyrosinemia Type 1 by Raymond D. Hickey, Clara T. Nicolas, Kari Allen, Shennen Mao, Faysal Elgilani, Jaime Glorioso, Bruce Amiot, Caitlin VanLith, Rebekah Guthman, Zeji Du, Harvey Chen, Cary O. Harding, Robert A. Kaiser, Scott L. Nyberg, and Joseph B. Lillegard in Cell Transplantation
Supplemental Material, Supp_Figure_3JPG for Autologous Gene and Cell Therapy Provides Safe and Long-Term Curative Therapy in A Large Pig Model of Hereditary Tyrosinemia Type 1 by Raymond D. Hickey, Clara T. Nicolas, Kari Allen, Shennen Mao, Faysal Elgilani, Jaime Glorioso, Bruce Amiot, Caitlin VanLith, Rebekah Guthman, Zeji Du, Harvey Chen, Cary O. Harding, Robert A. Kaiser, Scott L. Nyberg, and Joseph B. Lillegard in Cell Transplantation