Abstract
Type 1 diabetes (T1D) is characterized by the loss of insulin-producing β-cells in the pancreas. T1D can be treated using cadaveric islet transplantation, but this therapy is severely limited by a lack of pancreas donors. To develop an alternative cell source for transplantation therapy, we carried out the epigenetic characterization in nine different adult mouse tissues and identified visceral adipose-derived progenitors as a candidate cell population. Chromatin conformation, assessed using chromatin immunoprecipitation (ChIP) sequencing and validated by ChIP-polymerase chain reaction (PCR) at key endocrine pancreatic gene promoters, revealed similarities between visceral fat and endocrine pancreas. Multiple techniques involving quantitative PCR, in-situ PCR, confocal microscopy, and flow cytometry confirmed the presence of measurable (2–1000-fold over detectable limits) pancreatic gene transcripts and mesenchymal progenitor cell markers (CD73, CD90 and CD105; >98%) in visceral adipose tissue-derived mesenchymal cells (AMCs). The differentiation potential of AMCs was explored in transgenic reporter mice expressing green fluorescent protein (GFP) under the regulation of the Pdx1 (pancreatic and duodenal homeobox-1) gene promoter. GFP expression was measured as an index of Pdx1 promoter activity to optimize culture conditions for endocrine pancreatic differentiation. Differentiated AMCs demonstrated their capacity to induce pancreatic endocrine genes as evidenced by increased GFP expression and validated using TaqMan real-time PCR (at least 2–200-fold relative to undifferentiated AMCs). Human AMCs differentiated using optimized protocols continued to produce insulin following transplantation in NOD/SCID mice. Our studies provide a systematic analysis of potential islet progenitor populations using genome-wide profiling studies and characterize visceral adipose-derived cells for replacement therapy in diabetes.
Keywords: Visceral adipose tissue, insulin, type 1 diabetes, histone modifications, ChIP-seq and RNA-seq
Introduction
Diabetes mellitus is a chronic metabolic disease defined by an inability to regulate circulating glucose concentrations. Type 1 diabetes (T1D) is characterized by the selective autoimmune-mediated destruction of pancreatic islet β-cells. This pathological loss of β-cell mass, results in a failure to produce insulin, in response to changes in blood glucose concentrations. If untreated, the resultant hyperglycemia can lead to serious microvascular (retinopathy, nephropathy, and neuropathy) and/or macrovascular complications (coronary/peripheral artery disease and stroke). Individuals with T1D require stringent monitoring of blood glucose levels and treatment with exogenous insulin administered through regular injections or through continuous monitoring insulin pumps1.
Current management plans for T1D patients are generally effective and can achieve good glycemic control with intensive insulin therapy. However, these pharmacological approaches, while effective, fail to completely recapitulate the true biology of a healthy pancreas. This can be achieved either through total pancreas transplantation or through islet transplantation, so as to replace the dying/lost insulin-producing β-cells. In 2000, Shapiro et al.2 first described a procedure involving a steroid-free immunosuppressive regimen for the transplantation of allogeneic islets isolated from the human cadaveric pancreas (the Edmonton protocol). This study demonstrated that for patients with T1D, islet transplantation can result in insulin independence with excellent metabolic control when glucocorticoid-free immunosuppression is combined with the infusion of an adequate islet mass. Unfortunately, this technique is limited by low numbers of cadaveric donors and the restricted yield of viable cells for transplantation3. Therefore, many research groups across the world are in search of alternative cell sources that have the potential to differentiate into insulin-producing cells for transplantation4.
Cells isolated from pancreatic tissues (endocrine, acinar or ductal) or other endodermal sources have potential as alternative sources for the generation of insulin-producing cells. There is now a growing body of evidence to suggest that it is possible to induce the trans-differentiation of endodermal cells such as hepatocytes5–7, intra-/extra-hepatic biliary epithelial cells8 and gallbladder epithelium9,10 to express markers characteristic of the pancreatic lineage. Adipose tissue has been one of the more widely explored sources of tissues for assessing its potential to differentiate into insulin-producing cells11–16. However, adipocytes are located in distinct depots, which demonstrate significantly different gene expression patterns17–19. Thus, progenitor/precursor cells residing in the visceral or the subcutaneous fat depots possess inherent and specific metabolic characteristics that may guide the differentiation of progenitor cells derived from such tissues. We theorize that epigenetic mechanisms that dictate the chromatin conformation of DNA in adipose tissue depots govern their cell fate during differentiation and that understanding the epigenetic conformation (histone modifications) in adipose tissue depots would aid in assessing their differentiation potential to an endocrine pancreatic fate.
Genetic and epigenetic mechanisms are thought to represent the primary control for cellular differentiation and developmental biology. Epigenetic mechanisms principally involve histone post-translational modifications and DNA methylation20, but can also include noncoding RNAs21–23 and nuclear dynamics24,25. Combining chromatin structure analysis using chromatin immunoprecipitation (ChIP) with comprehensive sequencing provides genome-wide measurements of chromatin structure, which can be integrated with RNA transcriptome sequencing to provide a detailed analysis of transcriptional networks in a given tissue. Here we show that the profiling of histone modifications and gene expression in multiple mouse tissues using ChIP-seq and RNA-seq platforms identified visceral adipose tissue as a source of progenitor cells that are epigenetically poised to differentiate into an endocrine pancreatic lineage. We present the characterization and differentiation of these cells using a variety of imaging and molecular techniques. Finally, we present data on their potential to produce insulin, following transplantation in mice.
Materials and Methods
All animal and human work presented herein was approved by specific Animal and Human Ethics Committees at the St. Vincent’s Hospital, Melbourne, Australia. Animals were handled in accordance with the National Health and Medical Research Council (NHMRC) guidelines for the care and maintenance of experimental animals.
Chromatin Immunoprecipitation Analysis and Sequencing
Healthy 12-week-old male C57/Bl-6 mice were euthanized and organs (skin, peritoneum, heart, visceral adipose, subcutaneous adipose, liver, stomach, brain, and pancreas) were harvested immediately. Organs were maintained in chilled low glucose Dulbecco’s modified Eagle’s medium (DMEM; Gibco, Thermo Fisher Scientific, Waltham, MA, USA) containing 5 mM glucose and 2 mM L-glutamine (GlutaMAX-1; Gibco, Thermo Fisher Scientific) on ice. Single cell suspensions were generated using mechanical and enzymatic dissociation. Pelleted cells were collected for chromatin immunoprecipitation using a protocol described previously26. Briefly, cells were cross-linked using 1% formaldehyde (Sigma, St. Louis, MO, USA) and DNA sheared using a Bioruptor to generate fragments of 200–500 base pairs (bp) required for ChIP. Chromatin was immunoprecipitated using 2 µg of specific antibodies for H3K4 trimethyl, H3K9 trimethyl and H3Ac (Merck Millipore, Burlington, MA, USA), with rabbit immunoglobulin (Ig)G (Merck Millipore), used as an isotype control. Chromatin was precipitated using A/G plus beads (Pierce™, Thermo Fisher Scientific), eluted and reverse cross-linked using an overnight incubation with 4 M sodium chloride (NaCl) at 65°C. Following proteinase-K (Sigma) digestion, DNA was precipitated using phenol-chloroform-isoamyl alcohol and dissolved in nuclease-free water. Quantitative real-time polymerase chain reaction (PCR) was performed as 10 µl reactions in a 96-well optical clear plate using 1 µl of precipitated DNA and custom promoter-specific primers (Sigma; sequences obtained from Xu et al.27) and SYBRPCR master mix (Applied Biosystems, Thermo Fisher Scientific, Waltham, MA, USA). The results were presented as the percent of input.
Ion ChIP-seq libraries were prepared from the ChIP DNA using an Ion Plus Fragment Library Kit (Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA). Briefly, purified ChIP DNA was end-repaired and purified with two rounds of AMPure XP bead (Beckman Coulter, Brea, CA, USA) capture to size-select fragments approximately 100–250 bp in length. The end-repaired DNA was ligated to Ion-compatible adapters. The library was amplified using PCR with a limited number of cycles to generate sufficient material for downstream template preparation. The final library was between 170–220 bp in length. The purified, amplified ChIP DNA library was diluted and an Ion OneTouch 200 Template Kit v2 DL (Invitrogen) was used to prepare template-positive Ion OneTouch 200 Ion Sphere Particles (ISPs) for 200 base-read sequencing of the ChIP DNA. Template-positive ISPs were enriched using an Ion OneTouch ES instrument and sequenced using an Ion PGM 200 Sequencing Kit (Invitrogen) and the Ion Torrent PGM Instrument using Ion 316 chips (Thermo Fisher Scientific). Data were analyzed using the Avadis NGS system (Bangalore, India). Sequencing data were validated using promoter-specific primers (Sigma; sequences obtained from Xu et al.27) as described above.
RNA Isolation, Poly(A)-Enrichment and Transcriptome Sequencing
Organs were harvested, homogenized, dissolved in TRIzol Reagent (Thermo Fisher Scientific) and RNA extracted following the manufacturer’s instructions. The Ambion MicroPoly(A)Purist™ kit (Ambion, Thermo Fisher Scientific) was used to enrich the mRNA transcripts from the previously isolated total RNA, which contains oligo(dT) cellulose to isolate the poly(A) RNA fraction. The poly(A) RNA was eluted using pre-warmed RNA storage solution, re-precipitated and quantitated on Qubit (Thermo Fisher Scientific). Ion RNA-Seq libraries were prepared from this poly-A RNA using an Ion Total RNA-Seq Kit v2 (Invitrogen). Briefly, the re-precipitated poly(A)-enriched RNA was fragmented using RNase III and purified using nucleic acid binding beads. The purified, fragmented RNA was ligated to Ion-compatible adaptors and subsequently amplified using PCR with a limited number of cycles to generate sufficient material for downstream template preparation. The library was purified and analyzed using an Agilent 2100 Bioanalyzer instrument (Agilent Technologies, Mulgrave, VIC, Australia). The purified, amplified poly(A)-enriched library was diluted and an Ion OneTouch 200 Template Kit v2 DL was used to prepare template-positive Ion OneTouch 200 Ion Sphere Particles (ISPs) for 200 base-read sequencing of the enriched RNA. Template-positive ISPs were enriched using an Ion OneTouch ES instrument and sequenced using an Ion PGM 200 Sequencing Kit and the Ion Torrent PGM Instrument using Ion 316 chips. Data were analyzed using the Avadis NGS system (Bangalore, India).
Isolation of Human Adipose-Derived Mesenchymal Cells
Human subcutaneous and visceral adipose tissues were available as surgical excess following elective abdominoplasty at St Vincent’s Private Hospital (Melbourne, Australia). Cells were isolated from these tissues using a protocol previously described by Zuk et al.28, with modifications. Briefly, adipose tissue was mechanically minced into fine pieces and cells were dissociated using an enzymatic digestion (0.2% w/v) collagenase 1 for 60 min at 37°C. Cells were washed, centrifuged to remove adipocytes and pelleted cells were cultured under standard conditions in low glucose DMEM containing 5 mM glucose, 2 mM L-glutamine (Sigma) and 100 U/ml antibiotic/antimycotic cocktail (Gibco) at 37°C in a humidified atmosphere containing 5% CO2. Adherent populations were grown until they became confluent under standard conditions, and sub-cultured at a ratio of 1:2. The cultured populations of cells were termed adipose-derived mesenchymal cells (AMCs).
Gene Expression Characterization Using Quantitative PCR
Fresh adipose tissue from donor samples (Day 0) and AMCs cultured to different passages were harvested in TRIzol and RNA was extracted following the manufacturer’s instructions. cDNA was prepared using a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) and quantitative real-time PCR of samples was performed in 96-well optical clear plates using an Assay-on-Demand probe and primer mix (Applied Biosystems; Supplementary Table 2) with TaqMan Fast Universal PCR Master Mix, with 40 ng/100 ng DNA inputs. PCR was performed using the ViiA7 Real-Time PCR platform and data analyzed using Applied Biosystems ViiA7 Software. The results were normalized to housekeeping genes (18 S). Data are presented as fold difference over the detectable Ct value, which was calculated using the ΔΔCt method (Ct value of 39 was considered to be undetectable)29.
Gene Expression Characterization Using In-situ PCR
Sub-cultured AMCs were characterized using an in-situ PCR protocol previously described by Ranjan et al.30. Briefly, AMCs were sub-cultured onto eight-well chamber slides (Thermo Fisher Scientific) and cultured to semi-confluence. Cells were fixed in freshly prepared 4% paraformaldehyde (PFA; Sigma) and permeabilized using a freeze–thaw technique. Cells were incubated with 20 µl 0.1 M ammonium chloride solution and in-situ cDNA was prepared using a High Capacity cDNA Reverse Transcription Kit. In-situ PCR was performed for 25 cycles using Assay-on-Demand probe (Supplementary Table 2) and a primer mix with TaqMan Fast Universal PCR Master Mix, with independent reactions carried out in each well of the eight-well chamber slide. Slides were fixed again in freshly prepared 4% PFA and mounted in Vectashield mountant (Vector Laboratories, Burlingame, CA, USA) containing Hoechst 33342 (Invitrogen). Images were captured using a Zeiss LSM 510 laser scanning microscope. Magnification, laser power and detector gains were set below saturation and were identical across samples.
Immunocytochemistry Characterization
Sub-cultured AMCs were characterized using an immunocytochemistry protocol previously described by Joglekar et al.26 Briefly, AMCs were sub-cultured onto eight-well chamber slides (Thermo Fisher Scientific) and grown to confluence. Cells were fixed in freshly prepared 4% PFA and permeabilized using chilled 0.1% Triton X-100 (Sigma) for 30 min at room temperature. Cells were exposed to blocking buffer composed of 4% normal donkey serum (NDS; Sigma) for 30 min at room temperature. Primary antibodies were diluted (1:100) in blocking buffer (4% NDS) and incubated with cells overnight at 4°C. Secondary antibodies were diluted (1:200) in blocking buffer (4% NDS) and incubated with cells for 1 h at 37°C and mounted in Vectashield mountant containing Hoechst 33342 (Sigma). Images were captured using a Zeiss LSM 510 laser scanning microscope. Magnification, laser power and detector gains were set below saturation and were identical across samples. Details of primary and secondary antibodies used herein are provided in Supplementary Table 2.
Flow Cytometry Characterization
Sub-cultured AMCs were trypsinized into a single cell suspension and maintained in serum-containing media at 37oC for 60 min to allow the cells time to regenerate the surface markers damaged or lost during the trypsinization. Non-specific epitopes on cells were blocked using 4% NDS in phosphate-buffered saline for 30 min at room temperature followed by labeling cells in suspension with fluorescent-conjugated primary antibodies at 4oC for 60 min in the dark. Primary antibodies were purchased from BD Biosciences and used at the concentrations indicated; anti-human CD29-PE (1:100), CD34-FITC (1.5:100), CD45-FITC (2.5:100), CD90-PE (1:200), CD31-FITC (1.5:100), CD44-PE (1:100), CD73-FITC (1:100) and CD105-PE (1:100) with their respective isotype controls (2.5:100). Cells were imaged on a fluorescence-activated cell sorter Calibur (Becton Dickinson, Franklin Lakes, NJ, USA). Gating was implemented after profiling unstained and isotype control cell samples.
Differentiation of Mouse AMCs Into an Endocrine Pancreatic Lineage
Transgenic mice contained a green fluorescent protein (GFP) reporter under the control of essential pancreatic promoters; the pancreatic and duodenal homeobox 1 (Pdx1). Pdx1-GFP mice were provided by Prof. Edouard Stanley and Prof. Andrew Elefanty (MCRI, Melbourne, Australia) and were maintained at our animal facility. Adipose-derived cells were isolated and cultured using the protocol described above for the isolation and culture of human cells. The differentiation potential of mouse AMCs was confirmed using the protocol described previously31–35. Mouse AMCs were sub-cultured as a single cell suspension in serum-free media into low adhesion tissue culture six-well plates, with each well containing 1 × 106 cells. During differentiation, the expression of GFP was visualized and imaged using the FLoid Cell Imaging Station (ThermoFisher Scientific, Melbourne, VIC, Australia). Further differentiation experiments were performed in black opaque 96-well flat bottom microplates for luminescence and fluorescence assays. Differentiation assays used an amended protocol to address the limitations of the small volumes and well sizes used in the experiments. Adipose-derived cells were sub-cultured into serum-free DMEM/F12 medium containing 17.5 mM glucose, 1.0% (w/v) bovine serum albumin and insulin-transferrin-selenium. GFP signals in each well were quantitated using POLARstar OPTIMA (BMG Labtech Pty Ltd, Mornington, VIC, Australia) multi-detection microplate reader in real time. GFP intensity was used as an index of promoter activity36. The level of fluorescence in the green channel was automatically recorded and analyzed using MARS Data Analysis software (BMG Labtech Pty Ltd, Mornington, VIC, Australia) for each of the 96-wells in the microplate. All values generated from these data were corrected with respect to the blank (wells containing media but no cells) and presented corrected to the fluorescence detected at day 1, which were presented as ‘0’ fluorescence (this did not indicate that no GFP expression was detected). This provided a standard against which changes in GFP intensity could be measured. The GFP intensity was therefore expected to diminish further when exposed to an antagonist or to increase in intensity when exposed to a known agonist of differentiation.
Transplantation of Adipose-Derived Islet-like Cell Aggregates
TheraCytes™ (TheraCyte Inc, Laguna Hills, CA, USA) were loaded with 300 to 500 islet-like cell aggregates (ICAs) differentiated from human AMC cultures. The sealed TheraCytes™ containing adipose-derived ICAs were transplanted in the peritoneal cavity and orientated such that they laid flush against the peritoneum above the left kidney in 12-week-old male NOD/SCID mice, obtained from the Animal Resource Centre (WA, Australia). Animals were made diabetic following a partial pancreatectomy procedure34,35 and euthanized after 90 days and TheraCytes™ were retrieved. The contents of the TheraCytes™ were scraped and collected using fresh scalpel blades and solubilized in TRIzol Reagent. RNA was extracted and cDNA was generated using the High Capacity cDNA Reverse Transcription Kits. Quantitative PCR characterization of samples was performed as 5 µl reactions in a 96-well optical clear plate using 40 ng equivalent of cDNA input and TaqMan Fast Universal PCR Master Mix, as described previously. All values generated from these data were represented as cycle threshold (Ct-) values and presented as fold difference over the detectable Ct value, which was calculated using the ΔΔCt method (Ct value of 39 considered to be undetectable)29.
Human Insulin Enzyme-linked Immunosorbent Assay
Mice were administered 2 g/kg glucose at T=0 and blood samples were taken at 30 min. Insulin was measured in serum following the centrifugation of blood samples using a human ultra-sensitive insulin enzyme-linked immunosorbent assay (ELISA) kit (Mercodia, Uppsala, Sweden) and values were normalized to the protein content.
Statistics
All data were obtained from at least three or more independent experiments as stated in the methods and/or legends. A two-way analysis of variance (ANOVA; ordinary with Fisher’s least significant difference test) or a one-way ANOVA (ordinary with Holm–Sidak’s correction) were used for data analysis using Prism (Sigma Software Distribution, Devon, UK). Statistical significance was taken at p<0.05.
Results
Epigenetic and Transcriptomic Discovery Analyses
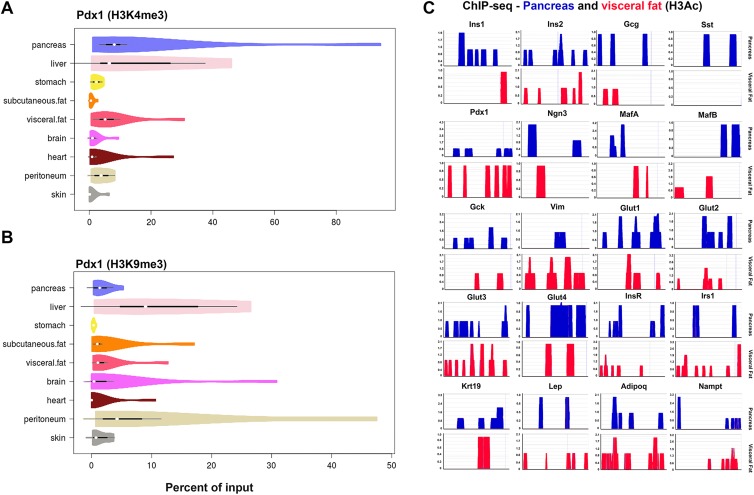
The chromatin structure of cells isolated from nine different types of mouse tissue (pancreas, liver, stomach, subcutaneous fat depot, visceral fat depot, brain, heart, peritoneal layer, and skin) was characterized using chromatin immunoprecipitation (ChIP). Two distinct histone modifications at the Pdx1 gene promoter region were examined (see methods for details). Pancreatic tissue demonstrated the highest level of open chromatin conformation for this master regulatory endocrine pancreatic transcription factor as evidenced by a higher level of H3K4 trimethylation as compared with H3K9 trimethylation (Fig. 1A, B). Interestingly, similar comparisons across other tissues indicated that the visceral fat depot was very different from the subcutaneous fat depot and contained cells that had a more ‘open’ conformation at the Pdx1 gene region. Most of the other tissues showed either very low levels of open chromatin modifications (stomach, subcutaneous fat, brain, skin) or showed significantly higher levels of inactive mark (H3K9me3) at the Pdx1 gene promoter region (e.g. liver, peritoneal layer; Fig. 1A, B). We then assessed open chromatin conformation in visceral and pancreatic tissues using a ChIP-sequencing approach (Fig. 1C). Indeed, the relative abundance of DNA that was pulled down for this open chromatin modification (H3-acetylation) was comparable between the pancreas and the visceral adipose tissue for multiple endocrine genes including ins2, Pdx1, mafB, gck, glut1, glut3 and insR. This inherent epigenetic similarity with the pancreas suggested that cells derived from visceral adipose tissue could represent a potential source of endocrine progenitors.
Fig 1.
Histone modifications at (pro-) endocrine gene promoters in mouse tissues. (A) and (B) represent trimethylation (me3) at the H3K4 and H3K9 regions respectively; representing an open (active; H3K4me3) or closed (inactive; H3K9me3) chromosomal conformation at the Pdx1 promoter (N=6 animals/group). All measurements are normalized to respective immunoprecipitated input DNA and presented as % of input. White dots/circles within these violin plots presented in (A) and (B) represent the medians; the black box limits indicate the 25th and 75th percentiles as determined by R software; whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles, while the polygons represent density estimates of data and extend to extreme values. (C) ChIP was carried for H3-acetylation; an active mark of gene expression, and immunoprecipitated DNA was sequenced using the Ion Torrent sequencing platform (N=6 animals/group). A comparative analysis of H3 acetylation through different gene promoters is presented in the mouse pancreatic (upper: blue) and visceral adipose tissues (lower: red). The y-axis indicates the abundance of H3ac-modified DNA within 1000 bases upstream of the transcription start site for each gene compared here.
ChIP: chromatin immunoprecipitation.
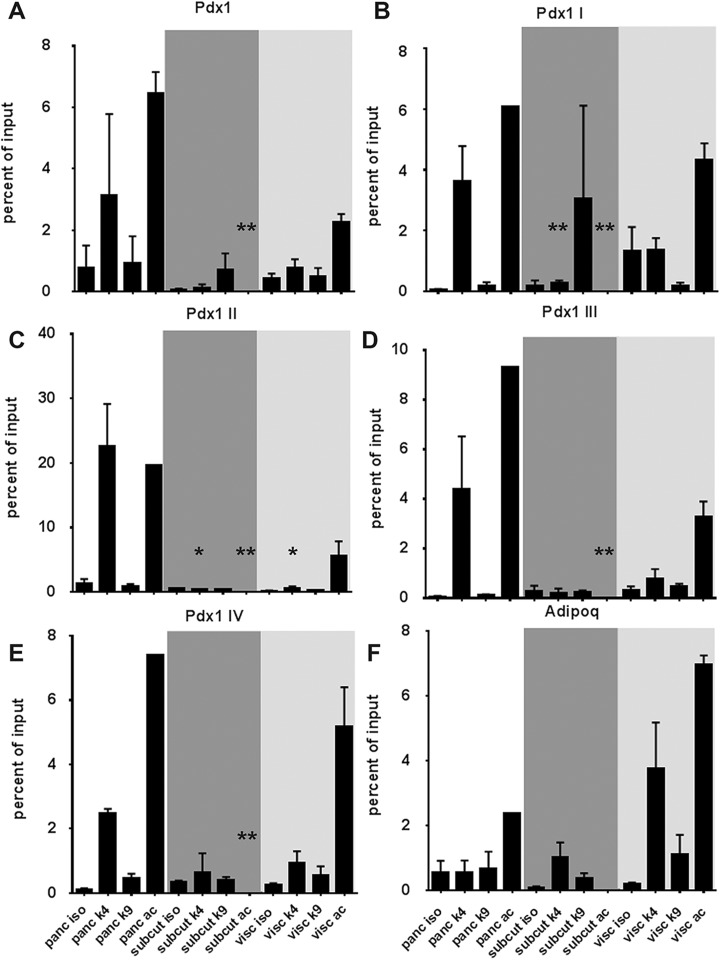
The results of the sequencing study were validated using real-time quantitative PCR on multiple samples for H3K4 and H3K9 trimethylation as well as H3 acetylation at selected regions of the Pdx1 gene and its promoter (Fig. 2A–E). Due to its importance in pancreas development and β-cell specification, five different primer pairs targeting Pdx1 promoter and upstream elements27, together with the gene promoter region for adiponectin (Fig. 2F) were used. As depicted in Fig. 2, pancreatic tissue samples showed an abundance of H3K4 trimethylation and H3 acetylation across all Pdx1 promoter regions, as compared with the subcutaneous adipose tissue samples. Interestingly, visceral adipose tissue samples showed a similar enrichment of methylation and acetylation signatures, especially in the upstream elements (regions I through IV) of the Pdx1 gene promoter. These data confirmed that the visceral adipose tissue samples were enriched for histone post-translational modifications that favor Pdx1 gene transcription. Collectively, these data demonstrate the epigenetic similarities between the pancreas and visceral adipose tissues, providing evidence to support the assertion that visceral adipose tissue-derived cells may contain an intrinsic capacity to differentiate into Pdx1-expressing endocrine progenitor cells.
Fig 2.
Validation of ChIP sequencing using quantitative real-time PCR. Ct values for isotype (control), H3K4 trimethylation (me3), H3K9 trimethylation (me3) and H3 ac pulldown were normalized to input DNA and presented as percent of input for the Pdx1 promoter region (A) as well as upstream regions I–IV (B–E) of the Pdx1 gene, which are recognized to be the pre-patterning modifications for pancreatic cell fate (Xu et al.27). Adipose-specific epigenetic modifications were assessed at the adiponectin (Adipoq) gene promoter region. Data presented as mean and SD from at least N=5 or 6 tissues in each group. Shaded background regions indicate pancreatic (white backdrop), subcutaneous adipose tissue (dark grey backdrop) and visceral adipose (light grey backdrop) tissue samples. Significant differences are represented by a *(p<0.05) or **(p<0.01) as compared with the respective pancreatic tissue modification (K4, K9 or ac).
ChIP: chromatin immunoprecipitation; Ct: cycle threshold; PCR: polymerase chain reaction; SD: standard deviation.
Gene expression analysis of pancreatic and adipose tissue samples was achieved using RNA sequencing. The relative transcript abundance of multiple genes is presented in Fig. S1. Pancreatic tissue samples contained relatively higher levels of transcripts for endocrine hormones (Ins1, Ins2, Gcg), which were not detected in visceral adipose tissue samples. Key pancreatic transcription factors in β-cell fate determination; Pdx1, MafB, Gata6, as well as important pancreatic function-related transcripts; glucokinase, glut1 and glut 2, were detected in both tissues, albeit at lower levels in visceral adipose samples. The characteristic mesenchymal intermediate filament vimentin (Vim) and adipokines; leptin (Lep), adiponectin (Adipoq), resistin (Retn) and visfatin (Nampt), were detected at significantly high levels in adipose tissue samples relative to those detected in the pancreas (Fig. S1). We therefore decided to further study adipose tissue-derived cells as a potential source for in-vitro differentiation to insulin-producing cells.
Isolation and Characterization of Adipose Tissue-derived Cells
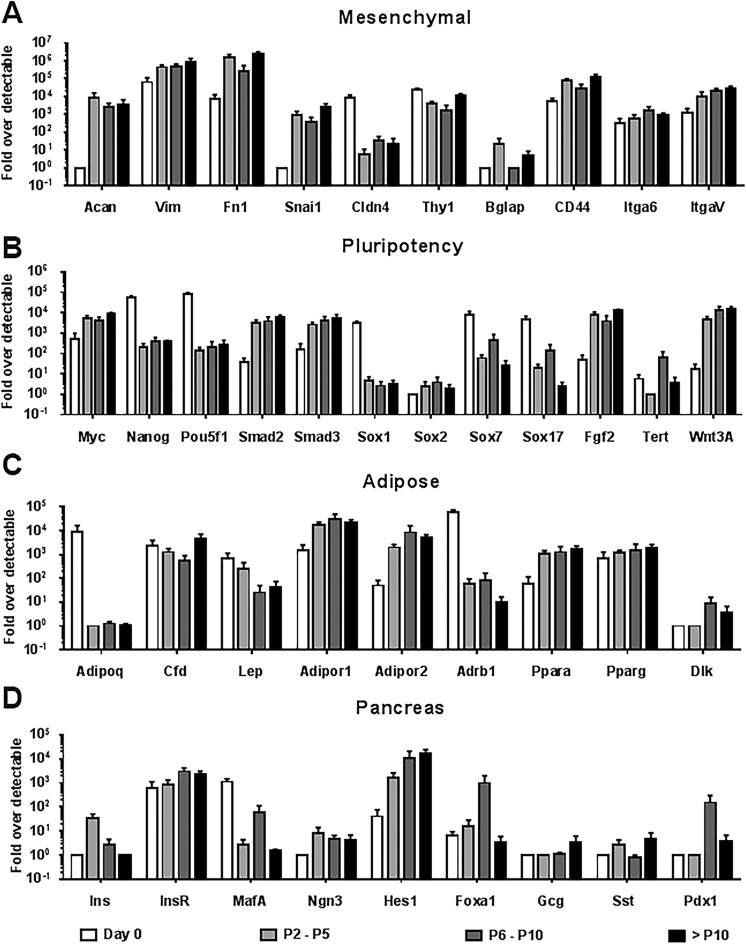
Human visceral adipose-derived progenitor cells were cultured as an adherent population of fibroblast-like cells. These cells mostly exhibited a bipolar morphology. Owing to their mesenchymal-like appearance in culture, we termed this population as AMCs. Cultured AMCs were characterized with respect to a selected panel of mesenchymal-, pluripotency-, adipose- and pancreas-associated genes at periodic stages of culture, using TaqMan-based real-time quantitative PCR (Fig. 3). Increased subculture was typically associated with either increases or stable expression of mesenchymal markers in culture (Fig. 3A). This data corroborates the visual observation of cultures achieving an increasingly homogenous mesenchymal-like morphology when cultured for multiple passages. Changes to the expression of pluripotency-associated genes in cultured cells were inconsistent, with AMCs observed to be enriched for the expression of certain genes (Myc, Smad2/3, Fgf2, Wnt3A) while the expression of others was decreased (Fig. 3B). The elevated expression of some of these markers was not indicative of the population achieving a pluripotent phenotype but rather suggested a much broader differentiation potential (increased plasticity) with prolonged passaging. The expression of adipose or preadipocyte markers in AMCs offered information regarding the retention of any adipose characteristics of the source tissue and a measure of their propensity to spontaneously differentiate into this phenotype (Fig. 3C). The expression of key adipokines (adiponectin and leptin) decreased in abundance in the AMCs as compared with the day 0 samples, whereas other adipose-associated transcripts were typically expressed at consistent levels throughout the culture.
Fig 3.
Gene expression characterization of cultured human AMCs. A comprehensive panel of genes were profiled for their expression in visceral adipose tissue (day 0) and AMCs (passage 2/P2 through passage 10/P10 or more) generated from the same preparations. TaqMan-based qPCR was performed and normalized to 18 s rRNA (housekeeping gene) and presented as fold difference over the detectable Ct limit (Ct value of 39 was considered to be undetectable) using the ΔΔCt method (Hardikar et al.29). Samples were characterized with respect to selected panel of (A) mesenchymal-, (B) pluripotency-, (C) adipose- and (D) pancreas-associated genes at different stages of culture. Data are generated from at least 3 to 5 different human preparations and presented as mean ± SEM.
AMC: adipose tissue-derived mesenchymal cell; Ct: cycle threshold; qPCR: quantitative polymerase chain reaction; SEM: standard error of the mean.
AMCs were profiled for a panel of pancreas-associated markers to investigate the expression of key endocrine pancreatic transcripts during sub-culturing (Fig. 3D). Transcripts encoding endocrine hormones (Ins, Gcg, and Sst) were undetectable or expressed at very low levels in a majority of AMC preparations. When transcription factors essential to the pancreas and β-cell development were assessed, we observed limited expression of Ngn3 and MafA, while Pdx1 and Foxa1 expression increased following 6 to 10 passages in vitro. Hes1, the negative regulator of Ngn3, was seen to be enriched following prolonged in-vitro expansion.
We carried out in-situ PCR using protocols optimized in our lab30, to confirm the expression of adipose and pancreatic gene transcripts. Visfatin mRNA (positive control), was observed to be almost ubiquitously expressed in cultured cells (Fig. S2). The fluorescent signal (FAM) of amplified visfatin mRNA was seen to be concentrated in the perinuclear region, as is expected for most mRNAs. Similarly, the fluorescent signal of amplified smooth muscle actin mRNA was observed to be present in a majority of cultured cells. However, the expression of MafA or Ngn3 was observed to be significantly limited, with only a small fraction of the cells within AMC cultures expressing these transcripts. These observations suggest that the AMCs contained discrete sub-populations of MafA- and Ngn3-expressing cells.
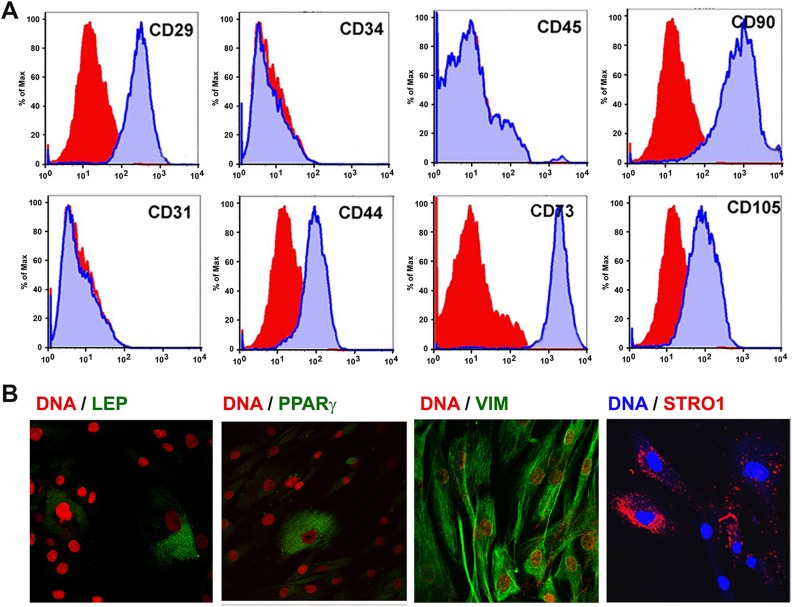
The cell surface antigens expressed by AMCs were quantified using flow cytometry (Fig. 4A). AMCs represent a fairly homogeneous population of mesenchymal-like cells immunopositive for CD29 (95.2 ± 1.4%), CD44 (96.2 ± 4.4%), CD73 (97.8%), CD90 (98.4%) and CD105 (98.6 ± 1.9%), and immunonegative for CD31 (0.8 ± 0.3%), CD34 (0.7 ± 0.4%) and CD45 (0.9 ± 0.3%). Expression of leptin, peroxisome proliferator-activated receptor gamma (PPARγ), and vimentin were assessed using immunocytochemistry. Adipose-specific leptin and PPARγ were present only in few cells while vimentin was observed to be ubiquitously expressed in cultured AMCs (Fig. 4B). The stromal precursor antigen-1 (Stro-1), a potential biomarker of multipotent progenitors37, was present in a majority of AMCs indicating multilineage differentiation potential (Fig. 4B).
Fig 4.
Surface and intracellular antigen profile of human AMCs. Human AMCs were characterized using flow cytometry (A) for the expression of different mesenchymal and hematopoietic surface antigens. Isotype controls are shown in red, while the specific antibody that was used is in blue; N=4/group. (B) AMCs exhibit presence of adipose markers (leptin and PPARγ) in fewer cells and are more homogeneous for the expression of the mesenchymal marker; vimentin and the pluripotency marker Stro1. Nuclei are stained with Hoechst and are presented here in red or blue, while other antigens are shown in green or red as indicated. Images were obtained using LSM 510 Meta confocal microscope on an Axio Observer Z1 platform. Laser intensities, gains and offsets were maintained similar, and below saturation levels for each scan. Bar represents 20 μm.
AMC: adipose tissue-derived mesenchymal cell; PPARγ: peroxisome proliferator-activated receptor gamma.
Differentiation of AMCs into ICAs
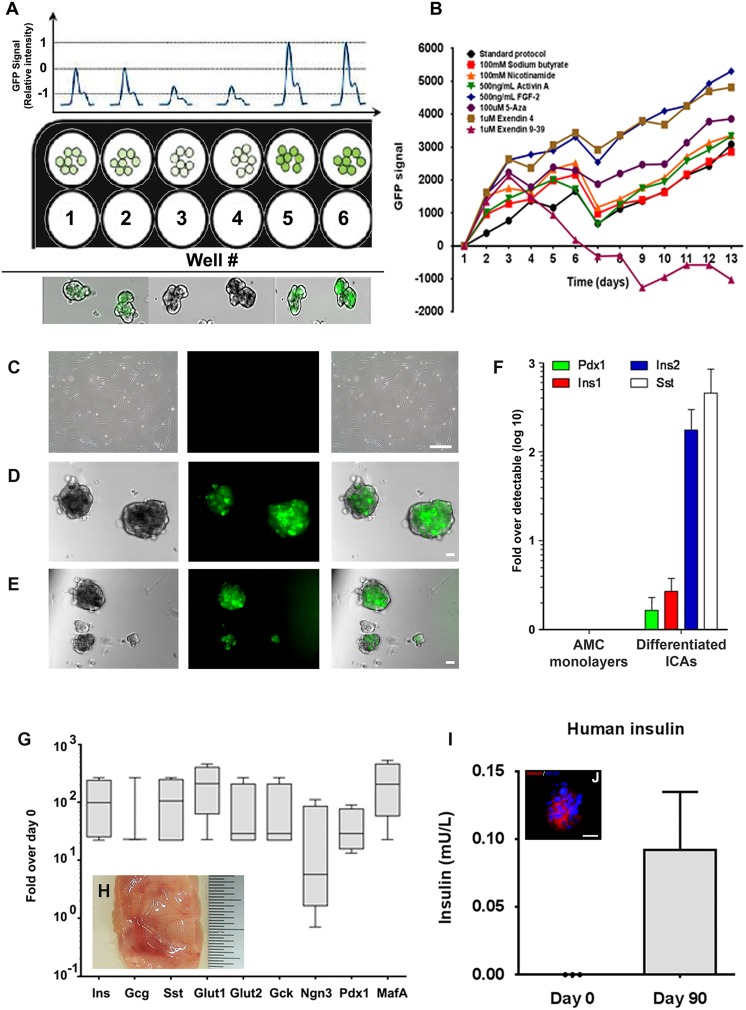
The differentiation process was investigated using visceral AMCs from a transgenic mouse model. These reporter mice contained a GFP transgene under the control of the Pdx1 gene promoter38. AMCs isolated from this transgenic mouse strain were analogous to equivalent cultures generated from human adipose tissue (not shown). When exposed to serum-free conditions31,32,34,35,39, AMCs formed ICAs that expressed the GFP reporter, indicating Pdx1 promoter activation (Fig. 5A). The fluorescent signal generated by these ICAs was quantitatively measured using the 96-well plate reader, where GFP intensity was quantified over time as a measure of endocrine pancreatic commitment. Differentiation was induced in the presence of multiple growth and differentiation factors at the concentrations indicated in Supplementary Table 1, and the effect on Pdx1 promoter activity was determined and presented in Fig. 5B. AMCs derived from this transgenic animal failed to show a detectable expression of GFP, indicating sub-optimal Pdx1 gene promoter activity under standard culture conditions (Fig. 5C). However, exposure to exendin-4 (Fig. 5D) and fibroblast growth factor 2 (FGF2; Fig. 5E) increased the GFP signal with time, reflecting increased promoter activity (Fig. 5B). These data indicated that FGF2, as well as the exendin-4 agonist, promoted the differentiation of mouse AMCs into the endocrine pancreatic lineage. Conversely, the antagonist exendin-(9-39) was found to suppress GFP expression, indicating decreased promoter activity and retardation of endocrine differentiation. Mouse AMC-derived ICAs were harvested and analyzed using TaqMan-based real-time quantitative PCR. Undifferentiated monolayer cultures contained undetectable endocrine transcripts, but differentiated ICAs contained increased pancreatic transcripts, including Pdx1, insulin 1, insulin 2 and somatostatin (Fig. 5F).
Fig 5.
Differentiation and function of AMCs. (A) Visceral adipose-derived AMCs from Pdx1-GFP mice were exposed to various growth and differentiation factors. Panel (A) represents the experimental setup where GFP reporter (Pdx1-GFP) AMCs were plated in (black) 96-well plates and fluorescent intensities recorded as detailed in methods. Note that some (minimal) level of GFP seen in the cells (wells 1 and 2), represented as ‘0’ intensity, diminished when exposed to the GLP1 antagonist (exendin 9-39; wells 3 and 4) or to increase in intensity when exposed to an agonist (wells 5 and 6). All conditions were set up in triplicate. (B) Cultures were stimulated to differentiate in the presence of the small molecules described in Supplementary Table 2 and the intensity of GFP fluorescence was measured at the same time of the day and at regular (24 h) intervals. Data were corrected with respect to the blank (well containing media but no cells) and presented relative to the GFP signal measured on day 1. Values generated for day 1 are therefore presented as ‘0’ fluorescence, while subsequent measurements are presented relative to day 1. AMCs generated from Pdx1-GFP transgenic mice do not show GFP expression as monolayers (C). However, when stimulated to differentiate into an endocrine pancreatic lineage, AMCs formed multiple, small ICAs that express GFP in presence of exendin-4 (D) or FGF2 (E). Bar = 20μm. GFP-expressing ICAs were analyzed using TaqMan-based quantitative PCR for the expression of endocrine pancreatic gene transcripts. Data are presented as fold difference over the detectable Ct value was calculated using the ΔΔCt method (Hardikar A et al.29) (F). Human AMCs were induced to differentiate into endocrine pancreatic lineage using FGF2 and exendin-4 agonist and transplanted in NOD/SCID mice using TheraCyte™ devices. Human AMC-derived ICAs showed detectable expression of pro-endocrine gene transcripts (G). Box and whisker plot showing the abundance of transcript at day 90 relative to expression in day 0 (undifferentiated) AMCs. The line in the box represents the median and the box represents the upper and lower quartile (N=4 mice). ICA-containing TheraCytes™ showed evidence of vascularization (H), showed detectable levels of human insulin (mean ± SD; N=3) in mouse circulation (I) and contained insulin-producing cells after 90 days of in-vivo studies (J); red: insulin, blue: DAPI: nuclei; bar = 20μm.
AMC: adipose tissue-derived mesenchymal cell; ICA: islet-like cell aggregate; AMC: Adipose-derived mesenchymal cells; ICAs: Islet-like Cell Aggregates; Pdx1: Pancreatic and duodenal homeobox 1; GLP1: Glucagon-like Peptide 1; GFP: Green Fluoroscence Protein; FGF2: Fibroblast Growth Factor 2; PCR: Polymerase Chain Reaction; Ct: Cycle threshold; NOD/SCID: Non-obese Diabetic/Severe Combined Immunodeficiency; DAPI: 4′,6-diamidino-2-phenylindole; SD: Standard Deviation.
Transplantation Studies of Adipose-derived ICAs
We then adapted the differentiation conditions identified using GFP-expressing mouse AMCs to differentiate human visceral AMCs in vitro. Human AMC-derived ICAs were transplanted into the abdominal cavity of NOD/SCID mice using TheraCyte™ devices. These animals were subsequently rendered diabetic following partial pancreatectomy. Grafts retrieved 90 days post-transplantation showed a significant increase in pro-insulin gene transcripts (Fig. 5G). The transcripts for the glucose transporters Glut1 and Glut2 were increased compared with day 0 samples, which is particularly interesting given that Glut1 expression is significantly higher in human β-cells. Pdx1, Ngn3, and MafA expression were also elevated by around 10- to 100-fold, as compared with their day 0 levels. Analysis of the retrieved graft from TheraCyte™ devices demonstrated efficient vascularization (Fig. 5 H). Blood samples collected from peripheral circulation of these mice following a glucose injection (30 minutes before retrieval) showed that differentiated and engrafted human AMC-derived ICAs could secrete detectable levels of insulin in response to a glucose stimulus (Fig. 5I) and contained insulin-immunopositive cells (Fig. 5J, red).
Discussion
Any study involving the conversion of stem or progenitor cells to an insulin-producing phenotype involves overwriting and reprogramming elements of a profoundly complex and carefully orchestrated biological system. The discovery study presented in this manuscript was initiated to understand the histone modifications at the pro-pancreatic gene promoter region, in cells derived from multiple tissues. This unbiased approach provided the opportunity to examine and characterize the epigenome of a wide variety of potential cell populations, and to provide information regarding their differentiation potential into an endocrine pancreatic lineage. The similarities identified at representative promoter regions of genes relevant to the β-cell development and insulin expression provided evidence of an underlying predisposition of visceral adipose-derived cells to differentiate into an endocrine pancreatic lineage. As we summarized earlier40, populations of lineage-committed cells represent a source that would predictably require fewer manipulations, to achieve differentiation into the desired endocrine pancreatic phenotype. We theorized that cells derived from a tissue that already exhibited several key epigenetic traits similar to a pancreatic β-cell would represent a viable candidate population for differentiation studies.
We characterized and compared the abundance of different histone modifications that confer an ‘open’ or active chromatin conformation in different tissues using ChIP-seq analysis followed by validation using real-time quantitative PCR. Analysis of immunoprecipitated DNA from visceral adipose tissue demonstrated an epigenetic signature (H3K4me3, H3K9me3, and H3Ac) at islet-specific gene promoters, similar to those in the pancreas. However, this did not imply that these similarities manifest at the transcriptome level. Subsequent transcriptome analysis demonstrated distinct differences in the mRNA profiles of the respective tissues, as expected for two developmentally distinct tissues with such separate functional and histological phenotypes. Nonetheless, the epigenetic similarities between adipose and pancreatic tissues demanded further investigation into the understanding the differentiation potential of visceral adipose-derived cells.
We generated a fibroblast-like cell population from adipose tissue and referred to them as AMCs. Early passage (P0–P2) AMCs did not represent a uniform population of cells, however, following further in-vitro culture, these AMCs tended to become more homogeneous and exhibit a characteristic mesenchymal phenotype, indicating a shift in cell composition during culture41. AMCs demonstrated an increasing abundance of mesenchymal (vim, fn1, snai1, CD44, ItgaV) as well as pluripotency (myc, smad2/3, fgf2, wnt3A) markers during in-vitro culture (Fig. 3A, B). The abundance of adipogenic or preadipocyte transcripts (Adipoq, cfd, Lep, Adrb1) progressively decreased during P0–P10 while pancreatic or pro-endocrine gene transcripts (insR, Ngn3, FoxA1, Pdx1) increased during this time (Fig. 3C, D). Although this gene expression profile indicates a pancreatic lineage prone precursor population, passaging of AMCs was also associated with progressive increases in the abundance of Hes1; a negative regulator of the pro-endocrine gene Ngn3. Since such gene expression data represent population analysis, which averages the variation amongst the millions of single cells in a population, we used in-situ PCR to capture low copy number transcripts in AMC cultures. Using in-situ PCR established in our lab30, we observed that a small subset of AMCs contain pancreas-specific transcripts in high abundance. However, despite the inherent heterogeneity observed in cells derived from adipose tissue, the propensity to differentiate into an endocrine pancreatic lineage is detected in our initial discovery and cultured cell characterization studies.
We used transgenic AMCs isolated from Pdx1-GFP mice to provide real-time monitoring of GFP intensity as an index of differentiation to endocrine pancreatic lineage36. The differentiation process was adapted from a protocol used in our lab for the differentiation of human pancreatic precursor cells to an endocrine pancreas lineage31,32,34,35,39. Cultured AMCs did not express the reporter due to inactivity of the conjugate Pdx1 promoter (Fig. 5C). However, when stimulated to differentiate, promoter activity increases in response to the differentiation process, which is reflected by an increase in GFP expression proportional to promoter activity (Fig. 5B, D, E). This technique provided a means to measure promoter activity in real time, to extrapolate gene transcription under different conditions. By quantifying the intensity of the GFP reporter signal at regular intervals, the activity of the associated promoter was measured and the commitment to a specific phenotype inferred. Increasing expression of the GFP reporter in differentiating AMCs generated from Pdx1-GFP animals indicated that they had achieved a state of commitment to an endocrine pancreatic lineage.
Cultured transgenic cells were stimulated to differentiate when treated with a variety of small molecules intended to promote the differentiation of mouse AMCs into an endocrine pancreatic lineage. These agents were specifically chosen for their documented effects on chromatin structure, differentiation and pancreas biology (see references 56–63 in Supplementary Table 1). We observed enhanced GFP expression in the presence of FGF2 and exendin-4. FGF2 is involved in many biological processes including angiogenesis, embryonic development, and wound healing. It has been shown to promote the differentiation of adipose-derived cells42 and loss of FGF signaling has been implicated in β-cell loss and the development of diabetes43. We31,44 and others45,46 have also demonstrated a role of FGF2 in the early stages of endocrine pancreas differentiation. Exendin-4, a glucagon-like peptide-1 (GLP-1) receptor agonist promotes the differentiation of pancreatic progenitor populations (such as ductal progenitor cells) and increases proliferation and maturation in β-cells during development47,48. Exendin-(9-39), a truncated version of exendin-4 that acts as a competitive antagonist at the GLP-1 receptor49,50, was chosen as a negative control and was seen to suppress differentiation (Fig. 5B).
Animal studies were carried out using the TheraCyte™ devices for transplantation. TheraCyte™ devices were used to facilitate easy and clean retrieval of human AMCs following transplantation. After optimal engraftment (determined as 2 weeks post-transplant, through previous studies from our lab34,35), these mice underwent partial pancreatectomy to remove the distal portion of the pancreas, rendering them surgically diabetic. This protocol38 is well described by our lab and has been shown to promote the differentiation of transplanted cells potentially through the paracrine factors released from the regenerating pancreas28,30. The results of the transplantation studies suggested that transplanted cells continued to mature and differentiate following surgery. Tissue grafts were recovered after 90 days and profiled for major pancreatic hormones, transcription factors and receptors using quantitative PCR. Analysis of the circulating human insulin concentrations in mouse plasma of transplanted animals demonstrated that the transplanted cells were capable of producing and secreting insulin (Fig. 5I, J). These studies confirmed that AMCs could not only differentiate to an endocrine pancreatic lineage in vitro but potentially act as a source of insulin for treatment of diabetes in vivo.
Over the past several years, we have assessed the potential of multiple human stem/progenitor cells including those from pancreatic ducts, bone marrow and umbilical cord blood-derived mononuclear cells. In order to assess differentiation efficiency of human AMCs, we compared the levels of insulin transcripts following differentiation of various biological replicates (and different passages) of the above stem/progenitor cell types. Intriguingly, differentiated human AMCs contained a significantly higher number of insulin transcripts than any of the other stem/progenitor cells tested in our lab (Fig. S3), although these were all significantly lower as compared with insulin transcripts detected in human islets. The present study therefore has merits in identification of a cell type (visceral AMCs) and a protocol (based on mouse AMC Pdx1 expression) for efficient differentiation of progenitor cells. As compared with several other therapies, such as those involving islet transplantation, the major advantage is that the source of AMCs can be autologous and potentially more abundant.
Strengths and Limitations of This Study
The study strengths are (1) a genome-wide epigenetic and transcriptomic profiling of multiple mouse tissues; and (2) identification of visceral, but not subcutaneous adipose tissue as a preferred source of progenitor cells with the potential to differentiate into an endocrine pancreatic cell lineage. Characterization of AMCs demonstrates that these could be expanded and differentiated into an endocrine pancreatic lineage in vitro and that they would continue to differentiate and produce human insulin in vivo. These cells appear to retain their differentiated characteristics in vivo, indicating that this cell population may prove to be an important candidate for cell replacement therapy in diabetes.
One of the study limitations is the depth of sequencing. We employed an Ion Torrent PGM platform for ChIP-seq and RNA-seq studies using a 316 chip that can provide up to 3 million clean reads. Although multiple samples were pooled from different sequencing runs, the overall low sequencing depth indicates that some of the lower abundance transcripts or DNA species may not have been captured into the representative libraries that we screened. However, it is intriguing to note that in spite of this low depth, several of the pancreatic pro-endocrine genes were seen to be accessible for transcription (Fig. 1C) and that several of these genes were also transcribed (Supplementary Fig. 1) in visceral adipose tissue. These observations present the first evidence that pro-endocrine gene transcripts are expressed in a subset of AMCs.
Another potential limitation is the applicability of in-vitro differentiation protocols optimized in mice, to the differentiation of human AMCs. Even though the broader functions of human and mouse tissues are similar, they can differ from each other in aspects including tissue architecture51,52, response to different enzymes53, cytokines and pathway activation54,55 and islet cell proliferation39. We identified a protocol that facilitates maximal activation of the mouse Pdx1 reporter gene during differentiation of mouse AMCs (Fig. 5B). While this knowledge generated from the in-vitro mouse AMC reporter system, may or may not be the most efficient protocol to differentiate human AMCs to insulin-producing cells, these studies provide us with a starting point to test the differentiation potential of human AMCs. Further studies to explore these possibilities are merited.
The current study opens up several avenues for future exploration. Although pluripotency and pro-pancreatic gene transcripts increased in abundance with progressive passaging, Hes1 expression increased concomitantly. Strategies targeting Hes1 repression, either through site-directed mutagenesis or by inhibition of Hes1 via regulatory RNAs or small molecules may facilitate the differentiation of this easily derivable as well as highly abundant source of pancreatic progenitor cells.
Supplemental Material
Supplemental Material, FigureS1_Hardikar for Epigenetic and Transcriptome Profiling Identifies a Population of Visceral Adipose-Derived Progenitor Cells with the Potential to Differentiate into an Endocrine Pancreatic Lineage by Michael D. Williams, Mugdha V. Joglekar, Sarang N. Satoor, Wilson Wong, Effie Keramidaris, Amanda Rixon, Philip O’Connell, Wayne J. Hawthorne, Geraldine M. Mitchell, and Anandwardhan A. Hardikar in Cell Transplantation
Supplemental Material
Supplemental Material, FigureS2_Hardikar for Epigenetic and Transcriptome Profiling Identifies a Population of Visceral Adipose-Derived Progenitor Cells with the Potential to Differentiate into an Endocrine Pancreatic Lineage by Michael D. Williams, Mugdha V. Joglekar, Sarang N. Satoor, Wilson Wong, Effie Keramidaris, Amanda Rixon, Philip O’Connell, Wayne J. Hawthorne, Geraldine M. Mitchell, and Anandwardhan A. Hardikar in Cell Transplantation
Supplemental Material
Supplemental Material, Figure_S3 for Epigenetic and Transcriptome Profiling Identifies a Population of Visceral Adipose-Derived Progenitor Cells with the Potential to Differentiate into an Endocrine Pancreatic Lineage by Michael D. Williams, Mugdha V. Joglekar, Sarang N. Satoor, Wilson Wong, Effie Keramidaris, Amanda Rixon, Philip O’Connell, Wayne J. Hawthorne, Geraldine M. Mitchell, and Anandwardhan A. Hardikar in Cell Transplantation
Supplemental Material
Supplemental Material, Supplementary_Table_1_new for Epigenetic and Transcriptome Profiling Identifies a Population of Visceral Adipose-Derived Progenitor Cells with the Potential to Differentiate into an Endocrine Pancreatic Lineage by Michael D. Williams, Mugdha V. Joglekar, Sarang N. Satoor, Wilson Wong, Effie Keramidaris, Amanda Rixon, Philip O’Connell, Wayne J. Hawthorne, Geraldine M. Mitchell, and Anandwardhan A. Hardikar in Cell Transplantation
Acknowledgments
The support provided to MDW and WW through the Australian Government’s post-graduate awards, MVJ through JDRF International post-doctoral fellowship, SNS through an NHMRC post-doctoral grant, EK and AR through the O’Brien Institute, GM through the NHMRC and O’Brien Institute and AAH through the ARC Future Fellowship and currently through the JDRF Australia Career Development Award is highly acknowledged. The authors acknowledge the surgical team at SVHM, Melbourne for providing the adipose tissues and Prof. Edouard G. Stanley and Prof. Andrew Elefanty (CMRI, Melbourne) for the generous gift of Pdx1-GFP reporter mice. The infrastructure support provided to AAH through the NHMRC Clinical Trials Center, The University of Sydney, The Rebecca Cooper Medical Research Foundation and the O’Brien Institute/St Vincent’s Hospital, Melbourne is highly acknowledged.
Footnotes
Author Contributions: MDW performed majority of all experiments, analyzed the data and wrote the manuscript draft. MVJ performed flow cytometry, ChIP, ELISAs and analyzed the data. SNS and WW assisted in sequencing studies and analysis. AR and EK provided assistance in mouse studies and derivation of AMCs. PO’C and WJH contributed human islets for RNA analysis. GM co-supervised MDW with AAH and provided AMCs and input for the study. AAH and MVJ designed and planned the study and wrote the final draft with all authors who agreed on the final draft.
Ethical Approval: This study was approved by our institutional review board.
Statement of Human and Animal Rights: This article describes studies carried out with animals and adipose tissues obtained from human subjects as per the respective approvals from the institutional animal and human ethics committees.
Statement of Informed Consent: All human tissues were obtained following a written informed consent as per the ethical committees' guidelines/approvals.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Einhorn D. Advances in diabetes for the millennium: insulin treatment and glucose monitoring. MedGenMed. 2004;6(suppl 3):8. [PMC free article] [PubMed] [Google Scholar]
- 2. Shapiro AM, Lakey JR, Ryan EA, Korbutt GS, Toth E, Warnock GL, Kneteman NM, Rajotte RV. Islet transplantation in seven patients with type 1 diabetes mellitus using a glucocorticoid-free immunosuppressive regimen. N Engl J Med. 2000;343(4):230–238. [DOI] [PubMed] [Google Scholar]
- 3. Croon AC, Karlsson R, Bergstrom C, Bjorklund E, Moller C, Tyden L, Tibell A. Lack of donors limits the use of islet transplantation as treatment for diabetes. Transplant Proc. 2003;35(2):764. [DOI] [PubMed] [Google Scholar]
- 4. Gangaram-Panday ST, Faas MM, de Vos P. Towards stem-cell therapy in the endocrine pancreas. Trends Mol Med. 2007;13(4):164–173. [DOI] [PubMed] [Google Scholar]
- 5. Ferber S, Halkin A, Cohen H, Ber I, Einav Y, Goldberg I, Barshack I, Seijffers R, Kopolovic J, Kaiser N, Karasik A. Pancreatic and duodenal homeobox gene 1 induces expression of insulin genes in liver and ameliorates streptozotocin-induced hyperglycemia. Nat Med. 2000;6(5):568–572. [DOI] [PubMed] [Google Scholar]
- 6. Motoyama H, Ogawa S, Kubo A, Miwa S, Nakayama J, Tagawa Y, Miyagawa S. In vitro reprogramming of adult hepatocytes into insulin-producing cells without viral vectors. Biochem Biophys Res Commun. 2009;385(1):123–128. [DOI] [PubMed] [Google Scholar]
- 7. Zalzman M, Anker-Kitai L, Efrat S. Differentiation of human liver-derived, insulin-producing cells toward the beta-cell phenotype. Diabetes. 2005;54(9):2568–2575. [DOI] [PubMed] [Google Scholar]
- 8. Nagaya M, Katsuta H, Kaneto H, Bonner-Weir S, Weir GC. Adult mouse intrahepatic biliary epithelial cells induced in vitro to become insulin-producing cells. J Endocrinol. 2009;201(1):37–47. [DOI] [PubMed] [Google Scholar]
- 9. Sahu S, Joglekar MV, Dumbre R, Phadnis SM, Tosh D, Hardikar AA. Islet-like cell clusters occur naturally in human gall bladder and are retained in diabetic conditions. J Cell Mol Med. 2009;13(5):999–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Galivo F, Benedetti E, Wang Y, Pelz C, Schug J, Kaestner KH, Grompe M. Reprogramming human gallbladder cells into insulin-producing beta-like cells. Plos One. 2017;12(8):e0181812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Timper K, Seboek D, Eberhardt M, Linscheid P, Christ-Crain M, Keller U, Muller B, Zulewski H. Human adipose tissue-derived mesenchymal stem cells differentiate into insulin, somatostatin, and glucagon expressing cells. Biochem Biophys Res Commun. 2006;341(4):1135–1140. [DOI] [PubMed] [Google Scholar]
- 12. Chandra V, G S, Phadnis S, Nair PD, Bhonde RR. Generation of pancreatic hormone-expressing islet-like cell aggregates from murine adipose tissue-derived stem cells. Stem Cells. 2009;27(8):1941–1953. [DOI] [PubMed] [Google Scholar]
- 13. Moshtagh PR, Emami SH, Sharifi AM. Differentiation of human adipose-derived mesenchymal stem cell into insulin-producing cells: an in vitro study. J Physiol Biochem. 2013;69(3):451–458. [DOI] [PubMed] [Google Scholar]
- 14. Dave SD, Trivedi HL, Chooramani SG, Chandra T. Management of type 1 diabetes mellitus using in vitro autologous adipose tissue trans-differentiated insulin-making cells. BMJ Case Rep. 2013;2013. doi: 10.1136/bcr-2013-200226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gabr MM, Zakaria MM, Refaie AF, Abdel-Rahman EA, Reda AM, Ali SS, Khater SM, Ashamallah SA, Ismail AM, Ismail HEA, El-Badri N, Ghoneim MA. . From human mesenchymal stem cells to insulin-producing cells: comparison between bone marrow- and adipose tissue-derived cells. Biomed Res Int. 2017;2017:3854232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Enderami SE, Soleimani M, Mortazavi Y, Nadri S, Salimi A. Generation of insulin-producing cells from human adipose-derived mesenchymal stem cells on pva scaffold by optimized differentiation protocol. J Cell Physiol. 2018;233(5):4327–4337. [DOI] [PubMed] [Google Scholar]
- 17. Satoor SN, Puranik AS, Kumar S, Williams MD, Ghale M, Rahalkar A, Karandikar MS, Shouche Y, Patole M, Bhonde R, Yajnik CS, Hardikar AA. Location, location, location: beneficial effects of autologous fat transplantation. Sci Rep. 2011;1:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Perrini S, Laviola L, Cignarelli A, Melchiorre M, De Stefano F, Caccioppoli C, Natalicchio A, Orlando MR, Garruti G, De Fazio M, Catalano G, Memeo V, Giorgino R, Giorgino F. Fat depot-related differences in gene expression, adiponectin secretion, and insulin action and signalling in human adipocytes differentiated in vitro from precursor stromal cells. Diabetologia. 2008;51(1):155–164. [DOI] [PubMed] [Google Scholar]
- 19. Pellegrinelli V, Carobbio S, Vidal-Puig A. Adipose tissue plasticity: how fat depots respond differently to pathophysiological cues. Diabetologia. 2016;59(6):1075–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. D’Urso A, Brickner JH. Mechanisms of epigenetic memory. Trends Genet. 2014;30(6):230–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wilusz JE, Sunwoo H, Spector DL. Long noncoding rnas: functional surprises from the rna world. Genes Dev. 2009;23(13):1494–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kim VN. Microrna biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6(5):376–385. [DOI] [PubMed] [Google Scholar]
- 23. Joglekar MV, Parekh VS, Hardikar AA. New pancreas from old: microregulators of pancreas regeneration. Trends Endocrinol Metab. 2007;18(10):393–400. [DOI] [PubMed] [Google Scholar]
- 24. Rajapakse I, Groudine M. On emerging nuclear order. J Cell Biol. 2011;192(5):711–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jayani RS, Ramanujam PL, Galande S. Studying histone modifications and their genomic functions by employing chromatin immunoprecipitation and immunoblotting. Methods Cell Biol. 2010;98:35–56. [DOI] [PubMed] [Google Scholar]
- 26. Joglekar MV, Hardikar AA. Isolation, expansion, and characterization of human islet-derived progenitor cells. Methods Mol Biol. 2012;879:351–366. [DOI] [PubMed] [Google Scholar]
- 27. Xu CR, Cole PA, Meyers DJ, Kormish J, Dent S, Zaret KS. Chromatin “prepattern” and histone modifiers in a fate choice for liver and pancreas. Science. 2011;332(6032):963–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zuk PA, Zhu M, Mizuno H, Huang J, Futrell JW, Katz AJ, Benhaim P, Lorenz HP, Hedrick MH. Multilineage cells from human adipose tissue: implications for cell-based therapies. Tissue Eng. 2001;7(2):211–228. [DOI] [PubMed] [Google Scholar]
- 29. Hardikar AA, Farr RJ, Joglekar MV. Circulating micrornas: understanding the limits for quantitative measurement by real-time pcr. J Am Heart Assoc. 2014;3(1):e000792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ranjan AK, Joglekar MV, Atre AN, Patole M, Bhonde RR, Hardikar AA. Simultaneous imaging of microrna or mrna territories with protein territory in mammalian cells at single cell resolution. RNA Biol. 2012;9(7):949–953. [DOI] [PubMed] [Google Scholar]
- 31. Hardikar AA, Marcus-Samuels B, Geras-Raaka E, Raaka BM, Gershengorn MC. Human pancreatic precursor cells secrete fgf2 to stimulate clustering into hormone-expressing islet-like cell aggregates. Proc Natl Acad Sci U S A. 2003;100(12):7117–7122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gershengorn MC, Hardikar AA, Wei C, Geras-Raaka E, Marcus-Samuels B, Raaka BM. Epithelial-to-mesenchymal transition generates proliferative human islet precursor cells. Science. 2004;306(5705):2261–2264. [DOI] [PubMed] [Google Scholar]
- 33. Joglekar MV, Patil D, Joglekar VM, Rao GV, Reddy DN, Mitnala S, Shouche Y, Hardikar AA. The mir-30 family micrornas confer epithelial phenotype to human pancreatic cells. Islets. 2009;1(2):137–147. [DOI] [PubMed] [Google Scholar]
- 34. Parekh VS, Joglekar MV, Hardikar AA. Differentiation of human umbilical cord blood-derived mononuclear cells to endocrine pancreatic lineage. Differentiation. 2009;78(4):232–240. [DOI] [PubMed] [Google Scholar]
- 35. Phadnis SM, Joglekar MV, Dalvi MP, Muthyala S, Nair PD, Ghaskadbi SM, Bhonde RR, Hardikar AA. Human bone marrow-derived mesenchymal cells differentiate and mature into endocrine pancreatic lineage in vivo. Cytotherapy. 2011;13(3):279–293. [DOI] [PubMed] [Google Scholar]
- 36. Williams MD, Wong W, Rixon A, Satoor SN, Hardikar AA, Joglekar MV. Pdx1 (gfp/w) mice for isolation, characterization, and differentiation of pancreatic progenitor cells. Methods Mol Biol. 2014;1194:271–288. [DOI] [PubMed] [Google Scholar]
- 37. Psaltis PJ, Paton S, See F, Arthur A, Martin S, Itescu S, Worthley SG, Gronthos S, Zannettino AC. Enrichment for stro-1 expression enhances the cardiovascular paracrine activity of human bone marrow-derived mesenchymal cell populations. J Cell Physiol. 2010;223(2):530–540. [DOI] [PubMed] [Google Scholar]
- 38. Holland AM, Micallef SJ, Li X, Elefanty AG, Stanley EG. A mouse carrying the green fluorescent protein gene targeted to the pdx1 locus facilitates the study of pancreas development and function. Genesis. 2006;44(6):304–307. [DOI] [PubMed] [Google Scholar]
- 39. Joglekar MV, Hardikar AA. Epithelial-to-mesenchymal transition in pancreatic islet beta cells. Cell Cycle. 2010;9(20):4077–4079. [DOI] [PubMed] [Google Scholar]
- 40. Gershengorn MC, Geras-Raaka E, Hardikar AA, Raaka BM. Are better islet cell precursors generated by epithelial-to-mesenchymal transition? Cell Cycle. 2005;4(3):380–382. [DOI] [PubMed] [Google Scholar]
- 41. Schellenberg A, Stiehl T, Horn P, Joussen S, Pallua N, Ho AD, Wagner W. Population dynamics of mesenchymal stromal cells during culture expansion. Cytotherapy. 2012;14(4):401–411. [DOI] [PubMed] [Google Scholar]
- 42. Hutley LJ, Newell FS, Kim YH, Luo X, Widberg CH, Shurety W, Prins JB, Whitehead JP. A putative role for endogenous fgf-2 in fgf-1 mediated differentiation of human preadipocytes. Mol Cell Endocrinol. 2011;339(1–2):165–171. [DOI] [PubMed] [Google Scholar]
- 43. Hart AW, Baeza N, Apelqvist A, Edlund H. Attenuation of fgf signalling in mouse beta-cells leads to diabetes. Nature. 2000;408(6814):864–868. [DOI] [PubMed] [Google Scholar]
- 44. Dalvi MP, Umrani MR, Joglekar MV, Hardikar AA. Human pancreatic islet progenitor cells demonstrate phenotypic plasticity in vitro. J Biosci. 2009;34(4):523–528. [DOI] [PubMed] [Google Scholar]
- 45. Ameri J, Stahlberg A, Pedersen J, Johansson JK, Johannesson MM, Artner I, Semb H. Fgf2 specifies hesc-derived definitive endoderm into foregut/midgut cell lineages in a concentration-dependent manner. Stem Cells. 2010;28(1):45–56. [DOI] [PubMed] [Google Scholar]
- 46. Tang W, Qin J, Tang J, Zhang H, Zhou Z, Li B, Geng Q, Wu W, Xia Y, Xu X. Aberrant reduction of mir-141 increased cd47/cul3 in hirschsprung’s disease. Cell Physiol Biochem. 2013;32(6):1655–1667. [DOI] [PubMed] [Google Scholar]
- 47. Xu G, Stoffers DA, Habener JF, Bonner-Weir S. Exendin-4 stimulates both beta-cell replication and neogenesis, resulting in increased beta-cell mass and improved glucose tolerance in diabetic rats. Diabetes. 1999;48(12):2270–2276. [DOI] [PubMed] [Google Scholar]
- 48. Abraham EJ, Leech CA, Lin JC, Zulewski H, Habener JF. Insulinotropic hormone glucagon-like peptide-1 differentiation of human pancreatic islet-derived progenitor cells into insulin-producing cells. Endocrinology. 2002;143(8):3152–3161. [DOI] [PubMed] [Google Scholar]
- 49. Goke R, Fehmann HC, Linn T, Schmidt H, Krause M, Eng J, Goke B. Exendin-4 is a high potency agonist and truncated exendin-(9-39)-amide an antagonist at the glucagon-like peptide 1-(7-36)-amide receptor of insulin-secreting beta-cells. J Biol Chem. 1993;268(26):19650–19655. [PubMed] [Google Scholar]
- 50. Serre V, Dolci W, Schaerer E, Scrocchi L, Drucker D, Efrat S, Thorens B. Exendin-(9-39) is an inverse agonist of the murine glucagon-like peptide-1 receptor: Implications for basal intracellular cyclic adenosine 3’,5’-monophosphate levels and beta-cell glucose competence. Endocrinology. 1998;139(11):4448–4454. [DOI] [PubMed] [Google Scholar]
- 51. Kilimnik G, Jo J, Periwal V, Zielinski MC, Hara M. Quantification of islet size and architecture. Islets. 2012;4(2):167–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kim A, Miller K, Jo J, Kilimnik G, Wojcik P, Hara M. Islet architecture: a comparative study. Islets. 2009;1(2):129–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Joglekar MV, Wong W, Maynard CL, Umrani MR, Martin D, Loudovaris T, Thomas HE, Dalgaard LT, Hardikar AA. Expression of mir-206 in human islets and its role in glucokinase regulation. Am J Physiol Endocrinol Metab. 2018. [DOI] [PubMed] [Google Scholar]
- 54. Joglekar MV, Trivedi PM, Kay TW, Hawthorne WJ, O’Connell PJ, Jenkins AJ, Hardikar AA, Thomas HE. Human islet cells are killed by bid-independent mechanisms in response to fas ligand. Apoptosis. 2016;21(4):379–389. [DOI] [PubMed] [Google Scholar]
- 55. McGlone ER, Tan TM. Of mice not men? Actions of interleukin-6 on glucose tolerance. Cell Metab. 2018;27(6):1157–1158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material, FigureS1_Hardikar for Epigenetic and Transcriptome Profiling Identifies a Population of Visceral Adipose-Derived Progenitor Cells with the Potential to Differentiate into an Endocrine Pancreatic Lineage by Michael D. Williams, Mugdha V. Joglekar, Sarang N. Satoor, Wilson Wong, Effie Keramidaris, Amanda Rixon, Philip O’Connell, Wayne J. Hawthorne, Geraldine M. Mitchell, and Anandwardhan A. Hardikar in Cell Transplantation
Supplemental Material, FigureS2_Hardikar for Epigenetic and Transcriptome Profiling Identifies a Population of Visceral Adipose-Derived Progenitor Cells with the Potential to Differentiate into an Endocrine Pancreatic Lineage by Michael D. Williams, Mugdha V. Joglekar, Sarang N. Satoor, Wilson Wong, Effie Keramidaris, Amanda Rixon, Philip O’Connell, Wayne J. Hawthorne, Geraldine M. Mitchell, and Anandwardhan A. Hardikar in Cell Transplantation
Supplemental Material, Figure_S3 for Epigenetic and Transcriptome Profiling Identifies a Population of Visceral Adipose-Derived Progenitor Cells with the Potential to Differentiate into an Endocrine Pancreatic Lineage by Michael D. Williams, Mugdha V. Joglekar, Sarang N. Satoor, Wilson Wong, Effie Keramidaris, Amanda Rixon, Philip O’Connell, Wayne J. Hawthorne, Geraldine M. Mitchell, and Anandwardhan A. Hardikar in Cell Transplantation
Supplemental Material, Supplementary_Table_1_new for Epigenetic and Transcriptome Profiling Identifies a Population of Visceral Adipose-Derived Progenitor Cells with the Potential to Differentiate into an Endocrine Pancreatic Lineage by Michael D. Williams, Mugdha V. Joglekar, Sarang N. Satoor, Wilson Wong, Effie Keramidaris, Amanda Rixon, Philip O’Connell, Wayne J. Hawthorne, Geraldine M. Mitchell, and Anandwardhan A. Hardikar in Cell Transplantation