Abstract
Chronic lymphocytic leukemia (CLL) is the most frequent leukemia type in which the genetic alterations influencing the clinico‐biological course are not entirely understood. CLL has a heterogeneous course, with some patients showing an indolent course and others experiencing an aggressive course. Whole‐genome sequencing and whole‐exome sequencing studies identified recurrently mutated genes in CLL and profiled its clonal evolution patterns. However, more recent whole‐genome sequencing studies also identified variants in non‐coding sequences of the CLL genome, revealing important lesions outside the protein‐coding regions. Here we describe the most representative non‐coding lesion of the CLL genome, including lesions in the 3′‐UTR region of NOTCH1 which result in the truncation of the NOTCH1 protein PEST domain, and non‐coding mutations in an enhancer region on chromosome 9p13 which result in reduced expression of the PAX5 transcription factor. In addition, we describe the role of microRNA in CLL, in particular the miR15a/miR16‐1 microRNA recurrently affected by deletions of chromosome 13q14. Together, new findings in non‐coding genome genetic lesions provide a more complete portrait of the genomic landscape of CLL with clinical implications.
Keywords: chronic lymphocytic leukemia, mutational analysis, non‐coding region
Abbreviations
- AID
activation‐induced deaminase
- BCR
B‐cell receptor
- BCL2
B‐cell lymphoma 2
- CLL
chronic lymphocytic leukemia
- DLBCL
diffuse large B‐cell lymphoma
- FL
follicular lymphoma
- IGHV
immunoglobulin heavy variable
- IL
interleukin
- MCL
mantle cell lymphoma
- M‐CLL
mutated chronic lymphocytic leukemia
- NF‐κB
nuclear factor‐κB
- NGS
next generation sequencing
- PFS
progression‐free survival
- SHM
somatic hypermutation
- U‐CLL
unmutated chronic lymphocytic leukemia
- WES
whole‐exome sequencing
- WGS
whole‐genome sequencing
1. Introduction
Chronic lymphocytic leukemia (CLL) is a common B‐cell tumor of adults; at diagnosis, the median age of patients is 72 years. Patients have been classically categorized depending on B‐cell receptor (BCR) immunoglobulin expression: the immunoglobulin heavy‐chain variable region gene (IGHV) harboring somatic hypermutation (SHM; IGHV‐mutated) group and the immunoglobulin heavy‐chain variable region gene harboring unmutated (IGHV‐unmutated) group (Damle et al., 1999; Hamblin et al., 1999). Patients with tumor clones with < 2% difference from germline or no mutation in the IGHV‐unmutated gene have a poorer prognosis compared with patients with the IGHV‐mutated gene (Damle et al., 1999; Hamblin et al., 1999).
Deletion of chromosome 13q14, del(13q14), is the most frequent cytogenetic aberration in CLL, occurring in ~ 55% of newly presented cases; if occurring as the sole genetic abnormality, it is associated with a benign course. Deletion of chromosome 11q, del(11q), occurs in ~ 25% of progressive but previously untreated patients, and in ~ 10% of early‐stage patients (Quesada et al., 2011; Zenz et al., 2010). Deletion 11q targets the ATM gene, which encodes for the proximal DNA damage response kinase ATM. Trisomy 12 occurs in ~ 15% of newly presented cases (Seiffert et al., 2012). Deletion of chromosome 17p, del(17p), occurs in ~ 7% of newly presented cases. Deletion of chromosome 17p targets the TP53 gene and is associated with chemoresistance (Hallek et al., 2010). Genomic studies have disclosed the complexity of cancer clonal architecture and identified several genetic prognostic biomarkers that are significantly associated with CLL overall survival, time to first treatment in cases managed with watch‐and‐wait, or progression‐free survival (PFS) in treated cases (Crespo et al., 2003; Damle et al., 1999; Döhner et al., 2000; Malek, 2013).
Whole genome/exome sequencing (WES/WGS) studies in CLL revealed recurrently mutated driver genes such as NOTCH1, MYD88, TP53, ATM, SF3B1, FBXW7, POT1, CHD2, RPS15, IKZF3, ZNF292, ZMYM3, ARID1A and PTPN11 (Fabbri et al., 2011; Landau et al., 2015; Puente et al., 2011, 2015; Quesada et al., 2011; Ramsay et al., 2013; Rossi et al., 2012). Important benefits of WGS studies included the identification of variants in non‐coding sequences of the CLL genome, revealing important lesions outside the protein‐coding regions which could help to disclose, together with the coding genetic lesion, the complexity of the CLL genetic landscape.
2. The miR‐15a and miR16‐1 in CLL
The identification of the epicenter of the minimal deleted region loss, ~ 30 kb on chromosome 13q, revealed the first example of non‐coding region alteration in CLL. The del13q14 leads to the monoallelic loss of the microRNA, miR15a and miR16‐1 (Fig. 1) (Calin et al., 2002). In normal cells, miR15a and miR16‐1 downmodulate at the post‐transcriptional level the expression of key regulators of apoptosis and cell cycle. The notion that miR15a/16‐1 and BCL2 expression levels are inversely correlated in CLL, and that downregulation of miR15a/16‐1 results in an increase of B‐cell lymphoma 2 (BCL2) expression, with consequent inhibition of apoptosis, led to the identification of BCL2 as the primary target of miR15a/16‐1 (Table 1) (Cimmino et al., 2005; Cory and Adams 2002, 2005; Sanchez‐Beato et al., 2003). Consistently, the miR15a/16‐1 consensus regions on the BCL2 mRNA disclosed that these two microRNA species are direct negative regulators of BCL2 at the post‐transcriptional level (Fig. 1) (Calin et al., 2008). The miR15a/16‐1 conditional deletion in mouse B‐cells results in the development of a CLL‐like monoclonal CD5+ lymphocyte proliferation in 40% of mice, proof of its involvement in CLL pathogenesis (Klein et al., 2010).
Figure 1.
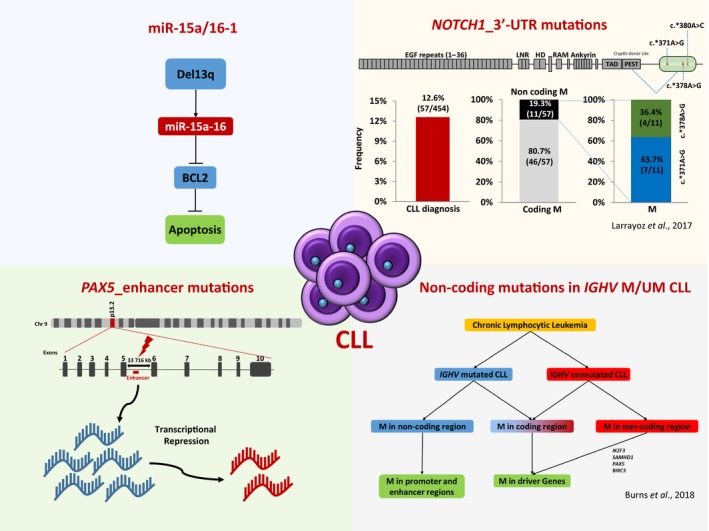
Non‐coding mutations in CLL. Portrayal of the most representative non‐coding lesion of the CLL genome.
Table 1.
Summary of non‐coding lesions in CLL
| Reference | Genomic region | Genes | Mutations | Functional consequences | Pathways |
|---|---|---|---|---|---|
| Cimmino et al. (2005) | del13q14 | miR‐15a miR16‐1 | – | miR‐15a/16‐1 downregulation in leukemic cell lines resulted in an increase of BCL2 expression with consequent inhibition of apoptosis | Cell cycle |
| Klein et al. (2010) | del13q14 | miR‐15a miR16‐1 | – | miR‐15a/16‐1 deletion in mice developed a CLL‐like monoclonal CD5+ lymphocyte proliferation | Cell cycle |
| Puente et al. (2015) | 3′‐UTR | NOTCH1 | c.*371A>G | This mutation is predicted to remove a PEST domain of NOTCH1 and to increase protein stability | NOTCH |
| Puente et al. (2015) | 9p13 | PAX5 | – | Decrease in PAX5 expression | BCR |
3. NOTCH1 coding and non‐coding mutations
NOTCH1 encodes a class I transmembrane protein which acts as a ligand‐activated transcription factor that plays a key role in cell proliferation, differentiation and apoptosis (Paganin and Ferrando, 2011). NOTCH1 binds to its ligand and then undergoes proteolytic cleavages which enable its intracellular domain to translocate into the nucleus to mediate transcriptional activation of multiple target genes, including TP53, MYC and genes which encode components of the nuclear factor κB (NF‐κB) pathway. Most CLL mutations which affect NOTCH1 are frameshift or nonsense events which are clustered within exon 34 (including a highly recurrent c.7544_7545delCT deletion) and are selected to disrupt the PEST domain of the protein (Fabbri et al., 2011; Rossi et al., 2012; Sportoletti et al., 2010). As the PEST domain is required to limit both the intensity and duration of NOTCH1 signaling activation, its removal is predicted to impair NOTCH1 degradation, thereby resulting in the accumulation of an active NOTCH1 isoform and subsequent deregulation of signaling (Fabbri et al., 2011, 2017; Sportoletti et al., 2010). Consistent with this prediction, multiple cellular pathways, including those controlling cell metabolism and cell cycle progression, are deregulated in CLL harboring NOTCH1 mutations (Del Giudice et al., 2012; Fabbri et al., 2017; Puente et al., 2011). NOTCH1 is preferentially targeted in specific clinical and biological groups of CLL (Sportoletti et al., 2010), including cases that have developed into Richter syndrome (Rossi et al., 2012; Villamor et al., 2013), cases harboring unmutated‐IGHV genes, cases harboring a subset eight BCR configuration, and cases harboring trisomy of chromosome 12 (Del Giudice et al., 2012; Fabbri et al., 2011; Rossi et al., 2012, 2013; Sportoletti et al., 2010).
Among CLL harboring NOTCH1 mutations, treatment with FCR, R‐Clb or O‐Clb does not result in the expected increase in PFS compared with treatment with FC or with chlorambucil alone (Stilgenbauer et al., 2014). These observations point to NOTCH1 mutations as a biomarker of resistance to the anti‐CD20 antibodies rituximab and ofatumumab in CLL. The outcome of CLL patients treated with obinutuzumab combined with chlorambucil improves independently of NOTCH1 mutation status, suggesting that the augmented cytotoxicity of obinutuzumab or the increased dose of the anti‐(CD20) IgG1 antibody used in the obinutuzumab‐chlorambucil schema overcomes NOTCH1 mutation‐associated resistance to rituximab (Estenfelder et al., 2016). The mechanism underlying the anti‐CD20 refractoriness associated with NOTCH1 mutations remains obscure.
NOTCH1 is broadly activated in CLL, where most of the cases express the intracellular active portion of the NOTCH1 protein, despite the absence of coding gene mutations. A proportion of cases lacking coding gene mutations but having biochemical clues of NOTCH1 activation is now justified by the occurrence of 3′‐UTR mutations in the non‐coding region of the NOTCH1 exon 34 (Fabbri et al., 2017). Non‐coding mutations in the 3′‐UTR of NOTCH1 (c.*371A>G) have been also described in 2–4% of cases of CLL (Fig. 1) (Bittolo et al., 2017; Nadeu et al., 2016; Puente et al., 2015). The 3′‐UTR of NOTCH1 mutation leads to a novel splicing event between a cryptic donor site located in the coding region of NOTCH1 exon 34 and a newly created acceptor site in the 3′‐UTR, resulting in a deletion that includes the last 158 coding bases. This within‐exon splicing is predicted to remove a PEST domain of NOTCH1 and to increase protein stability, as previously described for NOTCH1 mutation affecting exon 34 (Table 1) (Rossi et al., 2012). The 3′‐UTR mutations are mutually exclusive of other NOTCH1 somatic variants, consistent with the notion that they are selected by the tumor as an alternative genetic mechanism of the PEST domain deletion. Indeed, CLL cells having the 3′‐UTR mutations or the exon 34 coding mutations display constitutive levels of cleaved and active NOTCH1 protein (D'Agaro et al., 2017). Besides mimicking the biological effect of exon 34 mutations, the 3′‐UTR non‐coding mutation of NOTCH1 also has the same clinical consequences (Puente et al., 2015).
3.1. Clinical impact of NOTCH1 non‐coding mutations
NOTCH1 coding mutations identified in CLL affect exon 34 and include the highly recurrent c.7544_7545delCT deletion (Arruga et al., 2014; Puente et al., 2011; Rosati et al., 2009). At diagnosis, these mutations occur in ~ 8% of cases and have a high prevalence in advanced disease stages, in treatment‐refractory disease and after transformation to Richter syndrome (Baliakas et al., 2015; Oscier et al., 2013). The prognostic value of NOTCH1 non‐coding mutations was validated in chemotherapy first‐line treatment patients by the UK CLL4 trial study (Larrayoz et al., 2017) (Fig. 1). That study also showed that NOTCH1 non‐coding mutations together with coding mutation increase the power to predict outcomes in CLL patients. Taken together, these studies support analysis of NOTCH1 non‐coding region in order to stratify reduced survival patients better and to identify patients predestined to respond poorly to rituximab treatments (Stilgenbauer et al., 2014).
4. Non‐coding mutations affecting PAX5 gene
The B‐cell specific activator protein, also known as PAX5, is a transcription factor and is an important B‐cell precursor for normal B‐cell differentiation and maturation (Nutt et al., 2001). PAX5 gene expression is involved in IGHV gene rearrangement, BCR signal transduction and B‐cell survival, so deletion or inactivation of PAX5 gene led to cell arrest in Pro‐B‐cell stage. PAX5 heterozygous mice showed an accumulation of interleukin (IL)7‐dependent proB cells and developed B‐ALL when endangered by infections (Martin‐Lorenzo et al., 2015). Furthermore, PAX5 translocations and mutations have been observed in B‐cell lymphomas and B‐ALL (Mullighan et al., 2007; Poppe et al., 2005).
PAX5 recurrent mutations in enhancer non‐coding regions have been recently associated to activity alterations of the gene (Fig. 1) (Puente et al., 2015). A large WGS study of 150 CLL patients identified non‐coding mutations in the PAX5 enhancer region on chromosome 9p13. Patients harboring non‐coding mutations in this region showed a pronounced decrease in PAX5 expression compared with PAX5 wild‐type patients (Table 1) (Puente et al., 2015). By using the CRISPR/Cas9 approach to target the PAX5 enhancer region in an indicative cell line, they also showed a decrease in PAX5 expression. The study identified 42/506 (8%) CLL‐mutated samples in other B‐cell lymphomas such as diffuse large B‐cell lymphoma (DLBCL) in 29% of cases, follicular lymphoma (FL) in 23% of cases and mantle cell lymphoma (MCL) in 5% of cases. CLL patients harboring PAX5 non‐coding mutations were preferentially associated to the IGHV‐mutated subgroup but were not related to other recurrent CLL mutations, with the exception of del13q14, suggesting that PAX5 non‐coding mutations are early events in the development of the disease. A second study identified non‐coding PAX5 enhancer mutations in 3/13 (23%) of CLL cases. In contrast to the first description of PAX5 mutations, in this study the identified PAX5 mutations co‐existed with MYD88, ATM, NOTCH1, SF3B1 and ZMYM3 mutations (Rose‐Zerilli et al., 2016). Finally, a recent study of 46 CLL cases identified PAX5 non‐coding mutations in 17.4% of CLL cases, which correlated with non‐coding elements of transcription elongation sites, and promoter and enhancer regions. These PAX5 promoter mutations were found in 22% of IGHV unmutated patients, confirming previous studies (Burns et al., 2018).
5. Immunoglobulin gene mutations
In normal B lymphocytes, activation‐induced deaminase (AID) is required for the productive generation of antibody diversity by inducing SHM of IGV region and by mediating IGH class‐switch recombination during the development of protective effector mechanisms (Peled et al., 2008; Stavnezer et al., 2008). The on‐target AID activities consist in the conversion of cytidine to uridine on single‐stranded DNA at the IG locus during germinal center reaction. Functional evidence indicates that BCR pathway activation in CLL derives from contacts between tumor cells and antigens, which are influenced, among other factors, by the SHM action of the rearranged IGHV genes (Vardi et al., 2014). The IGHV genes of CLL can accumulate variations as a consequence of the SHM process. The prevalence of mutated IGHV genes is higher among newly diagnosed and asymptomatic CLL patients (~ 60%), whereas the prevalence of IGHV unmutated genes is higher among progressive (~ 50–60%) and relapsed/refractory (~ 70–80%) CLL patients.
Whole‐genome sequencing analysis of a variety of human tumors revealed a new type of off‐target AID activity and related deaminases, revealing multiple mutation clusters of < 10 kb in most of the tumors analyzed (Alexandrov et al., 2013; Nik‐Zainal et al., 2012). Localized regions of increased mutation density from random substitutions are called kataegis sites and are typically scattered across tumor genomes at a distance of ~ 0.1–1 Mb from each other, revealing 70% multiple mutation cytidine to thymidine transitions clusters. Kataegis mutations occurred in the same DNA strand, by catalytic processivity, and have frequently been associated with genomic rearrangements (Stephens et al., 2011). On these bases, kataegis was proposed to be the result of processive cytidine deamination of single‐strand DNA exposed by the resection of double‐strand breaks during DNA repair (Sakofsky et al., 2014).
A recent WGS study of 46 CLL patients provided a complete description of non‐coding mutation landscapes of both mutated and unmutated IGHV CLL (Burns et al., 2018) (Fig. 1) The study demonstrated that ~ 25% of kataegis non‐coding mutations outside the immunoglobulin loci occurred in genes relevant to CLL. They identified non‐coding mutations in the ATM gene that may negatively impact on ATM expression and found non‐coding mutations in the regulatory region of TCL1A gene. In particular, analysis of IGHV unmutated CLL cases revealed additional non‐coding mutations in CLL driver genes such as IKZF3, SAMHD1, PAX5 and BIRC3. Finally, they found that IGHV unmutated CLL harbored coding mutations in driver genes, whereas IGHV‐mutated CLL harbored non‐coding promoter and enhancer mutations caused by aberrant AID activity (Burns et al., 2018). Finally, they observed that recurrently non‐coding mutated regions harbored in CLL patients were associated with the concomitant presence of coding mutations, suggesting a functional relevance in CLL pathogenesis.
6. Concluding remarks
Current therapeutic approaches involve target proteins; for this reason, non‐coding mutations have only been studied for research purposes and not for medicine cancer care in the clinic. Although non‐coding mutations are related to the protein‐coding gene expressions they regulate, studies of these mutations might help to identify suitable therapeutic approaches to target linked proteins. In CLL, the presence of different genomic lesions demonstrated the enormous biological heterogeneity of this tumor. WGS studies identified non‐coding recurrent mutations, including the 3′‐UTR of NOTCH1 and a PAX5 enhancer, resulting in significant activity alterations of these transcription factors genes of well‐known importance in leukemia and other malignancies (Lobry et al., 2011; O'Brien et al., 2011). Previous studies have shown the effect of NOTCH1 mutations in CLL prognosis (Puente et al., 2011; Villamor et al., 2013). However, these studies may seriously underestimate the true incidence of NOTCH1 deregulation in CLL, considering that ~ 20% of NOTCH1‐mutated tumors were also mutated in the 3′‐UTR region. The function of microRNA to regulate gene expression is essential to provide fine control of several cell processes, and deregulation of microRNA may be involved in CLL development/progression. However, more studies are necessary to determine whether microRNA from CLL cells can be used in clinical practice. These independent CLL cohort studies have revealed new driver lesions involved in CLL evolution, helping to clarify the clinical impact of the heterogeneous molecular composition of the disease, resulting in new opportunities for improving the clinical management and personalized treatment of CLL patients.
Authors contributions
Both authors contributed to the writing of this review article.
Conflict of interest
The authors have no conflicts of interest to declare.
References
- Alexandrov LB, Nik‐Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Børresen‐Dale AL et al (2013) Signatures of mutational processes in human cancer. Nature 500, 415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arruga F, Gizdic B, Serra S, Vaisitti T, Ciardullo C, Coscia M, Laurenti L, D'Arena G, Jaksic O, Inghirami G et al (2014) Functional impact of NOTCH1 mutations in chronic lymphocytic leukemia. Leukemia 28, 1060–1070. [DOI] [PubMed] [Google Scholar]
- Baliakas P, Hadzidimitriou A, Sutton L‐A, Rossi D, Minga E, Villamor N, Larrayoz M, Kminkova J, Agathangelidis A, Davis Z et al (2015) Recurrent mutations refine prognosis in chronic lymphocytic leukemia. Leukemia 29, 329–336. [DOI] [PubMed] [Google Scholar]
- Bittolo T, Pozzo F, Bomben R, D'Agaro T, Bravin V, Bulian P, Rossi FM, Zucchetto A, Degan M, Macor P et al (2017) Mutations in the 3′ untranslated region of NOTCH1 are associated with low CD20 expression levels chronic lymphocytic leukemia. Haematologica 102, e305–e309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns A, Alsolami R, Becq J, Stamatopoulos B, Timbs A, Bruce D, Robbe P, Vavoulis D, Clifford R, Cabes M et al (2018) Whole‐genome sequencing of chronic lymphocytic leukaemia reveals distinct differences in the mutational landscape between IgHVmut and IgHVunmut subgroups. Leukemia 32, 573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K et al (2002) Frequent deletions and down‐regulation of micro‐RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA 99, 15524–15529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Cimmino A, Fabbri M, Ferracin M, Wojcik SE, Shimizu M, Taccioli C, Zanesi N, Garzon R, Aqeilan RI et al (2008) MiR‐15a and miR‐16‐1 cluster functions in human leukemia. Proc Natl Acad Sci U S A 105, 5166–5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M et al (2005) miR‐15 and miR‐16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA 102, 13944–13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cory S and Adams JM (2002) The Bcl2 family: regulators of the cellular life‐or‐death switch. Nat Rev Cancer 2, 647–656. [DOI] [PubMed] [Google Scholar]
- Cory S and Adams JM (2005) Killing cancer cells by flipping the Bcl‐2/Bax switch. Cancer Cell 8, 5–6. [DOI] [PubMed] [Google Scholar]
- Crespo M, Bosch F, Villamor N, Bellosillo B, Colomer D, Rozman M, Marcé S, López‐Guillermo A, Campo E, Montserrat E (2003) ZAP‐70 expression as a surrogate for immunoglobulin‐variable region mutations in chronic lymphocytic leukemia. N Engl J Med 348, 1764–1775. [DOI] [PubMed] [Google Scholar]
- D'Agaro T, Bittolo T, Bravin V, Dal Bo M, Pozzo F, Bulian P, Rossi FM, Zucchetto A, Degan M, D'Arena G et al (2017) NOTCH1 mutational status in chronic lymphocytic leukaemia: clinical relevance of subclonal mutations and mutation types. Br J Haematol 182, 597–602. [DOI] [PubMed] [Google Scholar]
- Damle RN, Wasil T, Fais F, Ghiotto F, Valetto A, Allen SL, Buchbinder A, Budman D, Dittmar K, Kolitz J et al (1999) Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood 94, 1840–1847. [PubMed] [Google Scholar]
- Del Giudice I, Rossi D, Chiaretti S, Marinelli M, Tavolaro S, Gabrielli S, Laurenti L, Marasca R, Rasi S, Fangazio M et al (2012) NOTCH1 mutations in +12 chronic lymphocytic leukemia (CLL) confer an unfavorable prognosis, induce a distinctive transcriptional profiling and refine the intermediate prognosis of +12 CLL. Haematologica 97, 437–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Döhner H, Stilgenbauer S, Benner A, Leupolt E, Kröber A, Bullinger L, Döhner K, Bentz M and Lichter P (2000) Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med 343, 1910–1916. [DOI] [PubMed] [Google Scholar]
- Estenfelder S, Tausch E, Robrecht S, Bahlo J, Goede V, Ritgen M, van Dongen JJM, Langerak AW, Fingerle‐Rowson G, Kneba M et al (2016) Gene mutations and treatment outcome in the context of chlorambucil (Clb) without or with the addition of rituximab (R) or obinutuzumab (GA‐101, G): results of an extensive analysis of the phase III study CLL11 of the German CLL Study Group. Blood 128, Abstract 3227. [Google Scholar]
- Fabbri G, Rasi S, Rossi D, Trifonov V, Khiabanian H, Ma J, Grunn A, Fangazio M, Capello D, Monti S et al (2011) Analysis of the chronic lymphocytic leukemia coding genome: role of NOTCH1 mutational activation. J Exp Med 208, 1389–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabbri G, Holmes AB, Viganotti M, Scuoppo C, Belver L, Herranz D, Yan XJ, Kieso Y, Rossi D, Gaidano G et al (2017) Common non mutational NOTCH1 activation in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A 114, E2911–E2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallek M, Fischer K, Fingerle‐Rowson G, Fink AM, Busch R, Mayer J, Hensel M, Hopfinger G, Hess G, von Grünhagen U et al (2010) Addition of rituximab to fludarabine and cyclophosphamide in patients with chronic lymphocytic leukaemia: a randomised, open‐label, phase 3 trial. Lancet 376, 1164–1174. [DOI] [PubMed] [Google Scholar]
- Hamblin TJ, Davis Z, Gardiner A, Oscier DG and Stevenson FK (1999) Unmutated Ig V(H) genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood 94, 1848–1854. [PubMed] [Google Scholar]
- Klein U, Lia M, Crespo M, Siegel R, Shen Q, Mo T, Ambesi‐Impiombato A, Califano A, Migliazza A, Bhagat G et al (2010) The DLEU2/miR‐15a/16‐1 cluster controls B cell proliferation and its deletion leads to chronic lymphocytic leukemia. Cancer Cell 17, 28–40. [DOI] [PubMed] [Google Scholar]
- Landau DA, Tausch E, Taylor‐Weiner AN, Stewart C, Reiter JG, Bahlo J, Kluth S, Bozic I, Lawrence M, Böttcher S et al (2015) Mutations driving CLL and their evolution in progression and relapse. Nature 526, 525–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larrayoz M, Rose‐Zerilli MJ, Kadalayil L, Parker H, Blakemore S, Forster J, Davis Z, Steele AJ, Collins A, Else M et al (2017) Non‐coding NOTCH1 mutations in chronic lymphocytic leukemia; their clinical impact in the UK CLL4 trial. Leukemia 31, 510–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobry C, Oh P and Aifantis I (2011) Oncogenic and tumor suppressor functions of Notch in cancer: it's NOTCH what you think. J Exp Med 208, 1931–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malek SN (2013) The biology and clinical significance of acquired genomic copy number aberrations and recurrent gene mutations in chronic lymphocytic leukemia. Oncogene 32, 2805–2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin‐Lorenzo A, Hauer J, Vicente‐Dueñas C, Auer F, González‐Herrero I, García‐Ramírez I, Ginzel S, Thiele R, Constantinescu SN, Bartenhagen C et al (2015) Infection exposure is a causal factor in B precursor acute lymphoblastic leukemia as a result of Pax5 inherited susceptibility. Cancer Discov 5, 1328–1343. [DOI] [PubMed] [Google Scholar]
- Mullighan CG, Goorha S, Radtke I, Miller CB, Coustan‐Smith E, Dalton JD, Girtman K, Mathew S, Ma J, Pounds SB et al (2007) Genome‐wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature 446, 7582764. [DOI] [PubMed] [Google Scholar]
- Nadeu F, Delgado J, Royo C, Baumann T, Stankovic T, Pinyol M, Jares P, Navarro A, Martín‐García D, Beà S et al (2016) Clinical impact of clonal and subclonal TP53, SF3B1, BIRC3, NOTCH1, and ATM mutations in chronic lymphocytic leukemia. Blood 127, 2122–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nik‐Zainal S, Alexandrov LB, Wedge DC, Van Loo P, Greenman CD, Raine K, Jones D, Hinton J, Marshall J, Stebbings LA et al (2012) Mutational processes molding the genomes of 21 breast cancers. Cell 149, 979–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutt SL, Eberhard D, Horcher M, Rolink AG and Busslinger M (2001) Pax5 determines the identity of B cells from the beginning to the end of B‐lymphopoiesis. Int Rev Immunol. 20, 65–82. [DOI] [PubMed] [Google Scholar]
- O'Brien P, Morin P Jr, Ouellette RJ and Robichaud GA (2011) The Pax‐5 gene: a pluripotent regulator of B‐cell differentiation and cancer disease. Cancer Res 71, 7345–7350. [DOI] [PubMed] [Google Scholar]
- Oscier DG, Rose‐Zerilli MJJ, Winkelmann N, Gonzalez de Castro D, Gomez B, Forster J, Parker H, Parker A, Gardiner A, Collins A et al (2013) The clinical significance of NOTCH1 and SF3B1 mutations in the UK LRF CLL4 trial. Blood 120, 4441–4443. [DOI] [PubMed] [Google Scholar]
- Paganin M and Ferrando A (2011) Molecular pathogenesis and targeted therapies for NOTCH1‐induced T‐cell acute lymphoblastic leukemia. Blood Rev 25, 83–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peled JU, Kuang FL, Iglesias‐Ussel MD, Roa S, Kalis SL, Goodman MF and Scharff MD (2008) The biochemistry of somatic hypermutation. Annu Rev Immunol 26, 481–511. [DOI] [PubMed] [Google Scholar]
- Poppe B, De Paepe P, Michaux L, Dastugue N, Bastard C, Herens C, Moreau E, Cavazzini F, Yigit N, Van Limbergen H et al (2005) PAX5/IGH rearrangement is a recurrent finding in a subset of aggressive B‐NHL with complex chromosomal rearrangements. Genes Chromosom Cancer 44, 2182223. [DOI] [PubMed] [Google Scholar]
- Puente XS, Beà S, Valdés‐Mas R, Villamor N, Gutiérrez‐Abril J, Martín‐Subero JI, Munar M, Rubio‐Pérez C, Jares P, Aymerich M et al (2015) Non‐coding recurrent mutations in chronic lymphocytic leukaemia. Nature 526, 519–524. [DOI] [PubMed] [Google Scholar]
- Puente XS, Pinyol M, Quesada V, Conde L, Ordóñez GR, Villamor N, Escaramis G, Jares P, Beà S, González‐Díaz M et al (2011) Whole‐genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature 475, 101–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada V, Conde L, Villamor N, Ordóñez GR, Jares P, Bassaganyas L, Ramsay AJ, Beà S, Pinyol M, Martínez‐Trillos A et al (2011) Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat Genet 44, 47–52. [DOI] [PubMed] [Google Scholar]
- Ramsay AJ, Quesada V, Foronda M, Conde L, Martínez‐Trillos A, Villamor N, Rodríguez D, Kwarciak A, Garabaya C, Gallardo M et al (2013) POT1 mutations cause telomere dysfunction in chronic lymphocytic leukemia. Nat Genet 45, 526–530. [DOI] [PubMed] [Google Scholar]
- Rosati E, Sabatini R, Rampino G, Tabilio A, Di Ianni M, Fettucciari K, Bartoli S, Coaccioli S, Screpanti I and Marconi P (2009) Constitutively activated Notch signaling is involved in survival and apoptosis resistance of B‐CLL cells. Blood 113, 856–865. [DOI] [PubMed] [Google Scholar]
- Rose‐Zerilli MJ, Gibson J, Wang J, Tapper W, Davis Z, Parker H, Larrayoz M, McCarthy H, Walewska R, Forster J et al (2016) Longitudinal copy number, whole exome and targeted deep sequencing of ‘good risk’ IGHV‐mutated CLL patients with progressive disease. Leukemia 30, 1301–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi D, Rasi S, Fabbri G, Spina V, Fangazio M, Forconi F, Marasca R, Laurenti L, Bruscaggin A, Cerri M et al (2012) Mutations of NOTCH1 are an independent predictor of survival in chronic lymphocytic leukemia. Blood 119, 521–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi D, Spina V, Bomben R, Rasi S, Dal‐Bo M, Bruscaggin A, Rossi FM, Monti S, Degan M, Ciardullo C et al (2013) Association between molecular lesions and specific B‐cell receptor subsets in chronic lymphocytic leukemia. Blood 121, 4902–4905. [DOI] [PubMed] [Google Scholar]
- Sakofsky CJ, Roberts SA, Malc E, Mieczkowski PA, Resnick MA, Gordenin DA and Malkova A (2014) Break‐induced replication is a source of mutation clusters underlying kataegis. Cell Rep 7, 1640–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez‐Beato M, Sanchez‐Aguilera A and Piris MA (2003) Cell cycle deregulation in B‐cell lymphomas. Blood 101, 1220–1235. [DOI] [PubMed] [Google Scholar]
- Seiffert M, Dietrich S, Jethwa A, Glimm H, Lichter P and Zenz T (2012) Exploiting biological diversity and genomic aberrations in chronic lymphocytic leukemia. Leuk Lymphoma 53, 1023–1031. [DOI] [PubMed] [Google Scholar]
- Sportoletti P, Baldoni S, Cavalli L, Del Papa B, Bonifacio E, Ciurnelli R, Bell AS, Di Tommaso A, Rosati E, Crescenzi B et al (2010) NOTCH1 PEST domain mutation is an adverse prognostic factor in B‐CLL. Br J Haematol 151, 404–406. [DOI] [PubMed] [Google Scholar]
- Stavnezer J, Guikema JE and Schrader CE (2008) Mechanism and regulation of class switch recombination. Annu Rev Immunol 26, 261–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens PJ, Greenman CD, Fu B, Yang F, Bignell GR, Mudie LJ, Pleasance ED, Lau KW, Beare D, Stebbings LA et al (2011) Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell 144, 27–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stilgenbauer S, Schnaiter A, Paschka P, Zenz T, Rossi M, Döhner K, Bühler A, Böttcher S, Ritgen M, Kneba M et al (2014) Gene mutations and treatment outcome in chronic lymphocytic leukemia: results from the CLL8 trial. Blood 123, 3247–3254. [DOI] [PubMed] [Google Scholar]
- Vardi A, Agathangelidis A, Sutton LA, Ghia P, Rosenquist R and Stamatopoulos K (2014) Immunogenetic studies of chronic lymphocytic leukemia: revelations and speculations about ontogeny and clinical evolution. Cancer Res 74, 4211–4216. [DOI] [PubMed] [Google Scholar]
- Villamor N, Conde L, Martínez‐Trillos A, Cazorla M, Navarro A, Beà S, López C, Colomer D, Pinyol M, Aymerich M et al (2013) NOTCH1 mutations identify a genetic subgroup of chronic lymphocytic leukemia patients with high risk of transformation and poor outcome. Leukemia 27, 1100–1106. [DOI] [PubMed] [Google Scholar]
- Zenz T, Mertens D, Küppers R, Döhner H and Stilgenbauer S (2010) From pathogenesis to treatment of chronic lymphocytic leukaemia. Nat Rev Cancer 10, 37–50. [DOI] [PubMed] [Google Scholar]


