Abstract
Purpose:
Poly (ADP-ribose) polymerase (PARP) inhibitors are undergoing clinical evaluation for cancer therapy. Because PARP inhibition has been shown to enhance tumor cell sensitivity to radiation, we investigated the in vitro and in vivo effects of the novel PARP inhibitor E7016.
Experimental Design:
The effect of E7016 on the in vitro radiosensitivity of tumor cell lines was evaluated using clonogenic survival. DNA damage and repair were measured using γH2AX foci and neutral comet assay. Mitotic catastrophe was determined by immunostaining. Tumor growth delay was evaluated in mice for the effect of E7016 on in vivo (U251) tumor radiosensitivity.
Results:
Cell lines exposed to E7016 preirradiation yielded an increase in radiosensitivity with dose enhancement factors at a surviving fraction of 0.1 from 1.4 to 1.7. To assess DNA doublestrand breaks repair, γH2AX measured at 24 hours postirradiation had significantly more foci per cell in the E7016/irradiation group versus irradiation alone. Neutral comet assay further suggested unrepaired double-strand breaks with significantly greater DNA damage at 6 hours postirradiation in the combination group versus irradiation alone. Mitotic catastrophe staining revealed a significantly greater number of cells staining positive at 24 hours postirradiation in the combination group. In vivo, mice treated with E7016/irradiation/temozolomide had an additional growth delay of six days compared with the combination of temozolomide and irradiation.
Conclusions:
These results indicate that E7016 can enhance tumor cell radiosensitivity in vitro and in vivo through the inhibition of DNA repair. Moreover, enhanced growth delay with the addition of E7016 to temozolomide and radiotherapy in a glioma mouse model suggests a potential role for this drug in the treatment of glioblastoma multiforme.
Glioblastoma multiforme continues to be the most common primary malignant brain tumor in adults and carries a dismal survival rate of only 3.3% at two years (1). Recent advances in the treatment of glioblastoma multiforme published in the landmark study by Stupp et al. changed the standard of care with the discovery that temozolomide given concurrently with radiotherapy significantly increased overall survival from 12.1 to 14.6 months compared with radiotherapy alone (2). Despite this success, survival remains low, prompting investigators to seek alternate treatments through a better understanding of the cell biology and through new molecular targets that may enhance current treatments.
Poly (ADP-ribose) polymerase (PARP)-1 is a nuclear protein involved in sensing and signaling the presence of DNA damage (3). Activation of PARP-1 leads to the addition of poly (ADP-ribose) branched chains from nicotinamide adenine dinucleotide onto damaged DNA, particularly at the histone H1 and H2B tails (4). This ribosylation leads to relaxation of the chromatin structure and recruitment of additional repair proteins, including XRCC1, Pol β, and DNA ligase (5). If the DNA single-strand breaks are not properly repaired they can be converted to double-strand breaks (DSB) during DNA replication leading to cell death (5). Studies with pharmacologic inhibitors of PARP activity have shown them to enhance the sensitivity of tumor cells to agents that cause DNA damage including both chemotherapy and radiation (6, 7). PARP-1 activity is particularly important in the base excision repair of N7-methyl guanine damage typically caused by alkylating agents, including temozolomide (8).
Although the chemical 3-aminobenzamide has been shown to inhibit DNA repair and enhance tumor cell killing by irradiation, it has a relatively low potency and nonspecific effects independent of PARP inhibition (9, 10). Consequently, a number of PARP inhibitors have recently been developed with the goal of enhancing the cytotoxic effects of both chemotherapy and radiotherapy (11). E7016 (formerly known as GPI21016) is a novel orally available PARP inhibitor that has been shown to have antitumor efficacy in murine xenograft studies.8 Moreover, in studies using an intracranial xenograft model with a similar compound, GPI 15427, this class of inhibitors was shown to potentiate the activity of temozolomide in an intracranial mouse model suggesting adequate brain penetration (12). As an initial step in testing E7016 as a clinically applicable PARP inhibitor for use in combination with radiotherapy, we investigated the effects of E7016 on the radiosensitivity of human tumor cell lines. The data presented indicate that E7016 enhances tumor cell radiosensitivity both in vitro and in vivo. Moreover, E7016 had a greater than additive effect in vivo when used in combination with temozolomide and radiation, the current standard of care in glioblastoma multiforme therapy.
Materials and Methods
Cell lines and treatment.
The U251 human glioblastoma cell line was obtained from the National Cancer Institute Frederick Tumor Repository and was grown in DMEM (Invitrogen) with 10% fetal bovine serum. MiaPaca and DU145 (human pancreatic and prostate carcinoma, respectively) cell lines were obtained from the American Type Culture Collection and grown in RPMI1640 media (Quality Biological) containing 2 mmol/L L-glutamine and 5% fetal bovine serum. Cultures were maintained at 37°C in an atmosphere of 5% CO2 and 95% room air. E7016, provided by Eisai, was reconstituted in DMSO (10 mmol/L) and stored at −80°C. Cultures were irradiated using a Pantak X-ray source at a dose rate of 2.28 Gy/min.
PARP inhibitor assay.
The enzymatic inhibition of poly (ADP-ribose) at varying concentrations of E7016 was compared with a standard, commercially available PARP inhibitor using a chemiluminescent PARP assay (Trevigen). Protocol was according to the manufacturer’s instructions without changes. 3-Aminobenzamide serially diluted from 200 mmol/L was used as a control inhibitor.
PARP activity in the U251 cell line treated with E7016 was also assessed using Trevigen chemiluminescent PARP assay. Briefly, at specified times posttreatment, cells were collected in PBS by cell scraping, transferred to 15-mL conical and centrifuged for 10 min at 1,500 rpm at 4°C. Pellet was resuspended in 40 μL Ripa buffer, sonicated for 10 s and centrifuged for 10 min at 13,000 rpm at 4°C. Supernatant was collected and protein quantification was done using Bradford assay. ADP-ribosylation reaction was carried out according to manufacturer’s protocol to determine activity of PARP in each sample.
Clonogenic assay.
Cultures were trypsinized to generate a single-cell suspension and a specified number of cells were seeded into each well of 6-well tissue culture plates. After allowing cells time to attach (16 h), E7016 or the vehicle control was added at specified concentrations and the plates were irradiated 6 h later. Ten to twelve days after seeding, colonies were stained with crystal violet, the number of colonies containing at least 50 cells was determined, and the surviving fractions were calculated. Survival curves were generated after normalizing for the cytotoxicity generated by E7016 alone. Data presented are the mean ± SE from at least three independent experiments.
Mitotic catastrophe.
The presence of fragmented nuclei was used as the criterion for defining cells undergoing mitotic catastrophe. To visualize mitotic catastrophe, cells grown in chamber slides were fixed with methanol for 15 min. at −20°C, stained with mouse monoclonal anti-α-tubulin (Sigma), followed by staining with Texas Red conjugated goat antimouse antibody (Jackson ImmunoResearch). Nuclei were counterstained with 4´,6-diamidino-2-phenylindole (Sigma). Cells were visualized on Leica microscope. Two fields selected at random contained 100 cells for each treatment group. The experiment was repeated thrice for a total of 300 cells per treatment group. Two or more distinct nuclear lobes within a single cell were scored as positive for mitotic catastrophe.
Apoptotic cell death.
To evaluate apoptosis as a mechanism of cell death, Guava Nexin assay was done. Cells were treated with E7016 (3 μmol/L) 6 h prior to irradiation and were stained at 24 and 72 h postirradiation. Staining was per manufacturer’s protocol, and apoptosis buffer, annexin V-PE, and 7-AAD were used. Samples were then analyzed on the Guava EasyCyte System.
Cell cycle analysis.
Evaluation of the cell cycle distribution was done using flow cytometry. Cells were seeded in 10-cm dishes and after 24 h treated with 3 μmol/L E7016, 2 Gy, or the combination. For a positive control, cells were treated with 0.2 μg/mL nocodazole for 24 h prior to analysis. Cells were fixed, stained with phospho-H3 (to distinguish mitotic cells) and cell cycle reagent (Guava Technologies), and analyzed on flow cytometer (Guava Technologies).
Immunofluorescent staining for γH2AX.
Cells were grown and treated in chamber slides at a concentration of 0.5 to 1.0 × 105 cells/ mL. At specified times, media were aspirated and cells were fixed with 2% paraformaldehyde for 15 min at room temperature. Cells were washed thrice with PBS, and permeablized with 1% Triton-X-100 for 5 min on ice. Cells were then washed thrice with PBS and then thrice with 1% bovine serum albumin (BSA). Primary antibody (anti-γH2AX; Upstate) was added at a dilution of 1:500 in 1% BSA and incubated for 1 h at room temperature. Cells were washed thrice in 1% BSA before adding FITC-conjugated secondary antibody (goat antimouse IgG; Jackson ImmunoResearch) in a 1:100 dilution in 1% BSA. Cells were washed thrice in 1% BSA before adding 1 ug/mL 4’,6-diamidino-2-phenylindole (Sigma) in PBS for 30 min. Cells were washed twice with PBS and coverslips were mounted with antifade solution (VectaShield). Slides were examined on a Leica DMRXA fluorescent microscope (Wetzlar). Images were captured by a QImaging CCD camera, imported into QCapture, and analyzed using Microsoft PowerPoint. For each treatment condition, γH2AX foci were determined in at least 150 cells.
Neutral comet assay detection of DNA DSBs.
U251 cells grown in 100-mm3 dishes were exposed to E7016 (3 umol/L) for 6 h, then irradiated (10 Gy) and returned to the incubator. At specified times; cells were washed with PBS, trypsinized on ice, and collected. Cells were centrifuged at 200 relative centrifugal force for 5 min and resuspended at 1 × 105 cells/mL. Cells were combined with melted LMAgarose (Trevigen) at a 1:10 ratio and immediately pipetted onto Comet slide (Trevigen). Slides were placed at 4°C for 30 min then immersed in Lysis Solution (Trevigen) for 1 h at 4°C. Slides were washed thrice in 1 × Tris-Borate-EDTA buffer and then run on a horizontal electrophoresis apparatus for 10 min at 20 V. Slides were rinsed twice in dH2O, once in ethanol, and air-dried for 4 h. Propidium iodide (2.5 μg/mL; Becton Dickinson) was added to each slide. Slides were visualized and images captured on Leica microscope as described above. Comet software was used to record Olive Tail Moment for each cell. Assay was completed three separate times evaluating 50 cells per treatment per experiment, for a total of 150 cells per treatment group.
In vivo s.c. tumor growth delay.
All animal studies were conducted in accordance with the principles and procedures outlined in the NIH Guide for the Care and Use of Animals. Four- to six-week-old female nude mice (Frederick Labs) were used for all in vivo studies. Animals were caged in groups of five or less and were fed animal chow and water ad libitum. A single cell suspension (1 × 106) of U251 cells was implanted on the lateral aspect of the rear leg. Tumors were staged when 172 mm3 was reached, at which point animals were randomized into four groups and treated with either vehicle control, E7016 alone (40 mg/kg by oral gavage), temozolomide followed by irradiation (temozolomide, 3 mg/kg orally, 6 h prior to irradiation; Sigma), or temozolomide followed by E7016 20 min later, followed by 4 Gy 6 h later. Tumors were monitored and measured three times per week using perpendicular diameter measurements. Volumes were calculated using the formula (L × W × W)/2. Tumors were followed until they reached a minimum of 1,000 mm3. Absolute tumor growth delay was calculated for each animal as the mean number of days for the treated tumors to grow to 1,000 mm3 minus the mean number of days for the control group to reach 1,000 mm3. SE in days was calculated for each group. Student’s t-test was used to compare the temozolomide/ radiotherapy group with the triple-therapy group.
Results
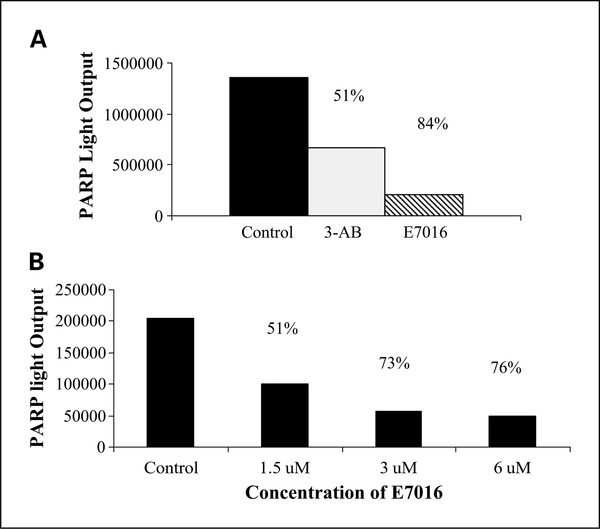
To determine if E7016 was inhibiting PARP at suitable concentrations for use in clonogenic survival analysis, a chemiluminescent PARP immunoassay was used. E7016 was used in a cell-free assay, and compared with the control (PARP enzyme without an inhibitor) E7016 (3 μmol/L) inhibited PARP activity by 84% and the known PARP inhibitor 3-aminobenzamide at 2 μmol/L (vendor-supplied positive control) inhibited PARP activity by 51% (Fig. 1A). To evaluate PARP inhibition in U251 cells, cells treated for 6 hours with 1.5 μmol/L, 3 μmol/L, and 6 μmol/L E7016 were collected and assayed. In U251 cells, the increasing concentration of E7016 produced increasing PARP inhibition up to 3 μmol/L (Fig. 1B).
Fig. 1.
Influence of E7016 on enzymatic activity of PARP. PARP inhibition was measured using a chemiluminescent immunoassay in a cell-free extract (A), 3 μmol/L, or in (B), U251 cells at 1.5, 3, and 6 μmol/L 6 h after E7016 treatment. Percent PARP inhibition posttreatment is indicated above the columns.
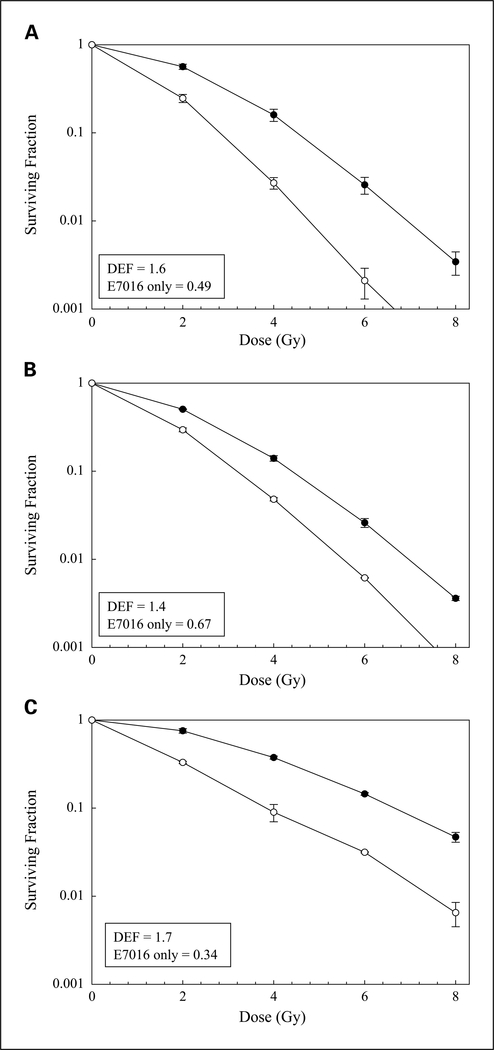
To determine whether radiosensitivity is enhanced by E7016, clonogenic survival analysis was done on the glioma cell line U251. For these studies, cells were treated with 3 μmol/L of E7016 for 6 hours prior to irradiation, which resulted in a surviving fraction of 49% (see Fig. 2A), an appropriate value for evaluating clonogenic survival with radiation. As shown in Fig. 2A, E7016 increased U251 radiosensitivity by a dose enhancement factor of 1.6 at a surviving fraction of 0.10. To determine whether this radiosensitizing effect could be extended to cell lines corresponding to other tumor types, the experiment was carried out in a pancreatic carcinoma cell line (MiaPaCa2) and prostate carcinoma cell line (DU145). Treatment of MiaPaCa2 cells with E7016 (3 μmol/L) prior to irradiation resulted in a drug-only surviving fraction of 67% and a dose enhancement factor of 1.4 at a surviving fraction of 0.10 (Fig. 2B). The DU145 cell line using a pretreatment E7016 concentration of 5 μmol/L had a drug-only surviving fraction of 34% and yielded a dose enhancement factor of 1.7 at a surviving fraction of 0.10 (Fig. 2C). These data indicate that E7016 exposure enhances radiation-induced cell killing in three separate tumor lines at drug doses shown to inhibit PARP activity.
Fig. 2.
The effects of E7016 on tumor cell radiosensitivity. Cells were seeded as a single-cell suspension and with a specified number of cells. After allowing cells time to attach (16 h), E7016 or the vehicle control was added at specified concentrations and the plates were irradiated 6 h later. Ten to twelve days after seeding, survival curves were generated after normalizing for the cytotoxicity generated by E7016 alone. A, U251 (3 μmol/L). B, MiaPaCa (3 μmol/L). C, DU145 (5 μmol/L).
Data presented are the mean ± SE from at least three independent experiments. ● = irradiation alone; ○ = irradiation plus E7016; DEF, dose enhancement factor.
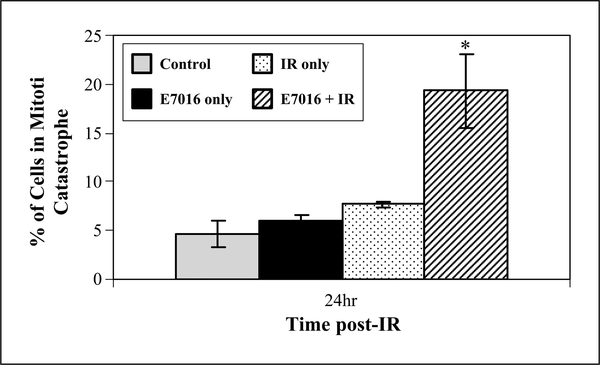
To begin to evaluate the mode of cell death through which E7016 enhanced radiosensitivity we focused on the U251 cell line and measured the possible E7016 enhancement in mitotic catastrophe and apoptosis. Following 6 hours of exposure to E7016 (3 μmol/L), cells were irradiated and evaluated 24 hours later for mitotic catastrophe, which was defined as the presence of two or more nuclear lobes within a single cell. As shown in Fig. 3, the number of cells in mitotic catastrophe was significantly greater in the E7016-treated irradiated cells than in cells that received radiation only at 24 hours postirradiation. Exposure of U251 cells to E7016 alone had no effect on mitotic catastrophe. To evaluate apoptosis, U251 cells were treated with E7016 6 hours prior to irradiation and then stained for annexin V-PE (positive in early and late apoptosis) and 7-AAD (positive in late apoptosis only) at 24 and 72 hours postirradiation. As expected for the glioma cell line (for radiation alone), minimal apoptosis of 4% to 6% was seen in the E7016, radiation, and combination groups at either 24 or 72 hours postirradiation (data not shown). Taken together, these data suggest that E7016-mediated radiosensitization occurs through an increase in the number of cells undergoing mitotic catastrophe and not an increase in the number of cells undergoing apoptosis.
Fig. 3.
Influence of E7016 on radiation-induced mitotic catastrophe. U251 cells growing in chamber slides were exposed to E7016 (3 μmol/L) for 6 h, irradiated (2 Gy), and fixed at 24 h postirradiation for immunocytochemical analysis of mitotic catastrophe. Nuclear fragmentation (defined as the presence of two or more distinct lobes within a single cell) was evaluated in 100 cells per treatment per experiment. Gray column, vehicle-treated cells; black columns, cells treated with E7016 alone; dotted columns, cells treated with 2 Gy alone; striped columns, cells treated with the combination of E7016 and 2 Gy. The data represent the mean of three independent experiments; bars, ± SE. *, P = 2.6 × 10−5 according to Student’s t test (2 Gy versus E7016 plus 2 Gy).
To determine whether the E7016-induced radiosensitization was the result of accumulation of cells in a more radiosensitive phase of the cell cycle, flow cytometry was used to define the cell cycle phase distribution of U251 cells after 6 hours of drug exposure (3 μmol/L). No difference in the cell cycle phase distributions of untreated and E7016 treated cells were detected (data not shown). Because activation of G2 checkpoint contributes to cell survival after irradiation, this parameter was defined in E7016-treated U251 cells. Cultures were exposed to E7016 (3 μmol/L) for 6 hours, irradiated (2Gy), and collected at times from 1 to 24 hours later for analysis of mitotic index according to phospho-H3 expression in 4N cells (13). Results showed that E7016 had no effect on the radiation-induced reduction and subsequent recovery in the mitotic index (data not shown). These results indicate that an abrogation of the G2 checkpoint does not mediate E7016-induced radiosensitization.
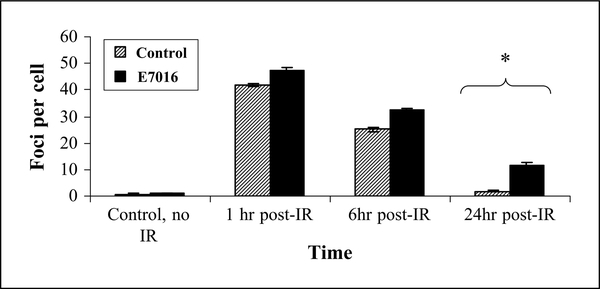
As it is known that PARP is involved in the repair of DNA damage, we next determined the effect on DSB repair in cells treated with E7016 (3). γH2AX foci staining is known to correlate with DSBs and that the number of foci per cell corresponds to the approximate number of DSB per cell (14). To define γH2AX foci, U251 cells were treated with E7016 for 6 hours prior to irradiation, collected, and stained at 1, 6, and 24 hours postirradiation. Immunocytochemistry revealed that at 1 hour postirradiation both the radiation only and the combination group had ~ 45 foci per cell, which did not differ significantly between groups (see Fig. 4). By 6 hours postirradiation, the number of foci per cell in each group had decreased to ~25 to 30 foci/cell and by 24 hours postirradiation the irradiation alone group had returned to baseline (<5 foci/cell) whereas the E7016-treated cells still had significantly more foci per cell at 24 hours (Fig. 4). This data suggested that E7016 inhibits the repair of radiation-induced DSBs.
Fig. 4.
Influence of E7016 on radiation-induced γH2AX foci. U251 cells growing in chamber slides were exposed to E7016 (3 μmol/L) for 6 h, then irradiated (2 Gy), returned to incubator, and fixed at the specified times for immunocytochemical analysis of nuclear γH2AX foci. Striped columns, cells treated with 2 Gy alone; black columns, cells treated with both E7016 and 2 Gy. Foci were evaluated three times in 50 nuclei per treatment per experiment; columns, mean of three independent experiments; bars, SE. *, P < 0.0001 according to Students t-test (E7016 plus 2 Gy versus 2 Gy alone).
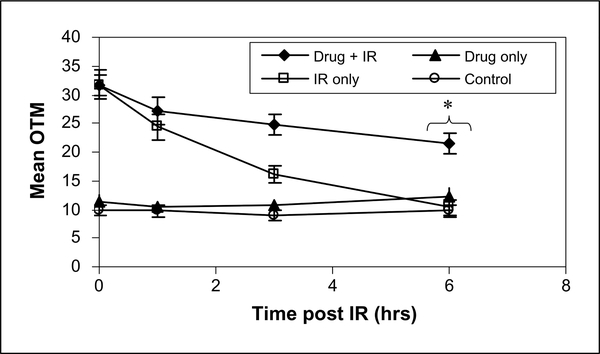
γH2AX expression at 24 hours after irradiation has been shown to correlate with radiosensitivity (15), but it reflects a chromatin level response. As an additional measure of the effects of E7016 on radiation-induced DSBs, the neutral comet assay was done. For the comet assay, cells were treated with 3 μmol/L E7016, 6 hours later irradiated (10 Gy), and collected out 6 hours later. As shown in Fig. 5, in cells exposed to E7016 plus radiation there was a significant increase in the amount of DNA damage remaining compared with cells receiving only radiation. Exposure of U251 cells to E7016 alone had no effect on DNA damage, nor did E7016 affect the initial level of DNA damage induced by radiation. Thus, consistent with the γH2AX data, these results indicate that E7016 inhibits the repair of radiation-induced DSBs.
Fig. 5.
Influence of E7016 on radiation-induced DNA damage by comet assay. U251 cells growing in 100-mm3 dishes were exposed to E7016 (3 μmol/L) for 6 h, then irradiated (10 Gy), returned to incubator, and comet assay was done under neutral conditions at specified times postirradiation. Open square, cells treated with 10 Gy alone; black diamond, cells treated with both E7016 and 10 Gy. Control refers to unirradiated cells exposed to vehicle; drug only refers to cells treated with E7016 only for specified time prior to assay. Assay was completed three separate times evaluating 50 cells per treatment per experiment. *, P = 0.0092 according to Students t-test (E7016 plus 10 Gy versus 10 Gy alone).
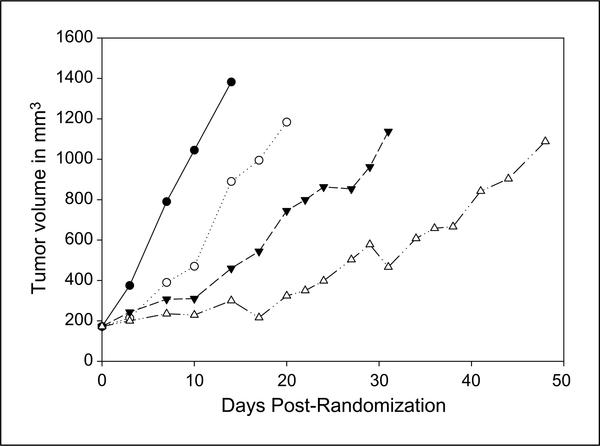
The in vitro data presented above indicate that E7016 enhances tumor cell radiosensitivity and that the mechanism involves the inhibition of DSB repair. However, the potential clinical application of this orally active agent in glioblastoma multiforme treatment will depend on its interaction with the current standard of care, which is temozolomide plus radiotherapy. Therefore, as an initial test of its potential clinical relevance in an in vivo glioblastoma multiforme model, E7016 was combined with temozolomide and radiation in a U251 tumor xenograft model. Mice bearing s.c. tumors (172 mm3) were randomized into four groups: vehicle, E7016 (40 mg/kg), temozolomide (3 mg/kg) prior to radiation (4 Gy), and E7016/ temozolomide/irradiation. The growth rates for the U251 tumors exposed to each treatment are shown in Fig. 6. For each group, the time to grow from 172 mm3 to 1,000 mm3 (a 5-fold increase in tumor size) was calculated using the tumor volumes from the individual mice in each group (mean + SE). The time for the tumor to grow from 172 to 1,000 mm3 increased from 10 + 2.1 days for vehicle-treated mice to 16.6 + 3.3 days for E7016-treated mice. Irradiation plus temozolomide increased the time to reach 1,000 mm3 to 30.2 + 4.8 days. When E7016 (40 mg) and temozolomide (3 mg/kg) were given prior to irradiation, there was a tumor growth delay of 41.0 + 9.5 days. The absolute growth delays (the time in days for tumors in treated mice to grow from 172 to 1,000 mm3 minus the time in days for tumors to reach the same size in vehicle- treated mice) were 6.6 days for E7016 alone, 20.2 days for the temozolomide/radiotherapy group, and of 31 days for E7016 plus the standard of care (radiotherapy/temozolomide). Therefore the triple therapy of E7016, temozolomide, and radiation produced a tumor growth delay greater than the individual E7016 treatment plus the standard of care treatment of radiotherapy plus temozolomide (P = 0.03, temozolomide/ radiotherapy versus E7016/temozolomide/radiotherapy). These data indicate that E7016 enhances the radiation/temozolomide-induced tumor growth delay of U251 xenografts.
Fig. 6.
Influence of E7016 on radiation-induced tumor growth delay. Mice were implanted with 1 × 106 U251 cells on the lateral aspect of the rear leg. Tumors were randomized at 172 mm3 into four groups and treated with either vehicle control, E7016 alone (40 mg/kg by oral gavage), temozolomide (3 mg/kg orally) followed by radiotherapy, or E7016/temozolomide/4Gy. Tumors were measured three times per week and followed until they reached at least 1,000 mm3. Volumes were calculated using the formula (L × W × W)/2. ● = vehicle alone; ○ = E7016 alone; ▼ = temozolomide/radiotherapy, ◇ = E7016/temozolomide/radiotherapy.
Discussion
Previous studies using 3-aminobenzamide have shown that PARP inhibition enhanced the in vitro sensitivity of tumor cell lines to radiation (6, 16–18). Those reports suggest that PARP inhibition might serve as a target for the development of radiation modifiers. To our knowledge, there are currently six PARP inhibitors that have been developed for the clinic, most of which are in either late preclinical studies or early clinical trials (3). E7016 has potent PARP inhibitory activity, is orally available, and crosses the blood brain barrier, suggesting that E7016 may be effective in glioblastoma multiforme therapy.
Given that the newer PARP inhibitors are derived from diverse chemical backbones including benzimidazoles, pyrrolocarbazoles, and phthalazinones, it is likely that the process of PARP inhibition will vary among compounds (11). Preclinical data show an enhancement of the response of tumor cells to radiation using some but not all the PARP inhibitors AG14361 (19), ABT-888 (20, 21), and INO-1001 (22), but no data have been published for KU-0059436, BSI-201, and E7016. The previous data, however, were derived in numerous cell lines, with various end points and diverse protocols. Thus, it may be difficult to generalize the radiation-modifying properties among PARP inhibitors, requiring the need to investigate the individual compounds. As shown in Fig. 2, however, E7016 significantly enhances the in vitro radiosensitivity of a variety of tumor cells.
A critical event in determining radiosensitivity is the repair of DNA DSBs. In the data presented here, γH2AX, a marker of DNA DSBs induced by clinically relevant doses of ionizing radiation (14), was elevated in the E7016-treated cells at 24 hours compared with those that were only irradiated (Fig. 4). The finding that PARP inhibition after radiation can lead to an inhibition of DNA DSBs has been reported previously (20, 21). In Liu et al., an increase in γH2AX was reported in cells treated with ABT-888 and irradiation versus irradiated cells only at 24 hours. However, in Albert et al., the increase in γH2AX was reported at 6 hours. This may have been due to the use by Albert et al. of a higher drug concentration and a longer exposure time than was used in either Liu et al. or in our work. Clearly, additional investigations are needed to further define the molecular processes of PARP-induced enhancement of radiosensitivity; however, the retention of γH2AX foci in combination with our comet data would suggest the effect is related to an inhibition of DNA repair.
Current treatment for glioblastoma involves maximal surgery followed by a combination of temozolomide and radiotherapy. Accordingly, the clinical potential of a novel radiosensitizing agent must be evaluated not simply in combination with radiation, but in combination with temozolomide/radiation. That is, the development of radiation modifiers for glioblastoma multiforme must account for the potential effects of temozolomide. Along these lines, whereas data presented here clearly showed that E7016 enhances tumor cell radiosensitivity in vitro consistent with previous reports of other PARP inhibitors, to generate insight into the potential clinical relevance, the critical in vivo experiment was to combine the PARP inhibitor with the current standard of care - temozolomide/radiation. As shown, the administration of E7016 to mice bearing U251 xenografts enhanced the effectiveness of the temozolomide/radiation combination. Thus, these results provide support for the addition of E7016 to the standard regimen of temozolomide and radiotherapy that is currently used in the clinic.
Translational Relevance.
Radiation therapy remains one of the mainstays of cancer treatment. The use of radiation is integral in achieving maximal local control and treatment with curative intent for various types of cancers. Some cancers, however, such as glioblastoma multiforme, are particularly more radioresistant and chemoresistant, and single-modality treatment offers little benefit to these patients. It is through the combination of treatments that greater local control and potential survival can be achieved. This preclinical study is important in helping to define systemic agents that can be used with radiation to enhance tumor cell kill. Our data show the poly (ADP-ribose) polymerase inhibitor E7016 sensitizes tumor cells to radiation and more importantly is tested in vivo in a model using both temozolomide and radiation.
Acknowledgments
Grant support: Intramural Research Program of the NIH, National Cancer Institute. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Disclosure of Potential Conflicts of Interest
X.Weizhung, J. Zhang, B.S. Slusher, employment, Eisai Research Institute.
J.Z., unpublished data.
References
- 1.Ohgaki H, Dessen P, Jourde B, et al. Genetic pathways to glioblastoma: a population-based study. Cancer Res 2004;64:6892–9. [DOI] [PubMed] [Google Scholar]
- 2.Stupp R, Mason W, Van Den Bent M, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med 2005;352:987–96. [DOI] [PubMed] [Google Scholar]
- 3.Ratnam K, Low JA. Current development of clinical inhibitors of poly(ADP-ribose) polymerase in oncology. Clin Cancer Res 2007;13:1383–8. [DOI] [PubMed] [Google Scholar]
- 4.Zaremba T, Curtin NJ. PARP inhibitor development for systemic cancer targeting. Anticancer Agents Med Chem 2007;7:515–23. [DOI] [PubMed] [Google Scholar]
- 5.Schreiber V, Dantzer F, Ame J, de Murcia G. Poly-(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol 2006;7:517–28. [DOI] [PubMed] [Google Scholar]
- 6.Durkacz BW, Omidiji O, Gray DA, Shall S. (ADP-ribose)n participates in DNA excision repair. Nature 1980;283:593–6. [DOI] [PubMed] [Google Scholar]
- 7.Bowman K, White A, Golding B, Griffin R, Curtin N. Potentiation of anti-cancer agent cytotoxicity by the potent poly(ADP-ribose) polymerase inhibitors NU1025and NU1064. BrJCancer1998;78:1269–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu L, Gerson S. Targeted modulation of MGMT: clinical implications. Clin Cancer Res 2006;12:328–31. [DOI] [PubMed] [Google Scholar]
- 9.Clark J, Ferris G, Pinder S. Inhibition of nuclear NAD nucleosidase and polyADP-ribose polymerase activity from rat liver by nicotinamide and 5’-methyl nicotinamide. Biochem Biophys Acta 1971;238:82–5. [DOI] [PubMed] [Google Scholar]
- 10.Milam K, Cleaver J. Inhibitors of poly(adenosine diphosphate-ribose) synthesis: effect on other metabolic processes. Science 1984;223:589–91. [DOI] [PubMed] [Google Scholar]
- 11.Southan G, Szabo C. Poly(ADP-ribose) polymerase inhibitors. Curr Med Chem 2003;10:321–40. [DOI] [PubMed] [Google Scholar]
- 12.Tentori L, Leonetti C, Scarsella M, et al. Systemic administration of GPI 15427, a novel poly(ADP-ribose) polymerase-1 inhibitor, increases the antitumor activity of temozolomide against intracranial melanoma, glioma, lymphoma. Clin Cancer Res 2003;9: 5370–9. [PubMed] [Google Scholar]
- 13.Xu B, Kim ST, Lim DS, Kastan MB. Two molecularly distinct G(2)/M checkpoints are induced by ionizing irradiation. Mol Cell Biol 2002;22:1049–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pilch DR, Sedelnikova OA, Redon C, Celeste A, Nussenzweig A, Bonner WM. Characteristics of γ-H2AX foci at DNA double-strand breaks sites. Biochem Cell Biol 2003;81:123–9. [DOI] [PubMed] [Google Scholar]
- 15.Banath J, Olive P. Expression of phosphorylated histone H2AX as a surrogate of cell killing by drugs that create DNA double-strand breaks. Cancer Res 2003; 63:4347–50. [PubMed] [Google Scholar]
- 16.Ueno A,Tanaka O, Matsudaira H. Inhibition of γ-ray dose-rate effects by D2O and inhibitors of poly(ADP-ribose) synthetase in cultured mammalian L5178Y cells. Radiat Res 1984;98:574–82. [PubMed] [Google Scholar]
- 17.Thraves P, Mossman K, Brennan T, Dritschilo A. Radiosensitization of human fibroblasts by 3-amino-benzamide: an inhibitor of poly(ADP-ribosylation). Radiat Res 1985;104:119–27. [PubMed] [Google Scholar]
- 18.Kelland L, Tonkin K. The effect of 3-aminobenzamide in the radiation response of three human cervix carcinoma xenografts. Radiother Oncol 1989;15: 363–9. [DOI] [PubMed] [Google Scholar]
- 19.Calabrese CR, Almassy R, Barton S, et al. Anticancer chemosensitization and radiosensitization by the novel poly(ADP-ribose) polymerase-1 inhibitor AG14361. J Natl Cancer Inst 2004;96:56–67. [DOI] [PubMed] [Google Scholar]
- 20.Liu S, Coackley C, Krause M, Jalali F, Chan N, Bristow R. A novel poly(ADP-ribose) polymerase inhibitor, ABT-888, radiosensitizes malignant human cell lines under hypoxia. Radiother Oncol 2008;88: 258–68. Epub 2008 May 2. [DOI] [PubMed] [Google Scholar]
- 21.Albert JM, Cao C, Kim KW, et al. Inhibition of poly-(ADP-ribose) polymerase enhances cell death and improves tumor growth delay in irradiated lung cancer models. Clin Cancer Res 2007;13:3033–42. [DOI] [PubMed] [Google Scholar]
- 22.Brock W, Milas L, Bergh S, Lo R, Szabo C, Mason K. Radiosensitization of human and rodent cell lines by INO-1001, a novel inhibitor of poly(ADP-ribose) polymerase. Cancer Lett 2004;205:155–60 [DOI] [PubMed] [Google Scholar]