Abstract
Background
Klinefelter syndrome (KS) is characterized by the presence of at least one supernumerary X chromosome. KS typical symptoms include tall stature, gynecomastia, hypogonadism and azoospermia. KS patients show a higher risk of developing metabolic and cardiovascular diseases, inflammatory and autoimmune disorders, osteoporosis and cancer. Long non-coding RNA (lncRNA) growth arrest-specific 5 (GAS5) has been shown to be involved in several biologic processes, including inflammatory and autoimmune diseases, vascular endothelial cells apoptosis and atherosclerosis, as well as cellular growth and proliferation, cellular development and cell-to-cell signaling and interaction. The lncRNA GAS5 expression profile in KS patients has never been evaluated so far.
Methods
To accomplish this, GAS5 mRNA levels were evaluated by Next Generation Sequencing (NGS) technology and qRT-PCR assay in 10 patients with KS and 10 age-matched controls.
Results
NGS results showed a significantly lncRNAGAS5up-regulation by 5.171-fold in patients with KS. Theresults of qRT-PCR confirmed the NGS data.
Conclusions
These findings showed the occurrence of lncRNA GAS5 over-expression in KS patients. Whether this lncRNA is involved in the pathogenesis of inflammation and autoimmune diseases, atherogenesis or germ cell depletion in KS patients is not known. Further studies are needed.
Keywords: Klinefelter syndrome, Rare disease, Cognitive deficits;NGS, mRNA, qRT-PCR
Background
Klinefelter syndrome (KS) is the most common sex-chromosome disorder in men, with an estimated prevalence of 1:660 newborns [1, 2]. The most common karyotype is the classic 47,XXY one, which accounts for the 80–90% of all cases [3]. It is a consequence of a non-disjunction of paired X-chromosomes during the first or second meiotic division [3], equally due to a paternal or maternal meiotic mal segregation event [4]. The remaining 10% of KS are due to chromosome mosaicisms (e.g. 46,XY/47,XXY) or to more complex karyotypes (X chromosome structural abnormalities such as 47,XX, der(Y), 47,X, der(X),Y, or other numeric sex chromosome abnormalities such as 48, XXXY, 48, XXYY and 49, XXXXY) [3, 5].
The chromosomal abnormality leads to a progressive germ cell degeneration starting from mid-puberty, impaired Sertoli cell function [6], total tubular atrophy or hyalinizing fibrosis and Leydig cell hyperplasia [7], clinically causing hypergonadotropic hypogonadism [8], small testis with increased consistency and infertility. Occasionally, foci of spermatogenesis have been observed in testis of KS patients [7]. Clinically, azoospermia is present in the 90% of non-mosaic KS, whereas severe oligozoospermia in the remaining 10% [7].
Several clinical manifestations associate with the syndrome. These include learning and language disability, reduction in intelligence quotient (IQ) scores of 10 to 15 points, but not into the intellectual disability range [6], increased risk for mitral valve prolapse, lower-extremity varicose veins, venous stasis ulcers, deep vein thrombosis and pulmonary embolism, autoimmune diseases, 20-fold-higher risk of developing breast cancer, type II diabetes mellitus (T2 DM) and metabolic syndrome [9], osteoporosis [10], extragonadal germ cell tumors and non-Hodgkin lymphoma [11, 12].
Molecular mechanisms underlying the variability in KS phenotype have not been clearly understood yet. Epigenetic mechanisms have been suggested to play a role [13]. Growth arrest-specific 5 (GAS5) gene, mapping on the 1q25.1 chromosome, encodes for a long non-coding RNA (lncRNA) which is involved in the modulation of gene expression, targeting many different downstream miRNAs [14, 15].
LncRNA GAS5 was initially identified as a tumor suppressor gene [16–21]. However, it has also been shown to support female germline stem cell (FGSC) survival [22] and adipocyte differentiation [15]. In addition, recent evidence pointed to lncRNA GAS5 a role in atherosclerosis [23] and autoimmune diseases [24], which are widely recognized as KS comorbidities.
No study evaluated the lncRNA GAS5 expression profile in KS patients so far. On this account, the present study was undertaken to investigate differently expressed genes and, specifically, lncRNA GAS5 expression in patients with KS and controls.
Methods
The present study belongs to a broad project investigating transcriptome differences between patients with KS and normal control subjects (NC). To accomplish this, 10 patients with non-mosaic KS and 10 healthy controls of similar age were selected. All men with KS (mean age 32.5) had 47,XXY karyotype, as confirmed by cytogenetic investigation performed on at least 50 metaphases. NC (mean age 32) had negative history of genetic diseases, normal testis volume and normal reproductive hormones (LH, FSH, total testosterone) levels. All men enrolled in this study (KS patients and controls) were Italians.
They underwent to transcriptome and qRT-PCR analysis which were performed on peripheral blood mononuclear cells (PBMCs) isolated with ficoll gradient.
RNA sequencing
RNA Sequencing, Next Generation Sequencing (NGS) and Data Analysis of NGS methods have been detailed in previous articles presenting NGS data from the same project [13, 25].
Indexed libraries were prepared from 1 μg/ea. purified RNA with TruSeq Stranded Total RNA (Illumina) Library Prep Kit according to the manufacturer’s instructions. Libraries were quantified using the Agilent 2100 Bioanalyzer (Agilent Technologies) and pooled such that each index-tagged sample was present in equimolar amounts, with final concentration of the pooled samples of 2bnM. The pooled samples underwent cluster generation and sequencing using an IlluminaHiSeq 2500 System (Illumina) in a 2 × 100 paired-end (RNA-seq) format. The raw sequence files generated (.fastq files) underwent quality control analysis using FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/).
Data analysis
Bioinformatics analysis were performed by Genomix4Life srl (Baronissi(SA), Italy). The quality checked reads were trimmed with cutadapt v.1.10 and then aligned to the human genome (hg19 assembly) using STAR v.2.5.2 [26], with standard parameters. Differentially expressed mRNAs were identified using DESeq2 v.1.12 [27].
Gene annotation, as provided by Ensembl (GRCh37), was obtained for all known genes in the human genome. We calculated the number of reads mapping to each transcript with HTSeq-count v.0.6.1 [28]. These raw read counts were then used as input to DESeq2 for calculation of normalized signal for each transcript in the samples, and differential expression was reported as fold change along with associated adjusted p-values (computed according to Benjamini-Hochberg). Differential expression data were further confirmed using Cuffdiff [28]. Functional analysis was performed using Ingenuity Pathway Analysis (IPA) with standard parameters.
Availability of data and materials
Raw data are available in ArrayExpress database repository (https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-6107/) with accession number E-MTAB-6107.
Validation whit qRT-PCR
To validate the results obtained by NGS analysis, we compared RT-PCR in 10 KS patients and 10 NC. Retro-transcription was performed with QuantiTect Reverse Transcription Kit (QIAGEN Sciences, Germantown, PA). Briefly, 600 ng of total RNA from each sample was then reconstituted in a final volume of 20 μl and the generated cDNA was used as a template for real-time quantitative PCR analysis using gene expression products. Quantification with qRT-PCR was performed using the 2-ΔΔCt method [29] by matching each KS patient to a respective NC aged ±3 years.
The target GAS5 and the reference gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH) TaqMan assays (Assay ID:Hs99999905_m1) were obtained from Applied Biosystems (Carlsbad,CA, USA).
The GAS5 TaqMan assays (Assay ID: Hs03464472_m1) used amplifies an amplicon between exon 10 and exon 11 (it is possible to check the characteristics of the probe used at the following link: https://www.thermofisher.com/taqman-gene-expression/product/Hs03464472_m1?CID = &ICID = &subtype=). The mean was obtained with the Software Version 1.5 supplied with the LightCycler 480, as previously reported [30].
Distribution analysis of measured gene transcript levels was performed using Shapiro – Wilk test and inferential statistical analysis of results was carried out using paired two tailed t-test and bivariate linear regression analysis. Graph Pad Prism 5 software was used for statistical analysis. A p value < 0.05 was accepted as significant.
Results
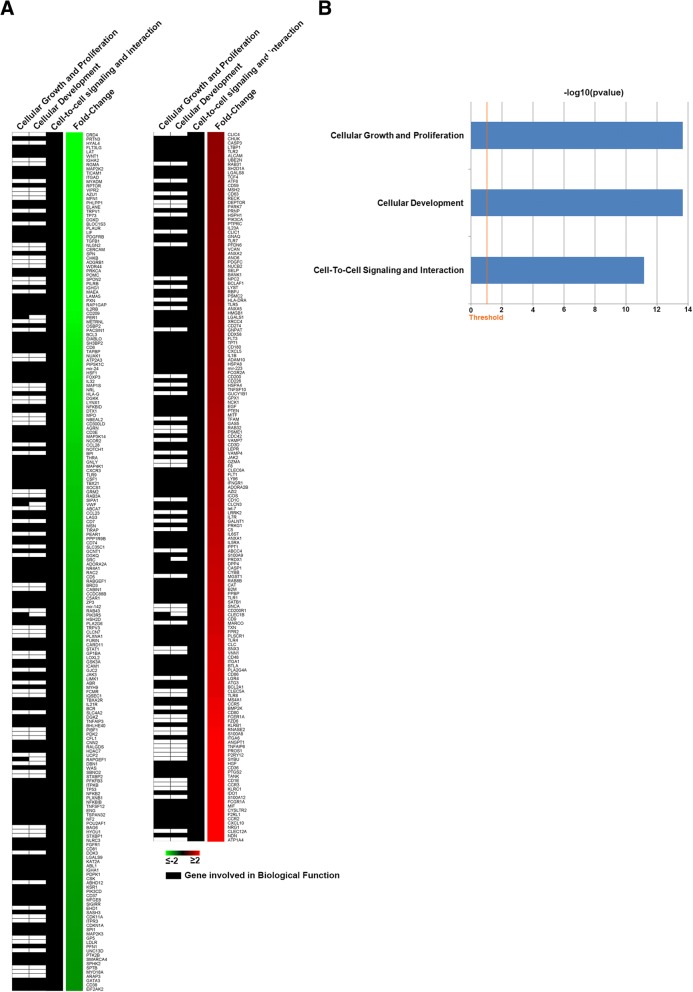
Clinical features and biochemical data of KS patients and controls have been reported elsewhere [13, 25]. Differential expression analysis showed 4448 genes differentially expressed with |fold-change| ≥2 and p-adj ≤ 0.05 comparing KS patients versus the 10 controls. Among these, 2698 were down-regulated and 1750 were up-regulated and Ingenuity Pathway Analysis (IPA) highlighted their involvement in several biological functions, including cellular growth and proliferation, cellular development and cell-to-cell signaling and interaction, where the transcription of GAS5 (locus 1:173832385–173,872,687) was up-regulated in the patients with KS by 5.171-fold (q-value< 0.0001) (Fig. 1).
Fig. 1.
Heatmap of gene functions. a. Heatmap showing the differentially expressed genes (|FC| ≥ 2 and p-adj ≤ 0.05) in whose function GAS5 is involved. b. Histogram showing the statistical significance of the functions where GAS5 is involved. The length of the bar is inversely proportional to the p-value
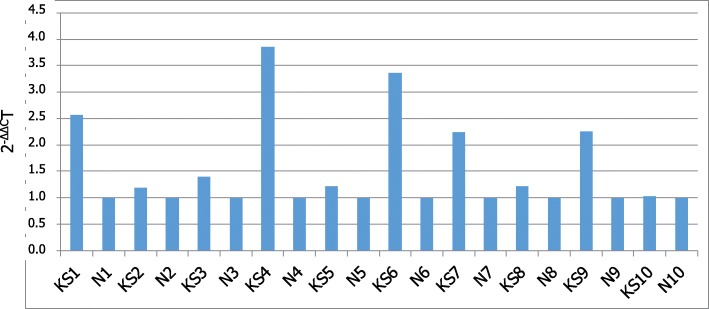
qRT-PCR was performed to validate these findings. Accordingly, the high sensitivity of this assay is optimal for ensuring accuracy in the evaluation of gene transcription levels. We examined lncRNA levels of GAS5 in 10 KS patients and 10 age-matched controls.
Distribution of expression values was normal in both KS patients and NC (p > 0.01). Therefore, we carried out a statistical inferential analysis using the parametric test paired two tailed t-test. Data acquired from this analysis revealed a significant increase of GAS5mRNA levels in KS patients compared to relative controls (p < 0.01), as shown in Fig. 2. In addition, we investigated the expression of the two groups by evaluating the relative expression value in terms of -ΔCt (Fig. 3). In this analysis, the mean expression of NC was 6.53 (value range 6.31–6.8; SD = 0.2; CV = 0.02; IC 95% 6.21–6.85), whereas the mean expression of the KS subjects was 7.28 (value range 6.55–8.28; SD = 0.65; CV = 0.085; IC 95% 6.81–7.75). Inferential statistical analysis reveals significant difference between two means expression value (p < 0.05). Linear regression analysis reveals no significant correlation between GAS5 mRNA levels and age of subjects involved in our study (p > 0.05).
Fig. 2.
GAS5 lncRNA expression in men with Klinefelter syndrome and normal controls. Data obtained by qRT-PCR. N, controls
Fig. 3.
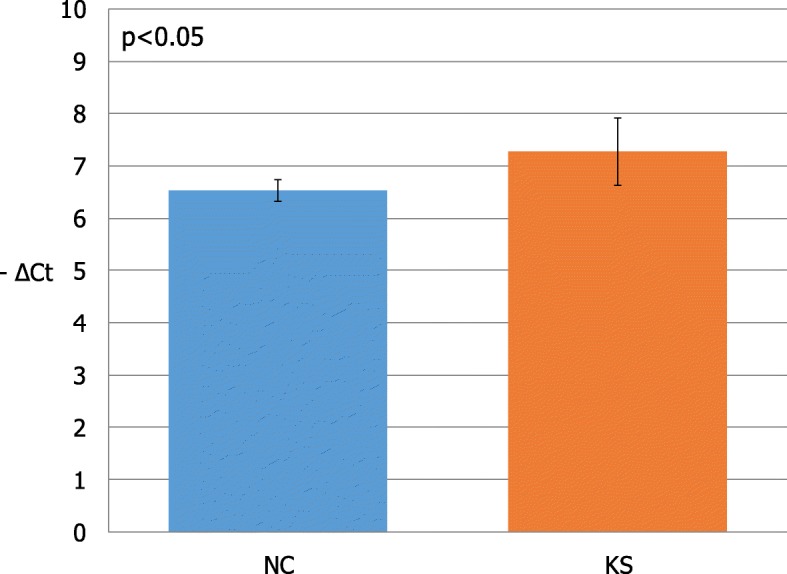
Mean of lncRNA GAS5 expression values in patients with Klinefelter syndrome and normal controls. KS, Klinefelter syndrome; NC, normal controls
Table 1 shows the main NGS findings validated by qRT-PCR presented so far that belong to the same project [13, 25]. The results confirm the data obtained by the NGS analysis and differences in values reflect the diversity of methods.
Table 1.
Differently-expressed transcripts in patients with Klinefelter syndrome (KS) and controls
Finally, fitting with the suppressing role of lncRNA GAS5 on IGF1R expression [31], our NGS data show that the IGF1R gene is down-regulated in KS subjects (FC = − 1.873 and p-adj = 0.000126) compared to normal subjects.
Discussion
We recently reported a decreased miRNAs and mitochondrial subunits expression in KS patients compared to controls [13, 25]. In the present study, we showed, for the first time, the occurrence of a higher lncRNA GAS5 expression in patients with KS compared to controls.
LncRNA GAS5 is involved in several biologic processes, including glucocorticoid (GC) actions, inflammatory and autoimmune diseases, vascular endothelial cells apoptosis and atherosclerosis, FGSC proliferation.
LncRNA GAS5 could directly interact with the DNA binding domain of the glucocorticoid receptor (GR) [32]. This prevents receptor binding to the GC responsive element (GRE) in target genes, blocking their transcription [32]. Because of its competition with GRE, lncRNA GAS5 is considered to act as a repressor of the GR action [33]. Accordingly, poor responders to GCs showed higher lncRNA GAS5 levels compared to normal ones [34]. GCs have a regulatory action on the immune system and are employed in inflammatory and autoimmune diseases. Altered lncRNA GAS5 levels in whole blood or leucocytes of patients with rheumatoid arthritis, systemic lupus erythematosus, multiple sclerosis, sarcoidosis, osteoarthritis and inflammatory bowel disease have already been reported [24, 35, 36], thus suggesting that lncRNA GAS5 may play a role in the pathogenesis of inflammatory and autoimmune diseases (probably modulating responsiveness to endogenous GCs), which are more frequent in KS patients.
LncRNA GAS5 expression is involved in atherogenesis. Its expression is significantly higher in plaques of atherosclerosis both from human and from animal models. Its over-expression enhances vascular endothelial cells apoptosis after lipoperoxidation. Therefore, lncRNA GAS5 has been suggested as a target for the therapy of atherosclerosis [23]. KS patients have an increased risk of atherogenesis, due to hypogonadism and metabolic syndrome [37]. Since the aforementioned evidence, a role for lncRNA GAS5 in atherogenesis in KS patients cannot be excluded.
Due to the involvement of lncGAS5 in several biological functions required for normal spermatogenesis, such as cellular growth and proliferation, cellular development and cell-to-cell signaling and interaction (Fig. 1), we speculate that lncGAS5 deregulation may play a role in germ cell loss in KS patients.
Very recently, lncRNA GAS5 has been regarded as a promoter of proliferation and survival of FGSC [22]. In greater detail, it has been found highly expressed in FGSCs and oocytes from neonatal mice and to facilitate cultured FGSC survival through the inhibition of their apoptosis in-vitro [22]. No study explored lncRNA GAS5 expression in male germ cells so far. This knowledge may help to understand its role, if any, in germ cell loss in KS patients. Indeed, lncRNA GAS5 has been shown to down-regulate the IGF1R expression [31]. Accordingly, we found that IGF1R expression was down-regulated in KS patients compared to controls. Growing evidence addressed to IGF1R a role in human testicular function [38] and in Sertoli cell proliferation and function [39]. Indeed, its knock-out (both the single one and in combination with the Insr) in Sertoli cells from mice affected the final testicular size, Sertoli cell number and the sperm output [40]. Therefore, a down-regulation of IGF1R in Sertoli cells might hypothetically explain germ cell loss in KS patients.
Finally, apoptosis is a mechanism responsible for the normal regulation of germ-cell death during differentiation and maturation of normal human germ cells. It is a prerequisite for a normal spermatogenesis [41, 42]. Therefore, apoptosis may contribute to the excessive germ-cell demise in KS patients. According with Koldemir and colleagues (2017), GAS5 accumulation in exosomes during apoptotic induction promotes two different mechanisms, microtubule stabilization and DNA strand breaks [43]. Therefore, it is plausible to hypothesize that GAS5 is involved in communication of cells upon cell death-promoting signals.
Conclusions
In conclusion, the results of this study showed a significant over-expression of lncRNA GAS5 in patients with non-mosaic KS compared to controls. The lncRNA seems to play a role in various biologic processes, such as GC actions, inflammatory and autoimmune diseases, vascular endothelial cells apoptosis and atherosclerosis and in FGSC proliferation. Furthermore, its involvement in cellular growth and proliferation, cellular development and cell-to-cell signaling and interaction suggests a possible role in germ cell loss. However, whether the lncRNA GAS5 over-expression is really involved in the pathogenesis of some of the typical features of KS patients (particularly in inflammatory and autoimmune diseases, atherogenesis and germ cell loss) needs to be further investigated.
Acknowledgements
None.
Funding
None.
Availability of data and materials
Differential expression data were further confirmed using Cuffdiff36.Raw data are available in ArrayExpress database repository (https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-6107/) with accession number E-MTAB-6107.
Abbreviations
- FGSC
Female germline stem cell
- GAS5
Growth arrest-specific 5
- GC
Glucocorticoid
- GR
Glucocorticoid receptor
- GRE
Glucocorticoid responsive element
- IQ
Intelligence quotient
- KS
Klinefelter syndrome
- lncRNA
Long non-coding RNA
- NGS
Next Generation Sequencing
- PBMCs
Peripheral blood mononuclear cells
- T2DM
Type II diabetes mellitus
Authors’ contributions
Concept and Design: AEC, MS and RC. Acquisition of data or analysis: AEC, MS, RC, RAC, LC, FR, GG, CR, SLV. Final approval: AEC, MS and RC. All authors read and approved the final manuscript, and they have the accountability for all aspects of work.
Ethics approval and consent to participate
This study was approved by the Ethical Committee of the University Teaching Hospital “Policlinico-Vittorio Emanuele”, Catania, Italy: trial registration number 49/2015/PO (Register of the Ethics Committee opinions). All the participants in the study signed an informed consent to publish their medical information.
Consent for publication
All the participants in the study signed an informed consentto publish.
Competing interests
The authors declare that there are no conflicts of interest regarding the publication of this study. All authors do not have financial competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Michele Salemi, Phone: +39 935 936440, Email: micezia@tiscali.it.
Rossella Cannarella, Email: roxcannarella@gmail.com.
Rosita A. Condorelli, Email: rosita.condorelli@unict.it
Laura Cimino, Email: lauracimino@hotmail.it.
Federico Ridolfo, Email: federico.ridolfo@alice.it.
Giorgio Giurato, Email: ggiurato@unisa.it.
Corrado Romano, Email: cromano@oasi.en.it.
Sandro La Vignera, Email: sandrolavignera@unict.it.
Aldo E. Calogero, Email: acaloger@unict.it
References
- 1.Morris JK, Alberman E, Scott C, Jacobs P. Is the prevalence of Klinefelter syndrome increasing? Eur J Hum Genet. 2008;16:163–170. doi: 10.1038/sj.ejhg.5201956. [DOI] [PubMed] [Google Scholar]
- 2.Klinefelter HF, Reifenstein EC, Albright F. Syndrome characterized by gynaecomastia, aspermatogenesis without a-Leydigism, and increased excretion of follicle stimulating hormone. J Clin Endocrinol Metab. 1942;2:615–627. doi: 10.1210/jcem-2-11-615. [DOI] [Google Scholar]
- 3.Maiburg M, Repping S, Giltay J. The genetic origin of Klinefelter syndrome and its effect on spermatogenesis. Fertil Steril. 2012;98:253–260. doi: 10.1016/j.fertnstert.2012.06.019. [DOI] [PubMed] [Google Scholar]
- 4.Wikstrom AM, Dunkel L. Klinefelter syndrome. Best Pract Res Clin Endocrinol Metab. 2011;25:239–250. doi: 10.1016/j.beem.2010.09.006. [DOI] [PubMed] [Google Scholar]
- 5.Frühmesser A, Kotzot D. Chromosomal variants in klinefelter syndrome. Sex Dev. 2011;5(3):109–123. doi: 10.1159/000327324. [DOI] [PubMed] [Google Scholar]
- 6.Aksglaede L, Link K, Giwercman A, Jørgensen N, Skakkebaek NE, Juul A. 47, XXY Klinefelter syndrome: clinical characteristics and age-specific recommendations for medical management. Am J Med Genet C Semin Med Genet. 2013;163C:55–63. doi: 10.1002/ajmg.c.31349. [DOI] [PubMed] [Google Scholar]
- 7.De Sanctis V, Ciccone S. Fertility preservation in adolescents with Klinefelter’s syndrome. Pediatr Endocrinol Rev Suppl 2010;1:178–181. [PubMed]
- 8.Groth KA, Skakkebæk A, Høst C, Gravholt CH, review BAC. Klinefelter syndrome--a clinical update. J Clin Endocrinol Metab. 2013;98:20–30. doi: 10.1210/jc.2012-2382. [DOI] [PubMed] [Google Scholar]
- 9.Bojesen A, Kristensen K, Birkebaek NH, Fedder J, Mosekilde L, Bennett P, Laurberg P, Frystyk J, Flyvbjerg A, Christiansen JS, Gravholt CH. The metabolic syndrome is frequent in Klinefelter’s syndrome and is associated with abdominal obesity and hypogonadism. Diabetes Care. 2006;29:1591–1598. doi: 10.2337/dc06-0145. [DOI] [PubMed] [Google Scholar]
- 10.van den Bergh JP, Hermus AR, Spruyt AI, Sweep CG, Corstens FH, Smals AG. Bone mineral density and quantitative ultrasound parameters in patients with Klinefelter's syndrome after long-term testosterone substitution. Osteoporos Int. 2001;12(1):55–62. doi: 10.1007/s001980170158. [DOI] [PubMed] [Google Scholar]
- 11.Wu FC, Tajar A, Beynon JM, Pye SR, Silman AJ, Finn JD, O'Neill TW, Bartfai G, Casanueva FF, Forti G, Giwercman A, Han TS, Kula K, Lean ME, Pendleton N, Punab M, Boonen S, Vanderschueren D, Labrie F, Huhtaniemi IT, EMAS Group Identification of lateonset hypogonadism in middle-aged and elderly men. N Engl J Med. 2010;363(2):123–135. doi: 10.1056/NEJMoa0911101. [DOI] [PubMed] [Google Scholar]
- 12.Kelleher S, Conway AJ, Handelsman DJ. Blood testosterone threshold for androgen deficiency symptoms. J Clin Endocrinol Metab. 2004;89(8):3813–3817. doi: 10.1210/jc.2004-0143. [DOI] [PubMed] [Google Scholar]
- 13.Cimino L, Salemi M, Cannarella R, Condorelli RA, Giurato G, Marchese G, La Vignera S, Calogero AE. Decreased miRNA expression in Klinefelter syndrome. Sci Rep. 2017;7:16672. doi: 10.1038/s41598-017-16892-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gu J, Wang Y, Wang X, Zhou D, Shao C, Zhou M, He Z. Downregulation of lncRNA GAS5 confers tamoxifen resistance by activating miR-222 in breast cancer. Cancer Lett. 2018;pii: S0304–3835(18):30447–30446. doi: 10.1016/j.canlet.2018.06.039. [DOI] [PubMed] [Google Scholar]
- 15.Li M, Xie Z, Wang P, Li J, Liu W, Tang S, Liu Z, Wu X, Wu Y, Shen H. The long noncoding RNA GAS5 negatively regulates the adipogenic differentiation of MSCs by modulating the miR-18a/CTGF axis as a ceRNA. Cell Death Dis. 2018;9(5):554. doi: 10.1038/s41419-018-0627-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mourtada-Maarabouni M, Hedge VL, Kirkham L, Farzaneh F, Williams GT. Growth arrest in human T-cells is controlled by the non-coding RNA growth-arrest-specific transcript 5 (GAS5) J Cell Sci. 2008;121:939–946. doi: 10.1242/jcs.024646. [DOI] [PubMed] [Google Scholar]
- 17.Mourtada-Maarabouni M, Pickard MR, Hedge VL, Farzaneh F, Williams GT. GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene. 2009;28:195–208. doi: 10.1038/onc.2008.373. [DOI] [PubMed] [Google Scholar]
- 18.Schneider C, King RM, Philipson L. Genes specifically expressed at growth arrest of mammalian cells. Cell. 1988;54:787–793. doi: 10.1016/S0092-8674(88)91065-3. [DOI] [PubMed] [Google Scholar]
- 19.Renganathan A, Kresoja-Rakic J, Echeverry N, Ziltener G, Vrugt B, Opitz I, Stahel RA, Felley-Bosco E. GAS5 long non-coding RNA in malignant pleural mesothelioma. Mol Cancer. 2014;13:119. doi: 10.1186/1476-4598-13-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pickard MR, Mourtada-Maarabouni M, Williams GT. Long non-coding RNA GAS5regulates apoptosis in prostate cancer cell lines. Biochim Biophys Acta. 2013;1832:1613–1623. doi: 10.1016/j.bbadis.2013.05.005. [DOI] [PubMed] [Google Scholar]
- 21.Yacqub-Usman K, Pickard MR, Williams GT. Reciprocal regulation of GAS5 lncRNA levels and mTOR inhibitor action in prostate cancer cells. Prostate. 2015;75:693–705. doi: 10.1002/pros.22952. [DOI] [PubMed] [Google Scholar]
- 22.Wang J, Gong X, Tian GG, Hou C, Zhu X, Pei X, Wang Y, Wu J. Long noncoding RNA growth arrest-specific 5 promotes proliferation and survival of female germline stem cells in vitro. Gene. 2018;653:14–21. doi: 10.1016/j.gene.2018.02.021. [DOI] [PubMed] [Google Scholar]
- 23.Chen L, Yang W, Guo Y, Chen W, Zheng P, Zeng J, Tong W. Exosomal lncRNA GAS5 regulates the apoptosis of macrophages and vascular endothelial cells in atherosclerosis. PLoS One. 2017;12(9):e0185406. doi: 10.1371/journal.pone.0185406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mayama T, Marr AK, Kino T. Differential expression of glucocorticoid receptor noncoding RNA repressor Gas5 in autoimmune and inflammatory diseases. Horm Metab Res. 2016;48(8):550–557. doi: 10.1055/s-0042-106898. [DOI] [PubMed] [Google Scholar]
- 25.Salemi M, Cimino L, Marino M, Cannarella R, Condorelli RA, Romano C, LaVignera S, Calogero AE. Next generation sequencing expression profiling of mitochondrial subunits in men with Klinefelter syndrome. Int J Med Sci. 2018;15:31–35. doi: 10.7150/ijms.21075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;(5):511–5. [DOI] [PMC free article] [PubMed]
- 29.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 30.Salemi M, La Vignera S, Castiglione R, Condorelli RA, Cimino L, Bosco P, Romano C, Romano C, Calogero AE. Expression of STRBP mRNA in patients with cryptorchidism and Down's syndrome. J Endocrinol Investig. 2012;35:5–7. doi: 10.1007/BF03345414. [DOI] [PubMed] [Google Scholar]
- 31.Dong S, Qu X, Li W, Zhong X, Li P, Yang S, Chen X, Shao M, Zhang L. The long non-coding RNA, GAS5, enhances gefitinib-induced cell death in innate EGFR tyrosine kinase inhibitor-resistant lung adenocarcinoma cells with wide-type EGFR via downregulation of the IGF-1R expression. J Hematol Oncol. 2015;8:43. doi: 10.1186/s13045-015-0140-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal. 2010;3:ra8. doi: 10.1126/scisignal.2000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pickard MR, Williams GT. Molecular and cellular mechanisms of action of tumour suppressor GAS5 LncRNA. Genes (Basel) 2015;6(3):484–499. doi: 10.3390/genes6030484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lucafo M, De Iudicibus S, Di Silvestre A, Pelin M, Candussio L, Martelossi S, Tommasini A, Piscianz E, Ventura A, Decorti G. Long noncoding RNA GAS5: a novel marker involved in glucocorticoid response. Curr Mol Med. 2015;15(1):94–99. doi: 10.2174/1566524015666150114122354. [DOI] [PubMed] [Google Scholar]
- 35.Xing D, Liang JQ, Li Y, Lu J, Jia HB, Xu LY, Ma XL. Identification of long noncoding RNA associated with osteoarthritis in humans. Orthop Surg. 2014;6(4):288–293. doi: 10.1111/os.12147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lucafò M, Di Silvestre A, Romano M, Avian A, Antonelli R, Martelossi S, Naviglio S, Tommasini A, Stocco G, Ventura A, Decorti G, De Iudicibus S. Role of the long non-coding RNA growth arrest-specific 5 in glucocorticoid response in children with inflammatory bowel disease. Basic Clin Pharmacol Toxicol 2018;122(1):87–93.ù. [DOI] [PubMed]
- 37.Calogero AE, Giagulli VA, Mongioì LM, Triggiani V, Radicioni AF, Jannini EA, Pasquali D. Klinefelter ItaliaN group (KING). Klinefelter syndrome: cardiovascular abnormalities and metabolic disorders. J Endocrinol Investig. 2017;40(7):705–712. doi: 10.1007/s40618-017-0619-9. [DOI] [PubMed] [Google Scholar]
- 38.Cannarella R, Mattina T, Condorelli RA, Mongioì LM, Pandini G, La Vignera S, Calogero AE. Chromosome 15 structural abnormalities: effect on IGF1R gene expression and function. Endocr Connect. 2017;6(7):528–539. doi: 10.1530/EC-17-0158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cannarella R, Condorelli RA, La Vignera S, Calogero AE. Effects of the insulin-like growth factor system on testicular differentiation and function: a review of the literature. Andrology. 2018;6(1):3–9. doi: 10.1111/andr.12444. [DOI] [PubMed] [Google Scholar]
- 40.Pitetti JL, Calvel P, Romero Y, Conne B, Truong V, Papaioannou MD, Schaad O, Docquier M, Herrera PL, Wilhelm D, Nef S. Insulin and IGF1 receptors are essential for XX and XY gonadal differentiation and adrenal development in mice. PLoS Genet. 2013;9:e1003160. doi: 10.1371/journal.pgen.1003160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grootegoed JA, Siep M, Baarends WM. Molecular and cellular mechanisms inspermatogenesis. Baillieres Best Pract Res Clin Endocrinol Metab. 2000;14:331–343. doi: 10.1053/beem.2000.0083. [DOI] [PubMed] [Google Scholar]
- 42.Print CG, Loveland KL. Germ cell suicide: new insights into apoptosis during spermatogenesis. BioEssays. 2000;22:423–430. doi: 10.1002/(SICI)1521-1878(200005)22:5<423::AID-BIES4>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 43.Koldemir O, Özgür E, Gezer U. Accumulation of GAS5 in exosomes is a marker of apoptosis induction. Biomed Rep. 2017;6(3):358–362. doi: 10.3892/br.2017.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Raw data are available in ArrayExpress database repository (https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-6107/) with accession number E-MTAB-6107.
Differential expression data were further confirmed using Cuffdiff36.Raw data are available in ArrayExpress database repository (https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-6107/) with accession number E-MTAB-6107.