Abstract
Background
The ability of MDR Gram-negative bacteria to evade even antibiotics of last resort is a severe global challenge. The development pipeline for conventional antibiotics cannot address this issue, but antimicrobial peptides (AMPs) offer an alternative solution.
Objectives
Two insect-derived AMPs (LS-sarcotoxin and LS-stomoxyn) were profiled to assess their suitability for systemic application in humans.
Methods
The peptides were tested against an extended panel of 114 clinical MDR Gram-negative bacterial isolates followed by time–kill analysis, interaction studies and assays to determine the likelihood of emerging resistance. In further in vitro studies we addressed cytotoxicity, cardiotoxicity and off-target interactions. In addition, an in vivo tolerability and pharmacokinetic study in mice was performed.
Results
LS-sarcotoxin and LS-stomoxyn showed potent and selective activity against Gram-negative bacteria and no cross-resistance with carbapenems, fluoroquinolones or aminoglycosides. Peptide concentrations of 4 or 8 mg/L inhibited 90% of the clinical MDR isolates of Escherichia coli, Enterobacter cloacae, Acinetobacter baumannii and Salmonella enterica isolates tested. The ‘all-d’ homologues of the peptides displayed markedly reduced activity, indicating a chiral target. Pharmacological profiling revealed a good in vitro therapeutic index, no cytotoxicity or cardiotoxicity, an inconspicuous broad-panel off-target profile, and no acute toxicity in mice at 10 mg/kg. In mouse pharmacokinetic experiments LS-sarcotoxin and LS-stomoxyn plasma levels above the lower limit of quantification (1 and 0.25 mg/mL, respectively) were detected after 5 and 15 min, respectively.
Conclusions
LS-sarcotoxin and LS-stomoxyn are suitable as lead candidates for the development of novel antibiotics; however, their pharmacokinetic properties need to be improved for systemic administration.
Introduction
The spread of antibiotic-resistant bacterial pathogens is a major challenge, with some strains now showing the worrying ability to overcome even antibiotics of last resort.1–4 The lack of antibiotics with novel targets or modes of action means that the development pipeline is insufficient to address MDR pathogens such as carbapenem-resistant Acinetobacter baumannii (CRAB), Pseudomonas aeruginosa (CRPA) and Enterobacteriaceae (CRE).5,6 Alternative treatment options include antibodies, probiotics and bacteriophages,7 but antimicrobial peptides (AMPs) are especially promising due to their potent antimicrobial activity and their ability to neutralize toxins.8,9
Insects produce the largest and most diverse repertoire of AMPs,10 and many insect AMPs have been assembled as a library of synthetic peptides.11–14 We previously described 23 AMPs from the medicinal maggots of the common green bottle fly Lucilia sericata, which colonizes habitats with remarkable microbial loads, such as carrion and infected wounds, and we tested these peptides against Escherichia coli and A. baumannii.13,15
Here we report the characterization of LS-sarcotoxin and LS-stomoxyn, two linear cationic AMPs from L. sericata, to assess their suitability as leads for systemic application in humans. Our experiments included in vitro and in vivo absorption, distribution, metabolism, excretion and toxicity (ADMET) analysis and activity profiling under physiological conditions against an extended panel of clinical MDR Gram-negative bacteria.
Materials and methods
Ethics
All procedures involving experimentation on animal subjects have been performed according to directive 63/2010 of the European Commission implemented in German animal welfare legislation and are registered by the competent authority (district government in Darmstadt, Hesse, Germany) under FH-1008. All procedures were in accordance with policies of Sanofi on the protection of animals and the responsible use of animals in research and production.16,17
Antimicrobial peptides
LS-sarcotoxin and LS-stomoxyn and the corresponding d-enantiomers were produced by solid-phase synthesis at ≥90% purity (GenScript, USA). The amino acid sequences and calculated physicochemical properties are listed in Table 1.
Table 1.
Properties of the synthetic L. sericata AMPs
| AMPa | Sequence | Size | MWb | pIb | Chargeb | Gc |
|---|---|---|---|---|---|---|
| LS-sarcotoxin | GWLKKIGKKIERVGQHTRDATIQTIGVAQQAANVAATLK-NH2 | 39 | 4199.86 | 11.63 | +6.1 | −0.321 |
| all-d LS-sarcotoxin | d-(GWLKKIGKKIERVGQHTRDATIQTIGVAQQAANVAATLK)-NH2 | 39 | 4199.86 | 11.63 | +6.1 | −0.321 |
| LS-stomoxyn | GFRKRFNKLVKKVKHTIKETANVSKDVAIVAGSGVAVGAAM-NH2 | 41 | 4326.13 | 11.72 | +8.1 | 0.059 |
| all-d LS-stomoxyn | d-(GFRKRFNKLVKKVKHTIKETANVSKDVAIVAGSGVAVGAAM)-NH2 | 41 | 4326.13 | 11.72 | +8.1 | 0.059 |
‘all-d’ signifies l-amino acids were replaced by the corresponding d-amino acids.
The AMP properties molecular weight (MW), isoelectric point (pI) and net charge at pH 7 (Charge) were calculated using software provided at http://pepcalc.com/.
G, GRAVY score, total hydropathy values of all the amino acids divided by the size.96
Bacterial isolates and culture conditions
The bacterial isolates (Tables S1 and S2, available as Supplementary data at JAC Online) were predominantly cultured in CAMHB (Becton Dickinson, Germany). Clinical isolates were provided by the Robert Koch Institute (Wernigerode, Germany), originating from hospitalized patients in Germany (Table S2).
Inhibition of microbial growth
MIC values were determined using initial cell populations and preparation methods appropriate for each test species (Supplementary Methods). MIC50 and MIC90 values (concentrations achieving no bacterial growth in 50%/90% of the tested isolates) were determined against a panel of MDR clinical isolates as previously defined.18
Chequerboard assay
Interactions between the peptides and colistin were investigated using the chequerboard assay,19 which we adapted to 384-well microtitre plates with an assay volume of 20 μL. The fractional inhibitory concentration (FIC) values for each AMP and colistin were calculated for each combination using the equations FICAMP = CAMP/MICAMP and FICCST = CCST/MICCST, where MICAMP and MICCST are the MICs of LS-sarcotoxin/LS-stomoxyn and colistin, respectively, and CAMP and CCST are the concentrations of LS-sarcotoxin/LS-stomoxyn and colistin in combination, respectively. Fractional inhibitory concentration index (FICI) values for each combination were calculated as follows: FICI = FICAMP + FICCST. Values ≤0.5 indicated synergy and values >4 indicated antagonism.20
Serial-passage mutagenesis
Serial-passage mutagenesis tests with E. coli and P. aeruginosa were conducted by a modification of the procedure described previously.21 Briefly, 10-fold-concentrated 1:2 dilution series of the peptides were prepared in 384-well microtitre plates over a concentration range of 5210–0.16 μg/mL in a volume of 50 μL, and 2 μL aliquots were transferred to new 384-well microtitre plates. These plates were sealed and stored at –80°C until further use. On each assay day 18 μL of the desired bacterial suspension (5 × 105 cfu/mL) was added. After incubation for 23 h, the content of the wells containing the second highest peptide concentration allowing bacterial growth was diluted 1:10000 in fresh CAMHB, and 18 μL aliquots were added to new plates containing the 10-fold concentrated peptide dilution series. This passaging was repeated for 30 days consecutively.
Time–kill kinetics
LS-sarcotoxin and LS-stomoxyn were added at 1×, 2×, 4× and 8× MIC.22 At time points 0, 0.5, 1, 2, 4 and 5 h after inoculation, 100 μL samples were diluted 1:10 in a 7-fold dilution series in PBS (pH 7.4) and plated on CAMHB agar using an Eddy Jet 2 (IUL, Spain). After overnight incubation at 37°C, the cultures were visualized using a Flash & Go camera (IUL) to determine the cfu/mL.
Haemolysis, cytotoxicity and potassium channel interaction assays
Haemolytic activity was assessed as previously described.23 Cytotoxicity was determined by measuring intracellular ATP levels of mycoplasma-free HepG2 cells (ATCC) using the CellTiter-Glo ATP monitoring kit (Promega).24 NOEC (no observed effect concentration) values were recorded, describing the highest peptide concentrations at which no cytotoxic effect (cell viability >80%) or precipitation of the test item was observed. AMPs were tested at concentrations of 1.56, 3.13, 6.25, 12.5, 25, 50, 100, 200 and 400 μM. AMP interaction with the human Ether-à-go-go-Related Gene (hERG) potassium channel was analysed using an automated patch-clamp method as previously described,25 with AMPs at concentrations of 0.12, 0.37, 1.1, 3.3, 10 and 30 μM.
Metabolic stability and plasma clearance
The metabolic stability of the AMPs was determined by measuring the half-life in human hepatocytes and calculating the human extraction ratio based on intrinsic, scaled and predicted hepatic clearance (Supplementary Methods).26 Scaled intrinsic hepatic clearance in humans was calculated based on a weight of 25.71 g liver/kg and a hepatocellularity of 99 × 106 cells/g liver. Scaled predicted hepatic clearance as well as the human extraction ratio were calculated based on a hepatic blood flow of 1.24 L/h/kg.27 Plasma clearance was determined as previously described.28
Pharmacological off-target profiling
Pharmacological off-target profiling was conducted by CEREP (Celle-Lévescault, France) to investigate inhibitory effects against the targets specified in Figure S1. All tests were conducted at a peptide concentration of 10 μM (42.0 and 43.3 mg/L for LS-sarcotoxin and LS-stomoxyn, respectively).
Handling of experimental mice
Six test-naive, healthy, 11-month-old, 34 ± 4 g male mice (RjOrl:SWISS, Janvier, France) were separated into two groups after delivery and kept in specific pathogen-free (SPF) facilities (open polycarbonate cages, EU TYP III, 820 cm2) on small animal litter composed of spruce granules (LIGNOCEL FS 14, cubic granulate, 3.5–4.5 mm). The housings were enriched with chew sticks (SAFE Block, SAFE, France), paper-based nesting material (SAFE crinklets, SAFE) and a mouse house/igloo. Each group of three animals was used as an experimental group in the study without any randomization. The mice were kept at 20–24°C, 45%–65% relative humidity and a light/dark cycle of 12 h/12 h in group housings and supplied with tap water and V1534-10 mm pellets ad libitum.
Acute toxicity and mouse pharmacokinetics
The peptides were resolved in Ampura® water (Fresenius, Germany) as stock solutions of 2.02 ± 0.02 mg/g and each was administered as a single intravenous dose of 0.19 ± 0.01 g through the tail vein to reach a final concentration of 10 mg/kg per mouse. Group 1 was treated with LS-sarcotoxin on 9 February 2017 [mouse (M) 1, 8:48:45 a.m.; M2, 11:07:48 a.m.; M3, 9:07:29 a.m.]. Group 2 was treated with LS-stomoxyn on 2 March 2017 (M1, 8:15:42 a.m.; M2, 8:19:23 a.m.; M3, 8:22:42 a.m.). No control groups were used. All procedures were performed in laboratory H826-304 (Industriepark Höchst). After administration, the mice were routinely evaluated for signs of toxicity. At 0.08, 0.25, 0.5, 1, 2, 4, 8 and 24 h after administration, 10 μL blood samples were transferred to heparin-coated tubes and then into ethanol containing 0.5% (v/v) ammonia and the plasma proteins were precipitated for 20 min at 1735 g. Supernatants were separated and analysed in triplicate by LC–MS/MS (Supplementary Methods) to evaluate the stability of the parent peptides. Three mice per group were used to calculate the standard deviation and the coefficient of variation in pharmacokinetic studies. After the experiment, the mice were euthanized with CO2.
Results
Antimicrobial activity against reference strains
LS-sarcotoxin and LS-stomoxyn lacked significant activity against Gram-positive bacteria and Candida albicans (MIC ≥1024 mg/L) as well as the Gram-negative bacterium Proteus mirabilis, which is intrinsically resistant to cationic peptides (Table S1).29,30 However, both peptides showed strong activity (MIC 4 mg/L) against the tested Enterobacteriaceae and A. baumannii. Furthermore, when tested against P. aeruginosa, LS-stomoxyn was considerably more active (MIC 8 mg/L) than LS-sarcotoxin (MIC 64 mg/L; Table S1).
Activity against an extended panel of Gram-negative clinical isolates
Next, we tested the AMPs against 114 MDR clinical isolates, which were selected based on their resistance phenotype, including but not limited to resistance to colistin and meropenem (Table S2). LS-stomoxyn was further tested against 52 isolates of P. aeruginosa. LS-sarcotoxin was active in the range 2–16 mg/L against isolates of E. coli, Enterobacter spp., Klebsiella spp., Salmonella enterica, Citrobacter freundii and Acinetobacter spp., resulting in MIC50 and MIC90 values of 4 and 8 mg/L, respectively (Table 2). The MIC profile of LS-stomoxyn ranged from 2 to >128 mg/L, with MIC50 and MIC90 values of 4 and 8 mg/L, respectively, except against K. pneumoniae (MIC90 32 mg/L). The activity of LS-stomoxyn differed strikingly from that of LS-sarcotoxin by the comparably high activity against P. aeruginosa, with MIC values in the range 4–64 mg/L and MIC50, respectively, and MIC90 values of 8 and 32 mg/L (Table 3). Both peptides were inactive or only weakly active (MIC ≥64 mg/L) against Stenotrophomonas maltophilia, Morganella morganii and Serratia fonticola, in accordance with previous findings, demonstrating that these species are intrinsically resistant to cationic peptides.29,30
Table 2.
MIC values of LS-sarcotoxin against a panel of Gram-negative clinical isolates
| Species and resistance phenotypea (number of isolates) | MIC of LS-sarcotoxin (mg/L)b |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| MIC50/90 (mg/L) | 2 | 4 | 8 | 16 | 32 | 64 | 128 | >128 | |
| E. coli (26) | 4/8 | 22 | 4 | ||||||
| CSTR MEMR (1) | 1 | ||||||||
| CSTR (9) | 8 | 1 | |||||||
| MEMR (4) | 3 | 1 | |||||||
| S (12) | 10 | 2 | |||||||
| E. cloacae (23) | 8/8 | 1 | 9 | 12 | 1 | ||||
| CSTR MEMR (1) | 1 | ||||||||
| CSTR (3) | 1 | 2 | |||||||
| MEMR (10) | 4 | 5 | 1 | ||||||
| S (9) | 5 | 4 | |||||||
| Enterobacter aerogenes (1) | 1 | ||||||||
| CSTR MEMR (1) | 1 | ||||||||
| K. pneumoniae (21) | 4/8 | 12 | 8 | 1 | |||||
| CSTR MEMR (6) | 4 | 2 | |||||||
| CSTR (9) | 4 | 4 | 1 | ||||||
| MEMR (2) | 2 | ||||||||
| S (4) | 2 | 2 | |||||||
| Klebsiella oxytoca (2) | 2 | ||||||||
| MEMR (2) | 2 | ||||||||
| S. enterica (10) | 4/8 | 5 | 5 | ||||||
| CSTR (2) | 1 | 1 | |||||||
| S (8) | 4 | 4 | |||||||
| C. freundii (1) | 1 | ||||||||
| MEMR (1) | 1 | ||||||||
| A. baumannii (20) | 4/8 | 15 | 5 | ||||||
| CSTR MEMR (3) | 2 | 1 | |||||||
| MEMR (16) | 12 | 4 | |||||||
| S (1) | 1 | ||||||||
| Acinetobacter pittii (1) | 1 | ||||||||
| S (1) | 1 | ||||||||
| P. aeruginosa (2) | 2 | ||||||||
| MEMR (2) | 2 | ||||||||
| S. maltophilia (2) | 1 | 1 | |||||||
| CSTR MEMR (2) | 1 | 1 | |||||||
| M. morganii (4) | 4 | ||||||||
| CSTR MEMR (1) | 1 | ||||||||
| CSTR (3) | 3 | ||||||||
| S. fonticola (1) | 1 | ||||||||
| CSTR MEMR (1) | 1 | ||||||||
CSTR, resistant to colistin; MEMR, resistant to meropenem; S, susceptible to colistin and meropenem.
The numbers of isolates for which the MIC value was determined are tabulated.
Table 3.
MIC values of LS-stomoxyn against a panel of Gram-negative clinical isolates
| Species and resistance phenotypea (number of isolates) | MIC of LS-stomoxyn (mg/L)b |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| MIC50/90 (mg/L) | 2 | 4 | 8 | 16 | 32 | 64 | 128 | >128 | |
| E. coli (26) | 4/8 | 15 | 9 | 1 | 1 | ||||
| CSTR MEMR (1) | 1 | ||||||||
| CSTR (9) | 5 | 3 | 1 | ||||||
| MEMR (4) | 2 | 2 | |||||||
| S (12) | 7 | 4 | 1 | ||||||
| E. cloacae (23) | 4/8 | 1 | 14 | 6 | 2 | ||||
| CSTR MEMR (1) | 1 | ||||||||
| CSTR (3) | 1 | 1 | 1 | ||||||
| MEMR (10) | 6 | 3 | 1 | ||||||
| S (9) | 7 | 1 | 1 | ||||||
| E. aerogenes (1) | 1 | ||||||||
| CSTR MEMR (1) | 1 | ||||||||
| K. pneumoniae (21) | 8/32 | 6 | 9 | 2 | 2 | 1 | 1 | ||
| CSTR MEMR (6) | 3 | 2 | 1 | ||||||
| CSTR (9) | 4 | 1 | 2 | 1 | 1 | ||||
| MEMR (2) | 2 | ||||||||
| S (4) | 1 | 3 | |||||||
| K. oxytoca (2) | 2 | ||||||||
| MEMR (2) | 2 | ||||||||
| S. enterica (10) | 4/8 | 6 | 4 | ||||||
| CSTR (2) | 2 | ||||||||
| S (8) | 4 | 4 | |||||||
| C. freundii (1) | 1 | ||||||||
| MEMR (1) | 1 | ||||||||
| A. baumannii (20) | 4/8 | 10 | 9 | 1 | |||||
| CSTR MEMR (3) | 1 | 2 | |||||||
| MEMR (16) | 9 | 6 | 1 | ||||||
| S (1) | 1 | ||||||||
| A. pittii (1) | 1 | ||||||||
| S (1) | 1 | ||||||||
| P. aeruginosa (54) | 8/32 | 3 | 28 | 14 | 8 | 1 | |||
| CSTR MEMR (1) | 1 | ||||||||
| MEMR (51) | 3 | 26 | 14 | 7 | 1 | ||||
| S (2) | 2 | ||||||||
| S. maltophilia (2) | 1 | 1 | |||||||
| CSTR MEMR (2) | 1 | 1 | |||||||
| M. morganii (4) | 2 | 2 | |||||||
| CSTR MEMR (1) | 1 | ||||||||
| CSTR (3) | 2 | 1 | |||||||
| S. fonticola (1) | 1 | ||||||||
| CSTR MEMR (1) | 1 | ||||||||
CSTR, resistant to colistin; MEMR, resistant to meropenem; S, susceptible to colistin and meropenem.
The numbers of isolates for which the MIC value was determined are tabulated.
To investigate potential correlation between colistin and/or meropenem resistance and a low susceptibility to the AMPs, we compared the signed-rank median MIC values of the two AMPs against the different resistance phenotypes (resistance to colistin and/or meropenem as well as isolates susceptible to both antibiotics) (Figure S2). No significant differences in AMP activity were observed against resistant and susceptible isolates, although the activity of LS-stomoxyn against the colistin-resistant but meropenem-susceptible strains was less pronounced (Figure S2).
Activity of all-d amino acid AMPs
To determine whether the AMPs recognize a specific target in a stereospecific manner or interact non-specifically with the lipid bilayer,31–35 we tested the natural all-l and corresponding all-d enantiomers of both AMPs against several Gram-negative reference strains and clinical isolates (Table 4). The all-d LS-sarcotoxin was virtually inactive against most strains, with MIC values ≥1024 mg/L. Some residual activity (MIC 128–256 mg/L) was observed against E. coli RKI 131/08, A. baumannii ATCC 19606 and A. baumannii RKI 19/09. The all-d LS-stomoxyn displayed a clear but less pronounced decline in activity, with MIC values of 16–64 mg/L.
Table 4.
Activity of all-d enantiomers of L. sericata AMPs compared with the native parental all-l enantiomers
| MIC (mg/L)a |
|||||
|---|---|---|---|---|---|
| Test strain | Phenotype | LS-sarcotoxin | all-d LS-sarcotoxinb | LS-stomoxyn | all-d LS-stomoxynb |
| E. coli | S | 4 | 1024 | 4 | 16 |
| E. coli | MEMR | 4 | 128 | 4 | 16 |
| K. pneumoniae | S | 4 | >1024 | 4 | 16 |
| K. pneumoniae | MEMR | 4 | >1024 | 4 | 16 |
| A. baumannii | S | 4 | 256 | 4 | 64 |
| A. baumannii | MEMR | 4 | 128 | 4 | 32 |
| P. aeruginosa | S | 64 | >1024 | 8 | 32 |
| P. aeruginosa | MEMR | 128 | >1024 | 8 | 32 |
| E. coli | CSTR | 4 | >1024 | 4 | 16 |
| K. pneumoniae | CSTR | 16 | >1024 | 8 | 32 |
MIC values were determined in CAMHB for the colistin/meropenem-susceptible (S) strains E. coli ATCC 25922, K. pneumoniae DSM 30104, A. baumannii ATCC 19606, P. aeruginosa ATCC 27853, the meropenem-resistant (MEMR) strains E. coli RKI 131/03, K. pneumoniae RKI 93/10, A. baumannii RKI 19/09, P. aeruginosa RKI 93/12, and the colistin-resistant (CSTR) strains E. coli RKI 6A-6 and K. pneumoniae RKI 19/16.
Enantiomer in which l-amino acids were replaced by the corresponding d-amino acids.
Activity under physiological conditions
Physiological conditions were approximated by supplementing the CAMHB medium with 150 mM NaCl or 1.25 mM CaCl2. A salt-dependent increase in MIC values was no higher than 2-fold in all experiments and there was little impact on the activity of LS-sarcotoxin (Table 5). The addition of 10% (v/v) human serum boosted the antibacterial activity (Table 5). The MIC values of LS-sarcotoxin decreased by 8- to 32-fold, to ∼0.25 mg/L for E. coli, K. pneumoniae and A. baumannii, whereas the MIC values for LS-stomoxyn decreased by 16- to 128-fold, to ∼0.063 mg/L. In contrast, the activity against P. aeruginosa declined by 2- to 4-fold. To replicate physiological conditions more accurately, we tested the peptides in CAMHB adjusted to 150 mM NaCl and also supplemented with 10% (v/v) human serum, which enhanced the activity of both AMPs even further (Table 5). The MIC values of LS-sarcotoxin against E. coli and A. baumannii were 128-fold lower (0.031 mg/L) and against K. pneumoniae and colistin-resistant E. coli they were 16- to 32-fold lower (0.13–1 mg/L). The MIC values of LS-stomoxyn were ∼0.016 mg/L for E. coli, K. pneumoniae and A. baumannii, reflecting at least a 256-fold increase in activity. A remarkable but less pronounced 64-fold increase in activity was also observed for the colistin-resistant E. coli and K. pneumoniae isolates. The addition of mouse serum rather than human serum achieved similar although somewhat less pronounced effects on the MIC values (Table S3).
Table 5.
Relative increase in activity under different approximated physiological conditions
| Fold decrease in MIC compared with CAMHBa |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| LS-sarcotoxin |
LS-stomoxyn |
||||||||
| Test strainb | Phenotype | N | C | S | S + N | N | C | S | S + N |
| E. coli | S | 1 | 1 | 16 | 128 | 1 | 1 | 64 | 256 |
| E. coli | MEMR | 1 | 1 | 32 | 128 | 1 | 0.5 | 128 | 256 |
| K. pneumoniae | S | 1 | 1 | 32 | 32 | 0.5 | 0.5 | 128 | 256 |
| K. pneumoniae | MEMR | 0.5 | 1 | 8 | 32 | 0.5 | 0.5 | 64 | 256 |
| A. baumannii | S | 1 | 1 | 16 | 128 | 0.5 | 0.5 | 32 | 256 |
| A. baumannii | MEMR | 1 | 1 | 16 | 128 | 1 | 0.5 | 64 | 256 |
| P. aeruginosa | S | ND | ND | 0.5 | 0.25 | 0.5 | 0.5 | 4 | 4 |
| P. aeruginosa | MEMR | ND | ND | 0.25 | 0.25 | 1 | 0.5 | 0.5 | 0.5 |
| E. coli | CSTR | ND | ND | 16 | 32 | ND | ND | 32 | 64 |
| K. pneumoniae | CSTR | ND | ND | 8 | 16 | ND | ND | 16 | 64 |
ND, not determined.
MICs were determined in CAMHB, CAMHB adjusted to 150 mM NaCl (N) or 1.25 mM CaCl2 (C), CAMHB supplemented with 10% human serum (S) and CAMHB supplemented with 10% human serum and adjusted to 150 mM NaCl (S + N). Fold changes in MIC values were calculated with respect to the MIC values obtained in CAMHB.
MIC values were determined for the colistin/meropenem-susceptible (S) strains E. coli ATCC 25922, K. pneumoniae DSM 30104, A. baumannii ATCC 19606, P. aeruginosa ATCC 27853, the meropenem-resistant (MEMR) strains E. coli RKI 131/03, K. pneumoniae RKI 93/10, A. baumannii RKI 19/09, P. aeruginosa RKI 93/12 and the colistin-resistant (CSTR) strains E. coli RKI 6A-6 and K. pneumoniae RKI 19/16.
Antibacterial activity in combination with colistin
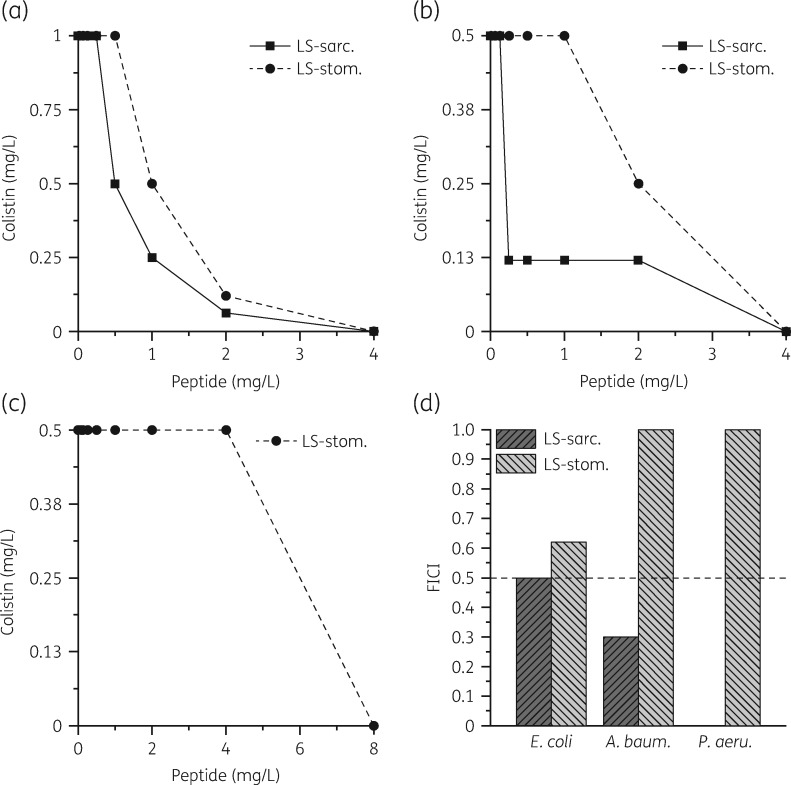
In preliminary experiments we observed 2-fold lower MIC values for LS-sarcotoxin and LS-stomoxyn when tested in the presence of sub-MIC concentrations of colistin (0.075 mg/L). For more detailed analysis, we performed chequerboard titration experiments (Figure 1). All experiments revealed a synergistic interaction between LS-sarcotoxin and colistin (FICI ≤0.5). No interaction was observed between LS-stomoxyn and colistin (FICI 1–0.6).
Figure 1.
Interaction of LS-sarcotoxin (LS-sarc.) and LS-stomoxyn (LS-stom.) with colistin tested on E. coli ATCC 25922 (a), A. baumannii ATCC 19606 (b) and P. aeruginosa ATCC 27853 (c). The data points of the isobolograms represent the concentrations of the two individual compounds in different combinations leading to complete bacterial growth inhibition. The calculated FICIs are displayed in (d). Values ≤0.5 indicate synergy.
Development of resistance to L. sericata AMPs
Serial passaging of E. coli and P. aeruginosa for 30 days in the presence of LS-sarcotoxin or LS-stomoxyn did not result in the emergence of any resistant mutants (Figure S3).
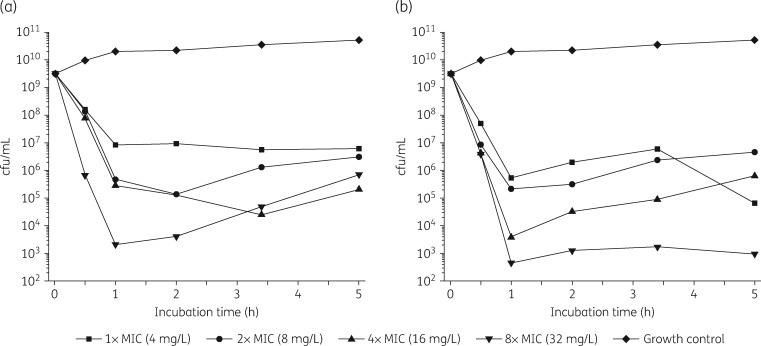
Time–kill kinetics
LS-sarcotoxin at 2× and 3× the MIC led to a rapid ≥3 log reduction in cfu/mL after incubation for 1 h (Figure 2). At a concentration corresponding to 8 × MIC, a ≥6 log reduction in cfu/mL was achieved after incubation for only 30 min. At 1 × MIC, LS-sarcotoxin reduced cfu/mL by 2.6 log after 1 h and did not achieve bactericidal activity. LS-stomoxyn was strongly bactericidal at all the concentrations we tested and caused a ≥3 log reduction after incubation for 1 h (Figure 2).
Figure 2.
Kill kinetics of LS-sarcotoxin (a) and LS-stomoxyn (b) against E. coli ATCC 25922 determined in CAMHB. The cfu/mL values are shown according to the incubation time for different peptide concentrations.
In vitro toxicity studies
For LS-sarcotoxin, haemolysis was observed at 1024 mg/L, whereas all-d LS-sarcotoxin, LS-stomoxyn and all-d LS-stomoxyn showed no haemolytic activity up to 1024 mg/L (Table 6). All four peptides showed minimal HepG2 toxicity, with NOECs of 100 μM (420 and 433 mg/L for LS-sarcotoxin and LS-stomoxyn, respectively; Table 6). Furthermore, as a model for cardiotoxicity, we investigated the effects of LS-sarcotoxin and LS-stomoxyn on the hERG potassium ion channel. No target-specific activity was observed at concentrations of up to 30 μM (126 and 130 mg/L, respectively), representing the highest test concentrations (Table 6).
Table 6.
In vitro toxicity and metabolic stability of the L. sericata peptides
| AMP | MHCa (mg/L) | NOECb (mg/L) | IC50 hERGc (mg/L) | t½ hepatocytesd (min) | Ehe (%) |
|---|---|---|---|---|---|
| LS-sarcotoxin | 1024 | 420 | >126 | 1060 | 13.9 |
| all-d LS-sarcotoxin | >1024 | 420 | ND | >5000 | <3 |
| LS-stomoxyn | >1024 | 433 | >130 | 77 | 68.8 |
| all-d LS-stomoxyn | >1024 | 433 | ND | 4110 | 3.9 |
ND, not determined.
Minimal haemolytic concentration determined for human erythrocytes.
Highest peptide concentration at which no cytotoxic effect (cell viability >80%) was observed for HepG2 cells.
Concentration at which the hERG channel was inhibited by 50%.
Half-life of AMPs determined in human cryopreserved hepatocytes.
Human extraction ratio: proportion of the compound that is eliminated by one passage through the liver.
In vitro stability studies
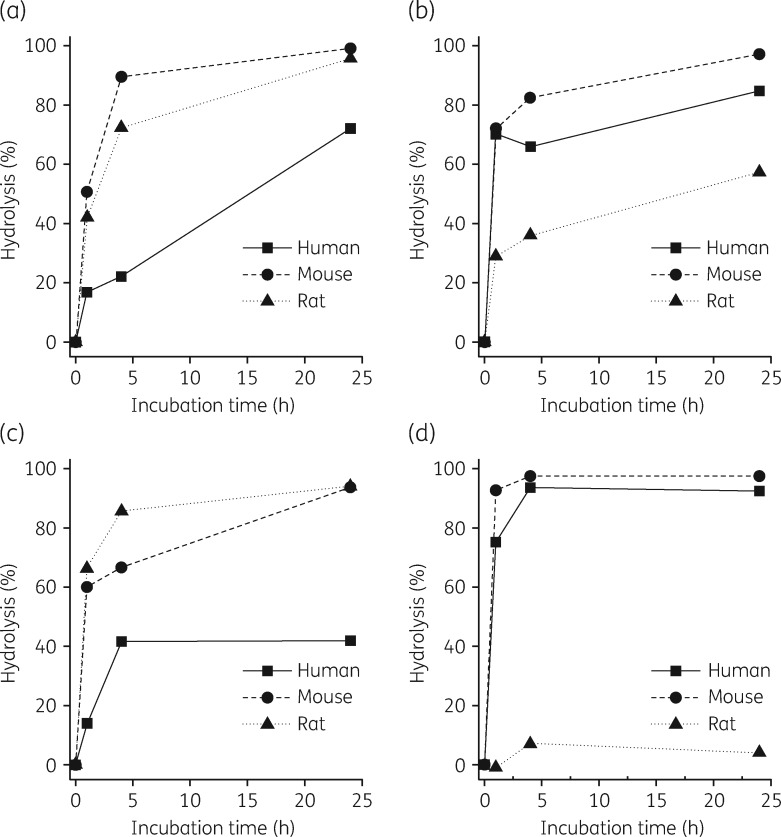
LS-sarcotoxin and all-d LS-sarcotoxin were stable, with half-lives >1000 min in human hepatocytes and calculated human extraction ratios <14% (Table 6). LS-stomoxyn was less stable, with a half-life of 77 min and a calculated human extraction ratio of 68.8%. In contrast, all-d LS-stomoxyn was stable, with a half-life of 4110 min and a calculated human extraction ratio of 3.9%. All four peptides were considered unstable in human, mouse and rat plasma after incubation for 24 h (Figure 3). Only all-d LS-stomoxyn was stable in rat plasma (4% hydrolysis). LS-sarcotoxin and its all-d enantiomer were stable in human plasma for 1 h with only 17% and 14% loss, respectively.
Figure 3.
Stability of LS-sarcotoxin (a), all-d LS-sarcotoxin (b), LS-stomoxyn (c) and all-d LS-stomoxyn (d) in plasma from different species. The extent of hydrolytic degradation is shown according to the incubation time in human, mouse and rat plasma.
Pharmacological off-target profiling
Pharmacological broad-spectrum off-target profiling of LS-sarcotoxin and LS-stomoxyn against 19 G-protein-coupled receptors (GPCRs), 8 ion channels, 2 transporters and 4 enzymes revealed few conspicuous interactions at a concentration of 10 μM (42.0 and 43.3 mg/L for LS-sarcotoxin and LS-stomoxyn, respectively) (Figure S1). We observed antagonistic inhibition of two human muscarinic acetylcholine receptors (M1 and M3) by LS-stomoxyn, reducing their activity by 49% and 44%, respectively, and antagonistic inhibition of the human M3 receptor by LS-sarcotoxin, reducing its activity by 31%.
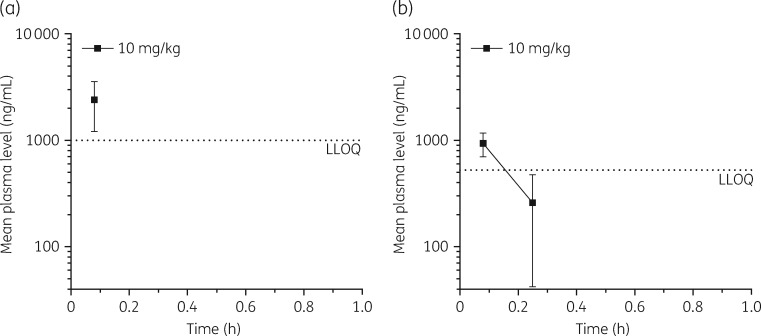
Tolerability and mouse pharmacokinetic profiling
LS-sarcotoxin and LS-stomoxyn were each administered to 3/6 test-naive, healthy male Swiss mice at a single intravenous dose of 10 mg/kg. All mice survived without signs of toxicity or adverse events. Both peptides were rapidly cleared from the plasma. For LS-sarcotoxin, plasma concentrations above the lower limit of quantification (LLOQ) were only detected 5 min after administration, the first sampling timepoint (Figure 4a). For LS-stomoxyn, plasma concentrations above the LLOQ were detected up to 15 min after administration (Figure 4b).
Figure 4.
Mean plasma levels (±SD) of LS-sarcotoxin (a) and LS-stomoxyn (b) after a single intravenous administration of 10 mg/kg to male Swiss mice. For LS-sarcotoxin and LS-stomoxyn, the LLOQ was determined as 1000 and 250 ng/mL, respectively.
Discussion
The selective activity of LS-sarcotoxin and LS-stomoxyn against Gram-negative bacteria was verified against a large panel of MDR clinical isolates, indicating the absence of cross-resistance to β-lactams, aminoglycosides, ciprofloxacin, chloramphenicol and sulfamerazine/trimethoprim. Selective activity against Gram-negative bacteria has been reported for other insect-derived AMPs36,37 and artificial AMPs.38 Among current clinical antibiotics, only colistin (polymyxin E) and polymyxin B display a similar activity profile,39 highlighting the reportedly similar mechanism of polymyxins and cationic α-helical AMPs.8,40–44
We also observed results that highlighted differences between the L. sericata AMPs and polymyxins. For example, the AMPs retained full (LS-sarcotoxin) or only moderately reduced (LS-stomoxyn) activity against colistin-resistant isolates of E. coli, Enterobacter cloacae, K. pneumoniae and A. baumannii (Figure S2). Furthermore, the synergistic activity of LS-sarcotoxin with colistin would not be observable if both compounds had an identical target. Cationic AMPs from various sources are active against bacterial isolates with acquired colistin resistance, including AMPs isolated from frog skin,45 cecropin A/melittin hybrid peptides,46 the artificial peptides WLBU2 and WR1247 and star-shaped peptide polymers composed of randomly polymerized lysine and valine residues, known as structurally nano-engineered antimicrobial peptide polymers (SNAPPs).48 Other cationic AMPs, including human cathelicidin LL-37,47 the insect cecropins A and B,49 the porcine AMP cecropin PI49 and the artificial tetra-branched peptide SET-M33L,38 show cross-resistance to colistin. Whereas E. coli ATCC 25922 and P. aeruginosa ATCC 27853 rapidly develop resistance (>100-fold change in MIC) to colistin in serial passaging experiments,50,51 we recovered no mutants resistant to LS-sarcotoxin or LS-stomoxyn during a 30 day passaging experiment at sub-MIC concentrations, also supporting a mode of action distinct from colistin.
Evidence for LS-sarcotoxin and LS-stomoxyn interacting with specific chiral targets rather than inducing detergent-like membrane lysis was provided by the low activity of their d-enantiomers.34,35,52 Complete loss of activity was previously observed for the all-d analogue of apidaecin, a proline-rich AMP from honeybees, which was recently shown to bind specifically to the bacterial heat shock protein DnaK and to block the assembly of the 50S ribosomal subunit in a stereospecific manner.53,54 In contrast, the l and d forms of the α-helical AMPs cecropin A, cecropin B and magainin-2 showed nearly identical antibacterial activity,31,52,55,56 indicating that the formation of pores in lipid bilayers is sufficient.31–35,56
AMPs with a large hydrophobic surface area or a high cationic charge can be toxic to human cells,57,58 but neither LS-sarcotoxin nor LS-stomoxyn showed evidence of haemolytic or cytotoxic effects. AMPs are typically less active in the presence of human serum due to salt-mediated charge repulsion, proteolytic degradation or interactions with plasma proteins.59–61 However, rather than the anticipated increase in MIC values, we observed a reduction for both peptides in the presence of human serum, as reported previously for polymyxin B, polymyxin B nonapeptide, colistin, magainin 2 and the synthetic polymyxin derivative SPR741, which may reflect interactions with the complement system.62–72 However, in preliminary experiments we also observed an increase in activity when we used serum in which the complement system was inactivated by heat treatment.
Despite the increasing number of AMP candidates,38,51,73,74 toxicity and unfavourable ADMET properties75,76 have thus far prevented the development of AMPs for systemic application.9,74,77 We found that LS-sarcotoxin and LS-stomoxyn were stable in the presence of hepatocytes (t½ 1060 and 77 min, respectively). Chemical analysis revealed the rapid loss of peptides exposed to human, mouse or rat plasma, but there was no difference in loss rate for the l and d forms, suggesting the AMPs are binding to plasma proteins such as albumin, apolipoproteins or glycoproteins.78–81 Although plasma protein binding of antibiotics affects pharmacological parameters,75,82–86 this does not rule out the suitability of the peptides for systemic application given that several approved antibiotics show >90% plasma protein binding.64,67,68,85,87 In contrast to other reported AMPs,88–90 LS-sarcotoxin and LS-stomoxyn presented inconspicuous off-target profiles. Mouse tolerability studies revealed that LS-stomoxyn or LS-stomoxyn was well tolerated, indeed better tolerated than colistin,38,51,91 which is approved for the clinic.92–94 Although both peptides were rapidly cleared from mouse plasma and in vitro stability studies showed similar results for rat and human plasma, several strategies to increase the half-life of AMPs have been described,38,48,60,95 which could be used to improve the observed pharmacokinetic profiles.
In conclusion, both LS-sarcotoxin and LS-stomoxyn are promising leads for the development of new antibiotics with activity against MDR Gram-negative bacteria and possibly a novel mode of action. However, the pharmacokinetic properties of the native insect AMPs need to be improved before proceeding to in vivo infection models.
Supplementary Material
Acknowledgements
We thank Kirsten-Susann Bommersheim, Sibylle Müller-Bertling and Kirstin Ganske for excellent technical assistance and Dr Richard M. Twyman for professional editing of the manuscript.
Funding
This work was supported by the Hesse State Ministry of Higher Education, Research and the Arts (HMWK) via a generous grant for the LOEWE Center for Insect Biotechnology and Bioresources and by the Federal Ministry of Education and Research (BMBF) in Germany via the project ‘Triple-In’.
Transparency declarations
None to declare.
References
- 1.CDC. Antibiotic Resistance Threats in the United States, 2013 2013. https://www.cdc.gov/drugresistance/pdf/ar-threats-2013-508.pdf.
- 2. O'Neill J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations 2016. https://amr-review.org/sites/default/files/160518_Final%20paper_with%20cover.pdf.
- 3. Stern S, Chorzelski S, Franken L. et al. Breaking Through the Wall: A Call for Concerted Action on Antibiotics Research and Development 2017. https://www.bundesgesundheitsministerium.de/fileadmin/Dateien/5_Publikationen/Gesundheit/Berichte/GUARD_Follow_Up_Report_Full_Report_final.pdf.
- 4.WHO. Prioritization of Pathogens to Guide Discovery, Research and Development of New Antibiotics for Drug-Resistant Bacterial Infections, Including Tuberculosis 2017. http://www.who.int/medicines/areas/rational_use/PPLreport_2017_09_19.pdf.
- 5. Taneja N, Kaur H.. Insights into newer antimicrobial agents against Gram-negative bacteria. Microbiol Insights 2016; 9: 9–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.WHO. Antibacterial Agents in Clinical Development: An Analysis of the Antibacterial Clinical Development Pipeline, Including Tuberculosis 2017. www.who.int/iris/bitstream/handle/10665/258965/WHO-EMP-IAU-2017.11-eng.pdf.
- 7. Czaplewski L, Bax R, Clokie M. et al. Alternatives to antibiotics—a pipeline portfolio review. Lancet Infect Dis 2016; 16: 239–51. [DOI] [PubMed] [Google Scholar]
- 8. Kang HK, Kim C, Seo CH. et al. The therapeutic applications of antimicrobial peptides (AMPs): a patent review. J Microbiol 2017; 55: 1–12. [DOI] [PubMed] [Google Scholar]
- 9. Kosikowska P, Lesner A.. Antimicrobial peptides (AMPs) as drug candidates: a patent review (2003-2015). Expert Opin Ther Pat 2016; 26: 689–702. [DOI] [PubMed] [Google Scholar]
- 10. Mylonakis E, Podsiadlowski L, Muhammed M. et al. Diversity, evolution and medical applications of insect antimicrobial peptides. Philos Trans R Soc Lond B Biol Sci 2016; 371: 20150290.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Altincicek B, Vilcinskas A.. Septic injury inducible genes in medicinal maggots of the blowfly Lucilia sericata. Insect Mol Biol 2009; 18: 119–25. [DOI] [PubMed] [Google Scholar]
- 12. Franta Z, Vogel H, Lehmann R. et al. Next generation sequencing identifies five major classes of potentially therapeutic enzymes secreted by Lucilia sericata medical maggots. Biomed Res Int 2016; 2016: 1–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Poppel AK, Vogel H, Wiesner J. et al. Antimicrobial peptides expressed in medicinal maggots of the blow fly Lucilia sericata show combinatorial activity against bacteria. Antimicrob Agents Chemother 2015; 59: 2508–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vogel H, Schmidtberg H, Vilcinskas A.. Comparative transcriptomics in three ladybird species supports a role for immunity in invasion biology. Dev Comp Immunol 2017; 67: 452–6. [DOI] [PubMed] [Google Scholar]
- 15. Jayamani E, Rajamuthiah R, Larkins-Ford J. et al. Insect-derived cecropins display activity against Acinetobacter baumannii in a whole-animal high-throughput Caenorhabditis elegans model. Antimicrob Agents Chemother 2015; 59: 1728–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanofi. Responsible Use of Animals in Research and Production 2016. https://www.sanofi.com/en/science-and-innovation/clinical-trials-and-results/responsible-use-of-animals-in-research-and-production/.
- 17.Sanofi. Sanofi Policy on the Protection of Animals 2015. https://www.sanofi.com/media/Project/One-Sanofi-Web/sanofi-com/common/docs/download-center/Sanofi_policy_protection_animals_EN_2015.pdf.
- 18. Magiorakos AP, Srinivasan A, Carey RB. et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 2012; 18: 268–81. [DOI] [PubMed] [Google Scholar]
- 19. Orhan G, Bayram A, Zer Y. et al. Synergy tests by Etest and checkerboard methods of antimicrobial combinations against Brucella melitensis. J Clin Microbiol 2005; 43: 140–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Johnson MD, MacDougall C, Ostrosky-Zeichner L. et al. Combination antifungal therapy. Antimicrob Agents Chemother 2004; 48: 693–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Silverman JA, Oliver N, Andrew T. et al. Resistance studies with daptomycin. Antimicrob Agents Chemother 2001; 45: 1799–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mascio CTM, Alder JD, Silverman JA.. Bactericidal action of daptomycin against stationary-phase and nondividing Staphylococcus aureus cells. Antimicrob Agents Chemother 2007; 51: 4255–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhu X, Shan A, Ma Z. et al. Bactericidal efficiency and modes of action of the novel antimicrobial peptide T9W against Pseudomonas aeruginosa. Antimicrob Agents Chemother 2015; 59: 3008–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hannah R, Beck M, Moravec R. et al. CellTiter-Glo™ Luminescent Cell Viability Assay: a sensitive and rapid method for determining cell viability. Promega Cell Notes 2001; 2: 11–3. [Google Scholar]
- 25. Houtmann S, Schombert B, Sanson C. et al. Automated patch-clamp methods for the hERG cardiac potassium channel. Methods Mol Biol 2017; 1641: 187–99. [DOI] [PubMed] [Google Scholar]
- 26. Ong V, Hough G, Schlosser M. et al. Preclinical evaluation of the stability, safety, and efficacy of CD101, a novel echinocandin. Antimicrob Agents Chemother 2016; 60: 6872–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Poulin P, Kenny JR, Hop CE. et al. In vitro-in vivo extrapolation of clearance: modeling hepatic metabolic clearance of highly bound drugs and comparative assessment with existing calculation methods. J Pharm Sci 2012; 101: 838–51. [DOI] [PubMed] [Google Scholar]
- 28. Ling LL, Schneider T, Peoples AJ. et al. A new antibiotic kills pathogens without detectable resistance. Nature 2015; 517: 455–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McCoy AJ, Liu H, Falla TJ. et al. Identification of Proteus mirabilis mutants with increased sensitivity to antimicrobial peptides. Antimicrob Agents Chemother 2001; 45: 2030–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Samonis G, Korbila IP, Maraki S. et al. Trends of isolation of intrinsically resistant to colistin Enterobacteriaceae and association with colistin use in a tertiary hospital. Eur J Clin Microbiol Infect Dis 2014; 33: 1505–10. [DOI] [PubMed] [Google Scholar]
- 31. Bland JM, De Lucca AJ, Jacks TJ. et al. All-d-cecropin B: synthesis, conformation, lipopolysaccharide binding, and antibacterial activity. Mol Cell Biochem 2001; 218: 105–11. [DOI] [PubMed] [Google Scholar]
- 32. Chen Y, Vasil AI, Rehaume L. et al. Comparison of biophysical and biologic properties of α-helical enantiomeric antimicrobial peptides. Chem Biol Drug Des 2006; 67: 162–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Huang J, Hao D, Chen Y. et al. Inhibitory effects and mechanisms of physiological conditions on the activity of enantiomeric forms of an alpha-helical antibacterial peptide against bacteria. Peptides 2011; 32: 1488–95. [DOI] [PubMed] [Google Scholar]
- 34. Manabe T, Kawasaki K.. d-form KLKLLLLLKLK-NH2 peptide exerts higher antimicrobial properties than its l-form counterpart via an association with bacterial cell wall components. Sci Rep 2017; 7: 43384.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhao Y, Zhang M, Qiu S. et al. Antimicrobial activity and stability of the d-amino acid substituted derivatives of antimicrobial peptide polybia-MPI. AMB Express 2016; 6: 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ebbensgaard A, Mordhorst H, Overgaard MT. et al. Comparative evaluation of the antimicrobial activity of different antimicrobial peptides against a range of pathogenic bacteria. PLoS One 2015; 10: e0144611.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tonk M, Vilcinskas A.. The medical potential of antimicrobial peptides from insects. Curr Top Med Chem 2017; 17: 554–75. [DOI] [PubMed] [Google Scholar]
- 38. Brunetti J, Falciani C, Roscia G. et al. In vitro and in vivo efficacy, toxicity, bio-distribution and resistance selection of a novel antibacterial drug candidate. Sci Rep 2016; 6: 26077.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Falagas ME, Kasiakou SK, Saravolatz LD.. Colistin: the revival of polymyxins for the management of multidrug-resistant Gram-negative bacterial infections. Clin Infect Dis 2005; 40: 1333–41. [DOI] [PubMed] [Google Scholar]
- 40. Brogden KA. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat Rev Microbiol 2005; 3: 238–50. [DOI] [PubMed] [Google Scholar]
- 41. Le CF, Fang CM, Sekaran SD.. Intracellular targeting mechanisms by antimicrobial peptides. Antimicrob Agents Chemother 2017; 61: e02340–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lohner K. Membrane-active antimicrobial peptides as template structures for novel antibiotic agents. Curr Top Med Chem 2017; 17: 508–19. [PubMed] [Google Scholar]
- 43. Nguyen LT, Haney EF, Vogel HJ.. The expanding scope of antimicrobial peptide structures and their modes of action. Trends Biotechnol 2011; 29: 464–72. [DOI] [PubMed] [Google Scholar]
- 44. Wimley WC. Describing the mechanism of antimicrobial peptide action with the interfacial activity model. ACS Chem Biol 2010; 5: 905–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Conlon JM, Sonnevend A, Pal T. et al. Efficacy of six frog skin-derived antimicrobial peptides against colistin-resistant strains of the Acinetobacter baumannii group. Int J Antimicrob Agents 2012; 39: 317–20. [DOI] [PubMed] [Google Scholar]
- 46. Rodriguez-Hernandez MJ, Saugar J, Docobo-Perez F. et al. Studies on the antimicrobial activity of cecropin A-melittin hybrid peptides in colistin-resistant clinical isolates of Acinetobacter baumannii. J Antimicrob Chemother 2006; 58: 95–100. [DOI] [PubMed] [Google Scholar]
- 47. Deslouches B, Steckbeck JD, Craigo JK. et al. Engineered cationic antimicrobial peptides to overcome multidrug resistance by ESKAPE pathogens. Antimicrob Agents Chemother 2015; 59: 1329–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lam SJ, O'Brien-Simpson NM, Pantarat N. et al. Combating multidrug-resistant Gram-negative bacteria with structurally nanoengineered antimicrobial peptide polymers. Nat Microbiol 2016; 1: 16162.. [DOI] [PubMed] [Google Scholar]
- 49. Vila-Farres X, Garcia de la Maria C, Lopez-Rojas R. et al. In vitro activity of several antimicrobial peptides against colistin-susceptible and colistin-resistant Acinetobacter baumannii. Clin Microbiol Infect 2012; 18: 383–7. [DOI] [PubMed] [Google Scholar]
- 50. Hashemi MM, Rovig J, Weber S. et al. Susceptibility of colistin-resistant, Gram-negative bacteria to antimicrobial peptides and ceragenins. Antimicrob Agents Chemother 2017; 61: e00292–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Uppu D, Konai MM, Sarkar P. et al. Membrane-active macromolecules kill antibiotic-tolerant bacteria and potentiate antibiotics towards Gram-negative bacteria. PLoS One 2017; 12: e0183263.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gad SC. Development of Therapeutic Agents Handbook. Hoboken, NJ, USA: John Wiley & Sons, 2012. [Google Scholar]
- 53. Casteels P, Tempst P.. Apidaecin-type peptide antibiotics function through a non-poreforming mechanism involving stereospecificity. Biochem Biophys Res Commun 1994; 199: 339–45. [DOI] [PubMed] [Google Scholar]
- 54. Krizsan A, Prahl C, Goldbach T. et al. Short proline-rich antimicrobial peptides inhibit either the bacterial 70S ribosome or the assembly of its large 50S subunit. ChemBioChem 2015; 16: 2304–8. [DOI] [PubMed] [Google Scholar]
- 55. Bessalle R, Kapitkovsky A, Gorea A. et al. All-D-magainin: chirality, antimicrobial activity and proteolytic resistance. FEBS Lett 1990; 274: 151–5. [DOI] [PubMed] [Google Scholar]
- 56. Wade D, Boman A, Wahlin B. et al. All-D amino acid-containing channel-forming antibiotic peptides. Proc Natl Acad Sci USA 1990; 87: 4761–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Teixeira V, Feio MJ, Bastos M.. Role of lipids in the interaction of antimicrobial peptides with membranes. Prog Lipid Res 2012; 51: 149–77. [DOI] [PubMed] [Google Scholar]
- 58. Zelezetsky I, Tossi A.. Alpha-helical antimicrobial peptides—using a sequence template to guide structure-activity relationship studies. Biochim Biophys Acta 2006; 1758: 1436–49. [DOI] [PubMed] [Google Scholar]
- 59. Johansson J, Gudmundsson GH, Rottenberg ME. et al. Conformation-dependent antibacterial activity of the naturally occurring human peptide LL-37. J Biol Chem 1998; 273: 3718–24. [DOI] [PubMed] [Google Scholar]
- 60. Knappe D, Henklein P, Hoffmann R. et al. Easy strategy to protect antimicrobial peptides from fast degradation in serum. Antimicrob Agents Chemother 2010; 54: 4003–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Maisetta G, Di Luca M, Esin S. et al. Evaluation of the inhibitory effects of human serum components on bactericidal activity of human beta defensin 3. Peptides 2008; 29: 1–6. [DOI] [PubMed] [Google Scholar]
- 62. Boswell FJ, Ashby JP, Andrews JM. et al. Effect of protein binding on the in vitro activity and pharmacodynamics of faropenem. J Antimicrob Chemother 2002; 50: 525–32. [DOI] [PubMed] [Google Scholar]
- 63. Cha R, Rybak MJ.. Influence of protein binding under controlled conditions on the bactericidal activity of daptomycin in an in vitro pharmacodynamic model. J Antimicrob Chemother 2004; 54: 259–62. [DOI] [PubMed] [Google Scholar]
- 64. Dalhoff A. Seventy-five years of research on protein binding—do we ever understand? Antimicrob Agents Chemother 2018; 62: pii: e01663–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Darveau RP, Cunningham MD, Seachord CL. et al. β-Lactam antibiotics potentiate magainin 2 antimicrobial activity in vitro and in vivo. Antimicrob Agents Chemother 1991; 35: 1153–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Davis SD, Iannetta A, Wedgwood RJ.. Paradoxical synergism and antagonism between serum and the antibacterial activity of colistin. J Infect Dis 1971; 123: 392–8. [DOI] [PubMed] [Google Scholar]
- 67. Dutcher BS, Reynard AM, Beck ME. et al. Potentiation of antibiotic bactericidal activity by normal human serum. Antimicrob Agents Chemother 1978; 13: 820–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Perl TM, Pfaller MA, Houston A. et al. Effect of serum on the in vitro activities of 11 broad-spectrum antibiotics. Antimicrob Agents Chemother 1990; 34: 2234–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Sweeney D, Cotroneo N, Lister T. et al. The impact of varied test conditions of the in vitro activity of the novel gyrase inhibitor SPR719 in combination with the potentiator SPR741 Poster. In: ASM 2017. Boston, MA, USA, 2017. [Google Scholar]
- 70. Traub WH, Kleber I.. Studies on the additive effect of polymyxin B and the bactericidal activity of human serum against Serratia marcescens. Chemotherapy 1975; 21: 189–204. [DOI] [PubMed] [Google Scholar]
- 71. Vaara M, Vaara T.. Sensitization of Gram-negative bacteria to antibiotics and complement by a nontoxic oligopeptide. Nature 1983; 303: 526–8. [DOI] [PubMed] [Google Scholar]
- 72. Vaara M, Viljanen P, Vaara T. et al. An outer membrane-disorganizing peptide PMBN sensitizes E. coli strains to serum bactericidal action. J Immunol 1984; 132: 2582–9. [PubMed] [Google Scholar]
- 73. de Breij A, Riool M, Cordfunke RA. et al. The antimicrobial peptide SAAP-148 combats drug-resistant bacteria and biofilms. Sci Transl Med 2018; 10: eaan4044.. [DOI] [PubMed] [Google Scholar]
- 74. Mandell JB, Deslouches B, Montelaro RC. et al. Elimination of antibiotic resistant surgical implant biofilms using an engineered cationic amphipathic peptide WLBU2. Sci Rep 2017; 7: 18098.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Chung TDY, Terry DB, Smith LH. et al. In vitro and in vivo assessment of ADME and PK properties during lead selection and lead optimization—guidelines, benchmarks and rules of thumb In: Sittampalam GS, Coussens NP, Brimacombe K, eds. Assay Guidance Manual. Bethesda, MD, USA: Eli Lilly & Company and the National Center for Advancing Translational Sciences, 2015; 1285–7. [PubMed] [Google Scholar]
- 76. Diao L, Meibohm B.. Pharmacokinetics and pharmacokinetic-pharmacodynamic correlations of therapeutic peptides. Clin Pharmacokinet 2013; 52: 855–68. [DOI] [PubMed] [Google Scholar]
- 77. Mahlapuu M, Håkansson J, Ringstad L. et al. Antimicrobial peptides: an emerging category of therapeutic agents. Front Cell Infect Microbiol 2016; 6: 194.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Bergogne-Berezin E. Clinical role of protein binding of quinolones. Clin Pharmacokinet 2002; 41: 741–50. [DOI] [PubMed] [Google Scholar]
- 79. Fan JC, Chen X, Wang Y. et al. Binding interactions of pefloxacin mesylate with bovine lactoferrin and human serum albumin. J Zhejiang Univ - Sci B 2006; 7: 452–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Kremer JM, Wilting J, Janssen LH.. Drug binding to human alpha-1-acid glycoprotein in health and disease. Pharmacol Rev 1988; 40: 1–47. [PubMed] [Google Scholar]
- 81. Sorensen O, Bratt T, Johnsen AH. et al. The human antibacterial cathelicidin, hCAP-18, is bound to lipoproteins in plasma. J Biol Chem 1999; 274: 22445–51. [DOI] [PubMed] [Google Scholar]
- 82. Beer J, Wagner CC, Zeitlinger M.. Protein binding of antimicrobials: methods for quantification and for investigation of its impact on bacterial killing. AAPS J 2009; 11: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Craig WA, Kunin CM.. Significance of serum protein and tissue binding of antimicrobial agents. Annu Rev Med 1976; 27: 287–300. [DOI] [PubMed] [Google Scholar]
- 84. Lindup WE, Orme MC.. Clinical pharmacology: plasma protein binding of drugs. Br Med J (Clin Res Ed) 1981; 282: 212–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. O’Neill AJ, Chopra I.. Preclinical evaluation of novel antibacterial agents by microbiological and molecular techniques. Expert Opin Investig Drugs 2004; 13: 1045–63. [DOI] [PubMed] [Google Scholar]
- 86. Wise R. The clinical relevance of protein binding and tissue concentrations in antimicrobial therapy. Clin Pharmacokinet 1986; 11: 470–82. [DOI] [PubMed] [Google Scholar]
- 87. Cheah S-E, Wang J, Nguyen VTT. et al. New pharmacokinetic/pharmacodynamic studies of systemically administered colistin against Pseudomonas aeruginosa and Acinetobacter baumannii in mouse thigh and lung infection models: smaller response in lung infection. J Antimicrob Chemother 2015; 70: 3291–7. [DOI] [PubMed] [Google Scholar]
- 88. Pundir P, Catalli A, Leggiadro C. et al. Pleurocidin, a novel antimicrobial peptide, induces human mast cell activation through the FPRL1 receptor. Mucosal Immunol 2013; 7: 177–87. [DOI] [PubMed] [Google Scholar]
- 89. Park YJ, Lee HY, Jung YS. et al. Antimicrobial peptide scolopendrasin VII, derived from the centipede Scolopendra subspinipes mutilans, stimulates macrophage chemotaxis via formyl peptide receptor 1. BMB Rep 2015; 48: 479–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Wang W, Jia J, Li C. et al. Antimicrobial peptide LL-37 promotes the proliferation and invasion of skin squamous cell carcinoma by upregulating DNA-binding protein A. Oncol Lett 2016; 12: 1745–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Korzybski T, Kowszyk-Gindifer Z, Kurylowicz W. et al. Colistin syn. Colimycin. In: Antibiotics Origin, Nature and Properties. Warsaw: PWN - Polish Scientific Publishers, 1967; 115–8. [Google Scholar]
- 92. Cheah SE, Li J, Tsuji BT. et al. Colistin and polymyxin B dosage regimens against Acinetobacter baumannii: differences in activity and the emergence of resistance. Antimicrob Agents Chemother 2016; 60: 3921–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Machuca I, Gutierrez-Gutierrez B, Gracia-Ahufinger I. et al. Mortality associated with bacteremia due to colistin-resistant Klebsiella pneumoniae with high-level meropenem resistance: importance of combination therapy without colistin and carbapenems. Antimicrob Agents Chemother 2017; 61: e00406–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Petrosillo N, Giannella M, Antonelli M. et al. Clinical experience of colistin-glycopeptide combination in critically ill patients infected with Gram-negative bacteria. Antimicrob Agents Chemother 2014; 58: 851–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Pini A, Giuliani A, Falciani C. et al. Antimicrobial activity of novel dendrimeric peptides obtained by phage display selection and rational modification. Antimicrob Agents Chemother 2005; 49: 2665–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Kyte J, Doolittle RF.. A simple method for displaying the hydropathic character of a protein. J Mol Biol 1982; 157: 105–32. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.