Abstract
Macromolecular condensation resulting from biologically regulated liquid–liquid phase separation is emerging as a mechanism to organize intracellular space in eukaryotes, with broad implications for cell physiology and pathology. Despite their small size, bacterial cells are also organized by proteins such as FtsZ, a tubulin homolog that assembles into a ring structure precisely at the cell midpoint and is required for cytokinesis. Here, we demonstrate that FtsZ can form crowding‐induced condensates, reminiscent of those observed for eukaryotic proteins. Formation of these FtsZ‐rich droplets occurs when FtsZ is bound to SlmA, a spatial regulator of FtsZ that antagonizes polymerization, while also binding to specific sites on chromosomal DNA. The resulting condensates are dynamic, allowing FtsZ to undergo GTP‐driven assembly to form protein fibers. They are sensitive to compartmentalization and to the presence of a membrane boundary in cell mimetic systems. This is a novel example of a bacterial nucleoprotein complex exhibiting condensation into liquid droplets, suggesting that phase separation may also play a functional role in the spatiotemporal organization of essential bacterial processes.
Keywords: bacterial division, biomolecular condensation, droplet microfluidics, macromolecular crowding, phase separation
Subject Categories: Cell Cycle; Microbiology, Virology & Host Pathogen Interaction; Protein Biosynthesis & Quality Control
Introduction
In eukaryotic cells, it is becoming clear that many protein systems can self‐organize into macromolecular condensates resulting from liquid–liquid phase separation 1, 2, 3. These dynamic condensates, which reversibly concentrate proteins and nucleic acids, play crucial roles in cell physiology (e.g., metabolic regulation, signaling, stress adaptation) and pathology (e.g., diseases related to protein aggregation, amyloid formation), thus emerging as a fundamental mechanism to organize the intracellular space 1, 2, 4, 5. Proteins that contain multivalent domains involved in protein–protein or protein–nucleic acid interactions and those containing intrinsically disordered regions are the ones more prone to form these condensates 1, 2, 3, 6. Nonspecific effects such as volume exclusion due to natural crowding can promote these phase separation processes 7, 8, 9, 10. Intriguingly, in bacterial cells such protein‐based liquid‐phase macromolecular condensates have not been described, although the bacterial nucleoid has been hypothesized to be a liquid phase 11, 12, 13.
In this work, we have explored this fundamental question analyzing the susceptibility of the FtsZ protein to reversibly partition into dynamic condensates. This protein is a soluble GTPase, ancestor of eukaryotic tubulin, that serves as a central element of the division ring in most bacteria 14. FtsZ contains and/or interacts with other companion division elements that display key molecular features similar to those that drive phase separation in eukaryotic cells. These include multivalency through protein–protein and protein–nucleic acid interactions and partially unstructured regions (FtsZ linker).
In the model Gram‐negative Escherichia coli, division is initiated by an FtsZ‐ring in which FtsZ localizes at midcell together with other proteins forming part of the divisome, the molecular assembly effecting cytokinesis 14. The initial pathway of ring formation is the GTP‐dependent polymerization of FtsZ 15, 16. In vitro, FtsZ self‐assembles into single‐stranded flexible filaments, following a concerted reaction linked to GTP and Mg2+ 15. These filaments tend to further associate into higher‐order structures, whose formation is favored by macromolecular crowding 17. Two additional proteins, FtsA and ZipA, are required to tether the FtsZ filaments to the cytoplasmic membrane, forming a proto‐ring, the first molecular assembly of the divisome 18, 19. As FtsZ is distributed along the bacterial length, the assembly at midcell of the FtsZ‐ring requires dedicated positioning mechanisms, including the Min system 20 and nucleoid occlusion mediated by the SlmA protein 21 that work to ensure assembly of the ring only at the correct location 22.
SlmA, the nucleoid occlusion effector, is a DNA‐binding protein that when bound as a dimer of dimers to its specifically recognized palindromic DNA sequences (SBSs; SlmA‐binding sequences) blocks the FtsZ‐ring formation in regions of the cell occupied by the nucleoid 23, 24. This protects the chromosome from scission upon septum formation 21. The SlmA·SBS complex has been found to accelerate the disassembly of the FtsZ filaments leading to their fragmentation into shorter species that are still able to hydrolyze GTP 25. Contrary to other antagonists that sequester FtsZ subunits decreasing the GTPase activity and blocking polymerization all over the cell 26, 27, 28, SlmA modulation has a spatial dimension. Thus, it occurs selectively nearby all chromosomal macrodomains except the SBS‐free Ter one 23, 29 without changing the GTPase activity of FtsZ single‐stranded filaments 25, 30.
SlmA, the nucleoid occlusion effector, is a DNA‐binding protein that when bound as a dimer of dimers to its specifically recognized palindromic DNA sequences (SBSs; SlmA‐binding sequences) blocks the FtsZ‐ring formation in regions of the cell occupied by the nucleoid 23, 24. This protects the chromosome from scission upon septum formation 21. The SlmA·SBS complex has been found to accelerate the disassembly of the FtsZ filaments leading to their fragmentation into shorter species that are still able to hydrolyze GTP 25. Contrary to other antagonists that sequester FtsZ subunits decreasing the GTPase activity and blocking polymerization all over the cell 26, 27, 28, SlmA modulation has a spatial dimension. Thus, it occurs selectively nearby all chromosomal macrodomains except the SBS‐free Ter one 23, 29 without changing the GTPase activity of FtsZ single‐stranded filaments 25, 30.
The bacterial interior in which these FtsZ‐mediated macromolecular interactions occur is a highly crowded and heterogeneous environment 31. Excluded volume and chemical effects due to crowding may significantly alter the equilibrium and kinetic properties of macromolecular reactions 32, 33 and, as previously mentioned, can also drive phase separation leading to the formation of membrane‐free compartments 8. Thus, in the presence of high concentrations of homogeneous or heterogeneous crowders, whether inert polymers, DNA, or proteins, the tendency of FtsZ to assemble into higher‐order fibers was found to be significantly enhanced 17, 34, 35. Besides, the reconstitution of FtsZ in crowded liquid–liquid phase separation (LLPS) systems mimicking microenvironments and compartmentalization, in bulk or encapsulated in microdroplets, demonstrated uneven distribution of FtsZ among preexisting phases and/or interfaces, which could be reversibly modulated by the association state of FtsZ 36, 37. These studies suggest that phase separation that may occur in the cytoplasm could affect the organization and reactivity of FtsZ, but phase separation of FtsZ itself was not observed.
Here we demonstrate that, when bound to SlmA under crowded conditions, FtsZ forms a dense droplet phase. We have analyzed how SlmA and its complexes with a short double‐stranded DNA containing the consensus sequence specifically recognized by the protein modulate the behavior of FtsZ in crowded solutions and LLPS systems. Our simple LLPS systems, models of cellular microenvironments, are composed of two polymers, one being PEG and the other one dextran or unspecific DNA. The FtsZ·SlmA division complexes were also reconstructed in microfluidics‐based microdroplets, comprising an LLPS system stabilized by a lipid mixture matching the composition in the E. coli inner membrane (EcL). These microdroplets constitute cell‐like environments that display, in addition to compartmentalization, a membrane boundary. Our results show that FtsZ·SlmA·SBS complexes form structures consistent with crowding‐driven liquid‐phase condensates that are dynamic. They evolve toward fibers with GTP addition and reassemble into condensates upon GTP exhaustion. When preexisting polymer‐rich phases are present, these dynamic FtsZ condensates preferentially accumulate into one of the phases and build up at lipid surfaces when the phase‐separated system is encapsulated in microdroplets. These findings provide a potential novel mechanism to modulate the functional reactivity of FtsZ during the cell cycle. Furthermore, they support that liquid‐phase biomolecular condensation can also act as an organizer of intracellular biochemistry in bacterial systems.
Results
FtsZ from Escherichia coli forms biomolecular condensates upon interaction with SlmA in cell‐like crowded conditions
FtsZ was found to form round structures compatible with liquid droplets in the presence of SlmA and a 24‐bp oligonucleotide containing the consensus sequence targeted by this protein (SBS), under crowding conditions, as revealed by confocal microscopy imaging (Fig 1A and B). Condensates (1–6 μm diameter) were observed in solutions containing FtsZ labeled with Alexa 647 (FtsZ‐Alexa 647), unlabeled SlmA, and fluorescein‐labeled SBS (SBS‐Fl), in which the two dyes colocalized, independently of the macromolecule used to crowd the solution (Ficoll, PEG, dextran; Fig 1B and Appendix Fig S1). These structures were not formed by FtsZ in the absence of SlmA·SBS (Appendix Fig S2) nor by FtsZ·SlmA·SBS complexes in the absence of crowding (Fig 1B). These findings were confirmed by turbidity experiments done in parallel (Fig 1C).
Figure 1. Formation of condensates of FtsZ·SlmA·SBS in crowding conditions.
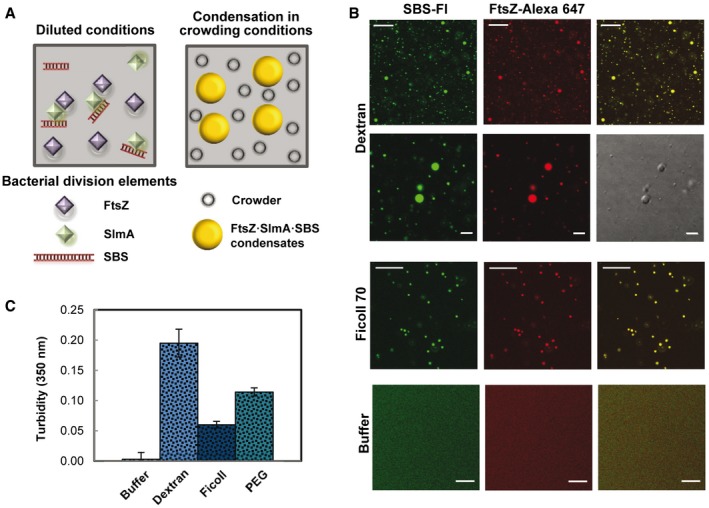
- Scheme of the Escherichia coli division elements involved in the formation of condensates. The concentrations of FtsZ, SlmA, and SBS used in this study were 12, 5, and 1 μM, unless indicated otherwise.
- Representative confocal images of the FtsZ·SlmA·SBS condensates formed in 150 g/l dextran 500 or Ficoll 70, and absence of condensates in dilute solution. Scale bars: 20 μm, except for images at higher magnification in dextran (5 μm, second row).
- Turbidity of FtsZ·SlmA·SBS in buffer (n = 3), in 150 g/l dextran 500 (n = 5) or Ficoll 70 (n = 3), and in 50 g/l PEG 8 (n = 3). Data correspond to the average ± SD.
To determine the factors influencing the formation of the condensates, confocal imaging and turbidity measurements were conducted on samples containing variable concentrations of the three elements (FtsZ, SlmA, and the SBS oligonucleotide), variable concentration of the crowding agent, and different ionic strengths. Condensation in dextran increased with the concentration of the two proteins and of the SBS oligonucleotide (Fig 2A). When SlmA, but not the SBS, was added to FtsZ, condensates in which the two labeled proteins colocalized were also observed, although in considerably lower amounts (Appendix Figs S3 and S4A). In the absence of FtsZ, scarce condensates were found in SlmA solutions, with or without the SBS sequence (Appendix Fig S4A and B). Only at high concentrations of SlmA (40 μM), far beyond the physiological concentration, abundant structures with variable morphology (including rounded‐like) were observed in the solutions (Appendix Fig S4C).
Figure 2. Dependence of the formation of FtsZ·SlmA·SBS condensates on protein and salt concentrations.
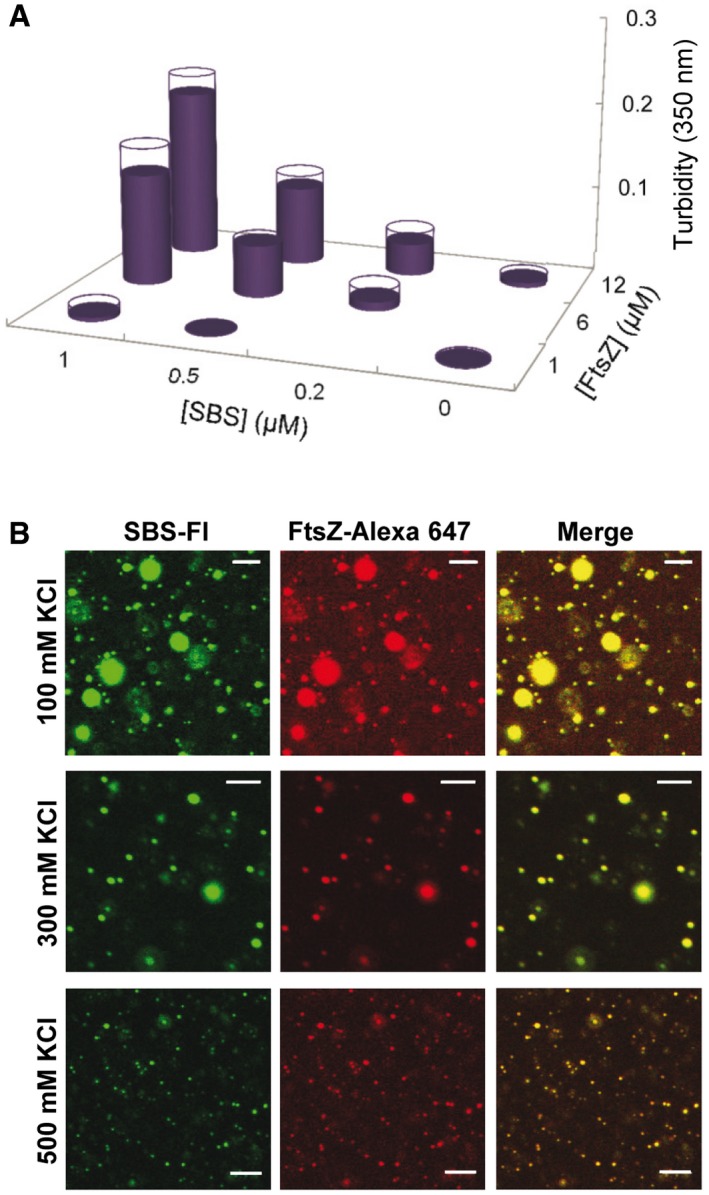
- Formation of condensates as a function of FtsZ and SlmA·SBS concentration, as measured by turbidity, in working buffer (300 mM KCl). SlmA concentration was fivefold that of SBS (except at 0.5 μM SBS, where SlmA concentration was 3 μM). Data are the average of two independent measurements. Errors (SD), symmetrical, are depicted as white disks.
- Representative confocal images of the FtsZ·SlmA·SBS condensates at the specified salt concentrations. The concentrations of FtsZ, SlmA, and SBS were 12, 5, and 1 μM, respectively. Scale bars: 5 μm.
As for other biomolecular condensates 9, FtsZ·SlmA·SBS condensation increased with the concentration of the crowding agent, being detectable by turbidity above 50 g/l dextran (Appendix Fig S5A). The formation of these condensates was dependent on ionic strength. Reducing the concentration of KCl with respect to that in the working buffer (300 mM KCl) resulted in larger condensates, whereas at 500 mM KCl, small structures were observed, although still abundant (Fig 2B and Appendix Fig S5B). This is likely related to the impact of ionic strength on the electrostatic interactions among positively charged SlmA and negatively charged FtsZ and SBS.
Therefore, condensation in this system was strongly favored by the simultaneous presence of the two proteins and the oligonucleotide, probably because this increases multivalency, a feature that has been related to condensation 1, 2, 3. Furthermore, crowding, by volume exclusion and/or other unspecific effects, and ionic strength, through modulation of electrostatic interactions, are among the factors determining the formation of these droplet‐like condensates.
FtsZ·SlmA·SBS condensates are dynamic and evolve toward fibers after addition of GTP
The round structures formed by FtsZ·SlmA·SBS were dynamic, a characteristic feature of liquid‐like droplets, as revealed by protein capture experiments similar to those carried out recently with other protein systems forming liquid‐phase condensates 9. Images show the final state and temporal evolution of FtsZ·SlmA·SBS nucleoprotein complexes containing FtsZ‐Alexa 647 in dextran, to which FtsZ‐Alexa 488 was subsequently added (Fig 3A). Colocalization of the two dyes indicated that newly added FtsZ was recruited into preformed condensates. These condensates remained dynamic and able to incorporate FtsZ after more than 3 h within similar timescales as the freshly prepared samples (Appendix Fig S6). Analogous behavior and comparable times of FtsZ uptake were found in other crowding solutions (Ficoll and PEG, Fig 3B and Appendix Fig S7). FtsZ·SlmA condensates (i.e., in the absence of the SBS oligonucleotide) behaved as permeable dynamic structures as well, despite the fewer droplets observed (Appendix Fig S8).
Figure 3. Dynamism of FtsZ·SlmA·SBS condensates.
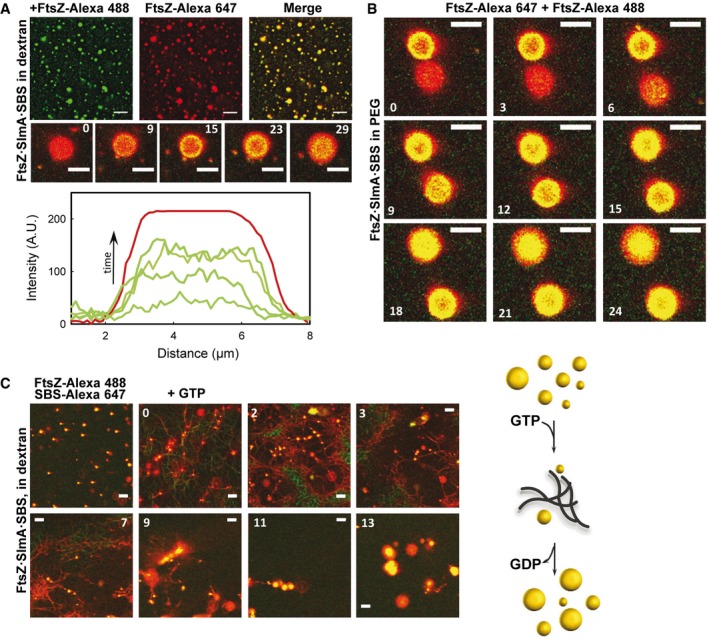
- Representative confocal images showing final state after addition of FtsZ‐Alexa 488 to FtsZ·SlmA·SBS complexes (FtsZ labeled with Alexa 647) in 150 g/l dextran. Below, images showing the stepwise diffusion of FtsZ‐Alexa 488 into the condensates containing FtsZ‐Alexa 647 at the indicated times in seconds (time zero, beginning of visualization for that particular condensate) and corresponding intensity profiles at selected times in the green channel. The profile in the red channel is shown as a reference and varies slightly among images. Scale bars: 5 μm (top row) and 4 μm (bottom row).
- Stepwise diffusion of FtsZ‐Alexa 488 into FtsZ·SlmA·SBS condensates (FtsZ labeled with Alexa 647) at the indicated times in seconds (time zero, beginning of visualization for those particular condensates) in 100 g/l PEG. Scale bars: 4 μm.
- Assembly of FtsZ fibers upon GTP addition (0.5 mM) to FtsZ·SlmA·SBS condensates and condensates formed after FtsZ fiber disassembly at the indicated times in minutes (time zero, GTP addition) in 150 g/l dextran. Scale bars: 5 μm. Scheme of the dynamic process on the right. The number of condensates decreases upon fibers formation, and they rearrange upon GTP depletion and fiber disassembly.
Next, we asked whether FtsZ within the condensates was active for assembly into fibers and how the GTP‐induced fibers were affected by SlmA·SBS. Addition of GTP on preformed FtsZ·SlmA·SBS condensates induced the formation of FtsZ fibers in which significant colocalization between FtsZ‐Alexa 488 and SBS‐Alexa 647 was observed (Fig 3C). Compared with control samples lacking SlmA·SBS (Fig EV1A), the fibers were thinner and their lifetime was appreciably shorter, as previously observed in dilute solution 25. Initially, the fibers coexisted with the round condensates, and then, the amount of fibers increased at the expense of the condensates. Upon GTP depletion, FtsZ fibers disassembled, and afterward, FtsZ, SlmA, and the oligonucleotide incorporated back into condensates larger than their original size (Fig 3C). FtsZ·SlmA·SBS condensates formed after GTP depletion were still dynamic, as newly added FtsZ rapidly incorporated into them (Appendix Fig S9). Preassembled GTP‐triggered FtsZ filaments were also sensitive to the action of SlmA·SBS. Addition of the nucleoprotein complex resulted in thinner fibers rapidly disassembling compared to the control without SlmA·SBS (Fig EV1B), as observed when eliciting polymerization of FtsZ·SlmA·SBS condensates (Fig 3C). Large condensates were also found in this case upon GTP exhaustion.
Figure EV1. Effect of SlmA·SBS on GTP‐triggered FtsZ fibers.
- Representative confocal images showing depolymerization of FtsZ fibers with time. Two independent experiments are shown in which FtsZ was labeled with either Alexa 647 or Alexa 488.
- Effect of SlmA·SBS added on preformed FtsZ fibers induced with GTP on time‐dependent disassembly.
These findings showed that the condensates formed by FtsZ and SlmA·SBS are dynamic entities in which FtsZ retains its GTP‐dependent assembly/disassembly properties.
Compartmentalization affects the distribution and localization of FtsZ·SlmA·SBS condensates in PEG/dextran phase‐separated systems
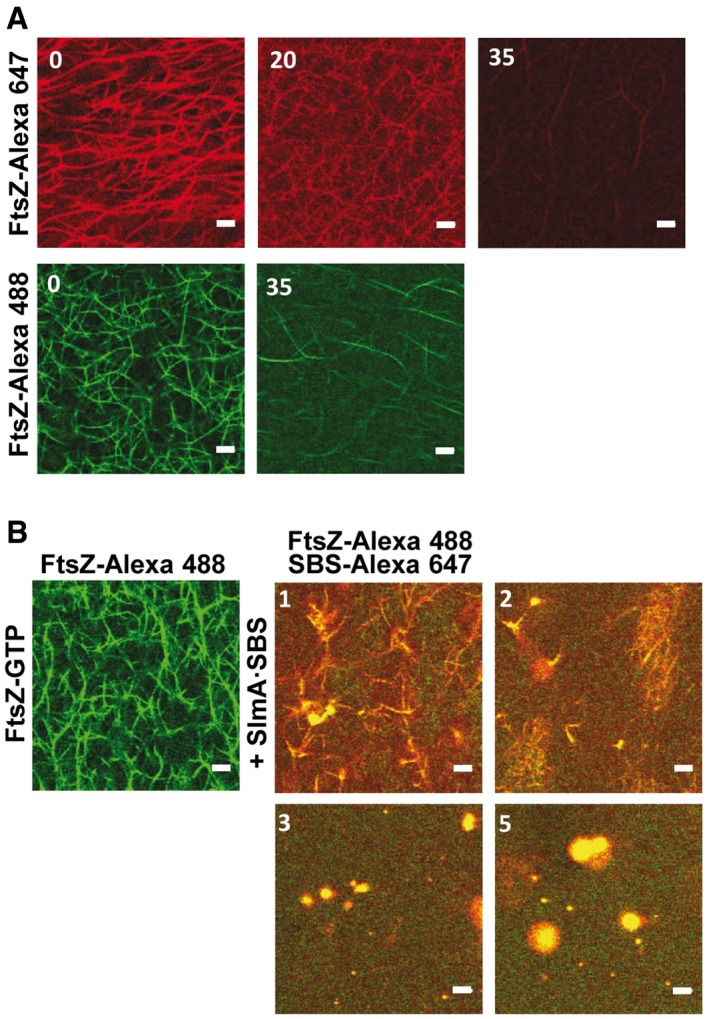
To determine the effect that microenvironments as those found in the cell might have on FtsZ·SlmA·SBS condensation, similar experiments to the ones described above were carried out in PEG/dextran LLPS systems, in which unassembled FtsZ distributes asymmetrically, partitioning preferentially into the dextran‐rich phase 36. Confocal microscopy images of an emulsion of PEG/dextran containing FtsZ and SlmA·SBS showed abundant condensates of similar size as the ones previously found in homogeneous crowders (see above), in which the two division proteins and SBS colocalized (Fig EV2A, Appendix Fig S10A–C). In this open, phase‐separated system, FtsZ·SlmA·SBS condensates distributed preferentially in the dextran phase, as shown by the lack of colocalization with labeled PEG (Appendix Fig S10B) and by fluorescence measurements (Appendix Fig S11A). This is likely because of the accumulation of the two proteins and the SBS in this phase (Appendix Fig S11A–D). As in the single crowder solutions, none or very few condensates were observed in this LLPS system for SlmA with or without the SBS (Appendix Fig S11B and C), and for FtsZ either alone 36 or in the presence of only SlmA (Appendix Fig S12) or only SBS (Appendix Fig S11E). Control experiments showed that these findings were highly reproducible, irrespectively of the fluorescent dyes used and of which element of the condensate was labeled (Appendix Figs S10 and S12).
Figure EV2. Formation of FtsZ·SlmA·SBS condensates in the PEG/dextran LLPS system and GTP‐induced FtsZ assembly into fibers.
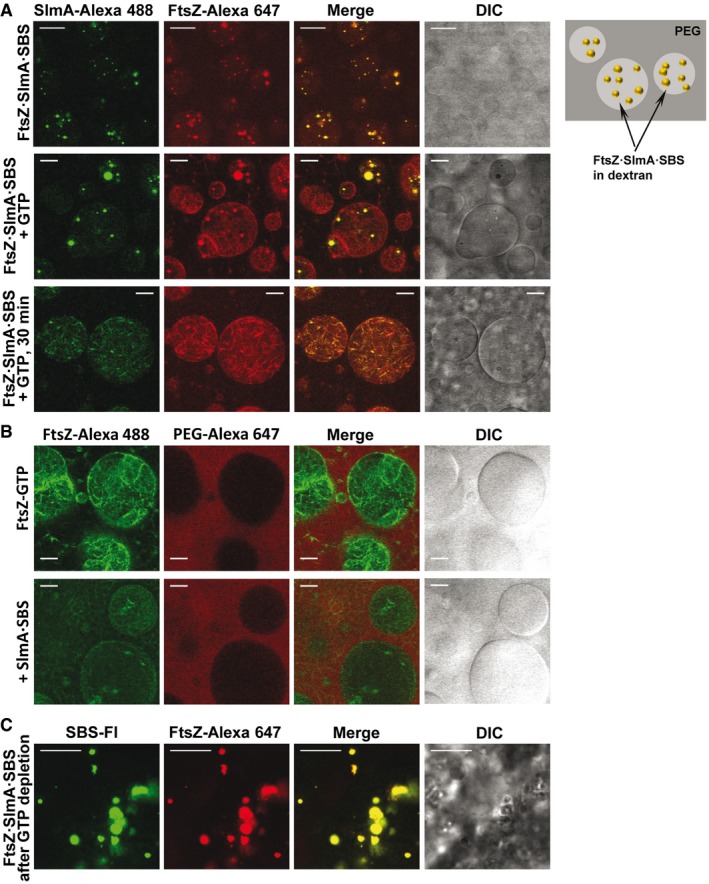
- Representative confocal and transmitted images of FtsZ·SlmA·SBS complexes and scheme of their distribution within the LLPS system on the right. Below, condensates in the presence of GTP (1 mM) at time zero and 30 min after addition of the nucleotide.
- Effect of addition of SlmA·SBS (bottom row) on preformed FtsZ‐GTP fibers induced with 1 mM GTP (top row).
- FtsZ·SlmA·SBS condensates formed after FtsZ fiber disassembly due to GTP (0.33 mM) depletion.
The impact of SlmA on the ability of FtsZ to assemble into fibers in the PEG/dextran LLPS system and on the distribution of the fibers was then analyzed. When GTP was added on the FtsZ·SlmA·SBS complexes, FtsZ filaments decorated with SlmA, distributed mostly within the dextran phase, were observed (Fig EV2A, middle and bottom row). The preference of these filaments for the interface was not as blatant as that of fibers formed solely by FtsZ (Fig EV2B, top row), likely due to their smaller size as a result of the action of SlmA·SBS. Depletion of GTP was found to reverse the process as, upon disassembly, liquid condensates were found again (Fig EV2C). Condensates located within the dextran phase were also found when SlmA·SBS was added to FtsZ fibers, previously elicited by GTP, after nucleotide exhaustion (Fig EV2B).
These experiments showed that SlmA bound to its specific nucleic acid sequence modifies the general arrangement and distribution of FtsZ fibers in LLPS systems mimicking compartmentalization. They also suggest that the formation of condensates in dynamic equilibrium with fibers, modulated by GTP binding and hydrolysis, is inherent to the FtsZ·SlmA·SBS system.
FtsZ·SlmA·SBS condensates contained in microdroplets with PEG/dextran phase‐separated systems accumulate at lipid surfaces
To determine how the membrane boundary and confinement in the E. coli cells may impact the formation of condensates by FtsZ and SlmA, the two proteins and the SBS sequence were encapsulated within phase‐separated microdroplets, picoliter cytomimetic systems surrounded by lipids, using a microfluidics approach we recently optimized 37 (Fig 4A). Encapsulation of FtsZ (with a tracer amount of FtsZ‐Alexa 647) in the stream containing PEG and SlmA·SBS (SBS labeled with fluorescein) in the stream with dextran rendered FtsZ·SlmA·SBS condensates located mostly at the lipid interface of the microdroplets (Fig 4B). FtsZ outside these condensates was mainly in one of the phases, presumably the dextran, the preference of SBS for this phase being more marked (Fig 4B).
Figure 4. Microfluidic encapsulation of FtsZ·SlmA·SBS in the PEG/dextran LLPS system inside microdroplets stabilized by the Escherichia coli lipid mixture.
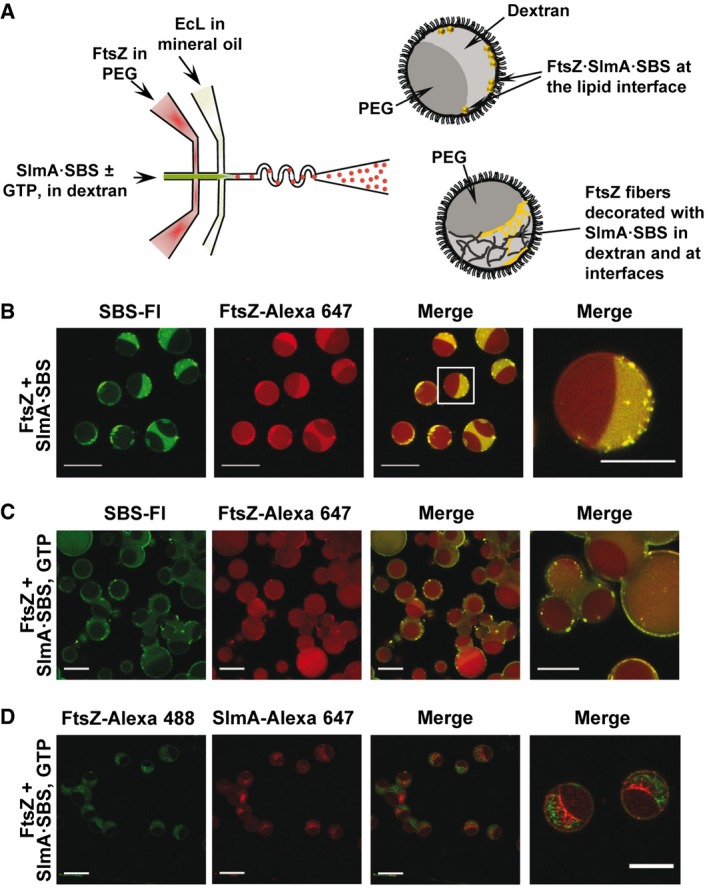
-
AScheme of the encapsulation setup and illustration, on the right, of the distribution of species within the encapsulated LLPS system.
-
B–DRepresentative confocal images of the microdroplets without (B) and with GTP (C and D). The concentrations of FtsZ, SlmA, and SBS were 12, 5, and 1 μM, respectively (B and C), or 6, 3, and 0.5 μM, respectively (D). 1 mM (C) or 2 mM GTP (D). Scale bars: 40 μm except in images on the far right (20 μm), which are either a magnification of the indicated region in the merged image (B) or independent images at higher magnification (C, D).
The same solutions were encapsulated including GTP in the stream with dextran, triggering the formation of FtsZ fibers shortly before encapsulation. Around 30 min after microdroplet production, FtsZ was almost completely disassembled and FtsZ·SlmA·SBS condensates appeared, again mainly at and nearby the lipid interface (Fig 4C and Movie EV1). To observe the fibers, solutions with higher GTP and lower FtsZ and SlmA·SBS concentrations were encapsulated, to increase the lifetime of the fibers (Fig 4D). FtsZ filaments were observed within the dextran phase, at the interface, and at the lipid boundary. In those areas with a higher local concentration of SlmA·SBS, the presence of the FtsZ filaments was less marked. This is compatible with the previously reported interference of SlmA·SBS with FtsZ filaments bundling 30, producing thinner structures more difficult to visualize by confocal microscopy. Interestingly enough, the size of the confined condensates, whether formed after depolymerization or not, was smaller (~ 1 μm diameter) than that observed under the same conditions in bulk solution (Appendix Fig S10C).
These experiments indicate that the FtsZ·SlmA·SBS system retains the tendency to reversibly form condensates when encapsulated inside micron‐sized containers mimicking the compartmentalization of the cytoplasm, and that they display a marked tendency toward the lipid boundary.
SlmA‐driven condensation of FtsZ in PEG/DNA as a model LLPS system closer to an intracellular environment
One LLPS system particularly relevant in the case of the DNA‐binding protein SlmA, in which we have previously studied the behavior of FtsZ 36, is that comprised by mixtures of PEG and nucleic acid phases. The nucleic acid phase consists of short salmon sperm DNA fragments prepared as previously described 38, allowing to reproduce some of the features of nucleic acid‐rich compartments in the bacterial cytoplasm as the charged nature.
In open, phase‐separated PEG/DNA systems, abundant FtsZ·SlmA·SBS condensates were found, mostly distributed in the DNA phase (Fig EV3A and Appendix Fig S13A), probably because of the preferential partition of the individual components (FtsZ, SlmA, and the SBS) into this phase (36 and Appendix Fig S13B). In this LLPS system, FtsZ·SlmA condensation (i.e., in the absence of the SBS sequence) was strongly favored with respect to that previously found in either single or biphasic crowder mixture (cf. Appendix Figs S13C, S3 and S12), likely because of the unspecific charge effects of the crowder DNA and/or unspecific binding of SlmA to these highly concentrated sequences. The simultaneous presence of SlmA, FtsZ, and DNA was determinant to achieve significant condensation in this LLPS system as no condensates were formed by FtsZ on its own 36, and a limited number were found for SlmA with or without SBS (Appendix Fig S13B). Like in the crowded systems previously described (see above), FtsZ·SlmA·SBS condensates obtained at high concentrations of the unspecific DNA were dynamic, as they captured FtsZ freshly added to the sample (Fig EV3B). GTP addition to these condensates induced the assembly of FtsZ into fibers coexisting with large liquid droplets (Appendix Fig S14). The fibers appeared mainly distributed within the DNA‐rich phase, as previously found for FtsZ fibers in the absence of SlmA 36, and also at the interface (Appendix Fig S14).
Figure EV3. FtsZ·SlmA·SBS condensates in the PEG/DNA LLPS system.
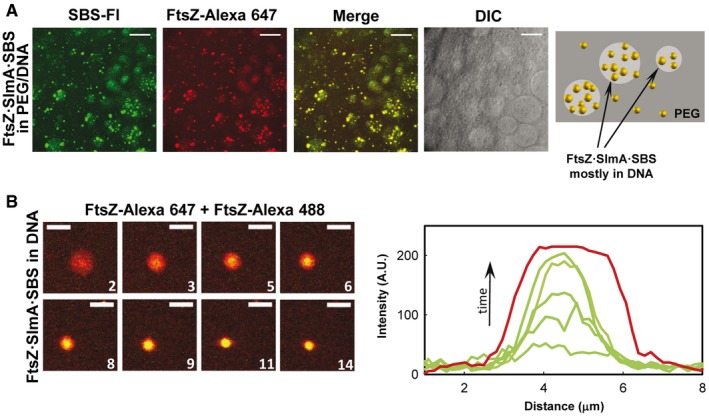
- Representative confocal images of FtsZ·SlmA·SBS complexes and schematic illustration of their disposition within the phases on the right.
- Stepwise diffusion of FtsZ‐Alexa 488 added on FtsZ·SlmA·SBS condensates (FtsZ labeled with Alexa 647) at the indicated times in seconds (time zero, beginning of visualization for this particular condensate) in 180 g/l DNA. On the right, representative intensity profiles in the green channel at different times. The profile in the red channel, shown as a reference, varies slightly within the images.
To reproduce charged and uncharged cellular compartments, FtsZ·SlmA·SBS condensates were also reconstructed in microfluidics‐based microdroplets stabilized by the E. coli lipid mixture containing the PEG/DNA LLPS system (Fig 5A). This required slight adaptation of the procedure earlier described to encapsulate FtsZ in microdroplets with the PEG/dextran LLPS system 37 (see Materials and Methods). Production of microdroplets with PEG/DNA and FtsZ was achieved by including the protein in the stream containing PEG, the other aqueous stream containing the unspecific DNA crowder and, when required, GTP. Generated microdroplets were of similar size as those previously produced with the PEG/dextran mixture, and the distribution of both phases among them was homogeneous (Appendix Fig S15). Simultaneous encapsulation of FtsZ and SlmA·SBS showed FtsZ·SlmA·SBS condensates located mostly at the lipid interface of the microdroplets (Fig 5B and Movie EV2), as with the PEG/dextran LLPS system. In the presence of GTP, fibers of FtsZ decorated by labeled SBS, presumably in complex with SlmA, were observed mainly in the DNA, and a large fraction of the protein and of the SBS located at the lipids (Fig 5C), similar to that found in the absence of SlmA·SBS (Appendix Fig S15).
Figure 5. Microfluidic encapsulation of FtsZ·SlmA·SBS in the PEG/DNA LLPS system inside microdroplets and GTP‐induced FtsZ fiber formation.
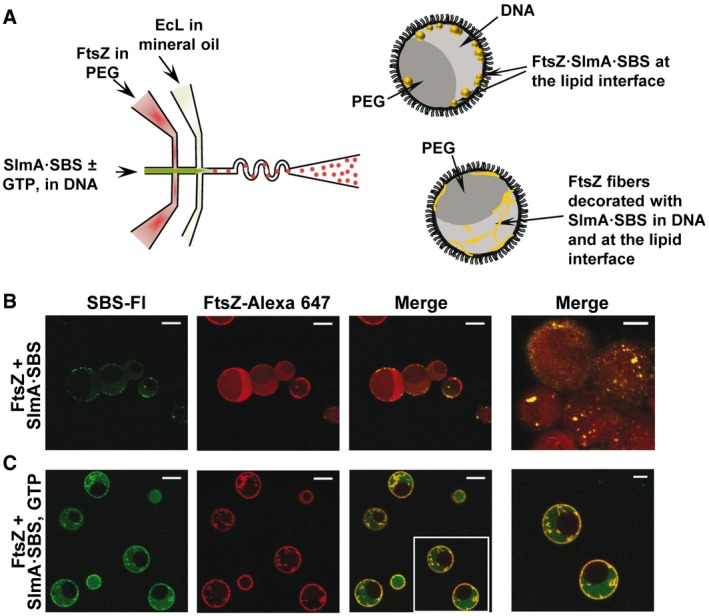
-
AScheme of the encapsulation procedure followed for the PEG/DNA LLPS system and illustration, on the right, of the distribution of species within the encapsulated LLPS system.
-
B, CRepresentative confocal images of the microdroplets stabilized by the Escherichia coli lipid mixture containing the biphasic PEG/DNA mixture and the FtsZ·SlmA·SBS complex without and with 1 mM GTP, respectively. Last image in (B) focuses in the lipid interface to show the high density of condensates. Scale bars: 20 μm except in images on the far right (10 μm), which are an independent image at higher magnification (B) or a magnification of the indicated region in the merged image (C).
These results show that the formation of biomolecular condensates by FtsZ and SlmA division proteins occurs not only in the presence of inert crowding agents but also in the presence of negatively charged ones mimicking the high nucleic acid content of the bacterial cytoplasm. Moreover, as in the PEG/dextran system, accumulation of the components in one of the phases, in this case the DNA‐rich one, resulted in asymmetrical distribution of the condensates in the open LLPS system and, when encapsulated in the microdroplets, condensates were principally assembled at the membrane boundary.
Discussion
In eukaryotes, formation of membrane‐less dynamic macromolecular condensates resulting from biologically regulated liquid–liquid phase separation is a recently discovered regulatory mechanism for spatiotemporal organization of essential intracellular processes. Here, we have shown that FtsZ reversibly forms condensates in the presence of SlmA, a nucleoid occlusion effector of division site selection, in complex with its specific SlmA‐binding sites on the chromosome (SBS), and that these condensates are consistent with crowding‐driven phase‐separated droplets. FtsZ SlmA·SBS condensates, in which FtsZ remains active for polymerization, were also found in cell‐like crowded phase‐separated systems revealing their preferential partition into one of the phases, and its accumulation at lipid surfaces of microdroplets generated by microfluidics.
The condensates found here are markedly different from the membrane‐free compartments previously described in prokaryotes, mostly involved in diverse metabolic pathways 39. Carboxysomes and Pdu and Eut microcompartments are examples of confined structural entities that consist of multiprotein complexes encased in a porous protein shell, thus allowing the limited exchange of substrates and reaction products with the surroundings 39, 40, 41, 42, 43. In contrast, FtsZ‐rich condensates only require a discrete number of components to be assembled and none of the molecules involved seem to provide a capsid encircling the whole structure, as they capture externally added protein.
The combined action of macromolecular crowding and specific multivalent protein–protein and protein–nucleic acid interactions is the molecular driver of FtsZ·SlmA·SBS condensation. Excluded volume effects in crowded solutions nonspecifically enhance macromolecular reactions, favoring macromolecular compaction and association 44. The expected magnitude of these effects is much more pronounced in the case of fibrous or large protein assemblies as the biomolecular condensates 1, 9, 10, 33, 45, 46, which is consistent with our findings for the division proteins. All of the crowders tested here induce the formation of FtsZ·SlmA·SBS condensates, and condensation correlates with crowder concentration and hence with volume exclusion effects. Notably, the abundance and size of the condensates seem to be dramatically enhanced at high concentrations of DNA, probably due to the additional exclusion provoked by electrostatic repulsion among DNA, FtsZ, and the SBS sequences, all negatively charged at the working pH. Large effects arising from high concentrations of unspecific DNA on FtsZ assembly and organization have been described before 35.
Weak transient interactions between multivalent proteins are known to drive liquid–liquid phase separation. Indeed, most biomolecular condensates described consist of various molecules containing multiple homo‐ or hetero‐association elements, such as nucleic acids, usually RNA, and proteins harboring various domains of interaction 1, 2, 3. Multivalency is clearly present in the FtsZ·SlmA·SBS complexes 47. Thus, FtsZ forms discrete oligomers in the absence of GTP 48, their size and number significantly enhanced by crowding 49, and the nucleic acid sequence anchors, in dilute solution, a dimer of dimers of SlmA 25, 30. The interaction of FtsZ with SlmA and with the SBS is required for liquid droplet formation, in line with previous observations that nucleic acids favor this type of structure 1, 50, 51. The concentrations of these three elements and ionic strength are also among the factors influencing condensation, as they modulate their mutual recognition.
The presence of unstructured regions in proteins also seems to promote condensation, and a number of intrinsically disordered proteins can form on their own, under crowding conditions in vitro and in vivo, liquid droplets that evolve toward solid aggregates in certain cases 10, 50, 51, 52, 53. Although FtsZ contains a flexible unstructured linker, of variable length depending upon the species 54, there is no reported evidence of crowding‐induced condensation of FtsZ itself. It cannot be discarded though that, since FtsZ self‐association can be finely tuned by solution conditions, certain combination of such conditions could lead to condensation. Furthermore, it remains to be determined if other FtsZ partners can elicit phase separation as we have observed in the presence of SlmA and its cognate SBS sequences.
Probably the most notable results derived from the reconstruction of FtsZ·SlmA·SBS complexes in phase‐separated systems are the marked partition of these condensates in DNA‐rich phases and the observed tendency of the condensates to concentrate at or nearby the lipid boundary when encapsulated inside microdroplets. Although it was initially proposed that disassembly of FtsZ filaments by SlmA would occur in the nucleoid 22, 55, different models suggest the antagonist activity hampering FtsZ ring formation falls at the membrane 22, 24, 30. One possibility is that many SlmA molecules are brought close to the membrane through transertion (coupled translation and membrane insertion) of proteins encoded by sequences near the SBS 30. This would be reminiscent of the proposed mechanism by which Noc protein functions as a nucleoid occlusion protein in Bacillus subtilis. By binding to the cytoplasmic membrane with an amphipathic helix, Noc physically connects the nucleoid to the membrane and crowds out FtsZ from those areas even if there is no evidence that Noc and FtsZ directly interact 56. Moreover, in that study, molecular crowding was invoked to explain exclusion of active FtsZ from non‐midcell positions, which is conceptually similar to what we propose.
Figure 6 illustrates the potential biological implications of our findings. In predivisional cells prior to chromosome segregation (stage 1), oligomers of SlmA are bound to their specific DNA target sequences over most of the nucleoid. These SlmA·SBS complexes recruit a portion of the cellular FtsZ, forming condensates in the crowded cytoplasm that cover most of the nucleoid, but which are biased toward the cell poles as the Ter macrodomains migrate to midcell 55, 57. The local concentration of the three elements in these condensates would be increased, likely enhancing their mutual recognition. Additionally, the nucleoprotein condensates may further accumulate in transient microenvironments resulting from crowding‐induced phase separation in the cytoplasm. Condensation into liquid droplets may aid in the localization of the elements in the vicinity of the membrane, where SlmA would compete with the membrane‐anchoring proteins for FtsZ binding 24, and reinforce the inhibition of FtsZ fiber formation by other systems operating at the membrane 20.
Figure 6. Scheme of the hypothetical influence of FtsZ·SlmA·SBS condensation on the action of SlmA over FtsZ in bacterial division.
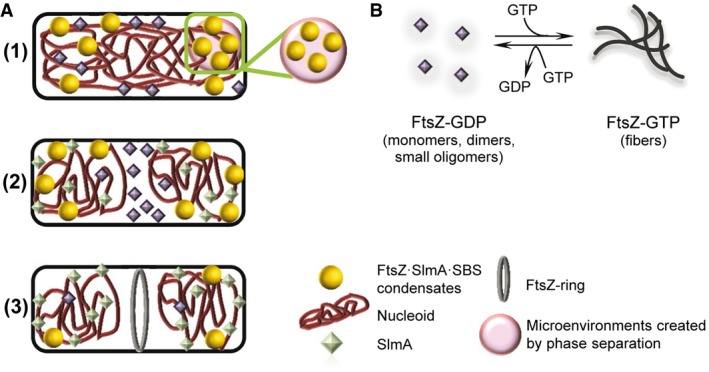
- (1) Under predivision conditions, SlmA·SBS may partially recruit FtsZ forming FtsZ·SlmA·SBS condensates, which may accumulate in transient microenvironments. Condensation into liquid droplets may aid in FtsZ·SlmA·SBS localization nearby the membrane. (2) Under division conditions, the amount of SlmA at midcell decreases and other elements would compete for the interaction with FtsZ that would leave the condensates and, (3) in the presence of GTP, would form a membrane‐anchored FtsZ ring. In non‐central regions, FtsZ would still be under the control of SlmA, which protects the chromosomes from scission by aberrant division ring formation.
- Scheme of the self‐association of FtsZ. In its GDP form, FtsZ is found as an ensemble of species of small size. GTP binding induces its assembly into fibers that disassemble upon depletion of the nucleotide by FtsZ GTPase activity.
After chromosome segregation begins and the midcell division site becomes available (stage 2), the migration of most of the nucleoid toward the poles and the localization of the Ter macrodomains to midcell decrease the amount of SlmA there 23. The Ter macrodomains and associated MatP protein then help to recruit ZapB and ZapA to the division site, where they form a large FtsZ‐independent structure that helps to recruit FtsZ at the correct time during the cell cycle 58. We postulate that this ZapB·ZapA·FtsZ complex, along with the FtsZ membrane anchors ZipA and FtsA, attracts additional FtsZ away from the FtsZ·SlmA·SBS complexes and, in the presence of GTP, forms a membrane‐anchored FtsZ ring (stage 3). In regions distal from midcell, SlmA would still sequester FtsZ in condensates, serving to protect the segregated chromosomes from scission by aberrant division ring formation in newborn cells.
The condensates of FtsZ and SlmA are dynamic, allowing the incorporation of additional protein, the rapid evolution of the integrated FtsZ toward filaments in the presence of GTP, and its recruitment back into the liquid droplets upon GTP depletion. The dynamism and reversibility are hallmarks of liquid droplets, and they appear to be particularly appropriate for the role of SlmA as a spatiotemporal regulator of the formation of FtsZ filaments. Enhancement of the mutual interactions between the two proteins and the target SBS by their local accumulation within these condensates would ensure that, although SlmA·SBS does not completely block FtsZ assembly, the fibers eventually formed under its control are smaller and rapidly disassemble to condense again into FtsZ·SlmA·SBS liquid droplets. FtsZ within these condensates would remain active for assembly when and where required and indeed, super‐resolution fluorescence imaging in vivo has provided evidence of non‐ring FtsZ persisting as patches that may act as precursors for its reassembly, also suggested to be involved in the formation of mobile complexes by recruitment of other binding partners 59.
The ability of FtsZ within the nucleoprotein condensates to form fibers can be explained as follows. According to Du and Lutkenhaus 24, SlmA accesses FtsZ by binding to its C‐terminus, and then severs FtsZ filaments by also binding to the core region of FtsZ. One could imagine that many SlmA molecules may not be always able to achieve both interaction steps, and only interact with the FtsZ C‐terminus. This would not block FtsZ polymerization. Moreover, if the interaction between FtsZ and SlmA·SBS depends upon the multivalency of the FtsZ C‐terminus as proposed 60, then as FtsZ filaments become shorter, they will be less likely to stay bound by SlmA·SBS. This in turn might result in some limited reassembly of FtsZ fibers, keeping them in a shortened equilibrium state. All this agrees with the fact that SlmA does not result in total disassembly of FtsZ filaments but only reduces their lifetime, as shown in Cabré et al 25. FtsZ mutants D86N and K190V (in the core domain) and K380M (at the C‐terminus) still form filaments when bound to SlmA, indicating that the loss of one charged residue in the target is enough to prevent SlmA from breaking FtsZ fibers. Given that formation of the FtsZ·SlmA·SBS condensates seems to be facilitated by interactions between the negatively charged FtsZ, positively charged SlmA, and negatively charged SBS, it is not surprising that changing a charged residue in FtsZ might have a large impact on the system.
Some of the next steps in understanding the potential role of the condensates we describe in this study will be to detect these condensates in vivo and to determine the effects of other FtsZ‐binding proteins such as FtsA, ZipA, and MinC on the condensates in vitro. It is intriguing that when SlmA is overproduced, FtsZ‐GFP is recruited to nucleoids 55, but no high‐resolution study has been done to determine whether there is a detectable phase separation in E. coli cells. Future work with mutants of FtsZ and other cell division proteins in vitro will be necessary to expand and refine our findings. On a broader level, our experimental approach can be extended to other systems undergoing liquid‐phase condensation to tackle the molecular mechanisms governing these processes in reconstituted cytomimetic systems with controlled composition.
Materials and Methods
Materials
GTP nucleotide, dextran 500, PEG 8, and other analytical grade chemicals were from Sigma. Ficoll 70 was from GE Healthcare. All crowders were dialyzed in 50 mM Tris–HCl, 300 mM KCl, pH 7.5, and their concentration measured as earlier described 36. Salmon sperm DNA was from Wako Pure Chemical Industries (Japan). Escherichia coli polar extract phospholipids, from Avanti Polar Lipids (Alabama, USA), were dissolved in spectroscopic grade chloroform (Merck) and kept at −20°C. Before use, a lipid film made by drying EcL in a Speed‐Vac device or under a nitrogen stream was resuspended in mineral oil to the final concentration by two cycles of vortex plus 15‐min sonication in a bath. Specific SBS oligonucleotides, either unlabeled or fluorescently labeled in the 5′ end with fluorescein or Alexa Fluor 647, were from IBA GmbH or Invitrogen.
Protein purification and labeling
FtsZ and SlmA were purified as described 23, 25, 48 and stored at −80°C until used. The proteins were covalently labeled in the amino groups with Alexa Fluor 488 or Alexa Fluor 647 carboxylic acid succinimidyl ester dyes (Molecular Probes/Invitrogen) as earlier stated 17, 25, 61, and stored at −80°C. The degree of labeling of FtsZ and SlmA, ranging between 0.2 and 0.9 moles of fluorophore per mole of protein, was estimated from their molar absorption coefficients.
Experiments were conducted in 50 mM Tris–HCl, 300 mM KCl, 1 mM MgCl2, pH 7.5 (working buffer), unless specified changes in KCl content. Protein solutions were equilibrated in the corresponding buffer, and the concentrations, except for the assays at variable concentrations or when otherwise indicated, of FtsZ, SlmA, and SBS were, respectively, 12, 5, and 1 μM (standard concentrations).
Specific SBS oligonucleotide hybridization
Complementary single‐stranded oligonucleotides were hybridized using a thermocycler as explained 25. The fluorescently labeled oligonucleotides were hybridized with a 10% excess of the complementary unlabeled one. The double‐stranded oligonucleotides generated in this way contained the SBS consensus sequence specifically targeted by SlmA (SBS, 5′‐AAGTAAGTGAGCGCTCACTTACGT‐3′, bases recognized by SlmA in bold) 23.
Unspecific DNA fragmentation and purification
For its use as crowder and in the PEG/DNA LLPS systems, salmon sperm DNA was fragmented and purified as described 36, following a slightly modified phenol:chloroform:isoamyl alcohol extraction method 38. The dried pellet was resuspended in 50 mM Tris–HCl, 300 mM KCl, pH 7.5. The DNA obtained, fragments of ≤ 300 bp 36, was kept at −20°C until used. DNA concentration was estimated from its dry weight after purification. Slight variations were found from batch to batch in the concentration at which phase separation with PEG was achieved, probably reflecting slight differences in DNA quantification, which did not affect the behavior of the division elements in this LLPS system.
Preparation of phases for LLPS systems and labeling of PEG
Enriched phases were prepared by mixing and subsequent isolation of PEG and dextran or unspecific DNA in 50 mM Tris–HCl, 300 mM KCl, pH 7.5, at concentrations rendering phase separation, as described in detail 36. Fluorescent labeling of PEG was done as described 36.
Turbidity measurements
Turbidity of samples containing 12 μM FtsZ, 5 μM SlmA, and 1 μM SBS in the presence and absence of Ficoll or dextran (150 g/l) or PEG (50 g/l) was determined at room temperature and 350 nm using a Varioskan Flash plate reader (Thermo). The absorbance of 200 μl solutions, measured every 10 min for 140 min, was stable during this time period. Reported values, average of 3–5 independent measurements ± SD, correspond to those recorded after 2‐h incubation. Additional turbidity measurements were conducted aimed at determining the effect of different concentrations of FtsZ, SlmA, and SBS or of KCl (in 150 g/l dextran) or the effect of dextran concentration (in the standard conditions of protein and KCl content). These samples were incubated at room temperature for 30 min. Absorbance values were stable for, at least, 10 min after incubation. Reported values correspond to those measured 10 min after incubation, and they are the average of 2–3 independent measurements ± SD.
Preparation of bulk emulsions of PEG/dextran and PEG/DNA LLPS systems
The bulk emulsions were formed by thoroughly mixing PEG‐rich and dextran‐rich or PEG‐rich and DNA‐rich phases in a 3:1 volume ratio as described 36. Proteins were directly added to this mixture, and, when required, polymerization of FtsZ was triggered by diffusion of GTP directly added over the mixture of the two phases. Localization of proteins and localization of the double‐stranded SBS oligonucleotide in the LLPS system phases were evaluated from the colocalization of the labeled element with a tracer amount (1 μM) of PEG‐Alexa 488 or PEG‐Alexa 647 depending on the dye attached to the protein or SBS. Images were acquired with different combinations of dyes (FtsZ‐Alexa 488, SlmA‐Alexa 488, and SBS‐Fl with PEG‐Alexa 647; FtsZ‐Alexa 647 and SlmA‐Alexa 647 with PEG‐Alexa 488) with equivalent results (Appendix Figs S10 and S12).
Diffusion of additional FtsZ into the preformed condensates with SlmA or SlmA·SBS
Samples with FtsZ containing 1 μM FtsZ labeled with Alexa 647 and SlmA ± SBS double‐stranded oligonucleotide, at the standard concentrations and in the specified crowding agents, were prepared and imaged before and after addition of 0.5–1 μM FtsZ‐Alexa 488. The diffusion of FtsZ‐Alexa 488 into the red labeled condensates was monitored with time.
Measurement of the partition of division elements in LLPS systems by fluorescence
Partition within the PEG/dextran mixture was calculated as described 36. Briefly, tracer (0.5 μM Alexa 488‐labeled proteins or fluorescein‐labeled SBS) and unlabeled species up to the standard concentration were gently added to the two phases in a 1:1 volume ratio. Mixture was allowed to phase separate and equilibrate for 30 min, and after centrifugation, phases were isolated and the fluorescence emission intensity of an aliquot of each phase was measured in PolarStar Galaxy (BMG Labtech GmbH, Germany) or Varioskan (Thermo) Plate Readers. Concentrations in the enriched phases were calculated by comparison with samples containing known amounts of tracer in the same phase. Control measurements proved tracer signals were in all cases linear with total concentration. Reported values correspond to the average of three independent measurements, six in the case of the samples with the three components, ± SD.
Microfluidic chip fabrication
Chips were constructed by conventional soft lithographic techniques as earlier explained 37. PDMS base Sylgard™ 184 was mixed in a 10:1 (w/w) with curing agent (Dow Corning GmbH, Germany), degassed, decanted onto masters (design details described elsewhere 62), and kept overnight at 65°C. Inlet and outlet holes were punched in the PDMS peeled from the master and channels sealed by a glass slide activating the surfaces by oxygen plasma (Diener electronic GmbH, Germany). For hydrophobic treatment of the chips, Aquapel (Pittsburgh Glass Works, LLC) was flushed in the channels and dried overnight at 65°C.
Microfluidic encapsulation of LLPS systems in microdroplets
Production of microdroplets by microfluidics was conducted basically as described 37. Briefly, encapsulation was achieved by mixing a PEG stream and another one with either dextran or salmon sperm DNA in an approximately 1:1 volume ratio prior to the droplet formation junction. When stated, Alexa 647‐ or Alexa 488‐labeled PEG (2 μM) was included in the PEG solution. FtsZ (12 or 25 μM) was added to one of the aqueous phases, and SlmA (6 or 10 μM) with or without SBS (1–2 μM) was added to the other. When required, tracer amounts (2 μM) of the proteins labeled with the specified dye and of the SBS oligonucleotide labeled with fluorescein (2 μM) were added. Comparable results were obtained with all dye combinations. When induction of FtsZ polymerization before encapsulation was required, the nucleotide GTP (2–4 mM) was included in the SlmA solution. The third stream supplied the mineral oil with the E. coli lipid mixture (20–25 g/l). In the particular case of the PEG/DNA LLPS system encapsulation, surfactant capability of the lipids seemed to be lower. Solutions were delivered at 120 μl/h (oil phase) and 5 and 7 μl/h (dextran and PEG aqueous phases, respectively), or 6 μl/h (both DNA and PEG aqueous phases), by automated syringe pumps (Cetoni GmbH, Germany). The sizes of generated microdroplets were similar (~ 15–20 μm diameter) as those previously reported for PEG/dextran 37. Droplet production in the microfluidic chip was monitored with an Axiovert 135 fluorescence microscope (Zeiss).
Confocal microscopy measurements and data analysis
The microdroplets generated by microfluidics were visualized either on chip or after collection in silicone chambers (Molecular probes/Invitrogen) glued to coverslips. These chambers were also used to visualize the division complexes in the presence of crowding agents or in the LLPS systems. Images were obtained with a Leica TCS SP2 inverted confocal microscope with a HCX PL APO 63× oil immersion objective (N.A. = 1.4–1.6; Leica, Mannheim, Germany). Ar (488 nm) and He‐Ne (633 nm) ion lasers were used to excite Alexa 488/Fluorescein and Alexa 647, respectively. Several images were registered across each sample, corresponding to different observation fields. Transmission light (DIC) and fluorescence images were taken simultaneously. Control images of unlabeled FtsZ·SlmA·SBS in working buffer (300 mM KCl) and 150 g/l dextran 500 clearly show the presence of condensates in the transmission light images (Appendix Fig S16).
ImageJ (National Institutes of Health, USA) was used to produce images and time‐lapse movies, to measure the distribution of sizes and to obtain the intensity profiles of the liquid droplets applying the straight line tool of the software through their equatorial section.
Author contributions
BM, SZ, and GR conceived the experimental work; BM and SZ analyzed results; BM, SZ, MS‐S, MAR‐R, and ML‐A performed experimental work; BM, SZ, WM, CDK, and GR discussed the results and wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Movie EV1
Movie EV2
Review Process File
Acknowledgements
Authors thank W.T.S. Huck and A. Piruska (Radboud University, Nijmegen) for kindly providing the chips designs and silicon masters for microfluidics; N. Ropero for technical assistance in protein purification and labeling; M.T. Seisdedos and G. Elvira (Confocal Laser and Multidimensional Microscopy Facility, CIB‐CSIC) for excellent support in imaging; and the Technical Support Facility (CIB‐CSIC) for invaluable input. This work was supported by the Fondo Europeo de Desarrollo Regional (FEDER) and the Agencia Estatal de Investigación (AEI); by the Spanish government (BFU2014‐52070‐C2‐2‐P and BFU2016‐75471‐C2‐1‐P, G. R.); by the National Institutes of Health (GM61074, W. M.); and by the National Science Foundation (MCB‐1715984, C. D. K.). M.L.‐A. was supported by the European Social Fund (ESF 2014‐2020).
EMBO Reports (2019) 20: e45946
Contributor Information
Begoña Monterroso, Email: monterroso@cib.csic.es.
Silvia Zorrilla, Email: silvia@cib.csic.es.
Germán Rivas, Email: grivas@cib.csic.es.
References
- 1. Banani SF, Lee HO, Hyman AA, Rosen MK (2017) Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol 18: 285–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Shin Y, Brangwynne CP (2017) Liquid phase condensation in cell physiology and disease. Science 357: eaaf4382 [DOI] [PubMed] [Google Scholar]
- 3. Alberti S (2017) Phase separation in biology. Curr Biol 27: R1097–R1102 [DOI] [PubMed] [Google Scholar]
- 4. Aguzzi A, Altmeyer M (2016) Phase separation: linking cellular compartmentalization to disease. Trends Cell Biol 26: 547–558 [DOI] [PubMed] [Google Scholar]
- 5. Alberti S (2017) The wisdom of crowds: regulating cell function through condensed states of living matter. J Cell Sci 130: 2789–2796 [DOI] [PubMed] [Google Scholar]
- 6. Shin Y, Berry J, Pannucci N, Haataja MP, Toettcher JE, Brangwynne CP (2017) Spatiotemporal control of intracellular phase transitions using light‐activated optoDroplets. Cell 168: 159–171 e14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Walter H, Brooks DE (1995) Phase separation in cytoplasm, due to macromolecular crowding, is the basis for microcompartmentation. FEBS Lett 361: 135–139 [DOI] [PubMed] [Google Scholar]
- 8. Keating CD (2012) Aqueous phase separation as a possible route to compartmentalization of biological molecules. Acc Chem Res 45: 2114–2124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Woodruff JB, Ferreira Gomes B, Widlund PO, Mahamid J, Honigmann A, Hyman AA (2017) The centrosome is a selective phase that nucleates microtubules by concentrating tubulin. Cell 169: 1066–1077 [DOI] [PubMed] [Google Scholar]
- 10. Hernandez‐Vega A, Braun M, Scharrel L, Jahnel M, Wegmann S, Hyman BT, Alberti S, Diez S, Hyman AA (2017) Local nucleation of microtubule bundles through tubulin concentration into a condensed tau phase. Cell Rep 20: 2304–2312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cunha S, Woldringh CL, Odijk T (2001) Polymer‐mediated compaction and internal dynamics of isolated Escherichia coli nucleoids. J Struct Biol 136: 53–66 [DOI] [PubMed] [Google Scholar]
- 12. de Vries R (2010) DNA condensation in bacteria: interplay between macromolecular crowding and nucleoid proteins. Biochimie 92: 1715–1721 [DOI] [PubMed] [Google Scholar]
- 13. Pelletier J, Halvorsen K, Ha BY, Paparcone R, Sandler SJ, Woldringh CL, Wong WP, Jun S (2012) Physical manipulation of the Escherichia coli chromosome reveals its soft nature. Proc Natl Acad Sci USA 109: E2649–E2656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Haeusser DP, Margolin W (2016) Splitsville: structural and functional insights into the dynamic bacterial Z ring. Nat Rev Microbiol 14: 305–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Erickson HP, Anderson DE, Osawa M (2010) FtsZ in bacterial cytokinesis: cytoskeleton and force generator all in one. Microbiol Mol Biol Rev 74: 504–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mingorance J, Rivas G, Vélez M, Gómez‐Puertas P, Vicente M (2010) Strong FtsZ is with the force: mechanisms to constrict bacteria. Trends Microbiol 18: 348–356 [DOI] [PubMed] [Google Scholar]
- 17. González JM, Jiménez M, Vélez M, Mingorance J, Andreu JM, Vicente M, Rivas G (2003) Essential cell division protein FtsZ assembles into one monomer‐thick ribbons under conditions resembling the crowded intracellular environment. J Biol Chem 278: 37664–37671 [DOI] [PubMed] [Google Scholar]
- 18. Rico AI, Krupka M, Vicente M (2013) In the beginning, Escherichia coli assembled the proto‐ring: an initial phase of division. J Biol Chem 288: 20830–20836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ortiz C, Natale P, Cueto L, Vicente M (2016) The keepers of the ring: regulators of FtsZ assembly. FEMS Microbiol Rev 40: 57–67 [DOI] [PubMed] [Google Scholar]
- 20. Rowlett VW, Margolin W (2015) The Min system and other nucleoid‐independent regulators of Z ring positioning. Front Microbiol 6: 478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Adams DW, Wu LJ, Errington J (2014) Cell cycle regulation by the bacterial nucleoid. Curr Opin Microbiol 22: 94–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mannik J, Bailey MW (2015) Spatial coordination between chromosomes and cell division proteins in Escherichia coli . Front Microbiol 6: 306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cho H, McManus HR, Dove SL, Bernhardt TG (2011) Nucleoid occlusion factor SlmA is a DNA‐activated FtsZ polymerization antagonist. Proc Natl Acad Sci USA 108: 3773–3778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Du S, Lutkenhaus J (2014) SlmA antagonism of FtsZ assembly employs a two‐pronged mechanism like MinCD. PLoS Genet 10: e1004460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cabre EJ, Monterroso B, Alfonso C, Sanchez‐Gorostiaga A, Reija B, Jimenez M, Vicente M, Zorrilla S, Rivas G (2015) The Nucleoid Occlusion SlmA protein accelerates the disassembly of the FtsZ protein polymers without affecting their GTPase activity. PLoS One 10: e0126434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen Y, Milam SL, Erickson HP (2012) SulA inhibits assembly of FtsZ by a simple sequestration mechanism. Biochemistry 51: 3100–3109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hill NS, Buske PJ, Shi Y, Levin PA (2013) A moonlighting enzyme links Escherichia coli cell size with central metabolism. PLoS Genet 9: e1003663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hernandez‐Rocamora VM, Alfonso C, Margolin W, Zorrilla S, Rivas G (2015) Evidence that bacteriophage lambda Kil peptide inhibits bacterial cell division by disrupting FtsZ protofilaments and sequestering protein subunits. J Biol Chem 290: 20325–20335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tonthat NK, Arold ST, Pickering BF, Van Dyke MW, Liang S, Lu Y, Beuria TK, Margolin W, Schumacher MA (2011) Molecular mechanism by which the nucleoid occlusion factor, SlmA, keeps cytokinesis in check. EMBO J 30: 154–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tonthat NK, Milam SL, Chinnam N, Whitfill T, Margolin W, Schumacher MA (2013) SlmA forms a higher‐order structure on DNA that inhibits cytokinetic Z‐ring formation over the nucleoid. Proc Natl Acad Sci USA 110: 10586–10591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vendeville A, Lariviere D, Fourmentin E (2011) An inventory of the bacterial macromolecular components and their spatial organization. FEMS Microbiol Rev 35: 395–414 [DOI] [PubMed] [Google Scholar]
- 32. Zhou HX, Rivas G, Minton AP (2008) Macromolecular crowding and confinement: biochemical, biophysical, and potential physiological consequences. Annu Rev Biophys 37: 375–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rivas G, Minton AP (2016) Macromolecular crowding in vitro, in vivo, and in between. Trends Biochem Sci 41: 970–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Popp D, Iwasa M, Narita A, Erickson HP, Maeda Y (2009) FtsZ condensates: an in vitro electron microscopy study. Biopolymers 91: 340–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Monterroso B, Reija B, Jimenez M, Zorrilla S, Rivas G (2016) Charged molecules modulate the volume exclusion effects exerted by crowders on FtsZ polymerization. PLoS One 11: e0149060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Monterroso B, Zorrilla S, Sobrinos‐Sanguino M, Keating CD, Rivas G (2016) Microenvironments created by liquid‐liquid phase transition control the dynamic distribution of bacterial division FtsZ protein. Sci Rep 6: 35140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sobrinos‐Sanguino M, Zorrilla S, Keating CD, Monterroso B, Rivas G (2017) Encapsulation of a compartmentalized cytoplasm mimic within a lipid membrane by microfluidics. Chem Commun 53: 4775–4778 [DOI] [PubMed] [Google Scholar]
- 38. Biswas N, Ichikawa M, Datta A, Sato YT, Yanagisawa M, Yoshikawa K (2012) Phase separation in crowded micro‐spheroids: DNA–PEG system. Chem Phys Lett 539–540: 157–162 [Google Scholar]
- 39. Holthuis JC, Ungermann C (2013) Cellular microcompartments constitute general suborganellar functional units in cells. Biol Chem 394: 151–161 [DOI] [PubMed] [Google Scholar]
- 40. Cheng S, Liu Y, Crowley CS, Yeates TO, Bobik TA (2008) Bacterial microcompartments: their properties and paradoxes. BioEssays 30: 1084–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Parsons JB, Frank S, Bhella D, Liang M, Prentice MB, Mulvihill DP, Warren MJ (2010) Synthesis of empty bacterial microcompartments, directed organelle protein incorporation, and evidence of filament‐associated organelle movement. Mol Cell 38: 305–315 [DOI] [PubMed] [Google Scholar]
- 42. Yeates TO, Kerfeld CA, Heinhorst S, Cannon GC, Shively JM (2008) Protein‐based organelles in bacteria: carboxysomes and related microcompartments. Nat Rev Microbiol 6: 681–691 [DOI] [PubMed] [Google Scholar]
- 43. Kerfeld CA, Aussignargues C, Zarzycki J, Cai F, Sutter M (2018) Bacterial microcompartments. Nat Rev Microbiol 16: 277–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rivas G, Ferrone F, Herzfeld J (2004) Life in a crowded world. EMBO Rep 5: 23–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ferrone FA, Rotter MA (2004) Crowding and the polymerization of sickle hemoglobin. J Mol Recognit 17: 497–504 [DOI] [PubMed] [Google Scholar]
- 46. Hatters DM, Minton AP, Howlett GJ (2002) Macromolecular crowding accelerates amyloid formation by human apolipoprotein C‐II. J Biol Chem 277: 7824–7830 [DOI] [PubMed] [Google Scholar]
- 47. Schumacher MA (2017) Bacterial nucleoid occlusion: multiple mechanisms for preventing chromosome bisection during cell division. Subcell Biochem 84: 267–298 [DOI] [PubMed] [Google Scholar]
- 48. Rivas G, López A, Mingorance J, Ferrándiz MJ, Zorrilla S, Minton AP, Vicente M, Andreu JM (2000) Magnesium‐induced linear self‐association of the FtsZ bacterial cell division protein monomer. The primary steps for FtsZ assembly. J Biol Chem 275: 11740–11749 [DOI] [PubMed] [Google Scholar]
- 49. Rivas G, Fernandez JA, Minton AP (2001) Direct observation of the enhancement of noncooperative protein self‐assembly by macromolecular crowding: indefinite linear self‐association of bacterial cell division protein FtsZ. Proc Natl Acad Sci USA 98: 3150–3155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lin Y, Protter DS, Rosen MK, Parker R (2015) Formation and maturation of phase‐separated liquid droplets by RNA‐binding proteins. Mol Cell 60: 208–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Patel A, Lee HO, Jawerth L, Maharana S, Jahnel M, Hein MY, Stoynov S, Mahamid J, Saha S, Franzmann TM et al (2015) A liquid‐to‐solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell 162: 1066–1077 [DOI] [PubMed] [Google Scholar]
- 52. Ambadipudi S, Biernat J, Riedel D, Mandelkow E, Zweckstetter M (2017) Liquid‐liquid phase separation of the microtubule‐binding repeats of the Alzheimer‐related protein Tau. Nat Commun 8: 275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, Mittag T, Taylor JP (2015) Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163: 123–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Buske PJ, Levin PA (2013) A flexible C‐terminal linker is required for proper FtsZ assembly in vitro and cytokinetic ring formation in vivo . Mol Microbiol 89: 249–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bernhardt TG, de Boer PA (2005) SlmA, a nucleoid‐associated, FtsZ binding protein required for blocking septal ring assembly over chromosomes in E. coli . Mol Cell 18: 555–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Adams DW, Wu LJ, Errington J (2015) Nucleoid occlusion protein Noc recruits DNA to the bacterial cell membrane. EMBO J 34: 491–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Espeli O, Borne R, Dupaigne P, Thiel A, Gigant E, Mercier R, Boccard F (2012) A MatP‐divisome interaction coordinates chromosome segregation with cell division in E. coli . EMBO J 31: 3198–3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Buss JA, Peters NT, Xiao J, Bernhardt TG (2017) ZapA and ZapB form an FtsZ‐independent structure at midcell. Mol Microbiol 104: 652–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Rowlett VW, Margolin W (2014) 3D‐SIM super‐resolution of FtsZ and its membrane tethers in Escherichia coli cells. Biophys J 107: L17–L20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Schumacher MA, Zeng W (2016) Structures of the nucleoid occlusion protein SlmA bound to DNA and the C‐terminal domain of the cytoskeletal protein FtsZ. Proc Natl Acad Sci USA 113: 4988–4993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Reija B, Monterroso B, Jimenez M, Vicente M, Rivas G, Zorrilla S (2011) Development of a homogeneous fluorescence anisotropy assay to monitor and measure FtsZ assembly in solution. Anal Biochem 418: 89–96 [DOI] [PubMed] [Google Scholar]
- 62. Mellouli S, Monterroso B, Vutukuri HR, te Brinke E, Chokkalingam V, Rivas G, Huck WTS (2013) Self‐organization of the bacterial cell‐division protein FtsZ in confined environments. Soft Matter 9: 10493–10500 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Movie EV1
Movie EV2
Review Process File


