Abstract
Background
This study investigated the diagnostic and prognostic values of kinesin superfamily proteins (KIFs) in breast cancer (BC) patients.
Material/Methods
All data were obtained from the Cancer Genome Atlas. DESeq was run to test for differentially expressed KIF genes. Patients were divided into high- and low-expression groups according to the median expression values of each KIF genes. Survival data were calculated using the Cox proportional hazard model. Comprehensive survival analysis was performed to evaluate the prognostic value of the prognostic signature. Gene set enrichment analysis (GSEA) was conducted to identify associated gene ontology and KEGG pathways.
Results
Bioinformatics analysis showed that all KIF genes were significantly enriched during DNA replication and the cell cycle, and co-expressed with each other. Thirteen KIF genes were differentially expressed in cancer and adjacent tissues, and high levels of KIF15, KIF20A, KIF23, KIF2C and KIF4A genes were significantly correlated with poor overall survival (OS). GSEA showed that BC patients with high expression of KIF15, KIF20A, KIF23, KIF2C and KIF4A were enriched in the cell cycle process, P53 regulation pathway and mismatch repair. Combinations of low expression of KIF15, KIF20A, KIF23, KIF2C and KIF4A were more highly correlated with favorable OS. Nomograms showed that the KIF4A risk score provided the maximum number of risk points (range 0–100), whereas other genes made a lower contribution.
Conclusions
We conclude that 13 KIF genes are differentially expressed in BC tumor tissues, and KIF15, KIF20A, KIF23, KIF2C and KIF4A are associated with prognostic factors in BC.
MeSH Keywords: Breast Neoplasms, Diagnosis, Kinesin, Prognosis, RNA
Background
Breast cancer (BC) remains the highest occurring cancer in women, in addition to being the third most frequent malignancy globally. In 2012, around 1.7 million persons worldwide had BC and nearly 500,000 died from the disease [1–4]. One among eight or 10 women will develop BC in their lifetime. BC mortality has decreased in North America as well as the European Union, but is still increasing in South America, Africa, and Asia. BC is the most common cause of cancer mortality in developing countries, compared with lung cancer in developed countries [5–7]. Genetic aspects and environmental exposure play a significant part in the etiology of BC [8,9]. The Human Genome Project has led to increasing attention being paid to cancer genetic susceptibility. A genetic factor which dysfunction amid normal tissues and tumors in the genome are the major potential sources of prognostic and diagnostic biomarkers [10]. Also, genes whose expression is interlinked with survival of BC might be prognostic biomarkers, as well as therapeutic targets [11–14]. As in other malignant neoplastic diseases, BC is considered to have dysfunction of numerous gene signaling pathways as well as networks that have an impact on tissue homeostasis.
There are 45 kinesin superfamily proteins (KIFs) with various functions in humans [15]. KIFs are involved in the molecular movements of axonal transportation. KIFs are ubiquitous in eukaryotes, and some are involved in transportation of vesicles and organelles inside cells [16,17]. KIF genes have been shown to play a crucial role in many tumors, and can be used to predict cancer diagnosis and prognosis [18,19] . It has been shown that the KIF family of genes is linked with BC [20,21], nevertheless, the joint analysis linked with multiple KIF family genes regarding BC have rarely been recorded. Complete examination of the diagnostic and prognostic values of KIF genes in BC requires additional investigation. The purpose of the present study was to explore the diagnostic and prognostic values of KIF gene expression in BC patients, on the basis of bioinformatics evaluation.
Material and Methods
Bioinformatics analysis of KIF genes
For analysis of the biological pathways and significance of the KIF family genes, a set of functional enrichment analysis for the KIF family was performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID 6.8, https://david.ncifcrf.gov/home.jsp, accessed August 3, 2018). Enriched P values <0.05 were statistically significant. Gene–gene interactions of KIF family genes were investigated via GeneMANIA (http://www.genemania.org/, accessed August 9, 2018) [22]. Protein–protein interactions were examined by the Search Instrument for the Retrieval of Interacting Genes/Proteins (STRING, https://string-db.org/, accessed August 9, 2018) [23]. We also applied the Cytoscape (version 3.6.1) Biological Networks Gene Ontology (BiNGO) instrument for performing Gene Ontology (GO) evaluation on the KIF gene family [24].
Data source
The knowledge of the clinical of BC patients and RNA sequence based on the patients were gathered by the Cancer Genome Atlas (TCGA) database (https://cancergenome.nih.gov/, accessed June 7, 2018). Using the edgeR package in R, we normalized mRNA sequencing data and examined mRNA expression in normal tissues and BC. Genes with an accustomed P value <0.01 and |log2 fold-change (FC)| >2 were considered to be significantly different in BC and adjacent tissues. We regarded the genes as being differentially expressed mRNA (DEM). Clinical characteristics of patients with BC included gender, ethnicity (Asian, black, white or other), age at diagnosis (<65 or ≥65 years), and tumor stage.
Survival analysis
For each KIF DEM, patients were divided into low- and high-expression groups according to the median expression values of each KIF genes. By utilizing survival curves by Kaplan-Meier analysis with log-rank test, we assessed the prognostic significance of every clinical aspect as well as DEM from a criterion of P<0.05. The Cox proportional hazards model was utilized for evaluating the comparative risk in such differentially expressed genes on overall survival (OS). The mRNAs significantly associated with OS in the Cox proportional hazards model were considered to be prognostic mRNAs.
Correlation analysis
Pearson correlation coefficient was assessed for identifying correlations between the prognostic mRNAs.
Joint-effects analysis and nomograms
To assess thoroughly the prognostic model, joint analysis and nomograms were performed on the KIF DEM prognostic signature. On the basis of previous survival analysis, we divided the combined genes into high-, intermediate- and low-risk groups, completed survival analysis on 3 groups of patients, and established a Cox regression model. In addition to the joint analysis, we examined the predictive prognostic value of the risk scoring using nomograms to assess the correlation among clinical status as well as risk score within BC OS. The possible implication of risk scoring on the basis of predicting clinical characteristics has similarly been discovered. C-index and calibration curve were considered with bootstrap self-sampling and internal verification.
Gene set enrichment analysis (GSEA)
The core concept in GSEA is to utilize a predefined group of genes (mainly by previous experimental outcomes or functional annotations) for ranking the genes in accordance with the extent of differential expression within the 2 types of samples, then verifying that the pre-established group of genes is supplemented at the bottom or top in the sorting table. To explore the differences in pathways as well as biological functions in the low- and high-expression sets of such prognostic KIF genes, GSEA (http://software.broadinstitute.org/gsea/index.jsp, accessed August 9, 2018) [25,26] was used to explore potential KEGG pathway and GO analysis within the Molecular Signatures Database (MSigDB) of c2 (curated gene sets) and c5 (GO gene sets) [27]. The criteria for significant enrichment gene sets in GSEA were: P<0.05 and false discovery rate (FDR) <0.25.
Statistical analysis
Log-rank assessment was utilized for comparing clinical aspects as well as univariate survival analysis of KIF genes. Clinicopathological parameters statistically linked to OS (P<0.05) were included in multivariate Cox proportional hazard regression models to adjust. Hazard ratios (HRs) and 95% confidence intervals (CIs) were used to assess the relative risk in many patients with BC. Multiple testing with the Benjamini–Hochberg procedure was used to control the FDR in GSEA. Statistical analysis was performed using SPSS 22.0 and R 3.5.1 software. P<0.05 was considered to be statistically significant.
Results
Bioinformatics analysis of the KIF family genes
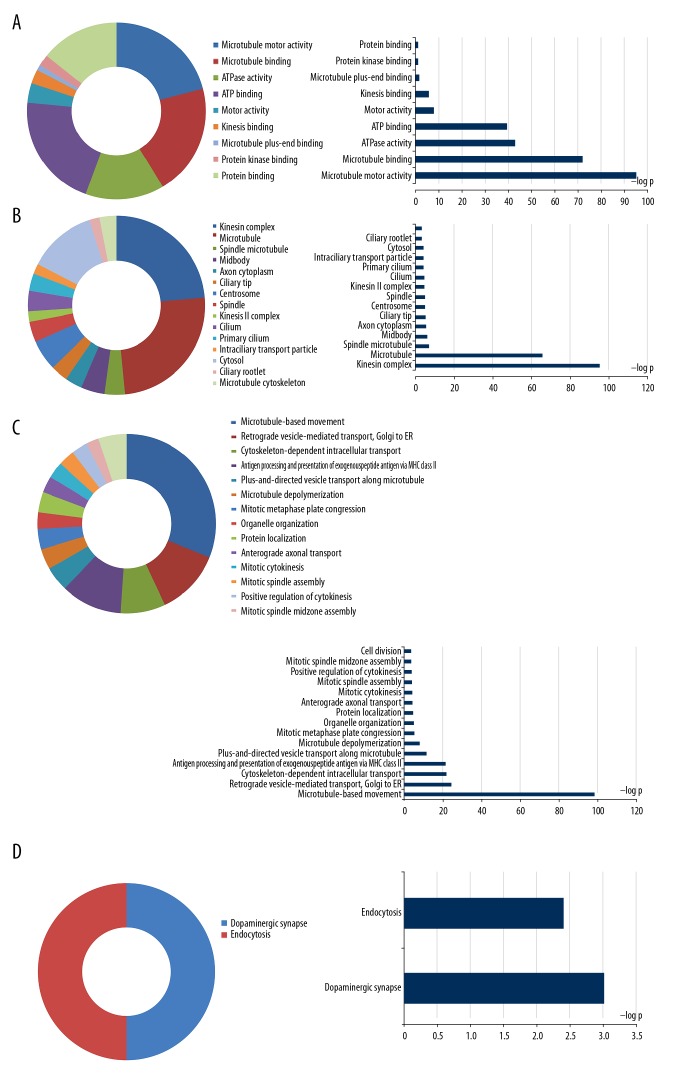
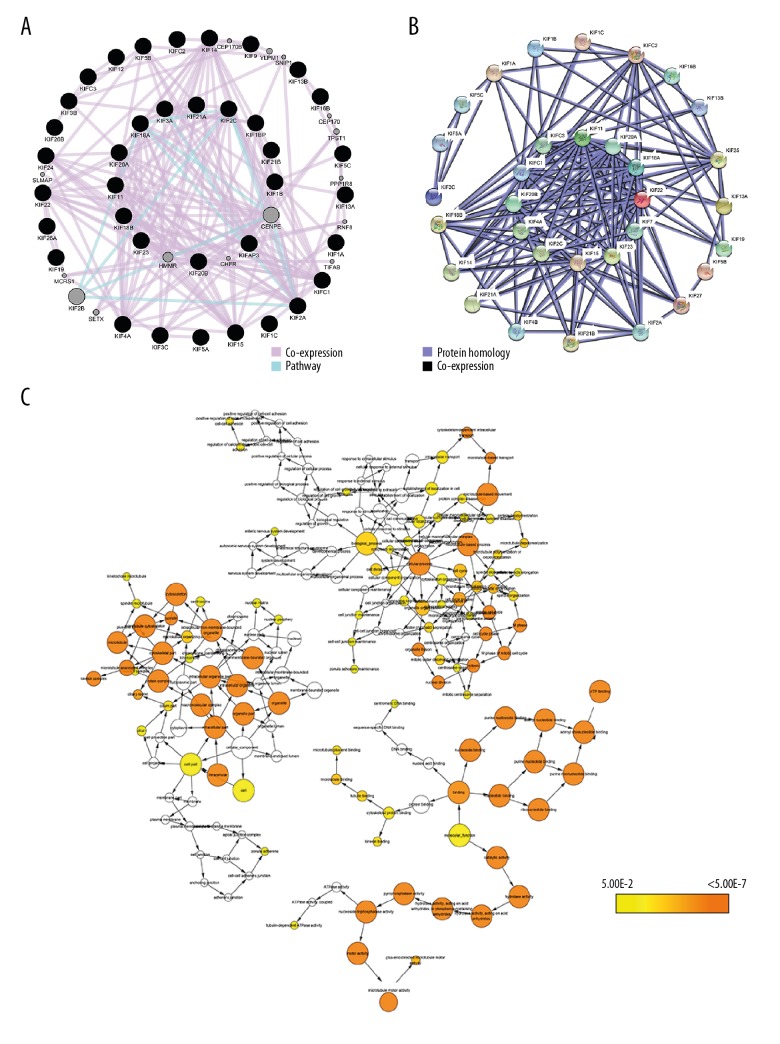
Enrichment analysis of GO terms for the KIF family genes, performed with DAVID, showed that KIF genes had suggestive enrichment for microtubule-based movement, and biological functions mainly included mitotic metaphase plate congression, mitotic cytokinesis, mitotic spindle assembly, cell division, mitotic spindle midzone assembly, and positive regulation of cytokinesis (Figure 1). Gene–gene and protein–protein interaction networks confirmed that the KIF genes had solid protein homology as well as co-expression with one another at the protein as well as gene levels (Figure 2A, 2B). The focused KIF genetic acyclic graph constructed by BiNGO in Cytoscape similarly showed that the main biological roles were cell cycle progression, cellular processes, and microtubule-based processes (Figure 2C).
Figure 1.
GO term and KEGG analysis of all the KIF family genes. GO term enrichments of KIF genes: (A) for MF; (B) for CC; (C) for BP. (D) KEGG enrichments of KIF genes. GO – gene ontology; KEGG – Kyoto Encyclopedia of Genes and Genomes; KIF – kinesin; MF – molecular function; CC – cellular component; BP – biological process.
Figure 2.
Protein–protein and gene–gene interaction networks of KIF genes. (A) GeneMANIA interaction networks. (B) Protein–protein interaction networks; (C) BiNGO analysis. KIF – kinesin; BiNGO – Biological Networks Gene Ontology tool.
Patient characteristics influencing differential KIF expression in BC
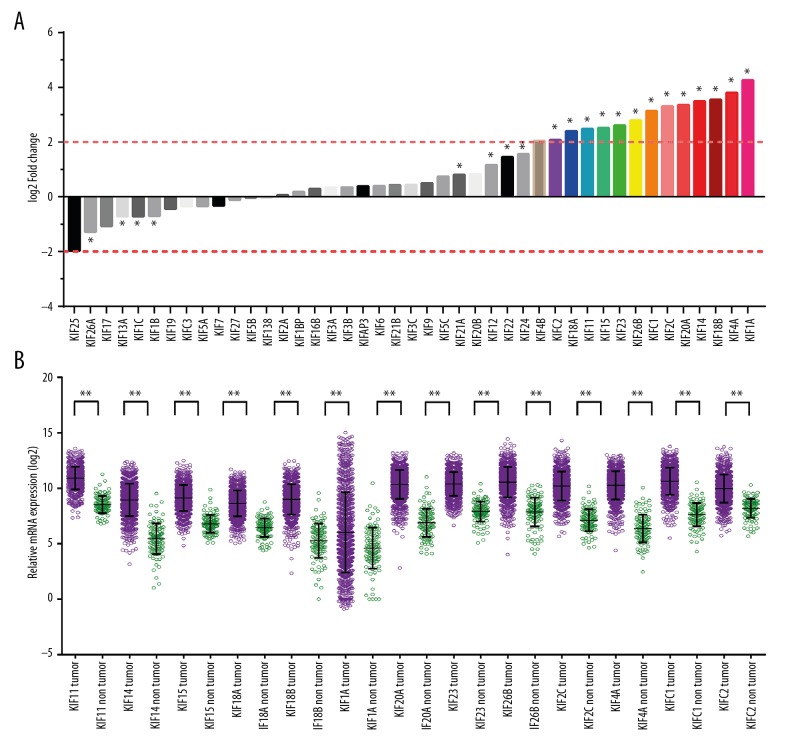
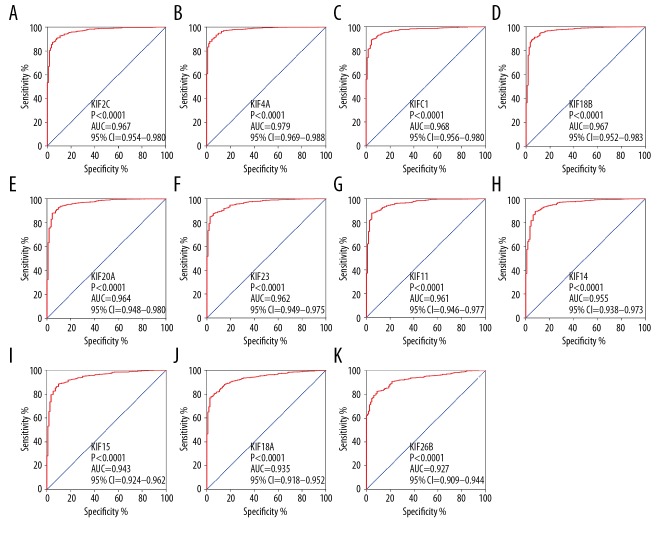
The |log2FC| of KIF family is shown in a histogram (Figure 3A). Thirteen KIF family genes met the standard of FDR <0.05 together with |log2FC| ≥2 (Table 1). A scatter plot produced using TCGA showed the difference in expression of the 13 KIF genes in invasive BC tissues compared with normal breast tissues (Figure 3B). Therefore, only the remaining 13 mRNAs were included in the step function screening to investigate the optimal combination, and all the mRNA expression data were log2 transformed for further analysis. The KIF genetic ROC analysis in the TCGA cohort specified that every KIF gene was highly accurate for discriminating normal breast and tumor tissues (area under the curve for the ROC curves in 11 KIF genes remained >0.9; Figure 4, Table 1).
Figure 3.
(A) Expression of KIF family genes. (B) Gene expression distribution of KIF genes in TCGA. * P<0.01. KIF – kinesin; TCGA – the Cancer Genome Atlas.
Table 1.
The difference expression between BC patients and normal breast tissues.
| Gene name | log2 fold change | P value | FDR | AUC | 95%CI | P value |
|---|---|---|---|---|---|---|
| KIF4A | 3.815 | 0.000 | 0.000 | 0.979 | 0.969–0.988 | 0.000 |
| KIFC1 | 3.134 | 0.000 | 0.000 | 0.968 | 0.956–0.980 | 0.000 |
| KIF18B | 3.545 | 0.000 | 0.000 | 0.967 | 0.951–0.983 | 0.000 |
| KIF2C | 3.302 | 0.000 | 0.000 | 0.967 | 0.954–0.980 | 0.000 |
| KIF20A | 3.352 | 0.000 | 0.000 | 0.964 | 0.948–0.980 | 0.000 |
| KIF23 | 2.609 | 0.000 | 0.000 | 0.962 | 0.949–0.975 | 0.000 |
| KIF11 | 2.488 | 0.000 | 0.000 | 0.961 | 0.945–0.977 | 0.000 |
| KIF14 | 3.489 | 0.000 | 0.000 | 0.955 | 0.938–0.973 | 0.000 |
| KIF22 | 1.450 | 0.000 | 0.000 | 0.950 | 0.936–0.965 | 0.000 |
| KIF15 | 2.519 | 0.000 | 0.000 | 0.943 | 0.924–0.962 | 0.000 |
| KIF18A | 2.405 | 0.000 | 0.000 | 0.935 | 0.917–0.952 | 0.000 |
| KIF24 | 1.563 | 0.002 | 0.013 | 0.929 | 0.911–0.948 | 0.000 |
| KIF26B | 2.793 | 0.000 | 0.000 | 0.927 | 0.909–0.944 | 0.000 |
| KIFC2 | 2.087 | 0.000 | 0.000 | 0.880 | 0.857–0.904 | 0.000 |
| KIF26A | −1.291 | 0.006 | 0.027 | 0.867 | 0.840–0.895 | 0.000 |
| KIF13A | −0.716 | 0.001 | 0.006 | 0.859 | 0.931–0.887 | 0.000 |
| KIF17 | −1.062 | 0.132 | 0.305 | 0.829 | 0.794–0.865 | 0.000 |
| KIF25 | −1.970 | 0.018 | 0.066 | 0.809 | 0.771–0.848 | 0.000 |
| KIF1B | −0.693 | 0.002 | 0.013 | 0.797 | 0.763–0.832 | 0.000 |
| KIF4B | 2.058 | 0.298 | 0.532 | 0.781 | 0.744–0.818 | 0.000 |
| KIF20B | 0.825 | 0.027 | 0.092 | 0.765 | 0.730–0.800 | 0.000 |
| KIF19 | −0.436 | 0.534 | 0.747 | 0.757 | 0.724–0.790 | 0.000 |
| KIF1C | −0.715 | 0.000 | 0.002 | 0.738 | 0.691–0.785 | 0.000 |
| KIF21A | 0.815 | 0.007 | 0.032 | 0.733 | 0.699–0.767 | 0.000 |
| KIFAP3 | 0.395 | 0.096 | 0.242 | 0.720 | 0.686–0.754 | 0.000 |
| KIF5A | −0.338 | 0.475 | 0.702 | 0.707 | 0.657–0.757 | 0.000 |
| KIFC3 | −0.346 | 0.261 | 0.486 | 0.704 | 0.664–0.743 | 0.000 |
| KIF9 | 0.507 | 0.367 | 0.605 | 0.684 | 0.645–0.724 | 0.000 |
| KIF7 | −0.322 | 0.377 | 0.616 | 0.679 | 0.637–0.721 | 0.000 |
| KIF3B | 0.352 | 0.071 | 0.194 | 0.665 | 0.629–0.700 | 0.000 |
| KIF13B | −0.027 | 0.815 | 0.927 | 0.626 | 0.592–0.659 | 0.000 |
| KIF3A | 0.348 | 0.343 | 0.580 | 0.617 | 0.569–0.664 | 0.000 |
| KIF1BP | 0.183 | 0.414 | 0.651 | 0.612 | 0.568–0.656 | 0.000 |
| KIF12 | 1.161 | 0.000 | 0.002 | 0.605 | 0.557–0.653 | 0.000 |
| KIF1A | 4.263 | 0.000 | 0.000 | 0.604 | 0.565–0.644 | 0.000 |
| KIF27 | −0.115 | 0.693 | 0.857 | 0.598 | 0.545–0.652 | 0.001 |
| KIF3C | 0.444 | 0.182 | 0.381 | 0.568 | 0.532–0.604 | 0.018 |
| KIF5B | −0.040 | 0.903 | 0.977 | 0.553 | 0.504–0.601 | 0.065 |
| KIF16B | 0.292 | 0.219 | 0.433 | 0.536 | 0.498–0.574 | 0.211 |
| KIF21B | 0.436 | 0.437 | 0.671 | 0.510 | 0.470–0.550 | 0.722 |
| KIF6 | 0.406 | 0.772 | 0.904 | 0.509 | 0.471–0.547 | 0.765 |
| KIF2A | 0.065 | 0.772 | 0.904 | 0.507 | 0.465–0.549 | 0.805 |
| KIF5C | 0.742 | 0.023 | 0.083 | 0.506 | 0.463–0.550 | 0.826 |
FDR – false discovery rate; AUC – area under the curve; 95%CI – 95% confidence interval.
Figure 4.
ROC curves (AUC>0.9) of KIF gens for distinguishing BC tumor tissue and adjacent normal tissues in TCGA. ROC curves of KIF2C (A), KIF4A (B), KIFC1 (C), KIF18B (D), KIF20A (E), KIF23 (F), KIF11 (G), KIF14 (H), KIF15 (I), KIF18A (J), and KIF26B (K). KIF – kinesin; TCGA – the Cancer Genome Atlas; BC – breast cancer; AUC – area under the curve; ROC – receiver operating characteristic.
Survival analysis and association analysis
In the TCGA invasive BC cohort, patients with advanced tumor stage and age ≥65 years had an increased risk of invasive BC mortality (Table 2). Other patient characteristics, including gender and race, within the TCGA cohort did not show a significant association with OS of invasive BC.
Table 2.
Demographic and clinical data for 1055 BC patients.
| Variables | Patients (n=1055) | No. of events | MST (days) | HR (95% CI) | Log-rank P |
|---|---|---|---|---|---|
| Race | 0.534 | ||||
| White | 732 | 109 | 3941 | Ref | |
| Others | 239 | 33 | 3873 | 1.132 (0.766–1.671) | |
| Missing | 84 | ||||
| Gender | 0.854 | ||||
| Female | 1043 | 148 | 3926 | Ref | |
| Male | 12 | 1 | NA | 0.832 (0.116–5.96) | |
| Age (years) | <0.001 | ||||
| ≥65 | 719 | 88 | 6456 | Ref | |
| <65 | 322 | 61 | 3418 | 2.18 (1.567–3.033) | |
| Missing | 14 | ||||
| Tumor stage | <0.001 | ||||
| I | 175 | 15 | 3959 | Ref | |
| II | 596 | 65 | 4267 | 1.71 (0.974–2.999) | |
| III | 241 | 43 | 3461 | 3.131 (1.738–5.641) | |
| IV | 20 | 15 | 1034 | 13.481 (6.572–27.654) | |
| Missing | 23 |
MST – median survival time; HR – hazard ratio; CI – confidence interval.
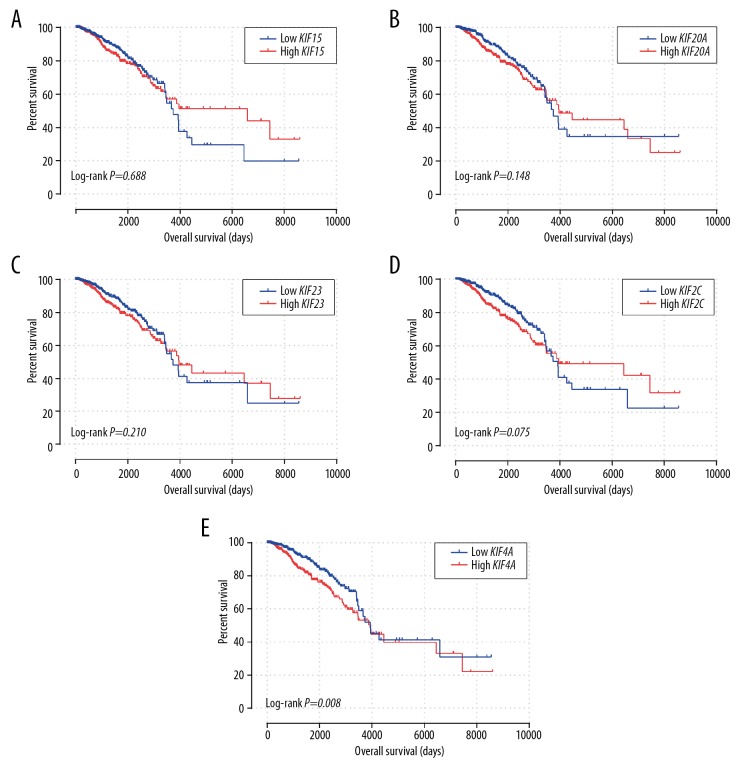
Survival analysis of the 13 differentially expressed KIF genes is shown in Table 3. Patients with low expression of KIF15, KIF20A, KIF23, KIF2C and KIF4A genes in the TCGA invasive BC cohort had an extended OS (Table 3, Figure 5A–5E). However, only the P values of KIF4A reached significance (P=0.008). It was suggested that elevated expression of KIF15 (adjusted P=0.045; adjusted HR=1.422; 95% CI=1.007–2.008), KIF20A (adjusted P=0.03; adjusted HR=1.467; 95% CI=1.038–2.072), KIF23 (adjusted P=0.014; adjusted HR=1.54; 95% CI=1.09–2.175), KIF2C (adjusted P=0.001; adjusted HR=1.805; 95% CI=1.276–2.553) and KIF4A (adjusted P=0.001; adjusted HR=1.805; 95% CI=1.276–2.553) (Table 3) was related to poor OS within invasive BC, after adjusting for tumor stage and age.
Table 3.
Prognostic survival analysis according to the high or low level of 13 diagnostic KIF genes and OS.
| Gene | Patients (n=1055) | Events | MST (days) | Crude HR (95% CI) | Crude P | Adjusted HR* (95% CI)* | Adjusted P* |
|---|---|---|---|---|---|---|---|
| KIF11 | |||||||
| High | 527 | 82 | 4456 | 1 | 1 | ||
| Low | 528 | 67 | 3736 | 1.106 (0.801–1.529) | 0.54 | 1.402 (0.991–1.982) | 0.056 |
| KIF14 | |||||||
| High | 527 | 76 | 7455 | 1 | 1 | ||
| Low | 528 | 73 | 3736 | 1.028 (0.745–1.417) | 0.869 | 1.187 (0.848–1.663) | 0.318 |
| KIF15 | |||||||
| High | 527 | 80 | 6593 | 1 | 1 | ||
| Low | 528 | 69 | 3736 | 1.068 (0.773–1.476) | 0.688 | 1.422 (1.007–2.008) | 0.045 |
| KIF18A | |||||||
| High | 527 | 82 | 3945 | 1 | 1 | ||
| Low | 528 | 67 | 3736 | 1.105 (0.799–1.527) | 0.546 | 1.367 (0.971–1.924) | 0.073 |
| KIF18B | |||||||
| High | 527 | 78 | 3959 | 1 | 1 | ||
| Low | 528 | 71 | 3736 | 1.143 (0.829–1.577) | 0.415 | 1.224 (0.875–1.713) | 0.238 |
| KIF1A | |||||||
| High | 527 | 74 | 3945 | 1 | 1 | ||
| Low | 528 | 75 | 3926 | 1.001 (0.726–1.38) | 0.996 | 1.046 (0.748–1.461) | 0.794 |
| KIF20A | |||||||
| High | 527 | 87 | 3959 | 1 | 1 | ||
| Low | 528 | 62 | 3736 | 1.273 (0.917–1.766) | 0.148 | 1.467 (1.038–2.072) | 0.03 |
| KIF23 | |||||||
| High | 527 | 83 | 3959 | 1 | 1 | ||
| Low | 528 | 66 | 3736 | 1.23 (0.889–1.701) | 0.21 | 1.54 (1.09–2.175) | 0.014 |
| KIF26B | |||||||
| High | 527 | 69 | 3472 | 1 | 1 | ||
| Low | 528 | 80 | 3959 | 1.067 (0.771–1.475) | 0.696 | 1.194 (0.848–1.682) | 0.309 |
| KIF2C | |||||||
| High | 527 | 84 | 3959 | 1 | 1 | ||
| Low | 528 | 65 | 3926 | 1.341 (0.97–1.855) | 0.075 | 1.805 (1.276–2.553) | 0.001 |
| KIF4A | |||||||
| High | 527 | 90 | 3941 | 1 | 1 | ||
| Low | 528 | 59 | 3926 | 1.557 (1.121–2.162) | 0.008 | 1.805 (1.276–2.553) | 0.001 |
| KIFC1 | |||||||
| High | 527 | 82 | 4456 | 1 | 1 | ||
| Low | 528 | 67 | 3492 | 1.106 (0.798–1.534) | 0.545 | 1.273 (0.902–1.796) | 0.17 |
| KIFC2 | |||||||
| High | 527 | 71 | 4456 | 1 | 1 | ||
| Low | 528 | 78 | 3669 | 0.983 (0.712–1.358) | 0.919 | 0.915 (0.653–1.281) | 0.603 |
Adjusted for age (stratified by 65 years) and tumor stage. KIF – kinesin; OS – overall survival; MST – median survival time; HR – hazard ratio; CI – confidence interval.
Figure 5.
Kaplan-Meier survival curves for KIF genes in BC of TCGA cohort. OS stratified by KIF15 (A), KIF20A (B), KIF23 (C), KIF2C (D), and KIF4A (E). KIF – kinesin; TCGA – the Cancer Genome Atlas; BC – breast cancer; OS – overall survival.
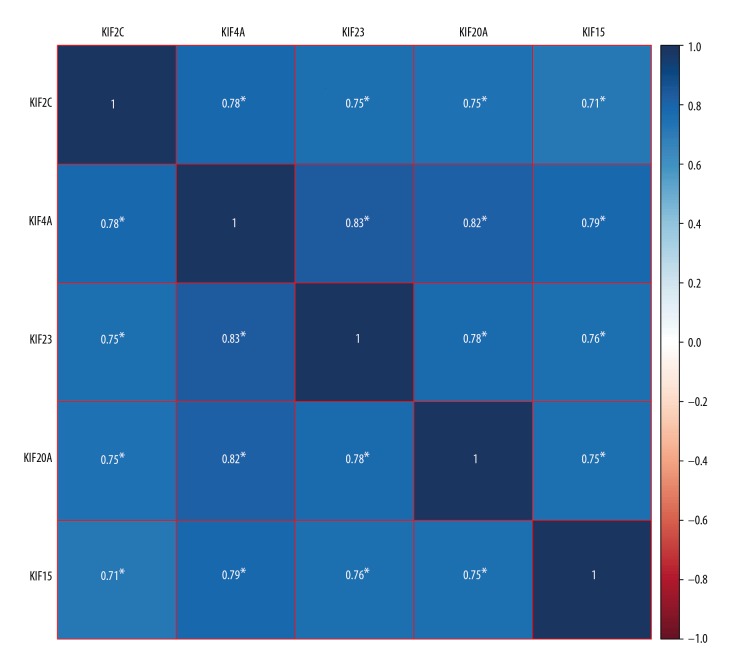
After performing survival analysis within the TCGA cohorts, co-expression analysis of KIF15, KIF20A, KIF23, KIF2C and KIF4A in BC malignant tissues was evaluated using Pearson’s correlation coefficient. The genes were co-expressed strongly with each other in the TCGA cohort (Figure 6).
Figure 6.
Co-expression heat map of KIF2C, KIF4A, KIF23, KIF20A and KIF15 in TCGA BC patients. KIF – kinesin; TCGA – the Cancer Genome Atlas; BC – breast cancer.
Effect of combinations of KIF gene expression on OS
Based on KIF gene survival analysis, KIF15, KIF20A, KIF23, KIF2C and KIF4A were screened as prognostic genes by multivariate survival analysis. A joint-effects model was utilized for determining the combined influence of the 5 KIF genes on OS of BC patients. The diverse groups for this analysis were generated in accordance with expression of KIF15, KIF20A, KIF23, KIF2C and KIF4A (Tables 4–7). The Kaplan-Meier estimator with a log-rank evaluation was administered to evaluate the prognostic significance of the gene expression combinations represented by each group. Two selected groups showed that the BC patients with high expression of KIF20A and KIF4A or high expression of KIF2C and KIF4A had poor OS (Table 8). Within the evaluation of low KIF15, KIF20A, KIF23, KIF2C and low KIF4A expression, the combinations in groups 4, 7, 10, 13, 16, 19, 22, 25 and 28 were highly correlated with favorable OS (all P<0.05; Table 8). In the analysis of high expression of KIF15, KIF20A, KIF23, KIF2C and KIF4A, the combinations in groups 3, 6, 9, 12, 15, 18, 21, 24, 27 and 30 were highly correlated with poor OS (all P<0.05; Table 8).
Table 4.
Grouping according to 2 selected genes.
| Group | Combination | Group | Combination |
|---|---|---|---|
| 1 | Low KIF15 + low KIF20A | 16 | Low KIF20A + low KIF2C |
| 2 | Low KIF15 + high KIF20A | 17 | Low KIF20A + high KIF2C |
| High KIF15 + low KIF20A | High KIF20A + low KIF2C | ||
| 3 | High KIF15 + high KIF20A | 18 | High KIF20A + high KIF2C |
| 4 | Low KIF15 + low KIF23 | 19 | Low KIF20A + low KIF4A |
| 5 | Low KIF15 + high KIF23 | 20 | Low KIF20A + high KIF4A |
| High KIF15 + low KIF23 | High KIF20A + low KIF4A | ||
| 6 | High KIF15 + high KIF23 | 21 | High KIF20A + high KIF4A |
| 7 | Low KIF15 + low KIF2C | 22 | Low KIF23 + low KIF2C |
| 8 | Low KIF15 + high KIF2C | 23 | Low KIF23 + high KIF2C |
| High KIF15 + low KIF2C | High KIF23 + low KIF2C | ||
| 9 | High KIF15 + high KIF2C | 24 | High KIF23 + high KIF2C |
| 10 | Low KIF15 + low KIF4A | 25 | Low KIF23 + low KIF4A |
| 11 | Low KIF15 + high KIF4A | 26 | Low KIF23 + high KIF4A |
| High KIF15 + low KIF4A | High KIF23 + low KIF4A | ||
| 12 | High KIF15 + high KIF4A | 27 | High KIF23 + high KIF4A |
| 13 | Low KIF20A + low KIF23 | 28 | Low KIF2C + low KIF4A |
| 14 | Low KIF20A + high KIF23 | 29 | Low KIF2C + high KIF4A |
| High KIF20A + low KIF23 | High KIF2C + low KIF4A | ||
| 15 | High KIF20A + high KIF23 | 30 | High KIF2C + high KIF4A |
KIF – kinesin.
Table 5.
Grouping according to 3 selected genes.
| Group | Combination | Group | Combination |
|---|---|---|---|
| a | Low KIF15 + low KIF20A + low KIF23 | A | Low KIF15 + low KIF2C + low KIF4A |
| b | Low KIF15 + high KIF20A + low KIF23 | B | High KIF15 + low KIF2C + low KIF4A |
| High KIF15 + low KIF20A + low KIF23 | Low KIF15 + high KIF2C + low KIF4A | ||
| Low KIF15 + low KIF20A + high KIF23 | Low KIF15 + low KIF2C + high KIF4A | ||
| High KIF15 + high KIF20A + low KIF23 | High KIF15 + high KIF2C + low KIF4A | ||
| Low KIF15 + high KIF20A + high KIF23 | High KIF15 + low KIF2C + high KIF4A | ||
| High KIF15 + low KIF20A + high KIF23 | Low KIF15 + high KIF2C + high KIF4A | ||
| c | High KIF15 + high KIF20A + high KIF23 | C | High KIF15 + high KIF2C + high KIF4A |
| d | Low KIF15 + low KIF20A + low KIF2C | D | Low KIF20A + low KIF23 + low KIF2C |
| e | High KIF15 + high KIF20A + low KIF2C | E | High KIF20A + low KIF23 + low KIF2C |
| Low KIF15 + high KIF20A + low KIF2C | Low KIF20A + high KIF23 + low KIF2C | ||
| High KIF15 + low KIF20A + low KIF2C | Low KIF20A + low KIF23 + high KIF2C | ||
| Low KIF15 + high KIF20A + high KIF2C | High KIF20A + high KIF23 + low KIF2C | ||
| High KIF15 + low KIF20A + high KIF2C | High KIF20A + low KIF23 + high KIF2C | ||
| Low KIF15 + low KIF20A + high KIF2C | Low KIF20A + high KIF23 + high KIF2C | ||
| f | High KIF15 + high KIF20A + high KIF2C | F | High KIF20A + high KIF23 + high KIF2C |
| g | Low KIF15 + low KIF20A + low KIF4A | G | Low KIF20A + low KIF23 + low KIF4A |
| h | Low KIF15 + high KIF20A + low KIF4A | H | High KIF20A + low KIF23 + low KIF4A |
| High KIF15 + low KIF20A + low KIF4A | Low KIF20A + high KIF23 + low KIF4A | ||
| Low KIF15 + high KIF20A + high KIF4A | Low KIF20A + low KIF23 + high KIF4A | ||
| High KIF15 + low KIF20A + high KIF4A | High KIF20A + high KIF23 + low KIF4A | ||
| High KIF15 + high KIF20A + low KIF4A | High KIF20A + low KIF23 + high KIF4A | ||
| Low KIF15 + low KIF20A + high KIF4A | Low KIF20A + high KIF23 + high KIF4A | ||
| i | High KIF15 + high KIF20A + high KIF4A | I | High KIF20A + high KIF23 + high KIF4A |
| j | Low KIF15 + low KIF23 + low KIF2C | J | Low KIF20A + low KIF2C + low KIF4A |
| k | High KIF15 + low KIF23 + low KIF2C | K | High KIF20A + low KIF2C + low KIF4A |
| Low KIF15 + high KIF23 + low KIF2C | Low KIF20A + high KIF2C + low KIF4A | ||
| Low KIF15 + low KIF23 + high KIF2C | Low KIF20A + low KIF2C + high KIF4A | ||
| High KIF15 + high KIF23 + low KIF2C | High KIF20A + high KIF2C + low KIF4A | ||
| High KIF15 + low KIF23 + high KIF2C | High KIF20A + low KIF2C + high KIF4A | ||
| Low KIF15 + high KIF23 + high KIF2C | Low KIF20A + high KIF2C + high KIF4A | ||
| l | High KIF15 + high KIF23 + high KIF2C | L | High KIF20A + high KIF2C + high KIF4A |
| m | Low KIF15 + low KIF23 + low KIF4A | M | Low KIF23 + low KIF2C + low KIF4A |
| n | High KIF15 + low KIF23 + low KIF4A | N | Low KIF23 + low KIF2C + high KIF4A |
| Low KIF15 + high KIF23 + low KIF4A | High KIF23 + low KIF2C + low KIF4A | ||
| Low KIF15 + low KIF23 + high KIF4A | Low KIF23 + high KIF2C + low KIF4A | ||
| High KIF15 + high KIF23 + low KIF4A | High KIF23 + high KIF2C + low KIF4A | ||
| High KIF15 + high KIF23 + high KIF4A | High KIF23 + low KIF2C + high KIF4A | ||
| Low KIF15 + high KIF23 + high KIF4A | Low KIF23 + high KIF2C + high KIF4A | ||
| o | High KIF15 + low KIF23 + high KIF4A | O | High KIF23 + high KIF2C + high KIF4A |
KIF – kinesin.
Table 6.
Grouping according to 4 selected genes.
| Group | Combination | Group | Combination |
|---|---|---|---|
| I | Low KIF15 + low KIF20A + low KIF23 + low KIF2C | X | Low KIF15 + low KIF23 + low KIF2C + low KIF4A |
| II | High KIF15 + high KIF20A + low KIF23 + low KIF2C | XI | High KIF15 + low KIF23 + low KIF2C + low KIF4A |
| Low KIF15 + high KIF20A + high KIF23 + high KIF2C | High KIF15 + low KIF23 + high KIF2C + high KIF4A | ||
| High KIF15 + low KIF20A + high KIF23 + high KIF2C | High KIF15 + high KIF23 + low KIF2C + high KIF4A | ||
| High KIF15 + high KIF20A + low KIF23 + high KIF2C | High KIF15 + high KIF23 + high KIF2C + low KIF4A | ||
| High KIF15 + high KIF20A + high KIF23 + low KIF2C | Low KIF15 + high KIF23 + low KIF2C + high KIF4A | ||
| Low KIF15 + high KIF20A + low KIF23 + high KIF2C | Low KIF15 + high KIF23 + high KIF2C + low KIF4A | ||
| Low KIF15 + high KIF20A + high KIF23 + low KIF2C | High KIF15 + low KIF23 + high KIF2C + low KIF4A | ||
| High KIF15 + low KIF20A + high KIF23 + low KIF2C | High KIF15 + low KIF23 + low KIF2C + high KIF4A | ||
| High KIF15 + low KIF20A + low KIF23 + high KIF2C | High KIF15 + high KIF23 + low KIF2C + low KIF4A | ||
| High KIF15 + low KIF20A + low KIF23 + low KIF2C | Low KIF15 + high KIF23 + low KIF2C + low KIF4A | ||
| Low KIF15 + high KIF20A + low KIF23 + low KIF2C | Low KIF15 + low KIF23 + high KIF2C + low KIF4A | ||
| Low KIF15 + low KIF20A + high KIF23 + low KIF2C | Low KIF15 + low KIF23 + low KIF2C + high KIF4A | ||
| Low KIF15 + low KIF20A + low KIF23 + high KIF2C | Low KIF15 + low KIF23 + high KIF2C + high KIF4A | ||
| Low KIF15 + low KIF20A + high KIF23 + high KIF2C | Low KIF15 + high KIF23 + high KIF2C + high KIF4A | ||
| III | High KIF15 + high KIF20A + high KIF23 + high KIF2C | XII | High KIF15 + high KIF23 + high KIF2C + high KIF4A |
| IV | Low KIF15 + low KIF20A + low KIF23 + low KIF4A | XIII | Low KIF20A + low KIF23 + low KIF2C + low KIF4A |
| V | High KIF15 + high KIF20A + low KIF23 + low KIF4A | XIV | High KIF20A + low KIF23 + low KIF2C + low KIF4A |
| Low KIF15 + high KIF20A + high KIF23 + high KIF4A | High KIF20A + low KIF23 + high KIF2C + high KIF4A | ||
| High KIF15 + low KIF20A + high KIF23 + high KIF4A | High KIF20A + high KIF23 + low KIF2C + high KIF4A | ||
| High KIF15 + high KIF20A + low KIF23 + high KIF4A | High KIF20A + high KIF23 + high KIF2C + low KIF4A | ||
| High KIF15 + high KIF20A + high KIF23 + low KIF4A | Low KIF20A + high KIF23 + low KIF2C + high KIF4A | ||
| Low KIF15 + high KIF20A + low KIF23 + high KIF4A | Low KIF20A + high KIF23 + high KIF2C + low KIF4A | ||
| Low KIF15 + high KIF20A + high KIF23 + low KIF4A | High KIF20A + low KIF23 + high KIF2C + low KIF4A | ||
| High KIF15 + low KIF20A + high KIF23 + low KIF4A | High KIF20A + low KIF23 + low KIF2C + high KIF4A | ||
| High KIF15 + low KIF20A + low KIF23 + high KIF4A | High KIF20A + high KIF23 + low KIF2C + low KIF4A | ||
| High KIF15 + low KIF20A + low KIF23 + low KIF4A | Low KIF20A + high KIF23 + low KIF2C + low KIF4A | ||
| Low KIF15 + high KIF20A + low KIF23 + low KIF4A | Low KIF20A + low KIF23 + high KIF2C + low KIF4A | ||
| Low KIF15 + low KIF20A + high KIF23 + low KIF4A | Low KIF20A + low KIF23 + low KIF2C + high KIF4A | ||
| Low KIF15 + low KIF20A + low KIF23 + high KIF4A | Low KIF20A + low KIF23 + high KIF2C + high KIF4A | ||
| Low KIF15 + low KIF20A + high KIF23 + high KIF4A | Low KIF20A + high KIF23 + high KIF2C + high KIF4A | ||
| VI | High KIF15 + high KIF20A + high KIF23 + high KIF4A | XV | High KIF20A + high KIF23 + high KIF2C + high KIF4A |
| VII | Low KIF15 + low KIF20A + low KIF2C + low KIF4A | ||
| VIII | High KIF15 + high KIF20A + low KIF2C + low KIF4A | ||
| Low KIF15 + high KIF20A + high KIF2C + high KIF4A | |||
| High KIF15 + low KIF20A + high KIF2C + high KIF4A | |||
| High KIF15 + high KIF20A + low KIF2C + high KIF4A | |||
| High KIF15 + high KIF20A + high KIF2C + low KIF4A | |||
| Low KIF15 + high KIF20A + low KIF2C + high KIF4A | |||
| Low KIF15 + high KIF20A + high KIF2C + low KIF4A | |||
| High KIF15 + low KIF20A + high KIF2C + low KIF4A | |||
| High KIF15 + low KIF20A + low KIF2C + high KIF4A | |||
| High KIF15 + low KIF20A + low KIF2C + low KIF4A | |||
| Low KIF15 + high KIF20A + low KIF2C + low KIF4A | |||
| Low KIF15 + low KIF20A + high KIF2C + low KIF4A | |||
| Low KIF15 + low KIF20A + low KIF2C + high KIF4A | |||
| Low KIF15 + low KIF20A + high KIF2C + high KIF4A | |||
| IX | High KIF15 + high KIF20A + high KIF2C + high KIF4A |
KIF – kinesin.
Table 7.
Grouping according to 5 selected genes.
| Group | Combination |
|---|---|
| ➀ | Low KIF15 + low KIF20A + low KIF23 + low KIF2C + low KIF4A |
| ➁ | High KIF15 + high KIF20A + low KIF23 + low KIF2C + low KIF4A |
| High KIF15 + low KIF20A + low KIF23 + low KIF2C + low KIF4A | |
| Low KIF15 + high KIF20A + low KIF23 + low KIF2C + low KIF4A | |
| Low KIF15 + low KIF20A + high KIF23 + low KIF2C + low KIF4A | |
| Low KIF15 + low KIF20A + low KIF23 + high KIF2C + low KIF4A | |
| Low KIF15 + low KIF20A + low KIF23 + low KIF2C + high KIF4A | |
| High KIF15 + low KIF20A + high KIF23 + low KIF2C + low KIF4A | |
| High KIF15 + low KIF20A + low KIF23 + high KIF2C + low KIF4A | |
| High KIF15 + low KIF20A + low KIF23 + low KIF2C + high KIF4A | |
| Low KIF15 + high KIF20A + high KIF23 + low KIF2C + low KIF4A | |
| Low KIF15 + high KIF20A + low KIF23 + high KIF2C + low KIF4A | |
| Low KIF15 + high KIF20A + low KIF23 + low KIF2C + high KIF4A | |
| Low KIF15 + low KIF20A + high KIF23 + high KIF2C + low KIF4A | |
| Low KIF15 + low KIF20A + high KIF23 + low KIF2C + high KIF4A | |
| Low KIF15 + low KIF20A + low KIF23 + high KIF2C + high KIF4A | |
| Low KIF15 + high KIF20A + low KIF23 + high KIF2C + high KIF4A | |
| Low KIF15 + high KIF20A + high KIF23 + low KIF2C + high KIF4A | |
| Low KIF15 + high KIF20A + high KIF23 + high KIF2C + low KIF4A | |
| High KIF15 + low KIF20A + low KIF23 + high KIF2C + high KIF4A | |
| High KIF15 + low KIF20A + high KIF23 + low KIF2C + high KIF4A | |
| High KIF15 + low KIF20A + high KIF23 + high KIF2C + low KIF4A | |
| High KIF15 + high KIF20A + low KIF23 + low KIF2C + high KIF4A | |
| High KIF15 + high KIF20A + low KIF23 + high KIF2C + low KIF4A | |
| High KIF15 + high KIF20A + high KIF23 + low KIF2C + low KIF4A | |
| Low KIF15 + high KIF20A + high KIF23 + high KIF2C + high KIF4A | |
| High KIF15 + low KIF20A + high KIF23 + high KIF2C + high KIF4A | |
| High KIF15 + high KIF20A + low KIF23 + high KIF2C + high KIF4A | |
| High KIF15 + high KIF20A + high KIF23 + low KIF2C + high KIF4A | |
| High KIF15 + high KIF20A + high KIF23 + high KIF2C + low KIF4A | |
| Low KIF15 + low KIF20A + high KIF23 + high KIF2C + high KIF4A | |
| ➂ | High KIF15 + high KIF20A + high KIF23 + high KIF2C + high KIF4A |
KIF – kinesin.
Table 8.
Joint analysis of the prognostic value of combination of KIF15, KIF20A, KIF23, KIF2C, and KIF4A expression of BC.
| Group | Patients | MST (days) | Crude p | Crude HR | Adjusted p | Adjusted HR(95% CI) * |
|---|---|---|---|---|---|---|
| 1 | 432 | 3736 | 0.367 | 1 | 0.065 | 1 |
| 2 | 192 | 3941 | 0.185 | 1.354 (0.865–2.121) | 0.16 | 1.419 (0.87–2.313) |
| 3 | 431 | 6593 | 0.291 | 1.219 (0.844–1.759) | 0.021 | 1.578 (1.073–2.322) |
| 4 | 444 | 3669 | 0.439 | 1 | 0.049 | 1 |
| 5 | 168 | 4456 | 0.229 | 1.328 (0.836–2.11) | 0.223 | 1.363 (0.828–2.245) |
| 6 | 443 | 3959 | 0.356 | 1.184 (0.827–1.695) | 0.014 | 1.615 (1.1–2.372) |
| 7 | 428 | 3736 | 0.398 | 1 | 0.008 | 1 |
| 8 | 200 | 6456 | 0.279 | 1.296 (0.811–2.071) | 0.021 | 1.825 (1.097–3.037) |
| 9 | 427 | 3959 | 0.23 | 1.244 (0.871–1.778) | 0.003 | 1.776 (1.211–2.604) |
| 10 | 436 | 3669 | 0.237 | 1 | 0.013 | 1 |
| 11 | 184 | 6456 | 0.396 | 1.223 (0.768–1.946) | 0.196 | 1.39 (0.843–2.29) |
| 12 | 435 | 3873 | 0.09 | 1.369 (0.952–1.967) | 0.003 | 1.787 (1.215–2.63) |
| 13 | 450 | 3736 | 0.292 | 1 | 0.038 | 1 |
| 14 | 156 | 3461 | 0.265 | 1.317 (0.812–2.137) | 0.116 | 1.508 (0.904–2.517) |
| 15 | 449 | 3959 | 0.14 | 1.308 (0.916–1.87) | 0.012 | 1.625 (1.112–2.374) |
| 16 | 441 | 3736 | 0.221 | 1 | 0.008 | 1 |
| 17 | 174 | 4456 | 0.546 | 1.169 (0.704–1.94) | 0.652 | 1.133 (0.659–1.949) |
| 18 | 440 | 3959 | 0.083 | 1.366 (0.96–1.944) | 0.003 | 1.76 (1.21–2.56) |
| 19 | 454 | 3736 | 0.051 | 1 | 0.011 | 1 |
| 20 | 148 | 6593 | 0.772 | 0.919 (0.518–1.631) | 0.735 | 1.114 (0.598–2.074) |
| 21 | 453 | 3941 | 0.032 | 1.463 (1.034–2.07) | 0.004 | 1.715 (1.188–2.475) |
| 22 | 445 | 3736 | 0.207 | 1 | 0.006 | 1 |
| 23 | 166 | 3941 | 0.197 | 1.379 (0.846–2.249) | 0.394 | 1.258 (0.742–2.131) |
| 24 | 444 | 3959 | 0.101 | 1.344 (0.944–1.914) | 0.002 | 1.844 (1.262–2.696) |
| 25 | 453 | 3736 | 0.070 | 1 | 0.004 | 1 |
| 26 | 150 | NUM | 0.805 | 0.93 (0.525–1.648) | 0.932 | 1.026 (0.573–1.837) |
| 27 | 452 | 3941 | 0.040 | 1.436 (1.017–2.027) | 0.002 | 1.787 (1.236–2.585) |
| 28 | 457 | 3736 | 0.033 | 1 | 0.001 | 1 |
| 29 | 142 | 4456 | 0.751 | 0.909 (0.505–1.636) | 0.457 | 0.785 (0.414–1.486) |
| 30 | 456 | 3873 | 0.020 | 1.503 (1.066–2.12) | 0.001 | 1.93 (1.34–2.78) |
| a | 402 | 3736 | 0.343 | 1 | 0.042 | 1 |
| b | 258 | 3941 | 0.157 | 1.354 (0.89–2.059) | 0.097 | 1.466 (0.933–2.302) |
| c | 395 | 3959 | 0.298 | 1.225 (0.836–1.794) | 0.013 | 1.674 (1.113–2.516) |
| d | 390 | 3736 | 0.37 | 1 | 0.04 | 1 |
| e | 283 | 3941 | 0.232 | 1.296 (0.847–1.983) | 0.129 | 1.426 (0.902–2.254) |
| f | 382 | 3959 | 0.211 | 1.274 (0.872–1.861) | 0.011 | 1.675 (1.123–2.497) |
| g | 399 | 3736 | 0.335 | 1 | 0.028 | 1 |
| h | 262 | 4456 | 0.45 | 1.181 (0.767–1.818) | 0.225 | 1.335 (0.837–2.131) |
| i | 394 | 3959 | 0.139 | 1.328 (0.912–1.934) | 0.008 | 1.713 (1.152–2.548) |
| j | 399 | 3669 | 0.34 | 1 | 0.018 | 1 |
| k | 267 | 4456 | 0.168 | 1.348 (0.882–2.06) | 0.088 | 1.487 (0.942–2.346) |
| l | 389 | 3959 | 0.262 | 1.24 (0.851–1.805) | 0.005 | 1.787 (1.193–2.675) |
| m | 403 | 3669 | 0.44 | 1 | 0.019 | 1 |
| n | 251 | 6456 | 0.519 | 1.154 (0.747–1.782) | 0.316 | 1.267 (0.798–2.011) |
| o | 401 | 3873 | 0.2 | 1.275 (0.879–1.849) | 0.006 | 1.763 (1.181–2.631) |
| A | 398 | 3669 | 0.334 | 1 | 0.018 | 1 |
| B | 263 | 6456 | 0.558 | 1.14 (0.735–1.767) | 0.315 | 1.273 (0.795–2.04) |
| C | 394 | 3959 | 0.141 | 1.321 (0.912–1.913) | 0.005 | 1.749 (1.18–2.593) |
| D | 411 | 3736 | 0.424 | 1 | 0.026 | 1 |
| E | 248 | 3941 | 0.312 | 1.252 (0.809–1.937) | 0.316 | 1.269 (0.797–2.023) |
| F | 396 | 6456 | 0.225 | 1.258 (0.869–1.821) | 0.007 | 1.721 (1.158–2.559) |
| G | 419 | 3736 | 0.2 | 1 | 0.023 | 1 |
| H | 227 | 6593 | 0.885 | 1.035 (0.649–1.651) | 0.46 | 1.204 (0.735–1.973) |
| I | 409 | 3941 | 0.096 | 1.358 (0.947–1.949) | 0.007 | 1.691 (1.151–2.484) |
| J | 414 | 3736 | 0.081 | 1 | 0.002 | 1 |
| K | 232 | 4456 | 0.964 | 0.989 (0.613–1.597) | 0.97 | 0.99 (0.591–1.658) |
| L | 409 | 3873 | 0.046 | 1.44 (1.006–2.061) | 0.002 | 1.823 (1.248–2.664) |
| M | 419 | 3736 | 0.218 | 1 | 0.002 | 1 |
| N | 229 | 4456 | 0.891 | 1.033 (0.646–1.653) | 0.965 | 0.989 (0.6–1.629) |
| O | 407 | 3959 | 0.103 | 1.346 (0.942–1.925) | 0.002 | 1.865 (1.268–2.742) |
| I | 375 | 3736 | 0.391 | 1 | 0.044 | 1 |
| II | 324 | 3941 | 0.179 | 1.324 (0.879–1.995) | 0.103 | 1.44 (0.929–2.231) |
| III | 356 | 3959 | 0.353 | 1.203 (0.815–1.777) | 0.013 | 1.693 (1.115–2.569) |
| IV | 381 | 3736 | 0.477 | 1 | 0.038 | 1 |
| V | 304 | 4456 | 0.338 | 1.224 (0.81–1.852) | 0.189 | 1.347 (0.864–2.099) |
| VI | 370 | 3959 | 0.254 | 1.252 (0.851–1.843) | 0.01 | 1.715 (1.135–2.593) |
| VII | 375 | 3736 | 0.381 | 1 | 0.037 | 1 |
| VIII | 315 | 4456 | 0.376 | 1.208 (0.795–1.837) | 0.186 | 1.355 (0.864–2.124) |
| IX | 365 | 3959 | 0.169 | 1.309 (0.892–1.921) | 0.01 | 1.701 (1.135–2.55) |
| X | 381 | 3669 | 0.544 | 1 | 0.026 | 1 |
| XI | 306 | 4456 | 0.441 | 1.178 (0.776–1.789) | 0.257 | 1.294 (0.829–2.02) |
| XII | 368 | 3959 | 0.285 | 1.231 (0.841–1.801) | 0.007 | 1.752 (1.163–2.638) |
| XIII | 394 | 3736 | 0.41 | 1 | 0.015 | 1 |
| XIV | 287 | 3941 | 0.553 | 1.138 (0.742–1.745) | 0.479 | 1.179 (0.747–1.859) |
| XV | 374 | 3959 | 0.182 | 1.29 (0.887–1.876) | 0.006 | 1.768 (1.182–2.645) |
| ➀ | 364 | 3736 | 0.455 | 1 | 0.045 | 1 |
| ➁ | 348 | 3941 | 0.24 | 1.275 (0.85–1.912) | 0.128 | 1.4 (0.908–2.158) |
| ➂ | 343 | 3959 | 0.316 | 1.224 (0.824–1.816) | 0.013 | 1.708 (1.119–2.606) |
Adjusted for age (stratified by 65 years) and tumor stage. KIF – kinesin; OS – overall survival; MST – median survival time; HR – hazard ratio; CI: confidence interval.
GSEA
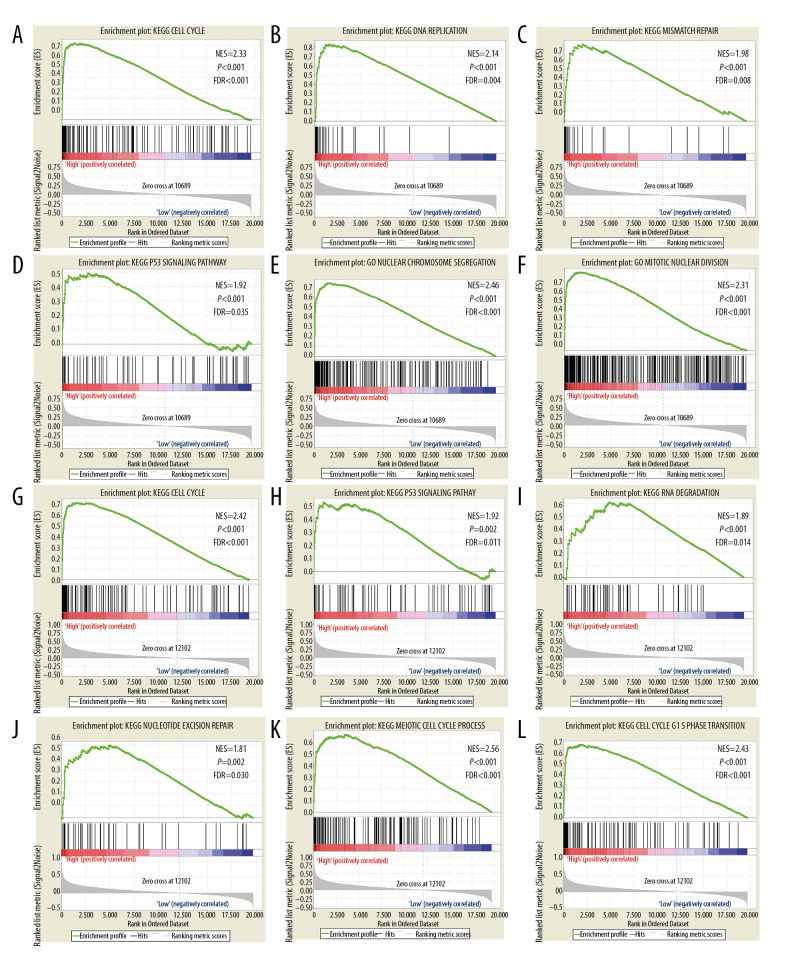
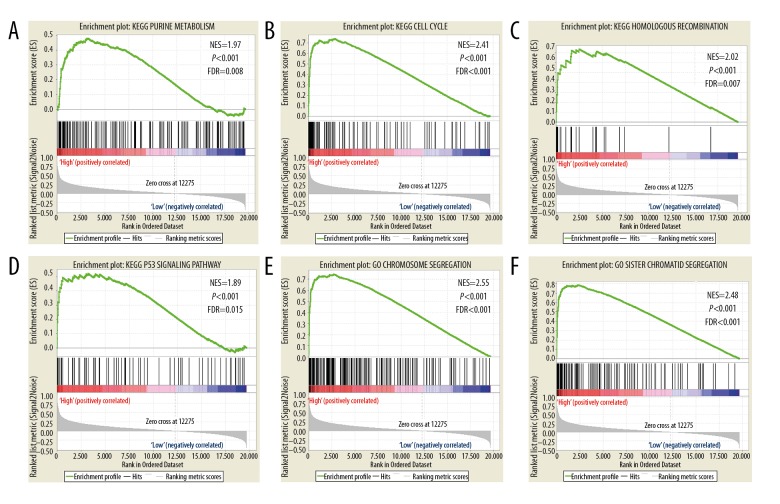
GSEA of the prognostic genes KIF15, KIF20A, KIF23, KIF2C and KIF4A was performed within the TCGA cohorts. The expression profiles of the genome-wide dataset in the TCGA-based cohorts were divided into 2 groups in accordance with the median prognostic KIF genetic values. GSEA outcomes of the TCGA cohort are shown in Figures 7A–7L, 8A–8L and 9A–9F), which suggested that their elevated expression remained linked with mismatch repair, P53 regulation pathway, and cell cycle progression.
Figure 7.
A–F shows GSEA of KIF2C in TCGA patients. (A–D) GSEA results of c2 reference gene sets for high KIF2C expression groups, (E–F) GSEA results of c5 reference gene sets for high KIF2C expression groups; G–L shows GSEA of KIF4A in TCGA patients. (G–J) GSEA results of c2 reference gene sets for high KIF4A expression groups; (K–L) GSEA results of c5 reference gene sets for high KIF4A expression groups. KIF – kinesin; GSEA – gene set enrichment analysis; TCGA – the Cancer Genome Atlas; BC – breast cancer.
Figure 8.
A–F shows GSEA of KIF15 in TCGA patients. (A–D) GSEA results of c2 reference gene sets for high KIF15 expression groups. (E–F) GSEA results of c5 reference gene sets for high KIF15 expression groups. G–L shows GSEA of KIF20A in TCGA patients. (G–J) GSEA results of c2 reference gene sets for high KIF20A expression groups. (K–L) GSEA results of c5 reference gene sets for high KIF20A expression groups. KIF – kinesin; GSEA – gene set enrichment analysis; TCGA – the Cancer Genome Atlas; BC – breast cancer.
Figure 9.
GSEA results of KIF23 in TCGA BC patients. (A–D) GSEA results of c2 reference gene sets for high KIF23 expression groups. (E–F) GSEA results of c5 reference gene sets for high KIF23 expression groups. KIF – kinesin; GSEA – gene set enrichment analysis; TCGA – the Cancer Genome Atlas; BC – breast cancer.
Nomogram analysis
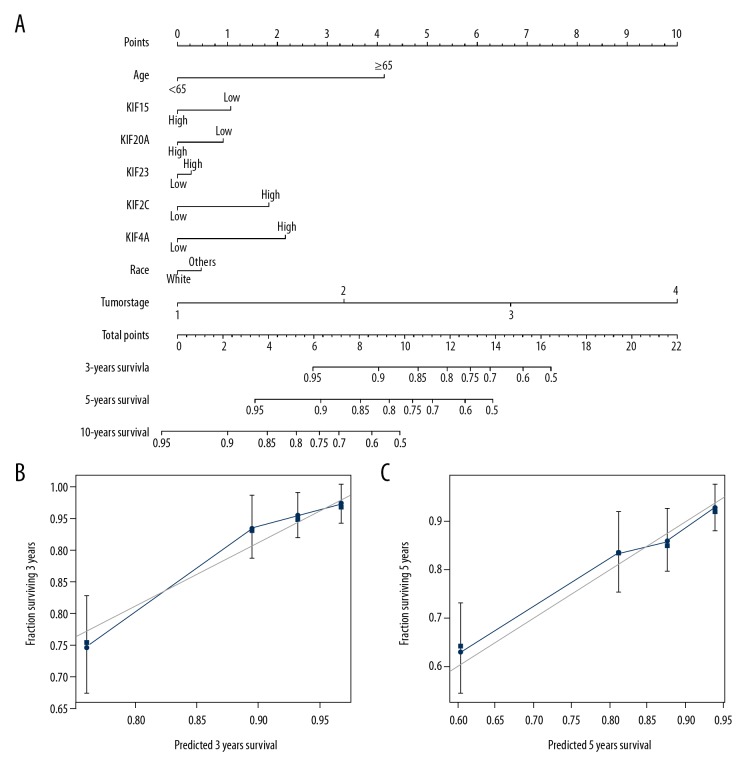
The nomogram was driven from rms as well as its supplementary packages on the base of information of patients having BC with comprehensive clinical evidence within TCGA. It showed that, among the 5 KIF genes, KIF4A had the greatest sum of risk points (ranging between 0 and 100), while the other genes made a considerably lower contribution (Figure 10A). By examining the conformity and discrimination using the nomogram model, bootstrap analysis on the bases of 1000 resampling tests had a C-index of 0.76 and 95% CI of 0.70–0.82. The discrimination is suitable. The calibration curve showed that the general point was close to the ideal curve of 45 degrees, indicating good compliance (Figure 10B, 10C).
Figure 10.
(A–C) Relationship between risk score and clinical information. Nomogram for predicting the 1-, 3-, and 5-year event (death) with risk score and clinical information. OS – overall survival.
Discussion
Kinesin motor activity is spatially as well as temporally controlled within mitosis to ensure that it occurs precisely with stable inward and outward forces. Nevertheless, over-expression of a few mitotic kinesins might produce further outward forces. This provokes a sequence of undesirable events, such as overshooting before anaphase, sister chromatid segregation before anaphase, increased spindle separation, and ultimately monopolar or bipolar spindle formation [28. Such things might cause imbalanced distribution of DNA, aneuploidy and a plethora of cancer phenotypes, together with metastatic and invasive behavior. Kinesin function might cause failed cytokinesis, imperfect spindle assembly and mitotic arrest, which stimulates apoptosis and killing of cancer cells [15]. Our evaluation of genetic function enrichment suggested that the KIF gene family is involved in biological processes of the cell cycle such as mitotic cytokinesis, mitotic spindle assembly, and positive regulation of cytokinesis. Our analysis established that KIF15, KIF20A, KIF23, KIF2C and KIF4A were co-expressed at the protein and gene levels.
We found 5 KIF genes of diagnostic and prognostic value. Extensive studies have reported these genes as potential diagnostic markers in multiple cancers. Among the 5 genes, KIF15 is likewise overexpressed in lung adenocarcinoma and might play a significant role in modifying the cell cycle [29]. Likewise, KIF15 promotes proliferation of pancreatic cancer cells via the MEK/ERK pathway [30]. KIF15 is overexpressed in BC cells and might have potential as a novel therapeutic target and a prognostic factor in endocrine-therapy-resistant BC [31]. We found that expression of KIF15 mRNA was significantly higher in BC than in adjacent tissues, and elevated expression of KIF15 in patients with BC was associated with poor OS. Our results agreed with previous studies that designated the KIF15 as an oncogene in BC.
In 2005, a study by Keisuke et al. [32] found that KIF20A was overexpressed in pancreatic cancer according to cDNA microarray analysis, and down-regulation of KIF20A significantly decreased tumor cell proliferation, confirming that KIF20A is carcinogenic in pancreatic cancer. Numerous studies have shown that KIF20A also has carcinogenic traits in various other cancers, such as nasopharyngeal carcinoma, liver cancer, melanoma, lung adenocarcinoma and glioma [33]. It has been suggested that the KIF20A gene is a potential diagnostic biomarker. We found that KIF20A has differential expression in BC and adjacent tissues, and high expression of KIF20A is related to poor OS in patients with BC, so it might also be a prognostic biomarker.
It has been found that KIF23 is up-regulated in patients with hepatocellular carcinoma, and it may be a marker for OS [34]. Zou et al. [31] showed that KIF4A, KIF15, KIF20A and KIF23 expression was significant in proliferating BC cells. They also showed that, among patients treated with tamoxifen, high expression of these 4 genes was highly correlated with poor recurrence-free survival. It has been suggested that over-expression of KIF23 is a valuable independent prognostic factor in lung tumors, particularly lung adenocarcinoma, and patients with p-stage I tumor stage and high expression of KIF23 have poorer survival than those with low expression [35]. In addition, the multivariate Cox proportional hazards model in our study, which was based on expression of KIF23, likewise divided patients in low- and high-expression groups, and patients with high expression had poor OS.
Nowadays, it is certain that KIF4A performs a significant role in cancer development and progression. Numerous studies have shown that KIF4A is a potential contributor to several malignant tumors, such as lung cancer [36], breast cancer [37], cervical cancer [38], hepatocellular carcinoma [39], and oral cancer [40]. Our results were consistent with previous studies. We also found that BC patients with high expression of KIF4A had poor OS compared with patients with low expression.
Shimo et al. have demonstrated that KIF2C is overexpressed in BC cells, and plays a major part in cytokinesis within these cells [41]. Additionally, they have discovered that down-regulation of KIF2C through treatment with siRNA suppresses development of BC cells [41]. We investigated the high expression of KIF4A, KIF15, KIF20A, KIF23 and KIF2C in patients in the TCGA database, which has been linked to poor OS. Our joint genetic analysis suggests that BC patients with high expression of 2–5 of these genes have poorer OS compared with patients with low gene expression. These findings suggest KIF4A, KIF15, KIF20A, KIF23 and KIF2C as potential prognostic biomarkers and therapeutic targets in BC.
GSEA showed that KIF4A, KIF15, KIF20A, KIF23 and KIF2C were significantly associated with the cell cycle, p53 and mismatch repair, that were associated with their biological functioning. It is well known that KIF genes play critical roles in DNA replication and cell cycle progression [15]. The results require additional experimental validation.
The present study had a few limitations. First, all the information was obtained from open databases, and the medical parameters were not complete. Therefore, we were not able to perform a far-reaching survival analysis of KIF genes, considering each latent prognostic variable of BC in the multivariate Cox proportional hazards regression model. Second, because of the varied origin of BC patients, together with the number of elements affecting BC prognosis, we were not able to construct a comprehensive hazard score model, which depended on the KIF genes articulation level for visualization forecast. Third, with the help of the correlation with the past research work, the constraint of our present investigation suggested that it just researched the relationship existing between the mRNA expression of the KIF genes and BC prognosis. Nonetheless, the connection between KIF protein level and BC requires additional investigation.
In spite of the above limitations, we established and validated the prognostic and diagnostic values of expression of KIF genes in BC patients, and similarly examined the potential mechanism linked with KIF4A, KIF15, KIF20A, KIF23 and KIF2C within BC prognosis by GSEA. When these outcomes are confirmed, the prognostic and diagnostic standards of KIF genetics on the extent of protein, such genes might hold a substantial clinical implication value in diagnosis of BC, as well as targeted therapy. Nevertheless, future verification with a larger study population is required to confirm that the KIF genes could be involved in diagnosis and prognostic monitoring of BC.
Conclusions
We revealed that 13 KIF genes were differentially expressed in BC tumor tissues, and may serve as latent diagnostic biomarkers in patients with BC. KIF15, KIF20A, KIF23, KIF2C and KIF4A have the potential to serve as prognostic biomarkers in patients with BC. Multivariate survival analysis, nomograms, and joint survival analysis showed high expression of these genes correlated with poor prognosis of BC. GO, KEGG and GSEA suggested that these genes affect the prognosis of BC by influencing the cell cycle. Our results need to be confirmed in further research.
Acknowledgements
We acknowledge the support of the National Key Clinical Specialty Programs (General Surgery & Oncology), in addition to the Key Laboratory of Early Prevention & Treatment for Regional High-Incidence Tumor (Guangxi Medical University), Ministry of Education, China. We also thank the contributors of the TCGA database (https://cancergenome.nih.gov/) for sharing the BC data in open access.
Footnotes
Source of support: The current research was supported by the Guangxi Zhuang Autonomous Region Higher Education Research Project (No.: KY2015ZD031)
Conflicts of interest
None.
References
- 1.Zhong ZB, Shan M, Wang J, et al. Decreased Wnt5a expression is a poor prognostic factor in triple-negative breast cancer. Med Sci Monit. 2016;22:1–7. doi: 10.12659/MSM.894821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–86. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 3.Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 4.Guo XH, Li JT, Zhang HW, et al. Relationship between ADAMTS8, ADAMTS18, and ADAMTS20 (a disintegrin and metalloproteinase with thrombospondin motifs) expressions and tumor molecular classification, clinical pathological parameters, and prognosis in breast invasive ductal carcinoma. Med Sci Monit. 2018;24:3726–35. doi: 10.12659/MSM.907310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harbeck N, Gnant M. Breast cancer. Lancet. 2017;389(10074):1134–50. doi: 10.1016/S0140-6736(16)31891-8. [DOI] [PubMed] [Google Scholar]
- 6.Ginsburg O, Bray F, Coleman MP, et al. The global burden of women’s cancers: A grand challenge in global health. Lancet. 2017;389(10071):847–60. doi: 10.1016/S0140-6736(16)31392-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu M, Zhang X, Li H, et al. MicroRNA-588 is downregulated and may have prognostic and functional roles in human breast cancer. Med Sci Monit. 2017;23:5690–96. doi: 10.12659/MSM.905126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu L, Shi W, Long J, et al. A transcriptome-wide association study of 229,000 women identifies new candidate susceptibility genes for breast cancer. Nat Genet. 2018;50(7):968–78. doi: 10.1038/s41588-018-0132-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nathanson KN, Wooster R, Weber BL. Breast cancer genetics: What we know and what we need. Nat Med. 2001;7(5):552–56. doi: 10.1038/87876. [DOI] [PubMed] [Google Scholar]
- 10.Yi H, Wang K, Jin H, et al. Overexpression of Rho-associated coiled-coil containing protein kinase 2 is correlated with clinical progression and poor prognosis in breast cancer. Med Sci Monit. 2018;24:4776–81. doi: 10.12659/MSM.908507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang N, Zuo L, Zheng H, et al. Increased expression of CD81 in breast cancer tissue is associated with reduced patient prognosis and increased cell migration and proliferation in MDA-MB-231 and MDA-MB-435S human breast cancer cell lines in vitro. Med Sci Monit. 2018;24:5739–47. doi: 10.12659/MSM.911612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kwan TT, Bardia A, Spring LM, et al. A digital RNA signature of circulating tumor cells predicting early therapeutic response in localized and metastatic breast cancer. Cancer Discov. 2018;8L:1286–99. doi: 10.1158/2159-8290.CD-18-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodriguez Bautista R, Ortega Gomez A, Hidalgo Miranda A, et al. Long non-coding RNAs: implications in targeted diagnoses, prognosis, and improved therapeutic strategies in human non- and triple-negative breast cancer. Clin Epigenetics. 2018;10:88. doi: 10.1186/s13148-018-0514-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liao XW, Zhu GZ, Huang R, et al. Identification of potential prognostic microRNA biomarkers for predicting survival in patients with hepatocellular carcinoma. Cancer Manag Res. 2018;10:787–803. doi: 10.2147/CMAR.S161334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lucanus AJ, Yip GW. Kinesin superfamily: Roles in breast cancer, patient prognosis and therapeutics. Oncogene. 2018;37(7):833–38. doi: 10.1038/onc.2017.406. [DOI] [PubMed] [Google Scholar]
- 16.Goldstein LS, Philp AV. The road less traveled: Emerging principles of kinesin motor utilization. Ann Rev Cell Dev Biol. 1999;15:141–83. doi: 10.1146/annurev.cellbio.15.1.141. [DOI] [PubMed] [Google Scholar]
- 17.Miki H, Setou M, Hirokawa N. All kinesin superfamily protein, KIF, genes in the mouse and human genome and transcripts. Mol Biol Cell. 2002;13:184a. doi: 10.1073/pnas.111145398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu XR, Gong H, Huang K. Oncogenic role of kinesin proteins and targeting kinesin therapy. Cancer Sci. 2013;104(6):651–56. doi: 10.1111/cas.12138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao HW, Chen XR, Cai QL, et al. Increased KIF4A expression is a potential prognostic factor in prostate cancer. Oncol Lett. 2018;15(5):7941–47. doi: 10.3892/ol.2018.8322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eskova A, Knapp B, Matelska D, et al. An RNAi screen identifies KIF15 as a novel regulator of the endocytic trafficking of integrin. J Cell Sci. 2014;127(11):2433–47. doi: 10.1242/jcs.137281. [DOI] [PubMed] [Google Scholar]
- 21.Groth-Pedersen L, Aits S, Corcelle-Termeau E, et al. Identification of cytoskeleton-associated proteins essential for lysosomal stability and survival of human cancer cells. PLoS One. 2012;7(10):e45381. doi: 10.1371/journal.pone.0045381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Warde-Farley D, Donaldson SL, Comes O, et al. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38:W214–20. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.von Mering C, Huynen M, Jaeggi D, et al. STRING: A database of predicted functional associations between proteins. Nucleic Acids Res. 2003;31(1):258–61. doi: 10.1093/nar/gkg034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maere S, Heymans K, Kuiper M. BiNGO: A Cytoscape plugin to assess overrepresentation of Gene Ontology categories in Biological Networks. Bioinformatics. 2005;21(16):3448–49. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 25.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mootha VK, Lindgren CM, Eriksson KF, et al. PGC-1 alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34(3):267–73. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 27.Liberzon A, Birger C, Thorvaldsdottir H, et al. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1(6):417–25. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wordeman L. How kinesin motor proteins drive mitotic spindle function: Lessons from molecular assays. Semin Cell Dev Biol. 2010;21(3):260–68. doi: 10.1016/j.semcdb.2010.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bidkhori G, Narimani Z, Hosseini Ashtiani S, et al. Reconstruction of an integrated genome-scale co-expression network reveals key modules involved in lung adenocarcinoma. PLoS One. 2013;8(7):e67552. doi: 10.1371/journal.pone.0067552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang J, Guo XJ, Xie CC, Jiang JX. KIF15 promotes pancreatic cancer proliferation via the MEK-ERK signalling pathway. Br J Cancer. 2017;117(2):245–55. doi: 10.1038/bjc.2017.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zou JX, Duan ZJ, Wang JJ, et al. Kinesin family deregulation coordinated by bromodomain protein ANCCA and histone methyltransferase MLL for breast cancer cell growth, survival, and tamoxifen resistance. Mol Cancer Res. 2014;12(4):539–49. doi: 10.1158/1541-7786.MCR-13-0459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taniuchi K, Nakagawa H, Nakamura T, et al. Down-regulation of RAB6KIFL/KIF20A, a kinesin involved with membrane trafficking of discs large homologue 5, can attenuate growth of pancreatic cancer cell. Cancer Res. 2005;65(1):105–12. [PubMed] [Google Scholar]
- 33.Zhao X, Zhou LL, Li X, et al. Overexpression of KIF20A confers malignant phenotype of lung adenocarcinoma by promoting cell proliferation and inhibiting apoptosis. Cancer Med. 2018;7(9):4678–89. doi: 10.1002/cam4.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun XT, Jin ZT, Song X, et al. Evaluation of KIF23 variant 1 expression and relevance as a novel prognostic factor in patients with hepatocellular carcinoma. BMC Cancer. 2015;15:961. doi: 10.1186/s12885-015-1987-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kato T, Wada H, Patel P, et al. Overexpression of KIF23 predicts clinical outcome in primary lung cancer patients. Lung Cancer. 2016;92:53–61. doi: 10.1016/j.lungcan.2015.11.018. [DOI] [PubMed] [Google Scholar]
- 36.Taniwaki M, Takano A, Ishikawa N, et al. Activation of KIF4A as a prognostic biomarker and therapeutic target for lung cancer. Clin Cancer Res. 2007;13(22):6624–31. doi: 10.1158/1078-0432.CCR-07-1328. [DOI] [PubMed] [Google Scholar]
- 37.Wang H, Lu CQ, Li Q, et al. The role of Kif4A in doxorubicin-induced apoptosis in breast cancer cells. Mol Cells. 2014;37(11):812–18. doi: 10.14348/molcells.2014.0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Narayan G, Bourdon V, Chaganti S, et al. Gene dosage alterations revealed by cDNA microarray analysis in cervical cancer: Identification of candidate amplified and overexpressed genes. Gene Chromosome Canc. 2007;46(4):373–84. doi: 10.1002/gcc.20418. [DOI] [PubMed] [Google Scholar]
- 39.Hou GJ, Dong CP, Dong ZH, et al. Upregulate KIF4A enhances proliferation, invasion of hepatocellular carcinoma and indicates poor prognosis across human cancer types. Sci Rep. 2017;7(1):4148. doi: 10.1038/s41598-017-04176-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Minakawa Y, Kasamatsu A, Koike H, et al. Kinesin family member 4A: A potential predictor for progression of human oral cancer. PLoS One. 2013;8(12):e85951. doi: 10.1371/journal.pone.0085951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shimo A, Tanikawa C, Nishidate T, et al. Involvement of kinesin family member 2C/mitotic centromere-associated kinesin overexpression in mammary carcinogenesis. Cancer Sci. 2008;99(1):62–70. doi: 10.1111/j.1349-7006.2007.00635.x. [DOI] [PMC free article] [PubMed] [Google Scholar]