Abstract
Peroxisomes are conserved organelles of eukaryotic cells with important roles in cellular metabolism, human health, redox homeostasis, as well as intracellular metabolite transfer and signaling. We review here the current status of the different co‐existing modes of biogenesis of peroxisomal membrane proteins demonstrating the fascinating adaptability in their targeting and sorting pathways. While earlier studies focused on peroxisomes as autonomous organelles, the necessity of the ER and potentially even mitochondria as sources of peroxisomal membrane proteins and lipids has come to light in recent years. Additionally, the intimate physical juxtaposition of peroxisomes with other organelles has transitioned from being viewed as random encounters to a growing appreciation of the expanding roles of such inter‐organellar membrane contact sites in metabolic and regulatory functions. Peroxisomal quality control mechanisms have also come of age with a variety of mechanisms operating both during biogenesis and in the cellular response to environmental cues.
Keywords: de novo peroxisome biogenesis, peroxisomal membrane contact sites, peroxisome growth and division, peroxisome quality control, peroxisomal membrane protein biogenesis
Subject Categories: Membrane & Intracellular Transport, Metabolism, Protein Biosynthesis & Quality Control
Glossary
- aa
amino acid
- ACBD
acyl‐CoA binding domain
- ADP
adenosine diphosphate
- APX
ascorbate peroxidase
- Arf
ADP‐ribosylation factors
- ATPase
adenosine triphosphatase
- BAK
BCL2 antagonist/killer
- BiFC
bimolecular fluorescence complementation
- CAML
calcium‐modulating cyclophilin ligand
- cAMP
cyclic adenosine monophosphate
- Cat
carnitine transferase
- CAT
catalase
- CERT
ceramide transfer protein
- CHO
Chinese hamster ovary
- Cit
citrate synthase
- CTD
C‐terminal domain
- Cys
cysteine
- DAG
diacylglycerol
- DHA
docosahexaenoic acid
- Dnm
dynamin
- DRP
dynamin‐related protein
- ERAD
endoplasmic reticulum‐associated degradation
- ER
endoplasmic reticulum
- ERMES
endoplasmic reticulum‐mitochondrial encounter structures
- ERppVs
endoplasmic reticulum‐derived pre‐peroxisomal vesicles
- ESCRT
endosomal sorting complexes required for transport
- Fis
fission
- GET
guided entry of tail‐anchor
- GFP
green fluorescent protein
- GTPase
guanosine triphosphatase
- GTP
guanosine triphosphate
- HEK
human embryonic kidney cells
- HSP
high‐speed pelletable
- ICL
isocitrate lyase
- Inp
inheritance of peroxisomes
- LD
lipid droplet
- LPMC
lysosome–peroxisome membrane contacts
- LSP
low‐speed pelletable
- MCS
membrane contact sites
- MCTP2
multiple C2 domain containing transmembrane protein
- MDppVs
mitochondrially derived pre‐peroxisomal vesicles
- MFF
mitochondrial fission factor
- mPTS
membrane peroxisomal targeting signals
- MTS
mitochondrial targeting signal
- Myo
myosin
- NTD
N‐terminal domain
- OSBP
oxysterol binding protein
- PBDs
peroxisome biogenesis disorders
- PE
phosphatidylethanolamine
- pER
pre‐peroxisomal endoplasmic reticulum
- Pex/PEX
peroxins from yeast/mammals
- Phe
phenylalanine
- PMPs
peroxisomal membrane proteins
- Pp
Pichia pastoris
- ppVs
pre‐peroxisomal vesicles
- Psd
phosphatidylserine decarboxylases
- PS
phosphatidylserine
- PTS
peroxisomal targeting signals
- QC
quality control
- RADAR
receptor accumulation and degradation in the absence of recycling
- RHD
reticulon homology domain
- RING
really interesting gene
- ROS
reactive oxygen species
- Sc
Saccharomyces cerevisiae
- SRP
signal recognition particle
- TA
tail‐anchored
- TMD
transmembrane domain
- TOMM20
translocator of outer mitochondrial membrane 20
- TRC
transmembrane recognition complex
- Ub
ubiquitin
- UPS
ubiquitin–proteasome system
- VAMP
vesicle‐associated membrane protein
- VAP
VAMP‐associated protein
- VDAC
voltage‐dependent anion channel
- WRB
tryptophan‐rich basic protein
- WT
wild type
- YFP
yellow fluorescent protein
Introduction
Peroxisomes are a conserved, intracellular organelle of eukaryotic cells and are involved in a range of metabolic functions that vary based on the organism in which they occur. General functions of metabolic pathways housed in peroxisomes include the β‐oxidation of fatty acids and the detoxification, by catalase, of hydrogen peroxide that is produced during fatty acid oxidation 1. Other metabolic functions and the role of peroxisomes in human disease are reviewed elsewhere 2, 3.
A characteristic feature of peroxisomes is that they proliferate or dissipate in response to external cues 4. In yeasts, peroxisome numbers, sizes, and enzyme repertoires can rapidly change by manipulating the carbon source in their growth medium. For example, Saccharomyces cerevisiae and Pichia pastoris will proliferate peroxisomes when grown in fatty acids, such as oleate, because the β‐oxidation of fatty acids occurs in peroxisomes. P. pastoris and Hansenula polymorpha also proliferate peroxisomes when grown in methanol, which is metabolized using peroxisomal enzymes. Conversely, when organisms are switched from peroxisome proliferation conditions to media that do not require peroxisomal metabolism, then the excess peroxisomes are degraded, typically by a selective form of autophagy called pexophagy 5. Similarly, excessive reactive oxygen species (ROS), hypoxia, or the depletion of iron can trigger pexophagy in different model organisms 6, 7, 8, 9.
The proteins implicated in peroxisome biogenesis are known as peroxins and the genes encoding them are dubbed PEX genes. More than half of these peroxins, referred to as Pex or PEX proteins in yeast and mammals, respectively, are required for the import of peroxisomal matrix proteins, and the rest are implicated in the targeting of the peroxisomal membrane proteins (PMPs) to the peroxisome membrane and in peroxisome proliferation. This review will mostly focus on exciting, new advances regarding peroxisome biogenesis, membrane contact sites (MCS) between peroxisomes and other organelles, and quality control (QC), while only a brief description of peroxisomal matrix import is provided for continuity. These topics will highlight the flexibility exploited by different model organisms in the relative use of redundant pathways for PMP and peroxisome biogenesis, the interconnectivity and communication between peroxisomes and other subcellular compartments, and the complex QC mechanisms associated with peroxisomes.
Brief overview of peroxisomal matrix protein import
Proteins destined for import into the peroxisome matrix or membrane possess peroxisomal targeting signals (PTSs) or membrane PTSs (mPTSs), respectively (Fig 1). The peroxisomal matrix proteins are synthesized in the cytosol and transported into the peroxisome matrix across translocons located in the peroxisome membrane (Fig 1A). Most peroxisomal matrix proteins have either a C‐terminal PTS1 or an N‐terminal PTS2. In yeast and mammals, these sequences are recognized by specific receptors, Pex5, for PTS1 and Pex7 for PTS2. The protein Pex9 is a Pex5‐related protein found in S. cerevisiae that acts on limited PTS1 cargos, such as malate synthase 1 and 2, as well as the glutathione transferase, making it a condition‐specific PTS receptor 10, 11. These receptors can either act alone (e.g., Pex5), or with co‐receptors (Pex7‐Pex18 or Pex7‐Pex20 in S. cerevisiae and P. pastoris, respectively, or PEX7‐PEX5L in mammals) to form receptor/cargo complexes, which dock at the peroxisome membrane with a docking complex (typically comprised in yeasts of Pex13, Pex14, and Pex17, but mammals lack Pex17). The minimal translocon in yeast involves Pex14 and Pex5 for PTS1 import 12, and likely Pex14/Pex17 and Pex18 for PTS2 import 13. Associated with the docking complex is another subcomplex comprised of three conserved RING (really interesting gene) domain proteins, Pex2, Pex10, and Pex12, that have E3 ligase activities. Together, the docking and RING subcomplexes form the importomer complex 14, 15.
Figure 1. Peroxisomal matrix and membrane protein import in yeast (an overview).
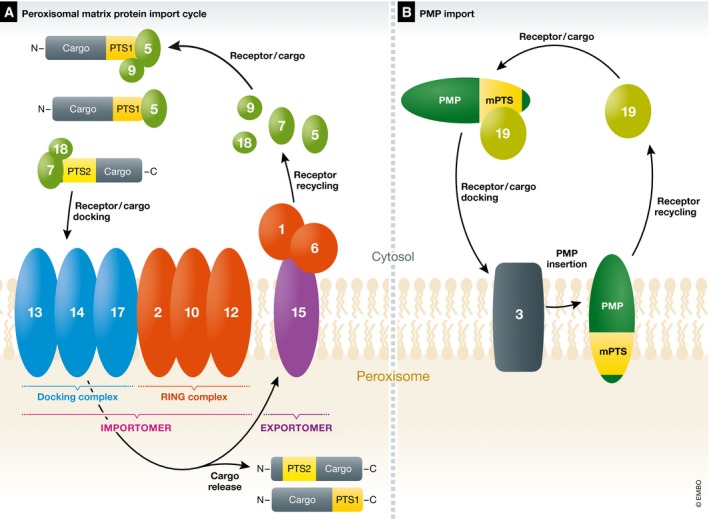
Most proteins destined for import into the peroxisome matrix possess either a C‐terminal PTS1 or an N‐terminal PTS2. (A) The peroxisomal matrix protein import cycle. These cargos synthesized in the cytosol are recognized by PTS receptors, Pex5 for PTS1 and Pex7 for PTS2, respectively. Pex7 generally works with a co‐receptor (Pex18/21 in S. cerevisiae or Pex20 in P. pastoris only Pex18 is shown). Pex9 is a Pex5‐related protein found in S. cerevisiae that acts on limited PTS1 cargos as described in the text 10, 11. The PTS receptor/cargo complex, along with the co‐receptor, where applicable, docks at the peroxisome membrane with the docking complex, comprised of Pex13, Pex14, and Pex17. PTS cargos are translocated into the peroxisome matrix across translocons in the peroxisome membrane. The minimal translocon in yeast involves Pex14 and Pex5 for PTS1 import 12, and likely Pex14/Pex17 and Pex18 for PTS2 import 13. Associated with the docking complex is the RING subcomplex comprised of Pex2, Pex10, and Pex12 that have E3 ligase activities involved in ubiquitin‐dependent, PTS‐receptor recycling and QC steps (sections Brief overview of peroxisomal matrix protein import and QC during peroxisomal matrix protein import). Together, the docking and RING subcomplexes form the importomer complex 14, 15. Following PTS cargo release in the peroxisome lumen, the PTS receptors, and co‐receptors where applicable, recycle from the peroxisomes back to the cytosol for another round of import, using components collectively called the exportomer, whose components are described in the text 16. (B) The PMP import cycle for the direct import of proteins into the peroxisome membrane (section The direct import of PMPs to peroxisomes). Each PMP has at least one mPTS that is bound to, and the PMP is chaperoned by, Pex19, which docks at the peroxisomes via interactions with Pex3. The PMP is inserted into the membrane and Pex19 recycles back to the cytosol for another round of PMP import.
The receptor/cargo complexes from the cytosol interact with the docking subcomplex, translocate into the peroxisome matrix or membrane and release their respective cargos in the peroxisome lumen. Then, the receptors, and co‐receptors where applicable, recycle from the peroxisomes back to the cytosol for another round of import, using components collectively called the exportomer 16, 17.
This export and recycling of the receptor and co‐receptor requires mono‐ubiquitination of a cysteine near the N‐terminus of Pex5 (in yeast and mammalian systems) 18, 19 and Pex20 (in P. pastoris) 20. Pex5 and Pex20 mono‐ubiquitination requires the typical ubiquitination enzymes—an E1 protein, an E2 in the form of Pex4 associated in yeast with the peroxisome membrane via the PMP, Pex22, and E3 ligase activity provided by one or more components of the peroxisomal RING subcomplex 21. The mono‐ubiquitinated PTS receptors or co‐receptors are recognized by peroxisome membrane‐associated AAA‐ATPases, Pex1 and Pex6 22, 23, which are associated with peroxisomes in an ATP‐dependent manner via interaction with specific PMPs (Pex15 in yeast or PEX26 in mammals). These ATPases are required to export and recycle mono‐ubiquitinated PTS receptors/co‐receptors 17, following which the PTS receptors/co‐receptors are deubiquitinated (by Ubp15 for the mono‐ubiquitinated Pex5 in yeast or by USP9X in mammals) and reused for subsequent rounds of import 24, 25.
When this mono‐ubiquitination is blocked, either by mutation of the ubiquitination site in the exported receptor or co‐receptor or by mutations in the receptor recycling machinery that recognizes this mono‐ubiquitin and exports the proteins to the cytosol, then an alternative pathway called receptor accumulation and degradation in the absence of recycling (RADAR) takes over 20. This is described later under quality control pathways.
Peroxisomal membrane proteins
Because many pex mutants (with the exception of pex3, pex16, and pex19) are defective only in peroxisome matrix protein import and still possess peroxisome remnants or ghosts containing PMPs, the sorting of PMPs requires components distinct from those involved in peroxisomal matrix protein import. PMPs fulfill a variety of functions such as serving as components of the peroxisomal translocon or the exportomer, membrane transporters for metabolites and ions, quality control or organelle division machineries, redox proteins, signaling molecules, organelle membrane tethers, and so on. PMPs have one or more mPTSs 26 that are sorted to the peroxisome membrane in either a Pex19‐dependent or Pex19‐independent manner 27.
Based on whether or not Pex19 is required for their membrane insertion step, these PMPs are broadly classified into two classes:
Class I or direct pathway—involving Pex19‐dependent membrane insertion of PMPs (most PMPs) 28, 29, 30 (Fig 1B).
Class II or indirect pathway—this alternative was proposed initially to address the PEX19‐independent membrane insertion of PEX3 in mammalian cells 30, 31, which traffics to peroxisomes via the endoplasmic reticulum (ER) 32, 33. However, since these early studies, many PMPs have been shown to traffic to the peroxisomes via the ER, we therefore prefer to call this the indirect pathway (i.e., via the ER) of PMP trafficking to peroxisomes 34.
It should also be noted that the same PMP may traffic to peroxisomes directly or indirectly. Thus, mammalian PEX3 can also be imported directly to peroxisomes in a PEX16‐ and PEX19‐dependent manner 35. Perhaps many (or even all) PMPs have the flexibility to be targeted to peroxisomes directly, or indirectly via the ER 34, with the latter being the only mode possible when there are no pre‐existing peroxisomes.
A subclass of PMPs is the tail‐anchored (TA) proteins—integral membrane proteins with a short, C‐terminal sequence adjoining their transmembrane domain (TMD) 36, whose insertion into the membranes (peroxisomal or ER) may be Pex19‐dependent or Pex19‐independent, either directly into pre‐existing peroxisomes or indirectly via prior insertion into membranes of other subcellular compartments, from which peroxisomes are subsequently derived. These topics are addressed later (sections The direct import of tail‐anchored proteins to peroxisomes and Insertion of tail‐anchored PMPs into the ER membrane).
Peroxisome biogenesis—divergent models ranging from growth and division to de novo mechanisms
Two models have co‐existed for decades regarding the biogenesis of peroxisomes and are likely to operate within the same cells in response to specific environmental cues. The older of these is the growth and division model 37, in which peroxisomes, like chloroplasts and mitochondria, arise from pre‐existing peroxisomes that grow to a certain size after acquiring their PMPs and matrix proteins directly from the cytosol. Then, upon activation by poorly characterized mechanisms, peroxisomes divide by fission to form a daughter peroxisome that then goes through this cycle again. The second model invokes de novo peroxisome biogenesis in which some PMPs are first inserted into the membrane of the ER, sorted to a region of the ER called the pre‐peroxisomal ER (pER), from where distinct pre‐peroxisomal vesicles (ppVs) containing the PMPs bud 38. Moreover, a recent study in mammals suggests that some ppVs might also originate from the mitochondria 39. The ppVs containing different subsets of PMPs then fuse, either in a heterotypic fashion 40 or with pre‐existing peroxisomes 41 to create mature or larger peroxisomes, respectively.
Finally, a third model blends and accommodates features of the PMP traffic envisioned in the growth and division model, as well as via the ER in the de novo biogenesis model 42. This third model invokes two routes for PMP insertion into peroxisomes—one involving direct insertion of PMPs into membranes of pre‐existing peroxisomes and the other invoking indirect traffic of PMPs to peroxisomes via the ER/mitochondria, followed by their subsequent sorting to the peroxisomes 37, 38, 39. In this review, we will mostly focus on the first two models, although the indirect PMP traffic via the ER invoked in this third model will be described in some detail in the de novo peroxisome biogenesis model (section The de novo peroxisome biogenesis model).
The growth and division model
The direct import of PMPs to peroxisomes
In the growth and division model, PMPs are inserted directly into the peroxisome membrane from the cytosol and the ER provides the lipids for membrane growth, most likely through organelle contact sites described later (section Peroxisome‐ER MCS) 41. PMPs are synthesized on free polyribosomes and post‐translationally imported into peroxisomes. Their hydrophobic TMDs have to be protected by chaperones soon after synthesis. Their mPTSs consist of a cluster of basic residues in a predicted α‐helical conformation with a minimal length of 11 amino acids and are generally flanked by one or two TMDs 43.
Pex19 is an acidic peroxin that associates with membranes through its C‐terminal farnesyl tail, and serves as a receptor and chaperone for Class I PMPs, recognizing and binding the mPTSs within these PMPs 28, 29. The binding of Pex19 near the TMDs of such PMPs facilitates the role of Pex19 as a chaperone 30. This role of Pex19 in stabilizing and chaperoning hydrophobic PMPs is underscored by the fact that several PMPs are unstable and degraded in cells lacking Pex19 44. Furthermore, the solubility of in vitro synthesized PMPs, such as PMP22, increases in the presence of Pex19 45.
Pex19 is a predominantly cytosolic protein that exhibits a characteristic domain organization 27, 46. A small but significant amount of the Pex19 population is also associated with the peroxisome membrane through the farnesylation of its C‐terminal end 47. The C‐terminal domain (CTD) of Pex19 participates in the recognition and binding of mPTS motifs in PMPs 48, 49, 50.
A role for the farnesylation of Pex19 is still unclear. Pex19 does not seem to require farnesylation to associate with membranes, and there are reports that it functions to allosterically modulate Pex19 function 51. Nuclear magnetic resonance data suggest that the C‐terminal residues of the CTD become rigid upon farnesylation, which in turn, might enhance the interactions of mammalian PEX19 with PMPs 51. In rats and mice, a splice variant of PEX19, called PEX19i, has been identified, which encodes a PEX19‐like protein with its C‐terminal farnesyl tail replaced by a hydrophobic region 52. The transcription of PEX19i was highly induced by the peroxisome proliferator, clofibrate, and this protein was functional in that it restored peroxisomes by complementation of PEX19‐deficient (ZP119) Chinese hamster ovary (CHO) cells and also bound several PMPs known to interact with PEX19. The ability of this protein to support peroxisome biogenesis also suggests that the farnesylation of PEX19 is not critical for its function.
Both Pex3 and Pex19 are involved in membrane insertion of the PMPs 53, 54. Pex19 directs the PMP to the peroxisomal membrane, where it docks with the transmembrane protein, Pex3, and thereby acts as a shuttling receptor (Fig 1B) 55. Surprisingly, only these two factors, Pex3 and Pex19, seem to be essential for the Class I pathway, independent of the topological complexity of the PMPs. It has been shown in mammals and Neurospora crassa that PMPs harboring one to six TMDs can be inserted into peroxisome membranes through this route 53, 56.
The N‐terminal region of Pex19 contains a high‐affinity Pex3‐binding site 48, 50, 55, 57. Pex3 possesses one TMD near its N‐terminus and exposes most of its polypeptide chain into the cytosol 58, 59, 60. The cytosolic domain of Pex3 serves as a docking factor for Pex19‐PMP complexes 55. Because lipid molecules can bind to Pex3 in competition with Pex19, such lipid binding may perturb the peroxisomal lipid bilayer to allow PMP insertion into the peroxisome membrane 61.
The Pex3 mPTS does not bind Pex19 directly, and therefore, its membrane insertion follows the indirect pathway 30, trafficking through the ER and possibly mitochondria (section ppVs derived from mitochondria (mammals)), rather than by direct import into peroxisome membranes. Nevertheless, mammalian PEX3 traffic to the peroxisome membrane depends on PEX16, which is a Class I PMP itself, and might also serve as a docking factor for PEX3‐PEX19 complexes at the peroxisome surface under conditions when PEX3 is forced to traffic to peroxisomes using the Class I pathway 35.
The direct import of tail‐anchored proteins to peroxisomes
At least two proteins implicated in peroxisome biogenesis in mammals are the TA PMPs (FIS1 and PEX26), which can be inserted directly by the Class I pathway 53. Evidence of the direct targeting of PEX26 in mammalian cells comes from a cell‐free reaction in which a complex containing PEX19 and PEX26 accumulates in a pex3 mutant cell line, and PEX26 from this complex can be targeted to peroxisomes in semi‐permeabilized cells in a PEX3‐dependent, but ASNA1/TRC40‐independent, manner 62. Either removal of the mPTS in PEX26 or the absence of PEX19 in mammalian cells impairs PEX26 targeting to peroxisomes. A ternary complex between PEX19, PEX26, and PEX3 has been detected 54. The yeast orthologue of PEX26 is Pex15 and it too is targeted to peroxisomes in a similar manner 63. Interestingly, a new function has been uncovered for Pex19 in S. cerevisiae, which is also apparently involved in the insertion of the TA proteins, Fis1 and Gem1, into mitochondria 64.
How is TMD binding and release mediated during direct insertion of PMPs into the peroxisome membrane? This role of Pex19 was addressed using Neurospora proteins 53. Pex19 was reported to bind Pex26, preventing it from aggregation followed by its insertion into peroxisome membranes in a Pex3‐dependent manner, mimicking the mammalian system. This chaperone‐like activity of Pex19 depends on hydrophobic contacts via an amphipathic helix in the CTD of Pex19 and the TA PMP. This study also identified an additional amphipathic helix in Pex19, lying between the N‐terminal, Pex3 binding region in Pex19, and its CTD. Hydrophobicity in this region of Pex19 is obligatory for the insertion of the TMD of the TA PMP, but not for chaperone activity or Pex3 binding. Another hydrophobic surface at the base of Pex3, adjacent to where it is anchored in the membrane, promotes an unconventional form of membrane association of the TA PMP and is also required for the membrane insertion of its TMD. Together, these data support a model in which hydrophobic moieties in Pex19 and Pex3 act in distinct capacities to promote TMD binding, release, and insertion.
However, PEX26 and its yeast orthologue, Pex15, are capable of also targeting to peroxisomes via the ER (probably a minor pathway) in mammalian cells, in what is reminiscent of the indirect pathway 65. This pathway is described later (section Insertion of tail‐anchored PMPs into the ER membrane).
Peroxisome fission in the growth and division model
According to the current model, during peroxisome growth and division, peroxisome fission happens in a 3‐step process involving peroxisome elongation, constriction, and scission (Fig 2, top panel) 41. Pex11 is essential for the first step (Fig 2, panel 1). Its overexpression causes peroxisome proliferation, and its deletion causes enlarged peroxisomes and a decrease in their number 66. Penicillium chrysogenum Pex11 was shown in vitro to bind, impart curvature, and tubulate liposome membranes, particularly those containing negatively charged phospholipids mimicking those in peroxisome membranes 67. This feature is conserved from yeast to human PEX11 isoforms.
Figure 2. Peroxisome fission in the growth and division model.
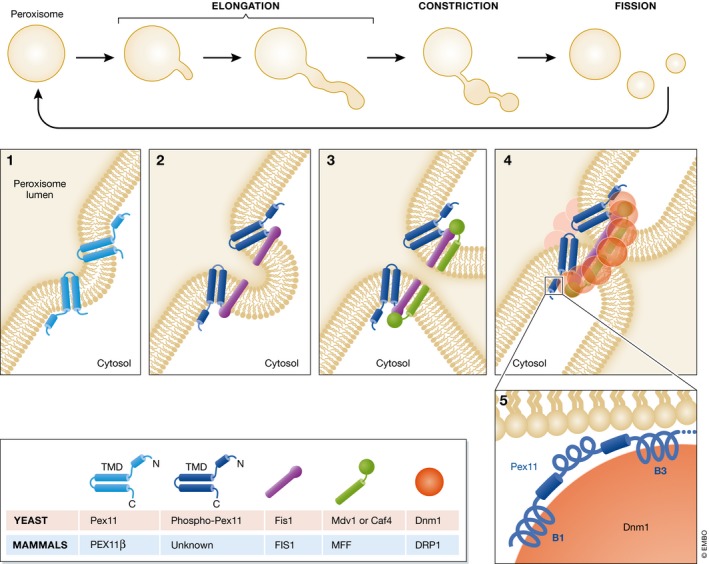
According to the growth and division model, peroxisome fission happens in a 3‐step process. During the first step of elongation, Pex11 (PEX11β in mammals), a transmembrane protein that imparts curvature to peroxisome membranes (panel 1), is essential for the elongation step. The topology shown here for Pex11 is based on studies in H. polymorpha 67. The second step, involving membrane constriction, is poorly understood and we do not know any proteins implicated in this step. The third step, peroxisome fission, starts in P. pastoris with the phosphorylation of Pex11(S173) that stimulates its interaction with the adaptor, Fis1 (panel 2) 78. Note that the topology of PpPex11 has not been documented, so it is unclear whether the phosphorylation is on the cytosolic or the peroxisome matrix side. Fis1 then recruits the peripheral receptors, Mdv1 and/or Caf4 (panel 3) 75. Mdv1 and/or Caf4 assemble a Dnm1 ring around the peroxisome constriction site (panel 4). Mammals do not have homologues for these proteins, and DRP1 is recruited to peroxisomes by MFF and FIS1 79. Yeast Dnm1 interacts with Fis1 and two Pex11 helices named B1 and B3 (panel 5). The hydrolysis of GTP by Dnm1, enhanced by the interaction with the B3 helix of Pex11, leads to a constriction that divides the peroxisome 82.
While Pex11 causes membrane tubulation in vitro, the cytoskeleton to which peroxisomes are attached in yeast and mammalian cells likely also plays a role in peroxisome tubulation and elongation, prior to division. Yeast peroxisomes are associated with an actin/myosin cytoskeleton, involving the Myo2 motor linked to peroxisomes via the proteins, Inp1 and Inp2 (inheritance of peroxisomes) 68, 69. In mammals, however, peroxisomes are associated with microtubules through the Ras GTPase, MIRO1, a potential adaptor linking mammalian peroxisomes to microtubules 70. MIRO1 localizes to both peroxisome and mitochondria. Distinct splice variants of MIRO1 are targeted specifically to peroxisomes and mitochondria in human embryonic kidney (HEK) cells, with the MIRO1‐variant 4 being more specific for peroxisomes in these cells. When MIRO1 is targeted exclusively to peroxisomes, it mediates pulling forces that contribute to peroxisome membrane elongation and proliferation in a cell type‐dependent manner 70. It should be noted, however, that in mammalian cells, peroxisomes can also elongate independently of microtubules, and peroxisome elongation is promoted by microtubule‐depolymerizing drugs 71, 72. This suggests that a PEX11 isoform, PEX11β, and motor forces such as those mediated by MIRO1 can independently promote peroxisome proliferation, but may cooperate under physiological conditions.
There is less information about peroxisome constriction, and actually, this step is poorly understood. However, given the fact that peroxisomes share their components of a common division machinery with mitochondria, some insights may be gleaned from the multiple constriction steps involved in mitochondrial division 73, 74.
In S. cerevisiae, the GTPase, Dnm1, accomplishes the final step of scission 75. In dnm1Δ cells, a single enlarged peroxisome protrudes from the mother cell into the bud, demonstrating that Dnm1 is required for the final step (scission), but not for the elongation step 76. Dnm1 forms a ring‐like structure around membranes, and the hydrolysis of GTP leads to a constriction that divides the organelle 77 (Fig 2, panel 4). Unlike canonical dynamins, yeast Dnm1 does not have pleckstrin‐homology domains for direct membrane binding. Instead, it binds to adaptors, such as Fis1, a TA protein localized to both peroxisomes and mitochondria 75. Fis1 interacts with phosphorylated Pex11 (as described later in this section) at peroxisome membranes 78 and recruits the yeast peripheral membrane receptors, Mdv1 and Caf4, which, in turn, assemble Dnm1 75 (Fig 2, panel 2–4). In higher eukaryotes that do not have Mdv1 and Caf4 homologues, the mitochondrial fission factor (MFF) recruits the dynamin‐related protein (DRP1) 79, 80.
Because DRP1 in mammalian cells is recruited to PEX11‐enriched peroxisomal membranes 81, evidence was sought for a functional link and/or a physical interaction between H. polymorpha Dnm11 and Pex11. Direct interaction was confirmed by co‐precipitation from wild‐type (WT) H. polymorpha cell lysates and interactions between Dnm1 expressed and purified from Escherichia coli and purified yeast Pex11 82. Similar interactions were also reported between PEX11β and DRP1 in mammalian cells 82. Using amino acid substitutions in peptide arrays corresponding to regions of H. polymorpha Pex11, two regions (named B1 and B3) in yeast Pex11 were identified, in which single amino acid substitutions abolished the ability of Pex11 to interact with Dnm1. Secondary structure predictions show that the B3 region of Pex11 is part of a larger amphipathic helix, whereas the B1 region folds into an alpha helical structure required for this interaction (Fig 2, panel 5).
Because Pex11 is required for Dnm1 function in H. polymorpha, kinetic experiments were performed to elucidate whether Pex11 binding alters the Dnm1 kinetic properties. Purified Dnm1 hydrolyzed GTP in a time‐dependent manner. The addition of purified Pex11 resulted in a small increase of GTPase activity. The addition of the complete B3 amphipathic helix of Pex11 significantly enhanced the catalytic activity and showed that the B3 region can act in vitro as a GTPase‐activating protein for Dnm1 82. This represents a novel function for Pex11 that is distinct from its membrane elongation activity.
Peroxisomes and mitochondria share a common organelle division machinery 75, 83, which must be activated differentially on peroxisomes and mitochondria in response to different cues. Studies in P. pastoris (Pp) shed light on how PpPex11 is activated to promote peroxisome division specifically in oleate. On growth of yeast cells in this medium, PEX11 gene expression increases 1,000‐fold as compared to its steady‐state levels in glucose medium 84. PpPex11 expression is coordinated with the initiation of peroxisome biogenesis and the protein is phosphorylated at Ser173 (S173) 78. P. pastoris mutants pex11(S173A) (unphosphorylated) and pex11(S173D) (constitutive phosphomimic) exhibit juxtaposed elongated peroxisomes and hyper‐divided forms, respectively, although protein levels remain unchanged. This phosphorylation occurs at the peroxisomes and the modification allows Pex11 to interact with Fis1, a key component of the peroxisome division machinery. Since Fis1 also interacts with Dnm1 85, it may aid the assembly of the peroxisome fission complex that encircles and constricts the peroxisome membrane, causing division. The coordinated action of phosphorylated Pex11 in recruiting Fis1, the binding of Dnm1 by both Fis1 and Pex11, and the activation of the GTPase of Dnm1 by Pex11 explain how peroxisome fission is mediated locally.
Analogous results were observed also with S. cerevisiae (Sc) ScPex11, which is also phosphorylated (at Ser165 and/or 167) 86. The phosphomimic form stimulates peroxisome division upon overexpression, whereas the non‐phosphorylated form mimics the phenotype of Pex11‐deficient cells. These mutant phenotypes were not caused by changes in the levels of the transcripts or the protein in a comparison of the WT and mutant cells expressing these proteins. However, the PEX11 transcription was rapidly destabilized in YPD medium relative to peroxisome‐inducing, oleate medium, but the Pex11 protein was stable in both media. The overproduction of the Pho85 kinase caused the hyperphosphorylation of Pex11 and peroxisome proliferation, and conversely in cells lacking Pho85 kinase, Pex11 was not phosphorylated. These data point to the Pho85 kinase in yeast as the regulator of Pex11 phosphoregulation.
Interestingly, the role of Pex11 and Fis1 in peroxisome division is dependent on the environment. Neither Pex11 nor Fis1 is necessary for peroxisome division in P. pastoris cells grown in methanol 78, showing that the proteins that control peroxisome division likely depend on the specific environmental conditions that trigger peroxisome division. Since most organisms, including yeasts, possess multiple Pex11‐family members, it is plausible that some other family member and a different, Fis1‐independent, dynamin‐family member are required for peroxisome division for P. pastoris cells grown in methanol 78. This function could be provided by the Vps1 protein, another dynamin‐like GTPase in yeast 87.
Yeast cells have a family of Pex11‐related proteins, such as Pex25 and Pex27, and these have been best studied in S. cerevisiae. While ScPex11 promotes the proliferation of pre‐existing peroxisomes, ScPex25 initiates remodeling at the peroxisomal membrane and ScPex27 acts to counter this activity 88.
Pex34 is a peroxisomal integral membrane protein that functions both independently and jointly with the Pex11‐family proteins (Pex11, Pex25, and Pex27 in S. cerevisiae) to regulate peroxisome populations under peroxisome‐induction and constitutive‐expression conditions 89. Pex34 interacts with these peroxins and its elevated expression causes peroxisome proliferation in both WT and pex34Δ cells. In view of the related functions of ScPex34 and mammalian PEX16, we speculate that Pex34 may enhance PMP and de novo peroxisome biogenesis.
Mammalian cells also have multiple PEX11‐family members, denoted as α, β, and γ that serve as peroxisome membrane elongation and division factors 90. As is the case in yeast, these proteins interact with mammalian FIS1, a limiting factor in peroxisome division 90. In the plant, Arabidopsis thaliana (At), the PEX11 protein family consists of the three phylogenetically distinct subfamilies PEX11a, PEX11b, and PEX11c to PEX11e 91. All five Arabidopsis PEX11 proteins are peroxisomal and PEX11a and PEX11c to PEX11e behave as peroxisomal integral membrane proteins. Overexpression of AtPEX11 genes in Arabidopsis induced peroxisome proliferation, whereas reduction in gene expression decreased peroxisome abundance 91.
Another S. cerevisiae PMP, Pex35, a distant homologue of several curvature‐generating human proteins, regulates the fission process 92. Its deletion causes a significant reduction in peroxisomes/cell, and conversely, its overexpression results in a multi‐lobular peroxisome phenotype due to enhanced peroxisome fission. A systematic complementation screen revealed that Pex35 is in the proximity of Pex11 and Arf1, a small GTPase. In S. cerevisiae, Arf1 and Arf3 are ADP‐ribosylation factors that upregulate and downregulate peroxisome fission, respectively 93, 94. The double mutant, arf1Δ pex35Δ, exhibited an increase in the size and a reduction in the number of peroxisomes, analogous to the phenotype seen in the single mutants arf1Δ, pex35Δ, or pex11Δ cells 92. The overexpression of Pex35 in arf1Δ cells restores the normal peroxisome number, suggesting a redundant role between Pex35 and Arf1. However, the authors did not investigate the effects of overexpression or deletion of the PEX35 gene in pex11Δ cells and the mechanism by which Pex35 and Arf1 modulate Pex11 function is not understood.
Both intrinsic and extrinsic signals activate peroxisome division. One example of an intrinsic signal comes from the finding that Yarrowia lipolytica Pex16 is involved in peroxisome division 95. As peroxisomes grow via the import of matrix proteins, there is a redistribution of the peroxisomal matrix enzyme, acyl‐CoA oxidase, from the matrix to the luminal leaflet of the peroxisome membrane, where it associates with Pex16, which negatively regulates peroxisome division in Y. lipolytica. This interaction relieves the inhibitory action of Pex16 95, thereby allowing mature peroxisomes to divide by allowing the biosynthesis of phosphatidic acid and diacylglycerol (DAG) in the membrane 96. The formation of these two lipids and the subsequent trans‐bilayer movement of DAG initiate the assembly of a complex between Pex10 and Pex19, the dynamin‐like GTPase Vps1, and several actin cytoskeletal proteins on the peroxisomal surface. This protein complex promotes membrane fission, which is the terminal step of peroxisome division 96.
There may also be peroxisome‐generated metabolites that signal division in human cells, based on the observation that both impairment in the peroxisomal matrix protein import of certain fatty acid β‐oxidation enzymes (acyl‐CoA oxidase and 2‐enoyl‐CoA hydratase/D‐3‐hydroxyacyl‐CoA dehydrogenase) or the loss of either of these enzymes causes a reduction in the number of peroxisomes 97. However, there must also be extrinsic signals coming from outside the peroxisome matrix because peroxisomes deficient in the import of peroxisomal matrix proteins can still divide.
In human cells, metabolites, like docosahexaenoic acid (DHA, C22:6n‐3), can also drive peroxisome division 98. In fibroblasts isolated from patients impaired in peroxisomal fatty acid β‐oxidation, peroxisomes were much less abundant than in normal cells. Treatment of these patient fibroblasts with DHA induced the proliferation of peroxisomes, in a DRP1‐dependent fashion, to the level seen in normal fibroblasts. Time‐lapse imaging analysis of peroxisomal morphogenesis performed in the presence of DHA revealed the sequence of steps involved in peroxisome division, including PEX11β‐dependent elongation followed by peroxisomal fission. DHA‐enhanced peroxisomal division was microtubule‐independent, suggesting that cytoskeletal proteins like MIRO1 might not be involved.
Taken together, it is clear that the import of PMPs and matrix proteins underlies the growth of peroxisomes that then divide in response to extrinsic and intrinsic stimuli.
The de novo peroxisome biogenesis model
The growth and division model had difficulty explaining how peroxisomes could arise in peroxisome biogenesis mutants that displayed no evidence of pre‐existing peroxisomes, and yet could be complemented by the missing gene to generate new peroxisomes. The alternative model invoked to address this issue involves de novo peroxisome biogenesis in which some PMPs are first inserted into the membrane of the ER, sorted to a region of the ER called the pER, from where ppVs containing the PMPs bud (Fig 3). Nuances of the de novo biogenesis model are whether one or multiple types of ppVs bud from pre‐existing membranous compartments, whether the ER alone or other organelles, such as mitochondria, contribute to peroxisome biogenesis, and finally where the lipids required for peroxisome membranes originate within the cells. The presence in peroxisomes of lipids that originate in other subcellular compartments, and of metabolites either shared or transferred between peroxisomes and other organelles, has led to recent interest in inter‐organelle MCS (section Membrane contact sites involving peroxisomes), where peroxisomes are one of the partners.
Figure 3. Schematic representation of de novo peroxisome biogenesis pathways in yeast and mammals.
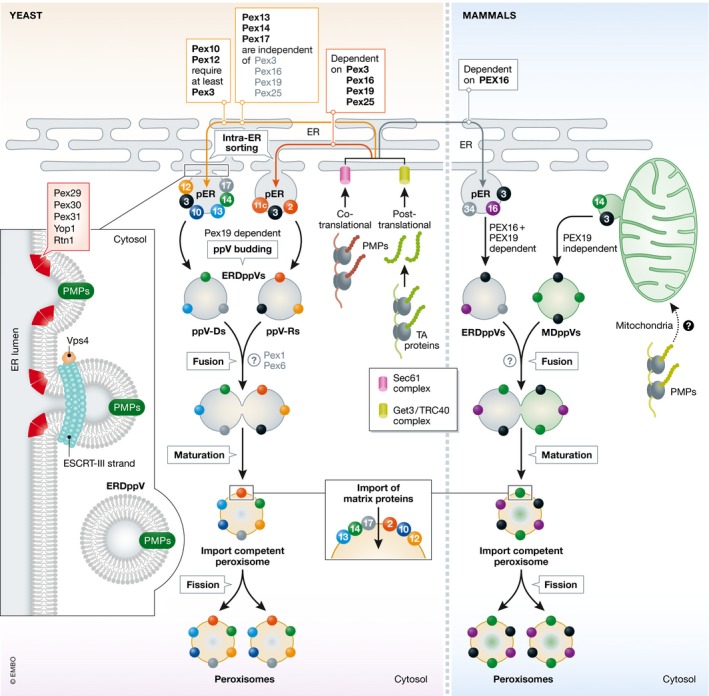
The first step in the de novo biogenesis is the indirect import of PMPs to the ER. Some PMPs are co‐translationally inserted into the ER membrane via the Sec61 complex 33, 108 and TA PMPs are post‐translationally incorporated into the ER membrane via the Get3 complex (yeast) 63 or ASNA1/TRC40 (mammals) 62. After PMP insertion into the ER, work in yeasts shows that an intra‐ER sorting step targets the PMPs to sub‐domains of the ER called the pER 32. Work in P. pastoris reveals that this routing of PMPs is either dependent or independent of Pex3, 16, 19, and 25 116, 117, 118. Studies from several yeasts define at least two modes of intra‐ER sorting of PMPs. One pathway is exemplified by the docking subcomplex proteins (Pex13, 14, and 17), which are independent of Pex3, 16, 19, and 25 118, 137. The other is exemplified by the RING‐domain PMPs (Pex10, 12, 2, 11c) and is dependent on Pex3, 16, 19, and 25 for intra‐ER sorting 118, 137. The exit sites for ppV budding are marked by the presence of several proteins (shown in inset on the left) including Pex29, 30, 31, which interact with Yop1 and Rtn1 and impart positive curvature in the ER 122, 124, 130. Subsequently, ESCRT‐III proteins (Vps20 and Snf7) are proposed to play a role in ppV scission 121 in an energy‐dependent manner, perhaps facilitated by Vps4 (stimulating disassembly of ESCRT‐III at the ER) 121. The ppVs bud from the pER in a Pex19‐dependent manner 115, 120. ERDppVs are of two distinct varieties—ppV‐R, containing Pex3, Pex2, and Pex11C and ppV‐D comprised of Pex13, Pex14, Pex17, Pex10, Pex12, and Pex3 116. Subsequently, these ppVs fuse heterotypically or with pre‐existing peroxisomes 40, 42, 119. In mammals, ppV formation is different in that several PMPs are sorted to the pER in a PEX16‐dependent manner 31, 146 and several other PMPs are routed to peroxisomes via mitochondria, from which MDppVs are formed in a PEX19‐independent manner 39. Subsequently, ERDppVs and MDppVs are proposed to fuse to form import‐competent peroxisomes, which subsequently import the matrix proteins and become metabolically active organelles. The question mark (?) represents uncertainty regarding either the known 42, 119, 144 or unknown proteins required for this fusion step.
The machinery responsible for PMP import in the direct and indirect peroxisome biogenesis pathways described above is still poorly understood and is incompletely characterized. In the various genetic screens conducted for defects in peroxisome biogenesis in multiple model organisms from yeast to plants to mammals, only three proteins, Pex3, Pex19, and Pex16 (whose functional orthologues in P. pastoris and S. cerevisiae are Pex36 and Pex34, respectively), have been described to play a clear role in PMP biogenesis.
The five specific steps in de novo peroxisome biogenesis are described next and consist of the following—(i) PMP insertion into the ER, (ii) intra‐ER sorting of PMPs to the pER, (iii) PMP exit from the pER in ppVs, (iv) ppV fusion with pre‐existing peroxisomes or heterotypic ppV fusion, and (v) potential involvement of ppVs derived from both the ER and mitochondrial membranes (Fig 3).
PMP insertion into the ER via the Sec61 complex during de novo peroxisome biogenesis
Integral membrane proteins of the cell surface and most intracellular compartments of eukaryotic cells are assembled at the ER. Several PMPs in yeast, plants, and mammals traffic through the ER, prior to being transported to the peroxisomes (Table 1). Two highly conserved and parallel pathways mediate membrane protein targeting to and insertion into this organelle. The classical co‐translational pathway, utilized by most membrane proteins, involves targeting by the signal recognition particle (SRP) followed by insertion via the Sec61 translocon 99. The second pathway is a post‐translational process, employed by many TA membrane proteins, and is composed of entirely different factors centered around a cytosolic ATPase termed ASNA1/TRC40 or Get3 100.
Table 1.
PMPs trafficking via the ER to peroxisomes in various model organisms
| Organism | PMPs at ER | Comments | Overexpression (Y/N) | References |
|---|---|---|---|---|
| Y. lipolytica | Pex2, Pex16 | N‐glycosylated in ER | N | 101 |
| S. cerevisiae | Pex3 | Artificial ER signal sequence; targeted to peroxisome | Y | 102 |
| S. cerevisiae | Pex1, Pex2, Pex4, Pex6, Pex8, Pex10‐15, Pex19, Pex25, Pex27, Ant1 | Traffics via the ER upon reintroduction of the WT PEX3 gene | N | 34 |
| S. cerevisiae | Pex15 | N‐glycosylated in ER | Y | 103 |
| A. thaliana | SSE1 (Pex16 homologue) | In peroxisomes, ER of roots, leaves, and suspension cells | Y | 104 |
| Cottonseed peroxisomal ascorbate peroxidase | APX | At ER domain and in peroxisomes in tobacco cells in suspension | Y | 105 |
| Monkey and human cells | Human PEX16 | Traffics via the ER to peroxisomes in normal and Pex16‐deficient human cells | Y | 31 |
| S. cerevisiae and mammals | 14 PMPs | Associated with ER‐localized ribosomes | N | 106 |
| Mammals | PEX3, PEX19 | Implicated in post‐translational insertion and sorting of a lipid droplet (LD) protein, UBXD8, to LDs |
Y (PEX19) N (PEX3) |
107 |
In Y. lipolytica, the ER‐to‐peroxisome traffic of Pex2 and Pex16 required Srp54, a subunit of the SRP 101, suggesting an involvement of the Sec61 complex in the insertion of these PMPs. In S. cerevisiae, Pex8, Pex13, and Pex14 were inserted into the ER in a manner dependent on the Sec61 complex 34.
By appending an artificial glycosylation signal at the N‐terminus of ScPex3, a substantial reduction in labeling (glycosylation) was observed when Sec61 mutants (either Sec61‐2, a temperature‐sensitive mutant, or a SEC61‐variant controlled by a doxycycline‐regulatable, TET promoter) were used, in comparison with the WT cells 108. In addition, using an in vitro system, a Pex3 construct with the glycosylation tag was glycosylated in the presence of yeast ER membranes and ER integration of Pex3 occurred post‐translationally. The authors also defined a conserved N‐terminal stretch of positively charged amino acids (5–6 aa) upstream of the TMD of Pex3, which might be a signal anchor sequence.
The targeting of mammalian PEX3 to the ER is also SRP‐dependent 33. Unlike the case in yeast, PEX3 is inserted into the ER co‐translationally, but it requires the Sec61 complex, similar to yeast. It is worth noting, however, that the mRNA encoding Pex3 in yeast is mostly localized to the ER, suggesting it could be inserted co‐translationally as well 109. An α‐helical region (HR) in PEX3 that partially overlaps with the N‐terminal stretch of positively charged amino acids described in yeast Pex3 108, and the TMD of PEX3, serve as the signal sequence for the in vitro insertion of PEX3 into ER microsomes 33. The HR is responsible for the ER membrane insertion of PEX3 and interacts with Sec61α and translocating chain‐associated membrane proteins, sequentially.
The first direct evidence of direct insertion of a PMP into the ER in higher eukaryotes was obtained in plants 105. When peroxisomal ascorbate peroxidase (APX) from cottonseed was transiently expressed in tobacco BY‐2 cells, it localized to the pER and to peroxisomes. In vitro experiments showed that APX integrates specifically into microsome‐derived ER membranes (93%), only 5% into peroxisomes membranes, and not into mitochondria, chloroplast, or plasma membranes.
In mammalian cells expressing endogenous levels of PEX16, this protein was observed mostly at the peroxisomes. However, when expressed in PEX16‐deficient cells lacking peroxisomes, PEX16 was observed at the ER. Similar to PEX3, mammalian PEX16 is inserted co‐translationally to the ER 31. Overexpression of PEX16 in WT cells caused the protein to exhibit dual localization at both the peroxisomes and ER.
Insertion of tail‐anchored PMPs into the ER membrane
Most TA proteins residing in the yeast ER are targeted by the GET (guided entry of tail‐anchors) complex, wherein Get3 binds the TMD of the TA protein and then following an interaction with the Get1/Get2 receptor complex, Get3 releases its cargo for insertion into the ER membrane 63, 110. This process is generally independent of Sec61 111. ASNA1/TRC40 is the mammalian homologue of Get3 112. In mammals, insertion of TA proteins into the ER is facilitated by the interaction of ASNA1/TRC40 with a membrane receptor complex formed by WRB (tryptophan‐rich basic protein) 113 and CAML (calcium‐modulating cyclophilin ligand) 114. The mechanism by which the Get3 complex directs TA proteins to their appropriate pathways (secretory, peroxisomes, mitochondria, etc.) is unknown, but it has been suggested that this targeting may be determined by the length and hydrophilicity of the TMD in the TA proteins 36.
In S. cerevisiae, Get3 interacts with a TA PMP, Pex15, and this interaction depends on the TMD of Pex15 63. Pex15 is then inserted into the ER through the Get3 complex, and independently of Sec61, before it is targeted finally to peroxisomes 34, 63. An ER targeting signal overlapping with its mPTS was found in Pex15 103. In the absence of the GET complex, Pex15 mis‐localized to the mitochondria 63, showing that Pex15 requires the GET complex for proper ER targeting. Additionally, in pex19Δ cells, overexpression of Pex15 caused it to remain in the ER, showing Pex19‐independent ER insertion followed by Pex19‐dependent targeting to peroxisomes. It is likely that the TA PMPs are inserted post‐translationally into the ER membrane because their C‐terminal TMD is occluded by the ribosomes until protein translation is complete. The mitochondrial mis‐localization of Pex15 in the absence of GET function was independent of Pex19, because although Pex15 remains in the ER in pex19Δ cells, it is mitochondrial in both get1Δ get2Δ cells and get1Δ get2Δ pex19Δ cells. Additionally, this result suggests that the ER membrane insertion of Pex15 by the GET complex precedes Pex19 function 63. It should be noted that the ER insertion of TA PMPs is distinct from the subsequent Pex19‐dependent budding of these PMPs from the ER 115.
Intra‐ER sorting of PMPs
Once PMPs are inserted into the ER, they must be sorted to specific sites (pER) in the ER from which ppVs exit. The signals responsible for the ER insertion and for intra‐ER sorting have been studied and shown to be distinct in yeast Pex3 32. The N‐terminal 17‐amino acid segment of Pex3 has two signals, conserved also in its human and Drosophila homologues, that are each sufficient for sorting to the pER. This was shown neatly by the finding that a chimeric protein containing the N‐terminal domain of Pex3 fused to the transmembrane and cytoplasmic segments of the ER protein, Sec66, sorts only to the pER in WT cells and does not colocalize with peroxisomes. Subsequent transport to existing peroxisomes requires the Pex3 TMD.
Two types of intra‐ER sorting of PMPs to the pER have been defined in P. pastoris 116, 117. One of these exemplified by the intra‐ER sorting of the docking subcomplex proteins (Pex13, Pex14, and Pex17) and Pex3, is independent of Pex3 and Pex19 116, 117. The other, illustrated by the RING‐domain PMPs (Pex2, Pex10, and Pex12) and Pex11C, requires both Pex3 and Pex19 in the form of a tripartite complex for intra‐ER sorting to the pER 116, 118. In cells lacking Pex3 and/or Pex19, these PMPs mis‐localized all over the ER, and consequently, this sorting pathway ultimately affects the ppVs that bud from the pER (Fig 3).
PEX16 is involved in the PEX19‐independent recruitment of PMPs, such as PEX3 and PMP34, to the ER or in their intra‐ER sorting in mammalian cells 65. A comprehensive mutational analysis of PEX16 was performed to elucidate the molecular targeting signals responsible for its ER‐to‐peroxisome trafficking and the domain(s) involved in PMP recruitment at the ER. The first TMD (aa 110–131) of PEX16, or a TMD from another ER protein, is both necessary and sufficient for its targeting to the ER. A separate region, comprising amino acids 71–81, serves as the ER‐to‐peroxisome targeting signal, as judged by the fact that the deletion of this sequence caused the PEX16(Δ66‐81)‐GFP to remain in the ER. PEX16 recruits multiple PMPs to the ER as shown for two TA PMPs (PEX26 and FIS1), as well as multi‐span PMPs (PMP34, PEX11β, and PEX10), a function that is conserved in plants 65. This recruitment depends on amino acids 66–103 in PEX16 and is independent of PEX3 and PEX19. Exactly how PEX16 recruits PMPs to the ER was not clear from these studies, but the role of P. pastoris Pex36, a functional homologue of PEX16, sheds some light on this process 118. Pex36 is a recently identified PMP in P. pastoris 118 that is required for cell growth in conditions that require peroxisomes for the metabolism of certain carbon sources. The growth defect in cells lacking Pex36 can be rescued by the expression of human PEX16, S. cerevisiae Pex34, or by overexpression of the endogenous P. pastoris Pex25. Pex36 is not an essential protein for peroxisome proliferation, but in the absence of the functionally redundant protein, Pex25, it becomes essential and < 20% of the pex25∆ pex36∆ cells show import‐incompetent, peroxisome remnants. In the absence of Pex25 and Pex36 proteins, peroxisome biogenesis and the intra‐ER sorting of Pex2 and Pex11C (a Pex11 family protein) are seriously impaired, likely by affecting Pex3 and Pex19 function.
Exit of ER‐associated PMPs, likely via ppVs
A key control feature of ppV production is that while PMPs reside in the ER, peroxisomal matrix protein import should not occur into the wrong subcellular compartment, namely the ER, so there must be some mechanism preventing this. The solution appears to be to segregate PMPs into distinct ER‐derived ppVs (ERDppVs) 116, 117, 119, and possibly also mitochondrially derived ppVs (MDppVs) 39.
Multiple functional epitope‐tagged PMPs have been used in vitro and in vivo to follow ppV budding in several model organisms (Table 2). Most of these studies show that in the absence of Pex19, little or no ppV budding occurs, showing a requirement for Pex19. Additional screens and assays have identified other proteins (Table 3) that are required, but this analysis is at its early stages.
Table 2.
Selected cargo proteins used to study ppV budding in vivo and in vitro
| Protein | Organism | Role of cargo protein | References |
|---|---|---|---|
| Pex15 | S. cerevisiae | Anchors Pex6 at the peroxisome membrane | 115 |
| Pex11 | P. pastoris | Involved in peroxisome division | 78 120 |
| Pex3 | S. cerevisiae P. pastoris | Required for PMP biogenesis of RING peroxins and PMP import into pre‐existing peroxisomes | 32 115 116 120 121 122 |
| Pex2 | P. pastoris | RING peroxin and part of peroxisomal E3 ligase complex | 116 118 |
| Pex17 | P. pastoris | Component of docking subcomplex in yeast | 116 |
Table 3.
Proteins implicated in ppV budding
| Proteins | Organism | Role in ppV budding | References |
|---|---|---|---|
| Pex29 and Pex30 | S. cerevisiae | ER resident proteins that physically interact with reticulon proteins (Rtn1 and Yop1); induce membrane curvature and facilitates formation of tubular structures | 122 |
| Pex30 and Pex31 | S. cerevisiae | Reticulon‐like ER‐shaping proteins predominantly localized at the pER | 124 |
| ESCRT‐III proteins (Vps20 and Snf7) | S. cerevisiae | Involved in release of ppVs from ER via membrane scissioning | 121 |
| Sec238 and Srp54 | Y. lipolytica | Implicated in the exit of Pex2 and Pex16 from ER | 101 |
| Sec16B | HeLa cells | Regulates the transport of peroxisome biogenesis factors from the ER | 125 126 |
| Pex19 | S. cerevisiae P. pastoris | Mediates ppV budding from ER and in intra‐ER sorting of RING peroxins | 115 120 |
| Sec20, Sec39, and Dsl1 | S. cerevisiae | Secretory proteins facilitate the exit of Pex3 from the ER | 127 |
| Seipin complex | S. cerevisiae | Facilitates ppV and LD budding | 128 129 |
Several studies using yeast revealed distinct types of ppVs containing different PMPs or subcomponents of the importomer complex. Seminal studies in Y. lipolytica provided the first report of biochemically distinct ppVs (P1 and P2) loaded with Pex2 and Pex16 123. Later work in S. cerevisiae and P. pastoris showed that two biochemically distinct ppVs arise from the ER 116, 119 (Fig 3). In S. cerevisiae, one type of ppV, that we call ppV‐D, has components of the docking subcomplex of the importomer (Pex13, Pex14, and Pex17), while the other has components of the peroxisomal RING subcomplex (Pex2, Pex10, and Pex12) 119 (Fig 3, left panel). However, work in P. pastoris showed that the ppV‐D vesicles contain, in addition to the docking subcomplex subcomponents, the PMPs, Pex3, and two RING PMPs, Pex10 and Pex12, whereas the ppV‐R vesicles contain Pex2, Pex3, and Pex11C 116, 118.
The proteins required for ppV budding are still poorly understood and incompletely characterized (Table 3). Their functions are described briefly next, but other proteins are also likely to be required for ppV budding and this remains an active area for ongoing research.
Interestingly, the sites of ppV budding, marked by the Seipin complex and Pex30 in yeast (and a similar protein called multiple C2 domain containing transmembrane protein MCTP2, in higher eukaryotes), also correspond to the sites of lipid droplet (LD) formation, suggesting a link between LDs and peroxisomes 128, 129.
Role of Pex19 in ppV budding
Although Pex19 has several roles described elsewhere in this manuscript (sections Insertion of tail‐anchored PMPs into the ER membrane, Intra‐ER sorting of PMPs, Exit of ER‐associated PMPs, likely via ppVs), it plays an essential in vitro and in vivo role in ppV budding in several organisms 34, 115, 120. In vitro assays demonstrating the budding of ppVs from the ER in S. cerevisiae and P. pastoris show that ppV budding, in which PMPs bud from membranes of permeabilized yeast cells into the cytosol, is an energy‐, cytosol‐ and Pex19‐dependent process 115, 120.
Pex19 consists of an intrinsically disordered region located between distinctive N‐terminal domain (NTD) and CTD 46, 47, 117. Only 3% of Pex19 associates with peroxisomal membranes through its C‐terminal farnesyl tail 47, but this association also requires Pex3 as a docking factor 55. Crystallographic studies reveal that the NTD (aa 1–44) of Pex19 binds Pex3 and its CTD (aa 160–300) possesses an mPTS binding site 49, 50, 57.
A recent study revealed the critical regions of Pex19 involved in de novo peroxisome biogenesis in P. pastoris pex19Δ cells under peroxisome proliferation conditions (methanol as sole source of carbon) 117. Cells devoid of either the N‐terminal, Pex3‐binding domain or the C‐terminal mPTS binding region of Pex19, which had been presumed to be essential, still formed import‐competent peroxisomes, but grew more slowly on methanol. Only a central domain of PpPex19 (aa 89–150) that retains binding sites for Pex11 and Pex25 (and perhaps other unknown proteins) was essential for de novo peroxisome biogenesis in cells lacking pre‐existing peroxisomes. Recently, a part of this central domain was identified as having an amphipathic helix, called alpha‐d in N. crassa, that is necessary for the insertion of certain TA PMPs into the peroxisome membrane 53. This alpha‐d segment is conserved and corresponds to aa 96–107 in PpPex19 and lies within the central domain required for de novo peroxisome biogenesis. It remains to be tested whether this alpha‐d region, and/or some other part of the central domain, of PpPex19 is required for ppV budding. Beyond this, further mechanisms await additional investigations, particularly the discovery of other proteins that interact with this domain.
Involvement of other proteins in ppV budding
Interestingly, Sec238, a protein involved in the secretory pathway in Y. lipolytica, is implicated in the exit of Pex2 and Pex16 from the ER 101. A subunit of the SRP, SRP54, is also involved in this process 101, which is surprising because one might have expected defective Pex2 and Pex16 insertion into the ER in the SRP54 knockout cells, rather than an intra‐ER sorting and or ER‐exit defect. In both mutants, the traffic of Pex2 and Pex16 was significantly delayed, but not completely blocked, and indirectly affected the number and sizes of the resulting peroxisomes. A possible explanation for the function of SRP54 in peroxisome biogenesis is that it does not play a direct role in Pex2 and Pex16 budding into ppVs, but rather indirectly affects the insertion into the ER of other protein factors required for ppV budding.
In the secretory pathway of yeast, the ER exit sites for ER to Golgi vesicular trafficking are marked by the presence of Sec16, which has two mammalian orthologues SEC16A and SEC16B. The C‐terminal region of SEC16B, which is not conserved in SEC16A, regulates the transport of peroxisomal biogenesis factors from the ER to peroxisomes in mammalian cells 125. Upon overexpression of SEC16B, PEX3 and PEX16 were redistributed from peroxisomes to SEC16B‐positive ER membranes in mammalian cells 126. Knockdown of SEC16B, but not SEC16A, by RNAi inhibited the transport of PEX16 from the ER to peroxisomes, and also suppressed expression of PEX3. These phenotypes were reversed by the expression of RNAi‐resistant Sec16B. These data suggest that SEC16B, located in ER areas other than ER exit sites (perhaps the pER), plays a role in the exit of PEX3 and PEX16 from the ER to peroxisomes (most likely via ppVs).
The formation of ppVs and their exit from the pER requires proteins (and likely lipids) that impart membrane curvature. The ER‐shaping reticulon proteins, through physical interaction with other reticulon homology domain (RHD)‐family proteins like Pex29 and Pex30, assist in regulating Pex3 sorting through the ER and releasing ppVs 122, 124, 130. Pex30 and its paralogue, Pex31, have membrane‐shaping capabilities like the reticulon proteins, which may help in defining and segregating the ppV exit site in the ER 124 (Fig 3). In fact, it has been suggested that Pex30‐containing protein complexes act as focal points (effectively the pER) from which peroxisomes form and that the tubular ER architecture organized by the RHD proteins controls this process 124, 130.
Certain subunits of the endosomal sorting complexes required for transport (ESCRT)‐III are required for ppV budding from the ER into the cytosol 121. The absence of ESCRT‐III proteins impedes de novo peroxisome formation and results in an aberrant peroxisome population in vivo. Using a cell‐free ppV budding assay in S. cerevisiae, it was shown that the ESCRT‐III subunits, Vps20 and Snf7, are necessary for ppV budding (Fig 3). The involvement of specific ESCRT‐III components in ppV budding has been explained in terms of a model wherein Vps20 is recruited to sites of ppV formation, which in turn recruits and activates the polymerization of Snf7 to drive membrane scission and release of the ppV to the cytosol 121. Other ESCRT‐III proteins like Did4 and Vps24 are also involved in ppV scission, but are not essential for this process, and may influence the rate of ppV formation by recruiting other proteins, like the AAA‐ATPase, Vps4, for disassembly of ESCRT‐III at the ER 131, 132.
Notably, ESCRT proteins are normally involved topologically in “reverse budding events” away from the cytosol 132, but in this case their role in ppV budding would have to be “normal” in that ppVs bud into the cytosol 121. However, there is some precedence for ESCRTs possibly being involved also in such “normal” topology budding 133, 134, but this is a matter requiring more careful investigation.
A study in S. cerevisiae showed that ER‐associated secretory proteins (Sec20, Sec39, and Dsl1), which form a complex at the ER, are involved in the early stages of peroxisome biogenesis 127. In cells in which these proteins were repressed, there was a relocalization of Pex3 to tubular vesicular structures and the cells lacked mature peroxisomes. Cells lacking only Sec39 affected the normal trafficking of Pex3 from ER to peroxisomes. Whether these proteins are involved in the intra‐ER sorting of PMPs or in ppV budding is unknown.
Distinct ppVs in peroxisome biogenesis
Several earlier studies had suggested the absence of functional peroxisomes and membrane remnants in yeast pex3Δ cells, but upon reintroduction of Pex3 in these cells peroxisomes re‐emerge by the de novo pathway from the ER membrane 34, 44, 135, 136. However, recent studies in H. polymorpha 137 and then in S. cerevisiae 138 show the existence of ppVs, as well as predominantly import‐incompetent, peroxisomal membrane structures, in pex3Δ atg1Δ cells. Previous studies may have missed the existence of such structures because they are degraded by selective autophagy in H. polymorpha, which requires the Atg1 kinase 137. Oleic acid‐induced pex3Δ and pex3Δ atg1Δ S. cerevisiae cells displayed characteristic fluorescent punctae for Pex14‐GFP, but unlike the situation in H. polymorpha, the number of Pex14‐GFP punctae detected in both these mutant strains was similar, suggesting a less prominent role of autophagy in degrading peroxisome remnants in S. cerevisiae 138. These fluorescent spots did not colocalize with the ER, but in some regions they were closely associated with the ER. In these cells, PMPs (especially Pex14) did not accumulate in the ER but were localized in membrane vesicles as revealed by electron, immunoelectron, and fluorescence microscopies and subcellular fractionation experiments 137, 138. Furthermore, cell fractionation analysis showed that in S. cerevisiae while most of the Pex14 co‐migrated with Por1 (a mitochondrial marker), a small fraction co‐migrated with Kar2 (ER marker). Flotation analysis showed the presence of Pex14 in gradient fractions of lighter density indicating its association with membranes.
The peroxisomal membrane structures observed in pex3Δ mutants in S. cerevisiae and H. polymorpha were similar to each other since these vesicles contain common PMPs such as Pex8, Pex13 and Pex14, but not Pex10, Pex11, and Ant1 and RING subcomplex components 137, 138. Peroxisomal matrix proteins were detected in lower amounts in these vesicles, but, without protease‐protection experiments, it is unclear if these were present on or within the membrane vesicles.
A major difference is that while these ppVs are stable in S. cerevisiae, they are degraded by autophagy in H. polymorpha, unless autophagy is blocked 137, 138. Upon the reintroduction of Pex3 in pex3Δ atg1Δ cells of H. polymorpha, the vesicles containing Pex14 were able to import peroxisomal matrix protein markers like GFP‐SKL and become mature peroxisomes, showing that they are peroxisome biogenesis intermediates 137.
These findings of ppVs in pex3Δ cells and the presence of several PMPs in these ppVs and not the ER contradict previous reports that may have misinterpreted an ER‐proximal location of the PMPs by fluorescence microscopy as being the ER itself 34, 44, 135, 136. However, this finding of the in vivo presence of ppVs in pex3Δ cells of H. polymorpha and S. cerevisiae is in accord with the observation in vitro that import‐incompetent ppVs can still be formed using P. pastoris components, and these contain components of the ppV‐D, and not the ppV‐R vesicles 116, 120.
Notably, Pex19 and Pex25 were not required for the formation of these ppVs in pex3Δ atg1Δ cells of H. polymorpha 137, which remains a puzzle given the reported requirement of Pex19 for ppV budding in P. pastoris and S. cerevisiae 115, 120. One possibility that remains unexplored is whether these ppVs seen in pex3Δ cells are MDppVs because some PMPs are targeted to mitochondria in the absence of pre‐existing peroxisomes 39. Obviously, further research into this Pex19 dependence of ppV formation is needed.
ppV fusion
Once ppVs are generated from the ER (or possibly also other subcellular compartments like the mitochondria), they appear to have either no, or only limited, import competence 40, 116, 119, 137, 138. This is because several studies show that, both in vitro and in vivo, the ppVs containing the components of the docking subcomplex do not contain some or all components of the RING subcomplex 116, 119, 137, 138. It should be noted that all three constituents (Pex2, Pex10 and Pex12) come together in the form of a subcomplex and are necessary for their mutual stabilities 139. Additionally, these proteins have RING E3 ligase activities, either individually or jointly, and these play a key role in PTS receptor recycling, a key step for the efficient import of peroxisome matrix proteins 21. The separation of one or more RING subcomplex constituents into different ppVs immediately provides an explanation for the lack of full import competence of the ppVs. Their subsequent acquisition of import competence is explained by membrane fusion events of which two versions exist in the literature.
One model suggested in Y. lipolytica and S. cerevisiae is that the ppV‐D and ppV‐R fuse in a manner that is dependent on the AAA‐ATPases, Pex1 and Pex6, to create import‐competent peroxisomes 40, 119 (Fig 3).
Using isopycnic density gradient centrifugation at 20,000 × g (low speed), a high‐speed pelletable (HSP) and a low‐speed pelletable (LSP) peroxisome fractions were found, with the first being the precursor of the second one 101. The HSP fraction can be subdivided by isopycnic density gradient centrifugation into six different vesicular subforms named P1–P6, representing different stages of immature peroxisomes harboring specific proteins 123. The key step in ppV fusion is that P1 and P2 fuse to create P3, which transitions in vivo to mature peroxisomes P4, P5, and P6 in a multi‐step manner 40, 123. The fusion of P1 and P2 is a multi‐step process subdivided into vesicle priming, docking, and fusion. At the beginning of the process, both Pex1 and Pex6 are associated with P2, while only Pex1 is associated with P1. Pex1 and Pex6 have been proposed to prime these vesicles asymmetrically. P1 peroxisomes are primed by cytosol‐dependent and ATP hydrolysis‐triggered release of Pex1, whereas P2 peroxisomes are primed by cytosol‐dependent and ATP hydrolysis‐triggered release of Pex6. This is followed by peroxisome docking, which requires P2‐associated Pex1, whereas neither Pex1 nor Pex6 needs to associate with primed P1 to achieve docking. The final step, the real fusion, is shown to be independent of Pex1, Pex6, cytosol, and ATP 40. The mechanisms proposed for these steps are however in conflict with the proposed double ring, hexa‐heteromeric structure of the Pex1‐Pex6 ATPase complex 140, 141 and the multiple roles of these ATPases in ppV fusion described here, in inhibiting pexophagy 16, 142, as well as in PTS‐receptor recycling (described later in section QC during peroxisomal matrix protein import) and peroxisomal matrix protein import 143.
An alternative model, also emanating from studies in S. cerevisiae, suggests that most peroxisome biogenesis in yeast with pre‐existing peroxisomes is by growth and division and any fusion of ppVs derived from the ER must occur with pre‐existing peroxisomes to allow lipid addition and membrane growth 42, 144. In these studies, no evidence was found for the localization of PMPs to distinct ppVs reported earlier 119, and the authors did not find support for the requirement of Pex1 and Pex6 for the formation of new peroxisomal membranes by fusion of ER‐derived vesicles. However, the authors do concede that there may be conditions (e.g., absence of pre‐existing peroxisomes) when de novo peroxisome biogenesis predominates 42, 144. The proteins involved in the fusion of ER‐derived vesicles with pre‐existing peroxisomes remain unknown in these studies.
ppVs derived from mitochondria (mammals)
We discussed earlier the trafficking of several PMPs via the ER during de novo peroxisome biogenesis in WT cells. In mammalian cells, many PMPs, such as PEX3, PEX12, PEX13, PEX14, PEX26, PMP34, and ALDP, are targeted to mitochondria in pex mutant cell lines lacking functional peroxisomes 39. The McBride group investigated whether the import of PMPs to the mitochondrial membranes in mammalian cells is an artifact and concluded that it was not.
They used mutant fibroblast cells from a patient lacking both PEX3 (called Pex3mut) and peroxisomes to examine peroxisome biogenesis. Adenoviral expression of PEX3‐YFP, followed by the use of fluorescence microscopy, showed that this exogenously expressed PEX3‐YFP and endogenous PEX14 were targeted to the mitochondrial outer membrane. Subcellular fractionation also confirmed the presence of PEX3‐YFP and PEX14 in the mitochondrial membrane fraction and cell‐free import experiments showed the targeting of PEX3‐YFP to mitochondria and not to ER microsomes 39. From this mitochondrially targeted PEX3‐YFP, they documented MDppV budding (stage 1), followed by the import of other PMPs, like PMP70, into these structures (stage II), and finally, import‐competent peroxisomes containing matrix markers (e.g., catalase) were observed (stage III). Upon acquisition of import competence by peroxisomes, PEX3‐YFP and Pex14 no longer targeted mitochondria and shifted exclusively to peroxisomes. The GTPase, DRP1, involved in peroxisome division, the retromer component, VPS35, necessary for the transport of other mitochondrially derived vesicles to peroxisomes 145 and PEX19, were not required for this MDppV budding (Fig 3). However, as of now, specific components required for this process have not been found.
Interestingly, they also used fibroblasts from a patient lacking PEX16, which traffics to mammalian peroxisomes via the ER 31. Confirming this observation, upon complementation of this Pex16mut cell line with ectopically expressed PEX16‐YFP, this protein was targeted to the ER and then formed ERDppVs, which were required to fuse with the MDppVs, to form normal import‐competent peroxisomes 39. PEX14 was initially absent from ER‐derived PEX16 vesicles, but they observed a second stage during which PEX14 was enriched within PEX16‐positive structures, which were in very close contact with mitochondria.
In Pex3mut cells overexpressing PEX16‐mRFP, a re‐routing of PEX3‐YFP was observed via the ER, rather than through mitochondria. However, under these conditions, fewer import‐competent peroxisomes were generated leading the authors to conclude that PEX3 must traffic via the mitochondria to efficiently generate functional peroxisomes, and that ERDppVs carrying PEX16 and PEX3 are insufficient to initiate the rapid import of PMPs. This conclusion, however, seems at odds with other studies showing that mammalian PEX3 is sorted to peroxisomes via the ER in a PEX16‐dependent fashion 146.
Using whole‐cell fusion experiments, the fusion between mitochondrially derived PEX3 vesicles and ER‐derived PEX16 vesicles was visualized 3 h after the cell fusion event. Based on these data, it was concluded that mitochondria are an essential part of the peroxisome de novo biogenesis pathway 39. This result recapitulates another remarkable conclusion made in S. cerevisiae, albeit under an artificial situation, showing that peroxisomes can arise from mitochondrial membranes 147. In WT yeast cells, an artificial, ectopically expressed Pex3‐GFP fusion was targeted to mitochondria when its N‐terminal ER and PTS were replaced by a mitochondrial targeting signal (MTS) from the mitochondrial membrane protein, Tom20, but peroxisome formation and matrix protein targeting were not affected. In contrast, no peroxisomes were formed in pex3Δ cells and specific peroxisomal membrane and matrix markers were mis‐localized instead to the cytosol. However, upon expression of this Tom20‐Pex3‐GFP fusion in pex3Δ cells, some peroxisomes were produced and they contained much of the peroxisomal membrane and matrix proteins analyzed, as well as a small, but significant amount, of the ectopically expressed construct. These results were interpreted to mean that peroxisomes could arise by complementation of the pex3Δ cells by the Tom20‐Pex3‐GFP fusion from mitochondria. However, the results would have been more convincing if the absence of any targeting of this fusion protein to the ER had been confirmed directly, without assuming that ER targeting had been completely eliminated by replacement of the ER targeting signal. Additionally, it is unclear if MDppVs play any role in the process.
There are also some caveats associated with the experiments of McBride group 39, which were performed using fibroblast cells that lacked the PEX3 or PEX16 proteins, which could behave differently from normal WT cells. Additionally, PEX3‐YFP was overexpressed and could have been driven to mitochondria. Countering this point, however, PEX16‐YFP was also overexpressed but was not observed at mitochondria. There is evidence that the overexpression of Pex15 in S. cerevisiae causes its accumulation in the ER 103, and its mammalian counterpart, PEX26, accumulates in particular cell lines in mitochondria 148. Despite these reservations, if this involvement of MDppVs in peroxisome biogenesis proves reproducible and generalizable to other organisms, this model would also explain how premature peroxisomal matrix import is prevented into the wrong subcellular compartment by the segregation of the peroxisomal matrix protein import machinery.
Membrane contact sites involving peroxisomes
In recent years, there has been increasing recognition that subcellular organelles communicate and interact with each other dynamically, and multiple MCSs have been defined involving peroxisomes and other subcellular compartments (Fig 4). Remarkably, multiple tethers have been discovered for the MCSs involving the same organelles, and most probably each of them plays different roles or is induced by different metabolic conditions. Membrane contact can be achieved by protein–protein and/or protein–lipid interactions. As a general rule of thumb, contact sites have been evoked for non‐vesicular transport (e.g., metabolites such as lipids) and for communication (e.g., signals, often involved in calcium exchange). Thus, this discussion of MCSs is relevant to the acquisition of lipids and for signaling events in both the growth and division, as well as the de novo peroxisome biogenesis, models. Recent studies also attribute additional functions to MCSs, such as the site of organelle fission 149, 150, 151, 152, or as the membrane source during (autophagy‐related) organelle degradation 153. Interestingly, a few studies have associated the MCS of ER–mitochondria with mitochondrial protein translocation complexes, suggesting a possible direct translocation of membrane proteins, and however, additional evidence is needed to confirm this suggestion 154, 155.
Figure 4. Peroxisome membrane contact sites.
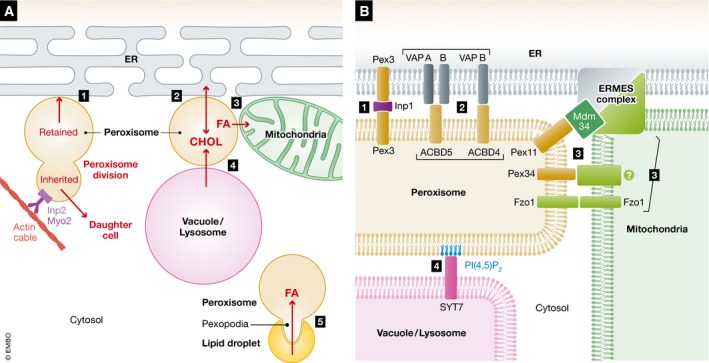
(A) MCSs of peroxisomes with several organelles (labeled 1–5), and their suggested functions. During cell division in yeast, some peroxisomes are retained in mother cells via tethering to the ER and the new peroxisomes, produced by division, are inherited (Inh) to daughter cells moving along actin cable 163. Yeast peroxisomes are pulled by the class V myosin motor, Myo2, which is attached to the peroxisomal membrane by the Inp2 protein 164. The functions of peroxisome–mitochondria MCSs linked either by Pex11‐Mdm34, Fzo1‐Fzo1, or Pex34 with an undefined (?) mitochondrial partner are unknown (panel B), but may play a role in peroxisome fission 150, 165. The other MCSs shown with other organelles are implicated in the transfer of lipids, such as fatty acids (FA) 65, 157, 158 and cholesterol (CHOL) from or to peroxisomes 170, respectively. Arrows indicate the direction of lipid traffic or organelle movement to daughter cell. (B) Known tether components of peroxisome MCSs from panel (A) represented in a single peroxisome for simplicity. However, most likely each peroxisome may not have all of these MCSs simultaneously and these sites are also dynamic in nature. 1 and 2, ER‐peroxisome tethers: 3, mitochondria‐peroxisome tethers; 4, lysosome‐peroxisome tether; 5, peroxisome–LD tether.
Peroxisome–ER MCS
In mammals, the ER has the most abundant contact sites for peroxisomes (more than 90% of peroxisomes contact the ER) followed by mitochondria (~ 20%) and LDs (~ 20%) 156. Recently, two independent groups identified the tethers between the ER and peroxisomes in mammals 65, 157, 158 (Fig 4B). The ER tethers are the vesicle‐associated membrane protein (VAMP)‐associated proteins (VAPs), which are implicated in tethering several different MCSs 159. For example, ER‐mitochondrial MCSs are promoted by VAPB‐PTPIP51 (protein tyrosine phosphatase interacting protein 51) interaction, while ER–Golgi MCSs are promoted by VAP interaction with OSBP (oxysterol binding protein), CERT (ceramide transfer protein), and Nir1‐3 (Pyk2 N‐terminal domain‐interacting receptor 1). A VAP homologue in yeast (Scs2) has been implicated in MCSs between the ER and the plasma membrane. VAPs are ER‐localized TA proteins with an N‐terminal, major sperm protein domain exposed to the cytosol and they act as protein receptors for partner proteins containing two Phe (F) residues in an acidic tract (FFAT) motif 160. The peroxisomal tethers are the TA PMPs with acyl‐CoA binding domains (ACBD), ACBD4 and ACBD5, both of which have FFAT motifs in their middle domains and an ACBD in each of their N‐terminal regions 65, 157, 158.
Disrupting the tether by silencing VAPs or ACBD5 increases peroxisome mobility in fibroblasts. Overexpression of ACBD5 in fibroblasts deficient in peroxisome fission (DRP1 or MFF‐deficient fibroblasts) induces peroxisome elongation, suggesting the tether functions in peroxisome growth. Conversely, overexpression of the tethers in WT cells (VAPB, ACBD4, and ACBD5) induces contact sites 157. The ACBD domain is not essential for peroxisome–ER tethering, but it is most probably required for the lipid exchange, as mutation in this domain affects β‐oxidation of very‐long‐chain fatty acids in peroxisomes 161. In agreement with this function, the total cellular levels of plasmalogens and cholesterol were reduced in the absence of these tethers 65.
The non‐vesicular traffic of lipids between the ER and peroxisomes has been previously proposed in S. cerevisiae using a novel in vitro assay 162. Glycerophospholipid biosynthesis occurs mostly at the ER, except for the conversion of phosphatidylserine (PS) to phosphatidylethanolamine (PE), which is catalyzed in mitochondria or in the Golgi apparatus, by phosphatidylserine decarboxylases (Psd1 and Psd2, respectively). A rapid conversion of PS to PE was observed when E. coli Psd was localized in the peroxisomal matrix in a yeast strain lacking the endogenous Psds, indicating an efficient traffic of phospholipids between the organelles. This transport was not blocked in sec mutants (required for secretory vesicular trafficking from the ER) and did not require cytosolic factors or ATP. However, the involvement of any MCS between peroxisomes and mitochondria in this process has not been tested.
In yeast, the peroxisome–ER MCS has also been implicated in peroxisome inheritance 163. During cell division, the Inp1 (inheritance of peroxisomes 1) protein forms a complex with two Pex3 molecules, one localized at the peroxisomes and the other localized at the cortical ER, which permits the preservation of a population of peroxisomes in the mother cell (Fig 4). Inp2, which is most probably a Class I PMP, interacts with the motor protein, Myo2 (myosin 2), to transport the non‐tethered peroxisomes to the budding cell 164.
Peroxisome–mitochondria MCS
The existence of sites of close proximity between the peroxisomes and mitochondria was confirmed by bimolecular fluorescence complementation (BiFC) in S. cerevisiae tagging three different peroxins (Pex3, Pex11, and Pex25) and two mitochondrial (Tom70 and Tom20) reporters 165. Overexpression of Pex34 (the closest homologue to P. pastoris Pex36 and mammalian PEX16) or Fzo1 (a yeast mitofusin protein) enhanced the number of contact sites. When expressed at endogenous levels, Pex34 was enriched in the contact sites. Mitofusins have been implicated in several contact sites between mitochondria and mitochondria, ER, melanosomes, and LDs 166. Interestingly, overexpressed Fzo1 was observed at the peroxisomes, suggesting that it could be also localized there. This is additionally supported by the fact that Fzo1 physically interacts with Pex19 and Pex14, suggesting that Fzo1 could be the tether in both organelles (Fig 4B). However, one concern is that Fzo1 was seen at peroxisomes only upon overexpression. In contrast, Pex34 induces and localizes to MCSs; however, it is not yet clear if it is a peroxisomal tether. If it is a tether, its mitochondrial counterpart is unknown (Fig 4B).
The rates of contact site formation increase when cells are induced in oleate suggesting that the transfer of β‐oxidation products might be the reason. Overexpression of Pex34 resulted in a marked increase in CO2 production (product of the Krebs cycle), indicating a stimulation in the transport of acetyl‐CoA from peroxisomes to mitochondria. Acetyl‐CoA can be transported from the peroxisome to mitochondria by two pathways 167. The first pathway is through conversion of acetyl‐CoA to citrate by the peroxisomal citrate synthase (Cit2). The second is through conversion of acetyl‐CoA to acetylcarnitine by carnitine transferase (Cat2). Overexpression of Pex34 did not cause CO2 production in cit2Δ cells and reduced CO2 levels were observed in cat2Δ cells, indicating that citrate is the most prominent molecule being transferred by this MCS. In conclusion, these data suggest that Pex34 functions in the transfer of β‐oxidation intermediates between peroxisomes and mitochondria. The presence of Fzo1 was not required for the overexpression of Pex34 to induce contact sites or acetyl‐CoA transfer indicating a different role for that tether.
Surprisingly, deletion of the yeast PEX34 and PEX11 genes did not alter the number of contact sites observed by BiFC 165. However, lack of Pex11, a PMP implicated in MCS with mitochondria, reduced the colocalization between a mitochondrial component of the ERMES (ER‐mitochondrial encounter structures) complex, Mdm34 (Mdm34‐mCherry), and Pex14‐GFP 150). Pex11 tethers the two organelles through direct interaction with Mdm34 (Fig 4B). Because Pex11 is implicated in peroxisome fission and interacts directly with Fis1, a shared component necessary for mitochondrial and peroxisomal fission, it is possible that the Pex11‐Mdm34 MCS functions in peroxisome fission.
In mouse Leydig tumor cells, contact sites between peroxisomes and mitochondria are strongly increased upon treatment of the cells with dibutyryl cyclic adenosine monophosphate (cAMP), a potent signaling molecule for steroidogenesis 168. A similar phenotype is observed when a splice variant of enoyl‐CoA δ isomerase 2 (ACBD2 isoform A) is overexpressed, leading the authors to speculate that the increase in MCSs results in an increase of both basal and hormone‐stimulated steroid formation, plausibly through favoring the inter‐organellar exchange of metabolites involved in steroid biosynthesis. The core component of the tether is ACBD2, an ACBD‐containing protein harboring a cleavable, N‐terminal MTS and a non‐canonical, C‐terminal, PTS1 (a tripeptide, PKL), but it lacks the TA normally present near the C‐terminus of most ACBD family proteins. Due to the dual targeting signal of ACBD2, it has been suggested that the MCS is promoted through the simultaneous binding of ACBD2 with PEX5 and TOMM20 (translocator of outer mitochondrial membrane 20), the receptors for the PTS and MTS, respectively. This topic however needs further investigation.
Peroxisome–LD MCS
Several organelles have contact sites with LDs. Because in most yeasts and in plants peroxisomes are the sole organelles that perform fatty acid β‐oxidation, it seemed reasonable to expect contact sites that transfer fatty acids. Indeed, close proximity between LDs and peroxisomes has been observed in mammals, yeast, and plants. In yeast cells grown in fatty acid (oleate), peroxisome–LD contact sites are more numerous and stable 169. At these contacts, peroxisomal protrusions termed pexopodia were found that extended into the LD core (Fig 4A). These protrusions might represent places of hemi‐fusion between the outer leaflet of the peroxisomal membrane with the phospholipid monolayer of LDs, while the inner leaflet invades the LD core. Pexopodia are enriched in proteins involved in β‐oxidation, indicating that they might be places where fatty acids are shuttled from LDs to peroxisomes. Consistently, the number of pexopodia was reduced in cells containing defective peroxisomes that lack β‐oxidation enzymes.
Peroxisome–lysosome MCS
A recent study has shown the existence of lysosome–peroxisome membrane contacts (LPMC) essential for the cellular trafficking of cholesterol (Fig 4A) 170. Tethering between the two organelles involves synaptotagmin VII (SYT7) on lysosomes and phosphatidylinositol‐4, 5‐bisphosphate [PI(4,5)P2] on peroxisomes (Fig 4B). PIP4K2A, a PI(5)P‐kinase, is implicated in the synthesis of PI(4,5)P2 from PI4P at the peroxisome surface 171. Disruption of PIP4K2A or depletion of peroxisomal PI(4,5)P2 caused robust cholesterol accumulation in lysosomes and reduced LPMC.
Quality control in peroxisome homeostasis
QC during de novo peroxisome biogenesis
We reviewed earlier the de novo pathway for peroxisome biogenesis. Because of the potential of mis‐sorting peroxisomal proteins to the wrong subcellular compartment or having misfolded, aberrant, or overexpressed peroxisomal proteins residing in the wrong compartment, the question of quality control pathways to prevent the detrimental consequences of such events has arisen. Although not shown explicitly, any PMPs that are missorted to, or misfolded in, the ER, are likely to be subjected to the ERAD (ER‐associated degradation) pathway 172.
In addition, the peroxisomal AAA complex and pexophagy play a role in QC of de novo peroxisome biogenesis intermediates 16, 142 (Fig 5). It has been reported that in the absence of the S. cerevisiae AAA complex components Pex1 or Pex6, or the PMP, Pex15, that anchors Pex6 at the peroxisome membrane, the PMPs present in ppVs are degraded by pexophagy 16. Evidence for this comes from the finding that in pex1Δ atg1Δ cells (deficient in Pex1 and all forms of autophagy), most peroxisomal membranes are associated with phagophore assembly sites involved in pexophagy. Degradation depended on Atg11 and the pexophagy receptor, Atg36, which are specific proteins required for pexophagy.
Figure 5. Peroxisomal quality control pathways.
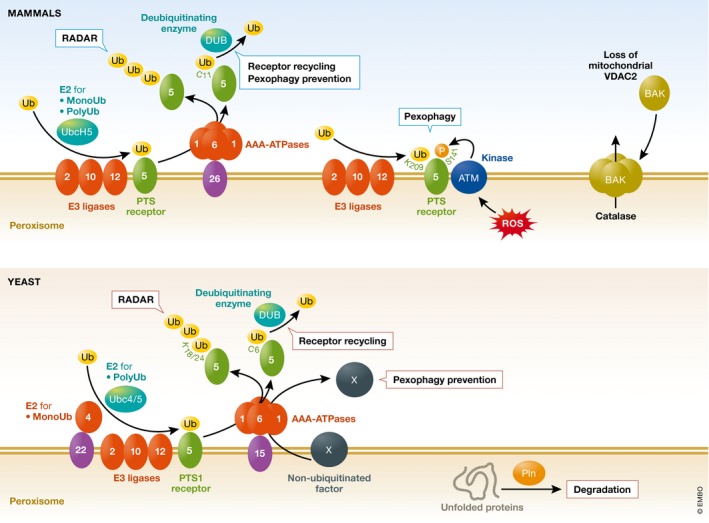
During peroxisomal matrix protein import, after the PTS receptor protein, Pex5, has released the cargo in the peroxisomal matrix, it follows different fates. Receptor Recycling: Mono‐ubiquitination (monoUb) of Pex5 occurs at a conserved cysteine residue (C6 in yeast and C11 in mammals) catalyzed by the E2‐enzyme complex (Pex4/Pex22) in yeast or UbcH5 in mammals, and the E3 ligases Pex2, Pex10, and Pex12 21. Next, mono‐ubiquitinated Pex5 is recycled to the cytosol, mediated by the AAA‐ATPases, Pex1, and Pex6 141. Finally, ubiquitin is removed by a deubiquitinating enzyme (DUB) and Pex5 becomes available for another round of import 24. RADAR: As a quality control mechanism, poly‐ubiquitination (polyUb) of Pex5 at conserved lysine residues in yeast by the E2‐enzyme Ubc4 and E3 ligases direct Pex5 for degradation by the proteasome (RADAR) 186. In mammals, the E2‐enzyme UbcH5 has been implicated in mono‐ and poly‐ubiquitination of PEX5. Pexophagy (and its prevention): As is the case in yeast, during receptor recycling in mammals, Pex1 and Pex6 are implicated in the recycling of PEX5, but in addition their presence prevents pexophagy 142. The mechanism in mammals is through recycling PEX5 from the peroxisomal membrane, as PEX5 is also target of ubiquitination which is recognized by autophagy factors driving pexophagy 186. For example, pexophagy is induced by high levels of peroxisomal reactive oxygen species (ROS), which recruits ATM to peroxisomes. ATM phosphorylates (P) PEX5 at Ser141 (S141), which then mediates the ubiquitination of PEX5 either at Lys209 (K209) 9. Alternatively, Cys11 is ubiquitinated 7. In yeast, the pexophagy prevention by the AAA‐ATPases is most probably independent of Pex5, which is not required for pexophagy. Instead, a different factor (X) that triggers pexophagy, which could be the yeast pexophagy receptor (Atg36), might need to be removed from the peroxisome surface by Pex1 and Pex6. Other QC mechanisms: The peroxisomal Lon type AAA‐protease, Pln, degrades damaged proteins in the peroxisomal matrix 182, 187, 188, 189. This protease is absent in S. cerevisiae but is present in other yeasts. Another QC mechanism may operate to recycle back to the cytosol damaged enzymes, which could be degraded by the proteasome. Such a mechanism has been described by the release of catalase to the cytosol by a peroxisome‐localized BAK, due to the lack of mitochondrial VDAC2, which normally retains BAK at mitochondria 179.
A similar observation that the presence of a functional AAA complex prevents pexophagy is also true in mammalian cells 142. Mutants of pex1, pex6, or pex26 accumulate ubiquitinated receptors at the peroxisomal membrane (Fig 5). However, while such ubiquitination triggers pexophagy in mammalian cells 142, preventing this accumulation does not abolish pexophagy of these structures in S. cerevisiae, consistent with the view that pexophagy in yeast is not ubiquitin‐dependent 16.
A good example of a QC pathway activated upon overexpression and mis‐sorting of a PMP comes from the overexpression of a TA PMP, Pex15, when it is mistargeted to mitochondria in S. cerevisiae 173. When this happens, a mitochondrial AAA protein, Msp1, engages in the destruction of Pex15. Interestingly, low levels of Msp1 are also found in the peroxisomal membrane, but here the Pex15 is rapidly converted from an Msp1‐susceptible to an Msp1‐resistant form, perhaps due to the association of Pex15 with Pex3, which protects Pex15 from degradation by Msp1.
Finally, we have evidence that several unimported peroxisomal matrix proteins, as well as the cytosolic pools of Pex5 and Pex7 in P. pastoris, are degraded by a pexophagy‐receptor‐independent form of selective autophagy (X. Wang, P. Wang, Z. Zhang, J.C. Farré, X. Li, R. Wang, Z. Xia, S. Subramani, & C. Ma, unpublished data). The degradation of unimported peroxisomal matrix proteins is of particular physiological relevance in patients with peroxisome biogenesis disorders (PBDs) where the mis‐localization of peroxisomal enzymes involved in metabolism to the cytosol might cause futile enzymatic reactions and the creation of toxic products, such as hydrogen peroxide.
QC to maintain peroxisome homeostasis and in response to environmental cues
QC also plays an important role in peroxisome homeostasis and particularly in cellular adaptation when cells are shifted from media requiring peroxisome metabolism to other media wherein these metabolic pathways can be bypassed. A good example of such adaptation is seen in fungi, wherein all fatty acid β‐oxidation occurs in peroxisomes. Not surprisingly, yeast cells grown in media such as oleate, induce peroxisomes, but when these cells are moved to glucose medium, they use glycolysis and do not need peroxisomal metabolism to function, resulting in the turnover of the excess and redundant peroxisomes. The signaling pathways, as well as the selective and general autophagy machinery, required for this type of organelle reprogramming are well known and have been reviewed elsewhere 174.
Along similar lines, when peroxisomes are either damaged by excessive ROS production, or when these organelles produce too much ROS due to their own metabolic pathways, such peroxisomes in mammalian cells are marked by ubiquitination, a common signal that triggers autophagy in mammals. Specifically, ROS activates ATM kinase in mammalian cells, causing it to translocate to the peroxisome membrane, where it ubiquitinates PEX5 at K209 by a phosphorylation‐dependent mechanism (Fig 5, upper panel) 9. Alternatively, ROS can also induce ubiquitination of PEX5 (Cys11) 175. These ubiquitination processes trigger pexophagy. It is unclear why the sites of PEX5 ubiquitination are different, but one possibility is that it depends on where and how the ROS is produced.
Another example is hypoxia, when cells are forced to reduce oxygen consumption by metabolic pathways, one of which is the use of oxygen in peroxisomes to produce hydrogen peroxide. In such instances, HIF2α induction promotes pexophagy in mammalian cells 8. However, whether this happens through the ubiquitination of PEX5 (K209 or Cys11) is unclear.
Peroxisomal metabolites, such as hydrogen peroxide, which are actively produced during plant photorespiration, can oxidize peroxisomes and trigger pexophagy 176. However, the mechanism of this pexophagy is unknown.
Developmental reprogramming of peroxisomes can also activate the degradation of peroxisomal proteins selectively. This is evident during the transition of glyoxysomes (a type of peroxisome in plants housing the glyoxylate pathway enzymes, such as isocitrate lyase‐ICL) to peroxisomes, when the content of the peroxisome matrix is altered drastically. A few days after germination, ICL, which is present in seed glyoxysomes, is degraded when photosynthesis begins. ICL and malate synthase are stabilized when a peroxisome‐associated, ubiquitin‐conjugating enzyme, Pex4, and its peroxisomal membrane anchor, Pex22, are both mutated, suggesting that matrix proteins might exit the peroxisome for ubiquitin‐dependent cytosolic degradation 177. A genetic screen for additional components needed for peroxisome‐associated matrix protein degradation of a GFP‐ICL fusion protein led to the identification in A. thaliana of three persistently fluorescent (pfl) GFP‐ICL mutants 178. One was defective in the PEROXIN14 (PEX14) gene, suggesting that ICL must enter the peroxisome for efficient degradation. A second mutant was missing the peroxisomal 3‐ketoacyl‐CoA thiolase encoded by the PEROXISOME DEFECTIVE1 (PED1) gene, suggesting that peroxisomal metabolism influences the rate of matrix protein degradation. The third pfl mutant that displayed normal matrix protein import carried a novel lesion in PEROXIN6 (PEX6), which encodes a peroxisome‐associated ATPase that is involved in recycling PTS receptors back to the cytosol. The isolation of pex6‐2 as a pfl mutant supports the hypothesis that matrix proteins can exit the peroxisome for cytosolic degradation. A model for how peroxisomal matrix proteins might be removed from peroxisomes and degraded via the proteasome has been described 178. Similar evidence for the selective export of a peroxisomal matrix protein, catalase, has emerged in mammalian cells 179 and will be described in the next section. In both plant and mammals, however, the mechanisms involved in this model are still obscure and ripe for further investigation.
QC during peroxisomal matrix protein import
As described earlier, the PTS receptors/co‐receptors shuttle relevant PTS cargos from the cytosol to the peroxisome, prior to their return to the cytosol for additional rounds of peroxisomal matrix protein import. An interesting ubiquitin–proteasome system (UPS)‐dependent, QC system similar to the ERAD pathway 172 also exists on peroxisomes.
During the peroxisomal matrix protein import cycle, Pex5 and Pex20 of P. pastoris have two alternative fates that are dependent on the types and specific sites of ubiquitination present on these proteins. In both cases, mono‐ubiquitination of P. pastoris Pex5 (at Cys11) or Pex20 (Cys8) signals recycling of these proteins back to the cytosol for another round of matrix protein import, in a process that depends on the presence of Pex1 and Pex6, the protein that anchors Pex6 to peroxisomes (ScPex15), as well as the E1, E2, and E3 enzymes involved in the reactions that conjugate ubiquitin (Ub) to the cysteines 21 (Fig 5). Pex1 and Pex6 recycle off the peroxisomes as they hydrolyze ATP. A de‐ubiquitination step is required in the cytosol to remove the mono‐ubiquitin from Pex5 in yeast and mammalian cells 24, 25.
In the absence of one or more of these proteins required for Pex5 or Pex20 recycling, the peroxisomal import machinery would get blocked, creating a logjam of unimported matrix proteins that could be detrimental for the cells. Under these conditions, an alternative UPS‐dependent RADAR pathway is activated 20 (Fig 5). In this process, P. pastoris Pex5(K22) and Pex20(K19) are poly‐ubiquitinated, followed by their extraction (independent of Pex1 and Pex6) and the delivery of these poly‐ubiquitinated proteins to the UPS for destruction 21. In the process, this QC system clears the logjam at the peroxisomes in an attempt to allow matrix protein import. The E1, E2, and E3 enzymes required for the mono‐ and poly‐ubiquitination of Pex5 and Pex20 have been described 21. Additionally, these mechanisms are conserved in Pex5 from yeast to plants to mammals, but the particular ubiquitination enzymes and the sites of ubiquitination may vary from system to system 21.
The PTS2 receptor, Pex7, also ferries cargos between the cytosol and peroxisomes, and recycles back to the cytosol 180. In P. pastoris, Pex7 is also subject to quality control and regulation 181, but its stability and dynamics are different from those of Pex5 and Pex20. Pex7 is constitutively degraded in WT cells but is stabilized in pex mutants affecting matrix protein import, suggesting a link to the peroxisomal matrix protein import cycle.
Degradation of Pex7 is more prevalent in cells grown in methanol, in which the PTS2 pathway is nonessential, in comparison with oleate, suggesting regulation of Pex7 turnover 181. Pex7 must shuttle into and out of peroxisomes before it is poly‐ubiquitinated and degraded by the UPS. The shuttling of Pex7, and consequently its degradation, is dependent on the receptor recycling pathways of Pex5 and Pex20 and relies on an interaction between Pex7 and Pex20.
Peroxisomal matrix proteins must also be properly folded and assembled to function properly, and are likely to be subject to QC when these processes malfunction. In H. polymorpha, a peroxisomal Lon protease, Pln, plays a role in degradation of unfolded and non‐assembled peroxisomal matrix proteins 182 (Fig 5, lower panel). In the absence of Pln, intracellular ROS levels increase. In A. thaliana, LON2 protease is involved in the degradation of glyoxysomal proteins inside the peroxisomes 183.
A novel form of QC for certain peroxisomal matrix proteins has been uncovered in mammalian CHO and HeLa cells in the form of a regulated permeabilization of peroxisomes to leak out some, but not all, matrix proteins. This discovery came from the analysis of a peroxisome‐deficient CHO cell line that was shown to be deficient in the mitochondrial, voltage‐dependent anion channel (VDAC2) 179. In these cells, the protein BAK (BCL2 antagonist/killer) was mis‐localized substantially from mitochondria to peroxisomes, where it caused enhanced peroxisomal permeability to catalase located normally in the peroxisomal matrix (Fig 5, upper panel). Overexpression of BAK, or its targeting to peroxisomes, or the use of BAK activators enhanced the level of catalase in the cytosol. Conversely, the knockdown of BAK, or the expression of a mutant form of BAK or the use of BAK inhibitors reduced the level of cytosolic catalase. This suggests, as described earlier in plant cells undergoing the glyoxysome to peroxisome transition 178, that some peroxisomal matrix proteins can be exported back to the cytosol. However, the mechanism of this process is even less studied in mammalian cells.
Last, but not the least, PMPs in the peroxisome membrane could also be misfolded or mis‐assembled and subject to turnover. Indeed, in H. polymorpha, Pex13 is subject to ubiquitination and degradation 184. This process is dependent both on Ub and the peroxisomal ubiquitination machinery (the E2 enzyme, Pex4, and the RING E3 complex component, Pex2), but a direct involvement of the ubiquitination machinery in Pex13 ubiquitination is still missing. In Arabidopsis, also Pex13 is degraded in a manner that is dependent on the RING E3 ligase SP1, which is a PMP that interacts with Pex13 and Pex2 185. SP1 promotes the ubiquitination of Pex13 in vitro. How Pex13 is extracted from the peroxisome membrane and degraded is not clear at present.
Summary and concluding remarks
Our understanding of the players and many of the mechanisms involved in targeting of proteins to peroxisomes, matrix protein import into peroxisomes, and the role of peroxins in PBDs has advanced dramatically in the last three decades. We hope it is obvious from this review that remarkable progress has also been made in the past few years regarding the mechanisms by which PMPs of different varieties are targeted to peroxisomes, both during growth and division, as well as during de novo peroxisome biogenesis. The flexibility and adaptability of PMP sorting between different peroxisome biogenesis pathways is astounding, especially since it is conserved across evolution. Yet, despite this progress, new players and mechanisms involved in the de novo biogenesis of peroxisomes are still being uncovered. The fact that earlier screens for peroxisome biogenesis mutants in multiple organisms had missed the identification of these genes is likely a reflection of the fact that these are either redundant or essential genes that are now being uncovered by more sophisticated genetic screens or through biochemical means. We now have a pretty comprehensive view of the involvement of QC at all steps of peroxisome biogenesis, but more work needs to be done in uncovering the underlying mechanisms, as well their role in disease. Likewise, we have just scratched the surface of the dynamic contact sites between peroxisomes and other organelles, and more importantly their physiological roles. Undoubtedly, the role of these MCSs in human health and disease will see greater progress. The future for research in the peroxisome biogenesis arena remains as exciting and relevant as ever, and we can look forward to additional important insights.
Conflict of interest
The authors declare that they have no conflict of interest.
In need of answers.
What are the proteins and mechanisms required for ppV budding and what are the roles of ppVs in delivering PMPs and lipids to peroxisomes?
What are the exact contributions of the ER and mitochondria to peroxisome biogenesis and how general is this in different organisms?
How do the same PMPs sort to peroxisomes by both the Class I or II pathways and what regulates these processes?
What is the mechanism by which some proteins, once imported into the peroxisome matrix, exit the peroxisomes?
What are the proteins, mechanisms, and signals regulating QC processes that maintain peroxisome homeostasis? What signals trigger these QC pathways?
What PBDs are caused by mutations in recently identified proteins required for de novo peroxisome biogenesis, ppV budding, and QC pathways?
Acknowledgements
This work was supported by an NIH grant 2RO1 DK41737 to SS, who holds a Tata Chancellor's Endowed Professorship in Molecular Biology at the University of California, San Diego.
EMBO Reports (2019) 20: e46864
See the Glossary for abbreviations used in this article.
References
- 1. Tabak HF, Braakman I, Distel B (1999) Peroxisomes: simple in function but complex in maintenance. Trends Cell Biol 9: 447–453 [DOI] [PubMed] [Google Scholar]
- 2. Walker CL, Pomatto LCD, Tripathi DN, Davies KJA (2018) Redox regulation of homeostasis and proteostasis in peroxisomes. Physiol Rev 98: 89–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Waterham HR, Ferdinandusse S, Wanders RJ (2016) Human disorders of peroxisome metabolism and biogenesis. Biochim Biophys Acta 1863: 922–933 [DOI] [PubMed] [Google Scholar]
- 4. Honsho M, Yamashita S, Fujiki Y (2016) Peroxisome homeostasis: mechanisms of division and selective degradation of peroxisomes in mammals. Biochim Biophys Acta 1863: 984–991 [DOI] [PubMed] [Google Scholar]
- 5. Till A, Lakhani R, Burnett SF, Subramani S (2012) Pexophagy: the selective degradation of peroxisomes. Int J Cell Biol 2012: 512721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jin A, Lee JN, Kim MS, Kwak S, Kim SJ, Song K, Choe SK, Park R (2016) 2,2′‐dipyridyl induces pexophagy. Biochem Biophys Res Commun 469: 941–947 [DOI] [PubMed] [Google Scholar]
- 7. Nordgren M, Francisco T, Lismont C, Hennebel L, Brees C, Wang B, Van Veldhoven PP, Azevedo JE, Fransen M (2015) Export‐deficient monoubiquitinated PEX5 triggers peroxisome removal in SV40 large T antigen‐transformed mouse embryonic fibroblasts. Autophagy 11: 1326–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Walter KM, Schonenberger MJ, Trotzmuller M, Horn M, Elsasser HP, Moser AB, Lucas MS, Schwarz T, Gerber PA, Faust PL et al (2014) Hif‐2alpha promotes degradation of mammalian peroxisomes by selective autophagy. Cell Metab 20: 882–897 [DOI] [PubMed] [Google Scholar]
- 9. Zhang J, Tripathi DN, Jing J, Alexander A, Kim J, Powell RT, Dere R, Tait‐Mulder J, Lee JH, Paull TT et al (2015) ATM functions at the peroxisome to induce pexophagy in response to ROS. Nat Cell Biol 17: 1259–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yifrach E, Chuartzman SG, Dahan N, Maskit S, Zada L, Weill U, Yofe I, Olender T, Schuldiner M, Zalckvar E (2016) Characterization of proteome dynamics during growth in oleate reveals a new peroxisome‐targeting receptor. J Cell Sci 129: 4067–4075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Effelsberg D, Cruz‐Zaragoza LD, Schliebs W, Erdmann R (2016) Pex9p is a new yeast peroxisomal import receptor for PTS1‐containing proteins. J Cell Sci 129: 4057–4066 [DOI] [PubMed] [Google Scholar]
- 12. Ma C, Schumann U, Rayapuram N, Subramani S (2009) The peroxisomal matrix import of Pex8p requires only PTS receptors and Pex14p. Mol Biol Cell 20: 3680–3689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Montilla‐Martinez M, Beck S, Klumper J, Meinecke M, Schliebs W, Wagner R, Erdmann R (2015) Distinct pores for peroxisomal import of PTS1 and PTS2 proteins. Cell Rep 13: 2126–2134 [DOI] [PubMed] [Google Scholar]
- 14. Meinecke M, Cizmowski C, Schliebs W, Kruger V, Beck S, Wagner R, Erdmann R (2010) The peroxisomal importomer constitutes a large and highly dynamic pore. Nat Cell Biol 12: 273–277 [DOI] [PubMed] [Google Scholar]
- 15. Rayapuram N, Subramani S (2006) The importomer–a peroxisomal membrane complex involved in protein translocation into the peroxisome matrix. Biochim Biophys Acta 1763: 1613–1619 [DOI] [PubMed] [Google Scholar]
- 16. Nuttall JM, Motley AM, Hettema EH (2014) Deficiency of the exportomer components Pex1, Pex6, and Pex15 causes enhanced pexophagy in Saccharomyces cerevisiae . Autophagy 10: 835–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Platta HW, Hagen S, Erdmann R (2013) The exportomer: the peroxisomal receptor export machinery. Cell Mol Life Sci 70: 1393–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Carvalho AF, Pinto MP, Grou CP, Alencastre IS, Fransen M, Sa‐Miranda C, Azevedo JE (2007) Ubiquitination of mammalian Pex5p, the peroxisomal import receptor. J Biol Chem 282: 31267–31272 [DOI] [PubMed] [Google Scholar]
- 19. Platta HW, El Magraoui F, Schlee D, Grunau S, Girzalsky W, Erdmann R (2007) Ubiquitination of the peroxisomal import receptor Pex5p is required for its recycling. J Cell Biol 177: 197–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Leon S, Zhang L, McDonald WH, Yates J III, Cregg JM, Subramani S (2006) Dynamics of the peroxisomal import cycle of PpPex20p: ubiquitin‐dependent localization and regulation. J Cell Biol 172: 67–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu X, Ma C, Subramani S (2012) Recent advances in peroxisomal matrix protein import. Curr Opin Cell Biol 24: 484–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Erdmann R, Schwerter DP, Grimm I, Girzalsky W (2018) Receptor recognition by the peroxisomal AAA‐complex depends on the presence of the ubiquitin moiety and is mediated by Pex1p. J Biol Chem 293: 15458–15470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pedrosa AG, Francisco T, Bicho D, Dias AF, Barros‐Barbosa A, Hagmann V, Dodt G, Rodrigues TA, Azevedo JE (2018) Peroxisomal monoubiquitinated PEX5 interacts with the AAA ATPases PEX1 and PEX6 and is unfolded during its dislocation into the cytosol. J Biol Chem 293: 11553–11563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Debelyy MO, Platta HW, Saffian D, Hensel A, Thoms S, Meyer HE, Warscheid B, Girzalsky W, Erdmann R (2011) Ubp15p, a ubiquitin hydrolase associated with the peroxisomal export machinery. J Biol Chem 286: 28223–28234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Grou CP, Francisco T, Rodrigues TA, Freitas MO, Pinto MP, Carvalho AF, Domingues P, Wood SA, Rodriguez‐Borges JE, Sa‐Miranda C et al (2012) Identification of ubiquitin‐specific protease 9X (USP9X) as a deubiquitinase acting on ubiquitin‐peroxin 5 (PEX5) thioester conjugate. J Biol Chem 287: 12815–12827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jones JM, Morrell JC, Gould SJ (2001) Multiple distinct targeting signals in integral peroxisomal membrane proteins. J Cell Biol 153: 1141–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Giannopoulou EA, Emmanouilidis L, Sattler M, Dodt G, Wilmanns M (2016) Towards the molecular mechanism of the integration of peroxisomal membrane proteins. Biochim Biophys Acta 1863: 863–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sacksteder KA, Jones JM, South ST, Li X, Liu Y, Gould SJ (2000) PEX19 binds multiple peroxisomal membrane proteins, is predominantly cytoplasmic, and is required for peroxisome membrane synthesis. J Cell Biol 148: 931–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Snyder WB, Koller A, Choy AJ, Subramani S (2000) The peroxin Pex19p interacts with multiple, integral membrane proteins at the peroxisomal membrane. J Cell Biol 149: 1171–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jones JM, Morrell JC, Gould SJ (2004) PEX19 is a predominantly cytosolic chaperone and import receptor for class 1 peroxisomal membrane proteins. J Cell Biol 164: 57–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kim PK, Mullen RT, Schumann U, Lippincott‐Schwartz J (2006) The origin and maintenance of mammalian peroxisomes involves a de novo PEX16‐dependent pathway from the ER. J Cell Biol 173: 521–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fakieh MH, Drake PJ, Lacey J, Munck JM, Motley AM, Hettema EH (2013) Intra‐ER sorting of the peroxisomal membrane protein Pex3 relies on its luminal domain. Biol Open 2: 829–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mayerhofer PU, Bano‐Polo M, Mingarro I, Johnson AE (2016) Human peroxin PEX3 Is co‐translationally integrated into the ER and exits the ER in budding vesicles. Traffic 17: 117–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. van der Zand A, Braakman I, Tabak HF (2010) Peroxisomal membrane proteins insert into the endoplasmic reticulum. Mol Biol Cell 21: 2057–2065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Matsuzaki T, Fujiki Y (2008) The peroxisomal membrane protein import receptor Pex3p is directly transported to peroxisomes by a novel Pex19p‐ and Pex16p‐dependent pathway. J Cell Biol 183: 1275–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Borgese N, Brambillasca S, Colombo S (2007) How tails guide tail‐anchored proteins to their destinations. Curr Opin Cell Biol 19: 368–375 [DOI] [PubMed] [Google Scholar]
- 37. Agrawal G, Subramani S (2013) Emerging role of the endoplasmic reticulum in peroxisome biogenesis. Front Physiol 4: 286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Agrawal G, Subramani S (2016) De novo peroxisome biogenesis: evolving concepts and conundrums. Biochim Biophys Acta 1863: 892–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sugiura A, Mattie S, Prudent J, McBride HM (2017) Newly born peroxisomes are a hybrid of mitochondrial and ER‐derived pre‐peroxisomes. Nature 542: 251–254 [DOI] [PubMed] [Google Scholar]
- 40. Titorenko VI, Rachubinski RA (2000) Peroxisomal membrane fusion requires two AAA family ATPases, Pex1p and Pex6p. J Cell Biol 150: 881–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Motley AM, Hettema EH (2007) Yeast peroxisomes multiply by growth and division. J Cell Biol 178: 399–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Motley AM, Galvin PC, Ekal L, Nuttall JM, Hettema EH (2015) Reevaluation of the role of Pex1 and dynamin‐related proteins in peroxisome membrane biogenesis. J Cell Biol 211: 1041–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rottensteiner H, Kramer A, Lorenzen S, Stein K, Landgraf C, Volkmer‐Engert R, Erdmann R (2004) Peroxisomal membrane proteins contain common Pex19p‐binding sites that are an integral part of their targeting signals. Mol Biol Cell 15: 3406–3417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hettema EH, Girzalsky W, van Den Berg M, Erdmann R, Distel B (2000) Saccharomyces cerevisiae Pex3p and Pex19p are required for proper localization and stability of peroxisomal membrane proteins. EMBO J 19: 223–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Shibata H, Kashiwayama Y, Imanaka T, Kato H (2004) Domain architecture and activity of human Pex19p, a chaperone‐like protein for intracellular trafficking of peroxisomal membrane proteins. J Biol Chem 279: 38486–38494 [DOI] [PubMed] [Google Scholar]
- 46. Sato Y, Shibata H, Nakatsu T, Nakano H, Kashiwayama Y, Imanaka T, Kato H (2010) Structural basis for docking of peroxisomal membrane protein carrier Pex19p onto its receptor Pex3p. EMBO J 29: 4083–4093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rucktaschel R, Thoms S, Sidorovitch V, Halbach A, Pechlivanis M, Volkmer R, Alexandrov K, Kuhlmann J, Rottensteiner H, Erdmann R (2009) Farnesylation of Pex19p is required for its structural integrity and function in peroxisome biogenesis. J Biol Chem 284: 20885–20896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Matsuzono Y, Matsuzaki T, Fujiki Y (2006) Functional domain mapping of peroxin Pex19p: interaction with Pex3p is essential for function and translocation. J Cell Sci 119: 3539–3550 [DOI] [PubMed] [Google Scholar]
- 49. Schueller N, Holton SJ, Fodor K, Milewski M, Konarev P, Stanley WA, Wolf J, Erdmann R, Schliebs W, Song YH et al (2010) The peroxisomal receptor Pex19p forms a helical mPTS recognition domain. EMBO J 29: 2491–2500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Snyder WB, Faber KN, Wenzel TJ, Koller A, Luers GH, Rangell L, Keller GA, Subramani S (1999) Pex19p interacts with Pex3p and Pex10p and is essential for peroxisome biogenesis in Pichia pastoris . Mol Biol Cell 10: 1745–1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Emmanouilidis L, Schutz U, Tripsianes K, Madl T, Radke J, Rucktaschel R, Wilmanns M, Schliebs W, Erdmann R, Sattler M (2017) Allosteric modulation of peroxisomal membrane protein recognition by farnesylation of the peroxisomal import receptor PEX19. Nat Commun 8: 14635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kinoshita N, Matsuura A, Fujiki Y (2017) Peroxisome biogenesis: a novel inducible PEX19 splicing variant is involved in early stages of peroxisome proliferation. J Biochem 161: 297–308 [DOI] [PubMed] [Google Scholar]
- 53. Chen Y, Pieuchot L, Loh RA, Yang J, Kari TM, Wong JY, Jedd G (2014) Hydrophobic handoff for direct delivery of peroxisome tail‐anchored proteins. Nat Commun 5: 5790 [DOI] [PubMed] [Google Scholar]
- 54. Matsuzono Y, Fujiki Y (2006) In vitro transport of membrane proteins to peroxisomes by shuttling receptor Pex19p. J Biol Chem 281: 36–42 [DOI] [PubMed] [Google Scholar]
- 55. Fang Y, Morrell JC, Jones JM, Gould SJ (2004) PEX3 functions as a PEX19 docking factor in the import of class I peroxisomal membrane proteins. J Cell Biol 164: 863–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Liu Y, Yagita Y, Fujiki Y (2016) Assembly of peroxisomal membrane proteins via the direct Pex19p‐Pex3p pathway. Traffic 17: 433–455 [DOI] [PubMed] [Google Scholar]
- 57. Schmidt F, Treiber N, Zocher G, Bjelic S, Steinmetz MO, Kalbacher H, Stehle T, Dodt G (2010) Insights into peroxisome function from the structure of PEX3 in complex with a soluble fragment of PEX19. J Biol Chem 285: 25410–25417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ghaedi K, Tamura S, Okumoto K, Matsuzono Y, Fujiki Y (2000) The peroxin Pex3p initiates membrane assembly in peroxisome biogenesis. Mol Biol Cell 11: 2085–2102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kammerer S, Holzinger A, Welsch U, Roscher AA (1998) Cloning and characterization of the gene encoding the human peroxisomal assembly protein Pex3p. FEBS Lett 429: 53–60 [DOI] [PubMed] [Google Scholar]
- 60. Soukupova M, Sprenger C, Gorgas K, Kunau WH, Dodt G (1999) Identification and characterization of the human peroxin PEX3. Eur J Cell Biol 78: 357–374 [DOI] [PubMed] [Google Scholar]
- 61. Pinto MP, Grou CP, Fransen M, Sa‐Miranda C, Azevedo JE (2009) The cytosolic domain of PEX3, a protein involved in the biogenesis of peroxisomes, binds membrane lipids. Biochim Biophys Acta 1793: 1669–1675 [DOI] [PubMed] [Google Scholar]
- 62. Yagita Y, Hiromasa T, Fujiki Y (2013) Tail‐anchored PEX26 targets peroxisomes via a PEX19‐dependent and TRC40‐independent class I pathway. J Cell Biol 200: 651–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Schuldiner M, Metz J, Schmid V, Denic V, Rakwalska M, Schmitt HD, Schwappach B, Weissman JS (2008) The GET complex mediates insertion of tail‐anchored proteins into the ER membrane. Cell 134: 634–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Cichocki BA, Krumpe K, Vitali DG, Rapaport D (2018) Pex19 is involved in importing dually targeted tail‐anchored proteins to both mitochondria and peroxisomes. Traffic 19: 770–785 [DOI] [PubMed] [Google Scholar]
- 65. Hua R, Cheng D, Coyaud E, Freeman S, Di Pietro E, Wang Y, Vissa A, Yip CM, Fairn GD, Braverman N et al (2017) VAPs and ACBD5 tether peroxisomes to the ER for peroxisome maintenance and lipid homeostasis. J Cell Biol 216: 367–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Erdmann R, Blobel G (1995) Giant peroxisomes in oleic acid‐induced Saccharomyces cerevisiae lacking the peroxisomal membrane protein Pmp27p. J Cell Biol 128: 509–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Opalinski L, Kiel JA, Williams C, Veenhuis M, van der Klei IJ (2011) Membrane curvature during peroxisome fission requires Pex11. EMBO J 30: 5–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Fagarasanu M, Fagarasanu A, Tam YY, Aitchison JD, Rachubinski RA (2005) Inp1p is a peroxisomal membrane protein required for peroxisome inheritance in Saccharomyces cerevisiae . J Cell Biol 169: 765–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Fagarasanu A, Mast FD, Knoblach B, Jin Y, Brunner MJ, Logan MR, Glover JN, Eitzen GA, Aitchison JD, Weisman LS et al (2009) Myosin‐driven peroxisome partitioning in S. cerevisiae . J Cell Biol 186: 541–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Castro IG, Richards DM, Metz J, Costello JL, Passmore JB, Schrader TA, Gouveia A, Ribeiro D, Schrader M (2018) A role for mitochondrial Rho GTPase 1 (MIRO1) in motility and membrane dynamics of peroxisomes. Traffic 19: 229–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Schrader M, Burkhardt JK, Baumgart E, Luers G, Spring H, Volkl A, Fahimi HD (1996) Interaction of microtubules with peroxisomes. Tubular and spherical peroxisomes in HepG2 cells and their alterations induced by microtubule‐active drugs. Eur J Cell Biol 69: 24–35 [PubMed] [Google Scholar]
- 72. Passmore JB, Pinho S, Gomez‐Lazaro M, Schrader M (2017) The respiratory chain inhibitor rotenone affects peroxisomal dynamics via its microtubule‐destabilising activity. Histochem Cell Biol 148: 331–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Kraus F, Ryan MT (2017) The constriction and scission machineries involved in mitochondrial fission. J Cell Sci 130: 2953–2960 [DOI] [PubMed] [Google Scholar]
- 74. Lee JE, Westrate LM, Wu H, Page C, Voeltz GK (2016) Multiple dynamin family members collaborate to drive mitochondrial division. Nature 540: 139–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Motley AM, Ward GP, Hettema EH (2008) Dnm1p‐dependent peroxisome fission requires Caf4p, Mdv1p and Fis1p. J Cell Sci 121: 1633–1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Thoms S, Erdmann R (2005) Dynamin‐related proteins and Pex11 proteins in peroxisome division and proliferation. FEBS J 272: 5169–5181 [DOI] [PubMed] [Google Scholar]
- 77. Mears JA, Lackner LL, Fang S, Ingerman E, Nunnari J, Hinshaw JE (2011) Conformational changes in Dnm1 support a contractile mechanism for mitochondrial fission. Nat Struct Mol Biol 18: 20–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Joshi S, Agrawal G, Subramani S (2012) Phosphorylation‐dependent Pex11p and Fis1p interaction regulates peroxisome division. Mol Biol Cell 23: 1307–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Loson OC, Song Z, Chen H, Chan DC (2013) Fis1, Mff, MiD49, and MiD51 mediate Drp1 recruitment in mitochondrial fission. Mol Biol Cell 24: 659–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Gandre‐Babbe S, van der Bliek AM (2008) The novel tail‐anchored membrane protein Mff controls mitochondrial and peroxisomal fission in mammalian cells. Mol Biol Cell 19: 2402–2412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Li X, Gould SJ (2003) The dynamin‐like GTPase DLP1 is essential for peroxisome division and is recruited to peroxisomes in part by PEX11. J Biol Chem 278: 17012–17020 [DOI] [PubMed] [Google Scholar]
- 82. Williams C, Opalinski L, Landgraf C, Costello J, Schrader M, Krikken AM, Knoops K, Kram AM, Volkmer R, van der Klei IJ (2015) The membrane remodeling protein Pex11p activates the GTPase Dnm1p during peroxisomal fission. Proc Natl Acad Sci USA 112: 6377–6382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Hu J (2010) Molecular basis of peroxisome division and proliferation in plants. Int Rev Cell Mol Biol 279: 79–99 [DOI] [PubMed] [Google Scholar]
- 84. Karpichev IV, Small GM (1998) Global regulatory functions of Oaf1p and Pip2p (Oaf2p), transcription factors that regulate genes encoding peroxisomal proteins in Saccharomyces cerevisiae . Mol Cell Biol 18: 6560–6570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Koppenol‐Raab M, Harwig MC, Posey AE, Egner JM, MacKenzie KR, Hill RB (2016) A targeted mutation identified through pKa measurements indicates a postrecruitment Role for Fis1 in yeast mitochondrial fission. J Biol Chem 291: 20329–20344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Knoblach B, Rachubinski RA (2010) Phosphorylation‐dependent activation of peroxisome proliferator protein PEX11 controls peroxisome abundance. J Biol Chem 285: 6670–6680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Hoepfner D, van den Berg M, Philippsen P, Tabak HF, Hettema EH (2001) A role for Vps1p, actin, and the Myo2p motor in peroxisome abundance and inheritance in Saccharomyces cerevisiae . J Cell Biol 155: 979–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Huber A, Koch J, Kragler F, Brocard C, Hartig A (2012) A subtle interplay between three Pex11 proteins shapes de novo formation and fission of peroxisomes. Traffic 13: 157–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Tower RJ, Fagarasanu A, Aitchison JD, Rachubinski RA (2011) The peroxin Pex34p functions with the Pex11 family of peroxisomal divisional proteins to regulate the peroxisome population in yeast. Mol Biol Cell 22: 1727–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Koch J, Pranjic K, Huber A, Ellinger A, Hartig A, Kragler F, Brocard C (2010) PEX11 family members are membrane elongation factors that coordinate peroxisome proliferation and maintenance. J Cell Sci 123: 3389–4300 [DOI] [PubMed] [Google Scholar]
- 91. Orth T, Reumann S, Zhang X, Fan J, Wenzel D, Quan S, Hu J (2007) The PEROXIN11 protein family controls peroxisome proliferation in Arabidopsis . Plant Cell 19: 333–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Yofe I, Soliman K, Chuartzman SG, Morgan B, Weill U, Yifrach E, Dick TP, Cooper SJ, Ejsing CS, Schuldiner M et al (2017) Pex35 is a regulator of peroxisome abundance. J Cell Sci 130: 791–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Anthonio EA, Brees C, Baumgart‐Vogt E, Hongu T, Huybrechts SJ, Van Dijck P, Mannaerts GP, Kanaho Y, Van Veldhoven PP, Fransen M (2009) Small G proteins in peroxisome biogenesis: the potential involvement of ADP‐ribosylation factor 6. BMC Cell Biol 10: 58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Lay D, Grosshans BL, Heid H, Gorgas K, Just WW (2005) Binding and functions of ADP‐ribosylation factor on mammalian and yeast peroxisomes. J Biol Chem 280: 34489–34499 [DOI] [PubMed] [Google Scholar]
- 95. Guo T, Kit YY, Nicaud JM, Le Dall MT, Sears SK, Vali H, Chan H, Rachubinski RA, Titorenko VI (2003) Peroxisome division in the yeast Yarrowia lipolytica is regulated by a signal from inside the peroxisome. J Cell Biol 162: 1255–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Guo T, Gregg C, Boukh‐Viner T, Kyryakov P, Goldberg A, Bourque S, Banu F, Haile S, Milijevic S, San KH et al (2007) A signal from inside the peroxisome initiates its division by promoting the remodeling of the peroxisomal membrane. J Cell Biol 177: 289–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Chang CC, South S, Warren D, Jones J, Moser AB, Moser HW, Gould SJ (1999) Metabolic control of peroxisome abundance. J Cell Sci 112(Pt 10): 1579–1590 [DOI] [PubMed] [Google Scholar]
- 98. Itoyama A, Honsho M, Abe Y, Moser A, Yoshida Y, Fujiki Y (2012) Docosahexaenoic acid mediates peroxisomal elongation, a prerequisite for peroxisome division. J Cell Sci 125: 589–602 [DOI] [PubMed] [Google Scholar]
- 99. Rapoport TA (2007) Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature 450: 663–669 [DOI] [PubMed] [Google Scholar]
- 100. Hegde RS, Keenan RJ (2011) Tail‐anchored membrane protein insertion into the endoplasmic reticulum. Nat Rev Mol Cell Biol 12: 787–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Titorenko VI, Rachubinski RA (1998) Mutants of the yeast Yarrowia lipolytica defective in protein exit from the endoplasmic reticulum are also defective in peroxisome biogenesis. Mol Cell Biol 18: 2789–2803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Kragt A, Voorn‐Brouwer T, van den Berg M, Distel B (2005) Endoplasmic reticulum‐directed Pex3p routes to peroxisomes and restores peroxisome formation in a Saccharomyces cerevisiae pex3Delta strain. J Biol Chem 280: 34350–34357 [DOI] [PubMed] [Google Scholar]
- 103. Elgersma Y, Kwast L, van den Berg M, Snyder WB, Distel B, Subramani S, Tabak HF (1997) Overexpression of Pex15p, a phosphorylated peroxisomal integral membrane protein required for peroxisome assembly in S. cerevisiae, causes proliferation of the endoplasmic reticulum membrane. EMBO J 16: 7326–7341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Karnik SK, Trelease RN (2005) Arabidopsis peroxin 16 coexists at steady state in peroxisomes and endoplasmic reticulum. Plant Physiol 138: 1967–1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Mullen RT, Lisenbee CS, Miernyk JA, Trelease RN (1999) Peroxisomal membrane ascorbate peroxidase is sorted to a membranous network that resembles a subdomain of the endoplasmic reticulum. Plant Cell 11: 2167–2185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Jan CH, Williams CC, Weissman JS (2014) Principles of ER cotranslational translocation revealed by proximity‐specific ribosome profiling. Science 346: 1257521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Schrul B, Kopito RR (2016) Peroxin‐dependent targeting of a lipid‐droplet‐destined membrane protein to ER subdomains. Nat Cell Biol 18: 740–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Thoms S, Harms I, Kalies KU, Gartner J (2012) Peroxisome formation requires the endoplasmic reticulum channel protein Sec61. Traffic 13: 599–609 [DOI] [PubMed] [Google Scholar]
- 109. Zipor G, Haim‐Vilmovsky L, Gelin‐Licht R, Gadir N, Brocard C, Gerst JE (2009) Localization of mRNAs coding for peroxisomal proteins in the yeast, Saccharomyces cerevisiae . Proc Natl Acad Sci USA 106: 19848–19853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Wang F, Chan C, Weir NR, Denic V (2014) The Get1/2 transmembrane complex is an endoplasmic‐reticulum membrane protein insertase. Nature 512: 441–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Steel GJ, Brownsword J, Stirling CJ (2002) Tail‐anchored protein insertion into yeast ER requires a novel posttranslational mechanism which is independent of the SEC machinery. Biochemistry 41: 11914–11920 [DOI] [PubMed] [Google Scholar]
- 112. Stefanovic S, Hegde RS (2007) Identification of a targeting factor for posttranslational membrane protein insertion into the ER. Cell 128: 1147–1159 [DOI] [PubMed] [Google Scholar]
- 113. Vilardi F, Lorenz H, Dobberstein B (2011) WRB is the receptor for TRC40/Asna1‐mediated insertion of tail‐anchored proteins into the ER membrane. J Cell Sci 124: 1301–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Vilardi F, Stephan M, Clancy A, Janshoff A, Schwappach B (2014) WRB and CAML are necessary and sufficient to mediate tail‐anchored protein targeting to the ER membrane. PLoS One 9: e85033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Lam SK, Yoda N, Schekman R (2011) A vesicle carrier that mediates peroxisome protein traffic from the endoplasmic reticulum. Proc Natl Acad Sci USA 108: E51–E52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Agrawal G, Fassas SN, Xia ZJ, Subramani S (2016) Distinct requirements for intra‐ER sorting and budding of peroxisomal membrane proteins from the ER. J Cell Biol 212: 335–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Agrawal G, Shang HH, Xia ZJ, Subramani S (2017) Functional regions of the peroxin Pex19 necessary for peroxisome biogenesis. J Biol Chem 292: 11547–11560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Farré JC, Carolino K, Stasyk OV, Stasyk OG, Hodzic Z, Agrawal G, Till A, Proietto M, Cregg J, Sibirny AA et al (2017) A new yeast peroxin, Pex36, a functional homolog of mammalian PEX16, functions in the ER‐to‐Peroxisome traffic of peroxisomal membrane proteins. J Mol Biol 429: 3743–3762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. van der Zand A, Gent J, Braakman I, Tabak HF (2012) Biochemically distinct vesicles from the endoplasmic reticulum fuse to form peroxisomes. Cell 149: 397–409 [DOI] [PubMed] [Google Scholar]
- 120. Agrawal G, Joshi S, Subramani S (2011) Cell‐free sorting of peroxisomal membrane proteins from the endoplasmic reticulum. Proc Natl Acad Sci USA 108: 9113–9118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Mast FD, Herricks T, Strehler KM, Miller LR, Saleem RA, Rachubinski RA, Aitchison JD (2018) ESCRT‐III is required for scissioning new peroxisomes from the endoplasmic reticulum. J Cell Biol 217: 2087–2102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Mast FD, Jamakhandi A, Saleem RA, Dilworth DJ, Rogers RS, Rachubinski RA, Aitchison JD (2016) Peroxins Pex30 and Pex29 dynamically associate with reticulons to regulate peroxisome biogenesis from the endoplasmic reticulum. J Biol Chem 291: 15408–15427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Titorenko VI, Chan H, Rachubinski RA (2000) Fusion of small peroxisomal vesicles in vitro reconstructs an early step in the in vivo multistep peroxisome assembly pathway of Yarrowia lipolytica . J Cell Biol 148: 29–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Joshi AS, Huang X, Choudhary V, Levine TP, Hu J, Prinz WA (2016) A family of membrane‐shaping proteins at ER subdomains regulates pre‐peroxisomal vesicle biogenesis. J Cell Biol 215: 515–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Tani K, Tagaya M, Yonekawa S, Baba T (2011) Dual function of Sec16B: endoplasmic reticulum‐derived protein secretion and peroxisome biogenesis in mammalian cells. Cell Logist 1: 164–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Yonekawa S, Furuno A, Baba T, Fujiki Y, Ogasawara Y, Yamamoto A, Tagaya M, Tani K (2011) Sec16B is involved in the endoplasmic reticulum export of the peroxisomal membrane biogenesis factor peroxin 16 (Pex16) in mammalian cells. Proc Natl Acad Sci USA 108: 12746–12751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Perry RJ, Mast FD, Rachubinski RA (2009) Endoplasmic reticulum‐associated secretory proteins Sec20p, Sec39p, and Dsl1p are involved in peroxisome biogenesis. Eukaryot Cell 8: 830–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Joshi AS, Nebenfuehr B, Choudhary V, Satpute‐Krishnan P, Levine TP, Golden A, Prinz WA (2018) Lipid droplet and peroxisome biogenesis occur at the same ER subdomains. Nat Commun 9: 2940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Wang S, Idrissi FZ, Hermansson M, Grippa A, Ejsing CS, Carvalho P (2018) Seipin and the membrane‐shaping protein Pex30 cooperate in organelle budding from the endoplasmic reticulum. Nat Commun 9: 2939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. David C, Koch J, Oeljeklaus S, Laernsack A, Melchior S, Wiese S, Schummer A, Erdmann R, Warscheid B, Brocard C (2013) A combined approach of quantitative interaction proteomics and live‐cell imaging reveals a regulatory role for endoplasmic reticulum (ER) reticulon homology proteins in peroxisome biogenesis. Mol Cell Proteomics 12: 2408–2425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Henne WM, Buchkovich NJ, Emr SD (2011) The ESCRT pathway. Dev Cell 21: 77–91 [DOI] [PubMed] [Google Scholar]
- 132. Schoneberg J, Lee IH, Iwasa JH, Hurley JH (2017) Reverse‐topology membrane scission by the ESCRT proteins. Nat Rev Mol Cell Biol 18: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. McCullough J, Clippinger AK, Talledge N, Skowyra ML, Saunders MG, Naismith TV, Colf LA, Afonine P, Arthur C, Sundquist WI et al (2015) Structure and membrane remodeling activity of ESCRT‐III helical polymers. Science 350: 1548–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. McCullough J, Frost A, Sundquist WI (2018) Structures, functions, and dynamics of ESCRT‐III/Vps4 membrane remodeling and fission complexes. Annu Rev Cell Dev Biol 34: 85–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. South ST, Sacksteder KA, Li X, Liu Y, Gould SJ (2000) Inhibitors of COPI and COPII do not block PEX3‐mediated peroxisome synthesis. J Cell Biol 149: 1345–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Haan GJ, Baerends RJ, Krikken AM, Otzen M, Veenhuis M, van der Klei IJ (2006) Reassembly of peroxisomes in Hansenula polymorpha pex3 cells on reintroduction of Pex3p involves the nuclear envelope. FEMS Yeast Res 6: 186–194 [DOI] [PubMed] [Google Scholar]
- 137. Knoops K, Manivannan S, Cepinska MN, Krikken AM, Kram AM, Veenhuis M, van der Klei IJ (2014) Preperoxisomal vesicles can form in the absence of Pex3. J Cell Biol 204: 659–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Wroblewska JP, Cruz‐Zaragoza LD, Yuan W, Schummer A, Chuartzman SG, de Boer R, Oeljeklaus S, Schuldiner M, Zalckvar E, Warscheid B et al (2017) Saccharomyces cerevisiae cells lacking Pex3 contain membrane vesicles that harbor a subset of peroxisomal membrane proteins. Biochim Biophys Acta 1864: 1656–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Hazra PP, Suriapranata I, Snyder WB, Subramani S (2002) Peroxisome remnants in pex3delta cells and the requirement of Pex3p for interactions between the peroxisomal docking and translocation subcomplexes. Traffic 3: 560–754 [DOI] [PubMed] [Google Scholar]
- 140. Blok NB, Tan D, Wang RY, Penczek PA, Baker D, DiMaio F, Rapoport TA, Walz T (2015) Unique double‐ring structure of the peroxisomal Pex1/Pex6 ATPase complex revealed by cryo‐electron microscopy. Proc Natl Acad Sci USA 112: E4017–E4025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Grimm I, Saffian D, Platta HW, Erdmann R (2012) The AAA‐type ATPases Pex1p and Pex6p and their role in peroxisomal matrix protein import in Saccharomyces cerevisiae . Biochim Biophys Acta 1823: 150–158 [DOI] [PubMed] [Google Scholar]
- 142. Law KB, Bronte‐Tinkew D, Di Pietro E, Snowden A, Jones RO, Moser A, Brumell JH, Braverman N, Kim PK (2017) The peroxisomal AAA ATPase complex prevents pexophagy and development of peroxisome biogenesis disorders. Autophagy 13: 868–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Saffian D, Grimm I, Girzalsky W, Erdmann R (2012) ATP‐dependent assembly of the heteromeric Pex1p‐Pex6p‐complex of the peroxisomal matrix protein import machinery. J Struct Biol 179: 126–132 [DOI] [PubMed] [Google Scholar]
- 144. Knoops K, de Boer R, Kram A, van der Klei IJ (2015) Yeast pex1 cells contain peroxisomal ghosts that import matrix proteins upon reintroduction of Pex1. J Cell Biol 211: 955–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Sugiura A, McLelland GL, Fon EA, McBride HM (2014) A new pathway for mitochondrial quality control: mitochondrial‐derived vesicles. EMBO J 33: 2142–2156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Aranovich A, Hua R, Rutenberg AD, Kim PK (2014) PEX16 contributes to peroxisome maintenance by constantly trafficking PEX3 via the ER. J Cell Sci 127: 3675–3686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Rucktaschel R, Halbach A, Girzalsky W, Rottensteiner H, Erdmann R (2010) De novo synthesis of peroxisomes upon mitochondrial targeting of Pex3p. Eur J Cell Biol 89: 947–954 [DOI] [PubMed] [Google Scholar]
- 148. Kim PK, Hettema EH (2015) Multiple pathways for protein transport to peroxisomes. J Mol Biol 427: 1176–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Friedman JR, Lackner LL, West M, DiBenedetto JR, Nunnari J, Voeltz GK (2011) ER tubules mark sites of mitochondrial division. Science 334: 358–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Mattiazzi Usaj M, Brloznik M, Kaferle P, Zitnik M, Wolinski H, Leitner F, Kohlwein SD, Zupan B, Petrovic U (2015) Genome‐wide localization study of yeast Pex11 identifies peroxisome‐mitochondria interactions through the ERMES complex. J Mol Biol 427: 2072–2087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Hoyer MJ, Chitwood PJ, Ebmeier CC, Striepen JF, Qi RZ, Old WM, Voeltz GK (2018) A novel class of ER membrane proteins regulates ER‐associated endosome fission. Cell 175: 254–265 e14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Wu H, Carvalho P, Voeltz GK (2018) Here, there, and everywhere: the importance of ER membrane contact sites. Science 361: eaan5835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Yoboue ED, Sitia R, Simmen T (2018) Redox crosstalk at endoplasmic reticulum (ER) membrane contact sites (MCS) uses toxic waste to deliver messages. Cell Death Dis 9: 331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Meisinger C, Rissler M, Chacinska A, Szklarz LK, Milenkovic D, Kozjak V, Schonfisch B, Lohaus C, Meyer HE, Yaffe MP et al (2004) The mitochondrial morphology protein Mdm10 functions in assembly of the preprotein translocase of the outer membrane. Dev Cell 7: 61–71 [DOI] [PubMed] [Google Scholar]
- 155. Lahiri S, Chao JT, Tavassoli S, Wong AK, Choudhary V, Young BP, Loewen CJ, Prinz WA (2014) A conserved endoplasmic reticulum membrane protein complex (EMC) facilitates phospholipid transfer from the ER to mitochondria. PLoS Biol 12: e1001969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Valm AM, Cohen S, Legant WR, Melunis J, Hershberg U, Wait E, Cohen AR, Davidson MW, Betzig E, Lippincott‐Schwartz J (2017) Applying systems‐level spectral imaging and analysis to reveal the organelle interactome. Nature 546: 162–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Costello JL, Castro IG, Hacker C, Schrader TA, Metz J, Zeuschner D, Azadi AS, Godinho LF, Costina V, Findeisen P et al (2017) ACBD5 and VAPB mediate membrane associations between peroxisomes and the ER. J Cell Biol 216: 331–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Costello JL, Castro IG, Schrader TA, Islinger M, Schrader M (2017) Peroxisomal ACBD4 interacts with VAPB and promotes ER‐peroxisome associations. Cell Cycle 16: 1039–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Murphy SE, Levine TP (2016) VAP, a versatile access point for the endoplasmic reticulum: review and analysis of FFAT‐like motifs in the VAPome. Biochim Biophys Acta 1861: 952–961 [DOI] [PubMed] [Google Scholar]
- 160. Loewen CJ, Roy A, Levine TP (2003) A conserved ER targeting motif in three families of lipid binding proteins and in Opi1p binds VAP. EMBO J 22: 2025–2035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Yagita Y, Shinohara K, Abe Y, Nakagawa K, Al‐Owain M, Alkuraya FS, Fujiki Y (2017) Deficiency of a retinal dystrophy protein, Acyl‐CoA binding domain‐containing 5 (ACBD5), impairs peroxisomal beta‐oxidation of very‐long‐chain fatty acids. J Biol Chem 292: 691–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Raychaudhuri S, Prinz WA (2008) Nonvesicular phospholipid transfer between peroxisomes and the endoplasmic reticulum. Proc Natl Acad Sci USA 105: 15785–15790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Knoblach B, Sun X, Coquelle N, Fagarasanu A, Poirier RL, Rachubinski RA (2013) An ER‐peroxisome tether exerts peroxisome population control in yeast. EMBO J 32: 2439–2453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Saraya R, Cepinska MN, Kiel JA, Veenhuis M, van der Klei IJ (2010) A conserved function for Inp2 in peroxisome inheritance. Biochim Biophys Acta 1803: 617–622 [DOI] [PubMed] [Google Scholar]
- 165. Shai N, Yifrach E, van Roermund CWT, Cohen N, Bibi C, IJlst L, Cavellini L, Meurisse J, Schuster R, Zada L et al (2018) Systematic mapping of contact sites reveals tethers and a function for the peroxisome‐mitochondria contact. Nat Commun 9: 1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. de Brito OM, Scorrano L (2008) Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature 456: 605–610 [DOI] [PubMed] [Google Scholar]
- 167. van Roermund CW, Elgersma Y, Singh N, Wanders RJ, Tabak HF (1995) The membrane of peroxisomes in Saccharomyces cerevisiae is impermeable to NAD(H) and acetyl‐CoA under in vivo conditions. EMBO J 14: 3480–3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Fan J, Li X, Issop L, Culty M, Papadopoulos V (2016) ACBD2/ECI2‐mediated peroxisome‐mitochondria interactions in Leydig cell steroid biosynthesis. Mol Endocrinol 30: 763–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Binns D, Januszewski T, Chen Y, Hill J, Markin VS, Zhao Y, Gilpin C, Chapman KD, Anderson RG, Goodman JM (2006) An intimate collaboration between peroxisomes and lipid bodies. J Cell Biol 173: 719–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Chu BB, Liao YC, Qi W, Xie C, Du X, Wang J, Yang H, Miao HH, Li BL, Song BL (2015) Cholesterol transport through lysosome‐peroxisome membrane contacts. Cell 161: 291–306 [DOI] [PubMed] [Google Scholar]
- 171. Hu A, Zhao XT, Tu H, Xiao T, Fu T, Wang Y, Liu Y, Shi XJ, Luo J, Song BL (2018) PIP4K2A regulates intracellular cholesterol transport through modulating PI(4,5)P2 homeostasis. J Lipid Res 59: 507–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Neal S, Jaeger PA, Duttke SH, Benner C, Glass CK, Ideker T, Hampton RY (2018) The Dfm1 Derlin is required for ERAD retrotranslocation of integral membrane proteins. Mol Cell 69: 915 [DOI] [PubMed] [Google Scholar]
- 173. Weir NR, Kamber RA, Martenson JS, Denic V (2017) The AAA protein Msp1 mediates clearance of excess tail‐anchored proteins from the peroxisomal membrane. Elife 6: e28507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Farré JC, Subramani S (2016) Mechanistic insights into selective autophagy pathways: lessons from yeast. Nat Rev Mol Cell Biol 17: 537–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Fransen M, Lismont C, Walton P (2017) The peroxisome‐mitochondria connection: how and why? Int J Mol Sci 18: E1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Shibata M, Oikawa K, Yoshimoto K, Kondo M, Mano S, Yamada K, Hayashi M, Sakamoto W, Ohsumi Y, Nishimura M (2013) Highly oxidized peroxisomes are selectively degraded via autophagy in Arabidopsis . Plant Cell 25: 4967–4983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Zolman BK, Monroe‐Augustus M, Silva ID, Bartel B (2005) Identification and functional characterization of Arabidopsis PEROXIN4 and the interacting protein PEROXIN22. Plant Cell 17: 3422–3435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Burkhart SE, Lingard MJ, Bartel B (2013) Genetic dissection of peroxisome‐associated matrix protein degradation in Arabidopsis thaliana . Genetics 193: 125–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Hosoi KI, Miyata N, Mukai S, Furuki S, Okumoto K, Cheng EH, Fujiki Y (2017) The VDAC2‐BAK axis regulates peroxisomal membrane permeability. J Cell Biol 216: 709–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Nair DM, Purdue PE, Lazarow PB (2004) Pex7p translocates in and out of peroxisomes in Saccharomyces cerevisiae . J Cell Biol 167: 599–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Hagstrom D, Ma C, Guha‐Polley S, Subramani S (2014) The unique degradation pathway of the PTS2 receptor, Pex7, is dependent on the PTS receptor/coreceptor, Pex5 and Pex20. Mol Biol Cell 25: 2634–2643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Aksam EB, Koek A, Kiel JA, Jourdan S, Veenhuis M, van der Klei IJ (2007) A peroxisomal lon protease and peroxisome degradation by autophagy play key roles in vitality of Hansenula polymorpha cells. Autophagy 3: 96–105 [DOI] [PubMed] [Google Scholar]
- 183. Goto‐Yamada S, Mano S, Oikawa K, Shibata M, Nishimura M (2014) Interaction between chaperone and protease functions of LON2, and autophagy during the functional transition of peroxisomes. Plant Signal Behav 9: e28838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Chen X, Devarajan S, Danda N, Williams C (2018) Insights into the role of the peroxisomal ubiquitination machinery in Pex13p degradation in the yeast Hansenula polymorpha . J Mol Biol 430: 1545–1558 [DOI] [PubMed] [Google Scholar]
- 185. Pan R, Satkovich J, Hu J (2016) E3 ubiquitin ligase SP1 regulates peroxisome biogenesis in Arabidopsis . Proc Natl Acad Sci USA 113: E7307–E7316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Wang W, Subramani S (2017) Role of PEX5 ubiquitination in maintaining peroxisome dynamics and homeostasis. Cell Cycle 16: 2037–2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Bartel B, Farmer LM, Rinaldi MA, Young PG, Danan CH, Burkhart SE (2014) Mutation of the Arabidopsis LON2 peroxisomal protease enhances pexophagy. Autophagy 10: 518–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Bartoszewska M, Williams C, Kikhney A, Opalinski L, van Roermund CW, de Boer R, Veenhuis M, van der Klei IJ (2012) Peroxisomal proteostasis involves a Lon family protein that functions as protease and chaperone. J Biol Chem 287: 27380–27395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Goto‐Yamada S, Mano S, Nakamori C, Kondo M, Yamawaki R, Kato A, Nishimura M (2014) Chaperone and protease functions of LON protease 2 modulate the peroxisomal transition and degradation with autophagy. Plant Cell Physiol 55: 482–496 [DOI] [PubMed] [Google Scholar]


