Abstract
CDC14A codes for a conserved proline‐directed phosphatase, and mutations in the gene are associated with autosomal‐recessive severe to profound deafness, due to defective kinocilia. A role of CDC14A in cilia formation has also been described in other organisms. However, how human CDC14A impacts on cilia formation remains unclear. Here, we show that human RPE1 hCDC14APD cells, encoding a phosphatase dead version of hCDC14A, have longer cilia than wild‐type cells, while hCDC14A overexpression reduces cilia formation. Phospho‐proteome analysis of ciliated RPE1 cells identified actin‐associated and microtubule binding proteins regulating cilia length as hCDC14A substrates, including the actin‐binding protein drebrin. Indeed, we find that hCDC14A counteracts the CDK5‐dependent phosphorylation of drebrin at S142 during ciliogenesis. Further, we show that drebrin and hCDC14A regulate the recruitment of the actin organizer Arp2 to centrosomes. In addition, during ciliogenesis hCDC14A also regulates endocytosis and targeting of myosin Va vesicles to the basal body in a drebrin‐independent manner, indicating that it impacts primary cilia formation in a multilayered manner.
Keywords: actin, CDK5, DBN1, hCDC14A, primary cilium
Subject Categories: Cell Adhesion, Polarity & Cytoskeleton; Membrane & Intracellular Transport; Signal Transduction
Introduction
Cilia are dynamic, microtubule‐based organelles with functions in cell motility, transport of fluids (motile cilia) as well as sensory signaling (primary cilia) 1, 2. The steady state length of a cilium is determined by a series of events. These include the docking of vesicles derived from the Golgi or cytoplasmic membrane to the base of the cilium, the transport of cargo to the cilia tip by the microtubule anterograde intraflagellar transport (IFT) system, and the retrograde IFT‐mediated transport of turnover products back to the base 3. Modulation of these events can affect cilia length in the one or other direction. In addition, shorter cilia can also be the consequence of premature activation of proteins that normally promote cilia disassembly upon cell cycle entry. An example for this is the kinase CDK5 that induces, as one mechanism, proteolytic destabilization of the cilia disassembly factor “Nuclear Distribution protein nudE homolog 1” (NDE1) during ciliogenesis 4.
Importantly, the length of the cilium is a characteristic parameter that varies between cell types and probably has implications for cilia function. Indication for this notion comes from the observation that mutations in the genes coding for the CDK10/cyclin M protein kinase complex, which are the cause of STAR syndrome, affect cilia length 5, 6.
The actin cytoskeleton has an ill‐defined role in ciliation. Addition of low concentrations of the F‐actin‐depolymerizing drug cytochalasin D promotes cilia elongation partly by the regulation of vesicle trafficking, the inhibition of cilia disassembly factors, and the recruitment of actin‐binding proteins to the cilium 7, 8, 9. Recently, it was suggested that a branched, Arp2/3 organized actin network surrounds the centrosome and promotes transport of preciliary vesicles to the basal body via the action of the myosin Va motor protein 10. An additional function of the actin network and the actin regulator drebrin (DBN1) in ciliogenesis is the release of vesicles, so‐called ectosomes, from the tip of cilia 11.
The modulation of cilia length by the actin cytoskeleton raises the question about kinases and phosphatases that regulate the actin network. The CDK10/cyclin M protein kinase was proposed to negatively regulate ciliogenesis by inducing actin polymerization 5, 6. The proline‐directed phosphatase hCDC14A has the potential to counteract kinases that control the actin network during ciliogenesis because it is associated with actin filaments and in addition plays a role in ciliogenesis 12, 13, 14. Recently, it has been reported that nonsense mutations in human hCDC14A gene cause autosomal‐recessive severe to profound deafness. Deafness is caused by defective kinocilia of developing cochlear hair cells. In addition, the persistent kinocilia of vestibular hair cells in the organ of corti are also defective 15, 16. Furthermore, shorter cilia have been reported in zebrafish upon cdc14a or cdc14b depletion with morpholinos 14, 15. However, it is still an open question whether CDC14A regulates primary cilia in human cells and if so, how CDC14A modulates ciliogenesis on a molecular level.
Here, we have addressed the function of hCDC14A in ciliogenesis using serum‐starved human retinal pigmented epithelial (hTERT‐RPE1; herein, RPE1) cells as a model. We show that RPE1 cells that express a phosphatase dead version of hCDC14A (hCDC14A PD) have longer cilia than WT control cells because cilia continue to elongate in hCDC14A PD cells while they stop growing in WT cells. Phospho‐proteome and proximity‐dependent biotin identification (BioID) analyses of ciliated cells identified hCDC14A substrates including the actin bundler drebrin (DBN1) with functions in ciliogenesis 11. We show the counteracting phospho‐regulation of DBN1 at serine residue 142 by the proline‐directed kinase CDK5 17 and hCDC14A phosphatase during ciliogenesis. DBN1 and hCDC14A contribute to cilia length control by regulating the actin nucleator Arp2 at the basal body. hCDC14A has additional, DBN1 independent functions in endocytosis and transport of myosin Va marked vesicles to the basal body.
Results
hCDC14A PD RPE1 cells grow longer cilia than WT cells
To understand the function of hCDC14A in ciliogenesis, we analyzed cilia formation in serum‐starved hCDC14A PD RPE1 cells in comparison with WT. hCDC14A PD codes for an inactive hCDC14A phosphatase that carries a deletion of 77 amino acids (203–279) around the active cysteine 13, 18. Upon serum starvation, hCDC14A PD cells formed cilia with similar efficiency as WT cells independent of the cell's confluency that has an impact on ciliogenesis (Fig 1A and B) 7. However, 48 h after serum starvation, cilia of hCDC14A PD cells were longer in comparison with those of WT (Fig 1A and C). siRNA‐mediated depletion of hCDC14A also caused longer cilia compared to non‐specific control (NSC)‐treated RPE1 cells (Fig 1D). In addition, hCDC14 PD/+ cells carrying only one WT hCDC14A gene displayed an intermediate cilia length phenotype (Fig EV1A). Conversely, elongated cilia were not observed in RPE1 cells lacking the hCDC14A paralogue hCDC14B (hCDC14B −/−) suggesting that the cilia phenotype is specific to hCDC14A (Fig EV1B) 19.
Figure 1. Ablation of hCDC14A phosphatase activity in human RPE1 cells leads to the elongation of the primary cilium.
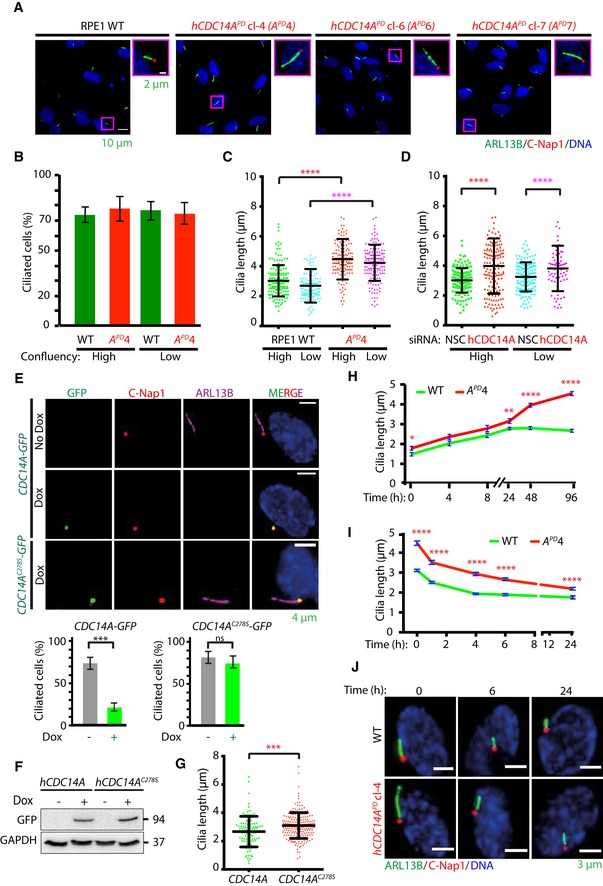
-
AWT and hCDC14A PD RPE1 cells were serum starved for 48 h for inducing ciliogenesis prior to fixation. Cilia were stained with Arl13B (green), while the basal bodies were marked with C‐Nap1 (red). DNA was stained with DAPI. Cilia in the magenta box are shown enlarged on the right picture.
-
B, CPercent of ciliated cells, as well as the length of cilia, was quantified from (A). Cilia length was measured by a semi‐automated ImageJ macro as described in Materials and Methods. Data points are the average of three independent experiments with N = 150 cilia for each experiment and condition. Error bars represent the mean ± SD. ****P ≤ 0.0001. High means 80–90% and low 40–50% confluency of RPE1 cells.
-
DElongated cilia phenotype was confirmed by siRNA‐mediated knockdown of hCDC14A. Three independent experiments, N = 150 cilia for each experiment and condition. Mean ± SD. ****P ≤ 0.0001. For definition of confluency see (C).
-
EhCDC14A‐GFP and hCDC14A C278S ‐GFP under control of the TetON promoter were stably integrated into RPE1 cells. hCDC14A‐GFP and hCDC14AC278S‐GFP localized to the C‐Nap1 marked basal body upon Dox induction. In addition, expression of hCDC14A‐GFP but not the phosphatase dead hCDC14A C278S ‐GFP mutant inhibited cilia formation. Ciliated cells were quantified 48 h after serum starvation with and without Dox addition. Three independent experiments, N = 150 cilia for each experiment and condition. Mean ± SD. ***P ≤ 0.001; ns: not significant.
-
FExpression of hCDC14A‐GFP and hCDC14A C278S ‐GFP was validated by immunoblot analysis with anti‐GFP antibodies. GAPDH was the loading control.
-
GRPE1 cells overexpressing hCDC14A C278S have longer cilia than those overexpressing hCDC14A. Three independent experiments, N = 100 cilia for hCDC14A expressing cells and 200 for hCDC14A C278S expressing cells. Mean ± SD. ***P ≤ 0.001.
-
HThe dynamics of ciliogenesis was determined by serum starving WT and hCDC14A PD cells for different time points. The cilia from hCDC14A PD cells continue to elongate even after 48 h of starvation, whereas those from WT cells reached constant length within 12–24 h. Three independent experiments, N = 150 cilia for each experiment and condition. Mean ± SEM. *P ≤ 0.05, **P ≤ 0.01; ****P ≤ 0.0001.
-
IIn a time course experiment, the cilia disassembly kinetics was measured by first inducing ciliation through 48 h of serum starvation prior to incubation with serum containing media for different time points. Measurement of cilia length indicated the comparable rates of cilia disassembly in RPE1 WT and hCDC14A PD cells. Cilia disassembly rates are shown in Fig EV1D. Three independent experiments, N = 150 cilia for each experiment and condition. Mean ± SEM. ****P < 0.0001.
-
JImages of cilia of WT RPE1 and hCDC14A PD RPE1 cells from (I), 0, 6, and 24 h after serum addition. Fixed cells were analyzed for cilia length.
Figure EV1. hCDC14A localizes to basal body and regulates cilia length.
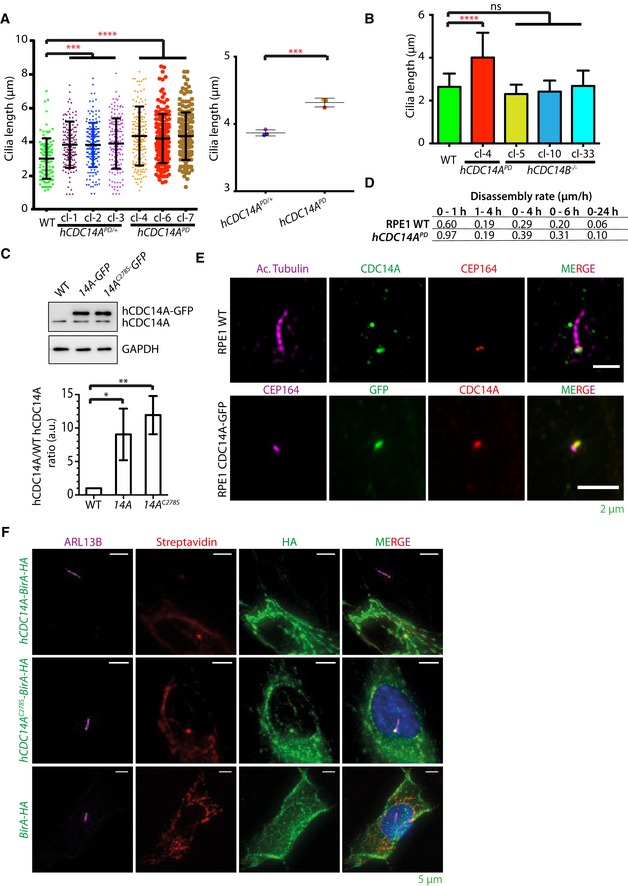
- One allele targeted hCDC14 PD/+ cells displayed an intermediate cilia length phenotype compared to WT and hCDC14 PD cells. Three independent clones of hCDC14 PD/+ and hCDC14A PD cells were used for this experiment. Three independent experiments, 150 cilia per condition and experiment. Mean ± SD. ***P ≤ 0.001; ****P ≤ 0.0001. One‐way ANOVA. The right side of the subfigure shows the comparison of the average cilia length of three independent hCDC14 PD/+ clonal cell lines with three independent hCDC14 PD clones. ***P ≤ 0.001. Unpaired Student's t‐test.
- Elongated cilia were not observed in hCDC14B −/− RPE1 cells suggesting that the cilia phenotype is specific to hCDC14A. Three independent hCDC14B −/− RPE1 cell lines were analyzed. Three independent experiments, 150 cilia per condition and experiment. Mean ± SD. ****P ≤ 0.0001, ns: not significant.
- Around 10‐fold overexpression of hCDC14A‐GFP was observed compared to endogenous hCDC14A. Three independent experiments. Mean ± SD. *P ≤ 0.05, **P ≤ 0.01.
- Cilia disassembly rates from Fig 1I.
- Endogenous hCDC14A could be detected at the basal body of RPE1 WT cells under ciliogenic condition (upper panel). Endogenous hCDC14A signal co‐localizes with the GFP signal from hCDC14A‐GFP expressing cells (lower panel).
- hCDC14A‐BirA and hCDC14AC278S‐BirA could be detected at the basal body, whereas BirA showed a diffused cellular signal without any basal body enrichment.
We next tested whether hCDC14A overexpression had the reverse phenotype of hCDC14A inactivation. hCDC14A and the phosphatase dead version hCDC14A C278S were expressed under control of the doxycycline (Dox)‐inducible TetON promoter. hCDC14A and hCDC14A C278S were expressed about ~10‐fold higher than endogenous WT CDC14A (Fig EV1C). This level of overexpression does not affect viability or the cell cycle profile in cycling cells 12. Overexpression of hCDC14A but not hCDC14A C278S inhibited cilia formation in most RPE1 cells (Fig 1E and F). Of the residual ~20% TetON‐hCDC14A cells with cilia, cilia were shorter than those in cells overexpressing hCDC14A C278S (Fig 1G). Taken together, hCDC14A regulates the length of cilia and elevated hCDC14A levels impair cilia formation.
Mutations can affect cilia length because of the impairment of cilia disassembly when cells re‐enter the cell cycle upon growth factor addition 4. Alternatively, continuous cilia elongation over the normal steady state point may be responsible for cilia elongation in CDC14APD cells. To evaluate these possibilities, we compared cilia formation and cilia disassembly of WT and hCDC14A PD cells. At the time of serum starvation, cilia of hCDC14A PD cells were slightly longer in comparison with RPE1 WT cells (Fig 1H). Elongation of cilia in response to serum starvation plateaued in WT cells after 12 h with an average cilium length of 2.8 μm. In contrast, cilia continued to elongate in hCDC14A PD cells over 96 h reaching a length of 4.6 μm (Fig 1H). Cilia disassembly rates were even slightly higher in hCDC14A PD cells in comparison with WT cells (Figs 1I and J, and EV1D). This suggests that it is the adjustment of cilia length but not cilia formation or disassembly, which is defective in hCDC14A PD cells.
We next characterized the structure of hCDC14A PD cilia in comparison with WT. Electron microscopy did not show obvious defects in the basal body and axoneme organization of hCDC14A PD cilia (Fig EV2A–C). In addition, distal appendage protein CEP164, the subdistal appendage proteins CEP170, ODF2, and ninein, the transition zone protein NPHP1, the cilia shaft protein ARL13B, the transport protein IFT88, the linker proteins C‐Nap1 and rootletin, and the pericentriolar proteins γ‐tubulin and pericentrin were similarly associated with cilia in WT and hCDC14A PD cells (Fig EV3A–G). Thus, cilia of hCDC14A PD cells do not show obvious structural defects.
Figure EV2. Electron micrograph of cilia from RPE1 WT and hCDC14 APD cells.
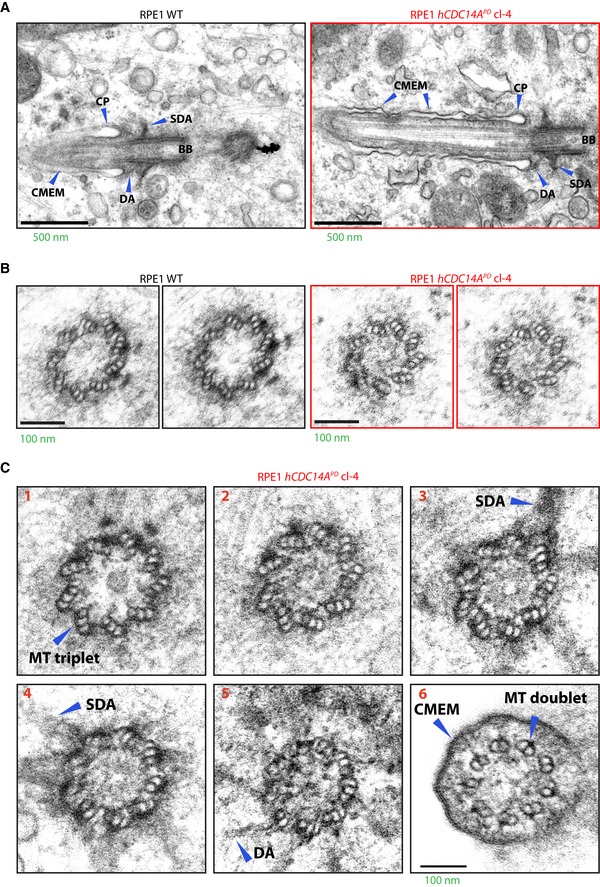
- Electron micrograph of cilia from 48 h serum‐starved RPE1 WT and hCDC14A PD cells. Basal body and the ciliary axoneme showed no gross structural defects in the longitudinal ciliary sections from hCDC14A PD cells.
- Similarly, the cross‐sections of the basal body from WT and hCDC14A PD cells represented a well‐organized triplet microtubule structure.
- Serial cross‐sections from a cilium of a hCDC14A PD cell. These sections indicate the normal organization of microtubules in the basal body (1‐4), transition zone (5), and axoneme (6).
Figure EV3. Association of ciliary components in RPE1 WT and hCDC14 APD cells.
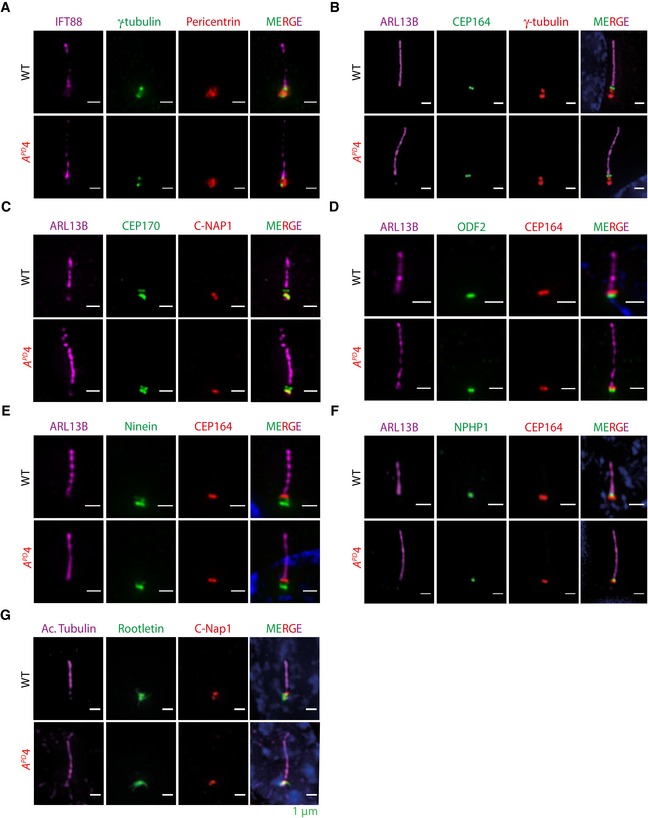
-
A–GCilia from hCDC14A PD cells are structurally similar to those from WT cells. Markers representing various basal body localizations showed no obvious localization defects for (A) pericentrin, (B, E and F) the distal appendage protein CEP164, (C) the subdistal appendage proteins CEP170, (D) ODF2, (E) ninein, (C and G) the proximal end protein C‐Nap1, (F) the transition zone protein NPHP1, and (G) the ciliary rootlet component rootletin. Scale bars: 1 μm.
hCDC14A localizes to the basal body and the actin cytoskeleton of ciliated cells
CDC14A localizes to the axoneme of kinocilia in inner ear cells 16. The localization of hCDC14A in cells with a primary cilium is, however, unclear. We first analyzed hCDC14A localization with an established antibody 13. Endogenous hCDC14A localizes with the actin cytoskeleton and was detected at the basal body of serum‐starved, ciliated RPE1 cells (Fig EV1E, top) 13. Because the hCDC14A antibody signal was relatively weak, we continued the localization analysis in RPE1 cells that stably expressed hCDC14A‐GFP or the phosphatase dead version hCDC14A C278S ‐GFP under the control of TetON promoter. Consistent with the antibody result, hCDC14A‐GFP was detected along actin cables in ciliated cells (Fig 2A and B). In addition, hCDC14A‐GFP and hCDC14AC278S‐GFP both associated with the basal body of cilia (Fig 1E). Similarly, hCDC14A‐GFP was detected at the basal body with an anti‐hCDC14A antibody (Fig EV1E, bottom). Because hCDC14A‐GFP expression reduced the number of cilia (Fig 1E), we fine mapped hCDC14A localization in ciliated cells expressing hCDC14A C278S ‐GFP. hCDC14AC278S‐GFP localized at two sites of the basal body. The most intense signal was at the proximal end of the basal body that contains the linker protein C‐Nap1 (encoded by CEP250) 20, 21 (Fig 2C and D). The second pool, relatively weaker in intensity, was close to the distal end of the basal body (Fig 2C and D). hCDC14A localized underneath the distal appendage marker CEP164 and overlapped in its localization with the subdistal appendage proteins ODF2 and ninein 22, 23. Thus, in ciliated cells, hCDC14A associates with the actin cytoskeleton and the basal body.
Figure 2. hCDC14A localizes to actin cytoskeleton and the basal body during ciliogenesis.
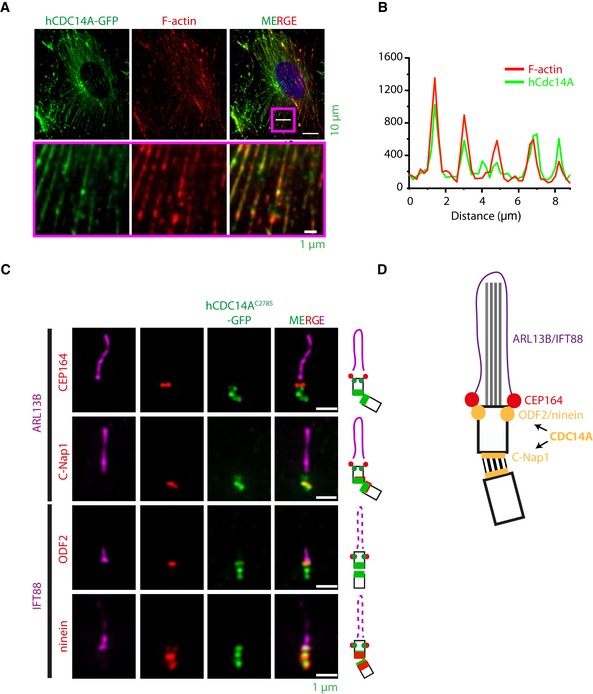
- TetON‐hCDC14A‐GFP RPE1 cells were serum starved for 48 h prior to fixation with paraformaldehyde and immunofluorescence microscopy. hCDC14A‐GFP co‐localizes with Phalloidin 555 conjugated dye that marks F‐actin fibers. The boxed area was 10‐fold enlarged and displayed below the corresponding images to indicate co‐localization of hCDC14A‐GFP and F‐actin.
- Line scan inside the magenta colored box showed in (A).
- Sub‐centrosomal localization of hCDC14AC278S‐GFP of methanol‐fixed cells was determined by correlative positioning of the GFP signal with the proteins Arl13B and IFT88 (cilia), C‐Nap1/ninein (proximal end), CEP164 (distal appendage), and ODF2/ninein (subdistal appendage). Results are fitted in the cartoons adjacent to the respective panels for better visualization.
- The cartoon summarizes the localization of hCDC14A on cilia. The distal appendage protein CEP164 marks the mother basal body. The C‐Nap1 organized centrosome linker connects the mother with the daughter basal body (black lines). ODF2 and ninein mark the subdistal appendages. Note, ninein has a second localization at the proximal end of basal bodies. hCDC14A localized to the subdistal appendages and the C‐Nap1 marked proximal end of the basal bodies.
hCDC14A dephosphorylates actin‐associated proteins during ciliogenesis
We applied two approaches, stable isotope labeling with amino acids in cell culture (SILAC) 24 and proximity BioID analysis 25, to identify substrates and proximity neighbors of hCDC14A during ciliogenesis. Comparison of the phospho‐proteomes of non‐induced and induced hCDC14A‐GFP expressing cells by mass spectrometric analysis identified actin‐associated proteins such as drebrin (DBN1), synaptopodin (SYNPO), LIM domain and actin‐binding 1 protein (LIMA1) and the microtubule‐associated protein MAP4 that functions in cilia length control 26 (Fig 3A–C and Dataset EV1). Interestingly, most of the phospho‐sites that were dephosphorylated by hCDC14A followed the pSP consensus of CDC14 phosphatases (Fig 3D) 12, 27, 28. However, compared to human cycling cells, in which hCDC14A dephosphorylated in ~40% of the cases pSPxK/R sites 12, the preference for positively charged amino acids at position pS+3 was reduced to 28% in cells under serum starvation (Fig 3D).
Figure 3. Identification of substrates and proximity neighbors of hCDC14A during ciliogenesis.
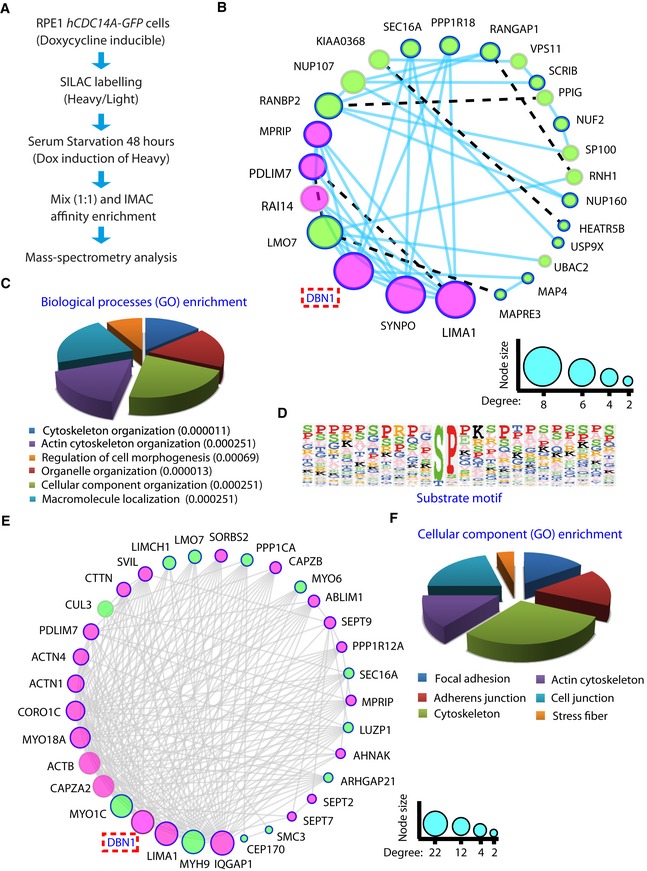
- Experimental scheme for hCDC14A‐GFP expression combined with phospho‐proteome (SILAC) analysis.
- Molecular network of the hypo‐phosphorylated proteins in ciliated hCDC14A‐GFP RPE1 cells. Gene network was constructed in Cytoscape using the function prediction GeneMANIA app. GeneMANIA identifies the most related genes to a query gene set (hits) using a guilt‐by‐association approach. The plugin uses a large database of functional interaction networks from multiple organisms to construct a molecular network using the query genes (hits). Physical interactions are denoted by solid lines (edges), whereas the shared protein domain interactions are represented by broken lines. The extent of interactions (degree) is indicated by the node size. The query proteins are marked with thick borders and the actin‐related proteins are marked with magenta colored nodes.
- Gene Ontology (GO) analysis of the proteins that are at least twofold hypo‐phosphorylated in the heavy (hCDC14A overexpressed) samples. Values inside the parentheses indicate P‐values.
- Substrate motif identified by analyzing the sequences of hypo‐phosphorylated peptides in Perseus.
- Molecular network of the neighbors of hCDC14A and hCDC14AC278S identified by proximity‐dependent biotin identification (BioID) during ciliogenesis. Gene network was constructed in Cytoscape as described in (B) by considering the physical interactions only.
- Gene Ontology (GO) analysis of the proteins from which at least five peptides were twofold enriched in the hCDC14A‐BirA and hCDC14AC278S‐BirA samples compared to BirA control.
The proximity BioID analysis 25 was performed with both hCDC14A WT and the phosphatase dead hCDC14AC278S N‐terminally tagged to the promiscuous biotin protein ligase (BirA). Consistent with the localization of the protein, BirA fused to hCDC14A or hCDC14AC278S biotinylated proteins at the basal body and along actin cables (Fig EV1F) 13. During ciliogenesis, hCDC14A and hCDC14AC278S showed proximity interactions with the actin‐associated proteins DBN1 and LIMA1, and the subdistal appendage protein CEP170 supporting the phospho‐proteome analysis (Fig 3E and F, and Dataset EV2; only BioID hits that were identified by hCDC14A and hCDC14AC278S are shown).
As a secondary screen, we tested proteins that were identified by the phospho‐proteome and BioID analyses for their impact on cilia length control. Proteins were depleted by siRNA during ciliogenesis of RPE1 hCDC14A WT and hCDC14A PD cells (Fig EV4A). Cilia length was subsequently measured by indirect immunofluorescence microscopy. Depletion of a number of proteins such as the subdistal appendage protein CEP170 had no impact on the cilia length in hCDC14A WT and hCDC14A PD cells (Fig EV4B and C). Depletion of DBN1, microtubule crosslinking factor 1 (MTCL1), or ubiquitin‐associated protein 2‐like (UBAP2L) significantly increased the length of cilia in WT cells without affecting those in hCDC14A PD cells (Fig EV4B–E). On the contrary, siRNA of “KN motif and ankyrin repeat domain‐containing protein 2” (KANK2), MAP4, PDZ and LIM Domain 7 (PDLIM7) or 40S ribosomal protein S2 (RPS2) reduced the length of hCDC14A PD cilia without having a significant impact on WT cilia (Fig EV4B–E). We reasoned that the group including DBN1 contains proteins whose loss of function or inhibition by either siRNA depletion in WT cells or hyper‐phosphorylation in hCDC14A PD cells promotes cilia elongation. In the second, MAP4 containing group, the hyper‐phosphorylated protein of hCDC14A PD cells would be hyperactive promoting longer cilia. Finally, knockdown of the actin regulator cortactin (CTTN) and the actin‐binding protein “LIM domain only protein 7” (LMO7) was found to increase cilia length in both WT and hCDC14A PD cells indicating that their functions in cilia length control do not require hCDC14A. In summary, hCDC14A phospho‐proteome and BioID analyses identified putative CDC14A substrates with functions in cilia length control.
Figure EV4. siRNA‐based secondary screen to validate substrates.

-
AExperimental scheme for siRNA screen for hCDC14A substrate(s) essential for cilia length control.
-
B–EAltogether we analyzed 24 proteins in this secondary screen. 14 proteins were chosen because they were hits in all four experiments (two replicates each for overexpression and BioID MS). Ten proteins appeared in at least two experiments (one replicate each for overexpression and BioID). Indicated proteins were depleted by siRNA during ciliogenesis in RPE1 WT and hCDC14A PD cells. Cilia length was subsequently measured by indirect immunofluorescence microscopy. The experiment was repeated twice, and 70–150 cilia were measured for each experiment; Mean ± SD. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001. One‐way ANOVA. All the other depletions did not significantly affect cilia length compared to the NSC control. Red dotted lines indicate the mean values of NSC.
hCDC14A regulates phosphorylation of DBN1 during ciliogenesis
Because DBN1 is a known phosphoprotein with functions in ciliogenesis, we analyzed the phospho‐regulation of this protein by hCDC14A in ciliated cells 11, 17. In cycling cells, serine 142 (S142) next to a proline residue of DBN1 is phosphorylated by the kinase CDK5 17, 29. This is the DBN1 phospho‐site that was dephosphorylated by hCDC14A in ciliated cells (Dataset EV1). To confirm dephosphorylation of pS142 in DBN1 by hCDC14A during ciliogenesis, we followed DBN1pS142 with a phospho‐specific antibody. Expression of Dox‐induced hCDC14A‐GFP but not the inactive hCDC14A C278S ‐GFP or GFP dephosphorylated DBN1pS142 (Fig 4A and B). In addition, S142 of DBN1 was hyper‐phosphorylated in hCDC14A PD RPE1 cell lines during ciliogenesis (independent clones 4, 6, and 7) compared to the WT control while DBN1 proteins levels were not reduced (Fig 4C). The identity of the DBN1pS142 and DBN1 bands was confirmed by their reduction in response to DBN1 siRNA (Fig 4C). Thus, DBN1pS142 is hyper‐phosphorylated in hCDC14A PD RPE1 cell during ciliogenesis in comparison with the RPE1 WT cells.
Figure 4. The phosphoprotein DBN1 is a substrate for hCDC14A.
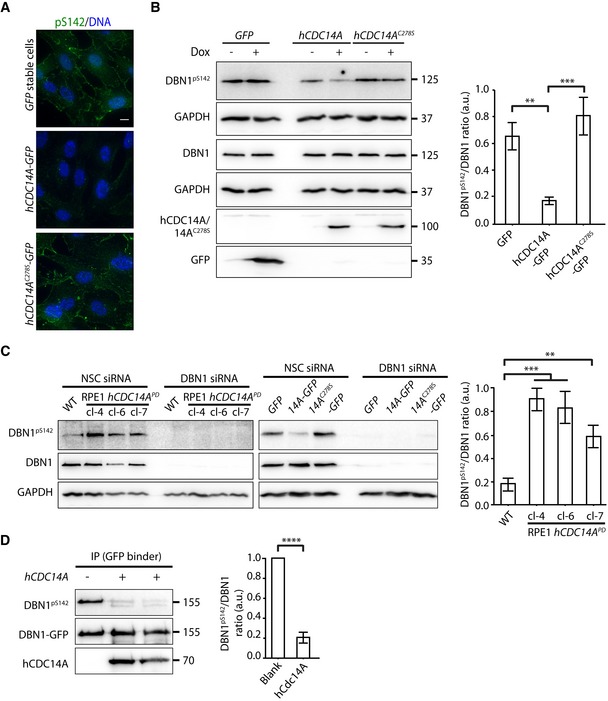
- hCDC14A, not the inactive hCDC14AC278S, dephosphorylates the serine residue 142 of DBN1 in vivo. RPE1 cells expressing GFP, hCDC14A‐GFP, and hCDC14A C278S ‐GFP under TetON promoter were serum starved in presence or absence of Dox for 48 h prior to fixation for immunofluorescence. DBN1pS142 was detected with a phospho‐specific antibody. DNA was stained with DAPI. Scale bar = 10 μm.
- Immunoblot analysis of samples from (A). The DBN1pS142/DBN1 ratio was densitometrically measured from the Dox treated samples and represented in the graph next to the immunoblot. In this experiment, two GAPDH blots were used to normalize protein levels because DBN1 and DBN1pS142 were analyzed in separated blots. Three independent experiments. Mean ± SD. **P ≤ 0.01, ***P ≤ 0.001.
- DBN1S142 was hyper‐phosphorylated in hCDC14A PD cell lines during ciliogenesis compared to the WT control. In this experiment, we sequentially analyzed DBN1pS142 and DBN1 on the same blot. The specificity of the DBN1pS142 antibody was validated by siRNA‐mediated knockdown of DBN1 in both hCDC14A PD cells and GFP/hCDC14A‐GFP/hCDC14AC278S‐GFP stable cell lines. Three independent experiments. Mean ± SD. **P ≤ 0.01, ***P ≤ 0.001.
- In vitro dephosphorylation of DBN1 by purified hCDC14A. DBN1‐GFP was transfected into HEK293T cells and expressed for 48 h. DBN1‐GFP was immunoprecipitated from cell lysates with GFP binder. The immunoprecipitated phospho‐DBN1 was then incubated with and without recombinant hCDC14A in vitro. The DBN1pS142/DBN1 ratio was densitometrically measured. Three independent experiments. Mean ± SD. ****P ≤ 0.0001.
To confirm the direct dephosphorylation of DBN1pS142 by hCDC14A, we incubated immunoprecipitated, phosphorylated DBN1 with recombinant and purified hCDC14A. DBN1pS412 was efficiently dephosphorylated by recombinant hCDC14A indicating that hCDC14A directly dephosphorylates DBN1 (Fig 4D). Taken together, these data suggest that DBN1pS142 is a direct substrate of hCDC14A during ciliogenesis.
Phospho‐regulation of DBN1 by hCDC14A contributes to cilia length control
To confirm the function of DBN1 in cilia length regulation, we constructed DBN1 RPE1 knockout (KO) cell lines using CRISPR/Cas9 technology (Fig 5A). Three independent gRNAs targeting exon 3, 5, or 7 of DBN1 were used to construct DBN1 KO cells (Fig 5A; gRNA1‐3). We have designed the gRNAs in such a way that even a possible truncated N‐terminal version of DBN1 would lack the critical domains AB1 (actin‐binding region 1) and AB2 (actin‐binding region 2; Fig 5A). Analysis of the DNA sequences from several clonal cell lines confirmed disrupting mutations in both DBN1 alleles (Fig 5B). As expected from gene disruption, DBN1 was no longer detected in DBN1 KO cell lines by immunoblotting with monoclonal antibody generated using a synthetic C‐terminal peptide (aa 632–649; Fig 5C). One clone from each gRNA (1.16, 2.18, and 3.3) was used for further ciliogenesis experiments and found to assemble longer cilia than WT cells (Fig 5D). Clone 2.19 did not affect cilia length; however, this cell line did not carry a mutation in DBN1 and the DBN1 protein was detected by immunoblotting (Fig 5C). This confirms the role of DBN1 in cilia length regulation.
Figure 5. Phospho‐regulation of DBN1 is critical for cilia length control.
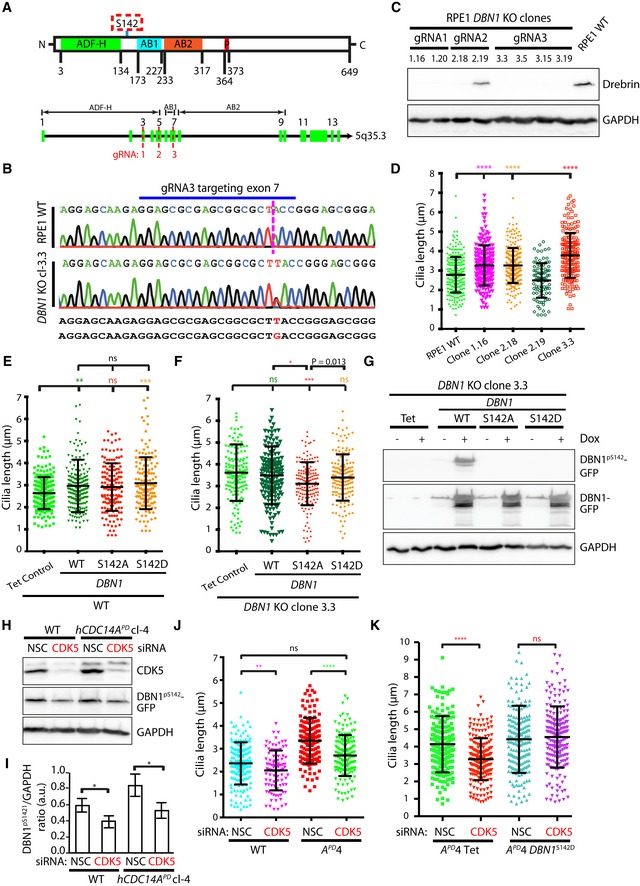
- Schematic representation of different domains of DBN1 (ADF‐H: ADF homology domain, AB1: actin‐binding region 1, AB2: actin‐binding region 2, P: proline‐rich region) 44. The numbers represent the amino acid positions of the denoted domains. Three independent gRNAs targeting different exons (3, 5, 7) of DBN1 were used to generate DBN1 KO cells.
- Successful targeting of DBN1 was confirmed by sequencing the PCR amplicons surrounding the locus. For example, in clone 3.3, there was a single base insertion in both DBN1 alleles (T in the one, G in the other) to cause disruption of protein expression through the introduction of a premature stop codon.
- The absence of DBN1 protein in the targeted clones was further verified by immunoblotting of the whole cell lysate. GAPDH is the loading control.
- One targeted clone from each gRNA (1.16, 2.18, 3.3) including the non‐targeted clone 2.19 and WT control was serum starved for 48 h to induce ciliogenesis. Subsequently, cilia length was measured. The cilia from DBN1 KO cells were significantly longer than those from WT or non‐targeted controls. Three independent experiments, N = 150 cilia for each experiment and condition. Mean ± SD. ****P ≤ 0.0001.
- Overexpression of DBN1, DBN1 S142A, and DBN1 S142D in RPE1 WT cells. Three independent experiments, N = 150 cilia for each experiment and condition. Mean ± SD. **P ≤ 0.01; ***P ≤ 0.001; ns: not specific.
- The phospho‐inhibitory (S142A) version of DBN1 rescued the elongated cilia of DBN1 KO cells. Three independent experiments, N = 150 cilia for each experiment and condition. Mean ± SD. *P ≤ 0.05; ***P ≤ 0.001; ns: not significant. Pairwise comparison was done with unpaired Student's t‐test and the P‐value was written in the figure. Global comparison was done with one‐way ANOVA and the P‐values were marked with stars.
- The overexpressed WT DBN1 was phosphorylated in cells as detected by phospho‐specific DBN1pS142 antibodies. The DBN1pS142 antibodies did not detect the overexpressed DBN1 S142A or DBN1 S142D.
- Immunoblot analysis confirming the successful siRNA depletion of CDK5 and indicating hypo‐phosphorylation of DBN1S142 in response to CDK5 depletion.
- The DBN1pS142/GAPDH ratio from (H) was densitometrically measured. Three independent experiments. Mean ± SD. *P ≤ 0.05.
- Depletion of CDK5 decreased the cilia length in WT cells. It restored cilia length in hCDC14A PD cells. Three independent experiments, N = 150 cilia for each experiment and condition. Mean ± SD. **P ≤ 0.01; ****P ≤ 0.0001; ns: not significant.
- CDK5‐mediated cilia length decrease can be completely blocked by expression of the phospho‐mimetic DBN1 S142D in hCDC14A PD cells. Three independent experiments; 150 cells per experiment and condition. Mean ± SD; ns = not significant; ****P ≤ 0.0001.
To test the impact of S142 phospho‐site in DBN1 on cilia length control, we expressed DBN1 versions of WT, phospho‐inhibitory (S142A), and phospho‐mimetic (S142D) in RPE1 WT and DBN1 KO RPE1 cells under control of the TetON promoter. All TetON‐DBN1 constructs were overexpressed about 10‐fold in WT (Fig EV5A) and DBN1 KO (Fig EV5B) cells. Overexpression of DBN1 or DBN1 S142D in RPE1 WT cells significantly increased cilia length, while overexpression of DBN1 S142A did not impact cilia length compared to the Tet control cells (Figs 5E and EV5A). Because of this overexpression effect, we did not observe rescue of the cilia length phenotype by TetON‐DBN1 in DBN1 KO cells (Figs 5F and EV5B). Interestingly, however, the non‐phosphorylated DBN1S142A 17 significantly reduced cilia length in comparison with DBN1WT or the phospho‐mimetic DBN1S142D in DBN1 KO cells (Fig 5F).
Figure EV5. DBN1 co‐localizes with actin fibers.

-
A, BSimilar extent of expression of all three DBN1 versions (WT, S142A, S142D) were observed in both WT (A) and DBN1 KO (B) cells (around 10‐fold compared to WT cells). Note that the WT and WT with DBN1 WT, DBN1 S142A, and DBN1 S142D samples were analyzed on the same blotting membrane. The DBN1/GAPDH ratios were densitometrically measured. The results are mean ± SD of two independent experiments. *P ≤ 0.05. One‐way ANOVA.
-
CDBN1 could not be detected in cilia. Antibody localization of DBN1 in ciliated RPE1 cells. CEP164 was used as a marker for the mother centriole and ARL13B for the cilium. The line scan on the right shows signal distribution along the cilium in the boxed on the left area for CEP164, ARL13B, and DBN1 (green).
-
DTetON‐DBN1‐GFP was expressed in ciliating RPE1 cells upon addition of Dox. The line scan on the right reflects ARL113B, CEP164, and DBN1‐GFP (green) intensity along the cilium shown in the boxed area on the left.
-
EDBN1‐GFP and F‐actin co‐localized in RPE1 cells during ciliogenesis. The intensities of both signals (right) were measured along the white line in the merge image on the left of ciliating RPE1 TetON‐DBN1‐GFP cells.
Similar cilia length in DBN1 KO cells expressing either DBN1 WT or phospho‐mimetic DBN1 S142D has led us to speculate that the TetON‐expressed DBN1 might be strongly phosphorylated in cells. Indeed, immunoblotting of the cell lysate with phospho‐specific DBN1pS142 antibodies confirmed that in TetON‐DBN1 cells, S142 was strongly phosphorylated (Fig 5G). No DBN1p142S signal was observed in cells overexpressing the S142A and S142D versions of DBN1, confirming the specificity of the DBN1p142 antibodies (Fig 5G). These data together suggest that the phosphorylated DBN1pS142 promotes elongation of cilia.
The kinase CDK5 and hCDC14A phosphatase counteract phosphorylation of DBN1 during ciliogenesis
In cycling cells, CDK5 kinase phosphorylates DBN1 at serine residue 142 17, 29, 30. If CDK5 regulates DBN1S142 during ciliogenesis, we would expect that CDK5 depletion by siRNA leads to hypo‐phosphorylation of DBN1S142 and the formation of shorter cilia as this was observed in DBN1 S142A cells (Fig 5E and F). Consistent with this, phosphorylation of DBN1pS142 was slightly reduced upon siRNA‐mediated depletion of CDK5 (Fig 5H and I). Moreover, CDK5 depletion restored the cilia length of hCDC14A PD RPE1 cells to the length of the WT control cells (Fig 5J).
To test the regulation of DBN1S142 by CDK5 further, we overexpressed the phospho‐mimetic version of DBN1 S142D in hCDC14A PD RPE1 cells and subsequently depleted CDK5 by siRNA. Cilia length reduction by CDK5 depletion was prevented by the expression of DBN1 S142D providing independent support for the notion that DBN1pS142 increases the length of cilia (Fig 5K). Thus, the opposing activities of CDC14A and CDK5 control phospho‐regulation of DBN1 at S142.
hCDC14A regulates endocytosis as well as myosin Va and Arp2 localization to the basal body during ciliogenesis
How do hCDC14A and DBN1 contribute to cilia length control? In Inner Medullar Collecting Duct (IMCD3) kidney (IMCD3) cells lacking a functional IFT27 gene, which is involved in intraflagellar transport, DBN1 localizes at the tip of the cilium 11. In these cells, DBN1 was identified as a member of a cilia‐associated actin network that promotes ectocytosis of vesicles from the cilia tip. Loss of DBN1 from cilia of hCDC14A PD RPE1 cells could explain the cilia length increase. However, in RPE1 WT cells DBN1 was not detected within the cilium using the antibodies that have been used for the IMCD3 cell study (Fig EV5C) 23. Similarly, Dox‐induced DBN1‐GFP did not localize along the cilium of RPE1 WT cells (Fig EV5D). However, DBN1‐GFP partially co‐localizes with actin fibers in RPE1 cells (Fig EV5E). Thus, in RPE1 cells DBN1 is associated with the actin cytoskeleton but not with the cilium. Cilia length control regulation via a DBN1 pool inside cilia is, therefore, unlikely for RPE1 cells.
Enhanced endocytic recycling to the pericentrosomal preciliary compartment could elongate cilia in hCDC14 PD RPE1 and DBN1 KO RPE1 cells 8, 31, 32. Interestingly, DBN1 KO HEK293 cells were reported to display enhanced endocytosis of dynamin‐dependent cargos 17. Consequently, we first tested the uptake of transferrin as a marker for endocytic recycling. Fluorescent transferrin was added to RPE1 cells simultaneously with serum starvation. After 45 min, cells were washed and analyzed for transferrin at the CEP164‐marked basal body. Transferrin at the basal body was higher in hCDC14A PD cells compared to WT cells or DBN1 KO cells (Fig 6A and B). We repeated the transferrin uptake experiment after 4 h of serum starvation as described in Fig 6A. At this later time point, the transferrin signal at basal bodies was still higher in hCDC14A PD cells than in WT cells or DBN1 KO cells (Fig 6A and B). Thus, hCDC14A inhibits endocytic uptake in RPE1 cells probably without the involvement of DBN1.
Figure 6. hCDC14 APD cells show enhanced endosome recycling as well as elevated docking of myosin Va vesicles and Arp2 recruitment to the basal body.

- Transferrin uptake was measured with serum starvation as described 17. Fluorescent transferrin was added for 45 min to the cells with serum starvation. A second sample was serum starved for 4 h followed by 45 min transferrin uptake with serum starvation. Cells were analyzed after washing. The red circle around the mother centriole marker CEP164 indicates the fluorescence intensity that was determined for (B). The encircled area in MERGE is shown 2‐fold enlarged on the right hand side of the figure.
- Quantification of (A). The fluorescence intensities of transferrin within a 2‐μm‐radius circle surrounding the CEP164 signal were quantified. An ImageJ macro was used to measure the intensity as described in Materials and Methods. Data were calculated from a total of 120 cells pooled from two independent experiments. Mean ± SD. ****P ≤ 0.0001.
- hCDC14A PD cells showed an increased localization of myosin Va containing vesicles within the 1 μm radius of the basal body (red circle). The encircled area in MERGE is shown 4‐fold enlarged on the right hand side of the figure.
- Quantification of (C). Data were calculated from a total of 180 cells pooled from three independent experiments. Mean ± SD. *P ≤ 0.05, ***P ≤ 0.001, ****P ≤ 0.0001.
- Arp2 localization in REP1 WT, hCDC14A PD, and DBN1 KO cells. Signal intensity was measured 30 min after serum starvation. The encircled area in MERGE is shown 4‐fold enlarged on the right hand side of the figure.
- Quantification of (E). Sub‐centrosomal (1 μm radius; red circle in E) Arp2 was measured by quantifying the intensity of Arp2 in WT, hCDC14A PD, and DBN1 KO cells. Both hCDC14A PD and DBN1 KO cells contain significantly higher Arp2 in the pericentrosomal area (1 μm radius) compared to WT cells after 30 min of serum starvation. Data were calculated from a total of 120 cells pooled from three independent experiments. Mean ± SD. ****P ≤ 0.0001.
- Pericentrosomal ARP2 enrichment was comparable in the RPE1 DBN1 KO cells (Tet Control) with DBN1 KO cells expressing different DBN1 versions (WT, S142A, S142D). Data were calculated from a total of 200 cells each pooled from two independent experiments. Mean ± SD. ns: not specific.
- Proposed model for primary cilia length regulation by hCDC14A, CDK5, and DBN1 in RPE1 cells.
Next, we used myosin Va accumulation at the CEP164‐marked mother centriole as an indication of preciliary vesicle transportation 10. In hCDC14A PD cells without serum starvation, the intensity of myosin Va vesicles surrounding the CEP164‐marked basal body was slightly higher in comparison with DBN1 KO RPE1 and RPE1 WT cells (Fig 6C and D). The myosin Va signal at the basal body increased within 30 min of serum starvation in all cell types but was highest in hCDC14A PD cells. After 24 h of ciliation, all three cell lines showed similar amounts of myosin Va at basal bodies. Thus, myosin Va recruitment to the basal body is negatively regulated by hCDC14A. This regulation probably does not involve DBN1.
Low dose of actin‐depolymerizing drug cytochalasin D was reported to promote ciliogenesis by stabilizing the Arp2‐associated pericentrosomal actin network 10. We observed enhanced Arp2 intensity surrounding the 1 μm radius of the basal body in hCDC14A PD and DBN1 KO cells 30 min after induction of ciliation through serum starvation (Fig 6E and F). Nevertheless, similar pericentrosomal Arp2 enrichment was observed in DBN1 KO cells expressing no DBN1 construct or DBN1 (WT, S142A, S142D) (Fig 6G). Such failure to detect differential Arp2 enrichment among DBN1 mutant overexpressing cells could be due to the disruption of delicate molecular balance associated with the overexpression of DBN1. These data denote that hCDC14A inhibits Arp2 accumulation at the basal body. DBN1 overlaps with hCDC14A in this inhibition.
Discussion
CDC14 phosphatases regulate ciliogenesis in a number of organisms including zebrafish, mouse, and human cells. CDC14A activity in human and mouse is needed for the formation of the kinocilia in the organ of corti 16. These kinocilia are motile 9+2 cilia with a special function in the organization of actin‐based stereocilia. Data presented in this study now indicate that hCDC14A plays a role in cilia length control of 9+0, non‐motile primary cilia. A phosphatase dead version of hCDC14A or hCDC14A depletion resulted in longer cilia, while elevated hCDC14A levels prevented cilia formation. The cilia length phenotype was specific to hCDC14A and was not observed in cells lacking the paralogue hCDC14B. In contrast to RPE1 cells, cdc14A or cdc14B depletion in zebrafish led to shorter cilia 14, 33, Presently, it is unclear why inactivation of CDC14A in zebrafish and human cells impacts cilia length in opposite ways. Variations in cilia phenotypes between organisms upon inactivation of orthologs have been reported before 34.
Whereas overexpression of CDC14A reduced cilium formation, inactivation of the protein did not affect cilia formation efficiency. This is probable due to the fact that the elongated cilia in phosphatase dead cells are the outcome of changed phospho‐status of hCDC14A substrates. In the overexpression situation, we possibly inhibit the initial steps leading to ciliogenesis. Latter conclusion is supported by the finding that overexpression of hCDC14A did not affect already assembled cilia (B. Uddin, unpublished observation).
Phospho‐proteome and BioID analyses identified hCDC14A substrates with a function in cilia length control. Importantly, most hCDC14A substrates during ciliogenesis were dephosphorylated on pSP sites, which is the signature of CDC14A phosphatases from yeast to human cells 12, 28. A secondary siRNA screen identified, among these hCDC14A substrates, proteins with a function in cilia length control. These include CTTN, LMO7, DBN1, MTCL1, UBAP2L, KANK2, MAP4, PDLIM7, and RPS2 (Fig EV4B–E). Some of them have been implicated in ciliogenesis 11, 26, 35.
Because DBN1 has been implicated in the regulation of ciliogenesis 11, we studied the phospho‐regulation of DBN1 in greater detail. Analysis of DBN1pS142 with a phospho‐specific antibody confirmed that DBN1 was hyper‐phosphorylated in hCDC14A PD RPE1 cells during ciliogenesis in comparison with CDC14A WT cells, while overexpression of hCDC14A reduced pS142 phosphorylation. Purified, recombinant hCDC14A dephosphorylated DBN1 at S142 indicating direct regulation by hCDC14A. Thus, DBN1pS142 is a substrate of hCDC14A during ciliogenesis. Moreover, the observation that depletion of CDK5 in ciliated RPE1 cells reduced DBN1pS142 phosphorylation suggests that S142 of DBN1 is phosphorylated by the proline‐directed kinase CDK5 during ciliogenesis. This is in line with reports on the phosphorylation of DBN1 by CDK5 in growth cones of neurons 29, 30.
Analysis of DBN1 mutants was complicated by the impact of DBN1 overexpression on cilia length. Despite this drawback, data presented here suggest that the hyper‐phosphorylated DBN1pS142 promotes cilia elongation, while the hypo‐phosphorylated DBN1 restricts cilia length. This became apparent from elongated cilia in DBN1 KO cells overexpressing DBN1 S142D in comparison with DBN1 S142A (Fig 5F) and the observation that DBN1 S142D overexpression prevented a reduction in cilium length in response to the depletion of CDK5 that phosphorylates DBN1 at S142 (Fig 5K).
A function of DBN1 in ectocytosis directly at the cilium has been proposed recently 11. However, because we could not detect DBN1 at the cilia tip or along cilia with published antibodies or overproduced DBN1‐GFP, we presently disfavor the model that DBN1 functions in an ectocytosis‐like process in WT RPE1 cells. An indication for actin‐remodeling function of DBN1 in ciliogenesis comes from the observation of an increased recruitment of the actin polymerizer Arp2 to centrosomes in DBN1 KO cells 10 (Fig 6F). This Arp2 pool has been implicated in the docking of Golgi derived, myosin Va‐associated preciliary vesicles to the basal body 10. However, overexpression of a phospho‐mimetic DBN1 S142D did not affect the Arp2 pool at centrosomes (Fig 6G). Presently, it is unclear whether overexpression of DBN1 S142D masked the role of DBN1 phospho‐regulation on the local Arp2 organization or simply DBN1 phosphorylation does not have a function in this process. It will be interesting to study how chromosomal DBN1 S142A and DBN1 S142D mutants affect the length of cilia.
CDK5 also affects cilia length by the phosphorylation and destabilization of the cilia disassembly factor NDE1 4. NDE1 disassembles cilia upon re‐entry of cells into the cell cycle but becomes inactivated during ciliation by CDK5‐mediated phosphorylation that induces NDE1 degradation via ubiquitination by the ubiquitin E3 ligase SCFFBW7. We did not detect NDE1 in the hCDC14A phospho‐proteome and BioID screens suggesting that the CDK5‐NDE1‐cilia length regulation functions without the involvement of hCDC14A. Maskey et al 4 noticed that NDE1 is not the only substrate of CDK5 in cilia length control. We propose that the phosphorylation of DBN1S142 by CDK5 provides a second way how CDK5 impacts cilia length.
During ciliation, hCDC14A PD RPE1 cells showed a range of alterations in comparison with WT cells (Fig 6H) that probably contribute to the elongated cilia. Arp2 and myosin Va recruitment to basal bodies and endocytic uptake were enhanced in hCDC14A PD RPE1 cells. How an increase in myosin Va‐associated preciliary vesicles to the basal body early in ciliogenesis impacts on cilia length growth after 24 h of ciliation is difficult to say mainly because we do not understand the composition of these vesicles. Increased delivery of Golgi derived, myosin V vesicles or the enhanced endocytic recycling may change the composition of the ciliary membrane early in ciliogenesis which then affects cilia length control at later time points. Presently, it is unclear to what extent each of these hCDC14A‐regulated steps contributes to cilia length control and which of the hCDC14A substrates are involved. It will be interesting to see whether these functions of hCDC14A are also counteracted by CDK5 or whether the “SP” specific CDK10/cyclin M kinase, whose misfunction is the cause of STAR syndrome 5, 6, plays a regulatory role here.
Materials and Methods
Cell culture and siRNA reverse transfection
RPE1, HEK293T (human embryonic kidney), and HEK293‐GP (GP2‐293, Clontech) cells were cultured in Gibco DMEM/F‐12 (ThermoFisher) media containing 10% FBS, 1% l‐glutamine, and 1% penicillin/streptomycin at 37°C and 5% CO2. Lipofectamine RNAiMAX transfection reagent (ThermoFisher) was used according to the manufacturer's instructions with a final concentration of 20 nM siRNA (please see the Appendix Table S1 and S2 for details of used siRNA).
Generation of knockout and stable cell lines
RPE1 cells with inducible expression of proteins of interest were constructed as previously described 36. Generation of zinc finger nuclease targeted hCDC14A PD cells was described previously 13, 18. CRISPR/Cas9‐mediated drebrin knockout was generated by targeting three different exons with three guide RNAs (gRNA1: GTACGGCTTCTGCAGTGTCA for exon 3; gRNA2: GCAGCGGCTCTCTAACGGGC for exon 5; and gRNA3: GGAGCGCGAGCGGCGCTACC for exon 7). The gRNAs were designed using the web tool (https://crispr.cos.uni-heidelberg.de/), ordered as primers and inserted next to the U6 promoter in the scaffold vector pSpCas9(BB)‐2A‐GFP(PX458) (a gift from Feng Zhang, Broad Institute of MIT and Harvard, Cambridge, MA; Addgene plasmid #48138) 37, 38. The plasmid pX458 with gRNA was electroporated into RPE1 cells (Neon transfection system, Invitrogen; pulse voltage 1,050 V, pulse width 30 ms, and pulse number 2) and 48 h later, the transfected cells were selected for GFP expression through FACS sorting. The sorted cells were then subjected to clonal propagation through limiting dilution on 96‐well plate. The emerging clones were harvested and screened via sequencing the genomic PCR amplicons (see the Appendix Table S3 for primer list). The successfully targeted clones were further confirmed by immunoblot analysis using the antibody MX823 (Progen). The MX823 is a mouse monoclonal antibody generated using synthetic C‐terminal peptide (aa 632–649). However, we have designed the gRNAs in such that even a possible truncated N‐terminal version of DBN1 would lack the critical domains AB1 (actin‐binding region 1) and AB2 (actin‐binding region 2; Fig 5A).
Immunofluorescence and microscopy
Cells were seeded on coverslips ~24 h before the treatments (starvation +/− and/or doxycycline +/− depending on the experimental set up) and washed once with PBS prior to fixation. The fixation condition was decided depending on the primary antibody used (see Appendix Table S4 for the detailed list of antibodies and fixation conditions). 4% paraformaldehyde was used for 10 min at room temperature, whereas methanol fixation was carried out at −20°C for 5 min with ice‐cold methanol. Then, the cells were permeabilized with 0.1% Triton X‐100 in PBS (10 min at RT), incubated with 10% FBS to block unspecific binding, and incubated with primary antibodies inside a wet chamber for 1 h. Upon three times washing with PBS, the cells were incubated with fluorescently labeled secondary antibodies for 30 min at RT, washed three times and mounted with Mowiol and dried overnight at room temperature. DNA was counterstained by adding DAPI (4′,6‐diamidino‐2‐phenylindole) with the secondary antibody solutions. A DeltaVision RT system (Applied Precision) equipped with an Olympus IX71 microscope was used to image immunofluorescence samples using Softworx software (Applied Precision). Comparable samples were always imaged with same exposures and intensities as well as the background subtracted images were displayed with ImageJ using same minimum and maximum brightness.
Immunoblot analysis
Cells were seeded on 6‐ or 12‐well plate and lysed directly with Laemmli Buffer supplemented with protease inhibitor (Roche; 11 873 580 001), phosphatase inhibitor (Roche; 04 906 845 001), phenylmethylsulfonyl fluoride (PMSF), and Benzonase (Merck, 101656; 1:500). The lysates were heated at 95°C for 5 min and centrifuged at 20,000 g for 5 min. Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) was performed as previously described 39. The membranes were blocked in 5% non‐fat milk in TBS‐T and incubated with appropriate primary and secondary antibodies (Appendix Table S5).
Measuring cilia length and protein intensity within a defined region of interest (ROI)
Cilia lengths of the background subtracted images were measured by a semi‐automated ImageJ macro developed and optimized by ZMBH imaging facility (Macro EV1). In short, the macro finds the best line fit for the channel that represents cilia. The line is then skeletonized and inflated to measure the length. Upon mouse click, the macro visualizes the processing steps with 400× zoom allowing the poorly processed cilia measurements to be discarded. Another macro that allows to select the size of region of interest (ROI) as well as to define channels to be measured was used for intensity measurements of signals, e.g., transferrin, Arp2, myosin Va surrounding 1–2 μm radius of centrosomes (Macro EV2).
Quantitative phospho‐proteome and BioID mass spectrometry analyses
The quantitative phospho‐proteome and BioID mass spectrometry analyses were conducted as described before 12. For global phospho‐proteome analysis, RPE1 cells with inducible expression of hCDC14A‐YFP were cultured in light (Arg12C14N Lys12C14N) and heavy (Arg13C15N Lys13C15N) SILAC medium (Silantes) in separate flasks. After seven passages, cells were expanded to four T175 flasks for each condition and starved for 48 h. Doxycycline (10 ng/ml) was added only to the heavy samples along with starvation for inducing hCDC14A‐YFP expression. Proteins were extracted and mixed at a ratio of 1:1. Strong cation exchange chromatography/immobilized metal affinity chromatography (SCX/IMAC) technique was used to enrich phospho‐peptides 40. The resulted 48 fractions (24 phospho‐enriched and 24 phospho‐depleted) were analyzed by LC‐MS using a Dionex UltiMate 3000RSLCnano HPLC system (Thermo Scientific) coupled to a LTQ Orbitrap Elite mass spectrometer (Thermo Scientific). Peptide identification and quantification were achieved using the MaxQuant software package (1.5.3.8) with its built‐in Andromeda search algorithm 41. The results of the database search were analyzed using Perseus 42. For BioID mass spectrometry analysis, RPE1 hCDC14A‐BirA‐HA, hCDC14A C278S ‐BirA‐HA, and BirA‐HA cells were cultured in medium containing 50 μM biotin. Protein expression was induced by adding Dox (10 ng/ml) during the 48 h of serum starvation. The cells were harvested and lysed using RIPA buffer (10 mM Tris–Cl, 150 mM NaCl, 1% Triton X‐100, 0.1% SDS) supplemented with protease inhibitor (Roche; 11 873 580 001), phenylmethylsulfonyl fluoride (PMSF), and Benzonase (Merck, 101656; 1:500). The lysate was clarified by centrifuging for 15 min at 20,000 g and the supernatant was incubated with streptavidin Sepharose beads (GE 17‐5113‐01) at 4°C for 3 h. Then, the beads were washed three times with RIPA buffer and the bound proteins were eluted by heating at 95°C for 10 min with 2× Laemmli buffer containing 2 mM biotin. The samples were dimethyl labeled (Heavy – BirA control; Medium – hCDC14AC278S‐BirA; and Light – hCDC14A‐BirA prior to mass spectrometry analysis) 43. Two replicates of each quantitative phospho‐proteome and BioID mass spectrometry experiment were run, and relevant results are deposited in Dataset EV1 and EV2, respectively.
In vitro phosphatase assay
DBN1‐GFP construct was transfected into HEK293T cells using polyethylenimine (PEI). The cells were harvested by scrapping after 48 h of transfection and lysed with RIPA buffer supplemented with protease inhibitor (Roche; 11 873 580 001), phenylmethylsulfonyl fluoride (PMSF), and Benzonase (Merck, 101656; 1:500). The DBN1‐GFP was immunoprecipitated by GFP‐Trap®_A beads (Chromotek) following manufacturer's protocol. Equal amount of bead‐captured DBN1 protein was incubated with purified hCDC14A in phosphatase assay buffer (30 mM imidazole, 1 mM DTT, 1 mM EDTA, 150 mM KCl, 1 mM MgCl2, and 25 mM K‐Hepes) for 2 h at 30°C. The products were analyzed on immunoblots following SDS/PAGE. A buffer control was used to determine the extent of dephosphorylation by quantifying pS142‐DBN1/DBN1 ratio.
Electron microscopy
RPE1 cells were seeded on coverslips and cultured at 37°C and 5% CO2 till they reached a confluency of 70–80%. Cells were rinsed in 100 mM phosphate buffer (PBS) three times and then fixed with 2.5% glutaraldehyde (GA) in 50 mM cacodylate buffer and 2% sucrose for 30 min at room temperature. After 5 times washing for 5 min with 50 mM cacodylate buffer, cells were incubated for 40 min in 2% osmium/cacodylate buffer on ice in darkness, followed by 4 times rinsing with dH2O and overnight staining at 4°C in 0.5% uranyl acetate. On the following day, coverslips were rinsed again 4 times with dH2O and via dehydration row (40, 50, 70, 80, 90, 95, 100% ethanol). Water was removed by ethanol within the cells. Coverslips were immediately placed on capsules filled with Spurr‐resin and polymerized at 60°C for 24–48 h. Embedded cells were sectioned using a Reichert Ultracut S Microtome (Leica Instruments, Vienna, Austria) to a thickness of 70 nm. Post‐staining with 3% uranyl acetate and lead citrate was performed. Sections were imaged at a Jeol JE‐1400 (Jeol Ltd., Tokyo, Japan), operating at 80 kV, equipped with a 4 k × 4 k digital camera (F416, TVIPS, Gauting, Germany). Micrographs were adjusted in brightness and contrast using ImageJ.
Statistical analysis
If not otherwise mentioned, at least 150 cilia or centrosomes were quantified for each experiment and the findings were confirmed by three independent experiments. The results were expressed as mean ± SD (Standard deviation). Prism 7 software (GraphPad) was used for statistical analyses. Mean between two groups was compared by t‐tests, whereas multiple inter‐group differences were analyzed by one‐way ANOVA (analysis of variance) followed by Tukey's multiple comparison test for post hoc comparisons.
Author contributions
BU, PP, and N‐PC performed experiments. MW analyzed cilia length for secondary siRNA screen experiment. AN analyzed cilia by electron microscopy. RH, BH, and TR performed mass spec analysis of BioID and phospho‐proteome experiments. AJ and HL developed ImageJ macros. GP and ES designed experiments, supervised students and wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Dataset EV1
Dataset EV2
Macro EV1
Macro EV2
Review Process File
Acknowledgements
We acknowledge Dr. Monika Langlotz from the ZMBH FACS Facility for all the FACS sorting experiments. This work is supported by Deutsche Forschungsgemeinschaft Grant Schi295/3‐4 (to E.S.), PE1883‐3 (to G.P.) and CRC873, A14 (to G.P.).
EMBO Reports (2019) 20: e46544
Contributor Information
Gislene Pereira, Email: gislene.pereira@cos.uni-heidelberg.de.
Elmar Schiebel, Email: e.schiebel@zmbh.uni-heidelberg.de.
References
- 1. Goetz SC, Anderson KV (2010) The primary cilium: a signalling centre during vertebrate development. Nat Rev Genet 11: 331–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Drummond IA (2012) Cilia functions in development. Curr Opin Cell Biol 24: 24–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Keeling J, Tsiokas L, Maskey D (2016) Cellular mechanisms of ciliary length control. Cells 5: E6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Maskey D, Marlin MC, Kim S, Kim S, Ong EC, Li G, Tsiokas L (2015) Cell cycle‐dependent ubiquitylation and destruction of NDE1 by CDK5‐FBW7 regulates ciliary length. EMBO J 34: 2424–2440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Guen VJ, Gamble C, Perez DE, Bourassa S, Zappel H, Gartner J, Lees JA, Colas P (2016) STAR syndrome‐associated CDK10/Cyclin M regulates actin network architecture and ciliogenesis. Cell Cycle 15: 678–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Guen VJ, Edvardson S, Fraenkel ND, Fattal‐Valevski A, Jalas C, Anteby I, Shaag A, Dor T, Gillis D, Kerem E et al (2018) A homozygous deleterious CDK10 mutation in a patient with agenesis of corpus callosum, retinopathy, and deafness. Am J Med Genet A 176: 92–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kim J, Jo H, Hong H, Kim MH, Kim JM, Lee JK, Heo WD, Kim J (2015) Actin remodelling factors control ciliogenesis by regulating YAP/TAZ activity and vesicle trafficking. Nat Commun 6: 6781 [DOI] [PubMed] [Google Scholar]
- 8. Kim J, Lee JE, Heynen‐Genel S, Suyama E, Ono K, Lee K, Ideker T, Aza‐Blanc P, Gleeson JG (2010) Functional genomic screen for modulators of ciliogenesis and cilium length. Nature 464: 1048–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kohli P, Hohne M, Jungst C, Bertsch S, Ebert LK, Schauss AC, Benzing T, Rinschen MM, Schermer B (2017) The ciliary membrane‐associated proteome reveals actin‐binding proteins as key components of cilia. EMBO Rep 18: 1521–1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wu CT, Chen HY, Tang TK (2018) Myosin‐Va is required for preciliary vesicle transportation to the mother centriole during ciliogenesis. Nat Cell Biol 20: 175–185 [DOI] [PubMed] [Google Scholar]
- 11. Nager AR, Goldstein JS, Herranz‐Perez V, Portran D, Ye F, Garcia‐Verdugo JM, Nachury MV (2017) An actin network dispatches ciliary gpcrs into extracellular vesicles to modulate signaling. Cell 168: 252–263.e14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen NP, Uddin B, Hardt R, Ding W, Panic M, Lucibello I, Kammerer P, Ruppert T, Schiebel E (2017) Human phosphatase CDC14A regulates actin organization through dephosphorylation of epithelial protein lost in neoplasm. Proc Natl Acad Sci USA 114: 5201–5206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen NP, Uddin B, Voit R, Schiebel E (2016) Human phosphatase CDC14A is recruited to the cell leading edge to regulate cell migration and adhesion. Proc Natl Acad Sci USA 113: 990–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Clement A, Solnica‐Krezel L, Gould KL (2012) Functional redundancy between Cdc14 phosphatases in zebrafish ciliogenesis. Dev Dyn 241: 1911–1921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Delmaghani S, Aghaie A, Bouyacoub Y, El Hachmi H, Bonnet C, Riahi Z, Chardenoux S, Perfettini I, Hardelin JP, Houmeida A et al (2016) Mutations in CDC14A, encoding a protein phosphatase involved in hair cell ciliogenesis, cause autosomal‐recessive severe to profound deafness. Am J Hum Genet 98: 1266–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Imtiaz A, Belyantseva IA, Beirl AJ, Fenollar‐Ferrer C, Bashir R, Bukhari I, Bouzid A, Shaukat U, Azaiez H, Booth KT et al (2018) CDC14A phosphatase is essential for hearing and male fertility in mouse and human. Hum Mol Genet 27: 780–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li B, Ding S, Feng N, Mooney N, Ooi YS, Ren L, Diep J, Kelly MR, Yasukawa LL, Patton JT et al (2017) Drebrin restricts rotavirus entry by inhibiting dynamin‐mediated endocytosis. Proc Natl Acad Sci USA 114: E3642–E3651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Uddin B, Chen NP, Panic M, Schiebel E (2015) Genome editing through large insertion leads to the skipping of targeted exon. BMC Genom 16: 1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Berdougo E, Nachury MV, Jackson PK, Jallepalli PV (2008) The nucleolar phosphatase Cdc14B is dispensable for chromosome segregation and mitotic exit in human cells. Cell Cycle 7: 1184–1190 [DOI] [PubMed] [Google Scholar]
- 20. Fry AM, Mayor T, Meraldi P, Stierhof YD, Tanaka K, Nigg EA (1998) C‐Nap1, a novel centrosomal coiled‐coil protein and candidate substrate of the cell cycle‐regulated protein kinase Nek2. J Cell Biol 141: 1563–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mayor T, Stierhof YD, Tanaka K, Fry AM, Nigg EA (2000) The centrosomal protein C‐Nap1 is required for cell cycle‐regulated centrosome cohesion. J Cell Biol 151: 837–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ishikawa H, Kubo A, Tsukita S, Tsukita S (2005) Odf2‐deficient mother centrioles lack distal/subdistal appendages and the ability to generate primary cilia. Nat Cell Biol 7: 517–524 [DOI] [PubMed] [Google Scholar]
- 23. Mazo G, Soplop N, Wang WJ, Uryu K, Tsou MB (2016) Spatial control of primary ciliogenesis by subdistal appendages alters sensation‐associated properties of cilia. Dev Cell 39: 424–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics 1: 376–386 [DOI] [PubMed] [Google Scholar]
- 25. Roux KJ, Kim DI, Raida M, Burke B (2012) A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J Cell Biol 196: 801–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ghossoub R, Hu Q, Failler M, Rouyez MC, Spitzbarth B, Mostowy S, Wolfrum U, Saunier S, Cossart P, Jamesnelson W et al (2013) Septins 2, 7 and 9 and MAP4 colocalize along the axoneme in the primary cilium and control ciliary length. J Cell Sci 126: 2583–2594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Visintin R, Craig K, Hwang ES, Prinz S, Tyers M, Amon A (1998) The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk‐dependent phosphorylation. Mol Cell 2: 709–718 [DOI] [PubMed] [Google Scholar]
- 28. Eissler CL, Mazon G, Powers BL, Savinov SN, Symington LS, Hall MC (2014) The Cdk/Cdc14 module controls activation of the Yen1 holliday junction resolvase to promote genome stability. Mol Cell 54: 80–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Worth DC, Daly CN, Geraldo S, Oozeer F, Gordon‐Weeks PR (2013) Drebrin contains a cryptic F‐actin‐bundling activity regulated by Cdk5 phosphorylation. J Cell Biol 202: 793–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tanabe K, Yamazaki H, Inaguma Y, Asada A, Kimura T, Takahashi J, Taoka M, Ohshima T, Furuichi T, Isobe T et al (2014) Phosphorylation of drebrin by cyclin‐dependent kinase 5 and its role in neuronal migration. PLoS One 9: e92291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ye X, Zeng H, Ning G, Reiter JF, Liu A (2014) C2 cd3 is critical for centriolar distal appendage assembly and ciliary vesicle docking in mammals. Proc Natl Acad Sci USA 111: 2164–2169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Joo K, Kim CG, Lee MS, Moon HY, Lee SH, Kim MJ, Kweon HS, Park WY, Kim CH, Gleeson JG et al (2013) CCDC41 is required for ciliary vesicle docking to the mother centriole. Proc Natl Acad Sci USA 110: 5987–5992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Clement A, Solnica‐Krezel L, Gould KL (2011) The Cdc14B phosphatase contributes to ciliogenesis in zebrafish. Development 138: 291–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hamel V, Steib E, Hamelin R, Armand F, Borgers S, Fluckiger I, Busso C, Olieric N, Sorzano COS, Steinmetz MO et al (2017) Identification of chlamydomonas central core centriolar proteins reveals a role for human WDR90 in ciliogenesis. Curr Biol 27: 2486–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bershteyn M, Atwood SX, Woo WM, Li M, Oro AE (2010) MIM and cortactin antagonism regulates ciliogenesis and hedgehog signaling. Dev Cell 19: 270–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Vlijm R, Li X, Panic M, Ruthnick D, Hata S, Herrmannsdorfer F, Kuner T, Heilemann M, Engelhardt J, Hell SW et al (2018) STED nanoscopy of the centrosome linker reveals a CEP68‐organized, periodic rootletin network anchored to a C‐Nap1 ring at centrioles. Proc Natl Acad Sci USA 115: E2246–E2253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F (2013) Genome engineering using the CRISPR‐Cas9 system. Nat Protoc 8: 2281–2308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O et al (2013) DNA targeting specificity of RNA‐guided Cas9 nucleases. Nat Biotechnol 31: 827–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- 40. Villen J, Gygi SP (2008) The SCX/IMAC enrichment approach for global phosphorylation analysis by mass spectrometry. Nat Protoc 3: 1630–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cox J, Mann M (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.‐range mass accuracies and proteome‐wide protein quantification. Nat Biotechnol 26: 1367–1372 [DOI] [PubMed] [Google Scholar]
- 42. Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, Mann M, Cox J (2016) The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods 13: 731–740 [DOI] [PubMed] [Google Scholar]
- 43. Boersema PJ, Raijmakers R, Lemeer S, Mohammed S, Heck AJ (2009) Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat Protoc 4: 484–494 [DOI] [PubMed] [Google Scholar]
- 44. Shirao T, Sekino Y (2017) General introduction to drebrin. Adv Exp Med Biol 1006: 3–22 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Dataset EV1
Dataset EV2
Macro EV1
Macro EV2
Review Process File


