Abstract
The generation, alteration, recognition, and erasure of epigenetic modifications of DNA are fundamental to controlling gene expression in mammals. These covalent DNA modifications include cytosine methylation by AdoMet-dependent methyltransferases and 5-methylcytosine oxidation by Fe(II)-dependent and α-ketoglutarate-dependent dioxygenases. Sequence-specific transcription factors are responsible for interpreting the modification status of specific regions of chromatin. This review focuses on recent developments in characterizing the functional and structural links between the modification status of two DNA bases: 5-methylcytosine and 5-methyluracil (thymine).
Introduction
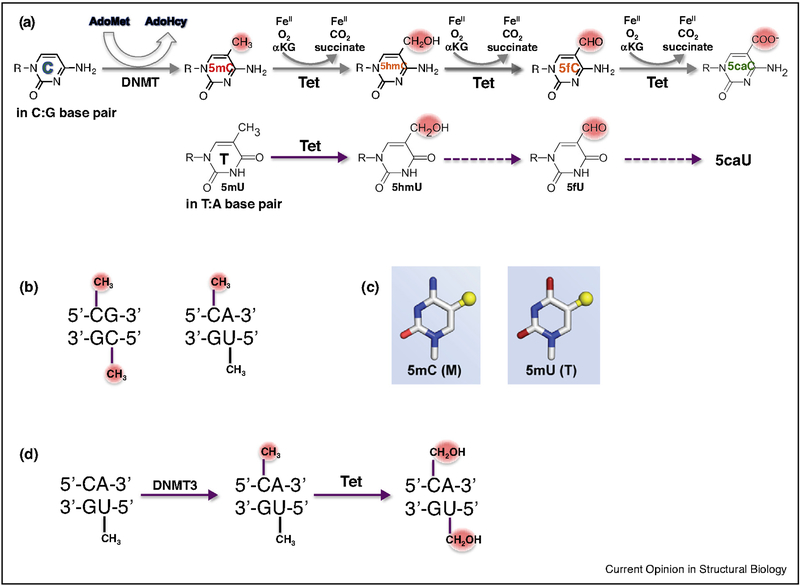
Five chemical forms of cytosine have been found in the DNA of higher organisms, including unmodified cytosine (C), 5-methylcytosine (5mC), 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC) (Figure 1a) [1–5]. While some of these modifications may affect the strength of base pairing C:G hydrogen (H) bonds [6–8], these chemical forms are equivalent in terms of base pairing specificity and protein coding. However, the various forms of cytosine can vary substantially in how they interact with transcription factors and other DNA-binding proteins, and in their influence on gene expression. There is great interest in the effects of these modifications in epigenetic regulation, development and differentiation, neuron function, and diseases [9,10]. In general, the post-synthetic modifications (or ‘epigenetic marks’) are added to cytosine in situ, following its incorporation into DNA in the unmodified form. DNA methyltransferases (Dnmts) modify specific cytosines to 5mC, usually within the sequence contexts CpG [11,12] or CpA [13,14•] (Figure 1b). A subset of these 5mC residues are then converted to 5hmC, 5fC, and 5caC in consecutive Fe(II)-dependent and α-ketoglutarate-dependent oxidation reactions by the ten-eleven translocation (Tet) dioxygenases [2–4].
Figure 1.
Methylation and oxidation in 5′-CpG-3′ and 5′-CpA-3′ dinucleotides. (a) DNA methyltransferases (DNMTs) convert C to 5-methylC (5mC). Tet dioxygenases then convert 5mC to 5-hydroxymethylC (5hmC), 5-formylC (5fC), and 5-carboxyC (5caC) in three consecutive Fe(II)-dependent and α-ketoglutarate-dependent oxidation reactions, without release of formaldehyde (Top). Tet dioxygenases can also convert thymine (5mU) to 5hmU, and potentially to 5fU and 5caU (Bottom). (b) CpG/CpG can be methylated on both strands; while TpG/CpA is intrinsically methylated on one strand (T, 5mU), and the C on the other strand may be methylated as well. (c) 5mC and 5mU (T) contain a methyl group (yellow sphere) in spatially equivalent positions at C5. (d) Dnmt3A and Dnmt3B can methylate cytosine in the context of a CpA/TpG dinucleotide. Tet dioxygenases can oxidize both 5mC and T (5mU).
Like 5mC, thymine contains a methyl group at C5 (Figure 1c). Thus CpA/TpG is an intrinsically hemimethylated DNA element. Cytosine methylation of CpA, by Dnmt3, generates a pseudo-symmetric fully-methylated 5mCpA/TpG dinucleotide. Further modification by Tet enzymes on both 5mC and T [15,16] can yield CpA/TpG sites having 5-hydroxymethyl (Figure 1d), 5-formyl or 5-carboxyl modifications on both 5mC and T.
In this review, we focus on the mechanisms for recognizing 5mCpG, 5mCpA, TpG, and their respective oxidative forms, in DNA. These modifications all protrude into the major groove of DNA, the primary recognition surface for proteins, and change its atomic shape and pattern of electrostatic charges. In principle, such changes can alter protein binding to specific recognition sequences in DNA, by strengthening interactions, weakening them, or abolishing them altogether. This, in turn, can modulate gene expression and thus affect cellular metabolism and differentiation and, on a broader scale, influence an organism’s development, aging and disease processes.
Several well-characterized classes of mammalian proteins interact with DNA in a methylation-responsive manner (Table 1). Two recent large-scale studies used SELEX-based approaches to reveal that DNA binding by proteins of the homeodomain, basic leucine zipper domain (bZIP), and tumor suppressor p53 families can be increased or decreased by cytosine methylation within the binding sequence [17••,18••]. There is thus a growing number of transcriptional regulators that have adapted to respond to different cytosine modification states, potentially acting as direct epigenetic sensors to instruct downstream events.
Table 1.
Structures of known DNA cytosine modification binding proteins
| TF family | Protein name | PDB | DNA | Mode of interaction |
|---|---|---|---|---|
| MBD | MeCP2 | 3C2I | 5mCpG | Methyl-Arg-G |
| MBD1 | 1IG4 | 5mCpG | ||
| MBD2 | 2KY8 | 5mCpG | ||
| MBD4 | 2MOE/4LG7/3VXX | 5mCpG | ||
| 3VYB | 5hmCpG | |||
| C2H2 ZF | Kaiso | 4F6M/4F6N | TpG/5mCpG | |
| Zfp57 | 4GZN | 5mCpG | ||
| Zfp57(E182Q) | 4M9V | 5caCpG | ||
| Wf4 | 2WBU/4M9E/5KE6 | CpG/5mCpG/TpG | ||
| Wf4 (E446D) | 5KEA | CpG | ||
| Egr1 | 4R2A/4X9J | 5mCpG | ||
| WT1 | 4R2E | 5mCpG | ||
| CTCF | 5T00 | 5mCpG and CpA | ||
| p53 bHLH-LZ (Other examples of 5caC binding) |
p53 | 3Q05 | TpG | |
| MAX | 5EYO | 5caCpG | Arginine | |
| WT1 | 4R2R | 5caCpG | Glutamine | |
| Pol II | 4Y7N | 5caC | Glutamine | |
| Tet3 (CXXC) | 5EXH | 5caC | Lysine | |
| bZIP | AP-1 (Jun) | 5T01 | T and 5mC | Van der Waals (vdw) |
| Zta (Epstein-Barr virus) | 5SZX | T and 5mC | vdw | |
| Homeodomain | HOBX13 | 5EGO | 5mCpG | vdw |
| CDX1 | 5LUX | 5mCpG | vdw | |
| CDX2 | 5LTY | 5mCpG | vdw | |
| LHX4 | 5HOD | TAA7TA | vdw | |
| Other candidates | MEIS1 | 5EGO/4XRM | TpG | Methyl-Arg-G |
| HOXB1-PBX1 | 1B72 | CpG and TpG | Vdw and methyl-R-G | |
| HOXA9-PBX1 | 1PUF | CpG and TpG | Vdw and methyl-R-G | |
| E2F8 | 4YO2 | CCCGCC | ||
| SRA | UHRF1 (SRA) | 2ZO0/3CLZ/2ZKD | 5mC | Base-flipping |
| UHRF2 (SRA) | 4PW7/4PW5 | 5mC/5hmC | ||
| SUVH5 (Arabidopsis) | 3Q0B/4YGI | 5mC/5hmC | ||
| TALE | dHax3 (X. campestris) | 3V6P/4GJP | T and 5mC | vdw (Glysine) |
| Unmodified CpG recognition | ||||
| TLR9 | 3WPC | CpG | H-bond | |
| CFP1 (CXXC1) | 3QMB | CpG | ||
| MBD1 (CXXC3) | 5W9Q | CpG | ||
| ID AX (CXXC4) | 5VC9 | CpG | ||
| RINF (CXXC5) | 5W9S | CpG | ||
| Tet1 (CXXC6) | 6ASD | CpG | ||
| Tet3 (CXXC) | 4HP1/4HP3 | CpC/CpG | ||
| MLL1 (CXXC7) | 2KKF/4NW3 | CpG | ||
| MLL2 (CXXC) | 4PZI | CpG | ||
| FBXL19(CXXC-PHD) | 6ASB | CpG | ||
| RNA N6mA binding | ||||
| YTH | YT521-B | 2 MTV | N6mA | Aromatic cage |
| YTH DC 1 | 4R3I | |||
| YTHDF1 | 4RCJ | |||
| MRB1 | 4U8T | |||
| Pho92 | 4RCM |
MBD proteins
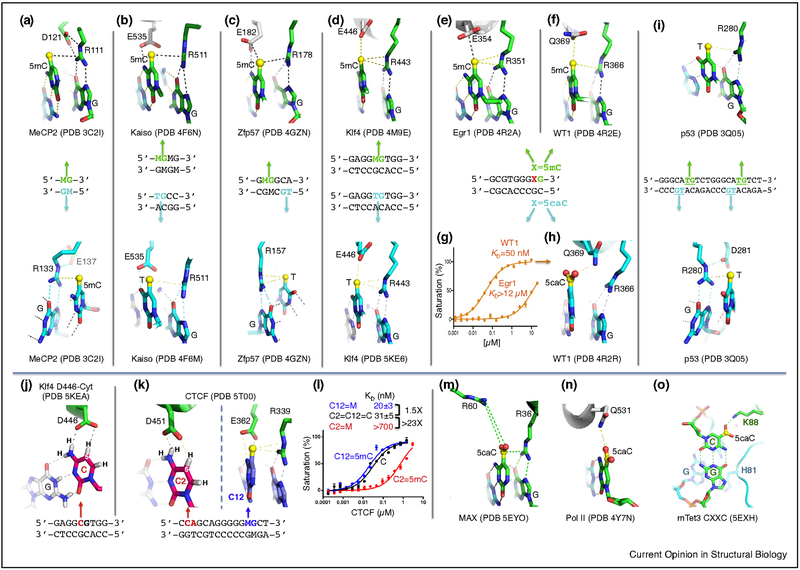
Methyl-binding domain proteins (MBDs) recognize fully-methylated CpG sequences, in which both DNA strands contain 5mC (5mCpG/5mCpG) [19]. The MBD domain of MeCP2 binds the symmetrically methylated CpG site using two arginines, each of which H-bonds to a guanine [20]. These two arginines also each make van der Waals contacts with the methyl group of the neighboring 5mC of the same DNA strand and form what we termed ‘5mC–Arg–G triads’ [21] (Figure 2a).
Figure 2.
Examples of methyl-Arg-Gua recognition and 5caC recognition. (a) MBD domain of MeCP2. PDB codes are given, DNA recognition sequences are shown with M = 5mC, and key interactions of each DNA strand are shown above and below the recognition sequence. (b) Kaiso (ZF), (c) Zfp57 (ZF), (d) Klf4 (ZF), (e) Egr1 (ZF) and (f) WT1 (ZF). (g) Effects of binding of DNA with 5caC against Egr1 and WT1. (h) WT1 (Q369) recognition of DNA with 5caC. (i) The p53 proteins recognize TpG, which can be replaced by 5mCpG. (j) Klf4 mutant (D446) interacts with unmodified cytosine. (k) In CTCF (ZF), D451 and E362 recognize, respectively, unmodified cytosine at position 2 and 5mC at position 12. (l) Effects of methylation at specific sites on binding by CTCF. (m) MAX (bHLH) recognition of 5caC. (n) Pol II recognition of 5caC. (o) Tet3 CXXC domain recognition of 5caC.
MeCP2 also binds methylated 5mCpA/TpG sites [22–25,26•,27••], with similar affinity to that of fully-methylated 5mCpG sites [28•], suggesting that ‘methyl–Arg–G triads’ would be a more appropriate name. Likewise, the MBD domain of MBD4 binds dinucleotides containing fully-methylated 5mCpG, and even to G:T mismatches occurring in the CpG context (5mCpG/TpG) [29]. In all three cases (5mCpG/5mCpG, 5mCpA/TpG, and 5mCpG/TpG), there are two symmetrically-positioned methyl groups, suggesting that the MBD proteins simultaneously recognize the two methyl groups on opposite strands. The 5mCpA/TpG binding may allow MeCP2 to control transcription in a ‘rheostat-like’ manner, in response to Dnmt3a activity, fine-tuning the cell-type-specific transcription of genes that are critical for brain function [14•].
C2H2 zinc finger (ZF) proteins
Two decades ago, Holliday argued that ‘sequences longer than CpG would be necessary for the regulation of gene expression by methylation’ [30]. Indeed, the recognition of some sequence-specific transcription factors is blocked by cytosine methylation only within the context of a longer specific sequence (e.g., the proteins CTCF [31,32] and Myc [33]). By contrast, certain Cys2-His2 (C2H2) zinc finger (ZF) proteins bind preferentially to DNA when CpG sites embedded within their recognition sequences are methylated [34]. The structures of several ZF domains bound to 5mC-containing DNA have been solved, including the transcription factors Kaiso, Zfp57, Klf4 (Krüppel-like factor 4), Egr1 (growth response protein 1), WT1 (Wilms tumor protein 1), and CTCF [35–39,40••]. Kaiso recognizes either methylated CpG dinucleotides [41] or an unmodified sequence with a TpG in the place of 5mCpG [42]. Arg511 of the Kaiso ZF domain interacts with the 5mCpG and TpG dinucleotides in a similar fashion [35], forming a methyl–Arg–G triad [21] (Figure 2b) like the two arginines of MBD. Similarly, Zfp57 uses a pair of methyl–Arg–G triads to recognize 5mCpG and TpG dinucleotides on two DNA strands (Figure 2c).
Like Kaiso, the consensus-binding sequence element for Klf4 contains either CpG, which can be methylated, or TpG, which is intrinsically methylated on one strand (Figure 2d). Klf4 is one of the four Yamanaka reprogramming factors [43], and like the other three (Myc, Oct4, and Sox2), contains TpG/CpA in its consensus recognition sequence [44]. These two different categories of 5-methyl-bearing sites may play distinct roles in development: non-CpG (mainly CpA) methylation disappears upon induced differentiation of embryonic stem cells, and is restored in induced pluripotent stem cells by reprogramming factors [45] that recognize CpA (or 5mCpA)-containing sequences.
Both Egr1 and WT1 bind similar consensus DNA sequence containing two CpG sites, each having high affinity for the methylated form of the sequence (Figure 2e,f), but exhibiting much-reduced affinity when either 5hmC or 5fC is present. This indicates that both Egr1 and WT1 differentiate primarily between the oxidized and unoxidized forms of 5mC, rather than between methylated and unmethylated C [38]. By contrast, 5caC affects the two proteins differently, greatly reducing binding by Egr1 but not by WT1 (Figure 2g). This difference can be ascribed to electrostatic interactions. In Egr1, a glutamate (E354) repels the negatively-charged carboxylate of 5caC, while the corresponding glutamine of WT1 (Q369) interacts favorably with the carboxylate (Figure 2h). Analogous residues to Egr1 E354 are found in MBD proteins (D121/E137 of MeCP2), Kaiso (E535), Zfp57 (E182), Klf4 (E446) (Figure 2a–d), and p53 (D281) (Figure 2i, see below). Acidic residues, aspartate and glutamate, are used sparingly in protein–DNA interactions [46], in part because of unfavorable electrostatic interaction with DNA phosphate groups, but juxtaposing an acidic residue with cytosine can specifically repel the negatively-charged carboxylate of 5caC.
Aspartate for cytosine and glutamate for 5-methlycytosine in C2H2 ZFs
A phage display study of Egr1 (also known as Zif268) revealed that aspartate (D), rather than glutamate (E) in the wild type, distinctly prefers binding to unmodified cytosine [47]. This observation led Choo and Klug to comment that ‘The physical basis for the interaction of aspartate/glutamate and cytosine is not yet clear, since hydrogen bonding contacts between these groups have yet to be observed in zinc finger cocrystal structure’ [48]. In bacterial one-hybrid experiments, where only unmodified bases were present, D was again found to preferentially juxtapose to cytosine [49].
We tested D/E interaction with cytosine/5mC in two important, 5mC-responsive regulatory proteins: Klf4 and CTCF [40••,50•]. Klf4 is one member of the specificity protein/Krüppel-like factor (Sp/Klf) family of ZF transcription factors, and the 5mC-interacting E446 of Klf4 is invariant among family members [37]. In the corresponding position of mouse Klf1, the neonatal anemia (Nan) mouse carries an E-to-D mutation (E339D) [51]. As with Klf4, ChIP-seq data for wild type Klf1 has the consensus-binding sequence containing either C or T as the base recognized by the wild type (E339), while the mutant (D339) displayed increased specificity for (unmodified) cytosine [52•]. We generated a corresponding E446D mutant in Klf4. The wild type (E446) has roughly equal binding affinity for 5mC or T, while the mutant (D446) has a preference for unmodified cytosine, due to decreased affinity for 5mC [50•]. The carboxylate group of D446 forms H-bonds with unmodified cytosine’s exocyclic N4 amino group and with its ring carbon C5 atom (Figure 2j). Both of these interactions would favor cytosine over thymine, and the latter one could explain the D446 preference for unmodified cytosine. By contrast, the wild-type E446 interacts with neither the 5mC N4 nitrogen nor the thymine O4 oxygen (Figure 2d).
The CTCF protein influences global chromatin architecture by sequence-specific DNA binding, via a tandem array of eleven C2H2 ZFs, and is present at ~80 000 sites in mammalian genomes [53,54]. ChIP experiments uncovered a broad CTCF-binding motif that contains a 12–15 bp consensus sequence [55–57]. Comparison to bisulfite sequencing data of various human cell types indicated that ~40% of variable CTCF binding is linked to differential DNA methylation, concentrated at the two conserved Cyt positions (2 and 12) within the 15-bp recognition sequence [58]. CTCF binding to the H19 imprinting control region sequence was inhibited by DNA methylation at a single CpG site [31,32,59], corresponding to position 2. Two negatively charged residues, D451 and E362 of CTCF, recognize, respectively, the two invariant cytosines at positions 2 and 12 [40••] (Figure 2k). The binding affinity for the oligonucleotide methylated at C2 was drastically reduced (by a factor of 23), while affinity was slightly increased when methylated at C12 (by a factor of 1.5) [40••] (Figure 2l). The distinct effects of methylation at positions 2 and 12 on binding affinity are due to the amino acids used in the interaction, with D452 preferring C and E362 preferring 5mC (Figure 2k).
p53 protein
The human tumor suppressor protein p53 binds as a tetramer to two repeated DNA sites, with each monomer recognizing a pentamer repeat TGCC(C/T) [60]. Each p53 monomer uses a methyl–Arg–G triad to recognize the TpG dinucleotide (Figure 2i). Thus three very different structural classes of DNA binding domains, in MBD, C2H2 ZFs, and p53, use the same methyl–Arg–G triad [21]. Substituting TpG with 5mCpG results in the largest observed increase of binding affinity by p53; by contrast, substitution with unmodified CpG reduces binding by an order of magnitude [18••].
Basic-helix–loop–helix (bHLH) transcription factors
The basic-helix–loop–helix (bHLH) transcription factor MYC, together with its binding partner MAX, regulates gene expression by binding to enhancer-box (E-box) elements (5′-CACGTG-3′) that contain a central CpG dinucleotide. Methylation of the central CpG greatly inhibits DNA binding by MAX [33]. However, unexpectedly, MAX exhibits the greatest affinity for an E-box containing 5caC (which, as noted above, is negatively charged), and much reduced affinities for the corresponding 5mC, 5hmC or 5fC forms [61•]. For context, 5caC is found at low levels (1–10% that of 5hmC), and preferentially accumulates at enhancers and other distal regulatory regions [62–67]. Interestingly, MAX Arg36 recognizes 5caC using a 5caC–Arg–G triad (Figure 2m), and the nearest other MAX residues to the carboxylate group are Arg60, Arg33 and Lys40, which yield a basic environment ideal for electrostatic attraction to the two carboxylate groups of symmetrically-modified 5caC:G base pairs. Given the similarities between the bHLH domains of MAX and its binding partners, and the structural conservation of the critical amino acids whether partnered with MYC or other proteins, it is likely that the ability to discriminate among 5mC oxidation states applies to most (if not all) MAX heterodimers, and to the extended MYC family of bHLH transcription factors. Thus, 5caC has the potential to function epigenetically by repelling acidic residues of MBDs and ZFs, but not of neutral (uncharged Gln in WT1) or positive side chains (MAX). In addition, 5fC and 5caC in DNA retard transcriptional elongation by Pol II; we note that the polymerase II subunit Rpb2 also uses a glutamine to hydrogen bond with the carboxylate of 5caC (Figure 2n) [68]. These observations suggest that transcription factors, as well as Pol II itself, may act as direct epigenetic sensors for DNA modifications.
One of the mammalian 5mC dioxygenases, Tet3, has its binding sites enriched for a sequence motif (5′-TCACGTGA-3′) at the transcription start sites of genes, particularly those involved in lysosome function [69]. Interestingly, this 8-bp sequence motif contains an E-box, and perfectly matches the recognition sequence for TFEB, another bHLH transcription factor and the master regulator of lysosomal genes. It is not yet known if the TFEB recognition sequences are modified (methylated or 5-carboxylated) in cells, particularly those sites associated with Tet3. It is conceivable that the DNA-binding domain of TFEB, which resembles that of MAX, binds to 5caC generated by Tet3 within this sequence motif. Besides the dioxygenase domain, Tet3 contains a DNA-binding CXXC domain, which binds 5caC (Figure 2o), though in a different sequence context [69,70]. The relationship of the transcription factor TFEB with Tet3 echoes that of WT1 with Tet2, in that WT1 displays high affinity for 5mC or 5caC but much reduced affinity for 5hmC or 5fC [38]. WT1 and Tet2 physically interact with one another [71,72], so WT1 may recruit Tet2 to its targets (containing 5mC) and/or Tet2 — together with its product (5caC) — could recruit WT1.
Basic leucine-zipper (bZIP) transcription factor family
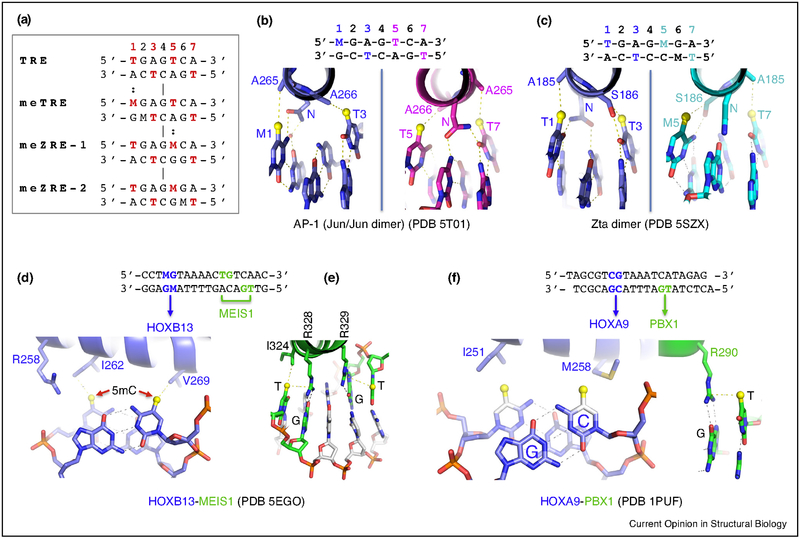
A classic basic leucine-zipper (bZIP) family transcription factor, activator protein 1 (AP-1), is substantially involved in gene regulation and controls such critical phenomena as oncogenesis, cell proliferation, and apoptosis [73,74]. Like MYC/MAX, AP-1 is a dimeric complex that comprises members of the Jun, Fos, ATF (activating transcription factor) and MAF (musculoaponeurotic fibrosarcoma) protein families [74]. AP-1 complexes include homodimers and heterodimers; for example, Jun/Jun and Jun/Fos activate a set of genes by binding a 7-bp element known as TRE (5′-TGAGTCA-3′), as well as a methylated response element known as meTRE (5′-MGAGTCA-3′ where M = 5mC) with a methylated CpG replacing the 5′ TpG dinucleotide [75–77]. Thus, the TRE and meTRE elements each contain two methyl groups at nucleotide positions 1 and 5 from the 5′ end, resulting in four methyl groups symmetrically positioned at base pair (bp) positions 1, 3, 5, and 7 (Figure 3a).
Figure 3.
Examples of methyl group recognition via van der Waals contacts. (a) Aligned DNA response elements with spatially equivalent methyl groups from T (5mU) or M (5mC). (b,c) Recognition of the four spatially constrained methyl groups of meTRE by Jun/Jun dimer (bZIP) or meZRE-2 by Zta dimer (bZIP). (d) Recognition of the two methyl groups of 5mCpG duplex by HOXB13 (homeodomain). (e) Homeodomain protein MEIS1 recognizes TpG via methyl–Arg–Gua triad. (f) HOXA9–PBX1 in complex with DNA. Both proteins could have preferential binding of methylated DNA. A methyl group (in yellow sphere) is modeled onto unmodified cytosine.
Epstein–Barr Virus (EBV) is a human-specific B cell-infecting gamma-herpesvirus [78,79]. An EBV transcription factor, Zta (also called EB1, BZLF1, or ZEBRA) was the first example of a sequence-specific transcription factor that preferentially binds methylated cytosine residues within a specific sequence, reverses epigenetic silencing and activates gene transcription [80]. The EBV virion genome is unmethylated, but becomes heavily methylated during the latent stage of the virus cycle [81–83]. Early lytic cycle activation depends upon the Zta homodimer preferentially recognizing methylated promoters containing meZREs, notable examples of which are meZRE1 (5′-TGAGMCA-3′) and meZRE2 (5′-TGAGMGA-3′) [80,84–86]. Significantly, this element contains two methyl groups at nucleotide positions 1 and 5 (Figure 3a), with a methylated cytosine in place of one of the inner thymine residues of the AP-1 element, resulting in the four spatially equivalent methyl groups. These methyl groups are in van der Waals contact with a conserved di-alanine in AP-1 dimer (Ala265 and Ala266 in Jun; Figure 3b), or with the corresponding Zta residues Ala185 and Ser186 (via its side chain carbon Cβ atom) (Figure 3c) [87•]. These analyses demonstrate a novel mechanism of 5mC/T recognition in a methylation-dependent, spatial and sequence-specific approach by bZIP transcription factors.
Besides binding 7-bp TRE elements (TGA-G-TCA), AP-1 Jun or Fos proteins can form heterodimers with ATF to recognize cAMP-response elements (CRE; TGA-CG-TCA) [74], which are also recognized by CRE-binding (CREB) transcription factors [88,89]. The difference between TRE and CRE elements is the one bp expansion in the central C:G bp to a CpG dinucleotide. Interestingly, CpG methylation decreased the affinity of ATF4, while replacing the outer T with 5mC led to greatly increased affinity [18••]. ChIP-seq data of CREB in hippocampal neurons identified a non-canonical CRE motif [90], where the inner A:T at bp-3 is replaced by G:C — methylation of the cytosine at bp-3 would restore the symmetrically positioned methyl groups. In addition, while methylation sensitivity was not seen for C/EBPβ binding to TTG-CG-CAA [18••], its binding was inhibited by 5hmC in the central CpG dinucleotide [91]. Indeed, the expanded central CpG dinucleotide potentially allows the CpG methylation/oxidation status to play a modulating role (e.g., see MAX above).
Homeodomain transcription factors
Homeodomain proteins are the family of transcription factors most-recently characterized as having preferential binding of methylated DNA [17••]. This class of proteins was enriched in factors playing central roles in embryonic and organismal development [92]. Crystal structures of the homeodomain proteins HOXB13, CDX1, CDX2, and LHX4 revealed a pair of hydrophobic residues making van der Waals contacts to the two methyl groups of duplex 5mCpG dinucleotides (Figure 3d) [17••]. HOXB13 also forms complexes with the TALE-class homeodomain protein MEIS1 [93], which recognizes a sequence motif containing two pairs of CpA/TpG sites (Figure 3e). MEIS1, like other proteins referred to above that recognize TpG, uses two neighboring arginines with each forming a methyl–Arg–G triad (Figure 3e). The observation made with HOXB13–MEIS1 complex could be extended to other TALE-mediated HOX complexes [18••], such as HOXB1–PBX1 [94] or HOXA9–PBX1 [95] (Figure 3f). Whether 5mCpG can replace the TpG, and the effects on MEIS1 or PBX1 binding, have yet to be tested.
SRA domains
The SET and RING finger associated (SRA) domains of UHRF1 and UHRF2 recognize hemimethylated CpG sequences, which contain 5mC in only one strand, such as arise during DNA replication [96–98]. UHRF1 (residues 124–628) binds to 5hmC DNA with >10-fold weaker affinity than to 5mC DNA [99], while UHRF2 (whether the SRA domain or full-length protein) showed small or no difference in binding affinity between the two modifications [100,131]. In addition, UHRF2 can directly and specifically bind A:5hmU in vitro (in contrast to A:5mU, which is A:T) [15]. The SRA domain uses a DNA recognition mode vastly different from that of other classes discussed above. Specifically, the SRA domain uses base flipping [101,102] to interact with DNA, similarly to DNA-modifying enzymes such as DNA methyltransferases, DNA glycosylases and Tet dioxygenases. Besides the SRA domain, UHRF1 contains a Tudor-PHD domain for binding one of three ligands: histone H3 methylated at Lys9 (H3K9, which is associated with methylated DNA) [103–107], an internal loop region of UHRF1 for allosteric regulation [108•], or DNA ligase 1 [109••] which is needed for the ligation of Okazaki fragments that are formed on the lagging template strand during DNA replication. UHRF1 also contains a C-terminal RING domain for E3 ligase activity that can ubiquitylate histone H3 [110,111], which is recognized by maintenance methyltransferase DNMT1 [112,113,114••]. Because UHRF1 is an important epigenetic regulator, maintaining DNA methylation and histone modifications in the cell, recent cancer research supports expression levels of UHRF1 serving as a universal diagnostic and prognostic biomarker [115], suggesting that better understanding of its activities is important.
N6-methyladenine in mammalian DNA and RNA
This section is brief due to space restrictions, but it has recently become clear that mammalian DNA can be methylated on adenines at the N6 position (N6mA) [116•,117•,118•]. This methylation is presumably subject to oxidative removal by Tet-like dioxygeneases [119]. Unlike the case for cytosine, however, Tet action on N6mA results in restoration of adenine (direct demethylation), involving formation of an N6-hydroxymethyl intermediate followed by spontaneous release of formal-dehyde. This differs from the series of oxidative states on 5mC, required because the cytosine C5 atom is an inert carbon. Very little is known about the ways in which N6mA DNA methyl marks are established, maintained, altered, or read in mammals, and which DNA-binding proteins are sensitive to those marks. We expect this to be a very active area of research over the next several years. However some hints as to a possible readout mechanism are provided by the better-studied detection of N6mA in RNA. This often involves an N6mA methyltransferase complex (METTL3–METTL14) [120,121], demethylases/dioxygenases (FTO and ALKBH5) [122,123], and N6mA-specific RNA-binding YTH domain proteins [124–127]. The YTH domain is a conserved 100–150-residue polypeptide [128], in which the N6mA methyl group is bound within a ‘cage’ of aromatic residues [129].
Summary
It is well established that methylation patterns are replicated, following semi-conservative DNA replication, via the selective recognition of hemi-methylated CpG dinucleotides at replication forks by DNMT1/UHRF1 complexes. This involves reading methyl marks on both DNA and associated histones, and mono-ubiquitination of histone H3. However, an unresolved fundamental question is how, and indeed whether, the pattern of oxidized 5mC derivatives is propagated at CpG and CpA sites. Many DNA binding proteins recognize consensus-binding elements, containing either CpG/CpG (which can be methylated on both strands), or TpG/CpA (which is intrinsically methylated on one strand, may be methylated on the other strand, and can be further modified on both strands). Because the methyl–Arg–G triad recognizes both 5mC and T, perhaps TpG dinucleotides are selected for when it is advantageous for a particular DNA sequence to be treated as if it is permanently methylated; this might even occur via targeted 5mC deamination [130] during development. On the other hand, the substitution of T by C (as in the cases of AP-1 and Zta) provides an opportunity for regulation by methylation and demethylation.
Transcription factors have adapted to respond to different states of cytosine modification, and gene activity is controlled on a finer ‘dimmer’ by these modifications than a simple ‘on’ or ‘off’ switch. For example, the DNA binding bHLH domain of MAX is responsive to all forms of modified CpG within its E-box recognition sequence, displaying the highest affinity for cognate sequences containing a central 5caC or unmodified C, with reduced affinity for 5fC and much lower affinity for 5mC or 5hmC. Progressive Tet-mediated oxidation of 5mC may thus be a way to titrate transcriptional activity in a graded and reversible fashion, with the 5mC form of such sites being the ‘off’ position and Tet-mediated oxidation steps a way of progressively increasing affinity for the binding site while moving towards 5caC. As many promoters are controlled by multiple transcription factors, several of which can have different patterns of methylation responsiveness, a very large number of combinatorial regulatory outputs is possible. In sum, a growing number of transcriptional regulators are being recognized as being responsive to different cytosine modification states, potentially acting as direct epigenetic sensors to instruct downstream events.
Acknowledgements
We thank Dr. Hideharu Hashimoto, Dr. Samuel Hong, Dr. Anamika Patel, and Dr. Dongxue Wang for discussion. The work in the Cheng Laboratory was supported in part by the National Institutes of Health (grant GM049245) and Cancer Prevention and Research Institute of Texas (RR160029).
Footnotes
Conflict of interest statement
The authors declare no conflict of interest.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as
• of special interest
•• of outstanding interest
- 1.Kriaucionis S, Heintz N: The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 2009, 324:929–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L et al. : Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 2009, 324:930–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y: Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 2011, 333:1300–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L et al. : Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 2011, 333:1303–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pfaffeneder T, Hackner B, Truss M, Munzel M, Muller M, Deiml CA, Hagemeier C, Carell T: The discovery of 5-formylcytosine in embryonic stem cell DNA. Angew Chem Int Ed Engl 2011, 50:7008–7012. [DOI] [PubMed] [Google Scholar]
- 6.Nakayama A, Yamazaki S, Taketsugu T: Quantum chemical investigations on the nonradiative deactivation pathways of cytosine derivatives. J Phys Chem A 2014, 118:9429–9437. [DOI] [PubMed] [Google Scholar]
- 7.Szulik MW, Pallan PS, Nocek B, Voehler M, Banerjee S, Brooks S, Joachimiak A, Egli M, Eichman BF, Stone MP: Differential stabilities and sequence-dependent base pair opening dynamics of Watson–Crick base pairs with 5-hydroxymethylcytosine, 5-formylcytosine, or 5-carboxylcytosine. Biochemistry 2015, 54:1294–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dai Q, Sanstead PJ, Peng CS, Han D, He C, Tokmakoff A: Weakened N3 hydrogen bonding by 5-formylcytosine and 5-carboxylcytosine reduces their base-pairing stability. ACS Chem Biol 2016, 11:470–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gasser SM, Li E: Epigenetics and disease: pharmaceutical opportunities. Prog Drug Res 2011, 67. [PubMed] [Google Scholar]
- 10.Huang Y, Rao A: Connections between TET proteins and aberrant DNA modification in cancer. Trends Genet 2014, 30:464–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bestor T, Laudano A, Mattaliano R, Ingram V: Cloning and sequencing of a cDNA encoding DNA methyltransferase of mouse cells. The carboxyl-terminal domain of the mammalian enzymes is related to bacterial restriction methyltransferases. J Mol Biol 1988, 203:971–983. [DOI] [PubMed] [Google Scholar]
- 12.Okano M, Xie S, Li E: Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat Genet 1998, 19:219–220. [DOI] [PubMed] [Google Scholar]
- 13.Gowher H, Jeltsch A: Enzymatic properties of recombinant Dnmt3a DNA methyltransferase from mouse: the enzyme modifies DNA in a non-processive manner and also methylates non-CpG [correction of non-CpA] sites. J Mol Biol 2001, 309:1201–1208. [DOI] [PubMed] [Google Scholar]
- 14.•.Stroud H, Su SC, Hrvatin S, Greben AW, Renthal W, Boxer LD, Nagy MA, Hochbaum DR, Kinde B, Gabel HW et al. : Early-life gene expression in neurons modulates lasting epigenetic states. Cell 2017, 171:1151–1164.The authors demonstrate that DNMT3A transiently binds transcribed regions of poorly-expressed genes in developing brain, initiating DNA methylation at CpA/TpG sequences, and that this methylation is inhibited by transcription. The 5mCpA/TpG is bound by the methyl-DNA-binding protein MECP2 and reduces transcription, reinforcing regions of low transcriptional activity.
- 15.Pfaffeneder T, Spada F, Wagner M, Brandmayr C, Laube SK, Eisen D, Truss M, Steinbacher J, Hackner B, Kotljarova O et al. : Tet oxidizes thymine to 5-hydroxymethyluracil in mouse embryonic stem cell DNA. Nat Chem Biol 2014, 10:574–581. [DOI] [PubMed] [Google Scholar]
- 16.Pais JE, Dai N, Tamanaha E, Vaisvila R, Fomenkov AI, Bitinaite J, Sun Z, Guan S, Correa IR Jr, Noren CJ et al. : Biochemical characterization of a Naegleria TET-like oxygenase and its application in single molecule sequencing of 5-methylcytosine. Proc Natl Acad Sci U S A 2015, 112:4316–4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.••.Yin Y, Morgunova E, Jolma A, Kaasinen E, Sahu B, Khund-Sayeed S, Das PK, Kivioja T, Dave K, Zhong F et al. : Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science 2017, 356.The authors use methylation-sensitive SELEX to assess over 500 human transcription factors, finding that many prefer methylated CpG sequences. The majority is in the extended homeodomain family, and play developmentally important roles.
- 18.••.Kribelbauer JF, Laptenko O, Chen S, Martini GD, Freed-Pastor WA, Prives C, Mann RS, Bussemaker HJ: Quantitative analysis of the DNA methylation sensitivity of transcription factor complexes. Cell Rep 2017, 19:2383–2395.The authors report a method, called EpiSELEX-seq, to assess the effects of 5mCpG on transcription factor binding free energy in a position-specific manner. They find that 5mCpG can both increase and decrease affinity, depending on where the modification occurs within the target site.
- 19.Lewis JD, Meehan RR, Henzel WJ, Maurer-Fogy I, Jeppesen P, Klein F, Bird A: Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell 1992, 69:905–914. [DOI] [PubMed] [Google Scholar]
- 20.Ho KL, McNae IW, Schmiedeberg L, Klose RJ, Bird AP, Walkinshaw MD: MeCP2 binding to DNA depends upon hydration at methyl-CpG. Mol Cell 2008, 29:525–531. [DOI] [PubMed] [Google Scholar]
- 21.Liu Y, Zhang X, Blumenthal RM, Cheng X: A common mode of recognition for methylated CpG. Trends Biochem Sci 2013, 38:177–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo JU, Su Y, Shin JH, Shin J, Li H, Xie B, Zhong C, Hu S, Le T, Fan G et al. : Distribution, recognition and regulation of non-CpG methylation in the adult mammalian brain. Nat Neurosci 2014, 17:215–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gabel HW, Kinde B, Stroud H, Gilbert CS, Harmin DA, Kastan NR, Hemberg M, Ebert DH, Greenberg ME: Disruption of DNA-methylation-dependent long gene repression in Rett syndrome. Nature 2015, 522:89–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen L, Chen K, Lavery LA, Baker SA, Shaw CA, Li W, Zoghbi HY: MeCP2 binds to non-CG methylated DNA as neurons mature, influencing transcription and the timing of onset for Rett syndrome. Proc Natl Acad Sci U S A 2015, 112:5509–5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kinde B, Gabel HW, Gilbert CS, Griffith EC, Greenberg ME: Reading the unique DNA methylation landscape of the brain: non-CpG methylation, hydroxymethylation, and MeCP2. Proc Natl Acad Sci U S A 2015, 112:6800–6806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.•.Kinde B, Wu DY, Greenberg ME, Gabel HW: DNA methylation in the gene body influences MeCP2-mediated gene repression. Proc Natl Acad Sci U S A 2016, 113:15114–15119.The authors find that MeCP2-repressed genes in brain are characterized by megabase regions of high methylation density, at both CpG and CpA sites. They suggest that the MeCP2 binding interferes with transcript elongation.
- 27.••.Lagger S, Connelly JC, Schweikert G, Webb S, Selfridge J, Ramsahoye BH, Yu M, He C, Sanguinetti G, Sowers LC et al. : MeCP2 recognizes cytosine methylated tri-nucleotide and di-nucleotide sequences to tune transcription in the mammalian brain. PLoS Genet 2017, 13:e1006793.The authors report that non-CpG binding of MeCP2 in brain is preferentially at the subset of CpA sites followed by C (5mCpApC), and occurs across long genomic domains. They find an inverse relationship between transcription and MeCP2 occupancy.
- 28.•.Sperlazza MJ, Bilinovich SM, Sinanan LM, Javier FR, Williams DC Jr: Structural Basis of MeCP2 Distribution on non-CpG methylated and hydroxymethylated DNA. J Mol Biol 2017, 429:1581–1594.The authors use biophysical methods to compare MeCP2, MBD2, and Rett syndrome-associated MeCP2 variant binding to 5mCpG and 5mCpA and also examine the effects of hydroxymethylation.
- 29.Otani J, Arita K, Kato T, Kinoshita M, Kimura H, Suetake I, Tajima S, Ariyoshi M, Shirakawa M: Structural basis of the versatile DNA recognition ability of the methyl-CpG binding domain of methyl-CpG binding domain protein 4. J Biol Chem 2013, 288:6351–6362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holliday R: DNA methylation in eukaryotes: 20 years on. Epigenetic Mech Gene Regulat 1996:5–27. [Google Scholar]
- 31.Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM: CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature 2000, 405:486–489. [DOI] [PubMed] [Google Scholar]
- 32.Renda M, Baglivo I, Burgess-Beusse B, Esposito S, Fattorusso R, Felsenfeld G, Pedone PV: Critical DNA binding interactions of the insulator protein CTCF: a small number of zinc fingers mediate strong binding, and a single finger–DNA interaction controls binding at imprinted loci. J Biol Chem 2007, 282:33336–33345. [DOI] [PubMed] [Google Scholar]
- 33.Prendergast GC, Lawe D, Ziff EB: Association of Myn, the murine homolog of max, with c-Myc stimulates methylation-sensitive DNA binding and ras cotransformation. Cell 1991, 65:395–407. [DOI] [PubMed] [Google Scholar]
- 34.Sasai N, Nakao M, Defossez PA: Sequence-specific recognition of methylated DNA by human zinc-finger proteins. Nucleic Acids Res 2010, 38:5015–5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Buck-Koehntop BA, Stanfield RL, Ekiert DC, Martinez-Yamout MA, Dyson HJ, Wilson IA, Wright PE: Molecular basis for recognition of methylated and specific DNA sequences by the zinc finger protein Kaiso. Proc Natl Acad Sci U S A 2012, 109:15229–15234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu Y, Toh H, Sasaki H, Zhang X, Cheng X: An atomic model of Zfp57 recognition of CpG methylation within a specific DNA sequence. Genes Dev 2012, 26:2374–2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu Y, Olanrewaju YO, Zheng Y, Hashimoto H, Blumenthal RM, Zhang X, Cheng X: Structural basis for Klf4 recognition of methylated DNA. Nucleic Acids Res 2014, 42:4859–4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hashimoto H, Olanrewaju YO, Zheng Y, Wilson GG, Zhang X, Cheng X: Wilms tumor protein recognizes 5-carboxylcytosine within a specific DNA sequence. Genes Dev 2014, 28:2304–2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zandarashvili L, White MA, Esadze A, Iwahara J: Structural impact of complete CpG methylation within target DNA on specific complex formation of the inducible transcription factor Egr-1. FEBS Lett 2015, 589:1748–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.••.Hashimoto H, Wang D, Horton JR, Zhang X, Corces VG, Cheng X: Structural basis for the versatile and methylation-dependent binding of CTCF to DNA. Mol Cell 2017, 66:711–720.The authors report the crystal structure of human CTCF DNA-binding domain bound to a known CTCF-binding site. The structure assigned roles to each of the 11 Zn fingers, and suggests a basis for CTCF sensitivity to 5mC.
- 41.Prokhortchouk A, Hendrich B, Jorgensen H, Ruzov A, Wilm M, Georgiev G, Bird A, Prokhortchouk E: The p120 catenin partner Kaiso is a DNA methylation-dependent transcriptional repressor. Genes Dev 2001, 15:1613–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Daniel JM, Spring CM, Crawford HC, Reynolds AB, Baig A: The p120(ctn)-binding partner Kaiso is a bi-modal DNA-binding protein that recognizes both a sequence-specific consensus and methylated CpG dinucleotides. Nucleic Acids Res 2002, 30:2911–2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takahashi K, Yamanaka S: Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126:663–676. [DOI] [PubMed] [Google Scholar]
- 44.Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J et al. : Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 2008, 133:1106–1117. [DOI] [PubMed] [Google Scholar]
- 45.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM et al. : Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 2009, 462:315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luscombe NM, Laskowski RA, Thornton JM: Amino acid–base interactions: a three-dimensional analysis of protein–DNA interactions at an atomic level. Nucleic Acids Res 2001, 29:2860–2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Choo Y, Klug A: Selection of DNA binding sites for zinc fingers using rationally randomized DNA reveals coded interactions. Proc Natl Acad Sci U S A 1994, 91:11168–11172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Choo Y, Klug A: Physical basis of a protein–DNA recognition code. Curr Opin Struct Biol 1997, 7:117–125. [DOI] [PubMed] [Google Scholar]
- 49.Gupta A, Christensen RG, Bell HA, Goodwin M, Patel RY, Pandey M, Enuameh MS, Rayla AL, Zhu C, Thibodeau-Beganny S et al. : An improved predictive recognition model for Cys2-His2 zinc finger proteins. Nucleic Acids Res 2014, 42:4800–4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.•.Hashimoto H, Wang D, Steves AN, Jin P, Blumenthal RM, Zhang X, Cheng X: Distinctive Klf4 mutants determine preference for DNA methylation status. Nucleic Acids Res 2016, 44:10177–10185.The authors examine mouse reprogramming factor Klf4, with focus on its 5-methyl preference (satisfied by either 5mC or T), and report on the key role of Glu446 in Zn finger 2.
- 51.Siatecka M, Sahr KE, Andersen SG, Mezei M, Bieker JJ, Peters LL: Severe anemia in the Nan mutant mouse caused by sequence-selective disruption of erythroid Kruppel-like factor. Proc Natl Acad Sci U S A 2010, 107:15151–15156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.•.Gillinder KR, Ilsley MD, Nebor D, Sachidanandam R, Lajoie M, Magor GW, Tallack MR, Bailey T, Landsberg MJ, Mackay JP et al. : Promiscuous DNA-binding of a mutant zinc finger protein corrupts the transcriptome and diminishes cell viability. Nucleic Acids Res 2017, 45:1130–1143.The authors examine the role of Zn finger 2 in mouse Klf1, looking genome-wide at binding and transcriptional effects of a missense mutation that leads to degenerate specificity and genetically dominant gene expression changes.
- 53.Chen H, Tian Y, Shu W, Bo X, Wang S: Comprehensive identification and annotation of cell type-specific and ubiquitous CTCF-binding sites in the human genome. PLoS One 2012, 7:e41374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maurano MT, Wang H, John S, Shafer A, Canfield T, Lee K, Stamatoyannopoulos JA: Role of DNA methylation in modulating transcription factor occupancy. Cell Rep 2015, 12:1184–1195. [DOI] [PubMed] [Google Scholar]
- 55.Nakahashi H, Kwon KR, Resch W, Vian L, Dose M, Stavreva D, Hakim O, Pruett N, Nelson S, Yamane A et al. : A genome-wide map of CTCF multivalency redefines the CTCF code. Cell Rep 2013, 3:1678–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rhee HS, Pugh BF: Comprehensive genome-wide protein–DNA interactions detected at single-nucleotide resolution. Cell 2011, 147:1408–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jothi R, Cuddapah S, Barski A, Cui K, Zhao K: Genome-wide identification of in vivo protein–DNA binding sites from ChIP-Seq data. Nucleic Acids Res 2008, 36:5221–5231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang H, Maurano MT, Qu H, Varley KE, Gertz J, Pauli F, Lee K, Canfield T, Weaver M, Sandstrom R et al. : Widespread plasticity in CTCF occupancy linked to DNA methylation. Genome Res 2012, 22:1680–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bell AC, Felsenfeld G: Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 2000, 405:482–485. [DOI] [PubMed] [Google Scholar]
- 60.Petty TJ, Emamzadah S, Costantino L, Petkova I, Stavridi ES, Saven JG, Vauthey E, Halazonetis TD: An induced fit mechanism regulates p53 DNA binding kinetics to confer sequence specificity. EMBO J 2011, 30:2167–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.•.Wang D, Hashimoto H, Zhang X, Barwick BG, Lonial S, Boise LH, Vertino PM, Cheng X: MAX is an epigenetic sensor of 5-carboxylcytosine and is altered in multiple myeloma. Nucleic Acids Res 2017, 45:2396–2407.The authors study the MYC binding partner MAX, which binds E-boxes in a modification-dependent manner. They find that MAX prefers the unmodified or 5-carboxy versions of CpG in E-boxes, and examine the effects of mutations associated with primary multiple myelomas.
- 62.Shen L, Wu H, Diep D, Yamaguchi S, D’Alessio AC, Fung HL, Zhang K, Zhang Y: Genome-wide analysis reveals TET- and TDG-dependent 5-methylcytosine oxidation dynamics. Cell 2013, 153:692–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Song CX, Szulwach KE, Dai Q, Fu Y, Mao SQ, Lin L, Street C, Li Y, Poidevin M, Wu H et al. : Genome-wide profiling of 5-formylcytosine reveals its roles in epigenetic priming. Cell 2013, 153:678–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu H, Wu X, Shen L, Zhang Y: Single-base resolution analysis of active DNA demethylation using methylase-assisted bisulfite sequencing. Nat Biotechnol 2014, 32:1231–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lu X, Han D, Zhao BS, Song CX, Zhang LS, Dore LC, He C: Base-resolution maps of 5-formylcytosine and 5-carboxylcytosine reveal genome-wide DNA demethylation dynamics. Cell Res 2015, 25:386–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sun Z, Dai N, Borgaro JG, Quimby A, Sun D, Correa IR Jr, Zheng Y, Zhu Z, Guan S: A sensitive approach to map genome-wide 5-hydroxymethylcytosine and 5-formylcytosine at single-base resolution. Mol Cell 2015, 57:750–761. [DOI] [PubMed] [Google Scholar]
- 67.Neri F, Incarnato D, Krepelova A, Rapelli S, Anselmi F, Parlato C, Medana C, Dal Bello F, Oliviero S: Single-base resolution analysis of 5-formyl and 5-carboxyl cytosine reveals promoter DNA methylation dynamics. Cell Rep 2015, 10:574–683. [DOI] [PubMed] [Google Scholar]
- 68.Wang L, Zhou Y, Xu L, Xiao R, Lu X, Chen L, Chong J, Li H, He C, Fu XD et al. : Molecular basis for 5-carboxycytosine recognition by RNA polymerase II elongation complex. Nature 2015, 523:621–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jin SG, Zhang ZM, Dunwell TL, Harter MR, Wu X, Johnson J, Li Z, Liu J, Szabo PE, Lu Q et al. : Tet3 reads 5-carboxylcytosine through its CXXC domain and is a potential guardian against neurodegeneration. Cell Rep 2016, 14:493–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xu Y, Xu C, Kato A, Tempel W, Abreu JG, Bian C, Hu Y, Hu D, Zhao B, Cerovina T et al. : Tet3 CXXC domain and dioxygenase activity cooperatively regulate key genes for Xenopus eye and neural development. Cell 2012, 151:1200–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rampal R, Alkalin A, Madzo J, Vasanthakumar A, Pronier E, Patel J, Li Y, Ahn J, Abdel-Wahab O, Shih A et al. : DNA hydroxymethylation profiling reveals that WT1 mutations result in loss of TET2 function in acute myeloid leukemia. Cell Rep 2014, 9:1841–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang Y, Xiao M, Chen X, Chen L, Xu Y, Lv L, Wang P, Yang H, Ma S, Lin H et al. : WT1 recruits TET2 to regulate its target gene expression and suppress leukemia cell proliferation. Mol Cell 2015, 57:662–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Karin M, Liu Z, Zandi E: AP-1 function and regulation. Curr Opin Cell Biol 1997, 9:240–246. [DOI] [PubMed] [Google Scholar]
- 74.Eferl R, Wagner EF: AP-1: a double-edged sword in tumorigenesis. Nat Rev Cancer 2003, 3:859–868. [DOI] [PubMed] [Google Scholar]
- 75.Glover JN, Harrison SC: Crystal structure of the heterodimeric bZIP transcription factor c-Fos-c-Jun bound to DNA. Nature 1995, 373:257–261. [DOI] [PubMed] [Google Scholar]
- 76.Tulchinsky EM, Georgiev GP, Lukanidin EM: Novel AP-1 binding site created by DNA-methylation. Oncogene 1996, 12:1737–1745. [PubMed] [Google Scholar]
- 77.Gustems M, Woellmer A, Rothbauer U, Eck SH, Wieland T, Lutter D, Hammerschmidt W: c-Jun/c-Fos heterodimers regulate cellular genes via a newly identified class of methylated DNA sequence motifs. Nucleic Acids Res 2014, 42:3059–3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chiu YF, Sugden B: Epstein–Barr virus: the path from latent to productive infection. Annu Rev Virol 2016, 3:359–372. [DOI] [PubMed] [Google Scholar]
- 79.Sugden B: Epstein–Barr virus: the path from association to causality for a ubiquitous human pathogen. PLoS Biol 2014, 12: e1001939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bhende PM, Seaman WT, Delecluse HJ, Kenney SC: The EBV lytic switch protein, Z, preferentially binds to and activates the methylated viral genome. Nat Genet 2004, 36:1099–1104. [DOI] [PubMed] [Google Scholar]
- 81.Paulson EJ, Speck SH: Differential methylation of Epstein–Barr virus latency promoters facilitates viral persistence in healthy seropositive individuals. J Virol 1999, 73:9959–9968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fernandez AF, Rosales C, Lopez-Nieva P, Grana O, Ballestar E, Ropero S, Espada J, Melo SA, Lujambio A, Fraga MF et al. : The dynamic DNA methylomes of double-stranded DNA viruses associated with human cancer. Genome Res 2009, 19:438–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kenney SC, Mertz JE: Regulation of the latent-lytic switch in Epstein–Barr virus. Semin Cancer Biol 2014, 26:60–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Farrell PJ, Rowe DT, Rooney CM, Kouzarides T: Epstein–Barr virus BZLF1 trans-activator specifically binds to a consensus AP-1 site and is related to c-fos. EMBO J 1989, 8:127–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bergbauer M, Kalla M, Schmeinck A, Gobel C, Rothbauer U, Eck S, Benet-Pages A, Strom TM, Hammerschmidt W: CpG-methylation regulates a class of Epstein–Barr virus promoters. PLoS Pathog 2010, 6:e1001114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yu KP, Heston L, Park R, Ding Z, Wang’ondu R, Delecluse HJ, Miller G: Latency of Epstein–Barr virus is disrupted by gain-of-function mutant cellular AP-1 proteins that preferentially bind methylated DNA. Proc Natl Acad Sci U S A 2013, 110:8176–8181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.•.Hong S, Wang D, Horton JR, Zhang X, Speck SH, Blumenthal RM, Cheng X: Methyl-dependent and spatial-specific DNA recognition by the orthologous transcription factors human AP-1 and Epstein–Barr virus Zta. Nucleic Acids Res 2017, 45:2503–2515.The authors examine AP-1 and its Epstein-Barr viral ortholog Zta, with focus on their 5-methyl preferences (satisfied by either 5mC or T). The distinct context dependencies of AP-1 and Zta for 5-methyl groups are explained by a key Ala-Ala vs. Ala-Ser dipeptide.
- 88.Montminy MR, Sevarino KA, Wagner JA, Mandel G, Goodman RH: Identification of a cyclic-AMP-responsive element within the rat somatostatin gene. Proc Natl Acad Sci U S A 1986, 83:6682–6686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Schumacher MA, Goodman RH, Brennan RG: The structure of a CREB bZIP.somatostatin CRE complex reveals the basis for selective dimerization and divalent cation-enhanced DNA binding. J Biol Chem 2000, 275:35242–35247. [DOI] [PubMed] [Google Scholar]
- 90.Lesiak A, Pelz C, Ando H, Zhu M, Davare M, Lambert TJ, Hansen KF, Obrietan K, Appleyard SM, Impey S et al. : A genome-wide screen of CREB occupancy identifies the RhoA inhibitors Par6C and Rnd3 as regulators of BDNF-induced synaptogenesis. PLoS One 2013, 8e64658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Khund Sayeed S, Zhao J, Sathyanarayana BK, Golla JP, Vinson C: C/EBPbeta (CEBPB) protein binding to the C/EBP|CRE DNA 8-mer TTGC|GTCA is inhibited by 5hmC and enhanced by 5mC, 5fC, and 5caC in the CG dinucleotide. Biochim Biophys Acta 2015, 1849:583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Burglin TR, Affolter M: Homeodomain proteins: an update. Chromosoma 2016, 125:497–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jolma A, Yin Y, Nitta KR, Dave K, Popov A, Taipale M, Enge M, Kivioja T, Morgunova E, Taipale J: DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature 2015, 527:384–388. [DOI] [PubMed] [Google Scholar]
- 94.Piper DE, Batchelor AH, Chang CP, Cleary ML, Wolberger C: Structure of a HoxB1–Pbx1 heterodimer bound to DNA: role of the hexapeptide and a fourth homeodomain helix in complex formation. Cell 1999, 96:587–597. [DOI] [PubMed] [Google Scholar]
- 95.LaRonde-LeBlanc NA, Wolberger C: Structure of HoxA9 and Pbx1 bound to DNA: Hox hexapeptide and DNA recognition anterior to posterior. Genes Dev 2003, 17:2060–2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hashimoto H, Horton JR, Zhang X, Bostick M, Jacobsen SE, Cheng X: The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix. Nature 2008, 455:826–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Arita K, Ariyoshi M, Tochio H, Nakamura Y, Shirakawa M: Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism. Nature 2008, 455:818–821. [DOI] [PubMed] [Google Scholar]
- 98.Avvakumov GV, Walker JR, Xue S, Li Y, Duan S, Bronner C, Arrowsmith CH, Dhe-Paganon S: Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1. Nature 2008, 455:822–825. [DOI] [PubMed] [Google Scholar]
- 99.Hashimoto H, Liu Y, Upadhyay AK, Chang Y, Howerton SB, Vertino PM, Zhang X, Cheng X: Recognition and potential mechanisms for replication and erasure of cytosine hydroxymethylation. Nucleic Acids Res 2012, 40:4841–4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhou T, Xiong J, Wang M, Yang N, Wong J, Zhu B, Xu RM: Structural basis for hydroxymethylcytosine recognition by the SRA domain of UHRF2. Mol Cell 2014, 54:879–886. [DOI] [PubMed] [Google Scholar]
- 101.Roberts RJ, Cheng X: Base flipping. Annu Rev Biochem 1998, 67:181–198. [DOI] [PubMed] [Google Scholar]
- 102.Hong S, Cheng X: DNA base flipping: a general mechanism for writing, reading, and erasing DNA modifications. Adv Exp Med Biol 2016, 945:321–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Nady N, Lemak A, Walker JR, Avvakumov GV, Kareta MS, Achour M, Xue S, Duan S, Allali-Hassani A, Zuo X et al. : Recognition of multivalent histone states associated with heterochromatin by UHRF1 protein. J Biol Chem 2011, 286:24300–24311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rajakumara E, Wang Z, Ma H, Hu L, Chen H, Lin Y, Guo R, Wu F, Li H, Lan F et al. : PHD finger recognition of unmodified Histone H3R2 links UHRF1 to regulation of euchromatic gene expression. Mol Cell 2011, 43:275–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Arita K, Isogai S, Oda T, Unoki M, Sugita K, Sekiyama N, Kuwata K, Hamamoto R, Tochio H, Sato M et al. : Recognition of modification status on a histone H3 tail by linked histone reader modules of the epigenetic regulator UHRF1. Proc Natl Acad Sci U S A 2012, 109:12950–12955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rothbart SB, Krajewski K, Nady N, Tempel W, Xue S, Badeaux AI, Barsyte-Lovejoy D, Martinez JY, Bedford MT, Fuchs SM et al. : Association of UHRF1 with methylated H3K9 directs the maintenance of DNA methylation. Nat Struct Mol Biol 2012, 19:1155–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rothbart SB, Dickson BM, Ong MS, Krajewski K, Houliston S, Kireev DB, Arrowsmith CH, Strahl BD: Multivalent histone engagement by the linked tandem Tudor and PHD domains of UHRF1 is required for the epigenetic inheritance of DNA methylation. Genes Dev 2013, 27:1288–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.•.Fang J, Cheng J, Wang J, Zhang Q, Liu M, Gong R, Wang P, Zhang X, Feng Y, Lan W et al. : Hemi-methylated DNA opens a closed conformation of UHRF1 to facilitate its histone recognition. Nat Commun 2016, 7:11197.The authors explore the structural basis for preferential binding of URHF1 to hemimethylated CpG sites, and how such binding potentiates URHF1 recognition of H3K9me3.
- 109.••.Ferry L, Fournier A, Tsusaka T, Adelmant G, Shimazu T, Matano S, Kirsh O, Amouroux R, Dohmae N, Suzuki T et al. : Methylation of DNA ligase 1 by G9a/GLP recruits UHRF1 to replicating DNA and regulates DNA methylation. Mol Cell 2017, 67:550–565 e555.The authors report that, in addition to preference for hemimethylated DNA and H3K9me3, UHRF1 is directly recruited to replication forks by DNA ligase (LIG1). This recruitment involves a lysine-methylated histone H3-like segment within LIG1.
- 110.Citterio E, Papait R, Nicassio F, Vecchi M, Gomiero P, Mantovani R, Di Fiore PP, Bonapace IM: Np95 is a histone-binding protein endowed with ubiquitin ligase activity. Mol Cell Biol 2004, 24:2526–2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Karagianni P, Amazit L, Qin J, Wong J: ICBP90, a novel methyl K9 H3 binding protein linking protein ubiquitination with heterochromatin formation. Mol Cell Biol 2008, 28:705–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Nishiyama A, Yamaguchi L, Sharif J, Johmura Y, Kawamura T, Nakanishi K, Shimamura S, Arita K, Kodama T, Ishikawa F et al. : Uhrf1-dependent H3K23 ubiquitylation couples maintenance DNA methylation and replication. Nature 2013, 502:249–253. [DOI] [PubMed] [Google Scholar]
- 113.Qin W, Wolf P, Liu N, Link S, Smets M, La Mastra F, Forne I, Pichler G, Horl D, Fellinger K et al. : DNA methylation requires a DNMT1 ubiquitin interacting motif (UIM) and histone ubiquitination. Cell Res 2015, 25:911–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.••.Ishiyama S, Nishiyama A, Saeki Y, Moritsugu K, Morimoto D, Yamaguchi L, Arai N, Matsumura R, Kawakami T, Mishima Y et al. : Structure of the Dnmt1 reader module complexed with a unique two-mono-ubiquitin mark on histone H3 reveals the basis for DNA methylation maintenance. Mol Cell 2017, 68:350–360.The authors study the replication foci targeting sequence (RFTS) of Dnmt1, and find that it has an unusual mechanism for simultaneously recognizing two monoubiquitinated lysines in H3 (H3-K18Ub/23Ub). They also find that incubation of Dnmt1 with H3-K18Ub/23Ub increases its catalytic activity in vitro.
- 115.Ashraf W, Ibrahim A, Alhosin M, Zaayter L, Ouararhni K, Papin C, Ahmad T, Hamiche A, Mely Y, Bronner C et al. : The epigenetic integrator UHRF1: on the road to become a universal biomarker for cancer. Oncotarget 2017, 8:51946–51962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.•.Koziol MJ, Bradshaw CR, Allen GE, Costa AS, Frezza C, Gurdon JB: Identification of methylated deoxyadenosines in vertebrates reveals diversity in DNA modifications. Nat Struct Mol Biol 2016, 23:24–30.The authors report a detailed protocol for N6mA DNA immunoprecipitation (DIP).
- 117.•.Wu TP, Wang T, Seetin MG, Lai Y, Zhu S, Lin K, Liu Y, Byrum SD, Mackintosh SG, Zhong M et al. : DNA methylation on N(6)-adenine in mammalian embryonic stem cells. Nature 2016, 532:329–333.The authors find N6mA in murine embryonic stem cells, and deficiency for the Tet ortholog Alkbh1 were associated with both increased N6mA levels and decreased transcription. The N6mA was preferentially associated with LINE-1 transposons, with the X-chromosome-located younger L1 elements being particularly-heavily modified.
- 118.•.Yao B, Cheng Y, Wang Z, Li Y, Chen L, Huang L, Zhang W, Chen D, Wu H, Tang B et al. : DNA N6-methyladenine is dynamically regulated in the mouse brain following environmental stress. Nat Commun 2017, 8:1122.The authors find N6mA levels in brain DNA increase in response to stress, with corresponding changes in gene expression. The genes showing changed expression are enriched for loci associated with neuropsychiatric disorders.
- 119.Zhang G, Huang H, Liu D, Cheng Y, Liu X, Zhang W, Yin R, Zhang D, Zhang P, Liu J et al. : N(6)-methyladenine DNA modification in Drosophila. Cell 2015, 161:893–906. [DOI] [PubMed] [Google Scholar]
- 120.Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, Jia G, Yu M, Lu Z, Deng X et al. : A METTL3–METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol 2014, 10:93–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wang Y, Li Y, Toth JI, Petroski MD, Zhang Z, Zhao JC: N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat Cell Biol 2014, 16:191–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, Yi C, Lindahl T, Pan T, Yang YG et al. : N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol 2011, 7:885–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, Vagbo CB, Shi Y, Wang WL, Song SH et al. : ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell 2013, 49:18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Theler D, Dominguez C, Blatter M, Boudet J, Allain FH: Solution structure of the YTH domain in complex with N6-methyladenosine RNA: a reader of methylated RNA. Nucleic Acids Res 2014, 42:13911–13919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zhu T, Roundtree IA, Wang P, Wang X, Wang L, Sun C, Tian Y, Li J, He C, Xu Y: Crystal structure of the YTH domain of YTHDF2 reveals mechanism for recognition of N6-methyladenosine. Cell Res 2014, 24:1493–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Li F, Zhao D, Wu J, Shi Y: Structure of the YTH domain of human YTHDF2 in complex with an m(6)A mononucleotide reveals an aromatic cage for m(6)A recognition. Cell Res 2014, 24:1490–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Xu C, Wang X, Liu K, Roundtree IA, Tempel W, Li Y, Lu Z, He C, Min J: Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat Chem Biol 2014, 10:927–929. [DOI] [PubMed] [Google Scholar]
- 128.Zhang Z, Theler D, Kaminska KH, Hiller M, de la Grange P, Pudimat R, Rafalska I, Heinrich B, Bujnicki JM, Allain FH et al. : The YTH domain is a novel RNA binding domain. J Biol Chem 2010, 285:14701–14710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Luo S, Tong L: Molecular basis for the recognition of methylated adenines in RNA by the eukaryotic YTH domain. Proc Natl Acad Sci U S A 2014, 111:13834–13839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wijesinghe P, Bhagwat AS: Efficient deamination of 5-methylcytosines in DNA by human APOBEC3A, but not by AID or APOBEC3G. Nucleic Acids Res 2012, 40:9206–9217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Vaughan RM, Dickson BM, Cornett EM, Harrison JS, Kuhlman B, Rothbart SB: Comparative biochemical analysis of UHRF proteins reveals molecular mechanisms that uncouple UHRF2 from DNA methylation maintenance. Nucleic Acids Res 2018, 46:4405–4416. [DOI] [PMC free article] [PubMed] [Google Scholar]