Abstract
Hydroxyl-radical footprinting (HRF) is a powerful method to probe structures of nucleic acid-protein complexes with single nucleotide resolution in solution. To tap the full quantitative potential of HRF, we describe a protocol HYDroxyl-Radical footprinting Interpretation for DNA (HYDROID) to quantify HRF data and integrate it with atomistic structural models. The stages of the HYDROID protocol include extraction of the lane profiles from gel images, quantification of the DNA cleavage frequency at every nucleotide, and theoretical estimation of the DNA cleavage frequency from atomistic structural models followed by comparison of experimental and theoretical results. Example scripts for every step of HRF data analysis and interpretation are provided for several nucleosome systems; they can be easily adapted to analyze user data. As input HYDROID requires polyacrylamide gel electrophoresis images of HRF products and optionally may use a molecular model of the DNA-protein complex. The HYDROID protocol can be used to quantify HRF over DNA regions of up to 100 nucleotides per gel image. In addition it can be applied to the analysis of RNA-protein complexes and free RNA or DNA molecules in solution. Compared to other methods, HYDROID is unique in its ability to simultaneously integrate HRF data with the analysis of atomistic structural models. HYDROID is freely available at https://github.com/ncbi/HYDROID. The complete protocol takes approximately ~ 3 hours. Users should be familiar with the command line interface, the Python scripting language and PDB file formats. A graphical user interface with basic functionality (HYDROID_GUI) is also available.
Keywords: nucleic acid footprinting, hydroxyl radicals, DNA-protein complexes, nucleic acids cleavage, PAGE, gel image quantification, molecular modeling, solvent accessible surface area
EDITORIAL SUMMARY
Hydroxyl-radical footprinting provides a wealth of information on the structure of nucleic acid-protein complexes. HYDROID is a software tool to quantify footprinting data from gel electrophoresis images and integrate it with structural models.
TWEET
HYDROID: a tool to quantify cleavage profiles of protein-nucleic acid complexes from hydroxyl-radical footprinting experiments
COVER TEASER
HYDROID quantifies hydroxyl-radical footprinting
INTRODUCTION
Structural characterization of nucleic acid-protein complexes with high precision is essential for our understanding of genome functioning including the regulation of gene expression, transcription, DNA replication and repair. While X-ray crystallography remains the gold standard for structural characterization at atomistic level, the technique is tedious and often falls short due to crystallization problems. DNA footprinting techniques have been traditionally used as coarse, yet versatile and easy ways to probe DNA-protein interactions in vitro1 and in vivo2,3. These methods use a variety of chemical or enzymatic agents (e.g. DNAse I and hydroxyl-radicals) to cleave DNA at solvent exposed positions. The location of these positions can be subsequently identified by gel electrophoresis provided that DNA strands are labeled with radioactive or fluorescent labels on one end. The precision of enzymatic DNA cleavage is limited by steric and sequence specific effects4. On the other hand, HRF is a powerful technique as hydroxyl-radicals mostly lack sequence specific bias and provide high resolution (up to single nucleotide) due to their small sizes (see ref. 5 for the detailed description of the HRF technique).
The basic interpretation of HRF data is often performed qualitatively by examining corresponding DNA polyacrylamide gel electrophoresis (PAGE) images, or gel lane intensity profiles derived from these images. However, such an approach usually does not lead to direct 3D structural interpretation of the data. To uncover the full potential of this high quality experimental data, two tasks have to be implemented: 1) quantification of actual DNA cleavage frequencies for every nucleotide position from PAGE data and 2) evaluation of DNA cleavage frequencies from corresponding atomistic molecular models. Combined together, these two analysis components allow the interpretation of HRF data at a more advanced level and enable the incorporation of the data into molecular modeling pipelines (Figure 1)6. Using such an approach high resolution atomistic structural models of DNA-protein complexes can be verified and/or refined by using HRF experimental data.
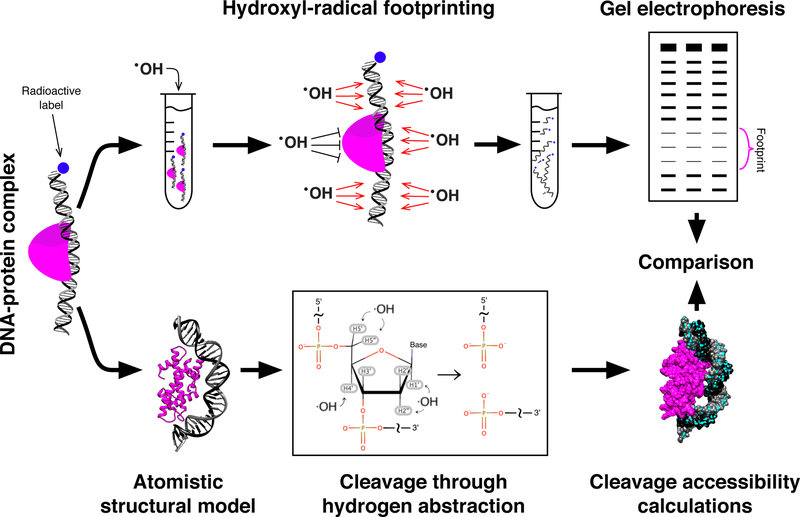
Figure 1. HRF of DNA-protein complexes and structural interpretation of the experimental data.
The schematic diagram outlines the basic principles of using HRF to elucidate the structure of DNA-protein complexes. Top: DNA-protein complexes are reconstituted on radiolabeled DNA and subjected to hydroxyl-radical cleavage; hydroxyl-radicals cleave DNA strands at sites not protected by protein in the regime of single hit kinetics (one cut per strand); the repertoire of cleaved DNA fragments is analyzed via denaturing gel electrophoresis and positions on the DNA sequence protected by protein become apparent. Bottom: atomistic structural models can be used to predict HRF lane profiles; the cleavage frequency depends on the accessibility of deoxyribose hydrogen atoms; comparison of predicted and experimental results may be used to refine or verify structural models.
Development of the Protocol
The theoretical basis for experimental HRF data quantification has been proposed previously7–10. However, while confronted with the practical need for structural interpretation of HRF data for DNA-protein complexes in our recent studies11,12 we realized that there is no easy-to-follow protocol and software for integrated HRF data quantification and atomistic structure analysis. Previously reported approaches8,9 that were used for PAGE gel quantification or solvent accessible surface area calculations for the 3D structure analysis have lost compatibility with the modern computer platforms or lack clear instructions on integrating HRF experimental data with 3D structural analysis. To complement this need, we developed a freely available software and here provide step-by-step instructions of our approach.
We present a protocol and software package HYDROID (HYDroxyl-Radical fOotprinting Interpretation for DNA). HYDROID provides quantitative analysis of HRF data at single base pair resolution, estimation of DNA cleavage frequencies from crystal structures or atomistic molecular models, and the comparison and integration of the two. HYDROID is written in Python and relies on free cross-platform components: ImageJ, for image analysis13 and FreeSASA library, for surface area calculations14. The cross-platform and robust nature of the HYDROID framework is supported by these well maintained, regularly updated and extensively tested core technologies. The HYDROID protocol consists of two libraries, HYDROIDexp (experimental data analysis and visualization) and HYDROIDpred (structure analysis) together with a set of examples and step-by-step instructions. HYDROID quantifies experimental gel lane profiles through data fitting with a variety of functions and fitting constraints. The package estimates the theoretical DNA cleavage frequency profiles from atomistic 3D structures through computing the solvent accessible surface area of deoxyribose hydrogen atoms. To familiarize oneself with the HYDROID protocol we have provided a video tutorial at https://ncbi.github.io/HYDROID/docs/video.html. The protocol has been successfully applied in two supporting studies11,12.
Applications of the method
HYDROID is a robust yet flexible tool to analyze and interpret DNA-footprinting experiments. It simultaneously combines the functionality to quantify HRF PAGE data (HYDROIDexp) and to estimate DNA cleavage frequency profiles from experimental structures or molecular models (HYDROIDpred).
Previously, we applied the HYDROID approach to study yeast centromeric nucleosomes and their complexes with the CENP-C protein11,12. High quality analysis of HRF experimental data using HYDROID allowed us to precisely determine DNA position on the histone octamer, construct an atomistic model of the centromeric nucleosome reconstituted on a specific DNA sequence and identify sites of CENP-C protein interactions with the nucleosome.
For this manuscript HYDROID approach was tested and validated using several experimental datasets on DNA-protein complexes. We analyzed the effects of different parameters on HYDROID performance and provided recommendations as to what parameters should be used when analyzing DNA-protein complexes. The Experimental Design section provides further details, corresponding examples and recommendations.
The core functionality of HYDROID modules can be easily adapted and used for a wider range of applications. Particularly, the HYDROIDexp module alone can be applied for the analysis of nucleic acids gel electrophoretic data of any type providing the exact quantification of band intensities. The band fitting algorithms developed in HYDROIDexp could in principle be adapted for the analysis of capillary electrophoresis data.
The HYDROIDpred module in its current form is capable of analyzing accessibility of ribose and deoxyribose hydrogen atoms using the H-SASA approach for either DNA or RNA alone or in complex with proteins. We have not used extensively the H-SASA approach to estimate cleavage frequency profiles in cases other than DNA-protein complexes. However, other studies previously suggested that estimating H-SASA profiles for certain hydrogen atoms in naked RNA or DNA could be analogously used to estimate HRF cleavage patterns10,15. For such cases, we suggest consulting the above mentioned references and specifying in the HYDROIDpred script which hydrogen atoms should be considered for analysis. Given the importance of the HRF method for RNA tertiary structure and RNA-protein complex analysis16–18, we expect that parts of the HYDROID package would be useful to this end and could be potentially incorporated into the RNA structure prediction pipelines19,20.
Comparison with other methods
A unique characteristic of HYDROID is the integration of HRF experimental data quantification and atomistic 3D structure analysis in one package. The advantages of the functionality provided by the individual components of HYDROID (HYDROIDexp and HYDROIDpred) are discussed below.
Certain standalone programs have been developed over the years to address gel quantification of HRF experiments, such as GelExplorer 8 or SAFA 9. However, one obstacle in using these programs currently is that they have not been updated for some time and lack compatibility with modern computer systems. Additional advantages of the HYDROIDexp library include the ability to choose between Gaussian or Lorentzian band shapes and a multitude of constrained fitting algorithms. The HYDROIDexp library also includes Python compatible tools for plotting profiles along the DNA sequence. It is free software (public domain) that may be reused in other projects.
With respect to the theoretical estimation of DNA cleavage frequency profiles from 3D structures, HYDROIDpred is currently the only available solution. It relies on calculating the accessibility hydrogen atoms via the FreeSASA library that is fast and free to use. HYDROIDprep extends FreeSASA by providing carefully selected sets of atomic radii for calculations. An alternative approach was reported previously (the RADACK approach) which tried to take into account the diffusion controlled nature of hydroxyl-radical attack reaction and the overall geometry of the molecule by means of stochastic simulations21. However, currently there is no publicly available software implementing the RADACK approach.
Advantages and limitations
The main advantage of HYDROID is its 2-in-1 design providing the user the ability to simultaneously quantify HRF experimental data and compare it to atomistic structural models of a protein-nucleic acid complex if available. Both components of HYDROID that implement these functionalities have their own advantages. Particularly, HYDROIDexp advantages include its cross-platform nature (runs on Linux, Windows, MacOS) and simultaneous implementation of a number of HRF profile fitting algorithms (Gaussian or Lorentzian band shapes combined with several constrained fitting algorithms). The constrained fitting algorithms perform well when deconvoluting the signal from partially overlapping gel bands. The advantages of HYDROIDpred include its free nature without any need for proprietary or licensed software components as well as a carefully selected and validated sets of parameters (probe radius, atom sizes) that can be used to obtain meaningful DNA cleavage frequency profile estimates from molecular models. The HYDROIDpred algorithm is based on estimating the solvent accessible area of the deoxyribose hydrogen atoms. This approach cannot directly account for the diffusion kinetics of hydroxyl-radicals (see the Experimental Design section for further discussion). Hence, care should be exercised when comparing the exact shapes of experimental and theoretical cleavage profiles.
The HYDROIDpred pipeline requires a PDB structure with hydrogen atoms. The suggested algorithm to add hydrogen atoms (Step 23) currently works only on Linux or Mac.
Overview of the HYDROID Protocol
HYDROID is composed of a protocol and a software package written in Python language that together with other components (ImageJ and FreeSASA) represents a robust framework for DNA-protein HRF experimental data analysis and interpretation. The protocol can be also applied for the analysis of RNA-protein complexes as well as free RNA or DNA molecules in solution. The HYDROID software package can be easily installed on Linux, Mac or Windows operating systems (see Box 1). It consists of two complementary pipelines (and corresponding software libraries): HYDROIDexp and HYDROIDpred. The HYDROIDexp pipeline is used to analyze PAGE gel images of HRF products to obtain DNA cleavage frequency values for every position on the studied DNA sequence (Figure 2, top). The HYDROIDpred pipeline is used to perform theoretical calculations of DNA cleavage frequency from a 3D atomistic structure or molecular model of a DNA-protein complex (Figure 2, bottom). The cleavage frequency calculations are done by calculating the solvent accessible surface area of deoxyribose hydrogen atoms (that are attacked by hydroxyl radicals and abstracted to induce DNA cleavage) for every nucleotide (H-SASA profiles). The H-SASA profiles estimate the expected DNA cleavage frequencies. Comparison and integration of experimental and theoretical profiles can be further used for experimental data interpretation and/or molecular model refinement. The main software functions implemented in HYDROID package are listed in Table 1. They are intended to be run from within a Python script, and certain functions have an interactive graphical user interface. Within the HYDROID package full-featured examples of HRF data and 3D structure analysis are provided in the form of thoroughly annotated Python scripts. These examples can be downloaded (see Box 1) and serve as modifiable templates for the analysis of the user data (see PROCEDURE and ANTICIPATED RESULTS). Below we outline the methodology behind the key stages of the HYDROID framework. Video illustrations of every stage are available at https://ncbi.github.io/HYDROID/docs/video.html.
BOX 1 | Installing software stack and downloading examples. TIMING 15 minutes.
This box provides instructions on software installation procedures for HYDROID, its supporting software programs Fiji ImageJ and REDUCE as well as downloading HYDROID examples Python scripts and data.
HYDROID installation
Detailed instructions are given at HYDROID GitHub repository (https://github.com/ncbi/HYDROID/blob/master/docs/INSTALL.md). The installation instructions for the prepackaged and precompiled version of HYDROID through the Conda package manager are given below (the FreeSASA library will be automatically installed too). This should install both HYDROIDexp and HYDROIDpred functionality on most modern versions of Linux, MacOS and Windows.
-
1
Install Miniconda Python distribution with Python2.7 for your platform from https://conda.io/miniconda.html.
-
2
Open default terminal on Linux or Mac, Anaconda Prompt (Start->Anaconda2->Anaconda Prompt) on Windows
-
3Run the following command to install HYDROID package
> conda install -c hydroid hydroid
-
4Test HYDROIDexp and HYDROIDpred functionality by issuing following commands in the terminal
> HYDROID_test_exp
An interactive window with a lane profile should open.> HYDROID_test_pred
A file test_H-SASA.csv should appear in the results folder of the current directory.
Downloading HYDROID examples
HYDROID examples provide data together with Python scripts that implement stages of the HYDROID analysis pipelines. Downloading and studying examples is the best way to learn how HYDROID works. Examples should be used as modifiable templates for user data analysis.
-
5
Open default terminal on Linux or Mac, Anaconda Prompt (Start->Anaconda2->Anaconda Prompt) on Windows
-
6Run the following command
> HYDROID_get_ex1
This will create ‘example1’ folder and download the contents of the Example 1. Detailed description of Example 1 and run instructions are available on GitHub at https://github.com/ncbi/HYDROID/tree/master/examples/example1> HYDROID_get_ex2
This will create ‘example2’ folder and download the contents of the Example 2. Detailed description of Example 1 and run instructions are available on GitHub at https://github.com/ncbi/HYDROID/tree/master/examples/example2
Both examples are also provided as a Supplementary data 1 (Example 1) and Supplementary data 2 (Example 2) zip-files.
ImageJ installation
ImageJ is a free and cross-platform image analysis software that can be used to extract 1D lane profiles from PAGE images of HRF or DNA sequencing reaction products (HYDROIDexp, Stage 1).
-
7
Download and install Fiji distribution of ImageJ from https://fiji.sc
-
8
Optional: Add the Workspace macro to be able to save and open workspaces.
-
9
Download Workspaces.txt from https://imagej.nih.gov/ij/macros/Workspaces.txt
-
10
Open Fiji ImageJ: Click Plugin->Macros->Install, point to the Workspaces.txt file
-
11
Optional: Open workspace from Example 2 (download instructions above): Click Plugin->Macros->Workspaces settings ..; Specify data folder from example2 folder; Click Plugin->Macros->Restore Workspace …; Click Ok; A PAGE image with highlighted lane profiles should open up.
REDUCE installation (Optional)
REDUCE is a program that can add hydrogen atoms to 3D structures in PDB format that otherwise lack determined hydrogen atoms’ positions. A version of REDUCE that provides consistent results for both DNA and protein can be installed as a part of AmberTools Package from http://ambermd.org/#AmberTools (also available as precompiled binaries through a Conda repository). AmberTools are currently available for Linux and Mac.
Figure 2. Data pipelines in HYDROID.
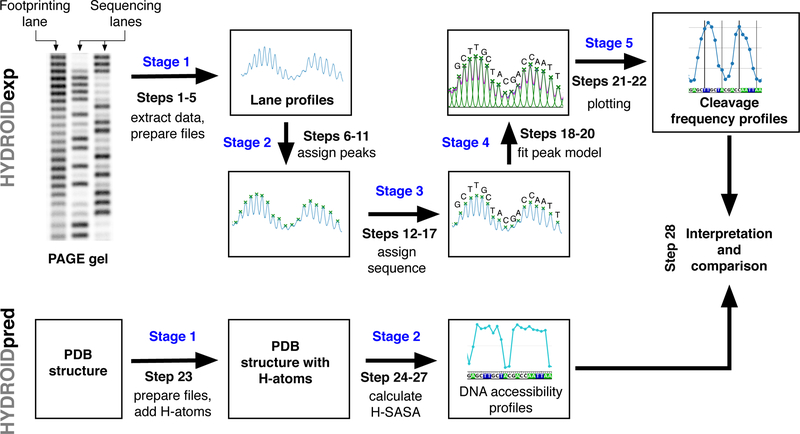
Procedure steps and stages are annotated on the schematic diagram for the analysis of hydroxyl-radical footprinting experimental data (top, HYDROIDexp) and theoretical estimation of DNA cleavage profiles from molecular structures (bottom, HYDROIDpred). The HYDROIDexp pipeline consists of five stages (annotated in blue): stage 1 – lane profiles in the form of data files are extracted from a PAGE image, stage 2 – locations of the bands (represented as peaks) are semi-automatically assigned on the lane profiles, stage 3 – correspondence between gel bands and DNA sequence is established, stage 4 – a mathematical model is fitted to describe the shape of the lane profile with Gaussian or Lorentzian functions, stage 5 – the individual band intensities (DNA cleavage frequencies) are extracted from the fitted model and plotted. The HYDROIDpred pipeline consists of two stages: stage 1 – a PDB file of the system of interest is obtained and hydrogen atoms are added to the atomistic structure if needed, stage 2 – DNA accessibility profiles are calculated by estimating the solvent accessible area of deoxyribose hydrogen atoms (H-SASA). Finally theoretical and experimental data sets can be compared.
Table 1.
Main HYDROID functions and their description.
| HYDROIDexp library | |
|---|---|
| assign_peaks_interactive | Provides interactive interface for a semi-automated algorithm used to identify positions of peaks on gel lane profiles. |
| call_peaks_interactive | Provides interactive interface to assign individual peaks with respect to DNA sequence position. |
| fit_peaks | Main function that fits a mathematical model to the gel lane profile and evaluates DNA cleavage intensities. |
| plot_prof_on_seq | Plots any data profile along the DNA sequence and allows for normalization and fitting of two or more profiles. |
| simulate_gel | Simulates the shape of gel lane profile and infers a gel lane image based on H-SASA profile. |
| HYDROIDpred library | |
| get_DNA_H_SASA | Estimates theoretical DNA cleavage profiles (H-SASA profiles) from a DNA-protein structure |
HYDROIDexp pipeline.
HYDROIDexp requires as an input a PAGE gel image in which HRF products and a sequencing reaction, both labeled on the same DNA strand, are run adjacent to each other. The pipeline yields relative DNA cleavage frequency values along the DNA sequence by quantifying the intensity of the bands on the PAGE gel. This pipeline can be applied in parallel to several samples that were run on the same gel. The pipeline consists of five stages. See Figure 2 for a workflow diagram.
Stage 1: Extracting lane profiles.
The experimental lane profiles are extracted from the gel image. Experimental gel lane intensity profile (lane profile) is hereafter defined as an array of intensity values on a gel image along a specified gel electrophoresis lane (in the direction of DNA migration on the gel). HYDROID functions read in experimental gel lane profile data saved as columns of numbers in the text files. To obtain “digital” experimental lane profiles from PAGE gel images we suggest using ImageJ software 13 with Bio-Formats plugin22 wherever necessary to open specific gel image file formats. Box 2 gives detailed instructions on how to extract lane profiles in a compatible tabular format text file from a PAGE image using ImageJ. In order for HYDROID to read in the data from the generated file, a separate configuration text file has to be specified, which specifies the lane names and corresponding column names in the generated file (see Box 3 for more details on input file formats for HYDROIDexp).
BOX 2 | Extracting lane profiles from PAGE images using ImageJ.
This box provides information on how to extract 1D lane profiles from PAGE images and save them in a tabular format (columns of numbers – one column per experimental gel lane) that can be read by HYDROID functions. More detailed instructions can be found within HYDROID at https://github.com/ncbi/HYDROID/blob/master/examples/example1/exp_s1_extract_lp.md.
-
Open Fiji ImageJ and open the PAGE gel image (example files “gel.png” in example/data/ directories). This can be done depending on the file type either through File->Open … or Plugins->Bio-Formats->Bio-Formats Importer ..
CRITICAL STEP The intensity of the pixels in the image should be proportional to the real signal measured by the gel scanner. Use BioFormats Plugin for ImageJ to read major gel scanner file formats in the original quality and format (e.g. gel-file format). Note: gel-file format may often be opened as tiff-file, but the signal intensities would be incorrect. Consult with the manual of your gel scanner.
-
Subtract image background intensity if necessary (Process-> Subtract background …) or (Process-> Math -> Subtract…).
CRITICAL STEP If substantial contribution from the background is present, it is important to subtract it for correct quantification.
Press Line tool in Area selection tools. Draw a line along the center of one lane from the start of the gel to the bottom. Note: The PAGE images often deviate from a rectangular grid-like structure; this complicates extracting lane profiles. If bands are more or less perpendicular to the lane path but the lane path could not be approximated by a straight line, using of segmented line to approximate the lane path is suggested (in ImageJ: right click on Line button, choose Segmented Line). The internal image straightening algorithm will then be automatically applied during the profile extraction.
Open ROI Manager: Analyze->Tools->ROI Manager.
Press Add [t] in the ROI Manager window to save the line position and geometry.
Repeat steps 4 and 5 for each gel lane. Try to start and stop each line at the equivalent positions, so their length is approximately the same. Or at least try to put the end positions of the lines at the equivalent positions on the gel lanes – this will facilitate alignment of HRF lane profiles with DNA sequencing profiles later.
Select lines in ROI Manager window and set their width to a reasonable value (ROI Manager: Properties …). Note: Since the PAGE gel image data is often noisy, the default 1 pixel width profile will be often very noisy too. Averaging it along the width of the lane is recommended. The width of averaging is recommended to be set to 25–50% of the lane width depending on the quality and shape of the bands.
Select all the saved lines in the ROI Manager window, choose More->Multi Plot. The lane profiles will be plotted. The segmented lines would be straightened automatically, check if they still follow lane paths.
-
In the plot window click Save … and save the data to an XLS file. The file will have data columns corresponding to each profile (X and Y values columns for each profile) in the same order as they were added to the ROI Manager. This file will be imported further by HYDROIDexp routines. An example of such file can be found at https://github.com/ncbi/HYDROID/blob/master/examples/example1/data/lane_profiles.xls.
CRITICAL STEP: Depending on the version of ImageJ the default file extension may be different. Always specify ‘xls’ as the file extension in order for the data to be saved in the correct format.
BOX 3 | Input file formats for HYDROIDexp.
The majority of HYDROIDexp functions read in two files: 1) a file with lane profiles data generated by ImageJ software, 2) lane profile configuration text file that specifies the names of the gel lanes and stores the information about the parameters used to process the lane profiles. Below we describe briefly their file format.
Lane profiles data file
This file is a tab or space separated data file saved by ImageJ with column names specified in the first line and looks as follows:
| X0 | Y0 | X1 | Y1 | X2 | Y2 |
| 0.00 | 1707.08 | 0.00 | 144.81 | 0.00 | 2515.93 |
| 0.000048 | 1786.45 | 0.000048 | 132.20 | 0.000048 | 2419.62 |
| 0.000095 | 1839.41 | 0.000095 | 134.25 | 0.000095 | 2253.74 |
The columns with Y in their name specify the actual intensity values in the lane profiles along the lanes and are further processed by HYDROID (it is assumed that the data points in the file are equally spaced with respect to their position along the lane – X value). A full example of such file can be found at https://github.com/ncbi/HYDROID/blob/master/examples/example1/data/lane_profiles.xls.
Lane profiles configuration file
This file is a comma separated file and looks as follows (spaces are ignored):
#Comments
| column, | lname, | leftlim, | rightlim, | peakthresh, | min_dist_left, | … |
| Y0, | GA_601TA_BS, | NaN, | NaN, | NaN, | NaN, | … |
| Y1, | CT_601TA_BS, | NaN, | NaN, | NaN, | NaN, | … |
| Y2, | scCSE4_601TA_BS, | 61, | 2172, | 0.031, | 5.164, | … |
| … |
Every line in the file describes parameters for a specific lane that can be analyzed by HYDROIDexp. The first parameter (column) references a specific column name in the lanes profile data file (see above), the second parameter (lname) assigns a name for that lane, all other columns provide specific values for the HYDROIDexp routines and should be set to NaN initially, they will be filled in interactively during Stages 2–3. A full example of such file that can be used as a starting point for new projects can be found at https://github.com/ncbi/HYDROID/blob/master/examples/example1/data/lane_config.csv. It further contains detailed description of every column meaning.
Stage 2: Assigning location of individual gel bands.
Positions of individual gel bands are identified on the gel lane profile via a semi-automated interactive algorithm. To deconvolute the lane profile into a set of contributions from individual bands, first one needs to know the number and approximate locations of bands on the lane profile. Each band usually corresponds to a local peak on the lane profile. The semi-automatic interactive algorithm implemented in assign_peaks_interactive function in HYDROID opens a graphical window and allows the user to vary parameters of the peak finding routine until all necessary peaks are correctly identified as judged by the user. The peak finding algorithm is based on the PeakUtils Python library. The algorithm is governed by two parameters: relative peak prominence (peakthresh parameter - only the peaks with amplitude higher than the threshold will be detected) and minimal distance between peaks. The peak finding algorithm splits the lane profile into several segments and allows the minimum distance between the peaks for the left most and right most segments to be chosen by the user (min_dist_left and min_dist_right parameters); values at the intermediate segments are then inferred using linear interpolation. If some bands overlap to the extent that no peaks can be identified on the lane profile or the intensity of the band is very low, a peak interpolation option is implemented in HYDROID to try to infer their positions from the known positions and spacing of the neighboring bands that are more prominent. Finally, the peak positions can be specified manually in the configuration file.
Stage 3: Assigning HRF peaks to DNA sequence.
Every band on the HRF gel lane (which is equivalent to a peak on the HRF lane profile) can be attributed to a specific position on the DNA sequence through comparison of HRF gel lanes to the lanes with the products of DNA sequencing reactions for the same DNA. In HYDROID the call_peaks_interactive function allows you to interactively plot the HRF lane profile together with the profiles of DNA sequencing reactions and the DNA sequence. By visually comparing the profiles with the DNA sequence, the user can specify the location of any single peak on the profile and its corresponding position on the DNA sequence. This allows you to establish the correspondence between all bands and positions on the DNA sequence.
Stage 4: Fitting lane profiles.
Mathematical models describing the intensity distribution of every band with Gaussian or Lorentzian functions are fitted to the lane profile data using fit_peaks function. This procedure allows you to deconvolute the raw experimental HRF profile into the contributions of individual gel bands, thus properly taking into account such effects as partial band signal overlap and unequal band width along the gel lane (see Box 4 for the mathematical details of the algorithm). This procedure is known to suffer from overfitting unless additional constraints are imposed on the fitting algorithm23. HYDROID implements a number of different constrained fitting options listed in Table 2. See section “Experimental HRF profiles quantification” for discussion of further details.
BOX 4 | Details of HRF profile deconvolution algorithm.
To extract DNA cleavage frequency values from gel lane intensity profiles of HRF experiments, several mathematical models can be fitted in HYDROID. The model describes the gel lane profile as a sum of individual contributions from every gel band. Distribution of intensity values of every gel band is in turn described as a bell-shaped function: Gaussian (formula 1) or Lorentzian (formula 2) with varying area (height) H, width σ and position D parameters. A Gaussian model function will be used as an example for further description.
| (1) |
| (2) |
After model fitting, the total area under each Gaussian (equal to the H parameter above) determines the overall intensity of the corresponding band and hence the DNA cleavage frequency represented by this band.
The profile deconvolution proceeds as follows. First, approximate locations of peaks are obtained from a semi-automated interactive algorithm (implemented via assign_peaks_interactive function, Figure 2, stage 2). Identified positions are used as starting values for the centers of Gaussian functions, D. Estimation of reasonable starting values for H and σ parameters and actual fitting are then performed via fit_peaks function.
In the first step the starting values for H and σ are determined by solving a simplified curve fitting problem. The width parameters of Gaussian functions are set to depend linearly on positions of peaks , while the H parameter of every Gaussian is set via the formula:
| (3) |
where is the value of experimental profile at position index i varies between 1 and M, where M is a number of gel bands. Optimal k and b, are then found by minimizing the sum of squared residuals between the experimental profile and the model.
In the second step, a non-linear least square fitting procedure using the Levenberg-Marquardt algorithm is performed with respect to , and of all individual peak functions. However, since we have many parameters, which can result in overfitting, a set of additional constraints is implemented in HYDROID. A list of constraints is shown in Table 2. For example, “dSIGMA>=0” constraints the widths of Gaussian functions so that they are not allowed to decrease with distance D from the start of the gel. The implementation of our fitting procedure is based on a substitution of variables, where the actual independent variables are the differences between the width parameters σ of the neighboring peaks so that the actual minimization occurs in a different parameter space, but via the same unconstrained minimization algorithm.
Table 2.
List of constraints in the fitting method available in HYDROID. (see Box 4 for parameter meaning)
| Constraint type (Abbreviation) | Description |
| dSIGMA>=0 | Peak width is not allowed to decrease with increasing D, position of the peak (i.e. from the start of the gel to the end). This dramatically improves stability of the solution and prevents overfitting. |
| SIGMA<2*dD | Peak width σ is not allowed to exceed twice the distance between a given peak/band and the following peak (dD). Implies automatically “dSIGMA>=0”. |
| SIGMA=k*D+b | Peak width σ is linearly related to the position of the peak (D). This effectively establishes a linear relationship between band width and its mobility. |
| log(SIGMA)=P2(log(M)) | Logarithm of peak width σ is proportional to the logarithm of the number of nucleotides in DNA, M, (or its molecular weight) via a second-degree polynomial. The optimal coefficients of polynomial are determined during fitting: |
| SAFA | Constraints as implemented in SAFA program 9, peak width is related to positions of neighboring peaks via |
To assess the quality of the experimental lane profile fitting by the model, HYDROID provides several goodness-of-fit measures: RMSD – root-mean-square deviation between experimental and predicted by the model values, relative RMSD – a variation of the latter, where deviation at every point is expressed as a fraction of the experimental value, and a Pearson correlation coefficient between experimental and predicted values.
DNA cleavage frequency values (total areas under the peaks on lane profile) are estimated by the non-linear least square fitting procedure. For every peak, the area is calculated under the segment of the curve that incorporates that peak. The segment is defined by the midpoints between the position of a given peak (position of its maximum) and peak positions of its neighbors to the left and to the right. The relative differences in the proxy area values determined for the experimental lane profile versus the fitted model profile are used as uncertainty estimates and referred to as “relative peak area errors”. Maximum relative peak area error among all peaks as well as root-mean-square average (“relative peak area RMSD”) are reported in HYDROID.
Stage 5: Calculating DNA cleavage frequency.
The DNA cleavage frequency for every position on the DNA is extracted as an area under the Gaussian or Lorentzian describing the corresponding band. The values of DNA cleavage frequency profile are plotted along the DNA sequence.
HYDROIDpred pipeline.
Theoretical calculation of DNA cleavage profiles by HYDROIDpred relies on the analysis of 3D atomistic structures of protein-DNA complexes and assessment of the accessibility of deoxyribose hydrogen atoms to hydroxyl radicals. 3D coordinates can be obtained via X-ray crystallography or molecular modeling (e.g. homology modeling, integrative modeling, docking, etc.). The solvent accessible surface area of deoxyribose hydrogen atoms is calculated for each nucleotide in the DNA molecule of the structural complex which results in two H-SASA profiles, one for each strand (in case if the protocol is applied to RNA, it will result in one H-SASA profile). These profiles yield estimated relative DNA cleavage frequency values. The pipeline consists of three key stages.
Stage 1: Preparation of an atomistic model of a DNA-protein complex with known positions of hydrogen atoms.
An important step is the inference of hydrogen atom positions if they are not available in the original structure. Small differences in positions of hydrogen atoms (e.g. different length parameters for the C-X bonds) may affect the magnitude of the estimated profiles (see section “Estimating theoretical DNA cleavage frequency profiles from atomistic structures” for a detailed discussion). REDUCE program24 from AmberTools 17 package25 with X-H bond distances derived from nuclear positions can be used when adding hydrogen atoms to the X-ray structures.
Stage 2: Estimating theoretical DNA cleavage frequencies.
Theoretical DNA cleavage frequency profiles are estimated by calculating the solvent accessible surface area of deoxyribose hydrogen atoms (H-SASA profiles) via the get_DNA_H_SASA function that uses FreeSASA library14. Lee- Richards algorithm is used26 in FreeSASA with the precision set to 200 slices per atom by default. Different probe radii for calculating SASA may be applied with 1.4 Å being the default. HYDROIDexp implements three sets of atomic radii for H-SASA calculations: a) default FreeSASA radii, b) radii values from CHARMM36 force field27, and c) radii values from AMBER ff10 (parm10) force field25. The latter two are derived from van der Waals rmin parameters (distance/radius at which Lennard-Jones energy has a minimum) of the corresponding force fields. Visualization of the calculated profiles is then performed (see Data visualization and comparison below).
Data visualization and comparison.
For DNA cleavage frequency profile visualization and comparison, HYDROID includes a plotting module (plot_prof_on_seq) based on Matplotlib library28. This module can plot several profiles simultaneously together with DNA sequence and allows several normalization techniques to be used (to bring profiles to the same scale). Some trivial normalization techniques are implemented, such as dividing each profile by its maximum value (“every_method”) or dividing both profiles by their mutual maximum value (“together_method”). In addition, the “fit method” performs a linear regression without the intercept between profile values and rescales both of them to the mutual maximum value. The use of a linear fit without intercept respects the physical requirement that positions inaccessible for cleavage should have values of zero on both profiles.
Additionally, we have implemented a technique to simulate gel lane profiles and gel images from cleavage frequency profiles (simulate_gel function). This functionality is useful to explore the expected shape of the gel lane profiles (and hence the resolution of the profile in the specific DNA region of interest) and design experiments. The Ogston DNA mobility model is used to simulate gel mobility29.
Experimental Design
HRF technique and data interpretation.
The HRF technique uses hydroxyl-radicals generated via the Fenton reaction or via irradiation to cleave DNA strands at solvent accessible sites5. The underlying chemistry is the following: a hydroxyl-radical attacks deoxyribose hydrogen atoms, which results in hydrogen abstraction and subsequent backbone cleavage (Figure 1). The main products of this chemical cleavage are two strands terminated by 3’ and 5’ phosphates that are adjacent to the attacked nucleotide in the original DNA strand. The analysis of DNA cleavage products is usually done using denaturing polyacrylamide gel electrophoresis (PAGE) of radio or fluorescently labeled DNA. Radiolabeling the 5’-end of DNA is generally recommended5 since during cleavage of 3′-labeled DNA strand, in addition to major 5′-phosphate-terminated product, a minor alternative product (a strand terminated by 5′-aldehyde group) is also produced. In the limit of single hit kinetics (one cut per DNA strand) the accessibility of every nucleotide for the hydroxyl-radical attack should be proportional to the intensity of the corresponding gel band5. The attribution of every band to a specific position on the DNA sequence is an important stage and is done by running products of various DNA sequencing reactions (such as Maxam-Gilbert reactions) on adjacent gel lanes (Figure 2), as a reference.
During HRF experiments, the nucleotide is attacked by hydroxyl-radicals and later is degraded yielding a corresponding DNA fragment that is one nucleotide shorter than the would-be DNA fragment terminated by the actually attacked nucleotide. The position on the DNA sequence specified by the user at this stage should correspond to the nucleotide that is actually attacked by hydroxyl-radicals (and degraded). This fact together with the nature of the employed DNA sequencing reactions should be taken into account when attributing a peak on the HRF profile to the position on the DNA sequence. Maxam-Gilbert sequencing reactions are convenient in this respect because they yield products that are also one nucleotide shorter that the would-be DNA fragments terminated by the attacked nucleotide 30.
The quantification of actual DNA cleavage frequencies requires measuring individual band intensities from gel lane intensity profiles. The partial overlap between bands and variation in their widths have to be taken into account31. To extract the total intensity of every band, a mathematical model was previously suggested which was used to fit the gel lane intensity profile8. In this approach the signal intensity of every band was described by a bell-shaped function and the resulting profile was modeled as a sum of the bell-shaped functions. Several modeling functions have been proposed so-far including Gaussian, Lorentzian8, simplified integrated Weibull function32 and more sophisticated functions33. While the Gaussian function is the default model to describe band broadening due to free diffusion of molecules, certain other factors (such as electric field, temperature gradients, non-homogeneities of the gel, etc.) should be also taken into account. Particularly, the auto radiographic detection methods might modify the original signal, making Lorentzian function a more appropriate model in certain cases34. From our experience the data obtained from gels using modern phosphorimaging setups is well fitted by Gaussian functions. Moreover, overfitting is a common issue for these types of problems and may be solved by imposing additional constraints on the solution23.
The quality of the gel image which is used to extract gel lane profiles is an important characteristic that might affect the quality of HRF results quantification. For good results it is expected that in the region of interest a complete ladder of electrophoretic bands representing DNA cleavage products differing by one nucleotide is clearly visible. However, HYDROID protocol may be also successful in deconvoluting lane profiles in the regions where some bands are poorly visible or have significant overlap. In this case the user should be able to specify the number of expected bands and their approximate locations. To this end the locations of the bands in other lanes of the gel may be used.
Computational structural modeling of DNA-protein complexes can be guided and verified by using high resolution DNA footprinting experimental methods. There is a pressing need for a software solution to perform these tasks and bridge the experimental data with 3D structural models. Previously Balasubramanian et al. performed a detailed study of the intensity of DNA strand scission through the attack on various deoxyribose hydrogen atoms7. They found that H5’ and H5’’ atoms had the highest reactivity followed by H4’, H3’, H2’+H2’’ and H1’. Moreover, the reactivity of hydrogen atoms was found to correlate well with their solvent accessible surface area (SASA), which can be estimated from X-ray structures or predicted from structural models. This finding forms the basis for the theoretical estimation of DNA cleavage frequencies from atomistic structures in HYDROID. However, as we discuss further, robust estimation of SASA for hydrogen atoms in a DNA-protein complex requires a careful choice of parameters, such as atomic radii, probe sphere radius and details of hydrogen atom placement algorithms.
Experimental HRF profile quantification.
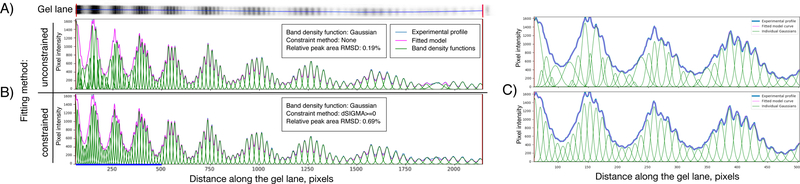
Quantification of experimental HRF lane profiles is done in HYDROIDexp by deconvoluting the shape of experimental profile as a sum of Gaussian (or Lorentzian) functions centered at band positions (see Box 4). The unconstrained fitting usually results in physically incorrect solutions (albeit having better goodness-of-fit characteristics) and clear irregularities in the positions and widths of bell curves fitting individual bands are seen (Figure 3C top panel as well as on the resulting DNA cleavage frequency plots Figure 4). To overcome this problem several constraints in fitting algorithms are implemented in HYDROIDexp (Table 1). The most flexible is the “dSIGMA>=0” algorithm, which does not allow for the widths of the Gaussian functions to decrease as one moves from the start of the gel to its end. This is based on a physical assumption that band broadening (higher dispersion coefficients) increases along with the mobility of DNA in the band. This constraint produced good fitted solutions in the majority of cases while imposing only minimal constraints (Figure 3). However, visual examination of the resulting solution is always recommended. Among constrained fit methods, the “dSIGMA>=0” algorithm usually provides the best fit as judged by goodness-of-fit characteristics. Other constraint algorithms also provide reasonable solutions with relative peak area RMSD values of less than 2% and almost identical resulting DNA cleavage frequency profiles (Supplementary Figure 1, 2).
Figure 3. Quantification of PAGE gel image by fitting a mathematical model.
A) PAGE image of the hydroxyl-radical footprinting products for one DNA strand of the nucleosome. The DNA migration direction on the image is from left to right. To obtain this image S. cerevisiae centromeric nucleosomes were reconstituted on a 601TA nucleosome-positioning DNA sequence with DNA radioactively labeled on 3’-end. Hydroxyl-radical footprinting of nucleosomes, DNA extraction, PAGE and radioactive signal acquisition were performed as described elsewhere11. B) Extracted lane profiles are fitted with a model that approximates every gel band as a Gaussian function; results of an unconstrained and constrained fitting are shown. The area under each Gaussian represents the intensity of the band and hence the DNA cleavage frequency. C) Zoomed version of the plots for the left part of the gel highlighted by blue line in panel B. The irregularities in the widths and positions of Gaussian for the unconstrained fitting method are clearly visible. Experimental and fitted curves cannot be discerned because they almost overlap. The Y-axis on the plots represents the lane profile intensity values.
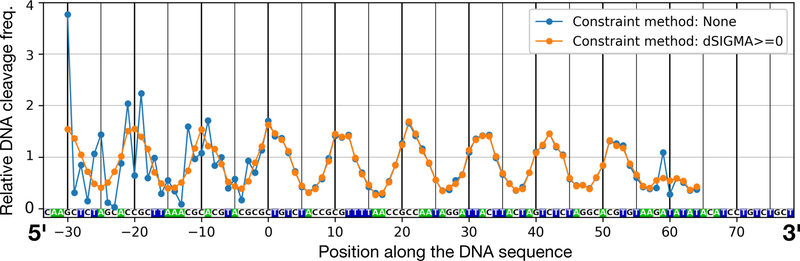
Figure 4. DNA cleavage frequency profiles extracted from HRF experiments.
According to our HRF data quantification methodology the lane profile curve in Figure 3B was fitted by a combination of Gaussian functions. The areas of these individual Gaussian functions represent the individual band intensities and hence DNA cleavage frequency values. These quantified DNA cleavage frequency values are plotted here for both unconstrained and constrained fitting results presented in Figure 3B. Correspondence between the DNA sequence and HRF profile was identified using Maxam-Gilbert sequencing reactions’ products run alongside the HRF products on the gel. Irregularities in the values resulting from the unconstrained fitting are visible. Zero on the x-axis marks the nucleotide positioned on the central dyad symmetry axis of nucleosome.
Gel lane profile deconvolution analysis in HYDROIDexp can be done using both Gaussian or Lorentzian functions to approximate band intensity distribution. Some previous studies have shown that Lorentzian may provide a better fit for DNA electrophoresis data8. For the experimental data sets used in this study Gaussian function provided a better fit than Lorentzian, although the differences in the quality of fitting did not exceed 2% estimated by relative RMSD (Supplementary Figure 1). However, the extracted DNA cleavage intensity profiles had a noticeable difference: using Lorentzian the maximum cleavage intensities tended to be higher and the minimum cleavage intensities lower than those produced by using Gaussian function(Supplementary Figure 2). This effect may be attributed to the slow decaying tails of the Lorentzian distribution, which causes peaks with higher intensity to contribute more to the signal amplitude at the neighboring low intensity bands.
Gel lane profiles sometimes may lack easily identifiable maxima for certain bands. This may happen due to noise, low intensity of some bands or band overlap. As long as the user can identify the number and approximate positions of these bands, the HYDROIDexp deconvolution algorithms can robustly quantify them. Figure 3C illustrates this for the left part of the gel profile where some bands overlap; Supplementary Figure 3 shows a case for the bands with low intensity and a certain level of noise. Supplementary Figure 4 presents the results of quantification of two independent HRF experiments for the same substrate and highlights good reproducibility of the results produced by our pipeline. Small variations can be attributed to experimental fluctuations during the footprinting process itself. Quantification of gel profiles obtained by electrophoresis of the same reaction products on two different gels yielded almost identical results (unpublished observations, AKS, HX).
Estimating theoretical DNA cleavage frequency profiles from atomistic structures.
Following the ideas in ref. 7 HYDROIDpred estimates theoretical DNA cleavage frequency profiles by hydroxyl-radicals via calculating the solvent accessible surface area (SASA) of deoxyribose hydrogen atoms (referred to as H-SASA profiles). In principle, the same method can be applied to calculating the solvent accessible surface area of ribose hydrogen atoms of RNA. To understand the influence of various parameters on the shape of H-SASA profiles we took the structure of a nucleosome as an example and calculated H-SASA profiles with different sets of parameters: probe radius, atomic radii sets, hydrogen atom addition methods, contributions from different hydrogen atoms. Interactive plots of H-SASA profiles calculated with different parameters are available at https://ncbi.github.io/HYDROID/examples/example1/results/nucl_H-SASA.html. The dependence of H-SASA profiles on the probe radius is rather straightforward: smaller probe sizes can discern finer details of DNA occlusion by protein but make the profile less smooth and more dependent on the local geometry of the DNA-protein interactions. Probe radius of 1.4 A is the usual choice, but calculating H-SASA profiles with different probe sizes might be useful to characterize DNA-protein interactions at different scales.
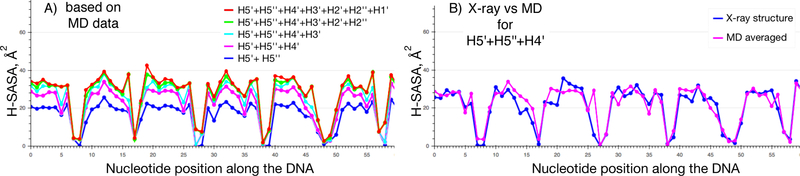
The H-SASA profiles depend rather significantly on the choice of atomic radii sets (FreeSASA default vs CHARMM36 vs AMBER10) and on the algorithm used to add hydrogen atoms (their positions in X-ray structures are usually not resolved). A methodologically consistent approach requires generating hydrogen atom positions using topology parameters from the same all-atom molecular mechanics force field used to estimate atomic radii. As shown previously, H-SASA profiles are sensitive to small fluctuations in DNA backbone geometry both due to X-ray structure inaccuracies and details of DNA conformation11. To address this problem, molecular dynamics (MD) simulations may be used to relax the X-ray structures and sample their dynamics. Supplementary Figure 5 shows differences between H-SASA profiles (based on all deoxyribose hydrogen atoms) obtained with and without MD relaxation and averaging. As we show next these differences are mainly due to contributions to H-SASA values from the least exposed H1’, H2’, H2’’, H3’ atoms, while the main contributions to H-SASA are made by the solvent accessible area values of H5’, H5’’ and H4’ atoms (Figure 5A). If H-SASA is calculated only for H5’, H5’’ and H4’ atoms, the correspondence between profiles calculated from MD and original X-ray structures is good (Figure 5B). Particularly, the locations of H-SASA minima (DNA nucleotides shielded by protein) are in agreement between the profiles (including the MD H-SASA profiles calculated with all deoxyribose hydrogen atoms considered). Hence, when using X-ray structures without MD relaxation, calculation of H-SASA profiles based on only H5’, H5’’ and H4’ atoms is recommended.
Figure 5. H-SASA profiles for DNA in nucleosome.
A) H-SASA (solvent accessible surface area of deoxyribose hydrogen atoms) profiles are calculated using contributions from different deoxyribose hydrogen atoms for each nucleotide along the DNA sequence starting from the center of the nucleosome. Profiles were calculated by averaging over 50 snapshots from MD simulations spaced 1 ns apart. B) Comparison of H-SASA profiles (based on H5’, H5’’ and H4’ atoms) as calculated from X-ray structure (with hydrogen atoms added via REDUCE) and MD trajectory. If only H5’, H5’’ and H4’ atoms are available, profiles do not differ much and X-ray structures can be used for reliable profile estimation. Zero on the x-axis marks the nucleotide positioned on the central dyad symmetry axis of nucleosome.
The differences in the shape of experimental and theoretical DNA cleavage profiles may be attributed to two facts. First, since HRF experiments are performed in solution, the dynamic nature of DNA conformation and interactions between DNA and protein would lead to averaging of the experimental profile. Such effects cannot be taken into account theoretically unless a dynamical ensemble of structures is considered and analyzed. Second, due to the diffusion controlled nature of the hydroxyl-radical attack reaction, the overall geometry of the molecule including the widths of the DNA grooves, can also modulate the attack probability10,21,35. The H-SASA profiles may only partially capture such effects. Hence, while comparing theoretical and experimental profiles the emphasis should be given to the comparison of the locations of the nucleotides that are maximally shielded from the hydroxyl-radical attack. The diffusion controlled effects are absent for the DNA positions completely protected by protein.
Example sets.
The HYDROID protocol uses two experimental data sets (termed “Example 1” and “Example 2”, available as Supplementary data 1 and Supplementary data 2, respectively, and also in the GitHub repository, see Box 1 on how to download them from command line) to illustrate the performance of HYDROID. Example 1 contains HRF data for S. cerevisiae centromeric nucleosomes reconstituted on 601TA DNA sequence with 3’-ends labeled radioactively and Example 2 contains HRF data of G. gallus nucleosomes reconstituted on 601 DNA sequence with 5’-ends labeled radioactively. The experimental details for the first and second sets can be found in refs. 11,36 and ref. 37, respectively. Both sets include the Python scripts for data analysis by HYDROID and the results files generated by HYDROID.
Example 1 further includes files with 3D coordinates – a molecular model of yeast centromeric nucleosome with 601TA DNA sequence38 not available in PDB that was built via homology modeling using Modeller39 and 3DNA40. Histone sequences were obtained from HistoneDB2.041 and from a corresponding X-ray structure of X. laevis nucleosome (PDB 3LZ0) as described previously 11. REDUCE program24 from AmberTools 17 package25 was used to generate hydrogen atoms positions with X-H bond distances derived from both nuclear and electronic cloud positions. In addition, a molecular dynamics approach was employed. Using VMD42 psfgen command with CHARMM36 force field 27 the hydrated structures were generated followed by minimization, relaxation and molecular dynamics simulations as described in our earlier studies 11.
Expertise needed to implement the protocol.
Users should be familiar with the command line interface on Linux, Mac or Windows. A basic knowledge of the Python scripting language would be beneficial in order to modify simple Python scripts using a text editor. For the HYDROIDpred part of the protocol understanding of the PDB-file format is beneficial.
A graphical user interface (GUI) wrapper is available for the basic functionality of HYDROIDexp part as an add-on module at https://github.com/intbio/HYDROID_GUI for those users that would prefer using GUI interface.
MATERIALS
EQUIPMENT AND DATA
A computer running a Linux, Mac or Windows environment.
PAGE images of HRF reaction products and DNA sequencing reaction products (typically obtained via phosphorimaging if DNA fragments are radiolabeled). The image files should be of good enough resolution so that in the region of interest the distance between the bands should be larger than 10 pixels. The image file format should be readable by ImageJ alone or through the BioFormats plugin.
(Optional) 3D atomistic structures or molecular models of the DNA-protein complex under study.
EQUIPMENT SETUP
Software installation.
Please refer to Box 1 for the installation instructions of HYDROID Python package and accompanying tools, Fiji ImageJ and REDUCE.
Downloading examples
The HYDROID package comes with a set of examples (protocol templates) available via the link https://github.com/ncbi/HYDROID/tree/master/examples. See Box 1 for the commands to automatically download them. The example files are also provided as Supplementary data 1 and 2.
PROCEDURE
CRITICAL Prior to following the procedure make sure HYDROID and accompanying software tools are installed following the instructions in Box 1.
Quantification of HRF PAGE images (HYDROIDexp pipeline and library).
-
1
Extracting lane profiles from PAGE image files and preparing data files for HYDROID. Follow the instructions in Box 2 to extract lane profile from a PAGE image using ImageJ software. Alternatively, if the user is more familiar with other biological image quantification software programs, including certain proprietary ones, an attempt can be made to replicate instructions in Box 2. In the latter case make sure that the resulting lane profile data file follows the format outlined in Box 3. Save the image file with the lines marking the extracted data ranges for further reference.
?TROUBLESHOOTING
-
2
Download sample HRF analysis example projects provided with HYDROID. See Box 2 for the download instructions. If DNA in HRF experiments was labeled at the 3’-end, use Example 1; if DNA in HRF experiments was labeled at the 5’-end, use Example 2. The example project folder contains the Python scripts for every stage of the pipeline and a data folder with several data files which will be used in the following steps.
CRITICAL STEP: To use your own data, modify the Python scripts corresponding to the example you choose to use in a text editor, as outlined in the subsequent steps of the Procedure.
-
3
Replace the data/lane_profiles.xls file with the lane profiles data file extracted from the PAGE image in Step 1.
-
4
Modify the data/lane_config.csv file to specify which columns from the data/lane_profiles.xls file should be analyzed by HYDROID and specify human understandable identifiers. Refer to Box 3 and description inside the file for more details. All other parameters should be set to NaN at this point (use the top line in the original file as a template).
-
5
Specify the DNA sequence under study in the data/DNA_seq.fasta file.
-
6
Identifying positions of peaks (electrophoretic band locations) on the gel lane profiles via a semi-automated interactive algorithm. Open exp_s2_assign_peaks.py script in a text editor and modify the lane_names variable (on lines 37–38) to include the names of the lanes from the data/lane_config.csv file that should be analyzed.
CRITICAL STEP: Identification of the peak positions is ultimately required only for the lanes with HRF data, but it can be used to examine profiles of the DNA sequencing lanes via the same procedure.
-
7Run the script via the following command in the terminal:
> python exp_s2_assign_peaks.py
For every lane specified in Step 6 an interactive window will open showing the plot of the lane profile and allowing to adjust parameters of the peak identification algorithm as well as a number of axillary parameters.
CRITICAL STEP: The interactive windows will open one after another. Close each window to proceed to the next one.
-
8
The goals of the next steps are to specify the data range for the analysis and to highlight each and every peak (band) in this range with an asterisk. Use the interactive sliders to specify the range of the values on the HRF profile that will be further analyzed (leftlim and rightlim parameters).
-
9Adjust the peakthresh (regulates the sensitivity of the algorithm to the magnitude of the peaks), min_dist_left (regulates the minimum allowed distance between the peaks on the left side of the data range), min_dist_right (regulates the minimum allowed distance between the peaks on the right side of the data range), Baseline (subtracts a linear baseline from the data before attempting to identify the peaks) and Segments (sets the number of segments to divide the data range to interpolate the minimum required distance between the peaks) parameters until each and every peak (band) location is highlighted by only one asterisk.CRITICAL STEP: Within the analyzed data range, each and every band location should be highlighted by only one asterisk. Otherwise the downstream analysis will be incorrect.CRITICAL STEP: Sometimes the intensity of the bands is very low, the data is noisy or the bands come so close together that no local maxima representing the band can be seen on lane profile. The deconvolution algorithm may still work well in such cases if the expected position of the band is marked by an asterisk. The latter may be achieved through turning on the Interpolate switch in the interactive window. The algorithm will try to guess the positions of the bands that are not represented by a definitive peak on the HRF profile. Additionally, the positions of the bands can be manually specified in the data/lane_config.csv file. To do so, enter their locations in the addpeaks column of the file.
?TROUBLESHOOTING
-
10
(Optional) The alignpos parameter allows to specify the position of any peak that will be used in the next stage to align the HRF profile with DNA sequencing profiles to call the DNA sequence. Usually this can be a prominent peak at the end of the gel that is unambiguously identified as a band representing the DNA product of the same length on HRF and DNA sequencing lanes.
-
11
Press the Save button. The data will be written to data/lane_config.csv file. Repeat the procedure for other lane profiles.
-
12
Calling DNA sequence on the HRF lane profile. Open exp_s3_call_peaks.py script in a text editor and modify the lane_sets list to include the names of the HRF lanes (footprinting_profile) together with the names of the corresponding DNA sequencing profiles (helper_profiles) from the data/lane_config.csv file.
CRITICAL STEP: Be sure to specify the correct DNA labeling end in the label variable (starting with line 35). Be sure that the DNA sequence supplied to every HRF lane profile (seq variable) corresponds to the specific DNA sequence of the DNA strand that was used in the experiment (written in 5’->3’ notation).
-
13Run the script via the following command:
> python exp_s3_call_peaks.py
An interactive window (see video tutorial) will open for every HRF lane profile specified in Step 12 where it will be plotted together with its corresponding DNA sequencing profiles. The sequence of the DNA strand under study will be also shown.
-
14
The goal of the following steps 14–17 is to manually assign any one peak on the HRF profile to the corresponding position on the DNA sequence by looking at the DNA sequencing profiles plotted on the same plot. Use the alignpos sliders if necessary to align the DNA sequencing profiles with each other and the HRF profile.
-
15
Use the sequence slider to select a position on the DNA sequence (the selected position will be highlighted as a capital letter).
-
16
Use the seqpeak slider to point the arrow to the peak on the HRF lane profile corresponding to the position on DNA sequence identified in the previous step
CRITICAL STEP: The user has to understand the nature of the chosen DNA sequencing reaction and its products in order to correctly accomplish this step (see experimental design).
-
17
Press the Save button. The data will be written to data/lane_config.csv file. Repeat the procedure for other HRF lane profiles.
-
18
Fitting a model to the HRF lane profile to quantify DNA cleavage frequencies. Open exp_s4_fit_model.py script in a text editor and modify the lane_data list to include the names of the HRF lanes that will be subjected to quantification. The parameters of the fit_peaks function can be also modified to tweak the results: peaktype can be set to Gaussian or Lorentzian depending on the desired band model (see Box 4), fitting_constraint can take one of the values specified in Table 2, maxfev controls the maximum number of optimization steps, set it to lower numbers to obtain quick preliminary results.
-
19Run the script via the following command:
> python exp_s4_fit_model.py
-
20
The script will output interactive graphical windows with the fitting results plots (Figure 3B). The results would be automatically saved to a CSV file specified in the script (results/LANE_NAME_fitted_intensities.csv). The script will be run sequentially for all lane profiles.
CRITICAL STEP: The user should examine the resulting plots (zoom in if necessary by using the zoom button) and confirm that there are no irregularities in the fitting results similar to ones shown in the upper panels of Figures 3B,3C. See also section “Challenges in experimental HRF profiles quantification” for additional guidance.
?TROUBLESHOOTING
-
21
Extract and plot cleavage frequencies along the DNA sequence. HYDROID provides a customizable function plot_prof_on_seq that may be used to produce plots of data profiles on top of the DNA sequence (Figure 4). Open exp_s5_plot_cl_freq.py script in a text editor and modify the lane_data list (near line 28) to include the names of the files containing the results of quantification from Step 20. The title of the plot and the output file name can be also specified (lines 40–43).
-
22Run the script via the following command:
> python exp_s5_plot_cl_freq.py
Interactive windows with plots will open and the plots will be saved in results/LANE_NAME_fitted_intensities.png.
Prediction of DNA cleavage frequencies from atomistic 3D structures (HYDROIDpred pipeline and library).
-
23Prepare a PDB file with a 3D structure of a DNA-protein complex. The PDB file representing your experimental system may be available in the PDB-database (www.pdb.org) or be a product of molecular modeling efforts. Save the resulting PDB-file in the data/structures subdirectory. As X-ray derived PDB structures often lack hydrogen atoms, you can use the following command to add hydrogen atoms:
> reduce -NUClear file.pdb > file_H.pdb
where file.pdb is the name of the original PDB file.
CRITICAL STEP: Make sure the REDUCE program is installed as detailed in Box 1.
-
24
Run SASA calculations to estimate DNA cleavage frequency profiles. Open the pred_s2_calc_H-SASA.py script in a text editor and modify the prof_data list to include the name(s) of the PDB structure file(s), the PDB identifier of the chain that represents the DNA strand under consideration, resids parameter should specify the range of residue numbers from the PDB-file that will be analyzed, seq parameter should provide the DNA sequence of the strand under study. Parameter Hcontrib is a list of seven numbers that specifies the weight of the contribution of each individual deoxyribose hydrogen atom in the following order [H1’ H2’ H2’’ H3’ H4’ H5’ H5’’]. For X-ray structures of low to moderate resolution (above 2.5–3 Å) taking into account only contributions from H4’, H5’and H5’’ atoms is suggested.
-
25Run the script via the following command:
> python pred_s2_calc_H-SASA.py
The estimated DNA cleavage profiles will be saved to CSV files results/PROF_NAME_H-SASA.csv, where PROF_NAME is the name specified earlier in the prof_data list.
-
26
Plot estimated DNA cleavage frequencies along DNA sequence Open the pred_s3_plot_H-SASA.py script in a text editor and modify the lane_data list to include the name of the CSV file with the estimated profile, DNA sequence of the strand under study and output file name.
-
2727Run the script via the following command:
> python pred_s3_plot_H-SASA.py
Interactive windows with plots will open, additionally the plots will be saved as PNG files.
?TROUBLESHOOTING
Comparison of data generated from HYDROIDexp and HYDROIDprep pipelines
-
28
The data from the CSV files obtained in steps 20 (experimental profiles) and 27 (theoretical profiles) can be open, plotted and compared using any plotting software preferred by the user (e.g. MS Excel). The CSV-file generated by the HYDROIDexp pipeline (see for example, results/scCSE4_601TA_BS_fitted_intensities.csv in Supplementary data 1) has columns named Site and Intensity corresponding to the DNA sequence position and quantified band intensity, which corresponds to DNA cleavage frequency, respectively. The CSV-file generated by the HYDROIDpred pipeline (see for example, results/scCSE4_601TA_BS_H-SASA.csv in Supplementary data 1) has columns named Site and H-SASA corresponding to the DNA sequence position and the theoretically estimated DNA cleavage probability, respectively.
For those with advanced Python skills we provide examples for manipulating and plotting results of Example 1 in the com_plot_ext_vs_pred.py file (Supplementary data 1).
TIMING
Time necessary for the protocol depends on the number of experimental samples to analyze and user’s familiarity with command line tools and Python scripting language. The estimates for a PAGE gel with two HRF lanes and four DNA sequencing lane based on Example 1 are provided below:
Step 1, Processing the image file with Fiji ImageJ: 30 min
Steps 2–11, Identifying the position of peaks (electrophoretic band locations): 30 min
Steps 12–17, Calling the DNA sequence on the HRF lane profile: 30 min
Steps 18–20, Fitting a model to the HRF lane profile: 30 min
Steps 21–22, Extracting and plotting DNA cleavage frequencies along the DNA sequence: 10 min
Step 23, Preparing a PDB file with a 3D structure of a DNA-protein complex: 15 min
Steps 24–25, Setting up and running SASA calculations to estimate DNA cleavage frequency profiles: 20 min
Steps 26–27, Plotting estimated DNA cleavage frequencies along the DNA sequence: 5 min
ANTICIPATED RESULTS
Case study: DNA-protein interactions in nucleosomes
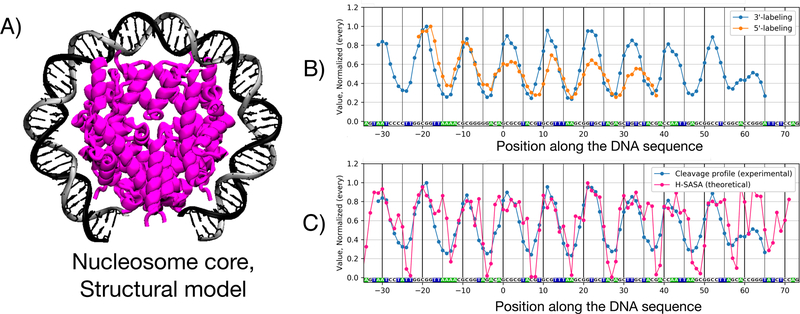
As a case study of the application of HYDROID to DNA-protein systems we analyzed and interpreted HRF experiments for several nucleosomes as they represent one of the most difficult systems to study and they manifest a periodic pattern of DNA-protein interactions. Two independent experimental data sets from different laboratories were obtained: a) HRF of nucleosomes reconstituted on 601 Widom sequence 43 using histones from G. gallus erythrocytes, b) HRF of nucleosomes reconstituted on a similar 601TA well-positioning sequence 38 using recombinant histones of S. cerevisiae centromere nucleosomes11. The two independent data sets also utilized different DNA labeling techniques: 5’-end was labeled in the first case and 3’-end in the second.
The results of the analyses of these experiments as well as comparisons to theoretical H-SASA profiles are presented in Figure 6. DNA in the nucleosome is known to be well positioned in the case of the 601/601TA sequence and is wound around the histone octamer in a superhelical fashion (Figure 6A). The DNA cleavage frequencies were previously shown to be very similar for the top and bottom strands due to the pseudo-symmetry of the nucleosome11. DNA cleavage profiles reflect the 10–11 bp periodicity of nucleosomal DNA as it rotates around the histone octamer surface by showing the corresponding minima and maxima (Figure 6B). The two experimental profiles produced using different end labeling match well but differ in certain aspects. This may reflect the reproducibility that one can expect from HRF analysis performed via different protocols. The key characteristic is the position of minima and maxima, while the exact magnitude of DNA cleavage may depend on experimental conditions, presence of partially assembled nucleosomal states44, DNA and protein dynamics45. The positions of minima and maxima match within 1 bp accuracy between the profiles (Figure 6B). For the 3’-end labeled DNA in the majority of cases minima are shifted by one base pair towards the 5’-end of the DNA when compared to the 5’-end labeled DNA experiment. A potential explanation for this subtle difference is that during hydroxyl-radical cleavage apart from major 5’- and 3’-phosphate terminated products other minor products are formed. Particularly, at the 5’-end a strand terminated by a 5’-aldehyde group can be formed7. It is one nucleotide longer than the 5’-phosphate-terminated strand, lacks the negative charge of the phosphate group, and has gel mobility 2–3 nucleotides slower than the regular product 46. On the 3’-end of the cleavage site the minor product is 3’-phosphoglycolate that is known to be less abundant and migrates close to the regular product 7. Thus 5’-end DNA labeling is generally recommended.
Figure 6. Experimental and theoretical DNA cleavage frequency profiles by hydroxyl-radicals in nucleosomes.
A) Structure of DNA-protein interactions in the nucleosome (from a homology model of S. cerevisiae nucleosome reconstituted on 601 DNA sequence based on PDB ID 3LZ0), central 70 bp of DNA are shown, histone tails are not shown. B) Comparison of two independently obtained experimental profiles for nucleosomes reconstituted on 601/601TA DNA sequence as quantified by HYDROIDexp. The 5’-end labeled profile corresponds to nucleosomes reconstituted on 601 nucleosome-positioning DNA sequence using histones from G. gallus erythrocytes (these data are available in HYDROID as Example 2, Supplementary data 2) DNA shows a periodic pattern of interactions with the histone octamer every 10–11 bp. The 3’-labeled profile corresponds to the nucleosomes reconstituted on a similar 601TA positioning sequence using recombinant histones of S. cerevisiae centromere nucleosomes (these data are available in HYDROID as Example 1, Supplementary data 1. C) Comparison of experimental and H-SASA profiles for centromeric S. cerevisiae nucleosome. The experimental profile is the same as 5’-end labeled profile from panel B. The H-SASA profile was obtained with HYDROIDpred using a homology model of centromeric S. cerevisiae nucleosome based on PDB 3LZ0 (also available in HYDROID Example 1, Supplementary data 1). In panels B) and C) zero on the x-axis marks the nucleotide positioned on the central dyad symmetry axis of nucleosome. The y-axis represents the normalized values of DNA cleavage frequency obtained either experimentally or theoretically. When plotting two profiles on one plot the “every” method from HYDORID package was used, which simply normalizes every profile to its maximum value.
An example comparison between theoretical DNA cleavage frequency profiles extracted from a structural model and experimental HRF data is shown in Figure 6C. The positions of minima of both experimental and theoretical profiles match within 1 bp accuracy. The shape of the theoretical profile is less smooth than for the experimental one, which reflects the limitations of the theoretical model. The smoother shape of experimental profiles is likely due to the dynamics of structures in solution as well as kinetic effects related to hydroxyl-radical diffusion. Interactive web-plots that compare H-SASA profiles derived with different parameters are available in the GitHub repository via the link https://ncbi.github.io/HYDROID/examples/example1/results/nucl_H-SASAvsEXP_BS.html.
Supplementary Material
Table 3.
Troubleshooting table.
| Steps | Problem | Possible reason | Solution |
|---|---|---|---|
| Step 1 | In Fiji ImageJ the gel image shows in negative colors. | Lookup table is set incorrectly. | Click Image->Lookup Tables-> Invert LUT |
| Step 9 | The semi-automatic algorithm cannot identify all needed band locations on HRF profile. | The data is too noisy or the magnitude of the peaks is very small. | 1) Try to adjust parameters so that only well resolved peaks are clearly identified. Click on the Interpolate switch to guess the missing positions. 2) If the previous option does not solve the problem: write down the positions of bands that are still missing by placing the mouse pointer at these locations. Refer to instruction in lane_config.csv file on how to specify these locations manually in the addpeaks column. |
| Step 20 | On the resulting plot the individual Gaussians describing bands at the end of the gel are too broad to represent a physically plausible solution. | Overfitting has occurred. | Use a more stringent fitting constraint option, for example, “SIGMA=k*D+b” |
| Step 27 | Theoretical DNA cleavage profiles look noisy | Small irregularities in 3D structure can affect deoxyribose hydrogen atom accessibility. | Use only contributions from H4’, H5’ and H5’’ deoxyribose hydrogen atoms to estimate cleavage profile. Set Hcontrib parameter to [0,0,0,0,1,1,1] |
ACKNOWLEDGEMENTS
This work was supported by the Intramural Research Programs of the National Library of Medicine (A.K.S., D.L., A.R.P.) and the National Cancer Institute (H.X.), National Institutes of Health; Russian Science Foundation [grant number 14–24-00031, agreement number 14–24-00031-p] (development of gel simulation algorithms and HYDROID_GUI package) (G.A.A., A.K.S.); Howard Hughes Medical Institute Janelia Research Campus and a Johns Hopkins University Bloomberg Distinguished Professorship (C.W.); US–Russia Collaboration in the Biomedical Sciences National Institutes of Health visiting fellows’ program (A.K.S.); NIH grants R01 GM119398 and R21CA220151 (V.M.S.), NIH grants R21DE025398 and P50DE019032 (D.A.G.). This work utilized the computational resources of the NIH HPC Biowulf cluster (http://hpc.nih.gov) and the shared research facilities of HPC computing resources at Lomonosov Moscow State University.
We thank T. Tullius for valuable suggestions, which helped to formulate the initial idea of this study, L. Zaslavsky for discussions on optimal data fitting algorithms and S. Mitternacht for modifications to the FreeSASA program.
Footnotes
COMPETING INTERESTS
The authors declare no competing interests.
DATA AVAILABILITY
All data used in this protocol are available at the HYDROID repository at https://github.com/ncbi/HYDROID.
REFERENCES
- 1.Tullius TD Physical studies of protein-DNA complexes by footprinting. Annual review of biophysics and biophysical chemistry 18, 213–237, doi: 10.1146/annurev.bb.18.060189.001241 (1989). [DOI] [PubMed] [Google Scholar]
- 2.Adilakshmi T, Lease RA & Woodson SA Hydroxyl radical footprinting in vivo: mapping macromolecular structures with synchrotron radiation. Nucleic acids research 34, e64, doi: 10.1093/nar/gkl291 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Song L & Crawford GE DNase-seq: a high-resolution technique for mapping active gene regulatory elements across the genome from mammalian cells. Cold Spring Harb Protoc, 10.1101/pdb.prot5384 (2010). [DOI] [PMC free article] [PubMed]
- 4.Koohy H, Down TA & Hubbard TJ Chromatin accessibility data sets show bias due to sequence specificity of the DNase I enzyme. PloS one 8, e69853, doi: 10.1371/journal.pone.0069853 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jain SS & Tullius TD Footprinting protein-DNA complexes using the hydroxyl radical. Nature protocols 3, 1092–1100 (2008). [DOI] [PubMed] [Google Scholar]
- 6.Armeev GA, Gorkovets TK, Efimova DA, Shaitan KV & Shaytan AK Modeling of the structure of protein–DNA complexes using the data from FRET and footprinting experiments. Moscow University Biological Sciences Bulletin 71, 29–33, doi: 10.3103/S0096392516010016 (2016). [DOI] [Google Scholar]
- 7.Balasubramanian B, Pogozelski WK & Tullius TD DNA strand breaking by the hydroxyl radical is governed by the accessible surface areas of the hydrogen atoms of the DNA backbone. Proceedings of the National Academy of Sciences of the United States of America 95, 9738–9743 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shadle SE et al. Quantitative analysis of electrophoresis data: novel curve fitting methodology and its application to the determination of a protein-DNA binding constant. Nucleic acids research 25, 850–860 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Das R, Laederach A, Pearlman SM, Herschlag D & Altman RB SAFA: semi-automated footprinting analysis software for high-throughput quantification of nucleic acid footprinting experiments. RNA (New York, N.Y.) 11, 344–354, doi: 10.1261/rna.7214405 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bishop EP et al. A map of minor groove shape and electrostatic potential from hydroxyl radical cleavage patterns of DNA. ACS chemical biology 6, 1314–1320, doi: 10.1021/cb200155t (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shaytan AK et al. Hydroxyl-radical footprinting combined with molecular modeling identifies unique features of DNA conformation and nucleosome positioning. Nucleic acids research 45, 9229–9243, doi: 10.1093/nar/gkx616 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiao H et al. Molecular basis of CENP-C association with the CENP-A nucleosome at yeast centromeres. Genes & development 31, 1958–1972, doi: 10.1101/gad.304782.117 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schneider CA, Rasband WS & Eliceiri KW NIH Image to ImageJ: 25 years of image analysis. Nature methods 9, 671–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mitternacht S FreeSASA: An open source C library for solvent accessible surface area calculations. F1000Research 5, 189 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ingle S, Azad RN, Jain SS & Tullius TD Chemical probing of RNA with the hydroxyl radical at single-atom resolution. Nucleic acids research 42, 12758–12767, doi: 10.1093/nar/gku934 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Costa M & Monachello D Probing RNA folding by hydroxyl radical footprinting. Methods in molecular biology 1086, 119–142, doi: 10.1007/978-1-62703-667-2_7 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Hampel KJ & Burke JM Time-resolved hydroxyl-radical footprinting of RNA using Fe(II)-EDTA. Methods 23, 233–239, doi: 10.1006/meth.2000.1134 (2001). [DOI] [PubMed] [Google Scholar]
- 18.Nilsen TW Mapping RNA-protein interactions using hydroxyl-radical footprinting. Cold Spring Harb Protoc 2014, 1333–1336, doi: 10.1101/pdb.prot080952 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Ding F, Lavender CA, Weeks KM & Dokholyan NV Three-dimensional RNA structure refinement by hydroxyl radical probing. Nature methods 9, 603–608, doi: 10.1038/nmeth.1976 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tian S & Das R RNA structure through multidimensional chemical mapping. Quarterly reviews of biophysics 49, e7, doi: 10.1017/S0033583516000020 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Begusova M, Spotheim-Maurizot M, Sy D, Michalik V & Charlier M RADACK, a stochastic simulation of hydroxyl radical attack to DNA. Journal of biomolecular structure & dynamics 19, 141–158, doi: 10.1080/07391102.2001.10506727 (2001). [DOI] [PubMed] [Google Scholar]
- 22.Linkert M et al. Metadata matters: access to image data in the real world. The Journal of cell biology 189, 777–782, doi: 10.1083/jcb.201004104 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takamoto K, Chance MR & Brenowitz M Semi-automated, single-band peak-fitting analysis of hydroxyl radical nucleic acid footprint autoradiograms for the quantitative analysis of transitions. Nucleic acids research 32, E119, doi: 10.1093/nar/gnh117 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Word JM, Lovell SC, Richardson JS & Richardson DC Asparagine and glutamine: using hydrogen atom contacts in the choice of side-chain amide orientation. Journal of molecular biology 285, 1735–1747, doi: 10.1006/jmbi.1998.2401 (1999). [DOI] [PubMed] [Google Scholar]
- 25.AMBER 17 (University of California, San Francisco: ). [Google Scholar]
- 26.Lee B & Richards FM The interpretation of protein structures: estimation of static accessibility. Journal of molecular biology 55, 379–400 (1971). [DOI] [PubMed] [Google Scholar]
- 27.Best RB et al. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone phi, psi and side-chain chi(1) and chi(2) dihedral angles. Journal of chemical theory and computation 8, 3257–3273, doi: 10.1021/ct300400x (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hunter JD Matplotlib: A 2D Graphics Environment. Computing in Science & Engineering 9, 90–95, doi: 10.1109/MCSE.2007.55 (2007). [DOI] [Google Scholar]
- 29.Slater GW, Desruisseaux C & Hubert SJ DNA Separation Mechanisms During Electrophoresis in Capillary Electrophoresis of Nucleic Acids: Volume I: Introduction to the Capillary Electrophoresis of Nucleic Acids (eds Mitchelson Keith R. & Cheng Jing) 27–41 (Humana Press, 2001). [DOI] [PubMed] [Google Scholar]
- 30.Maxam AM & Gilbert W A new method for sequencing DNA. Proceedings of the National Academy of Sciences of the United States of America 74, 560–564 (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brahmasandra SN, Burke DT, Mastrangelo CH & Burns MA Mobility, diffusion and dispersion of single-stranded DNA in sequencing gels. Electrophoresis 22, 1046–1062, doi:10.1002/1522–2683()22:6<1046::aid-elps1046>3.0.co;2-e (2001). [DOI] [PubMed] [Google Scholar]
- 32.Maramis CF & Delopoulos AN Improved Modeling of Lane Intensity Profiles on Gel Electrophoresis Images in XII Mediterranean Conference on Medical and Biological Engineering and Computing 2010: May 27 – 30, 2010 Chalkidiki, Greece (eds Bamidis Panagiotis D. & Pallikarakis Nicolas) 671–674 (Springer Berlin; Heidelberg, 2010). [Google Scholar]
- 33.Greenbaum JA, Pang B & Tullius TD Construction of a genome-scale structural map at single-nucleotide resolution. Genome research 17, 947–953, doi: 10.1101/gr.6073107 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berg JM APPENDIX: Lorentzian lineshapes are intrinsic to autoradiographic detection. Nucleic acids research 25, 850–860 (1997).9016637 [Google Scholar]
- 35.Spotheim-Maurizot M & Davidkova M Radiation damage to DNA in DNA-protein complexes. Mutation research 711, 41–48, doi: 10.1016/j.mrfmmm.2011.02.003 (2011). [DOI] [PubMed] [Google Scholar]
- 36.Xiao H et al. Molecular basis of CENP-C association with the CENP-A nucleosome at yeast centromeres. Genes and Development, in press (2017). [DOI] [PMC free article] [PubMed]
- 37.Morozov AV et al. Using DNA mechanics to predict in vitro nucleosome positions and formation energies. Nucleic acids research 37, 4707–4722, doi: 10.1093/nar/gkp475 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cloutier TE & Widom J DNA twisting flexibility and the formation of sharply looped protein-DNA complexes. Proceedings of the National Academy of Sciences of the United States of America 102, 3645–3650, doi: 10.1073/pnas.0409059102 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sali A & Blundell TL Comparative protein modelling by satisfaction of spatial restraints. Journal of molecular biology 234, 779–815, doi: 10.1006/jmbi.1993.1626 (1993). [DOI] [PubMed] [Google Scholar]
- 40.Lu XJ & Olson WK 3DNA: a versatile, integrated software system for the analysis, rebuilding and visualization of three-dimensional nucleic-acid structures. Nature protocols 3, 1213–1227, doi: 10.1038/nprot.2008.104 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Draizen EJ et al. HistoneDB 2.0: a histone database with variants--an integrated resource to explore histones and their variants. Database : the journal of biological databases and curation 2016, baw014, doi: 10.1093/database/baw014 (2016). [DOI] [PMC free article] [PubMed]
- 42.Humphrey W, Dalke A & Schulten K VMD: Visual molecular dynamics. Journal of Molecular Graphics 14, 33–38, doi: 10.1016/0263-7855(96)00018-5 (1996). [DOI] [PubMed] [Google Scholar]
- 43.Lowary PT & Widom J New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. Journal of molecular biology 276, 19–42, doi: 10.1006/jmbi.1997.1494 (1998). [DOI] [PubMed] [Google Scholar]
- 44.Rychkov GN et al. Partially Assembled Nucleosome Structures at Atomic Detail. Biophysical journal 112, 460–472, doi: 10.1016/j.bpj.2016.10.041 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eslami-Mossallam B, Schiessel H & van Noort J Nucleosome dynamics: Sequence matters. Adv Colloid Interface Sci 232, 101–113 (2016). [DOI] [PubMed] [Google Scholar]
- 46.Kappen LS & Goldberg IH Deoxyribonucleic acid damage by neocarzinostatin chromophore: strand breaks generated by selective oxidation of C-5’ of deoxyribose. Biochemistry 22, 4872–4878 (1983). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.