Summary
The recent surge in research on the intestinal microbiota has greatly changed our understanding of human biology. Significant technical advances in DNA sequencing analysis and its application to metagenomics and metatranscriptomics has profoundly enhanced our ability to quantify and track complex microbial communities and to begin understanding their impact on human health and disease. This has led to a better understanding of the relationships between the intestinal microbiome and renal physiology/ pathophysiology. In this review, we discuss the interactions between intestinal microbiota and kidney. We will focus on select aspects including the intestinal barrier, immunologic and soluble mediators of microbiome effects, and effects of dysbiosis on AKI. Relevant studies on microbiome changes in other renal diseases are highlighted. We also introduce potential mechanisms of intervention with regards to gut microbiota in renal diseases.
1. Dysbiosis of the microbiota causes disease
The human gut microbiome consists of at least 1000 genera of bacteria which build their own community [1]. Although there is a lack of clear understanding on what constitutes a ‘healthy’ gut microbiota, it is increasingly clear that imbalance of the gut microbiota, dysbiosis, is associated with disease development both in the intestine and in other organs. The intestinal epithelial barrier plays a key role in maintaining homeostasis of intestinal microbiota. In healthy individuals, this barrier largely prevents intestinal bacteria and toxins from crossing the intestinal mucosa into the circulatory system, or to other tissues and organs. As depicted in Figure 1, the intestinal epithelial barrier can be divided into three components – a biological barrier, a physical barrier, and an immune barrier – each of which can become dysfunctional.
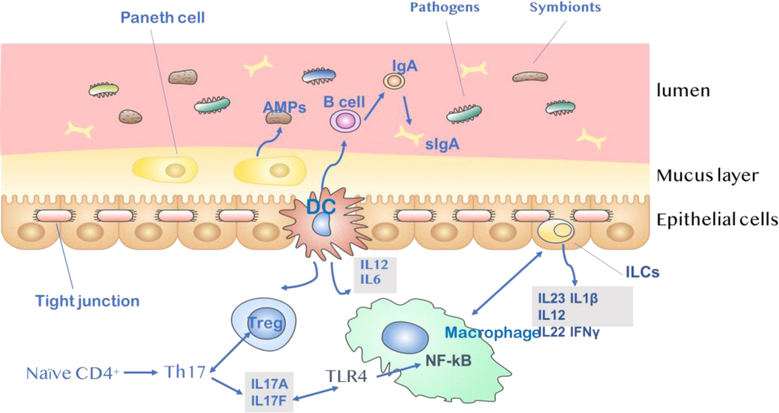
Fig.1. Intestinal barrier to bacteria.
A) Gut microbiota plays a direct protective role as a biological barrier. They are closely attached to the surface of the intestinal mucosa and compete with pathogenic bacteria and thus can inhibit the colonization and excessive growth of pathogens. B) The intestinal epithelial layer acts as a physical barrier. Tight junction proteins play an important role in the establishment and maintenance of cell polarity within tissues. They function as a fence that restricts the intermixing of lipid and protein components of apical and basolateral membrane domains, regulate the permeability of solutes and ions through the paracellular space and prevent pathogens from reaching the subepithelial tissue. C) Microbiota and epithelial cells interact with each other and regulate subsequent immune response. Translocation of bacterial products across the leaky intestinal barrier activates the immune system, resulting in systemic inflammation. IgA released from Paneth cells plays a fundamental role in mucosal immunity, mainly in response to colonization by specific commensal bacteria. Activation of DCs activated by intestinal microbes results in the secretion of proinflammatory cytokines such as IL-12, IL-6. These DCs promote the differentiation of naïve CD4+ T cells into regulatory T (Treg) cells and the maturation of B cells into IgA-secreting plasma cells. Th17-inducing bacteria may promote Th17 immunity via IL-17A/IL-17F induction, which may involve signaling mediated by the TLR ligands. Finnally, innate lymphoid cells(ILCs) function by limiting macrophage production of the pro-inflammatory cytokines IL-1β, IL-12, IL-23, IL22 and IFNγ.
The biological barrier is composed of bacterial and fungal symbionts. Large numbers of primarily anaerobic symbionts defend against pathogens by secreting antimicrobial proteins (AMPs) via Paneth cells to disrupt the surface structure of bacteria [2]. The commensal bacteria also yield short chain fatty acids (SCFAs); these end-products of carbohydrate digestion and fermentation are associated with pH reductions in the intestinal tract [3]. Symbionts are closely attached to the intestinal mucosal surface, and compete with pathogenic bacteria to inhibit the colonization and excessive growth of pathogens. This phenomenon, known as colonization resistance, plays a major role in maintaining intestinal homeostasis [4].
The physical barrier refers to the intestinal epithelial cells, whose apical tight junctions (TJs) form a defensive wall that prevents free diffusion from the intestinal lumen to the basolateral lamina propria. TJs of intestinal epithelial cells are mainly composed of transmembrane proteins and cytoplasmic proteins. These TJs serve two main functions. (1) They maintain the permeability barrier. Changes in TJs can lead to increased permeability through the physical barrier, ultimately allowing bacteria, endotoxins, and/or macromolecules to enter the circulatory system [5]. (2) TJs also maintain the polarity of intestinal epithelial cells. The fusion points between the plasma membranes of adjacent epithelial cells form a continuous apical surface and maintain cell polarity by restricting free diffusion of lipids and intact membrane proteins in different liquid spaces of cells [6].
The activation of the immune barrier response is the third strategy for maintaining microbial homeostasis. Dendritic cells (DC) in the lamina propria activate T cells to evoke an adaptive immune response. A key feature of the adaptive response is DC-mediated recruitment of regulatory T cells (Tregs), which critically depends on the activation of non-classical autophagy pathways [7–9]. Epithelial release of nuclear factor- κB (NF-κB) also plays a critical role in controlling proinflammatory response, as well as the nod-like receptor pyrin domain containing 3 (NLRP3) inflammasome [10]. Innate lymphoid cells (ILCs) located in the gut epithelium also have key defensive functions, producing or activating the release of immune-activating cytokines (i.e. IL- 22 [11], IFN-γ [11,12], IL-1β, IL-12 and IL-23 [13]) via adaptive and innate immune responses. Furthermore, the secretion of AMPs and IgA by Paneth cells protects mucosal surfaces and contributes to host-microbiota mutualism [14].
2. Microbiota-derived metabolites affect kidney function
Communication between gut and kidney occurs not only through the direct contact between host and microbiota, but also through indirect communication via microbiota-derived metabolites.
2.1. Short-chain fatty acids
As mentioned above, the gut microbiota produce SCFAs, including acetate, propionate, and butyrate, during the fermentation of dietary fibers. SCFAs are being increasingly recognized for their roles in regulating the dynamic balance between the microbes and host. The fact that SCFAs cannot be detected in germ-free (GF) mice suggests that SCFA production requires the presence of bacteria [15,16].
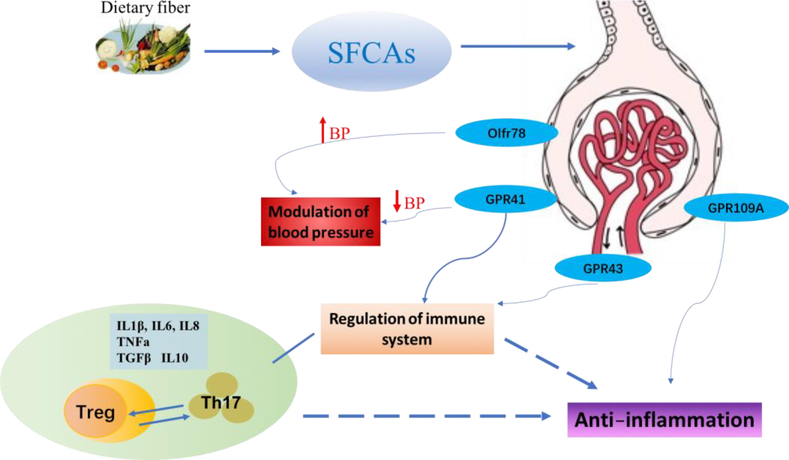
SCFAs primarily signal through G-protein-coupled receptors (GPCRs), a large protein family that detects molecules outside the cell and activates internal signal transduction pathways. GPR41, GPR43, and GPR109A are the major GPCRs activated by SCFAs. GPR41 is expressed in a variety of tissues and cell types, including the colon, kidneys, sympathetic nervous system, and blood vessels [17,18]. It is implicated in inhibiting cell proliferation and inducing apoptosis [19], energy homeostasis [20], T-cell differentiation, and immunity [21]. GPR43 knock-out (GPR43−/−) mice have decreased SCFA-induced release of glucagon-like peptide-1 (GLP-1), a key hormone that controls insulin release, and affects satiety and intestinal transit [22]. GPR43 mRNA and protein can be detected in eosinophils, basophils, neutrophils, monocytes, DCs, and mucosal mast cells [23,24], suggesting a broad role of GPR43-targeting SCFAs in immune responses. GPR109A is also expressed by immune cells and plays an important role in reducing the progression of atherosclerosis [25]. Additionally, another SCFA receptor named Olfr78 was recently found to play a functional role in the modulation of blood pressure [26]. Figure 2 shows a model of SCFAs and their GPRs in modulating kidney disease.
Fig.2. SCFAs and the receptors in kidney.
GPR41, GPR43, GPR109A and Olfr78 are four SCFA receptors identified in kidney. GPR41and GPR43 are activated by acetate, propionate, butyrate, and isobutyrate and function as regulators of the immune system. Luminal butyrate exerts anti-inflammatory effects via GPR109A and HDAC inhibition. SCFAs also regulate cytokine expression in T cells and generation of Tregs through HDAC inhibition. Effector Th17 cells have enhanced aerobic glycolysis, and inhibition of glycolysis promotes Treg cell generation. The activation of GPR41 can reduce blood pressure, whereas Olfr78, upon stimulation by acetate and propionate, increases blood pressure. SCFAs-mediated cytokines include IL-8, IL-6, IL-1β, TNFα, TGFβ, and IL-10.
SCFAs also act as histone deacetylase (HDAC) inhibitors that can regulate the epigenome by altering chromatin remodeling [27]. Pharmacologic HDAC inhibition has demonstrated anti-fibrotic, anti-inflammatory, and immunosuppressant effects in obstructed kidney and was found to reduce cyst formation in polycystic kidney disease [28]. By inhibiting HDAC activity, SCFAs increase the acetylation of histone, consequently modulating the expression of immune-regulating genes, including NF-κB, myogenic differentiation antigen, tumor protein p53, and nuclear factor of activated T cells (NFAT) [29]. These observations are consistent with studies showing that SCFAs, particularly acetate, play a crucial role in kidney disease. In a mouse model of kidney ischemia-reperfusion injury (IRI), acetate treatment administered in two separate intraperitoneal dosages (initially 30 minutes before ischemia induction and secondly at the moment of reperfusion) diminished serum levels of creatinine and urea [30]. Additionally, in a mouse model of experimental sepsis-induced AKI, acetate administration attenuated HDAC activity in T cells, resulting in restoration of oxidant-antioxidant balance [31]. Together, these data support a beneficial effect of SCFAs in kidney injury.
2.2. Trimethylamine-N-Oxide (TMAO)
Trimethylamine-N-oxide (TMAO), a small colorless amine oxide, is a by-product of phosphatidylcholine metabolism in the gut microbiota. It is directly derived from the metabolism of trimethylamine (TMA), which is synthesized exclusively by gut microbiota from dietary nutrients such as choline, an essential nutrient that forms the headgroup of the phospholipid phosphatidycholine, and carnitine, an amino acid with key metabolic functions. A clinical study of 283 individuals reported a correlation between elevated TMAO levels in plasma and an increased risk for major adverse cardiovascular events and death [32]. Gut microbiota-derived TMAO also contributes to renal interstitial fibrosis as shown in a mouse model of diet-induced obesity [33]. In C57BL/6J mice, a diet containing choline or TMAO led to tubulointerstitial fibrosis, collagen deposition, and phosphorylation through the TGFβ/Smad3 signaling pathway [34]. Chronic kidney disease (CKD) patients with increased plasma TMAO levels had lower long-term survival rate [35]. In hemodialysis patients, high TMAO level is associated with higher risk of sudden cardiac and all-cause mortality compared to those with lower TMAO level, especially in Caucasians [36] [37]. Cumulatively, these data suggest an important role for TMAO as a potential gut-derived toxin. To date, there is no published study on the effects of gut microbiota-derived TMAO levels in AKI, but limited CKD data and evidences from studies in related fields suggest that increased TMAO may worsen kidney function during AKI.
3. Gut microbiota and host immunity
Microbiota appear to promote the development of gut-associated lymphoid tissues (GALTs) and intestinal immune homeostasis through the regulation and induction of both adaptive and innate immune responses.
3.1. Tregs mediate intestinal immunity
Tregs exhibit diverse phenotypes depending on their origin (thymic vs. peripheral) and location (lymphoid vs. tissue-resident). DCs in the intestinal lamina propria induce the differentiation of gut-resident naïve T cells into intestinal Tregs following stimulation with lumen-derived antigens. These intestinal Tregs likely regulate effector T-cell responses to gut bacteria, including the expression of cell surface proteins and cytokine production [38]. Treg frequency is reduced several-fold in GF mice compared with mice housed in specific-pathogen free (SPF) facilities, indicating that gut microbiota regulate the Treg population [39]. Additional studies have shown that induced Tregs participate in maintaining tolerance to intestinal antigens [40]. Increasing evidence further demonstrates that the development of colonic Tregs occurs in response to species-specific bacterial antigens. Peripherally-derived Tregs (pTreg) have also been identified in a population of pathobiont-specific Tregs that selectively regulate pathobiont-driven inflammation in healthy mice [41]. In addition to their local effects on the intestinal Treg population, gut microbiota have also been found to affect peripheral Treg differentiation [42].
It appears that Treg induction serves as a strategy to establish commensalism, not only by helping the microorganisms to colonize their niche, but also by protecting the host from inflammation. Furthermore, Tregs appear to reinforce intestinal homeostasis by promoting tolerance to commensal bacterial and related-proteins [43]. These changes depend on the activity of T helper 17 (Th17) cells, a subset CD4 T helper (Th) cells that are defined by their production of the proinflammatory interleukin-17 (IL-17) [41]. The aryl hydrocarbon receptor/IL-22 axis in group 3 innate lymphoid cells (ILC3) has been shown to contribute to the suppression of inflammatory Th17 cell responses that maintains Treg-mediated gut homeostasis [44]. Thus, it is possible that ILC3 and Th cells share a number of features, and that ILCs may not only impair intestinal immunity against pathogen infection, but also induce autoimmunity in the gut by disturbing commensal flora [44].
3.2. IgA function in relation to intestinal microbiota
Immunoglobulin A (IgA) is essential for maintaining the symbiotic balance between the intestinal bacterial community and the host immune system [45]. The multiple functions of IgA in relation to intestinal microbiota include neutralizing toxins and viruses, blocking excessive live bacterial adherence or translocation, clearing unwanted macromolecular structures at the epithelial surface, and directed-sampling of luminal antigen [46]. IgA is produced by plasma cells in the small intestine by mechanisms that are independent of exogenous antigens and T cell contributions. In the steady state, IgA secreted by resident plasma cells act as natural polyreactive antibodies [47]. They possess innate-like recognition properties that may facilitate adaptation to the vast and dynamic array of exogenous microbiota and dietary antigens encountered at mucosal surfaces [48]. In the T cell transfer model of colitis, co-transfer of Tregs was required to limit pathology, maintain intestinal microbial diversity, and induce IgA [49]. In another study, depletion of Tregs reduced intestinal IgA level, suggesting that Tregs may directly induce IgA via secretion of TGFβ [50]. These findings indicate that there is an important Treg/IgA pathway at the host-microbiota interface.
3.3. Host immune response affects kidney disease
Emerging evidence from experimental models and human studies supports a key role of Th17 cells in renal injury. It has been shown that the induction and suppression of Th17 cells in the intestine leads to their migration and activation in kidney [51]. It appears that gut-derived Th17 cells can be recruited to inflamed kidney via a CCL20/CCR6 axis [52]. In addition, reduction of microorganisms by antibiotics reduces Th17 response and tissue damage in murine experimental crescentic glomerulonephritis [53]. Th17 cells are abundant in patients with autoimmune kidney diseases, such as anti-neutrophil cytoplasmic antibody (ANCA)-associated glomerulonephritis, and disease activity correlates with IL-17 serum levels [54]. Interestingly, cultured microbiota isolated from stools of systemic lupus erythematosus (SLE) patients, but not healthy controls‟ promoted lymphocyte activation and naïve CD4+ T cell differentiation into Th17 cells [55].
4. Microbiome and AKI
Acute kidney injury (AKI) involves multiple and overlapping immunological, biochemical, and hemodynamic mechanisms. Recent experimental data has revealed that the intestinal microbiota significantly affect outcomes in AKI [56]. Despite increasing data on microbiome and kidney diseases, there has been limited mechanistic or clinical data on AKI.
4.1. Gut microbiota and kidney inflammation
AKI is associated with intrarenal and systemic inflammation [57]. One mechanism by which gut microbiota reduces inflammation in kidney is through secretion of SCFAs. Four SCFA receptors have been reported in kidney: GPR41, GPR43, Olfr78, and GPR109a. GPR41 and GPR43 expression in the whole kidney and in the renal artery has been detected by RT-PCR [58]. In a mouse model of IRI, acetate treatment increased expression of GPR43 in kidney tissue [30,31]. Olfr78 is expressed in the renal juxtaglomerular apparatus, where it mediates renin secretion in response to SCFAs [58]. Finally, GPR109a has been reported in adipose tissue associated with the kidney at a relatively low level and its function in kidney disease remains unclear [59].
A number of findings suggest a beneficial effect of SCFAs in AKI. Recent data shows intraperitoneally administered SCFAs reduced kidney injury by modulating the inflammatory response, apoptosis and autophagy in a murine IRI model of AKI. Treatment with SCFAs not only decreased production of reactive oxygen species (ROS), but also diminished production of cytokines and chemokines such as IL-1β, IL6, TNF-α, and MCP-1. In addition, low levels of mRNA of the toll-like receptor 4 (TLR4) and its endogenous ligand were detected in kidney tissue, activation of NF-κB was inhibited in kidney epithelial cell line. Furthermore, in vitro analyses showed that SCFAs decreased maturation of DCs and inhibited the capacity of these cells to induce CD4 and CD8 T cell proliferation. SCFAs ameliorated the effects of hypoxia in kidney epithelial cells by improving mitochondrial biogenesis [30]. All these findings indicate that modulation of SCFAs in the gut could be a novel therapeutic approach against AKI. Mechanistic insights have also been gleaned from a rat model of contrast-induced nephropathy (CIN): administration of sodium butyrate decreased IL-6 levels in kidney tissue, preventing the translocation of NF-κB into the nucleus, and reducing inflammation and oxidative damage in the kidney after CIN [60]. This result suggests that SCFA-mediated HDAC inhibition may evoke therapeutic effects through a similar mechanism.
An emerging concept across biomedical research is that the use of “squeaky clean” mice can significantly alter research findings [61]. While searching for the reason at why murine kidney contains has abundant immune cells, including lymphocytes, it was hypothesized that GF mice may lack bacteria as well as kidney lymphocytes [62]. Contrary to expectations, ample kidney lymphocytes, including increased natural killer T (NKT) cells, and IL-4 cytokine, were found in kidneys of normal GF mice. Furthermore, the GF status worsens kidney histological and functional responses to IRI, with enhanced CD8 T cell trafficking. Conventionalized GF mice, in which normal gut bacteria have been reconstituted, appear to be protected from kidney injury: After IRI, 11-week-old conventionalized GF mice had less injury in the outer medulla and inner medulla compared with age-matched GF mice and 6-week-old GF mice, revealing the promise of therapeutic bacteria for AKI [62]. In another study, broad-spectrum antibiotics protected mice against IRI-induced kidney injury by reducing the maturation status of F4/80+ renal-resident macrophages and bone marrow-derived monocytes.
Furthermore, transplant of fecal material from untreated mice to microbiota depleted mice abolished this protective effect [63]. Thus, the relevance of antibiotic treatment and its effects on the intestinal microbiota are worth noting. Patient data suggests these effects are not limited to rodents: Long-term use of rifaximin, an oral broad-spectrum antibiotic that concentrates in the gastrointestinal tract, was found to increase glomerular filtration rate and natriuresis, thus improving systemic hemodynamics and renal function in patients with advanced cirrhosis [64,65].
4.2. Microbiome and uremic toxins
Normally, intestinal bacteria can generate uremic toxins that are absorbed into the blood and cleared by the kidney. However, changes in the microbiome composition can lead to excessive secretion of uremic toxins, which may damage renal tubular cells [66]. Indoxyl sulfate (IS) and p-cresyl sulfate (PCS) are protein-bound uremic toxins generated from colonic bacterial fermentation of dietary protein. Free serum levels of p-cresol (the precursor of PCS) are associated with increased mortality in hemodialysis patients [67]. The generation of p-cresol and indole (the precursor of IS) were also upregulated in patients on hemodialysis in comparison to healthy individuals [68]. TMA is another important urinary compound metabolized by gut microbiota and choline. It has been shown that the serum level of TMAO and related metabolites affect kidney functions and are associated with an increased risk of CKD and its complications, such as atherosclerosis [69]. Recent evidence from studies on AKI-to-CKD transition and end-stage renal disease (ESRD) indicate that changes in intestinal microbiota do occur in kidney diseases and reversing microbiota dysbiosis may prevent toxins and pathogens from permeating the intestinal barrier [56]. Hemodialysis or peritoneal dialysis may also induce intestinal barrier permeability in patients and contribute to the translocation of endotoxins from intestinal lumen to kidney [70].
4.3. Microbiome and blood pressure.
Arterial hypertension is a frequent finding in both acute and chronic kidney diseases. Recent studies have focused on the connection between the microbiome and hypertension. In GF mice, angiotensin II treatment was unable to induce hypertension [71]. In a model using chronic high salt intake, conventional mice developed early renal injury and hypertension. However, transplantation of fecal microbiota from conventional mice on chronic high salt diet to conventional mice on normal diet causes gut leakiness and renal injury. This indicates that high-salt-diet causes enteric dysbiosis that triggers renal injury and dysfunction [72]. Hydrogen sulfide (H2S) has been shown to be an important player in a variety of physiological functions, particularly in the renal vasculature. Deregulation of H2S biosynthesis causes or contributes to existing high blood pressure [73]. These limited results have highlighted the need for new research into the roles of gut microbiota in the development of hypertension and their influence on physiological levels of H2S, which could lead to novel strategies for exploiting microbiota for treatment of hypertension.
5. Modulation of gut microbiota in kidney disease
5.1. Probiotics and prebiotics
A number of therapeutic opportunities for targeting IS and PCS have been proposed, including inhibition of colonic bacterial biosynthesis, suppression of absorption, augmentation of clearance, and modulation of cellular pathways [74]. For example, orally administered AST-120, a highly potent activated charcoal adsorbent, reduces systemic toxin absorption through gastrointestinal sequestration, which may slow disease progression in CKD patients [75]. Probiotics consist of living bacteria, such as bifidobacterium species, lactobacilli, and streptococci can alter gut microbiota and affect the inflammatory response. Lactobacillus treatment can restore mucosal barrier function by increasing the expression of the TJ-associated protein zona occuldens-1 (ZO-1) [76]. Hemodialysis patients treated with oral Lactobacillus acidophilus showed decreased serum dimethylamine, a potential uremic toxin. A double-blind placebo-controlled randomized cross-over trial using synbiotics (co-administration of pre- and probiotics) showed beneficial effects on CKD stage 4–5 patients [77]. Table 1 summarizes recent advances in the use of probiotics and prebiotics as novel treatments for kidney disease [78,79][80] [81] [82] [83] [84] [85] [86].
Table.
Prebiotics and probiotics in kidney disease.
| Strain | Disease type | Effect | Year | Reference | |
|---|---|---|---|---|---|
| probiotics | Lactobacillus. acidophilus | CKD stage3,4 | Decrease Bun, Cr, uric acid, uremic toxins | 2009 2010 |
[78,79] |
| Diabetic + HD | Glucose homeostasis, Inflammation, oxidative stress | 2017 | [80] | ||
| Aging mice | Kidney klotho marker | 2012 | [81] | ||
| Lactobacillus. casei | LPS induced acute inflammatory model | Regulate immuno-coagulative response | 2018 | [82] | |
| Lactobacillus. rhamnosus | MRL/lpr lupus nephritis model | Anti-inflammatory, decreased IgG2a in kidney, adjust immunity | 2017 | [83] | |
| Bifidobacteria. bifidum | Mice model of primary hyperoxaluria | Limiting absorption across intestine | 2015 | [84] | |
| prebiotics | Oat and barley beta-glucans | Healthy individuals | Decrease p-cresyl sulfate | 2017 | [85] |
| xylooligosaccharide | Obese rats | Decrease renal oxidative stress and apoptosis | 2018 | [86] | |
Abbreviations: CKD: chronic kidney disease; Bun: blood urea nitrogen; Cr: creatinine; HD: hemodialysis; LPS: Lipopolysaccharide.
5.2. Targeting immune pathways
Direct targeting of immune responses downstream of microbial regulation has been explored as a strategy for limiting kidney damage. Kidney-infiltrating Th17 cells that mediate microbiome effects can be therapeutically targeted at multiple levels of the immune response. IL-23, a pro-inflammatory cytokine belonging to the IL-12 cytokine family, and Th17 cells identify a newly discovered axis for therapeutic intervention. Th17 cell expansion and maintenance can be disrupted using monoclonal antibodies against IL-23 (e.g. guselkumab [87]), and IL-23/IL-12 (e.g. ustekinumab [88]). IL-17A, secreted by Th17 cells, might be the most promising target and can be blocked with monoclonal anti-IL-17A antibodies, such as secukinumab and ixekizumab [89,90]. Alternatively, monoclonal antibodies such as brodalumab inhibit IL-17 receptor (IL-17R) signaling [91]. In addition to secretion of IL-17, Th17 cells are characterized by the expression of transcription factor RORγt. Several RORγt inhibitors are under development and might represent a promising approach for patients with immune-mediated kidney disease.
Strategies targeting the microbial source of immune regulation also show promise. The presence of lactic acid bacteria (Lactobacillales) in the gut microbiota can promote Treg cells and suppress disease-causing Th17 cells in kidney to attenuate kidney inflammation in lupus-prone mice [83]. Oral administration of Lactobacillus acidophilus ATCC 4356 led to attenuation of atherosclerotic progression through modulation of oxidative stress and inflammatory process in apoliprotein-E knockout (ApoE−/−) mice [92]. IgA produced in programmed cell death protein-1 (PD-1)-deficient (PD-1−/−) mice reduced bacterial-binding capacity and changed the microbial composition of the intestinal tract [93]. Additional data shows that intestinal microbiota enhances the efficacy of PD-1 and its ligand (PD-L1) therapy [94]. The ability of the intestinal microbes to rapidly respond to changes in the environment through dynamic alterations in gene expression renders microbial regulation of the immune barrier highly plastic. The use of functional metagenomics screening methods to study intestinal microbes is an exciting prospect for new therapeutic discovery [95].
Conclusion
The pathogenesis of AKI involves immunological, biochemical and hemodynamic mechanisms. Research on the microbiome has revealed new opportunities to better understand AKI. Figure 3 summarizes the key mechanisms by which microbiome dysbiosis can influence AKI. Experimental data in animal models has demonstrated the impact of intestinal microbiota on the process and outcome of AKI and highlighted the promise of treatment using prebiotics, probiotics or both. In the future, it will be important to determine whether targeted modulation of the intestinal microbiome can act as a potential therapeutic tool for AKI.
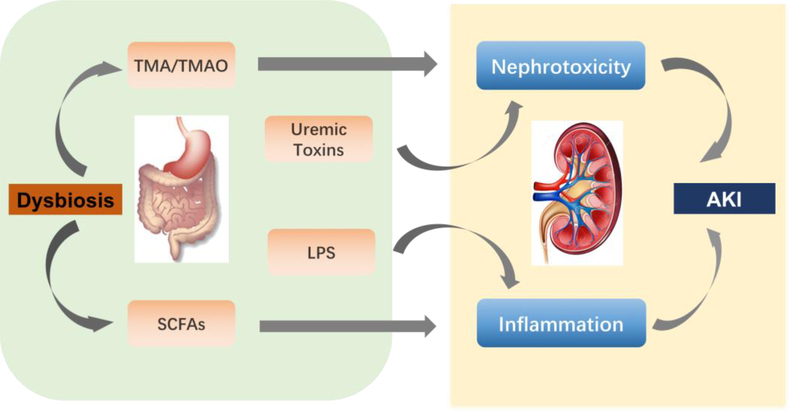
Fig.3. Dysbiosis during AKI.
AKI involves multiple and overlapping immunological, biochemical, and hemodynamic mechanisms. Gut dysbiosis generates SCFAs, which regulate systemic consequences of AKI. TMA/TMAO, combined with uremic toxins and other gut-derived toxins, contributes to nephrotoxicity-induced AKI.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gilbert JA, Blaser MJ, Caporaso JG et al. Current understanding of the human microbiome. Nature medicine 2018; 24: 392–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mergaert P Role of antimicrobial peptides in controlling symbiotic bacterial populations. Natural product reports 2018; February [DOI] [PubMed] [Google Scholar]
- 3.Cummings JH, Pomare EW, Branch WJ et al. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut 1987; 28: 1221–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kranich J, Maslowski KM, Mackay CR. Commensal flora and the regulation of inflammatory and autoimmune responses. Seminars in immunology 2011; 23: 139–145 [DOI] [PubMed] [Google Scholar]
- 5.Runkle EA, Mu D. Tight junction proteins: from barrier to tumorigenesis. Cancer letters 2013; 337: 41–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cereijido M, Contreras RG, Shoshani L et al. Tight junction and polarity interaction in the transporting epithelial phenotype. Biochimica et biophysica acta 2008; 1778: 770–793 [DOI] [PubMed] [Google Scholar]
- 7.Rescigno M, Urbano M, Valzasina B et al. Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat Immunol 2001; 2: 361–367 [DOI] [PubMed] [Google Scholar]
- 8.Colombo BM, Scalvenzi T, Benlamara S et al. Microbiota and mucosal immunity in amphibians. Frontiers in immunology 2015; 6: 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chu H, Khosravi A, Kusumawardhani IP et al. Gene-microbiota interactions contribute to the pathogenesis of inflammatory bowel disease. Science (New York, NY) 2016; 352: 1116–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Afonina IS, Zhong Z, Karin M et al. Limiting inflammation-the negative regulation of NF-kappaB and the NLRP3 inflammasome. Nat Immunol 2017; 18: 861–869 [DOI] [PubMed] [Google Scholar]
- 11.Zenewicz LA, Yancopoulos GD, Valenzuela DM et al. Innate and adaptive interleukin-22 protects mice from inflammatory bowel disease. Immunity 2008; 29: 947–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carasi P, Racedo SM, Jacquot C et al. Impact of kefir derived Lactobacillus kefiri on the mucosal immune response and gut microbiota. J Immunol Res 2015; 2015: 361604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bernink JH, Krabbendam L, Germar K et al. Interleukin-12 and −23 Control Plasticity of CD127(+) Group 1 and Group 3 Innate Lymphoid Cells in the Intestinal Lamina Propria. Immunity 2015; 43: 146–160 [DOI] [PubMed] [Google Scholar]
- 14.Ayabe T, Ashida T, Kohgo Y et al. The role of Paneth cells and their antimicrobial peptides in innate host defense. Trends Microbiol 2004; 12: 394–398 [DOI] [PubMed] [Google Scholar]
- 15.Hoverstad T, Midtvedt T. Short-chain fatty acids in germfree mice and rats. The Journal of nutrition 1986; 116: 1772–1776 [DOI] [PubMed] [Google Scholar]
- 16.Perry RJ, Peng L, Barry NA et al. Acetate mediates a microbiome-brain-beta-cell axis to promote metabolic syndrome. Nature 2016; 534: 213–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiong Y, Miyamoto N, Shibata K et al. Short-chain fatty acids stimulate leptin production in adipocytes through the G protein-coupled receptor GPR41. Proceedings of the National Academy of Sciences of the United States of America 2004; 101: 1045–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tazoe H, Otomo Y, Karaki S et al. Expression of short-chain fatty acid receptor GPR41 in the human colon. Biomedical research (Tokyo, Japan) 2009; 30: 149–156 [DOI] [PubMed] [Google Scholar]
- 19.Kimura M, Mizukami Y, Miura T et al. Orphan G protein-coupled receptor, GPR41, induces apoptosis via a p53/Bax pathway during ischemic hypoxia and reoxygenation. The Journal of biological chemistry 2001; 276: 26453–26460 [DOI] [PubMed] [Google Scholar]
- 20.Inoue D, Tsujimoto G, Kimura I. Regulation of Energy Homeostasis by GPR41. Frontiers in endocrinology 2014; 5: 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park J, Kim M, Kang SG et al. Short-chain fatty acids induce both effector and regulatory T cells by suppression of histone deacetylases and regulation of the mTOR-S6K pathway. Mucosal Immunol 2015; 8: 80–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tolhurst G, Heffron H, Lam YS et al. Short-chain fatty acids stimulate glucagon-like peptide-1 secretion via the G-protein-coupled receptor FFAR2. Diabetes 2012; 61: 364–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Le Poul E, Loison C, Struyf S et al. Functional characterization of human receptors for short chain fatty acids and their role in polymorphonuclear cell activation. The Journal of biological chemistry 2003; 278: 25481–25489 [DOI] [PubMed] [Google Scholar]
- 24.Karaki S, Tazoe H, Hayashi H et al. Expression of the short-chain fatty acid receptor, GPR43, in the human colon. Journal of molecular histology 2008; 39: 135–142 [DOI] [PubMed] [Google Scholar]
- 25.Lukasova M, Malaval C, Gille A et al. Nicotinic acid inhibits progression of atherosclerosis in mice through its receptor GPR109A expressed by immune cells. J Clin Invest 2011; 121: 1163–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pluznick J A novel SCFA receptor, the microbiota, and blood pressure regulation. Gut microbes 2014; 5: 202–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Waldecker M, Kautenburger T, Daumann H et al. Inhibition of histone-deacetylase activity by short-chain fatty acids and some polyphenol metabolites formed in the colon. The Journal of nutritional biochemistry 2008; 19: 587–593 [DOI] [PubMed] [Google Scholar]
- 28.Yanda MK, Liu Q, Cebotaru V et al. Histone deacetylase 6 inhibition reduces cysts by decreasing cAMP and Ca(2+) in knock-out mouse models of polycystic kidney disease. The Journal of biological chemistry 2017; 292: 17897–17908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schilderink R, Verseijden C, Seppen J et al. The SCFA butyrate stimulates the epithelial production of retinoic acid via inhibition of epithelial HDAC. Am J Physiol Gastrointest Liver Physiol 2016; 310: G1138–1146 [DOI] [PubMed] [Google Scholar]
- 30.Andrade-Oliveira V, Amano MT, Correa-Costa M et al. Gut Bacteria Products Prevent AKI Induced by Ischemia-Reperfusion. Journal of the American Society of Nephrology : JASN 2015; 26: 1877–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Al-Harbi NO, Nadeem A, Ahmad SF et al. Short chain fatty acid, acetate ameliorates sepsis-induced acute kidney injury by inhibition of NADPH oxidase signaling in T cells. International immunopharmacology 2018; 58: 24–31 [DOI] [PubMed] [Google Scholar]
- 32.Obeid R, Awwad HM, Rabagny Y et al. Plasma trimethylamine N-oxide concentration is associated with choline, phospholipids, and methyl metabolism. The American journal of clinical nutrition 2016; 103: 703–711 [DOI] [PubMed] [Google Scholar]
- 33.Sun G, Yin Z, Liu N et al. Gut microbial metabolite TMAO contributes to renal dysfunction in a mouse model of diet-induced obesity. Biochemical and biophysical research communications 2017; 493: 964–970 [DOI] [PubMed] [Google Scholar]
- 34.Wilson Tang WH, Wang Z, Kennedy DJ et al. Gut Microbiota-Dependent Trimethylamine N-oxide (TMAO) Pathway Contributes to Both Development of Renal Insufficiency and Mortality Risk in Chronic Kidney Disease. Circulation research 2015; 116: 448–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tomlinson JAP, Wheeler DC. The role of trimethylamine N-oxide as a mediator of cardiovascular complications in chronic kidney disease. Kidney international 2017; 92: 809–815 [DOI] [PubMed] [Google Scholar]
- 36.Mafune A, Iwamoto T, Tsutsumi Y et al. Associations among serum trimethylamine-N-oxide (TMAO) levels, kidney function and infarcted coronary artery number in patients undergoing cardiovascular surgery: a cross-sectional study. Clin Exp Nephrol 2016; 20: 731–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shafi T, Powe NR, Meyer TW et al. Trimethylamine N-Oxide and Cardiovascular Events in Hemodialysis Patients. Journal of the American Society of Nephrology : JASN 2017; 28: 321–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hall JA, Bouladoux N, Sun CM et al. Commensal DNA limits regulatory T cell conversion and is a natural adjuvant of intestinal immune responses. Immunity 2008; 29: 637–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu B, Tonkonogy SL, Sartor RB. Antigen-presenting cell production of IL-10 inhibits T-helper 1 and 17 cell responses and suppresses colitis in mice. Gastroenterology 2011; 141: 653–662, 662.e651–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cebula A, Seweryn M, Rempala GA et al. Thymus-derived regulatory T cells contribute to tolerance to commensal microbiota. Nature 2013; 497: 258–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu M, Pokrovskii M, Ding Y et al. c-MAF-dependent regulatory T cells mediate immunological tolerance to a gut pathobiont. Nature 2018; 554: 373–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim KS, Hong SW, Han D et al. Dietary antigens limit mucosal immunity by inducing regulatory T cells in the small intestine. Science (New York, NY) 2016; 351: 858–863 [DOI] [PubMed] [Google Scholar]
- 43.Barnes MJ, Powrie F. Regulatory T cells reinforce intestinal homeostasis. Immunity 2009; 31: 401–411 [DOI] [PubMed] [Google Scholar]
- 44.Qiu J, Guo X, Chen ZM et al. Group 3 innate lymphoid cells inhibit T-cell-mediated intestinal inflammation through aryl hydrocarbon receptor signaling and regulation of microflora. Immunity 2013; 39: 386–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hooper LV, Macpherson AJ. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nature reviews Immunology 2010; 10: 159–169 [DOI] [PubMed] [Google Scholar]
- 46.Strugnell RA, Wijburg OL. The role of secretory antibodies in infection immunity. Nature reviews Microbiology 2010; 8: 656–667 [DOI] [PubMed] [Google Scholar]
- 47.Bunker JJ, Erickson SA, Flynn TM et al. Natural polyreactive IgA antibodies coat the intestinal microbiota. Science (New York, NY) 2017; 358: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bunker JJ, Flynn TM, Koval JC et al. Innate and Adaptive Humoral Responses Coat Distinct Commensal Bacteria with Immunoglobulin A. Immunity 2015; 43: 541–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Russler-Germain EV, Rengarajan S, Hsieh CS. Antigen-specific regulatory T-cell responses to intestinal microbiota. Mucosal Immunol 2017; 10: 1375–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alexander KL, Katz J, Elson CO. CBirTox is a selective antigen-specific agonist of the Treg-IgA-microbiota homeostatic pathway. PloS one 2017; 12: e0181866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kitching AR, Holdsworth SR. The emergence of TH17 cells as effectors of renal injury. Journal of the American Society of Nephrology : JASN 2011; 22: 235–238 [DOI] [PubMed] [Google Scholar]
- 52.Krebs CF, Paust HJ, Krohn S et al. Autoimmune Renal Disease Is Exacerbated by S1P-Receptor-1-Dependent Intestinal Th17 Cell Migration to the Kidney. Immunity 2016; 45: 1078–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Krebs CF, Kapffer S, Paust HJ et al. MicroRNA-155 drives TH17 immune response and tissue injury in experimental crescentic GN. Journal of the American Society of Nephrology : JASN 2013; 24: 1955–1965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Velden J, Paust HJ, Hoxha E et al. Renal IL-17 expression in human ANCA-associated glomerulonephritis. Am J Physiol Renal Physiol 2012; 302: F1663–1673 [DOI] [PubMed] [Google Scholar]
- 55.Lopez P, de Paz B, Rodriguez-Carrio J et al. Th17 responses and natural IgM antibodies are related to gut microbiota composition in systemic lupus erythematosus patients. Scientific reports 2016; 6: 24072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Noel S, Martina-Lingua MN, Bandapalle S et al. Intestinal microbiota-kidney cross talk in acute kidney injury and chronic kidney disease. Nephron Clin Pract 2014; 127: 139–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rabb H, Griffin MD, McKay DB et al. Inflammation in AKI: Current Understanding, Key Questions, and Knowledge Gaps. Journal of the American Society of Nephrology : JASN 2016; 27: 371–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pluznick JL, Protzko RJ, Gevorgyan H et al. Olfactory receptor responding to gut microbiota-derived signals plays a role in renin secretion and blood pressure regulation. Proceedings of the National Academy of Sciences of the United States of America 2013; 110: 4410–4415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mielenz M, Kuhla B, Hammon HM. Abundance of adiponectin system and G-protein coupled receptor GPR109A mRNA in adipose tissue and liver of F2 offspring cows of Charolais x German Holstein crosses that differ in body fat accumulation. Journal of dairy science 2013; 96: 278–289 [DOI] [PubMed] [Google Scholar]
- 60.Machado RA, Constantino Lde S, Tomasi CD et al. Sodium butyrate decreases the activation of NF-kappaB reducing inflammation and oxidative damage in the kidney of rats subjected to contrast-induced nephropathy. Nephrology, dialysis, transplantation : official publication of the European Dialysis and Transplant Association - European Renal Association 2012; 27: 3136–3140 [DOI] [PubMed] [Google Scholar]
- 61.Willyard C Squeaky clean mice could be ruining research. Nature 2018; 556: 16–18 [DOI] [PubMed] [Google Scholar]
- 62.Jang HR, Gandolfo MT, Ko GJ et al. Early exposure to germs modifies kidney damage and inflammation after experimental ischemia-reperfusion injury. Am J Physiol Renal Physiol 2009; 297: F1457–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Emal D, Rampanelli E, Stroo I et al. Depletion of Gut Microbiota Protects against Renal Ischemia-Reperfusion Injury. Journal of the American Society of Nephrology : JASN 2017; 28: 1450–1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kalambokis GN, Mouzaki A, Rodi M et al. Rifaximin improves systemic hemodynamics and renal function in patients with alcohol-related cirrhosis and ascites. Clinical gastroenterology and hepatology : the official clinical practice journal of the American Gastroenterological Association 2012; 10: 815–818 [DOI] [PubMed] [Google Scholar]
- 65.Dong T, Aronsohn A, Gautham Reddy K et al. Rifaximin Decreases the Incidence and Severity of Acute Kidney Injury and Hepatorenal Syndrome in Cirrhosis. Digestive diseases and sciences 2016; 61: 3621–3626 [DOI] [PubMed] [Google Scholar]
- 66.Satoh M, Hayashi H, Watanabe M et al. Uremic toxins overload accelerates renal damage in a rat model of chronic renal failure. Nephron Exp Nephrol 2003; 95: e111–118 [DOI] [PubMed] [Google Scholar]
- 67.Bammens B. Urea and uremic solutes: how does peritoneal dialysis work? Seminars in nephrology 2011; 31: 127–137 [DOI] [PubMed] [Google Scholar]
- 68.Poesen R, Windey K, Neven E et al. The Influence of CKD on Colonic Microbial Metabolism. Journal of the American Society of Nephrology : JASN 2016; 27: 1389–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stubbs JR, House JA, Ocque AJ et al. Serum Trimethylamine-N-Oxide is Elevated in CKD and Correlates with Coronary Atherosclerosis Burden. Journal of the American Society of Nephrology : JASN 2016; 27: 305–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vaziri ND, Wong J, Pahl M et al. Chronic kidney disease alters intestinal microbial flora. Kidney international 2013; 83: 308–315 [DOI] [PubMed] [Google Scholar]
- 71.Karbach SH, Schonfelder T, Brandao I et al. Gut Microbiota Promote Angiotensin II-Induced Arterial Hypertension and Vascular Dysfunction. Journal of the American Heart Association 2016; 5: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hu J, Luo H, Wang J et al. Enteric dysbiosis-linked gut barrier disruption triggers early renal injury induced by chronic high salt feeding in mice. Exp Mol Med 2017; 49: e370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Weber GJ, Pushpakumar S, Tyagi SC et al. Homocysteine and hydrogen sulfide in epigenetic, metabolic and microbiota related renovascular hypertension. Pharmacol Res 2016; 113: 300–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yacoub R, Wyatt CM. Manipulating the gut microbiome to decrease uremic toxins. Kidney international 2017; 91: 521–523 [DOI] [PubMed] [Google Scholar]
- 75.Schulman G, Berl T, Beck GJ et al. Randomized Placebo-Controlled EPPIC Trials of AST-120 in CKD. Journal of the American Society of Nephrology : JASN 2015; 26: 1732–1746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Karczewski J, Troost FJ, Konings I et al. Regulation of human epithelial tight junction proteins by Lactobacillus plantarum in vivo and protective effects on the epithelial barrier. Am J Physiol Gastrointest Liver Physiol 2010; 298: G851–859 [DOI] [PubMed] [Google Scholar]
- 77.Rossi M, Johnson DW, Morrison M et al. SYNbiotics Easing Renal failure by improving Gut microbiologY (SYNERGY): a protocol of placebo-controlled randomised cross-over trial. BMC nephrology 2014; 15: 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ranganathan N, Friedman EA, Tam P et al. Probiotic dietary supplementation in patients with stage 3 and 4 chronic kidney disease: a 6-month pilot scale trial in Canada. Current medical research and opinion 2009; 25: 1919–1930 [DOI] [PubMed] [Google Scholar]
- 79.Ranganathan N, Ranganathan P, Friedman EA et al. Pilot study of probiotic dietary supplementation for promoting healthy kidney function in patients with chronic kidney disease. Advances in therapy 2010; 27: 634–647 [DOI] [PubMed] [Google Scholar]
- 80.Soleimani A, Zarrati Mojarrad M, Bahmani F et al. Probiotic supplementation in diabetic hemodialysis patients has beneficial metabolic effects. Kidney international 2017; 91: 435–442 [DOI] [PubMed] [Google Scholar]
- 81.Kaushal D, Kansal VK. Probiotic Dahi containing Lactobacillus acidophilus and Bifidobacterium bifidum alleviates age-inflicted oxidative stress and improves expression of biomarkers of ageing in mice. Molecular biology reports 2012; 39: 1791–1799 [DOI] [PubMed] [Google Scholar]
- 82.Haro C, Monaco ME, Medina M. Lactobacillus casei beneficially modulates immuno-coagulative response in an endotoxemia model. Blood coagulation & fibrinolysis : an international journal in haemostasis and thrombosis 2018; 29: 104–110 [DOI] [PubMed] [Google Scholar]
- 83.Mu Q, Zhang H, Liao X et al. Control of lupus nephritis by changes of gut microbiota. Microbiome 2017; 5: 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Klimesova K, Whittamore JM, Hatch M. Bifidobacterium animalis subsp. lactis decreases urinary oxalate excretion in a mouse model of primary hyperoxaluria. Urolithiasis 2015; 43: 107–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cosola C, De Angelis M, Rocchetti MT et al. Beta-Glucans Supplementation Associates with Reduction in P-Cresyl Sulfate Levels and Improved Endothelial Vascular Reactivity in Healthy Individuals. PloS one 2017; 12: e0169635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wanchai K, Yasom S, Tunapong W et al. Prebiotic prevents impaired kidney and renal Oat3 functions in obese rats. The Journal of endocrinology 2018; 237: 29–42 [DOI] [PubMed] [Google Scholar]
- 87.Sofen H, Smith S, Matheson RT et al. Guselkumab (an IL-23-specific mAb) demonstrates clinical and molecular response in patients with moderate-to-severe psoriasis. J Allergy Clin Immunol 2014; 133: 1032–1040 [DOI] [PubMed] [Google Scholar]
- 88.Jauregui-Amezaga A, Somers M, De Schepper H et al. Next generation of biologics for the treatment of Crohn’s disease: an evidence-based review on ustekinumab. Clinical and experimental gastroenterology 2017; 10: 293–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Patel DD, Lee DM, Kolbinger F et al. Effect of IL-17A blockade with secukinumab in autoimmune diseases. Ann Rheum Dis 2013; 72 Suppl 2: ii116–123 [DOI] [PubMed] [Google Scholar]
- 90.Leonardi C, Matheson R, Zachariae C et al. Anti-interleukin-17 monoclonal antibody ixekizumab in chronic plaque psoriasis. The New England journal of medicine 2012; 366: 1190–1199 [DOI] [PubMed] [Google Scholar]
- 91.Baker KF, Isaacs JD. Novel therapies for immune-mediated inflammatory diseases: What can we learn from their use in rheumatoid arthritis, spondyloarthritis, systemic lupus erythematosus, psoriasis, Crohn’s disease and ulcerative colitis? Ann Rheum Dis 2018; 77: 175–187 [DOI] [PubMed] [Google Scholar]
- 92.Chen L, Liu W, Li Y et al. Lactobacillus acidophilus ATCC 4356 attenuates the atherosclerotic progression through modulation of oxidative stress and inflammatory process. International immunopharmacology 2013; 17: 108–115 [DOI] [PubMed] [Google Scholar]
- 93.Kawamoto S, Tran TH, Maruya M et al. The inhibitory receptor PD-1 regulates IgA selection and bacterial composition in the gut. Science (New York, NY) 2012; 336: 485–489 [DOI] [PubMed] [Google Scholar]
- 94.Wang Y, Ma R, Liu F et al. Modulation of Gut Microbiota: A Novel Paradigm of Enhancing the Efficacy of Programmed Death-1 and Programmed Death Ligand-1 Blockade Therapy. Frontiers in immunology 2018; 9: 374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cohen LJ, Han S, Huang YH et al. Identification of the Colicin V Bacteriocin Gene Cluster by Functional Screening of a Human Microbiome Metagenomic Library. ACS infectious diseases 2018; 4: 27–32 [DOI] [PMC free article] [PubMed] [Google Scholar]