Abstract
Spermatozoon decondensation in the zygote leads to the initiation of chromatin remodeling during which protamines are removed and replaced with maternal histones. We hypothesize that damage to male germ cells induced by paternal exposure to cyclophosphamide may alter the timing of spermatozoal decondensation and the pattern of chromatin remodeling in the prepronuclear rat zygote. A specific order of sperm decondensation was observed, starting at the posterior end, proceeding to the ventral sides, followed by the tip, and finally the midbody region of the sperm head nucleus; subgroups of partially decondensed type a sperm nuclei were defined as types a1, a2, a3, and a4. Based on their frequencies relative to controls, paternal exposure to cyclophosphamide accelerated the timing of spermatozoal decondensation. Two distinct patterns of chromatin remodeling were observed for totally decondensed (type b) and recondensing (type c) sperm nuclei: H4K12ac showed a homogenous staining, whereas H3S10ph displayed a ring-like staining around the sperm nucleus; the distribution of these posttranslationally modified histones was not affected by cyclophosphamide exposure. In contrast, paternal cyclophosphamide treatment increased the number of gammaH2AX foci found in decondensing sperm nuclei. Small foci were significantly increased in type a2 and a3 nuclei, whereas a significant increase in the numbers of large foci was found in type b and c nuclei. This increase in gammaH2AX foci in the decondensing male genome suggests that damage recognition and repair pathways are initiated in prepronuclear rat zygotes. Thus, exposure of male rats to chronic low doses of cyclophosphamide accelerates spermatozoal decondensation and leads to the activation of gammaH2AX recognition of DNA damage in the male genome of the prepronuclear zygote.
Keywords: chromatin remodeling, cyclophosphamide, embryo, epigenetic marks, posttranslationally modified histones, prepronuclear zygote, sperm, sperm decondensation, toxicology
Introduction
The entry of the spermatozoon triggers the mature oocyte arrested at metaphase II to complete its second meiotic division. Once activated, stored oocyte proteins will block polyspermy and initiate major chromatin reorganization and reprogramming events in both the maternal and paternal genomes during an early window of embryonic development, defined as the zygotic prepronuclear (pre-PN) formation [1].
Chromatin decondensation of spermatozoa is a multistep process, starting with the induction of calcium influx by gamete membrane fusion and the resumption of meiosis by maternally activated protein kinase C [2–3] and the reduction of interprotamine sulfhydryl bonds by glutathione (GSH) [4]. In pre-PN zygotes, there are four stages of mouse sperm chromatin decondensation: condensed, partially decondensed (type a), totally decondensed (type b), and recondensing (type c); nuclei always decondense in the same order, from the posterior end, to the ventral sides, and then the tip of the sperm nucleus [5]. Chromatin remodeling is initiated when the spermatozoon chromatin begins to decondense; the male germ cell chromatin is packaged into a somatic cell chromatin by replacing sperm-specific protamines with somatic histones. Maternally provided cysteine protease degrades sperm-specific protamines, leaving the maternal somatic histones unaffected, while histone cell cycle regulation defective homolog A and nucleoplasmin chaperone proteins assemble nucleosomes, remodeling the paternal chromatin [6, 7]. Once this reprogramming is complete, the paternal chromatin recondenses and subsequently expands into the male pronucleus.
During sperm chromatin remodeling, the two parental sets of chromosomes exhibit striking epigenetic differences. The paternal chromosomal histones are mostly acetylated and devoid of methylation marks, while the maternal chromosomal histones are mostly methylated and lack acetylation marks [5, 8, 9]. Changes in histone modifications, induced by environmental and developmental cues, may play a role in regulating gene expression during early embryo development. The presence and localization of histones H4K12ac and H3S10ph were visualized in pre-PN zygotes and evaluated as indicators of the progression of chromatin remodeling in sperm from control and cyclophosphamide (CPA)-exposed rats. H4K12ac is one of the first maternally modified histones to be incorporated on the decondensing paternal chromatin [1], marking an open and active chromatin structure to support early embryonic events following fertilization [10]. H3S10ph marks (peri)centromeric heterochromatin important for homologous chromosome segregation during mouse oocyte maturation [11], the remodeling of paternal chromatin [1], and mitosis [12] in postfertilized oocytes. H3S10ph can be a marker of relaxed chromatin during transcription [13], or a marker of condensed chromatin during mitosis [14] and meiosis [11]. Thus, during pre-PN events, the epigenetic marks of the remodeling paternal chromatin are highly dynamic, potentially influencing the zygote as it progresses through the different sperm decondensation stages.
Histone modifications may also serve to mark DNA damage and recruit maternally stored DNA repair proteins following fertilization [15–17]. A histone H2A variant protein, H2AX (official symbol, H2AFX), is phosphorylated at serine 139 by phosphoinositide 3-kinase-related kinases to form γH2AX, which accumulates in foci to mark DNA double-strand breaks [18–20]. There are two types of γH2AX foci: a large population of small foci is important for the assembly of embryonic chromatin and mitotic cell cycle regulation, while a small population of large foci is indicative of the recruitment of DNA repair proteins [21–23]. In a previous study [24], we observed a striking increase in γH2AX focal numbers and volume in remodeled pronuclear-stage embryos in response to paternal CPA exposure, in support of a role for this modified histone in the processing of the DNA lesions by recruiting and maintaining repair factors in the vicinity of the damage during the first cell division in the zygote.
CPA (Cytoxan) is a chemotherapeutic [25, 26] and immunosuppressive agent used mainly in the treatment of breast cancer and non-Hodgkin disease. CPA is an alkylating agent, inducing DNA-DNA and DNA-protein cross-links, and single-strand breaks in rapidly dividing cells [26, 27]. Paternal exposure to CPA targets spermatogenesis in a dose- and time-specific manner [28]. The step of spermatogenesis most sensitive to CPA exposure is during spermiogenesis, at a time when most of the somatic histones are being replaced with sperm nuclear basic proteins, protamines, and histone variants [29–31].
The objectives of this study were to determine if paternal exposure to CPA impacts events during the development of early pre-PN zygotes. Since sperm DNA integrity and chromatin packaging are essential for the accurate and successful transmission of genetic information to progeny, we hypothesize that CPA-related alterations in sperm chromatin structure may affect the progression of sperm chromatin decondensation and remodeling after fertilization. If so, this could result in dysregulation of paternal and maternal gene expression, and, thereby, potentially account for the previously observed impaired pregnancy outcomes [28].
Materials and Methods
Drug Treatment and Collection of Rat Pre-PN Zygotes Generated In Vivo
Adult male (body weight, 350–400 g) and virgin female (body weight, 225–250 g) Sprague-Dawley rats were purchased from Charles River Canada (St. Constant, QC, Canada) and housed at the Animal Resources Centre, McIntyre Medical Building, McGill University (Montreal, QC, Canada). Animals received food and water ad libitum, and were maintained on a 0700–1900 h light/dark cycle. The drug treatment and zygote protocols previously described [32] were followed with minor modifications. After 1 wk of acclimatization, male rats were randomly assigned to one of two treatment groups (n = 11 per group) and gavaged with saline (vehicle) or CPA (CAS 6055-19-2; Sigma Chemical Co., St. Louis, MO), 6 mg/kg per day, six times per week for 4 wk [33, 34].
On the fifth week of treatment, control virgin females in proestrus were selected by a vaginal wash in the midafternoon, and were caged overnight in groups of two, with either a control or CPA-treated male. Pregnancies were confirmed with sperm-positive vaginal smears the following morning, designated as Gestation Day 0. Sperm-positive females were euthanized at 0900 h on Day 0 to collect pre-PN-stage zygotes. Since fertilization occurred after in vivo mating, the exact timing was not known. Thus, the progression of spermatozoal decondensation was assessed by analyzing the distribution of the stages of spermatozoa decondensation observed in the population of early pre-PN zygotes captured at one time point. Oviducts and proximal uteri were isolated and cleaned in prewarmed (37°) M2 culture medium (Sigma Chemical Co.). Zygotes were released from the ampullae into a drop of prewarmed (37°C) 1% hyaluronidase (Sigma Chemical Co.) in M2 medium to digest the cumulus cells. All animal protocols were carried out according to the guidelines of the Canadian Council on Animal Care, and were approved by the institutional animal care committee.
Immunofluorescence
The indirect immunofluorescence protocols used were described previously [18]. Zygotes fertilized by saline and CPA-exposed males were stained in parallel at room temperature unless otherwise stated. Briefly, zygotes were washed in 1× PBS (pH 7.4; Mg2+ and Ca2+ free), containing 1 mg/ml polyvinylpyrrolidone. Zona pellucidae were removed in prewarmed (37°C) acid Tyrode solution (pH 2.5) by pipetting up and down for 5 sec, and then washed in 1× PBS (pH 7.4; Mg2+ and Ca2+ free), containing 1 mg/ml polyvinylpyrrolidone. Zygotes were then fixed in 4% paraformaldehyde in Ca2+-/Mg2+-free PBS for 15 min, washed in 0.05% Tween 20 in PBS for 5 min, permeabilized in 0.2% Triton X-100 in PBS for 30 min, rewashed in 0.05% Tween 20 in PBS for 5 min, and blocked for 4–5 h in 10% goat serum, 3% BSA, and 0.1% Tween 20 in PBS.
To visualize chromatin remodeling of the paternal genome, we incubated the zygotes in rabbit polyclonal IgG anti-acetyl-histone H4 (Lys12) [1, 10] (1:200 dilution; catalog no. 06761; Upstate Cell Signaling Solutions, Billerica, MA) and rabbit polyclonal IgG anti-phosphorylated-histone H3 (S10ph) [1, 35] (1:200 dilution; catalog no. 06–570; Upstate Cell Signaling Solutions) overnight at 4°C in a humidified chamber. To visualize sites of DNA damage recognition and repair, we incubated the zygotes with mouse monoclonal IgG anti-γH2AX phospho-Serine-139 [20] (1:500 dilution; catalog no. 05–636, clone JBW103; Upstate Biotechnology, Charlottesville, VA) overnight at 4°C in a humidified chamber. Both primary and secondary antibodies were diluted in goat blocking solution (10% goat serum, 3% BSA, and 0.1% Tween 20 in PBS). Zygotes were then washed three times for 20 min in goat blocking solution, incubated for 1 h in goat fluorescein anti-rabbit IgG (H + L) (1:200 dilution; catalog no. F1–1000; Vector Laboratories, Burlington, ON, Canada) for H4K12ac and H3S10ph, and for 1 h in sheep fluorescein anti-mouse IgG (H + L) (1:200 dilution; catalog no. N1031V; Amersham Pharmacia Biosciences, Baie d'Urfe, QC, Canada) for γH2AX, and rewashed three times for 20 min in goat blocking solution. DNA was stained with propidium iodide (catalog no. P4864; Sigma Chemical Co.) at 10 μg/ml in goat blocking solution for 20 min, washed in 0.05% Tween 20 in PBS for 10 min, mounted in 3 μl of VectaShield mounting medium (Vector Laboratories) on a premarked pap pen slide, and covered with a cover slip. Slides were then stored at 4°C and visualized by confocal microcopy during the next 2 days. A total of 11 experimental replicates were done for each histone H4K12ac and H3S10ph marker, while 6 experimental replicates were done for the γH2AX study.
Confocal Microscopy
A Zeiss LSM 510 Axiovert 100M confocal microscope with a Plan-Apochromat ×63/1.4 oil differential interference contrast objective was used to visualize the fluorescence of early postfertilized zygotes. The best settings for laser scanning fluorescence imaging were determined experimentally for both primary H4K12ac and H3S10ph antibodies. All zygotes stained for H4K12ac and H3S10ph were scanned at a speed of 5, with an optical slice of 0.6 μm, zoom factor equal to 1, and a pinhole setting of 96 μm. Two scans of each optical section were compiled and averaged by the Zeiss LSM 510 computer software to give a final image that was 1024 × 1024 pixels in size. Due to significant variation in the H4K12ac signal intensity over the different paternal chromatin compaction levels, images of early pre-PN zygotes at the condensed and type a sperm nuclear decondensation stages were taken with a detector gain setting of 979, whereas the setting was 949 for type b and c sperm nuclei. Images of all zygotes stained with anti-H3S10ph were taken with a detector gain setting of 740. Images from Z-stacks were further analyzed and quantified using the Profile Program on the confocal microscope.
All zygotes stained for γH2AX were scanned at a speed of 6, with optical slice of <0.7μm, keeping the other settings unchanged. For qualitative purposes only, all γH2AX zygote images were taken at the optimized detector gain set for both the maternal nucleus and polar body. Images from Z-stacks were further reconstituted in three-dimensional images, and focal numbers and sizes were quantified using the overlay option on the confocal microscope.
Quantitative Analysis
H4K12ac and H3S10ph
Polyspermic zygotes (18 from the saline group, 14 from the CPA treatment group) were excluded from this analysis, leaving a total of 298 saline-sired zygotes and 300 CPA-sired zygotes. Quantitative analysis for all nonpolyspermic zygotes was done by drawing a single line, starting at the posterior end and proceeding to the anterior end of the sperm nucleus, across the longitudinal section of the sperm. The average fluorescence intensity was calculated from the middle two to three optical images across the longitudinal sperm section for each 10% of the sperm length. The number of zygotes analyzed for each sperm decondensation stage varied from 10 to 75 for each treatment group. The classification of pre-PN-stage zygotes as condensed, partially decondensed (type a), totally decondensed (type b), or recondensing (type c) sperm nuclei was described previously [5].
Representative staining patterns of the sperm chromatin remodeling in control and CPA-treated groups are shown above the graphs for each sperm decondensation stage. Each graph represents the average staining intensity of modified histones over 10% segments of the longitudinal section of the sperm nucleus, starting at the posterior end and proceeding to the tip region. The signal intensity was compared between adjacent sperm segments within the same treatment group and within the same sperm segment between treatment groups. The signal intensity was also qualitatively compared between sperm decondensation stages measured with the same detection settings.
γH2AX
Qualitative analysis of γH2AX foci number and size was done using a three-dimensional reconstitution of each zygote from the Z-stack sections of all individual zygotes. We then used the overlay option to measure the diameter of all foci and count the total number of foci in each size category: 0.01–0.79 or 0.80–2.5 μm in diameter. Unfertilized oocytes (54 saline, 44 CPA), polyspermic zygotes (1 saline, 9 CPA), and pronuclear zygotes (3 saline, 5 CPA) were excluded from this analysis, leaving a total of 79 zygotes sired by saline-treated males and 64 zygotes sired by CPA-treated males. The number of zygotes analyzed for each sperm decondensation stage varied from 2 to 30 for each treatment group.
Statistical Analyses
Chi-square analysis (Systat Version 10.2) and the Fisher exact test, with Bonferroni correction, were used to compare the progression of early zygotic development for all sperm decondensation stages observed in both treatment groups. Unbalanced two-way ANOVA with repeated measures and a logarithmic transformation of the data were done to detect any distance effect along the length of the sperm nucleus, any CPA drug effect, and any interaction between distance and treatment on chromatin remodeling with H4K12ac and H3S10ph for all sperm decondensation stages. Logarithmic transformation of the intensity was done because of the large variation in SEM. A post hoc contrast test for pair-wise comparison was done to confirm and complement the two-way ANOVA analysis by specifically identifying where along the sperm length segment chromatin remodeling with H4K12ac and H3S10ph differed between treatment groups and among specific adjacent sperm length segments within the same treatment group. These analyses were done with the SAS statistical analysis program (version 9.1). Comparison of the averaged intensity of DNA with the intensity values of H4K12ac is qualitative, since different dyes, propidium iodide, and FITC were used.
The Mann-Whitney U-test was used to compare the numbers of γH2AX foci between the control and CPA-treated groups for each individual sperm decondensation stage. This statistical analysis was done for both small (0.01–0.79 μm in diameter) and large (0.79–2.5 μm in diameter) foci. Both condensed and type a4 sperm nuclei were excluded from the analysis, since condensed sperm nuclei never had foci, and we did not obtain sufficient type a4 sperm nuclei for analysis due to the rapid rate of sperm decondensation at this stage. We did not compare stages for the same treatment group, since the data were dependent and zygotes were not obtained at all stages from at least three different males.
Results
Subclassification of Type a Sperm Nuclei in the Rat Zygote
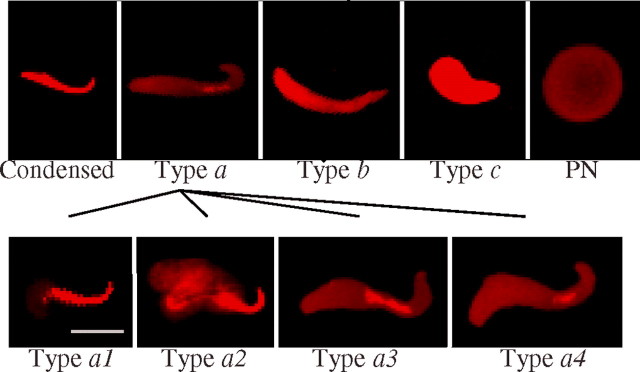
Following fertilization, sperm chromatin decondensation was divided into four stages based on the extent of chromatin compaction, as distinguished by the propidium iodide staining intensity. These sperm decondensation stages are: condensed sperm nuclei, type a partially decondensed sperm nuclei, type b totally decondensed sperm nuclei, and type c recondensing sperm nuclei. These stages are then followed by pronuclear formation (Fig. 1, top panel) [5]. Since we observed previously uncharacterized partially decondensed sperm nuclear patterns, type a sperm nuclei were further subdivided into four subgroups: 1) type a1, the sperm nucleus was starting to decondense at the posterior end; 2) type a2, half of the sperm nucleus was decondensed at the posterior end, and decondensation had progressed to a small extent on both ventral sides; 3) type a3, the posterior, ventral sides, and the tip of the sperm nucleus were decondensed; and 4) type a4, almost all of the sperm nucleus was decondensed, with the exception of a small condensed region in the midbody area (Fig. 1, bottom panel). This subclassification of type a sperm nuclei in the rat model facilitated our analysis of the sperm chromatin remodeling patterns described later, and highlights the unique sequential order of sperm decondensation in this species.
Fig. 1.
Types of paternal nuclei observed in early postfertilization zygotes. The DNA was stained with propidium iodide (red). Top panel) Condensed, type a, type b, type c and pronuclear sperm (PN) nuclei. Bottom panel) Subclassification of type a sperm nuclei: type a1, type a2, type a3, and type a4. Pictures were taken by confocal microscopy. Bar = 10 μm.
Paternal Exposure to CPA Alters the Progression of Sperm Chromatin Decondensation in Early Zygotes
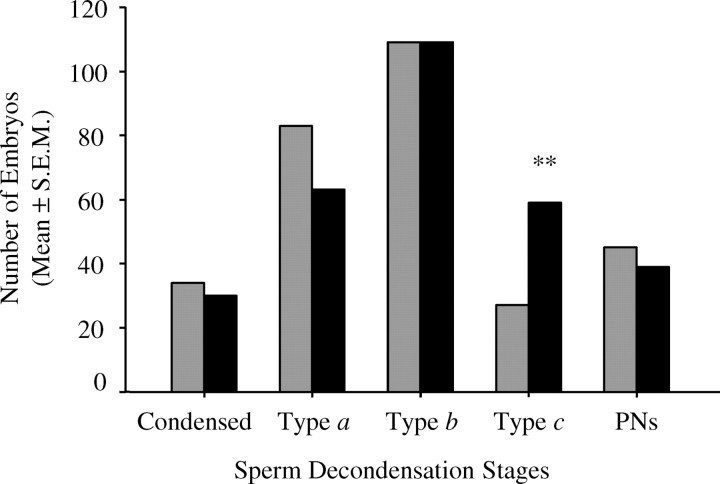
Treatment of male rats with CPA did not affect the fertilizing capacity of their spermatozoa, since the proportion of unfertilized to fertilized oocytes among females mated to saline- or CPA-treated males did not differ (saline, 92/316; CPA, 100/314). Since in vitro fertilization is not a commonly accessible technique in the rat, we collected zygotes at one time point and reported our data as the total number at each sperm nuclear stage at this time. The distribution of zygotes at the time of collection formed a bell-shaped curve, where most zygotes were found in the type b sperm nuclei decondensation stage (Fig. 2). Paternal exposure to CPA disturbed the distribution of these early zygotes by significantly increasing the number of type c sperm nuclei in the CPA group compared with controls (see type c sperm nuclei in Fig. 2) (P < 0.001).
Fig. 2.
The distribution of types of paternal nuclei in early pre-PN zygotes. The progression of early zygotes is accelerated following chronic paternal CPA treatment to type c sperm nuclei (n = 20 males for saline; n = 20 males for CPA). The gray bars represent the zygotes sired by sperm from saline-treated males, and the black bars represent zygotes sired by CPA-exposed sperm. Number of zygotes within each type of paternal nuclei: saline, 34, 83, 109, 27, 45; and CPA, 30, 63, 109, 59, 39 for condensed, type a, type b, type c, and PNs, respectively. Statistical analysis performed: χ2 analyses, Fisher exact test, with Bonferroni correction; **P < 0.001.
H4K12ac Sperm Chromatin Remodeling Pattern in Pre-PN Zygotes
The signal intensity of histone H4K12ac on the decondensing and remodeling sperm chromatin was quantified for both saline- and CPA-treated groups in all sperm decondensation stages (Fig. 3). In zygotes sired by saline-treated males, no visible staining with H4K12ac was seen across the whole sperm length in condensed sperm nuclei (Fig. 3a). A faint but visible H4K12ac mark initially appeared in type a1 sperm nuclei on the posterior end of the decondensing sperm chromatin, and gradually decreased from the midbody region to the tip of the sperm nucleus (Fig. 3b). In type a2 sperm nuclei, half of the sperm length had bright staining, with H4K12ac at the posterior end, and increased intensity to a small extent at the midbody and the tip of the sperm nucleus (Fig. 3c). In type a3 sperm nuclei, the posterior end, both ventral sides and tip showed positive H4K12ac staining, excluding the midbody region of the sperm head nucleus (Fig. 3d). The pattern of histone H4K12ac staining in type a4 sperm nuclei was very similar to type a3, except that the midbody of the sperm nucleus had started to decondense and stained with H4K12ac (Fig. 3e).
Fig. 3.
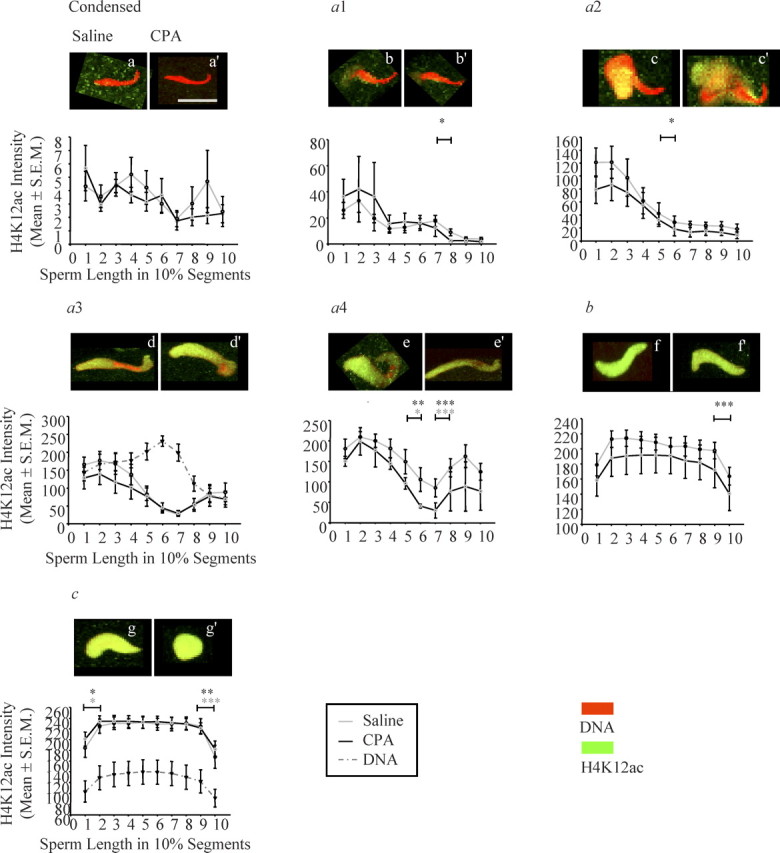
Quantitative analysis of the initial appearance, progression, and pattern of H4K12ac staining intensity on the paternal chromatin for all sperm decondensation stages of pre-PN zygotes. The sperm decondensation stage is on the top of each graph; below each stage, there is the best representative picture of saline-exposed sperm on the left (a) and CPA-exposed sperm on the right (a′) for each stage. DNA was stained with propidium iodide (red) and H4K12ac with FITC (green). The y axes represent H4K12ac intensity (mean ± SEM), and the x axes represent sperm length in 10% segments; error bars are the SEM. Light gray line, saline group; black line, CPA group; dashed dark gray line, DNA content. For each individual type of paternal nuclei, the staining intensity of histone H4K12ac differed significantly across the longitudinal section of the sperm length for almost all sperm decondensation stages with the exception of condensed sperm nuclei. CPA did not affect histone H4K12ac staining intensity and patterning in any of the sperm decondensation stages. Number of pre-PN zygotes analyzed in each sperm decondensation stage: saline, (a) n = 6, 11 zygotes, (b) n = 6, 12 zygotes, (c) n = 6, 8 zygotes, (d) n = 9, 18 zygotes, (e) n = 7, 10 zygotes, (f) n = 8, 39 zygotes, (g) n = 6, 10 zygotes; and CPA, (a′) n = 6, 15 zygotes, (b′) n = 5, 7 zygotes, (c′) n = 4, 7 zygotes, (d′) n = 8, 13 zygotes, (e′) n = 2, 2 zygotes, (f′) n = 9, 34 zygotes, (g′) n = 8, 26 zygotes for sperm decondensation stages: condensed, type a1, type a2, type a3, type a4, type b, and type c sperm nuclei, respectively. Statistical analyses were done using an unbalanced two-way ANOVA with repeated measures and a logarithmic transformation of the intensity followed by post hoc contrast tests with the SAS program. The asterisks are from the post hoc contrast test results; the color of the asterisks indicates treatment group, and the horizontal bars indicate between which sperm length segments there was a significant difference in the H4K12ac intensity; *P < 0.05, **P < 0.001, ***P < 0.0001. Bar in a = 10 μm for a–g′.
The initial appearance and progression of H4K12ac deposition followed the order of sperm chromatin decondensation. The signal intensity of H4K12ac between stages increased significantly as the paternal chromatin progressed through the earliest stages of sperm chromatin remodeling, where the greatest increase was observed at the posterior end, then at the tip, and then at the midbody region of the sperm head nucleus. In type b and c sperm nuclei, the entire sperm nucleus showed a bright and homogenous staining pattern with H4K12ac (Fig. 3, f and g). The specific pattern of staining observed in these three regions in type a subgroups was no longer distinguishable in type b and c sperm nuclei. The location along the longitudinal section of the sperm chromatin dictated the pattern of sperm chromatin remodeling with H4K12ac in nearly all sperm decondensation stages in both treatment groups (Fig. 3, Table 1; two-way ANOVA with post hoc contrast tests).
Table 1.
Quantification of the pattern of deposition of modified histones H4K12ac and H3S10ph in all sperm decondensation stages.a
| H4K12acb | H3S10phc | |||||
|---|---|---|---|---|---|---|
| Type of paternal nuclei | Drug P value | Distance P value | Interaction P value | Drug P value | Distance P value | Interaction P value |
| Condensed | 0.8201 | 0.1521 | 0.4277 | 0.0605 | 0.0017* | 0.3891 |
| Type a1 | 0.4847 | <0.0001*** | 0.285 | 0.3947 | <0.0001*** | 0.1 |
| Type a2 | 0.4352 | <0.0001*** | 0.6543 | 0.2277 | <0.0001*** | 0.5185 |
| Type a3 | 0.4647 | <0.0004** | 0.7738 | 0.3744 | <0.0001*** | 0.421 |
| Type a4 | 0.201 | <0.0001*** | 0.2361 | 0.7626 | <0.0001*** | 0.3307 |
| Type b | 0.3269 | <0.0001*** | 0.4832 | 0.5038 | <0.0001*** | 0.9855 |
| Type c | 0.7429 | <0.0001*** | 0.7771 | 0.1181 | <0.0001*** | 0.0297 |
Statistical analyses for H4K12ac and H3S10ph immunofluorescence by unbalanced two-way ANOVA with repeated measures and a logarithmic transformation of the intensity using SAS program; P values are adjusted: *P < 0.05, **P < 0.001, ***P < 0.0001.
The relationship between the pattern of chromatin remodeling with H4K12ac and the DNA content was assessed in type a3 sperm nuclei (Fig. 3d), because they were partially decondensed and clearly showed the three characteristic chromatin regions of the sperm head. Type c sperm nuclei (Fig. 3g) were selected because the remodeling of the sperm chromatin was completed by this stage [5]. In type a3 sperm nuclei, the level of DNA compaction was inversely related to the signal intensity with H4K12ac for all three regions of the sperm head (Fig. 3d). In type c sperm nuclei, the level of DNA compaction paralleled the staining intensity of H4K12ac (Fig. 3g).
The pattern of H4K12ac staining was also analyzed for the maternal nucleus and polar body of all sperm decondensation stages. The maternal chromosomes of unfertilized oocytes had a spot-like pattern of staining with H4K12ac. Following fertilization, in zygotes with either condensed or type a1 sperm nuclei, the maternal nuclear and polar body H4K12ac staining pattern and signal intensity were not distinguishable; both had many bright foci, mostly located in condensed or heterochromatin regions, while euchromatin regions had a low homogenous staining pattern (Supplemental Fig. S1a available online at www.biolreprod.org). The pattern of H4K12ac staining began to differ in zygotes with type a2 sperm nuclei. Here, the maternal nucleus appeared to have an increased number of foci, and the polar body had a reduced number, with four to five foci. In zygotes with type a3 and a4 sperm nuclei, there was a more marked difference in the number of foci between the maternal nucleus and the polar body (two to three foci) (Supplemental Fig. S1b available online at www.biolreprod.org). The pattern of H4K12ac staining became clearly distinguishable in zygotes with type b and c sperm nuclei, where the maternal nuclei had numerous, homogenously distributed foci, while the number of foci in the polar body was reduced to zero to one (Supplemental Fig. S1c available online at www.biolreprod.org).
Paternal exposure to CPA did not affect sperm chromatin remodeling, as depicted by the unchanged pattern of histone H4K12ac deposition for any of the sperm decondensation stages observed following fertilization (Fig. 3, a′–g′) and confirmed by the two-way ANOVA analysis (Table 1). In addition, paternal exposure to CPA did not affect the pattern of histone H4K12ac deposition in the maternal nucleus and polar body (data not shown).
H3S10ph Sperm Chromatin Remodeling Pattern in Pre-PN Zygotes
The pattern of histone H3S10ph deposition on the decondensing paternal chromatin was recorded for both saline- and CPA-treated groups in all sperm decondensation stages (Fig. 4). In zygotes sired by saline-treated males, no visible staining with histone H3S10ph was seen in condensed sperm nuclei, with a small peak in the midbody region of the sperm head nucleus (Fig. 4a). In type a1 sperm nuclei, a very faint staining at the posterior and ventral sides of the sperm head nucleus was observed (Fig. 4b). In type a2 sperm nuclei, H3S10ph signal intensity increased mostly at the posterior end, and stayed constant at the ventral sides of the sperm head nucleus (Fig. 4c). In type a3 sperm nuclei, a dramatic increase in H3S10ph signal intensity was observed at the posterior end, ventral sides, and the tip of the sperm head nucleus stained with H3S10ph (Fig. 4d). In type a4 sperm nuclei, H3S10ph pattern, and signal intensity resembled a3 sperm nuclei, except that H3S10ph localized to a greater extent in the midbody and tip regions of the sperm head nucleus (Fig. 4e). When comparing the staining intensity of histone H3S10ph in condensed, type a1, a2, a3, and a4 sperm nuclei, the greatest change was observed at the posterior end, then at the tip, and lastly at the midbody region of the sperm head nucleus. A ring-like pattern started to form in type a4 sperm nuclei at both extremities of the sperm head nucleus; this ring closed completely in type b and c sperm nuclei (Fig. 4, e–g). In type b and c sperm nuclei, the signal intensity with histone H3S10ph was the highest at both posterior and tip ends, and decreased to a small extent at the midbody region of the sperm head nucleus in type c sperm nuclei (Fig. 4, f and g). The pattern of sperm chromatin remodeling with H3S10ph was dependent on the location along the longitudinal section of the sperm chromatin in nearly all sperm decondensation stages in both treatment groups (Fig. 4 and Table 1; two-way ANOVA with post hoc contrast tests).
FIG. 4.
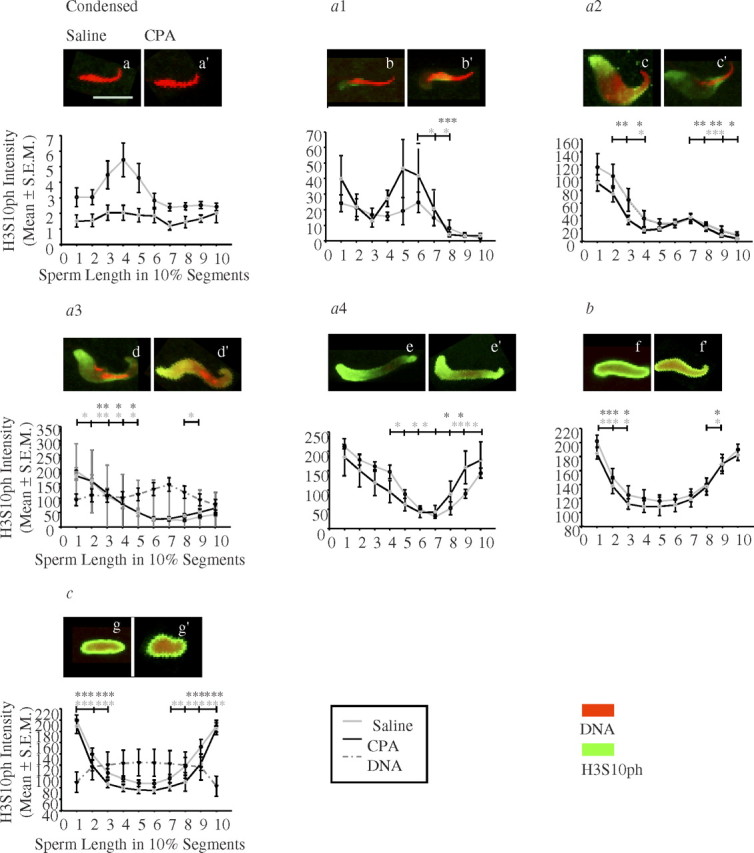
Quantitative analysis of the initial appearance, progression, and pattern of histone H3 phosphorylated at serine 10, H3S10ph, on the decondensing paternal nuclei in pre-PN zygotes. The sperm decondensation stage is on the top of each graph; below each stage, there is the best representative picture of saline-exposed sperm on the left (a) and CPA-exposed sperm on the right (a′) for each stage. DNA was stained with propidium: iodide (red) and H3S10ph with FITC (green). The y axes represent H3S10ph intensity (mean ± SEM), and the x axes represent sperm length in 10% segments. Error bars are the SEM. Light gray line, saline group; black line, CPA group; dashed dark gray line, DNA content. For each individual type of paternal nuclei, the localization and staining intensity of histone H3S10ph differed significantly along the different segments of the sperm length in all sperm decondensation stages. CPA exposure did not affect histone H3S10ph staining intensity or patterning for any of the sperm decondensation stages. Number of pre-PN zygotes analyzed at each sperm decondensation stage: saline, (a) n = 10, 33 zygotes, (b) n = 6, 7 zygotes, (c) n = 8, 15 zygotes, (d) n = 10, 17 zygotes, (e) n = 5, 6 zygotes, (f) n = 9, 70 zygotes, (g) n = 9, 17 zygotes; and CPA, (a′) n = 7, 17 zygotes, (b′) n = 4, 4 zygotes, (c′) n = 9, 15 zygotes, (d′) n = 8, 18 zygotes, (e′) n = 3, 3 zygotes, (f′) n = 9, 75 zygotes, (g′) n = 9, 33 zygotes for sperm decondensation stages: condensed, type a1, type a2, type a3, type a4, type b, and type c sperm nuclei, respectively. Statistical analyses, unbalanced two-way ANOVA with repeated measures, and a logarithmic transformation of the intensity followed by post hoc contrast tests were performed with the SAS program; the asterisks are from the post hoc contrast tests results, and the color refers to the appropriate treatment group. The horizontal bars indicate between which two adjacent sperm length segments there is a significant difference in H3S10ph intensity; *P < 0.05, **P < 0.001, ***P < 0.0001. Bar in a = 10 μm for a–g′.
The relationship between the level of DNA compaction and histone H3S10ph intensity was examined in both type a3 and type c sperm nuclei. In type a3 sperm nuclei, the level of DNA compaction was inversely related to histone H3S10ph staining intensity in all three sperm regions. When the sperm chromatin was decondensed, the signal for H3S10ph was high, and vice versa (Fig. 4d). In type c sperm nuclei, the level of DNA compaction was still inversely related to the staining intensity of histone H3S10ph; the signal intensity was high at the circumference of sperm nucleus, and low in the middle, while the DNA was decondensed around the circumference and condensed in the core of the sperm nucleus (Fig. 4g).
In zygotes from all the sperm decondensation stages analyzed, the maternal nucleus and polar body were not distinguishable (Supplemental Fig. S2 available online at www.biolreprod.org). They both had numerous fine, bright foci forming a ring pattern with histone H3S10ph at the circumference of the nucleus, with very low to no staining in the center. Occasionally, both the maternal and polar body had a single larger focus of the same intensity as the ring pattern. The ring observed in both maternal nuclei and polar bodies was very similar to the perinuclear ring observed on the sperm chromatin of type b and c sperm nuclei (Fig. 4, f and g).
The initial appearance, localization, and pattern of histone H3S10ph deposition on the remodeling sperm chromatin were not affected by paternal exposure to CPA (Fig. 4, a′–g′). The two-way ANOVA showed no effect of CPA treatment on the pattern of sperm chromatin remodeling with H3S10ph and no interaction between treatment and distance effect for any of the sperm decondensation stages analyzed (Table 1). In addition, paternal exposure to CPA did not affect histone H3S10ph pattern of deposition in either the maternal nucleus or the polar body (data not shown).
Characterization of the Distribution of γH2AX Foci in Pre-PN Zygotes
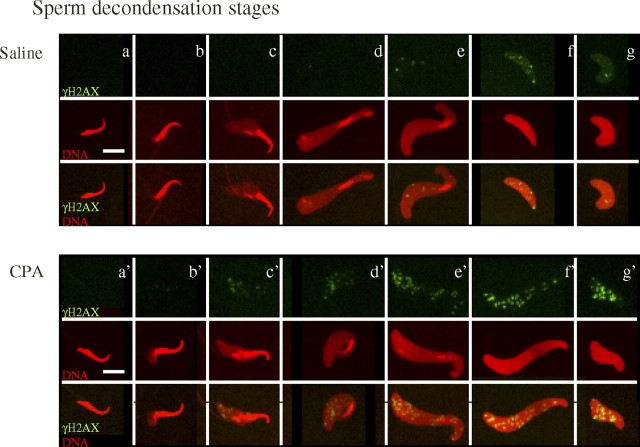
The appearance and pattern of distribution of γH2AX foci were analyzed in pre-PN zygotes at all stages of sperm decondensation (Fig. 5, a–g). In control zygotes, no γH2AX foci were observed in condensed and type a1 sperm nuclei (Fig. 5, a and b). A gradual increase in the population of small γH2AX foci was found in the later stages of sperm decondensation (Fig. 5, f and g), with a constant low occurrence of large γH2AX foci (Fig. 5, a–g).
Fig. 5.
The appearance and distribution of γH2AX foci on the decondensing sperm chromatin is altered in decondensing sperm nuclei after CPA exposure. The top panels are immunofluorescence projection images of saline-exposed sperm (a–g), and the bottom panels represent CPA-exposed sperm (a′–g′) at every sperm decondensation stage in pre-PN zygotes. The top row are projections of γH2AX with FITC (green) only, the middle row are projections of DNA stained with propidium iodide (red) only, and the last row represents the merged images of both γH2AX and DNA. Control group (a) condensed, (b) type a1, (c) type a2, (d) type a3 (still image from Supplemental Movie S1), (e) type a4, (f) type b, and (g) type c (still image from Supplemental Movie S2) sperm nuclei. CPA-treated group (a′) condensed, (b′) type a1, (c′) type a2, (d′) type a3 (still image from Supplemental Movie S3), (e′) type a4, (f′) type b, and (g′) type c (still image from Supplemental Movie S4) sperm nuclei. Foci were only distributed in regions of decondensed sperm chromatin. We observed an earlier appearance and an increase in the number and size of γH2AX foci, starting from type a2 sperm nuclei in our CPA treatment group. Bar = 10 μm.
In zygotes sired by sperm from CPA-exposed rats, there were a few small γH2AX foci in the posterior open sperm chromatin region in early decondensing sperm nuclei, such as the type a1 sperm nuclei (Fig. 5b′). γH2AX foci appeared on the decondensing posterior end of type a2, a3, and a4 sperm nuclei (Fig. 5, c′–e′). In type b and c sperm nuclei, an enhanced population of large γH2AX foci was detected (Fig. 5, f′ and g′).
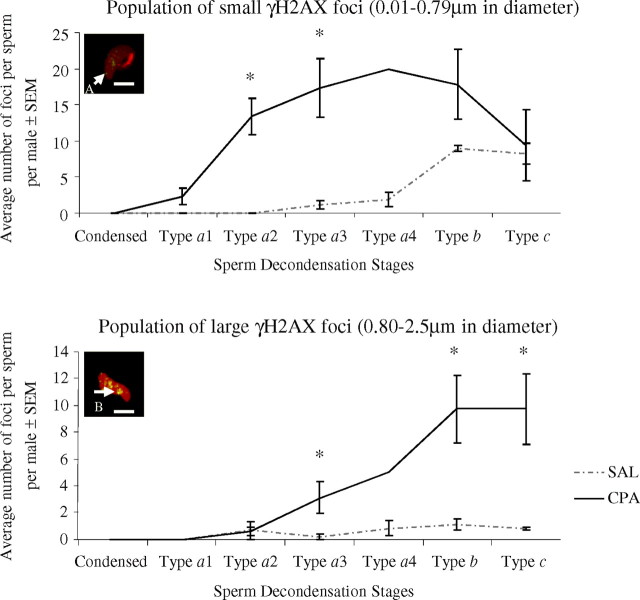
In the type a sperm nuclear subgroups of zygotes sired by CPA-exposed rats, the average number of small diameter (0.01–0.79 μm) foci significantly increased (Fig. 6, top panel). In addition, we observed a significant appearance of larger 0.80–2.5 μm diameter foci in type a3 and a4 sperm nuclei; this increase was enhanced in later stage b and c sperm nuclei (Fig. 6, bottom panel). Type a4 sperm nuclei were excluded from the analysis due to the very small number of zygotes within that stage.
Fig. 6.
Quantitative analysis of the average number of small (Top panel) and large (Bottom panel) γH2AX foci per sperm per male for all sperm decondensation stages. Bar = 10 μm. The y axis is the average number of foci per sperm per male ± SEM, and the x axis represents the sperm decondensation stages. Number of pre-PN zygotes analyzed at each sperm decondensation stage: saline, (a) n = 4, 16 zygotes, (b) n = 3, 5 zygotes, (c) n = 3, 4 zygotes, (d) n = 5, 9 zygotes, (e) n = 3, 4 zygotes, (f) n = 6, 30 zygotes, (g) n = 3, 11 zygotes; and CPA, (a′) n = 5, 17 zygotes, (b′) n = 3, 3 zygotes, (c′) n = 4, 8 zygotes, (d′) n = 4, 8 zygotes, (e′) n = 1, 2 zygotes, (f′) n = 6, 20 zygotes, (g′) n = 3, 6 zygotes for sperm decondensation stages: condensed, type a1, type a2, type a3, type a4, type b, and type c sperm nuclei, respectively. Mann-Whitney U-test was done to compare the average number of foci between CPA-treated and control group at all stages of sperm decondensation, independently and for both populations of γH2AX foci. *P < 0.05.
In both the maternal nucleus and polar body of control zygotes, we observed a perinuclear ring-like pattern with γH2AX staining formed by numerous small foci; occasionally, a large reaction was found on the ring in zygotes from all of the sperm decondensation stages analyzed (Supplemental Fig. S3 available online at www.biolreprod.org). Paternal exposure to CPA did not affect the pattern of γH2AX staining in the maternal nucleus or the polar body at any of the sperm decondensation stages (Supplemental Fig. S3 available online at www.biolreprod.org).
Using sperm 3D reconstruction, in type a subgroups γH2AX foci were always found in regions of open and loose sperm chromatin structure (Fig. 5 and Supplemental Movie S1 available online at www.biolreprod.org); in totally decondensed and recondensing type b and c sperm nuclei, foci were evenly distributed (Fig. 5 and Supplemental Movie S2 available online at www.biolreprod.org). Small γH2AX foci appeared precociously in the earliest sperm decondensation stages (Fig. 5 and Supplemental Movie S3 available online at www.biolreprod.org); a dramatic increase in the number of the larger γH2AX foci was observed in the later stages of sperm chromatin remodeling (Fig. 5 and Supplemental Movie S4 available online at www.biolreprod.org). This was not accompanied by a parallel impact on the maternal genome (data not shown).
Discussion
Chromatin remodeling of the paternal genome in early zygotes is not a random event: the pattern of deposition of maternally modified histones, H4K12ac and H3S10ph, on the remodeling of paternal chromatin is extremely precise and orderly. In almost all sperm decondensation stages, with the exception of condensed sperm nuclei, modified histone deposition varied significantly along the sperm and between sperm decondensation stages. These results suggest that sperm chromatin remodeling in the zygote is a highly dynamic process regulated by the order and level of DNA chromatin compaction.
Four phases of chromatin decondensation were observed in sperm nuclei during pre-PN zygotic development: condensed, type a partially decondensed, type b totally decondensed, and type c recondensing sperm nuclei [5]. In type a sperm nuclei, three sperm segments were clearly visible; the posterior, the anterior, and the midbody region of the sperm head. Decondensation always proceeded in the same order, from the posterior, to the ventral sides, to the tip and the midbody of the sperm head nucleus. Due to the novel segmental pattern of sperm chromatin decondensation in the rat, type a sperm nuclei were further subdivided into four subgroups: type a1, type a2, type a3, and type a4 sperm nuclei. Differences in the segmental pattern of sperm nuclear decondensation in rats from that previously described in mice [1] may be attributed to a species difference in sperm chromatin packaging, with two protamines in mice, but only one in rat sperm, or to the region-specific retention of nucleosomes during spermiogenesis [36–38].
Since the midbody region of the sperm head nucleus was the last segment of the sperm chromatin to decondense, we speculate that this may be the region of the sperm nucleus most protected against insult. The midbody, also referred to as the chromocenter, is a spermatozoal region enriched in modified nucleosomes and poor in protamine [1]. The presence of modified nucleosomes in the midbody may protect it against active paternal genome demethylation, and act as a barrier for sperm chromatin decondensation [1, 8, 38]. Alternatively, the midbody of the sperm nucleus may decondense last, due to its central location and higher protamine content.
The initial appearance and distribution of H4K12ac and H3S10ph on the remodeling sperm chromatin in type a subgroups followed the exact order of paternal chromatin decondensation: from the posterior, ventral sides, tip, and finally the midbody region of the sperm head nucleus. In type a1, a2, a3, and a4 sperm nuclei, the order and level of sperm DNA decondensation dictated the initial appearance and deposition pattern of modified histones H4K12ac and H3S10ph. The rare and distinct epigenetic marks retained in spermatozoa may also regulate the sequential order of sperm chromatin decondensation and the pattern of sperm chromatin remodeling in pre-PN zygotes, since spermatozoal nucleosomes have been found on genes guiding embryonic development [1, 39].
Posttranslational modifications on histone tails, such as H4K12ac and H3S10ph, are responsible for faithful chromatin compaction and cellular processes [40] in early postfertilized oocytes. We report two distinct patterns of chromatin remodeling in early postfertilized oocytes: one for H4K12ac, and a second for H3S10ph. We postulate that the high, homogenous pattern of staining with histone H4K12ac in type b and c sperm nuclei marks the open and active chromatin structure necessary for subsequent gene transcription and mitosis; this pattern was not affected by the level of DNA compaction. Furthermore, we speculate that the ring pattern of staining with histone H3S10ph in type b and c sperm nuclei marks the condensed chromatin structure characteristic of the early phase of mitosis [12], which is necessary for the recondensation of the paternal chromatin and the formation of the male pronucleus [41].
Previous work on the effects of paternal exposure to CPA on the formation and maturation of spermatozoa showed altered sperm chromatin packaging, decreased protamine, and thiol contents [42], changes in sperm nuclear matrix proteins [43], and altered in vitro nuclear decondensation patterns [27]. It has been suggested that these effects of CPA lead to a looser packaging of the sperm chromatin during spermiogenesis, perhaps linking these effects of drug exposure to an altered rate of nuclear decondensation during pre-PN zygotic development [44]. This suggestion is supported by our observation of an increase in the number of zygotes with type c sperm nuclei in the CPA group compared with controls. The absence of any effect of CPA on the pattern of sperm chromatin remodeling may be due to compensation mechanisms in the oocyte.
Our results suggest a cross-talk between the male and the female nucleus, since the zygote was able to sense and synchronize its development with the accelerated rate of the sperm chromatin decondensation. Previous work from our laboratory showed that paternal exposure to CPA accelerated the progression of later pronuclear-stage zygotes, and induced aberrant epigenetic reprogramming with histone H4K5ac and DNA methylation [32] and a biphasic DNA damage recognition response in the male genome [24, 32]. The time window of zygotic development, and the events taking place during this window, are distinct in the present study. In early pre-PN zygotes, the oocyte is completing its second cycle of meiosis, and the sperm chromatin is undergoing a complete protamine-to-histone remodeling. Here, we report that early events, such as the progression of sperm chromatin decondensation after fertilization, are disturbed without impacting sperm chromatin remodeling, as depicted by the patterns of two posttranslationally modified histones, H4K12ac and H3S10ph.
We then asked how early the zygote was able to recognize and respond to DNA damage induced by paternal CPA exposure. Small γH2AX foci were already visible in type a1 sperm nuclei. The increase in the number of small foci was significant in type a2 and a3 sperm nuclei, representing stages of rapid chromatin remodeling, when protamines are removed and replaced with histones [5]. In later stages of sperm decondensation, type c sperm nuclei, the average number of small foci in the CPA-treated group was comparable to control level. We observed an increase in the numbers of large γH2AX foci beginning in type a3 sperm nuclei and persisting through to type b and c sperm nuclei after paternal CPA exposure. The early induction of small γH2AX foci suggests a role for γH2AX during chromatin remodeling in the detection of CPA-damaged sperm chromatin during the removal and exchange of protamine for histones [23]. The subsequent disappearance of small γH2AX foci in remodeled sperm chromatin coincides with the increase in large foci, suggesting that these large foci may represent the accumulation of γH2AX marks at already existing sites of damage [23]. A progressive elevation in γH2AX focal volumes was observed in the male pronuclei of zygotes fertilized by CPA-exposed sperm, through S phase (PN3 and −4) into G2 (PN5) [24]. Previous studies showed that early pre-PN zygotes responded in a dose-dependent manner to DNA-damaging agents by activating the γH2AX signaling pathway following gamete fusion, and showed differences in parental DNA repair efficiency [22]. The pattern of γH2AX staining in the maternal nucleus and polar body was not affected by paternal CPA exposure, but may be sensitive to other DNA-damaging agents [21]. A dynamic interaction between small and large γH2AX foci in pre-PN zygotes fertilized by CPA-exposed sperm may indicate the presence of intricate control of damage recognition and repair mechanisms in the oocyte.
In summary, our data demonstrate that paternal exposure to CPA affects the progression of sperm chromatin decondensation, and activates a DNA damage response in the pre-PN rat zygote. The effects on sperm decondensation that are observed in pre-PN zygotes may represent the first manifestation of the detrimental effects previously observed among the progeny of CPA-treated males [28, 32, 34]. Both an undamaged sperm genome and an intact sperm nuclear matrix are necessary for the paternal genome to support embryonic development [45].
Supplementary Material
Acknowledgment
We thank Dr. H. J. Clarke (McGill University, Montreal, QC) for the gift of H4K12ac antibody and useful discussion, Ms. J. Laliberté (McGill University) for her assistance with confocal microscopy, and Jose Correa (McGill University) for his help with the statistical analysis of the H4K12ac and H3S10ph quantification data.
References
- 1. Van der Heijden GW, Derijck AAHA, Ramos L, Giele M, van der Vlag J, de Boer P. Transmission of modified nucleosomes from the mouse male germline to the zygote and subsequent remodeling of paternal chromatin. Dev Biol 2006; 298:458–469. [DOI] [PubMed] [Google Scholar]
- 2. Akabane H, Fan J, Xuehai Z, Zhu GZ. Protein kinase C activity in mouse eggs regulates gamete membrane interaction. Mol Reprod Dev 2007; 74:1465–1472. [DOI] [PubMed] [Google Scholar]
- 3. Yu Y, Halet G, Lai FA, Swann K. Regulation of diacylglycerol production and protein kinase C stimulation during sperm- and PLCζ-mediated mouse egg activation. Biol Cell 2008; 100:633–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Perreault SD, Barbee RR, Slott VL. Importance of glutathione in the acquisition and maintenance of sperm nuclear decondensing activity in maturing hamster oocyte. Dev Biol 1988; 125:181–186. [DOI] [PubMed] [Google Scholar]
- 5. Van der Heijden GW, Dieker JW, Derijck AAHA, Muller S, Berden JHM, Braat DDM, van der Vlag J, de Boer P. Asymmetry in histone H3 variants and lysine methylation between paternal and maternal chromatin of the early mouse zygote. Mech Dev 2005; 122:1008–1022. [DOI] [PubMed] [Google Scholar]
- 6. Leno GH, Mills AD, Philpott A, Laskey RA. Hyperphosphorylation of nucleoplasmin facilitates Xenopus sperm decondensation at fertilization. J Biol Chem 1996; 271:7253–7256. [DOI] [PubMed] [Google Scholar]
- 7. Loppin B, Bonnefoy E, Anselme C, Laurençon A, Karr TL, Couble P. The histone H3.3 chaperone HIRA is essential for chromatin assembly in the male pronucleus. Nature 2005; 437:1386–1390. [DOI] [PubMed] [Google Scholar]
- 8. Santos F, Peters AH, Otte AP, Reik W, Dean W. Dynamic chromatin modifications characterise the first cell cycle in mouse embryos. Dev Biol 2005; 280:225–236. [DOI] [PubMed] [Google Scholar]
- 9. Yoshida N, Brahmajosyula M, Shoji S, Amanai M, Perry ACF. Epigenetic discrimination by mouse metaphase II oocytes mediates asymmetric chromatin remodeling independently of meiotic exit. Dev Biol 2007; 301:464–477. [DOI] [PubMed] [Google Scholar]
- 10. Adenot PG, Mercier Y, Renard J-P, Thompson EM. Differential H4 acetylation of paternal and maternal chromatin precedes DNA replication and differential transcriptional activity in pronuclei of 1-cell mouse embryos. Development 1997; 124:4615–4625. [DOI] [PubMed] [Google Scholar]
- 11. Wang Q, Wang C-M, Ai J-S, Xiong B, Yin S, Hou Y, Chen D-Y, Schatten H, Sun Q-Y. Histone phosphorylation and pericentromeric histone modifications in oocyte meiosis. Cell Cycle 2006; 5:1974–1982. [DOI] [PubMed] [Google Scholar]
- 12. Song L, Li D, Liu R, Zhou H, Chen J, Huang X. Ser-10 phosphorylated histone H3 is involved in cytokinesis as a chromosomal passenger. Cell Biol Int 2007; 31:1184–1190. [DOI] [PubMed] [Google Scholar]
- 13. DeManno DA, Cottom JE, Kline MP, Peters CA, Maizels ET, Hunzicker-Dunn M. Follicle-stimulating hormone promotes histone H3 phosphorylation on serine-10. Mol Endocrinol 1999; 13:91–105. [DOI] [PubMed] [Google Scholar]
- 14. De la Barre AE, Gerson V, Gout S, Creaven M, Allis CD, Dimitrov S. Core histone N-termini play an essential role in mitotic chromosome condensation. EMBO J 2000; 19:379–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jaroudi S, SenGupta S. DNA repair in mammalian embryos. Mutat Res 2007; 635:53–77. [DOI] [PubMed] [Google Scholar]
- 16. Perry AC. Hijacking oocyte DNA repair machinery in transgenesis? Mol Reprod Dev 2000; 56:319–324. [DOI] [PubMed] [Google Scholar]
- 17. Generoso WM, Cain KT, Krishna M, Huff SW. Genetic lesions induced by chemicals in spermatozoa and spermatids of mice are repaired in the egg. Proc Natl Acad Sci U S A 1979; 76:435–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Stiff T, O'Driscoll M, Rief N, Iwabuchi K, Lobrich M, Jeggo PA. ATM and DNA-PK function redundantly to phosphorylate H2AX after exposure to ionizing radiation. Cancer Res 2004; 64:2390–2396. [DOI] [PubMed] [Google Scholar]
- 19. Rogakou EP, Boon C, Redon C, Bonner WM. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J Cell Biol 1999; 146:905–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rogakou EP, Pilch DR, Orr VS, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem 1998; 273:5858–5868. [DOI] [PubMed] [Google Scholar]
- 21. Ziegler-Birling C, Helmrich A, Tora L, Torres-Padilla M-E. Distribution of p53 binding protein 1 (53BP1) and phosphorylated H2A.X during mouse preimplantation development in the absence of DNA damage. Int J Dev Biol 2009; 53:1003–1011. [DOI] [PubMed] [Google Scholar]
- 22. Derijck AAHA, van der Heijden GW, Giele M, Philippens MEP, van Bavel CCAW, de Boer P. γH2AX signalling during sperm chromatin remodelling in the mouse zygote. DNA Repair (Amst)2006; 5:959–971. [DOI] [PubMed] [Google Scholar]
- 23. McManus KJ, Hendzel MJ. ATM-dependent DNA damage-independent mitotic phosphorylation of H2AX in normally growing mammalian cells. Mol Biol Cell 2005; 16:5013–5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Barton TS, Robaire B, Hales BF. DNA damage recognition in the rat zygote following chronic paternal cyclophosphamide exposure. Tox Sci 2007; 100:495–503. [DOI] [PubMed] [Google Scholar]
- 25. Thomson AB. Late reproductive sequelae following treatment of childhood cancer and options for fertility preservation. Best Pract Res Clin Endocrinol Metab 2002; 16:311–334. [DOI] [PubMed] [Google Scholar]
- 26. Colvin OM. An overview of cyclophosphamide development and clinical applications. Curr Pharm Des 1999; 5:555–560. [PubMed] [Google Scholar]
- 27. Qiu J, Hales BF, Robaire B. Effects of chronic low-dose cyclophosphamide exposure on the nuclei of rat spermatozoa. Biol Reprod 1995; 52:33–40. [DOI] [PubMed] [Google Scholar]
- 28. Trasler JM, Hales BF, Robaire B. A time-course study of chronic paternal cyclophosphamide treatment in rats: effects on pregnancy outcome and the male reproductive and hematologic systems. Biol Reprod 1987; 37:317–326. [DOI] [PubMed] [Google Scholar]
- 29. Lewis JD, Song Y, de Jong ME, Bagha SM, Ausió J. A walk through vertebrate and invertebrate protamines. Chromosoma 2003; 111:473–482. [DOI] [PubMed] [Google Scholar]
- 30. Hazzouri M, Pivot-Pajot C, Faure A-K, Usson Y, Pelletier R, Sèle B, Khochbin S, Rousseaux S. Regulated hyperacetylation of core histones during mouse spermatogenesis: involvement of histone deacetylases. Eur J Cell Biol 2000; 79:950–960. [DOI] [PubMed] [Google Scholar]
- 31. Codrington AM, Hales BF, Robaire B. Spermiogenic germ cell phase-specific DNA damage following cyclophosphamide exposure. J Androl 2004; 25:354–362. [DOI] [PubMed] [Google Scholar]
- 32. Barton TS, Robaire B, Hales BF. Epigenetic programming in the preimplantation rat embryo is disrupted by chronic paternal cyclophosphamide exposure. Proc Natl Acad Sci U S A 2005; 102:7865–7870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Trasler JM, Hales BF, Robaire B. Paternal cyclophosphamide treatment of rats causes fetal loss and malformations without affecting male fertility. Nature 1985; 316:144–146. [DOI] [PubMed] [Google Scholar]
- 34. Harrouk W, Khatabaksh S, Robaire B, Hales BF. Paternal exposure to cyclophosphamide dysregualtes the gene activation program in rat preimplantation embryos. Mol Reprod Dev 2000; 57:214–223. [DOI] [PubMed] [Google Scholar]
- 35. De la Barre AE, Angelov D, Molla A, Dimitrov S. The N-terminus of histone H2B, but not that of histone H3 or its phosphorylation, is essential for chromosome condensation. EMBO J 2001; 20:6383–6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Calvin HI. Comparative analysis of the nuclear basic proteins in rat, human, guinea pig, mouse and rabbit spermatozoa. Biochem Biophys Acta 1976; 434:377–389. [DOI] [PubMed] [Google Scholar]
- 37. Kistler WS, Keim PS, Heinriksom RL. Partial structural analysis of the basic chromosomal protein of rat spermatozoa. Biochem Biophys Acta 1976; 427:752–757. [DOI] [PubMed] [Google Scholar]
- 38. Balhorn R, Gledhill BL, Wyrobek AJ. Mouse sperm chromatin proteins: quantitative isolation and partial characterization. Biochemistry 1977; 16:4074–4080. [DOI] [PubMed] [Google Scholar]
- 39. Hammoud SS, Nix DA, Zhang H, Purwar J, Carrell DT, Cairns BR. Distinctive chromatin in human sperm packages genes for embryo development. Nature 2009; 460:473–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Eberlin A, Grauffel C, Oulad-Abdelghani M, Robert F, Torres-Padilla ME, Lambrot R, Spehmer D, Ponce-Perez L, Würtz JM, Stote RH, Kimmins S, Schultz P et al. Histone H3 tails containing dimethylated lysine and adjacent phosphorylated serine modifications adopt a specific conformation during mitosis and meiosis. Mol Cell Biol 2008; 28:1739–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Prigent C, Dimitrov S. Phosphorylation of serine 10 in histone H3, what for? J Cell Sci 2003; 116:3677–3685. [DOI] [PubMed] [Google Scholar]
- 42. Codrington AM, Hales BF, Robaire B. Exposure of male rats to cyclophosphamide alters the chromatin structure and basic proteome in spermatozoa. Hum Reprod 2007; 22:1431–1442. [DOI] [PubMed] [Google Scholar]
- 43. Codrington AM, Hales BF, Robaire B. Chronic cyclophosphamide exposure alters the profile of rat sperm nuclear matrix proteins. Biol Reprod 2007; 77:303–311. [DOI] [PubMed] [Google Scholar]
- 44. Perreault SD, Naish SJ, Zirkin BR. The timing of hamster sperm nuclear decondensation and male pronucleus formation is related to sperm nuclear disulfide bond content. Biol Reprod 1987; 36:239–244. [DOI] [PubMed] [Google Scholar]
- 45. Ward WS, Kimura Y, Yanagimachi R. An intact sperm nuclear matrix may be necessary for the mouse paternal genome to participate in embryonic development. Biol Reprod 1999; 60:702–706. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.