The emergence and spread of multidrug resistant (MDR) Mycobacterium tuberculosis complex (MTBC) strains is a critical global health problem. Between 2014 and 2018, 606 MTBC strains were isolated from 13,892 suspected pulmonary tuberculosis (TB) patients in Tehran, Iran, including 16 (2.6%) MDR-TB cases.
KEYWORDS: Mycobacterium tuberculosis, Tehran, extensive drug resistance, multidrug resistance, whole-genome sequencing
ABSTRACT
The emergence and spread of multidrug resistant (MDR) Mycobacterium tuberculosis complex (MTBC) strains is a critical global health problem. Between 2014 and 2018, 606 MTBC strains were isolated from 13,892 suspected pulmonary tuberculosis (TB) patients in Tehran, Iran, including 16 (2.6%) MDR-TB cases. A combination of phenotypic and genotypic methods (whole-genome sequencing) was employed for the identification of additional drug resistances and strain-to-strain genetic distances as a marker for recent transmission events. MDR and extensively drug-resistant (XDR) TB cases were almost exclusively infected by lineage 2/Beijing strains (14/16, P < 0.001). We further showed that recent transmission and/or recent introduction of lineage 2/Beijing strains contribute to high XDR-TB rates among all MDR-TB cases and should be considered an emerging threat for TB control in Tehran. In addition, the extensive pre-existing drug resistance profiles of MDR/XDR strains will further challenge TB diagnostics in the region.
INTRODUCTION
Nowadays, tuberculosis (TB) remains the number one cause of human death due to an infectious agent, causing an estimated 10.4 million new cases and 1.7 million deaths annually (1). The emergence and transmission of multidrug-resistant (MDR) Mycobacterium tuberculosis complex (MTBC) strains, defined as resistant to rifampin (RIF) and isoniazid (INH), is threatening worldwide TB control efforts (2). Resistance evolution in failing MDR-TB treatment regimens toward pre-extensively drug-resistant (pre-XDR) strains, defined as MDR plus either resistance to one fluoroquinolone (FQ) or one second-line injectable drug and XDR-TB (i.e., MDR plus resistance to both one fluoroquinolone and one injectable drug) is further aggravating the problem (3).
Since 1998, when the full-genome sequence of M. tuberculosis H37Rv was revealed (4), whole-genome sequencing (WGS) has been applied to a wide range of clinical aspects, including the prediction of drug susceptibility, epidemiological and transmission dynamics analysis, and research into the evolution of the MTBC on the local and global level (5). Several publications demonstrated a high sensitivity and specificity of WGS for the detection of drug resistance patterns of MTBC isolates, especially for the first-line drugs rifampin and isoniazid (6). Additionally, there is strong support for the utility of WGS in the detection of transmission events due to its unprecedented discriminatory power compared with other genotyping methods (IS6110 restriction fragment length polymorphism [RFLP], spoligotyping, and mycobacterial interspersed repetitive-unit–variable-number tandem-repeat [MIRU-VNTR]) (7, 8).
According to the latest report on the WHO’s global TB database, the incidence of TB in Iran is 14 (range, 11 to 18) cases per 100,000 inhabitants, and the estimated proportion of new TB cases with MDR- or rifampin-resistant TB is 1.3% (range, 0.6% to 2%) (http://www.who.int/tb/country/data/profiles/en/). These reports emphasize the need for an in-depth analysis to better understand and contain drug resistance development and transmission of MDR-TB in this region.
In our study, we analyzed MDR, pre-XDR, and XDR MTBC isolates from Tehran (the capital of Iran) by whole-genome sequencing in order to gain valuable information about the genetic diversity, drug resistance, and transmission dynamics of these strains in this region.
MATERIALS AND METHODS
Study population.
In this retrospective cross-sectional study, a total of 606 MTBC strains were retrieved from 13,892 suspected pulmonary TB patients from January 2014 to January 2018 at the Department of Mycobacteriology and Pulmonary Research, Pasteur Institute of Iran. All of the patients were residents in Tehran, the capital of Iran. Ethical reviews and informed written consent approval were granted by the Ethical Committee of the Pasteur Institute of Iran. This research has been performed in accordance with the ethical standards noted in the 1964 Declaration of Helsinki and its later amendments.
Drug susceptibility testing (DST) and determination of MIC.
DST was performed for all culture-positive specimens (n = 606) by the proportional method, using Lowenstein-Jensen (LJ) medium supplemented with isoniazid (INH) at 0.2 mg/liter, rifampin (RIF) at 40 mg/liter, streptomycin (STR) at 4 mg/liter, ethambutol (EMB) at 2 mg/liter, kanamycin (KAN) at 30 mg/liter, ofloxacin (OFX) at 2 mg/liter, and capreomycin (CAP) at 40 mg/liter (17). Bacteria grown on antimycobacterial agent-containing medium with growth exceeding 1% of the number of colonies on control medium were considered to be resistant to the anti-TB drug. To determine the MICs of the anti-TB drugs, an alamarBlue assay (Thermo Scientific, USA) was used as previously described (9). All tests were conducted in duplicate. For the MDR/pre-XDR/XDR strains, the MIC was determined for RIF/INH, and for monodrug-resistant strains, the MIC was determined for the respective anti-TB drug only.
Whole-genome sequencing and primary sequence data analysis.
For next-generation sequencing, genomic DNA was extracted from each LJ slant using the cetyltrimethylammonium bromide (CTAB) method, and sequencing libraries were prepared using the Illumina Nextera kit as described recently (10). Libraries were loaded onto a Illumina NextSeq 500 instrument in a paired-end 2 × 151-bp run. The resulting reads were mapped to the M. tuberculosis H37Rv genome (GenBank accession no. NC_000962.3) with the alignment program Burrow-Wheeler Aligner (BWA), and mappings were refined with the Genome Analysis Toolkit (GATK), sambamba, and SAMtools toolkits. For variant detection in mapped reads, we filtered SAMtools-derived mpileup files for minimum thresholds of at least 4 reads indicating a variant in both forward and reverse orientation, 4 reads calling the allele with at least a Phred score of 20, and 75% allele frequency. Detected variants included single nucleotide polymorphisms (SNPs), insertions, and deletions.
Comparative analysis of whole-genome sequence data.
For phylogenomic analysis, detected variant positions of all isolates were combined, supplementing the joint list with the respective information from the original mappings where necessary. SNP positions with a reliable base call in at least 95% of the isolates and covered by reads in all isolates were concatenated to a sequence alignment, excluding SNPs within a window of 12 bp from each other and those located in repetitive regions or resistance-associated genes. From the aligned sequences of concatenated SNPs, we built a maximum likelihood phylogenetic tree with FastTree version 2 in the Double precision built, with a general time reversible (GTR) substitution model, 1,000 resamples, and Gamma20 likelihood optimization, and visualized the resulting tree with the FigTree and EvolView tools. In addition, we constructed a maximum parsimony tree with the software BioNumerics version 7.6 (Applied Maths). The numbers on the branches indicate the number of distinct SNP positions between isolates. For the detection of clusters or related isolates likely derived from recent transmission, we employed the set of SNP positions to join isolates with a threshold of a maximum distance of 5 bp to the nearest group member.
Statistical analysis.
For assessing associations, the chi-square/Fisher exact test was applied. A P value of <0.05 was considered statistically significant. All analyses were done by using SPSS version 21 (SPSS, Inc., IL, USA) software.
RESULTS
Phenotypic resistance assays and phylogenetic analysis.
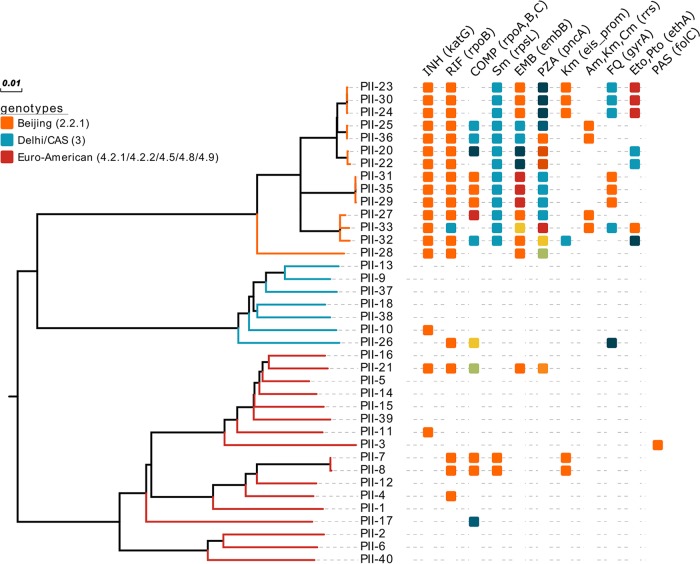
Phenotypic DST of 606 strains isolated between January 2014 to January 2018 detected a total of 16 (2.6%) MDR, pre-XDR, or XDR strains from single separate individuals. Among the 16 MDR strains, 8 had to be classified as pre-XDR, 5 strains as XDR, and 3 strains as MDR. In addition, 13 monodrug-resistant (3 mono-RIF, 3 mono-INH, 2 mono-STR, and 5 mono-EMB-resistant strains) and 9 pan-susceptible clinical strains were selected randomly for inclusion in this study. All 38 strains were analyzed by WGS, with all isolates reaching a mean coverage depth of at least 134-fold and a coverage breadth of at least 99.15% of the reference genome H37Rv. Classifying the strains by the detection of phylogenetically informative SNPs according to Coll et al. (11) assigned all isolates to one of three major lineages, lineage 2/Beijing (n = 14), lineage 4/Euro-American (n = 17), or lineage 3/Delhi-CAS (n = 7). Within the Beijing lineage, three strains belonged to the Central Asian outbreak type and seven strains to the W148 outbreak type (12). Interestingly, MDR, pre-XDR, and XDR-TB strains were significantly more likely to belong to the Beijing lineage (P < 0.001), with 14 out of the 16 MDR/pre-XDR/XDR strains belonging to the Beijing lineage, meaning that all Beijing strains in the study were at least found as being MDR-TB (Fig. 1; see Table S1 in the supplemental material).
FIG 1.
Maximum likelihood tree constructed from 5,660 SNP positions of the 38 isolates. Branches of the tree are colored according to major Mycobacterium tuberculosis lineages (Beijing, Delhi/CAS, and Euro-American). The boxes are color coded for distinct genetic drug resistance markers and bacterial fitness-associated (compensatory [COMP]) mutations (i.e., one color indicates a specific variant related). INH, isoniazid; RIF, rifampin; Sm, streptomycin; EMB, ethambutol; PZA, pyrazinamide; Km, Kanamycin; Am, amikacin; Cm, capreomycin; FQ, fluoroquinolone; Eto, ethionamide; Pto, prothionamide; PAS, para-aminosalicylic acid.
To determine possible MDR/XDR-TB transmission, we employed a maximum number of 5 SNP positions to the nearest cluster member to identify closely related isolates likely involved in recent transmission chains (13). This identifies four clusters, of which three are in the Beijing lineage comprising either two (one cluster) or three strains (two clusters) and one cluster comprises two Euro-American strains. Therefore, 8/14 lineage 2/Beijing strains and 8/16 MDR/pre-XDR/XDR strains are clustered (Fig. 1; see Fig. S1 in the supplemental material).
Polymorphisms in MDR/pre-XDR/XDR strains.
As summarized in Table S1, RIF resistance in MDR/pre-XDR/XDR strains (n = 16) was attributable to SNPs in the rpoB hot-spot region (S450L mutation), with an observed resistance by the proportional method, and was confirmed with the determination of the MIC by the alamarBlue assay. Out of 19 RIF-resistant strains (16 MDR and 3 mono-RIF-resistant strains), 13 (68.4%) also harbored additional mutations outside the rifampin resistance-determining region (RRDR) in rpoB, or in rpoA and rpoC, respectively. Of note, the mutation rpoC L516P was found in three lineage 2.2.1 (Beijing) strains and in two lineage 4.8 (mainly T) strains and was shown to be located in the interacting region of the RNA polymerase subunits RpoA and RpoC (14). The same was shown for the mutation rpoA T187A identified in two lineage 2 (Beijing W148) strains, suggesting that these strains indeed might have acquired a fitness (i.e., growth)-enhancing mutation.
INH resistance among all lineage 2 MDR strains was exclusively mediated by the mutation katG S315T. Two lineage 4.5 and one lineage 3 strains also had katG S315T. Another lineage 3 strain (PII-26) was identified with the combination katG N138H and ahpC t-76a and the highest MIC (64 mg/liter) compared with all other MDR strains (Table S1).
Furthermore, all MDR strains carried mutations or combinations of mutations in the embB gene and the embA promoter region, most likely conferring EMB resistance (e.g., embB M306I, M306V, Q497R, and embA -12c/t), although not all strains tested were phenotypically resistant to EMB at 2 mg/liter.
Resistance to STR in 13/16 (81.3%) MDR strains was mediated by rpsL K43R, and 15/16 (93.8%) of the MDR strains were observed with a mutation in the coding or upstream region of pncA associated with PZA resistance (Table S1).
Fluoroquinolone resistance in 6/16 (37.5%) strains was either linked to the mutation gyrA D94G or the combination of gyrA A90V and S91P. Four strains were found with the kanamycin resistance marker eis-8 c/a or eis-10 g/a, and four other strains were observed with an aminoglycoside and capreomycin cross-resistance mediated by rrs 1401 a/g.
In addition, 13 of these strains exhibited a mutation in the monooxygenase gene ethA which activates the drugs ethionamide and prothionamide. Especially in mutations leading to a premature stop codon, frameshifts and some reported mutations (i.e., ethA W256*, del 110 A, and T314I) might have an effect on the MIC of these drugs (15).
DISCUSSION
By employing WGS for drug resistance prediction and identification of strain clusters, we gained a better understanding of transmission dynamics and prevalent drug resistance profiles of M/XDR strains in Tehran. In particular, lineage 2/Beijing strains belonging to two previously defined MDR clades found in Russia, Eastern Europe, and Central Asia were also in Tehran associated with MDR/pre-XDR/XDR-TB (P < 0.001). These data are in concordance with our recent meta-analysis in which we showed that the prevalence of Beijing and Haarlem genotypes among MDR-TB isolates in this region was estimated to be 19.3% (95% confidence interval [CI], 13.1 to 27.5) and 18.7% (95% CI, 11.9 to 28.3), respectively (16). A previous publication reported that NEW-1 (47.8%) and the Beijing family (39.1%) were predominant in a period of 2011 to 2015 in Tehran (18). Surprisingly, our results indicate this scenario to be changing in this region; the significant increase of Beijing and reduction of NEW-1 genotypes (recently suggested as L4.5.1/Iran genotype by Mokrousov et al.) in MDR isolates detected in our study are crucial findings and are in contrast to recent reports for Iran and neighboring countries (19, 20).
Likewise, the clustering of closely related isolates identified with a maximum threshold of 5 SNPs, indicative of recent transmission, grouped the majority (8/14) of Beijing strains in clusters; therefore, also half (8/16) of the MDR/pre-XDR/XDR strains grouped into three clusters, while only two non-Beijing, non-MDR strains were found in a cluster. For our study, we included all 16 MDR/pre-XDR/XDR strains in our analysis and augmented the collection with randomly selected monodrug-resistant and susceptible strains. While this likely explains why there is only one non-MDR cluster identified, the crucial finding still remains that almost all of our MDR/pre-XDR/XDR strains are of the Beijing lineage (14/16) and more than half of them are probably from recent transmission (8/14). In addition, three of these MDR/pre-XDR/XDR Beijing strains match the Central Asia outbreak clone and seven match the Europe/Russian W148 outbreak type (12). Altogether, these findings might indicate that MDR-TB in the region may be at least partially driven by the introduction of MDR strains from neighboring high-burden countries, such as Azerbaijan, Armenia, Afghanistan, and Pakistan. Further investigations with a large number of strains are crucial to accredit these findings.
We also focused on SNPs in genes related to resistance to first- and second-line anti-TB drugs and compared the results with available phenotypic drug susceptibility testing. Beside the canonical mutation katG S315T, we observed one INH resistant lineage 3 (Delhi/CAS) strain with an unusual mutation combination (i.e., katG N138H and ahpC t-76a) translating into the highest MIC value (64 mg/liter) in our study cohort. The effect (MIC increase or compensatory effect) of the ahpC upstream mutation remains to be further investigated. All the RIF-resistant MTBC isolates (MDR/pre-XDR/XDR or mono-RIF resistant) in our study carried the rpoB S450L mutation, consistent with previous reports from other countries (21–23). Interestingly, compensatory mutations in rpoA/C are associated with the resistance mutation rpoB S450L (24), which is in concordance with our study. In a recent review by Gygli et al., the authors concluded that strains carrying this mutation (rpoB S450L) are able to outcompete strains with other RIF-resistant mutations because of its low fitness cost. This supposedly leads to an increase of strains with the rpoB S450L mutation in the MTBC population and the acquisition of additional compensatory mutations (24). According to our results, this holds true for RIF-resistant strains circulating in Tehran. As Li et al. showed recently, more investigations are needed to evaluate an epistatic interaction of these compensatory mutations related to RIF resistance and strain fitness and their role in the spread of MDR/pre-XDR/XDR or mono-RIF-resistant strains (25). In addition, we found the rpoA V59L mutation in one RIF-susceptible strain. The detection of compensatory mutations of RIF resistance in susceptible strains is not a rare phenomenon, as Li et al. reported two RIF-susceptible strains harboring the rpoC T721C mutation (25).
Regarding FQ resistance, two pre-XDR strains with gyrA D94G were susceptible to OFX by phenotypic DST which might be a test artifact, as this mutation is associated with the highest MIC values toward the fluoroquinolones (26). Overall the gyrA D94G, D94A, A90V, and S91P mutations found in our study are correlated with OFX resistance (P < 0.05), confirming the findings reported by Chein et al. and Li et al. (27, 28). Three strains were resistant to KAN linked with mutations in the eis promoter region (eis-10 g/a) (29), although the exact resistance level (MIC increase) remains to be confirmed. Another three strains were observed with the well-known marker rrs 1401 a/g, conferring cross-resistance to AMK, KAN, and CAP. This mutation was also found in one CAP-susceptible strain which was also reported in previous studies (30). More importantly, six strains were phenotypically resistant to CAP with no mutations detected; this phenomenon strengthens the role of efflux pump overexpression in MTBC drug resistance.
To the best of our knowledge, our work represents one of the most comprehensive epidemiological studies in this region. Between January 2014 and January 2018, 606 MTB strains were isolated from 13,892 suspected pulmonary TB patients, and the rate of MDR cases (2.6%) was twice as high as the estimation the WHO (1.3%) suggested for the region (http://www.who.int/tb/country/data/profiles/en/). We also found an alarming high rate of XDR cases among all MDR patients in Tehran plus additional 50% pre-XDR cases among all MDR patients. These findings could result from the high percentage of Afghani patients referred to the laboratory. Therefore, the ethnicity and origin of the studied patients must be considered an important issue in the current study. A further concern is the high number of MDR/pre-XDR/XDR strains with additional mutations in the genes ethA, embB, and pncA linked with resistance to ETO and PTO, EMB and PZA, respectively (31, 32) which further limits the treatment options for these patients.
In conclusion, two well-known lineage 2/Beijing MDR clades are the main cause for M/XDR-TB in Tehran, in parallel to high MDR-TB incidence settings in Eastern Europe, Russia, and Central Asia. Compared with previous reports, we see a reduction of NEW-1 genotypes (L4.5.1/Iran genotype) in MDR isolates, which suggests a likely import of resistant Beijing MDR/XDR-TB strains into Iran. Thus, even moderate incidence settings, such as Iran and its capital, are at risk to encounter rapidly increasing drug resistance rates via the introduction of already highly resistant and highly transmissible MTBC strains. The high-resolution data gathered in this study should be used to guide TB surveillance and containment measures in this region. More investigations with larger sample sizes from different provinces of the country will shed more light on the phylogenetic diversity and transmission dynamics of these strains.
Supplementary Material
ACKNOWLEDGMENTS
We thank all the personnel at the Mycobacteriology and Pulmonary Research Department, Pasteur Institute of Iran, for their assistance in this project, and Vanessa Mohr and Carina Hahn from the Research Center Borstel for their excellent technical assistance in the next-generation sequencing workflow.
This work was partially funded by the German Center for Infection Research (DZIF) and the Leibniz Science Campus Evolutionary Medicine of the Lung (EvoLUNG).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.01477-18.
REFERENCES
- 1.World Health Organization. 2017. Global tuberculosis control surveillance, planning, financing. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 2.Dheda K, Barry CE III, Maartens G. 2016. Tuberculosis. Lancet 387:1211–1226. doi: 10.1016/S0140-6736(15)00151-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nguyen L. 2016. Antibiotic resistance mechanisms in M. tuberculosis: an update. Arch Toxicol 90:1585–1604. doi: 10.1007/s00204-016-1727-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE 3rd, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail MA, Rajandream MA, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston JE, Taylor K, Whitehead S, Barrell BG. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 5.Satta G, Lipman M, Smith GP, Arnold C, Kon OM, McHugh TD. 2018. Mycobacterium tuberculosis and whole-genome sequencing: how close are we to unleashing its full potential? Clin Microbiol Infect 24:604–609. doi: 10.1016/j.cmi.2017.10.030. [DOI] [PubMed] [Google Scholar]
- 6.Papaventsis D, Casali N, Kontsevaya I, Drobniewski F, Cirillo DM, Nikolayevskyy V. 2017. Whole genome sequencing of Mycobacterium tuberculosis for detection of drug resistance: a systematic review. Clin Microbiol Infect 23:61. doi: 10.1016/j.cmi.2016.09.008. [DOI] [PubMed] [Google Scholar]
- 7.Roetzer A, Diel R, Kohl TA, Rückert C, Nübel U, Blom J, Wirth T, Jaenicke S, Schuback S, Rüsch-Gerdes S, Supply P, Kalinowski J, Niemann S. 2013. Whole genome sequencing versus traditional genotyping for investigation of a Mycobacterium tuberculosis outbreak: a longitudinal molecular epidemiological study. PLoS Med 10:e1001387. doi: 10.1371/journal.pmed.1001387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stucki D, Ballif M, Bodmer T, Coscolla M, Maurer AM, Droz S, Butz C, Borrell S, Längle C, Feldmann J, Furrer H, Mordasini C, Helbling P, Rieder HL, Egger M, Gagneux S, Fenner L. 2015. Tracking a tuberculosis outbreak over 21 years: strain-specific single-nucleotide polymorphism typing combined with targeted whole-genome sequencing. J Infect Dis 211:1306–1316. doi: 10.1093/infdis/jiu601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Palomino JC, Martin A, Camacho M, Guerra H, Swings J, Portaels F. 2002. Resazurin microtiter assay plate: simple and inexpensive method for detection of drug resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother 46:2720–2722. doi: 10.1128/AAC.46.8.2720-2722.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baym M, Kryazhimskiy S, Lieberman TD, Chung H, Desai MM, Kishony R. 2015. Inexpensive multiplexed library preparation for megabase-sized genomes. PLoS One 10:e0128036. doi: 10.1371/journal.pone.0128036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coll F, McNerney R, Guerra-Assunção JA, Glynn JR, Perdigão J, Viveiros M, Portugal I, Pain A, Martin N, Clark TG. 2014. A robust SNP barcode for typing Mycobacterium tuberculosis complex strains. Nat Commun 5:4812. doi: 10.1038/ncomms5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Merker M, Blin C, Mona S, Duforet-Frebourg N, Lecher S, Willery E, Blum MG, Rüsch-Gerdes S, Mokrousov I, Aleksic E, Allix-Béguec C, Antierens A, Augustynowicz-Kopeć E, Ballif M, Barletta F, Beck HP, Barry CE III, Bonnet M, Borroni E, Campos-Herrero I, Cirillo D, Cox H, Crowe S, Crudu V, Diel R, Drobniewski F, Fauville-Dufaux M, Gagneux S, Ghebremichael S, Hanekom M, Hoffner S, Jiao WW, Kalon S, Kohl TA, Kontsevaya I, Lillebæk T, Maeda S, Nikolayevskyy V, Rasmussen M, Rastogi N, Samper S, Sanchez-Padilla E, Savic B, Shamputa IC, Shen A, Sng LH, Stakenas P, Toit K, Varaine F, Vukovic D, Wahl C, Warren R, Supply P, Niemann S, Wirth T. 2015. Evolutionary history and global spread of the Mycobacterium tuberculosis Beijing lineage. Nat Genet 47:242–249. doi: 10.1038/ng.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walker TM, Ip CL, Harrell RH, Evans JT, Kapatai G, Dedicoat MJ, Eyre DW, Wilson DJ, Hawkey PM, Crook DW, Parkhill J, Harris D, Walker AS, Bowden R, Monk P, Smith EG, Peto TE. 2013. Whole-genome sequencing to delineate Mycobacterium tuberculosis outbreaks: a retrospective observational study. Lancet Infect Dis 13:137–146. doi: 10.1016/S1473-3099(12)70277-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Comas I, Borrell S, Roetzer A, Rose G, Malla B, Kato-Maeda M, Galagan J, Niemann S, Gagneux S. 2011. Whole-genome sequencing of rifampicin-resistant Mycobacterium tuberculosis strains identifies compensatory mutations in RNA polymerase genes. Nat Genet 44:106–110. doi: 10.1038/ng.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Casali N, Nikolayevskyy V, Balabanova Y, Harris SR, Ignatyeva O, Kontsevaya I, Corander J, Bryant J, Parkhill J, Nejentsev S, Horstmann RD, Brown T, Drobniewski F. 2014. Evolution and transmission of drug-resistant tuberculosis in a Russian population. Nat Genet 46:279–286. doi: 10.1038/ng.2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tarashi S, Fateh A, Jamnani FR, Siadat SD, Vaziri F. 2017. Prevalence of Beijing and Haarlem genotypes among multidrug-resistant Mycobacterium tuberculosis in Iran: systematic review and meta-analysis. Tuberculosis (Edinb) 107:31–37. doi: 10.1016/j.tube.2017.03.005. [DOI] [PubMed] [Google Scholar]
- 17.Drobniewski F, Rüsch-Gerdes S, Hoffner S. 2007. Subcommittee on Antimicrobial Susceptibility Testing of Mycobacterium tuberculosis of the European Committee for Antimicrobial Susceptibility Testing (EUCAST) of the European Society of Clinical Microbiology and Infectious Diseases (ESCMID). Antimicrobial susceptibility testing of Mycobacterium tuberculosis (EUCAST document E.DEF 8.1)—report of the Subcommittee on Antimicrobial Susceptibility Testing of Mycobacterium tuberculosis of the European Committee for Antimicrobial Susceptibility Testing (EUCAST) of the European Society of Clinical Microbiology and Infectious Diseases (ESCMID). Clin Microbiol Infect 13:1144–1156. doi: 10.1111/j.1469-0691.2007.01813.x. [DOI] [PubMed] [Google Scholar]
- 18.Khanipour S, Ebrahimzadeh N, Masoumi M, Sakhaei F, Alinezhad F, Safarpour E, Fateh A, Nour Nematollahi A, Hadizadeh Tasbiti A, Zolfaghari MR, Bahrmand AR, Mirsaeidi M, Rahimi Jamnani F, Vaziri F, Siadat SD. 2016. Haarlem 3 is the predominant genotype family in multidrug-resistant and extensively drug-resistant Mycobacterium tuberculosis in the capital of Iran: a 5-year survey. J Glob Antimicrob Resist 5:7–10. doi: 10.1016/j.jgar.2016.01.007. [DOI] [PubMed] [Google Scholar]
- 19.Mokrousov I. 2016. Emerging resistant clone of Mycobacterium tuberculosis in west Asia. Lancet Infect Dis 16:1326–1327. doi: 10.1016/S1473-3099(16)30460-1. [DOI] [PubMed] [Google Scholar]
- 20.Mokrousov I, Shitikov E, Skiba Y, Kolchenko S, Chernyaeva E, Vyazovaya A. 2017. Emerging peak on the phylogeographic landscape of Mycobacterium tuberculosis in West Asia: definitely smoke, likely fire. Mol Phylogenet Evol 116:202–212. doi: 10.1016/j.ympev.2017.09.002. [DOI] [PubMed] [Google Scholar]
- 21.Eldholm V, Monteserin J, Rieux A, Lopez B, Sobkowiak B, Ritacco V, Balloux F. 2015. Four decades of transmission of a multidrug-resistant Mycobacterium tuberculosis outbreak strain. Nat Commun 6:7119. doi: 10.1038/ncomms8119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ali A, Hasan Z, McNerney R, Mallard K, Hill-Cawthorne G, Coll F, Nair M, Pain A, Clark TG, Hasan R. 2015. Whole genome sequencing based characterization of extensively drug-resistant Mycobacterium tuberculosis isolates from Pakistan. PLoS One 10:e0117771. doi: 10.1371/journal.pone.0117771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar S, Jena L. 2014. Understanding rifampicin resistance in tuberculosis through a computational approach. Genomics Inform 12:276–282. doi: 10.5808/GI.2014.12.4.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gygli SM, Borrell S, Trauner A, Gagneux S. 2017. Antimicrobial resistance in Mycobacterium tuberculosis: mechanistic and evolutionary perspectives. FEMS Microbiol Rev 41:354–373. doi: 10.1093/femsre/fux011. [DOI] [PubMed] [Google Scholar]
- 25.Li QJ, Jiao WW, Yin QQ, Xu F, Li JQ, Sun L, Xiao J, Li YJ, Mokrousov I, Huang HR, Shen AD. 2016. Compensatory mutations of rifampin resistance are associated with transmission of multidrug-resistant Mycobacterium tuberculosis Beijing genotype strains in China. Antimicrob Agents Chemother 60:2807–2812. doi: 10.1128/AAC.02358-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nosova EY, Bukatina AA, Isaeva YD, Makarova MV, Galkina KY, Moroz AM. 2013. Analysis of mutations in the gyrA and gyrB genes and their association with the resistance of Mycobacterium tuberculosis to levofloxacin, moxifloxacin and gatifloxacin. J Med Microbiol 62:108–113. doi: 10.1099/jmm.0.046821-0. [DOI] [PubMed] [Google Scholar]
- 27.Chien JY, Chiu WY, Chien ST, Chiang CJ, Yu CJ, Hsueh PR. 2016. Mutations in gyrA and gyrB among fluoroquinolone- and multidrug-resistant Mycobacterium tuberculosis isolates. Antimicrob Agents Chemother 60:2090–2096. doi: 10.1128/AAC.01049-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li J, Gao X, Luo T, Wu J, Sun G, Liu Q, Jiang Y, Zhang Y, Mei J, Gao Q. 2014. Association of gyrA/B mutations and resistance levels to fluoroquinolones in clinical isolates of Mycobacterium tuberculosis. Emerg Microbes Infect 3:e19. doi: 10.1038/emi.2014.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zaunbrecher MA, Sikes RD Jr, Metchock B, Shinnick TM, Posey JE. 2009. Overexpression of the chromosomally encoded aminoglycoside acetyltransferase eis confers kanamycin resistance in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 106:20004–20009. doi: 10.1073/pnas.0907925106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Georghiou SB, Magana M, Garfein RS, Catanzaro DG, Catanzaro A, Rodwell TC. 2012. Evaluation of genetic mutations associated with Mycobacterium tuberculosis resistance to amikacin, kanamycin and capreomycin: a systematic review. PLoS One 7:e33275. doi: 10.1371/journal.pone.0033275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang F, Langley R, Gulten G, Dover LG, Besra GS, Jacobs WR Jr, Sacchettini JC. 2007. Mechanism of thioamide drug action against tuberculosis and leprosy. J Exp Med 204:73–78. doi: 10.1084/jem.20062100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Caminero JA, Sotgiu G, Zumla A, Migliori GB. 2010. Best drug treatment for multidrug-resistant and extensively drug-resistant tuberculosis. Lancet Infect Dis 10:621–629. doi: 10.1016/S1473-3099(10)70139-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.