This study aimed to validate a comprehensive diagnostic protocol based on real-time PCR for the rapid detection and identification of Bordetella pertussis, Bordetella parapertussis, and Bordetella holmesii, as well as its implementation in the diagnostic routine of a reference children’s hospital. The new algorithm included a triplex quantitative PCR (qPCR) targeting IS481 gene (in B. pertussis, B. holmesii, and some Bordetella bronchiseptica strains), pIS1001 (B. parapertussis-specific) and rnase P as the human internal control.
KEYWORDS: B. holmesii, B. parapertussis, Bordetella pertussis, real-time PCR, whooping cough
ABSTRACT
This study aimed to validate a comprehensive diagnostic protocol based on real-time PCR for the rapid detection and identification of Bordetella pertussis, Bordetella parapertussis, and Bordetella holmesii, as well as its implementation in the diagnostic routine of a reference children’s hospital. The new algorithm included a triplex quantitative PCR (qPCR) targeting IS481 gene (in B. pertussis, B. holmesii, and some Bordetella bronchiseptica strains), pIS1001 (B. parapertussis-specific) and rnase P as the human internal control. Two confirmatory singleplex tests for B. pertussis (ptxA-Pr) and B. holmesii (hIS1001) were performed if IS481 was positive. Analytical validation included determination of linear range, linearity, efficiency, precision, sensitivity, and a reference panel with clinical samples. Once validated, the new algorithm was prospectively implemented in children with clinical suspicion of whooping cough presenting to Hospital Sant Joan de Deu (Barcelona, Spain) over 12 months. Lower limits of detection obtained were 4.4, 13.9, and 27.3 genomic equivalents/ml of sample for IS481 (on B. pertussis), pIS1001 and hIS1001, and 777.9 for ptxA-Pr. qPCR efficiencies ranged from 86.0% to 96.9%. Intra- and interassay variabilities were <3% and <5%, respectively. Among 566 samples analyzed, B. pertussis, B. holmesii, and B. parapertussis were detected in 11.1%, 0.9% (only in females >4 years old), and 0.2% of samples, respectively. The new algorithm proved to be a useful microbiological diagnostic tool for whooping cough, demonstrating a low rate of other non-pertussis Bordetella species in our surveilled area.
INTRODUCTION
Pertussis is a vaccine-preventable acute respiratory disease primarily caused by Bordetella pertussis (1). Infants younger than 6 months are at higher risk to suffer from severe illness, hospitalization, and even fatal outcome (2, 3). Other, less prevalent Bordetella species, such as Bordetella parapertussis, Bordetella holmesii, and Bordetella bronchiseptica, can also produce pertussis-like illness.
In the last years and despite extensive vaccination programs, the resurgence of whooping cough has been documented worldwide. One possible explanation could be the replacement of B. pertussis by other Bordetella species (4, 5). This fact highlights the need of using precise diagnosis methods capable of identifying the etiological agent of the disease. An accurate identification at the species level is not only important from a clinical point of view to select the most appropriate antibiotic treatment, but also for health public purposes, since misdiagnosis of Bordetella species can lead to an incorrect assessment of pertussis vaccine effectiveness (6).
For this purpose, a number of microbiological techniques are readily available, such as culture, serology, and nucleic acid amplification tests (NAATs) (7). Although culture remains the gold standard, it has low sensitivity (8, 9). Serology is not an appropriate method to diagnose pertussis in pediatric populations, since it provides results that are difficult to interpret in immunized individuals and requires measuring antibody titers in the acute and convalescent phases of the disease, thus delaying time to result (10). Rapid, sensitive, and specific NAATs are being increasingly implemented to overcome the limitations of culture and serology (9, 11).
NAATs targeting IS481, at high copy number in the genome of B. pertussis, and IS1001 for B. parapertussis are commonly used (12, 13). However, both targets are also present in B. holmesii and some B. bronchiseptica strains (14). Several algorithms combine nonspecific targets (IS481, IS1001 or IS1002) and may include one or up to two specific targets for B. pertussis and B. holmesii (see Table S1). To our knowledge, only two published methods have reported the use of specific targets for the three most relevant Bordetella species (15, 16).
The present study aimed to adapt, optimize, and validate a diagnostic algorithm for the rapid detection and identification of B. pertussis, B. parapertussis, and B. holmesii. In addition, we sought to assess the disease burden caused by these species in our region by implementing the algorithm in pediatric patients suspected of pertussis.
(This work was presented in part at the 27th European Congress of Clinical Microbiology and Infectious Diseases, Vienna, Austria, 2017 [17].)
MATERIALS AND METHODS
Study design and setting.
Nasopharyngeal aspirates (NPAs) were prospectively collected from children and adolescents <18 years with clinical suspicion of whooping cough (according to CDC criteria), that were attended in Hospital Sant Joan de Deu (HSJD) between May 2016 and April 2017. This is a pediatric referral hospital that provides medical care services to more than 300,000 children in Catalonia (Spain). Information on age and sex variables of the patients was recorded for epidemiological purposes.
Sample collection and DNA extraction.
Nasopharyngeal aspirates were processed according to the protocol established at the clinical laboratory of the study site (18). Specimens showing poor quality (rnase P > 35 cycle thresholds [CT]) or weak IS481 positivity (40 > CT > 35) were subjected to additional DNA extraction using NucliSENS easyMag (bioMérieux, France), from an initial volume of 200 µl eluted into 25 µl.
qPCR reference method.
The standardized laboratory method of Hospital Sant Joan de Déu (HSJD) consisted of a duplex quantitative PCR (qPCR) that included hydrolysis probes (Roche Diagnostics GmbH, Germany) targeting IS481 and the human rnase P gene as a positive internal control for testing sample quality, as described in Brotons et al. (19) (Table 1). Delta Rn (dRn) CT values were manually set at 0.2 for both targets.
TABLE 1.
List of oligonucleotides and probes used in the multiplex and confirmatory qPCRs
| Target | Named | Sequencesc | Final concn (nM) | Reference or source |
|---|---|---|---|---|
| IS481 | IS481-Ref Fwd | TCCGAACCGGATTTGAGAAAC | 900 | 19 |
| IS481-Ref Rev | GTCGACGTAGGAAGGTCAATCG | 900 | ||
| IS481-Ref Probe | 6-FAM-CGCCAACCCCCCAGTTCACTCA-TAMRA | 300 | ||
| IS481 Fwd | CAAGGCCGAACGCTTCAT | 770 | 15 | |
| IS481 Rev | GAGTTCTGGTAGGTGTGAGCGTAA | 770 | ||
| IS481 Probea | 6-FAM-CAGTCGGCCTTGCGTGAGTGGG-BHQ1 | 260 | ||
| rnase P | rnase P Fwd | CCAAGTGTGAGGGCTGAAAAG | 150 | 15 |
| rnase P Rev | TGTTGTGGCTGATGAACTATAAAAGG | 150 | ||
| rnase P Probe | YAK-CCCCAGTCTCTGTCAGCACTCCCTTC-BHQ1 | 250 | ||
| IS1001 | pIS1001 Fwd | TCGAACGCGTGGAATGG | 600 | 15 |
| pIS1001 Rev | GGCCGTTGGCTTCAAATAGA | 600 | ||
| pIS1001 Probeb | Cy3-AGACCCAGGGCGCACGCTGTC-BHQ2 | 200 | ||
| hIS1001 Fwd | GGCGACAGCGAGACAGAATC | 900 | 15 | |
| hIS1001 Rev | GCCGCCTTGGCTCACTT | 900 | ||
| hIS1001 Probeb | Cy3-CGTGCAGATAGGCTTTTAGCTTGAGCGC-BHQ2 | 300 | ||
| ptxA-Pr | ptxA-Pr Fwd | CGCCAAGCTGAAGTAGCA | 900 | 11 |
| ptxA-Pr Rev | AAGGAGCGTTCATGCCG | 900 | ||
| ptxA-Pr Probe | 6-FAM-AGAATCGAGGGTTTTGTACGACGAATC-BBQ | 300 | This study |
Original fluorophore was modified for multiplexing.
Original fluorophore and quencher were modified for multiplexing.
Fluorophores and quenchers of hydrolysis probes are underlined. 6-FAM, 6-carboxyfluorescein; TAMRA, 6-carboxytetramethylrhodamine; BBQ, BlackBerry quencher; Cy3, cyanine 3; BHQ1, black hole quencher 1; BHQ2, black hole quencher 2; YAK, Yakima Yellow.
Ref, reference; Fwd, forward; Rev, reverse.
Samples yielding an IS481 CT of <35 were considered probable B. pertussis isolates, inferred from the high copy numbers of such targets in this species (estimated in 50 to 200) (20). Samples with a CT value of 35 to 40 were reported as Bordetella spp., and as negative if CT was >40.
New multiplex qPCR and diagnostic algorithm.
Previously published protocols for diagnosis of whooping cough were reviewed for designing the proposed algorithm (see Table S1). The method by Tatti et al. (15) was selected as the most complete and accurate basis for designing the new diagnostic algorithm, in relation to the number of species covered and the use of the human rnase P gene as the positive control (although not multiplexed). Its confirmatory target for B. pertussis, ptxS1, however, has been reported as cross-reactive with some strains of B. bronchiseptica (8, 21), so it was replaced by ptxA-Pr (11). The final design of the algorithm included three sequential qPCR assays for the specific identification of B. pertussis, B. parapertussis, and B. holmesii. The first triplex qPCR included the targets IS481, pIS1001 (B. parapertussis-specific) (13, 15), and the human rnase P (Table 1). If IS481 was positive, two confirmatory singleplex qPCRs were performed, ptxA-Pr for B. pertussis identification (21) and hIS1001 for B. holmesii (Table 2) (22).
TABLE 2.
Diagnostic algorithm used for the DNA detection and identification of Bordetella species
| Species | qPCR result for |
|||
|---|---|---|---|---|
| Multiplexa |
Singleplex ptxA-Pr | Singleplex hIS1001 | ||
| IS481 | pIS1001 | |||
| B. pertussis | + | − | + | − |
| B. parapertussis | − | + | − | − |
| B. holmesii | + | − | − | + |
| Bordetella spp. | + | − | − | − |
rnase P was used as the positive control. +, positive; −, negative.
Composition of the qPCR reactions only varied from the reference method in the concentrations and sequences of oligonucleotides, probes, and the reagents used for rnase P detection (Table 1). The ptxA-Pr probe was adapted by TIB-Molbiol (Berlin, Germany) to universal amplification conditions. dRn CT values were set at 0.2 for 6-carboxyfluorescein (6-FAM) or Yakima Yellow (YAK) probes and at 0.1 for cyanine 3 (Cy3).
Analytical validation.
Bacterial strains used were B. pertussis CECT 7974, B. parapertussis ATCC 15311, and B. holmesii ATCC 51541. DNA standards were prepared from bacterial suspensions in phosphate-buffered saline solution (PBS). DNA was extracted using NucliSENS easyMag, and concentrations were quantified with the UV-visible (UV-Vis) spectrophotometer Q500 (Quawell, USA). Genome equivalents (GE) of standards were estimated assuming the molecular size of B. pertussis Tohama I (Genbank accession number BX470248; 4,086,190 bp), B. parapertussis Bpp5 (Genbank accession number HE965803; 4,800,120 bp), and B. holmesii ATCC 51541 (Genbank accession number CP007494; 3,699,670 bp).
DNA standards were freshly prepared, by 10-fold dilutions ranging from 106 to 10° GE/ml of sample (104 to 10−2 GE/reaction and 105 to 10−1 fg DNA/reaction). Linear range and intra-assay variability were estimated by testing each dilution in triplicate on the same day, and consensus curves were used for calculating the efficiencies. Interassay variability was estimated with two additional replicates on successive days. Precision was acceptable if the mean coefficients of variation (CV) were ≤3% and ≤5% for intra-assay and interassay, respectively.
Analytical specificity was tested on a panel of 11 bacterial species, including Enterococcus faecalis, Escherichia coli, Haemophilus influenzae, Kingella kingae, Moraxella catarrhalis, Neisseria meningitidis, Pseudomonas aeruginosa, Staphylococcus aureus, Streptococcus agalactiae, Streptococcus mitis clinical isolates, and the Streptococcus pneumoniae R6.
In addition, a reference panel with clinical samples previously processed by the reference method was also tested by the new triplex qPCR. The purpose of this validation was to compare the performance of the two techniques targeting IS481 (the gene shared by the two methods) in nasopharyngeal matrices. NPAs were collected between December 2015 and April 2016 from children <1 year old with suspected pertussis. All IS481-positive samples by the reference duplex qPCR (n = 22), in addition to 22 negative samples gathered during this period, were subsequently analyzed by the proposed algorithm. Samples were stored frozen at −80°C between both analyses.
Statistical analysis.
Equations for the multiplex and singleplex qPCRs were calculated by plotting log10-transformed GE versus CT values. Lower limits of detection (LLOD) were estimated using probit regression analysis at 95% probability (see Table S2). Diagnostic sensitivity and specificity values were calculated as reported elsewhere (23).
Continuous variables were described as mean (standard deviation [SD]) or median values (interquartile range [IQR]). Parametric and nonparametric analyses were performed using a t test and the Mann-Whitney U test, respectively. Categorical data were analyzed by χ2 or Fisher’s exact test. Confidence intervals (CI) were set at 95% and significance at a two-sided P value of <0.05. All statistical analyses were performed with SPSS v.22 software (IBM Corp., USA), except for diagnostic sensitivity and specificity values, which were calculated with MedCalc Statistical Software v.17.6 (MedCalc Software Bvba, Belgium).
RESULTS
Analytical validation.
Linear ranges obtained spanned from 106 to 101 GE/ml of sample (r2 > 0.99). Efficiencies ranged from 86.0% to 96.9% (Table 3). LLOD for IS481 were 4.4 and 60.3 GE/ml of sample for B. pertussis and B. holmesii, respectively. Target pIS1001 showed a LLOD of 13.9 GE/ml of sample, and singleplex reactions hIS1001 and ptxA-Pr showed LLOD of 27.3 and 777.9 GE/ml of sample, respectively (Table 3 and Table S2).
TABLE 3.
New qPCR performances for each target and Bordetella species
| qPCR type | Target | Bacteria | Target copy no.b |
Curvea
|
Precision (CV %) |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Linear rangec | Sloped | Intercept | Efficiency (%) |
r2 | LLOD | Intra- assay |
Interassay | ||||
| Multiplex | IS481 | B. pertussis | 50 to 200 | 1.0 × 106 to 1.0 × 101 | –3.58 | 40.55 | 90.3 | 0.999 | 4.4 × 100 | 0.48 | 0.69 |
| B. holmesii | 8 to 10 | 0.8 × 106 to 0.8 × 102 | –3.71 | 44.36 | 86.0 | 0.999 | 6.0 × 101 | 0.63 | 1.75 | ||
| pIS1001 | B. parapertussis | ∼20 | 2.2 × 106 to 2.2 × 101 | –3.40 | 42.49 | 96.9 | 0.999 | 1.4 × 101 | 0.32 | 0.58 | |
| Singleplex | ptxA-Pr | B. pertussis | 1 | 1.0 × 106 to 1.0 × 103 | –3.51 | 49.01 | 92.8 | 1.000 | 7.8 × 102 | 0.94 | 1.35 |
| Singleplex | hIS1001 | B. holmesii | 3 to 5 | 0.6 × 106 to 0.6 × 102 | –3.50 | 43.72 | 92.9 | 0.998 | 2.7 × 101 | 0.67 | 0.81 |
Plotting CT versus log10 genome equivalents (GE) per ml of sample.
Number of copies present in the genomes of the different Bordetella species (10).
Genome equivalents of the specified bacteria per ml of sample. The DNA concentration range for each Bordetella species is as follows: B. pertussis, 0.9 × 105 to 0.9 × 10−1 fg/reaction (2.0 × 104 to 2.0 × 10−2 GE/reaction); B. parapertussis, 2.3 × 105 to 2.3 × 10−1 fg/reaction (4.4 × 104 to 4.4 × 10−2 GE/reaction); B. holmesii, 1.7 × 105 to 1.7·10−1 fg/reaction (1.6 × 104 to 1.6 × 10−2 GE/reaction) and 5.3 × 105 to 5.3 × 10−1 fg/reaction (1.3 × 104 to 1.3 × 10−2 GE/reaction) for multiplex and hIS1001, respectively.
The symbol (−) indicates negative values. LLOD, lower limit of detection; CV, coefficient of variation.
Precision estimates (CV) were <3% for all reactions, with 0.32% to 0.94% and 0.58% to 1.75% values for intra-assay and interassay, respectively (Table 3). Results of the specificity panel showed the four Bordetella targets to be genus specific, and the confirmatory targets pIS1001, ptxA-Pr, and hIS1001 were species specific.
The triplex qPCR correctly diagnosed all positive and negative samples of the reference panel of clinical samples. Sensitivity and specificity values for IS481 were both 100% (95% CI, 84.6 to 100.0% and 83.2 to 100.0%, respectively). Mean CT for IS481 in the reference and multiplex reactions were similar despite a freeze-thaw cycle between tests, with a mean difference between paired samples of 0.94 CT (95% CI, −0.40 to 2.78, P value = 0.159).
Burden of disease caused by B. pertussis, B. parapertussis, and B. holmesii.
During the study period, 578 NPAs were collected, from which 9 were excluded, as they were either processed by a different technique (n = 2), invalid samples (n = 1), or did not meet age inclusion criteria (n = 6). In addition, 3 samples showed poor quality (rnase P > 35 CT) and were also disregarded. A total of 566 samples was finally included in the study. Of them, 484 (85.5%) were negative for the targets IS481 or pIS1001. Eighty-two samples (14.5%) were Bordetella positive by either IS481 (n = 81, 98.8%) or pIS1001 (n = 1, 1.2%). The ptxA-Pr qPCR confirmed B. pertussis infection in 63 samples (76.8% of positives), and hIS1001 identified B. holmesii in five samples (6.1% of positives), one of them also coinfected with B. pertussis. The remaining 13 positive samples (15.9%) could not be identified at the species level and were reported as Bordetella sp. infection (CT values = 30.65 to 38.17).
A seasonal distribution of positive samples was observed, showing a higher incidence during warmer months (P value < 0.001) (Fig. 1), 62.2% of them identified within May to July (n = 51). B. pertussis was the most frequently detected species, while B. holmesii and B. parapertussis only circulated during the seasonal peak.
FIG 1.
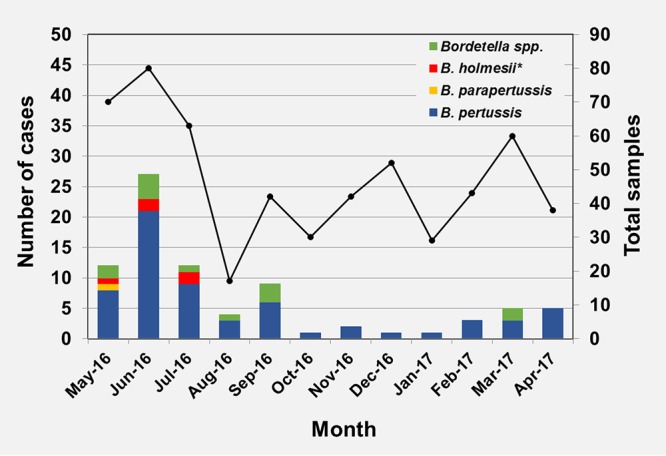
Seasonal distribution of microbiologically confirmed whooping cough cases by etiological agent (n = 566), includes one coinfection of B. holmesii and B. pertussis (*).
The median age of patients was 1.3 years (IQR, 0.24 to 5.85), and ages ranged from 7 days to 17.5 years. The distribution of B. pertussis was homogeneous across age groups, whereas B. holmesii was only detected in five children aged between 4.6 and 9.9 years, and B. parapertussis was only detected in an infant that was two months old (Fig. 2).
FIG 2.
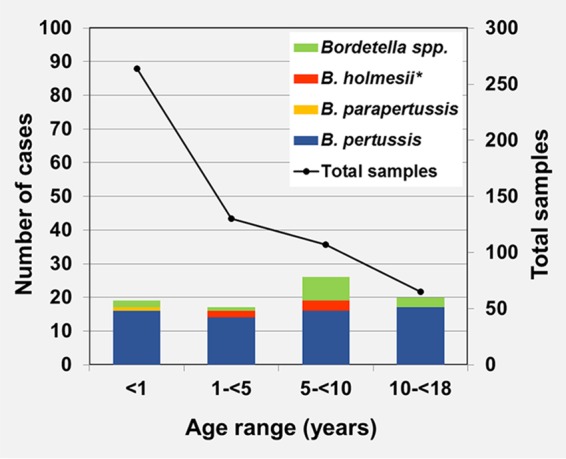
Age distribution of microbiologically confirmed cases and proportion of Bordetella species (n = 566). Includes one coinfection of B. holmesii and B. pertussis (*).
A remarkable difference in the positive rate was observed by age, with the infants younger than 1 year group showing the lowest positivity rate (7.2%). This rate increased with the age of patients (Fig. 2).
No differences in positivity rates were observed by gender, with proportions of 14.8% and 14.2% in males and females, respectively (P value = 0.857). In contrast, B. holmesii seemed to show a differential distribution, being only found in females (n = 5, P value = 0.06). B. parapertussis was only detected in a specimen taken from a male patient.
DISCUSSION
The present study proposed a rapid and easy-to-use protocol based on three qPCR assays for specific DNA detection of B. pertussis, B. holmesii, and B. parapertussis. The multicopy targets IS481, pIS1001, and hIS1001 were shown to be very sensitive, with LLOD values lower than 70 GE/ml of sample. The single-copy target ptxA-Pr had moderate but acceptable sensitivity, being able to detect <103 GE/ml of sample. qPCR efficiencies were above 90.0% for all targets except for hIS1001 (86%), all of them within the acceptability limits for qualitative methods (80.0% to 120.0%) (23). This simple and rapid multiplexed algorithm allowed the specific identification of the three Bordetella spp. in less than five hours.
The validated algorithm was implemented during 12 months at the study site. The most predominant species was B. pertussis, followed by B. holmesii and B. parapertussis. To date, there are few published European studies investigating the occurrence of other non-pertussis Bordetella infections. Retrospective studies performed in four European countries between 1992 and 2012 identified B. pertussis and B. parapertussis in 82.6 to 97.0% and 0.0 to 17.4% of the positive samples, respectively (5, 24–26). None of these investigations reported B. holmesii infections. However, two recent retrospective studies from 2013 to 2016 detected B. holmesii in 1.1 to 4.1% of positive samples and B. parapertussis in 0.3 to 8.2% of positive samples (27, 28). In addition, a study in France described a very high prevalence, up to 20%, of B. holmesii in adolescents and adults with pertussis-like symptoms (29). Our findings, in agreement with Mir-Cros et al. (28), confirm that B. holmesii is currently circulating in our region, and they could denote its increase as a causative agent of pertussis-like disease.
The incidence of pertussis in Spain has increased in all age groups despite the high levels of vaccination coverage (2). In our study, B. pertussis was shown to be evenly distributed among all ages, while B. holmesii was only detected in children >4 years old. This result is concordant with the higher prevalence of B. holmesii in symptomatic adolescents and adults that was previously reported in several studies (16, 29, 30).
A seasonal distribution of B. pertussis was observed, with higher occurrence in spring and summer, in line with the epidemiological trends of pertussis in Spain and Europe (31, 32). Although data of B. parapertussis and B. holmesii incidence rates presented in this study is limited, it appears to agree with literature suggesting cocirculation with B. pertussis, also supported by a remarkable number of B. pertussis-B. holmesii coinfections reported (16, 33, 34).
Bordetella infections were equally frequent in both sexes, as described by others (31). Interestingly, B. holmesii infection was only detected in females, although the low number of cases registered does not allow us to reach further conclusions on this potential association.
One study limitation was the lack of identification to species level in 15.9% of IS481-positive samples. Those Bordetella spp. likely corresponded to B. pertussis, in which ptxA-Pr qPCR results were negative due to low bacterial load or, less probably but possibly, to B. bronchiseptica strains for which no specific gene was investigated.
In conclusion, the new algorithm allowed improvement of accuracy of microbiological diagnosis of whooping cough at the study site by enhancing specificity while maintaining high sensitivity levels. In addition, we assessed the incidence of Bordetella spp. among the pediatric population of the geographical region of Catalonia during the algorithm implementation period. According to our data, the circulation of non-pertussis Bordetella species in this region seems to be minor and associated with seasonal increase of B. pertussis. Despite not representing a significant contribution to pertussis disease burden, it is essential to monitor the epidemiological patterns of these species to conduct an appropriate surveillance of the disease in our region. Nevertheless, our local data may not necessarily reflect the epidemiological status of Bordetella species in other areas, since differences in the species circulation can be highly influenced according to geographical and time variations (35). Therefore, local evaluations of diagnostic algorithms based on species-specific primers should carefully be undertaken before their implementation in any particular region.
Ethical considerations.
This study was approved by the ethics committee of HSJD, in conformity with the Helsinki Declaration of 1975 (revised in 2000); the Spanish Organic Law 15/1999, on December 13th, on data protection; and law 14/2007, on July 3rd, on biomedical research. For the present study, no informed consent was requested, as this is a population-based study in which there were no activities that could compromise laboratory performance, and samples were duly anonymized.
Supplementary Material
ACKNOWLEDGMENTS
A.V.-R. reports a grant received from Spanish Ministry of Economy and Competitiveness and the European Regional Development Fund (FEDER, Una manera de hacer Europa) (PI16/00247), and personal fees from European Centre for Disease Prevention and Control (“Pertinent” project). She also received financial support from bioMérieux S.A. for presenting part of this work at the 27th ECCMID Congress in Vienna (Austria), in 2017. C.M.-A. reports grants from European Centre for Disease Prevention and Control (“Pertinent” project), from Liliana Godia Foundation, and from CIBER of Epidemiology and Public Health (CIBERESP) during the conduct of the study. D.H., L.A., M.J., I.J., and P.G. have nothing to disclose.
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
We acknowledge the Department of Molecular Microbiology of HSJD for its significant cooperation and input into the implementation of the new diagnostic protocol. It is worth mentioning the valuable contributions of the “Pertinent” team (ECDC, EpiConcept and participant sites) during the project discussions when designing the diagnostic protocol. We also acknowledge Pedro Brotons for writing assistance and proofreading of the final text of the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.01231-18.
REFERENCES
- 1.Mattoo S, Cherry JD. 2005. Molecular pathogenesis, epidemiology, and clinical manifestations of respiratory infections due to Bordetella pertussis and other Bordetella subspecies. Clin Microbiol Rev 18:326–382. doi: 10.1128/CMR.18.2.326-382.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Centro Nacional de Epidemiología. 2016. Situación de la tos ferina en España, 2005–2016. Centro Nacional de Epidemiología, Madrid, Spain. [Google Scholar]
- 3.Centers for Disease Control and Prevention. 2015. Epidemiology and prevention of vaccine-preventable diseases, 13th ed Public Health Foundation, Washington, DC. [Google Scholar]
- 4.David S, van Furth R, Mooi FR. 2004. Efficacies of whole cell and acellular pertussis vaccines against Bordetella parapertussis in a mouse model. Vaccine 22:1892–1898. doi: 10.1016/j.vaccine.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 5.Pittet LF, Emonet S, François P, Bonetti E-J, Schrenzel J, Hug M, Altwegg M, Siegrist C-A, Posfay-Barbe KM. 2014. Diagnosis of whooping cough in Switzerland: differentiating Bordetella pertussis from Bordetella holmesii by polymerase chain reaction. PLoS One 9:e88936. doi: 10.1371/journal.pone.0088936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pittet LF, Emonet S, Schrenzel J, Siegrist C-A, Posfay-Barbe KM. 2014. Bordetella holmesii: an under-recognised Bordetella species. Lancet Infect Dis 14:510–519. doi: 10.1016/S1473-3099(14)70021-0. [DOI] [PubMed] [Google Scholar]
- 7.van der Zee A, Schellekens JFP, Mooi FR. 2015. Laboratory diagnosis of pertussis. Clin Microbiol Rev 28:1005–1026. doi: 10.1128/CMR.00031-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kolodkina V, Martinov V, Babenko A. 2014. Multiplex real-time PCR assay for detection and differentiation of Bordetella pertussis and Bordetella parapertussis. Iran J Microbiol 6:140–148. [PMC free article] [PubMed] [Google Scholar]
- 9.Lee AD, Cassiday PK, Pawloski LC, Tatti KM, Martin MD, Briere EC, Lucia Tondella M, Martin SW. 2018. Clinical evaluation and validation of laboratory methods for the diagnosis of Bordetella pertussis infection: Culture, polymerase chain reaction (PCR) and anti-pertussis toxin IgG serology (IgG-PT). PLoS One 13:e0195979. doi: 10.1371/journal.pone.0195979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guillot S, Guiso N, Riffelmann M, Wirsing Von König CH. 2014. Laboratory manual for the diagnosis of whooping cough caused by Bordetella pertussis/Bordetella parapertussis, update 2014. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 11.Fry NK, Duncan J, Wagner K, Tzivra O, Doshi N, Litt DJ, Crowcroft N, Miller E, George RC, Harrison TG. 2009. Role of PCR in the diagnosis of pertussis infection in infants: 5 years’ experience of provision of a same-day real-time PCR service in England and Wales from 2002 to 2007. J Med Microbiol 58:1023–1029. doi: 10.1099/jmm.0.009878-0. [DOI] [PubMed] [Google Scholar]
- 12.Glare EM, Paton JC, Premier RR, Lawrence AJ, Nisbet IT. 1990. Analysis of a repetitive DNA sequence from Bordetella pertussis and its application to the diagnosis of pertussis using the polymerase chain reaction. J Clin Microbiol 28:1982–1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van der Zee A, Agterberg C, van Agterveld M, Peeters M, Mooi FR. 1993. Characterization of IS1001, an insertion sequence element of Bordetella parapertussis. J Bacteriol 175:141–147. doi: 10.1128/jb.175.1.141-147.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tizolova A, Guiso N, Guillot S. 2013. Insertion sequences shared by Bordetella species and implications for the biological diagnosis of pertussis syndrome. Eur J Clin Microbiol Infect Dis 32:89–96. doi: 10.1007/s10096-012-1718-3. [DOI] [PubMed] [Google Scholar]
- 15.Tatti KM, Sparks KN, Boney KO, Tondella ML. 2011. Novel multitarget real-time PCR assay for rapid detection of Bordetella species in clinical specimens. J Clin Microbiol 49:4059–4066. doi: 10.1128/JCM.00601-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rodgers L, Martin SW, Cohn A, Budd J, Marcon M, Terranella A, Mandal S, Salamon D, Leber A, Tondella ML, Tatti K, Spicer K, Emanuel A, Koch E, McGlone L, Pawloski L, Lemaile-Williams M, Tucker N, Iyer R, Clark TA, Diorio M. 2013. Epidemiologic and laboratory features of a large outbreak of pertussis-like illnesses associated with cocirculating Bordetella holmesii and Bordetella pertussis—Ohio, 2010-2011. Clin Infect Dis 56:322–331. doi: 10.1093/cid/cis888. [DOI] [PubMed] [Google Scholar]
- 17.Valero-Rello A, Henares-Bonilla D, Jordan García Y, Carmona Parcerisa G, Perez-Argüello A, Acosta L, Jané MM-AC. 2017. Validation and implementation of a diagnostic algorithm for the detection of DNA from Bordetella pertussis, B. parapertussis and B. holmesii, based on sequential multiplex and singleplex qPCRs, abstr P0089. Abstr 27th European Congress of Clinical Microbiology and Infectious Diseases, Vienna, Austria, 22 to 25 April 2017. [Google Scholar]
- 18.Brotons P, De Paz HD, Esteva C, Latorre I, Muñoz-Almagro C. 2016. Validation of a loop-mediated isothermal amplification assay for rapid diagnosis of pertussis infection in nasopharyngeal samples. Expert Rev Mol Diagn 16:125–130. doi: 10.1586/14737159.2016.1112741. [DOI] [PubMed] [Google Scholar]
- 19.Brotons P, de Paz HD, Toledo D, Villanova M, Plans P, Jordan I, Dominguez A, Jane M, Godoy P, Muñoz-Almagro C, Alsedà M, Alvarez J, Arias C, Barrabeig I, Camps N, Carmona G, Carol M, Company M, Ferràs J, Ferrús G, Janè M, Minguell S, Rodríguez R, Sala MR, Torra R, Crespo I, Solano R, Caylà J, Lafuente S, Rius C, Domínguez A, del Amo E. 2016. Differences in Bordetella pertussis DNA load according to clinical and epidemiological characteristics of patients with whooping cough. J Infect 72:460–467. doi: 10.1016/j.jinf.2016.01.013. [DOI] [PubMed] [Google Scholar]
- 20.Arbefeville S, Levi MH, Ferrieri P. 2014. Development of a multiplex real-time PCR assay for the detection of Bordetella pertussis and Bordetella parapertussis in a single tube reaction. J Microbiol Methods 97:15–19. doi: 10.1016/j.mimet.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 21.European Centre for Disease Prevention and Control. 2012. Guidance and protocol for the use of real-time PCR in laboratory diagnosis of human infection with Bordetella pertussis and Bordetella parapertussis. European Centre for Disease Prevention and Control, Stockholm, Sweden. [Google Scholar]
- 22.Harvill ET, Goodfield LL, Ivanov Y, Smallridge WE, Meyer JA, Cassiday PK, Tondella ML, Brinkac L, Sanka R, Kim M, Losada L. 2014. Genome sequences of nine Bordetella holmesii strains isolated in the United States. Genome Announc 2:e00438-14. doi: 10.1128/genomeA.00438-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Broeders S, Huber I, Grohmann L, Berben G, Taverniers I, Mazzara M, Roosens N, Morisset D. 2014. Guidelines for validation of qualitative real-time PCR methods. Trends Food Sci Technol 37:115–126. doi: 10.1016/j.tifs.2014.03.008. [DOI] [Google Scholar]
- 24.Antila M, He Q, de Jong C, Aarts I, Verbakel H, Bruisten S, Keller S, Haanperä M, Mäkinen J, Eerola E, Viljanen MK, Mertsola J, van der Zee A. 2006. Bordetella holmesii DNA is not detected in nasopharyngeal swabs from Finnish and Dutch patients with suspected pertussis. J Med Microbiol 55:1043–1051. doi: 10.1099/jmm.0.46331-0. [DOI] [PubMed] [Google Scholar]
- 25.Templeton KE, Scheltinga SA, van der Zee A, Diederen BMW, van Kruijssen AM, Goossens H, Kuijper E, Claas ECJ. 2003. Evaluation of real-time PCR for detection of and discrimination between Bordetella pertussis, Bordetella parapertussis, and Bordetella holmesii for clinical diagnosis. J Clin Microbiol 41:4121–4126. doi: 10.1128/JCM.41.9.4121-4126.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van den Bossche D, De Bel A, De Smet D, Heylen O, Vekens E, Vandoorslaer K, Soetens O, Piérard D. 2013. Prevalence of Bordetella holmesii and Bordetella bronchiseptica in respiratory tract samples from Belgian patients with pertussis-like symptoms by sensitive culture method and mass spectrometry. Acta Clin Belg 68:341–348. doi: 10.2143/ACB.3341. [DOI] [PubMed] [Google Scholar]
- 27.Martini H, Detemmerman L, Soetens O, Yusuf E, Piérard D. 2017. Improving specificity of Bordetella pertussis detection using a four target real-time PCR. PLoS One 12:e0175587. doi: 10.1371/journal.pone.0175587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mir-Cros A, Codina G, Martín-Gómez MT, Fàbrega A, Martínez X, Jané M, Van Esso D, Cornejo T, Rodrigo C, Campins M, Pumarola T, González-López JJ. 2017. Emergence of Bordetella holmesii as a causative agent of whooping cough, Barcelona, Spain. Emerg Infect Dis 23:1856–1859. doi: 10.3201/eid2311.170960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Njamkepo E, Bonacorsi S, Debruyne M, Gibaud SA, Guillot S, Guiso N. 2011. Significant finding of Bordetella holmesii DNA in nasopharyngeal samples from French patients with suspected pertussis. J Clin Microbiol 49:4347–4348. doi: 10.1128/JCM.01272-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miranda C, Porte L, Garcia P. 2012. Bordetella holmesii in nasopharyngeal samples from Chilean patients with suspected Bordetella pertussis infection. J Clin Microbiol 50:1505–1505. doi: 10.1128/JCM.06747-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ciruela Navas P, Hernández Baeza S, ANP i VCM. 2015. Informe sobre els microorganismes declarats durant l’any 2015. Sistema de notificació microbiològica de Catalunya, Barcelona, Spain. [Google Scholar]
- 32.European Centre for Disease Prevention and Control. 2016. Pertussis—annual epidemiological report 2016 [2014 data]. European Centre for Disease Prevention and Control, Stockholm, Sweden. [Google Scholar]
- 33.Koepke R, Bartholomew ML, Eickhoff JC, Ayele RA, Rodd D, Kuennen J, Rosekrans J, Warshauer DM, Conway JH, Davis JP. 2015. Widespread Bordetella parapertussis infections—Wisconsin, 2011–2012: clinical and epidemiologic features and antibiotic use for treatment and prevention. Clin Infect Dis 61:1421–1431. doi: 10.1093/cid/civ514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mastrantonio P, Stefanelli P, Giuliano M, Herrera Rojas Y, Ciofi degli Atti M, Anemona A, Tozzi AE. 1998. Bordetella parapertussis infection in children: epidemiology, clinical symptoms, and molecular characteristics of isolates. J Clin Microbiol 36:999–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cox HC, Jacob K, Whiley DM, Bletchly C, Nimmo GR, Nissen MD, Sloots TP. 2013. Bordetella holmesii and pertussis diagnosis: authors’ reply. Pathology 45:532. doi: 10.1097/PAT.0b013e3283639c0d. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


