Abstract
Background and Objective:
Pseudomonas aeruginosa (P. aeruginosa) cause serious nosocomial and non-nosocomial infections. The blaOxacillinases (OXA)-23 and blaOXA24/40 induce resistance to carbapenems. The current study aimed at detecting blaOXA-23 and blaOXA-24/40 in P. aeruginosa strains isolated from patients with nosocomial and non-nosocomial infections. carbapenems. The current study aimed at detecting blaOXA-23 and blaOXA-24/40 in P. aeruginosa strains isolated from patients with nosocomial and non-nosocomial infections.
Methods:
The current descriptive cross sectional study was conducted in Sanandaj, Iran (Kurdistan Province) from December 2015 to August 2017, on 146 strains of Pseudomonas spp. isolated from patients’ specimens. Microbiological methods and polymerase chain reaction (PCR) for gyrB were applied to detect P. aeruginosa. Imipenem (IMP)-disk diffusion method and OXA-23-/OXA-24/40-multiplex PCR were used to identify resistant strains. Stata 12 using Fisher exact test and logistic regression were employed to analyze the data (P ≤0.05).
Results:
The gyrB-PCR results showed that 91.78% of isolates were P. aeruginosa. Nosocomial infection caused by P. aeruginosa was observed in 41.79% of the studied patients; however, 27.61% of P. aeruginosa strains were resistant to IMP; blaOXA-23 and blaOXA24/40 were detected in 11.19% and 2.24% of the strains, respectively; a co-existence of blaOXA-23 and blaOXA24/40 was also observed in 2.23% of P. aeruginosa strains. There were no significant relationships between antibiotic resistance and harboring resistance genes; in addition, between IMP resistance and age, gender, place of residence, inpatient/outpatient, and type of specimen no association was found (P≥ 0.05).
Conclusion:
Resistance to IMP and the detection of resistant genes in the current study were observed in the clinical samples. Antibiotics should be prescribed more cautiously in order to prevent antibiotic resistance in pathogens.
Introduction
Pseudomonas aeruginosa (P. aeruginosa) is the most common bacterial pathogen found in serious nosocomial and also non-nosocomial infections. Some of the infections caused by these bacteria are pneumonia, urinary tract infection (UTI), surgical site infections, and sepsis (1, 2). These bacteria are often associated with multi-drug resistance (MDR) and extensively drug resistance (XDR). Thus, different infections caused by P. aeruginosa are difficult to treat and lead to morbidity, mortality, and also high economic burden on the patients (1). P. aeruginosa strains are the common extended-spectrum beta-lactamase (ESBL) producers among pathogenic bacteria (3). According to Ambler Classification, beta-lactamases are classified into four groups of A, B, C, and D. Different genes encode class D beta-lactamases; in addition, they are poorly inhibited by clavulanic and ethylenediaminetetraacetic acids (EDTA), and have many differences in the amino acid sequences (4). The following types are examples of this enzyme: OXA (Oxacilinase)-1, 2, 10, 13, and 17 that are the prototype gene and cause resistance to oxacillin and cephalosporin (2). On the other hand, carbapenemhydrolyzing class D beta-lactamases such as OXA-23 and -24/40 induce resistance to carbapenems such as imipenem (IMP), meropenem (MEM), and doripenem (DRP). Hydrophobic bridge formed by Tyr112 and Met223 plays an important role in carbapenemase activity of this enzyme (5). OXA-23 was firstly reported in 1995 and OXA-24/40 was identified in Spain in 2000 (6, 7). Epidemiology of carbapenemresistant P. aeruginosa (CRPA) is studied in different geographical parts of the world and ranges from 10% to 50%. The lowest rates of CRPA are reported in Canada (3.3%) and the Dominican Republic (8%), and higher rates are observed in Brazil, Peru, Costa Rica, Russia, Greece, Poland, Iran, and Saudi Arabia (above 50%). The geographical distribution of CRPA is gradually increasing (8). Molecular and phenotypic techniques such as polymerase chain reaction (PCR) and disk diffusion method are used to study antibiotic resistant P. aeruginosa (9-11). According to the above mentioned contents, treatment of infections caused by P. aeruginosa harboring blaOXA-23 and blaOXA24/40 is very difficult. No study is conducted so far on the distribution of these genes in P. aeruginosa in Kurdistan Province, Iran. According to the increasing resistance, and failure of antibiotics to remove P. aeruginosa, the current study was conducted to investigate P. aeruginosa strains harboring blaOXA-23 and blaOXA24/40 and evaluate their characteristics. Such information can be used in healthcare centers and the hospital infection control practices advisory committees in order to make better decisions regarding the control and prevention of infectious diseases caused by these bacteria, which in turn can help to properly prescribe antibiotics. Therefore, the current study aimed at investigating the phenotypic and molecular detections of blaOXA-23 and blaOXA24/40 among P. aeruginosa strains isolated from patients with nosocomial and non-nosocomial infections in different tertiary hospitals in Kurdistan Province, Iran.
Materials and methods
The current descriptive cross sectional study was conducted at the Cellular and Molecular Research Center of Kurdistan University of Medical Sciences, Sanandaj, Iran from December 2015 to August 2017. All Pseudomonas spp. isolated from 49 females and 97 males admitted to tertiary hospitals were collected (Table 2). Inclusion criteria of the current study were admission to tertiary hospitals; having infections caused by P. aeruginosa, and isolation of P. aeruginosa from their clinical specimens. Strains that were not identified in molecular and phenotypic tests as P. aeruginosa and those that did not grow on bacterial cultures were excluded from the study. The study protocol was also approved by the Ethics Committee of the local university (ethical code: MUK.REC. 1394/337). Overall, 146 strains of Pseudomonas spp. were isolated. Patients’ demographic information was collected from the hospital information system (HIS). Nosocomial and non-nosocomial P. aeruginosa infections were also detected according to the definition of centers for disease control and prevention (CDC) (12). For P. aeruginosa species, phonotypic features were detected using microbiological methods (13). To extract DNA for PCR, single and pure colonies of overnight culture on Mueller-Hinton agar (MHA) (Merck, Germany) were dissolved in 500 µL of sterile deionized water in a 1.5-mL tube; then, powdered glass was added to them slightly; 500 µL of Tris-EDTA (ethylenediaminetetraacetic acid) (10 mM Tris, 1 mM EDTA, pH 8.0) was also added to them. After centrifugation (7000 rpm, five minutes), 3 µL of supernatant was used for PCR as DNA template. For P. aeruginosa molecular detection by PCR, gyrB (gyrase B) forward (F) and reverse (R) primers (SinaClon, Iran) in a final volume of 25 µL (7.5 µL deionized water, 3 µL DNA template, 1µL each F and R primers, and 12.5 µL Master mix) were used. P. aeruginosa ATCC 25922 (Darvash, Iran) and deionized water were applied as positive and negative controls, respectively (Table 1) (14). For antibiotic sensitivity testing, a suspension of P. aeruginosa adjusted to 0.5 McFarland turbidity standard was prepared, and then, was cultured on MHA. Kirby-Bauer disk diffusion method was applied according to Clinical and Laboratory Standards Institute (CLSI) guidelines with IMP (10 µg) (Rosco, Denmark) (12). Multiplex PCR for blaOXA-23 and blaOXA-24/40 using Fand R-primers (SinaClon, Iran) were performed. Acinetobacter baumannii strains harboring blaOXA-23 and blaOXA-24/40 and distilled water were respectively used as positive and negative controls in this assay (Table 1) (15). The PCR was amplified with the final volume of 21 µL (8 µL deionized water, 2 µL DNA template, 0.2 µL of each F and R primers for blaOXA-23 and blaOXA-24/40, and 10 µL Master mix) (7). Stata software version 12 using Fisher exact test and logistic regression analysis were used to analyze the data (P ≤0.05).
Table 2.
The Source of Pseudomonas aeruginosa in Hospitals
| Hospital | Outpatient, N (%) | Inpatient, N (%) |
Death,
N (%) |
Number of
Isolates (%) |
|---|---|---|---|---|
| Toohid | 9 (69.23) | 85 (71.42) | 1 (50) | 95 (70.89) |
| Besat | 3 (23.07) | 29 (24.36) | 1 (50) | 33 (24.62) |
| Imam Hossein | 1 (7.69) | 3 (2.52) | 0 | 4 (2.98) |
| Imam Khomeini | 0 | 1 (0.84) | 0 | 1 (0.74) |
| Fajr | 0 | 1 (0.84) | 0 | 1 (0.74) |
| Kowsar | 0 | 0 | 0 | 0 |
| Total | 13 | 119 | 2 | 134 |
Table 1.
Primers Sequences and PCR Conditions in the Current Study
| Primer and Gens Name |
Sequence | Product Size (bp) |
PCR Condition |
|---|---|---|---|
| gyr B | 5`-CCTGACCATCCGTCGCCACAAC-3` 5`-CGCAGCAGGATGCCGACGCC-3 |
222 | Initial denaturation at 95°C for 5 min 1 cycle followed by 35 cycles; denaturation at 94°C for 45 s, annealing at 66°C for 45 s, extension at 72°C for 1 min, and final extension at 72°C for 10 min 1 cycle |
| blaOXA-23 | 5-GAT CGG ATT GGA GAA CCAGA-3` 5`-ATT TCT GAC CGC ATT TCC AT-3` |
501 | Initial denaturation at 94˚C for 5 min 30 cycles, 94˚C for 25 s, 52˚C for 40 s, 72˚C for 50 s, and a final extension at 72˚C for 6 min |
| blaOXA-24/40 | 5`-GGT TAG TTG GCC CCC TTA AA-3` 5`-AGT TGA GCG AAA AGG GGA TT-3` |
246 |
Results
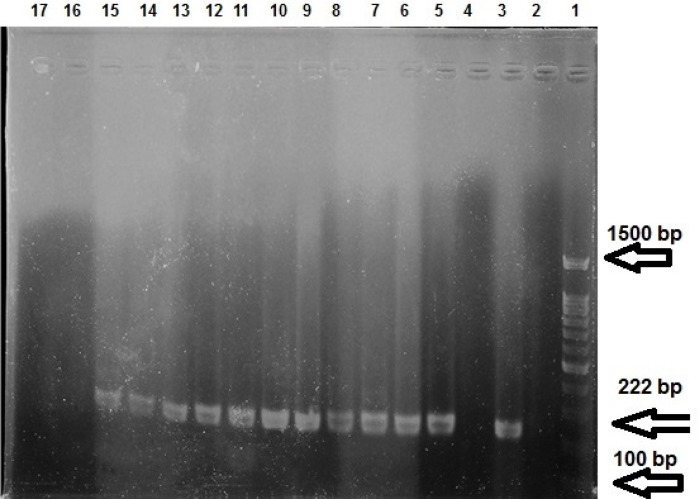
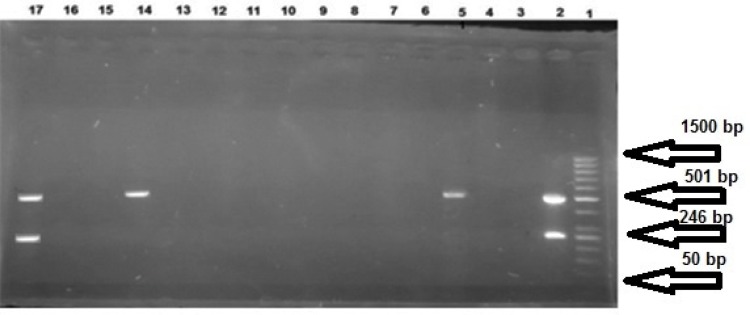
According to microbiological test results, 146 Pseudomonas spp. were detected. Phenotypic test showed 133 (91.09%) P. aeruginosa strains. But gyrB-PCR with 222 bp DNA fragments on gel electrophoresis determined 134 (91.78%) P. aeruginosa strains isolated from different hospitals (47 from females (35.07%) and 87 (64.93%) from males with the mean age of 50.35±20.19 years) (Figure1; Table 2). Nosocomial infection with P. aeruginosa was observed in 56 (41.79%) in-patients [14 (25%) females and 42 males (75%)] (Table 3). Results of the antibiotic sensitivity test showed IMP resistance in P. aeruginosa (Table 4) strains. Gel electrophoresis detected DNA fragments of blaOXA-23 on 501 bp and blaOXA-24/40 on 246 bp in 15 (11.19%) and three (2.24%) strains, respectively (Figure 2).Three (0.45%) of the 15 IMP-resistant isolates carried the blaOXA-23 and non-resistant isolates were the carriers of blaOXA-24/40. In addition, three (2.23%) strains of P. aeruginosa showed a co-existence of blaOXA-23 and blaOXA-24/40. Four (7.14%) P. aeruginosa strains isolated from nosocomial infections carried blaOXA-23, but none of them carried blaOXA-24/40.There was no significant relationship between antibiotic resistance and presence of genes, and between IMP-resistance and age, gender, place of residence, inpatient/ outpatient, and type of specimen (P ≥0.05).
Figure 1.
Gel electrophoresis of gyrB-PCR products; Line 1: marker, molecular weight 100–1500 bp; Line 2: negative control; Line 3: positive control; Lines 3, 16, 17: negative examples of gyrB; Line 5 to 15: positive examples of gyrB (222 bp lenght)
Table 3.
Source of Pseudomonas aeruginosa in Clinical Samples
| Source | No. (%) of Strains |
No. (%) of Strains related with Nosocomial infection |
|---|---|---|
| Total | 134 | 56 |
| Intensive Care Unit (ICU) | 40 (29.85) | 39 (69.64) |
| Women’s Internal | 13 (9.70) | 02 |
| Men’s Internal | 8 (5.97) | 1 (1.78) |
| Men’s Ward | 1 (0.75) | 0 |
| Women Heart | 3 (2.24) | 0 |
| Infectious | 15 (11.19) | 0 |
| Emergency | 12 (8.96) | 1(1.78) |
| Laboratory | 10 (7.46) | 0 |
| Men’s Surgery | 8 (5.97) | 4 (2.98) |
| Neurology | 7 (5.22) | 6 (4.47) |
| Respiratory | 4 (2.99) | 0 |
| Burn | 4 (2.98) | 2 (1.49) |
| Oncology | 3 (2.24) | 1 (1.78) |
| Women’s heart | 3 (2.23) | 1 (1.78) |
| Digestion | 2 (1.49) | 0 |
| Women’s Surgery | 1 (0.75) | 0 |
| Heart Surgery | 1 (0.75) | 0 |
| General Surgery | 1 (0.75) | 1 (1.78) |
| Orthopedic | 1 (0.75) | 0 |
| Total | 134 | 56 |
| Specimens | ||
| Urea | 61 (45.52) | 12 (21.42) |
| Tracheal | 29 (21.64) | 29 (51.78) |
| Wound | 15 (11.19) | 5 (8.92) |
| Blood | 16 (11.94) | 3 (5.35) |
| Lung Secretions | 4 (2.99) | 3 (5.35) |
| Pleural Fluid | 4 (2.98) | 2 (3.57) |
| Sputum | 2 (1.49) | 1 (1.78) |
| Intestines Biopsy | 1 (0.75) | 0 |
| Stool | 1 (0.75) | 0 |
| Abdominal Fluid | 1 (0.70) | 1 (1.78) |
* Men's internal ward was related to patients with internal diseases such as Endocrine disorders, liver disorders and rheumatism or diseases, Men's surgery ward was related to patients with surgery such as general medical surgery, urinary system, orthopedic surgery and neurosurgery
Table 4.
Results of IMP Sensitivity Testing for Pseudomonas aeruginosa
| Infection Type | Infection Type | ||
|---|---|---|---|
| Sensitive, N (%) | Intermediate, N (%) | Resistant, N (%) | |
| Total(134) | 90 (67.16) | 7(5.22) | 37 (27.61) |
| Nosocomial | 33 (36.66) | 2(2.22) | 21 (22.22) |
| Non-nosocomial | 57 (63.33) | 5(5.55) | 16 (18.88%) |
Discussion
P. aeruginosa cause infection in different hospital wards (2, 16). In the current study, among 146 Pseudomonas spp., 91.09% using phenotypic test and 91.78% using gyrB-PCR were identified as P. aeruginosa. Based on the results of phenotypic test and gyrB-PCR for P. aeruginosa, Farajzadeh Sheikh et al., reported all the 223 isolates as P. aeruginosa (17). PCR is a rapid technique; it is a gold standard with high sensitivity and specificity, and it is reliable to identify microbial pathogens (17, 18). In the current study, most P. aeruginosa strains were isolated from Toohid Hospital (70.89%), followed by Besat Hospital (24.62%). Since Toohid and Besat hospitals are the tertiary referral centers in Sanandaj, the center of Kurdistan Province, Iran, and most of the patients are referred to them from surrounding cities, the highest rates of infection were reported from these hospitals. Nosocomial infection caused by P. aeruginosa was observed in 41.79% of inpatients. The majority of P. aeruginosa strains were isolated from intensive care units (ICUs) (29.85%) of which 69.64% were related to nosocomial infections. Verma et al., in India reported 24% and 18% prevalence of P. aeruginosa respectively from burn ward and ICU in patients with nosocomial infection. In addition, resistance to antibiotics in these wards were high (19). Due to the fact that P. aeruginosa is an MDR and opportunistic pathogen; it can cause nosocomial infection, especially among inpatients admitted to ICU. Moreover, personal underlying risk factors such as nature and duration of invasive procedures used, length of ICU and hospital stay, and antibiotic treatment can affect the different rates of nosocomial infections in ICUs (19, 20). Results of the antibiotic sensitivity testing against IMP showed that bacteria isolated from nosocomial infections were more resistant to IMP (22.22%). Mohsenpour et al., reported that 134 isolates out of 374 were IMP-resistant, while 240 were IMP-sensitive; the resistance rates were higher in ICU and patients with nosocomial infections, which was similar to the results of the current study (21). Major factors leading to carbapenem-resistance are metallo-beta-lactamases (class D MBLs) and carbapenem-hydrolyzing oxacillinases (15). However, rates of antibiotic resistance in a study tend to differ according to certain factors such as type of antibiotics, genetic variations of bacteria and resistant strains, and differences in antibiotic consumption pattern at different locations (22) .In the current study, blaOXA-23 and blaOXA-24/40 were detected in 11.19% and 2.23% of the isolates, respectively. In addition, blaOXA-23 was detected in 7.14% of the patients with nosocomial infections. By performing multiplex PCR, Esenkaya Taşbent and Özdemir showed that of 184 IMPand/or MEM-resistant Pseudomonas spp. strains isolated from different clinical samples, 6.5% and 0.54% were positive for blaOXA-23 and blaOXA-24/40 , respectively (23). In the current study, 0.45% of 15 IMP-resistant isolates were the blaOXA-23 carriers, but none of the resistant isolates were blaOXA-24/40 carriers.
To justify this matter, the environment and/or genetic context can modify the phenotypic expression of resistant genes and thus, genotype does not always result in the expected phenotype (24). The results were similar to those of the current study. Based on the results of PCR, Odumosu et al., showed that the prevalence of blaOXA-10 in P. aeruginosa strains was 80% (10). Multiplex PCR results in a study by Farsiani and determine resistance patterns of bacteria, the employment of PCR method, and using easy-to-access and cost-effective methods. However, weaknesses and limitations of the current study were possible contamination of the laboratory environment, which may lead to false results and lack of access to the medical history and specimens of all the patients.
Conclusion
In the current study, IMP-resistant P. aeruginosa et al., showed harboring blaOXA-23 in all 36 isolates of A. baumannii and accordingly, the prevalence of strains were detected in different clinical samples taken from patients with nosocomial and non-nosocomi-blaOXA-24/40 was 64% (15). These rates in the studyal infections. Some of the isolates carried OXA genes.
by Odumosu were higher than those of the current Carbapenems are still the most important and effect study about bla OXA genes. According to the results of tive antibiotics against different infections caused PCR, Aghazadeh et al., detected OXA I, II, and III in P. aeruginosa species, and enterobacterial repetitive intergenic consensus-PCR (ERIC-PCR) proved high genetic diversity among these isolates (2). In a study by Saderi et al., 39.06% of 94 P. aeruginosa isolates were MBLs producers. They used the combination disk diffusion method to detect MBL-producing P. aeruginosa (25). Different mechanisms of gene transfer such as horizontal gene transfer (including transposable elements) can be the cause of transmission of class D carbapenem-hydrolyzing beta-lactamases genes among different bacterial strains; it is a global con¬cern threatening all the countries and communities (9, 15, 26, 27). Finally, the following can be considered as the strength points of the current study: clinical samples were gathered in the span of three years, employment of CLSI guidelines to diagnose by P. aeruginosa; therefore, according to the current study results, more effective planning and measures should be taken in order to determine the resistance and prevalence of such genes in the studied strains, and control and prevent the spread of these bacteria in hospital wards.
Acknowledgment
The article is part of Samaneh Rouhi’s PhD dissertation in molecular epidemiology of bacteria. The authors wish to extend their gratitude to the Research Deputy of Kurdistan University of Medical Sciences for financial supports.
Conflicts of interest
The authors declare that there are no conflicts of interest.
References
- 1.Skariyachan S, Sridhar VS, Packirisamy S, Kumargowda ST, Challapilli SB. Recent per- spectives on the molecular basis of biofilm formation by Pseudomonas aeruginosa and ap- proaches for treatment and biofilm dispersal. Folia microbiologica. 2018;19:1–20. doi: 10.1007/s12223-018-0585-4. [DOI] [PubMed] [Google Scholar]
- 2.Aghazadeh M, Samadi Kafil H, Ghotaslou R, Asgharzadeh M, Moghadami M, Akhi MT, Ho- jabri Z, Naghili B, Najafi K, Azimi S, Shokrian S. Prevalence of oxacillinase groups i, ii and iii in Pseudomonas aeruginosa isolates by poly- merase chain reaction and genotyping by ER- IC-PCR methods. Jundishapur Journal of Mi- crobiology. 2016;26:1–6. [Google Scholar]
- 3.Amirkamali S, Naserpour-Farivar T, Azarhoosh K, Peymani A. Distribution of the bla OXA, blaVEB-1, and bla GES-1 genes and resistance patterns of ESBL-producing Pseudomonas ae- ruginosa isolated from hospitals in Tehran and Qazvin, Iran. Rev Soc Bras Med Trop . 2017;50:315–20. doi: 10.1590/0037-8682-0478-2016. [DOI] [PubMed] [Google Scholar]
- 4.Queenan AM, Bush K. Carbapenemases: the versatile beta-lactamases. Clin Microbiol Rev. 2007;20(3):440–58. doi: 10.1128/CMR.00001-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.June CM, Vallier BC, Bonomo RA, Leonard DA, Powers RA. Structural origins of oxacillin- ase specificity in class D beta-lactamases. An- timicrob Agents Chemother . 2014;58(1):333–41. doi: 10.1128/AAC.01483-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaitany KC, Klinger NV, June CM, Ramey ME, Bonomo RA, Powers RA, Leonard DA. Struc- tures of the class D carbapenemases OXA-23 and OXA-146: Mechanistic basis of activity against carbapenems, extended-spectrum ceph- alosporins and aztreonam. Antimicrobial agents and chemotherapy. 2013 ;22:AAC–00762. doi: 10.1128/AAC.00762-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Woodford N, Ellington MJ, Coelho JM, Turton JF, Ward ME, Brown S, et al. Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int J Antimicrob Agents. 2006;27(4):351–3. doi: 10.1016/j.ijantimicag.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 8.Hong DJ, Bae K, Jang IH, Jeong SH, Kang HK, Lee K. Epidemiology and characteristics of metallo-β-lactamase-producing Pseudomo- nas aeruginosa. Infect Chemother. 2015;47:81–97. doi: 10.3947/ic.2015.47.2.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramazanzadeh R, Rouhi S, Shakib P, Shahbazi B, Bidarpour F, Karimi M. Molecular charac- terization of vibrio cholerae isolated from clini- cal samples in Kurdistan Province, Iran. Jundis- hapur journal of microbiology. 2015;8(5) doi: 10.5812/jjm.8(5)2015.18119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Odumosu BT, Adeniyi BA, Chandra R. First Detection of OXA-10 Extended-Spectrum Beta-Lactamases and the Occurrence of mexR and nfxB in Clinical Isolates of Pseu- domonas aeruginosa from Nigeria. Chemotherapy. 2016;61(2):87–92. doi: 10.1159/000441712. [DOI] [PubMed] [Google Scholar]
- 11.Smiljanic M, Kaase M, Ahmad-Nejad P, Ghe- bremedhin B. Comparison of in-house and com- mercial real time-PCR based carbapenemase gene detection methods in Enterobacteriaceae and non-fermenting gram-negative bacterial iso- lates. Ann Clin Microbiol Antimicrob. 2017;16(1):48. doi: 10.1186/s12941-017-0223-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lavakhamseh H, Shakib P, Rouhi S, Moham- madi B, Ramazanzadeh R. A survey on the prevalence and antibiotic sensitivity of nosoco- mial infections in the besat hospital, Sanandaj, Iran. Journal of Nosocomial Infection. 2014 Jan;1(2):1–8. [Google Scholar]
- 13.Ramazanzadeh R, Rouhi S, Hosainzadegan H, Shakib P, Nouri B. Co-occurrence of Extended- Spectrum Beta-Lactamases in isolated Entero- bacter spp From patients specimens. Archives of Clinical Infectious Diseases. 2016;11(3) [Google Scholar]
- 14.Mulamattathil SG, Bezuidenhout C, Mbewe M, Ateba CN. Isolation of environmental bacteria from surface and drinking water in Mafikeng, SouthAfrica, and characterization using their an- tibiotic resistance profiles. J Pathog. 2014;2014:1–11. doi: 10.1155/2014/371208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farsiani H, Mosavat A, Soleimanpour S, Nasab MN, Salimizand H, Jamehdar SA, et al. Limited genetic diversity and extensive antimicrobial resistance in clinical isolates of Acinetobacter baumannii in north-east Iran. J Med Microbiol . 2015;64:767–73. doi: 10.1099/jmm.0.000090. [DOI] [PubMed] [Google Scholar]
- 16.Canale FP, Davila SDV, Sasso CV, Pellarin NW, Mattar Dominguez MA. Immunization with Larrea divaricata Cav Proteins elicits op- sonic antibodies against Pseudomonas aerugi- nosa and induces phagocytic activity of murine macrophages. Microb Pathog. 2018;118:257–67. doi: 10.1016/j.micpath.2018.03.029. [DOI] [PubMed] [Google Scholar]
- 17.Sheikh AF, Rostami S, Jolodar A, Tabatabaie- far MA, Khorvash F, Saki A, Shoja S, Sheikhi R. Detection of metallo-beta lactamases among carbapenem-resistant Pseudomonas aeruginosa. Jundishapur journal of microbiology. 2014;7(11) doi: 10.5812/jjm.12289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gabar AM, Al-Daraghi WA. Use of PCR to De- tection Pseudomonas aeruginosa from Clinical Samples in Hilla Teaching Hospital. Journal of University of Babylon. 2016;24(5):1414–20. [Google Scholar]
- 19.Verma U, Kulshreshtha S, Khatri PK. MDR Pseudomonas aeruginosa in Nosocomial Infec- tion: Burden in ICU and Burn Units of a Ter- tiary Care Hospital. IJCMAS. 2018;7:1267–74. [Google Scholar]
- 20.Zhao GJ, Li D, Zhao Q, Song JX, Chen XR, Hong GL, et al. Incidence, risk factors and impact on outcomes of secondary infection in patients with septic shock: an 8-year retrospective study. Sci Rep . 2016;6:6–9. doi: 10.1038/srep38361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mohsenpour B, Rouhi S, Mehrdel R, Faraji T, Masaeli M, Ramazanzadeh R. Risk Factors Associated With Imipenem-Resistance Among Isolated Gram-Negative Bacteria From Patients in Sanandaj Hospitals, Iran. Avicenna Journal of Clinical Microbiology and Infection. 2016;3(1) [Google Scholar]
- 22.Ramazanzadeh R, Moradi Gh, Zandi S, Moham- madi S, Rouhi S, Pourzare M, et al. A survey of contamination rate and antibiotic resistant of Gram-negative bacteria isolated from patients in various wards of Toohid and Besat Hospitals of Sanandaj city during 2013-2014 years. PSJ. 2016;14:11–9. [Google Scholar]
- 23.Esenkaya Taşbent F, Özdemir M. The Presence of OXA Type Carbapenemases in Pseudomonas Strains: First Report from Turkey. MikrobiyolBul. 2015;49(1):26–34. doi: 10.5578/mb.8563. [DOI] [PubMed] [Google Scholar]
- 24.Hughes D, Andersson DI. Environmental and genetic modulation of the phenotypic ex- pression of antibiotic resistance. FEMS Microbiol Rev. 2017;41(3):374–91. doi: 10.1093/femsre/fux004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Owlia P, Saderi H, Karimi Z, Rad A, Bagher SM, Bahar MA. Phenotypic detection of Metal- lo-beta-Lactamase producing Pseudomonas aeruginosa strains isolated from burned pa- tients. Iranian Journal of Pathology. 2008 Jan;3(1):20–5. [Google Scholar]
- 26.Davoudi AR, Najafi N, Hoseini Shirazi M, Ah- angarkani F. Frequency of bacterial agents iso- lated from patients with nosocomial infection in teaching hospitals of Mazandaran University of Medical Sciences in 2012. Caspian J Intern Med . 2014;5:227–31. [PMC free article] [PubMed] [Google Scholar]
- 27.Gedik H. Antibiotic resistance status and its costs in hematological patients: A two-year analysis. Caspian journal of internal medicine. 2017;8(4):276. doi: 10.22088/cjim.8.4.276. [DOI] [PMC free article] [PubMed] [Google Scholar]