Auxin distribution revealed by DR5ver2::GUS and expression patterns of FveTARs and FveYUCs determined by the GUS reporter highlighted the endosperm as the main auxin biosynthesis site in strawberry achenes.
Keywords: Auxin biosynthesis, CRISPR/Cas9, DR5ver2::GUS, Fragaria vesca, fruit, root, spatial–temporal expression, strawberry, TARs, YUCs
Abstract
The critical role of auxin in strawberry fruit set and receptacle enlargement was demonstrated previously. While fertilization is known to trigger auxin biosynthesis, the specific tissue source of fertilization-induced auxin is not well understood. Here, the auxin reporter DR5ver2::GUS was introduced into wild strawberry (Fragaria vesca) to reveal auxin distribution in the seed and fruit receptacle pre- and post-fertilization as well as in the root. In addition, the expression of TAR and YUCCA genes coding for enzymes catalysing the two-step auxin biosynthesis pathway was investigated using their respective promoters fused to the β-glucuronidase (GUS) reporter. Two FveTARs and four FveYUCs were shown to be expressed primarily in the endosperm and embryo inside the achenes as well as in root tips and lateral root primordia. Expression of these reporters in dissected tissues provided more detailed and precise spatial (cell and tissue) and temporal (pre- and post-fertilization) information on where auxin is synthesized and accumulates than previous studies in strawberry. Moreover, we generated CRISPR-mediated knock-out mutants of FveYUC10, the most abundant YUC in seeds; the mutants had a lower free auxin level in young fruit, but displayed no obvious morphological phenotypes. However, overexpression of FveYUC10 resulted in elongated hypocotyls in Arabidopsis caused by elevated auxin level. Overall, the study revealed auxin accumulation in the chalazal seed coat, embryo, receptacle vasculature, root tip, and lateral root primordia and highlighted the endosperm as the main auxin biosynthesis site for fruit set.
Introduction
Cultivated strawberry (Fragaria ananassa) is one of the major fruit crops grown worldwide. Developmentally, strawberry fruit is unique in that the juicy flesh is developed from the receptacle, an enlarged stem tip, while the true fruit, called an achene, is developed from a pollinated ovary containing a single ovule (Darrow, 1966; Hollender et al., 2012). Because of the accessibility of the achenes on the fruit surface, strawberry fruit has long been used as a model system to study the roles of auxin in fruit development. When the achenes are removed after pollination, receptacle fruit fails to develop, but exogenous auxin application can substitute for the achene and restore the fruit development (Nitsch, 1950), suggesting the achene as the source of the auxin for promotion of receptacle fruit set. This early observation was supported later by RNA-seq data and measurement of free auxin. RNA-seq data of carefully staged achenes showed rapid up-regulation of auxin biosynthetic genes in the achenes soon after fertilization (Kang et al., 2013). Measurement of free auxin in the achene showed a dramatic increase starting from 3 d after fertilization and reaching a maximum of ~70 ng g−1 fresh weight at about 12 d after fertilization (Nitsch, 1950, 1955; Dreher and Poovaiah, 1982; Symons et al., 2012). By contrast, auxin biosynthesis genes were not expressed in the receptacle during early-stage fruit development (Kang et al., 2013) and free auxin in the fruit receptacle was about 5 ng g−1 fresh weight, significantly lower than that in the achenes (Dreher and Poovaiah, 1982; Symons et al., 2012; Estrada-Johnson et al., 2017). Together, these observations suggest that fertilization-induced auxin biosynthesis primarily occurs in the achenes of strawberry.
Most of our basic understanding of auxin synthesis and signaling was derived from molecular genetic studies in Arabidopsis, including the revelation of the two-step biosynthetic pathway of indole-3-acetic acid (IAA), the major auxin. The tryptophan aminotransferase of Arabidopsis 1 (TAA1)/TAR family of aminotransferases convert tryptophan to indole-3-pyruvic acid (IPyA), and the YUCCA (YUC) family of flavin-containing monooxygenases convert IPyA to IAA (Mashiguchi et al., 2011; Stepanova et al., 2011; Won et al., 2011). Multiple TARs and YUCs in Arabidopsis play similar roles in embryogenesis, vascular patterning, flower development, and shade avoidance (Zhao et al., 2001; Cheng et al., 2006, 2007; Stepanova et al., 2008; Tao et al., 2008; Yamada et al., 2009). The homologs of these TAA/TAR and YUC genes were identified in strawberry (Liu et al., 2012, 2014; Kang et al., 2013) and functional studies were previously conducted for FaYUC1 and FaYUC2 in cultivated strawberries and FvYUC6 in the wild strawberry, indicating roles in leaf, root, flower, and fruit development (Liu et al., 2012, 2014). During late-stage fruit development (green stage to ripe), FaYUC2, FaTAR2, and FaTAA1 were shown to be expressed in the receptacle fruit, and transient silencing of FaTAR2 in ripening fruit resulted in altered auxin response (Liu et al., 2014; Estrada-Johnson et al., 2017). These studies suggest that certain levels of auxin biosynthesis in the receptacle may contribute to fruit ripening, which is unexpected given the non-climacteric fruit ripening of strawberry and that auxin was previously thought of as an inhibitor of fruit ripening (Given et al., 1988; Chai et al., 2011; Jia et al., 2011; Li et al., 2011).
Here, we focus on understanding the role of auxin in early fruit development, specifically fruit set, taking advantage of existing transcriptome data of wild strawberry (Fragaria vesca) fruit tissues (Kang et al., 2013). The comprehensive dataset included five finely dissected fruit tissue types at five early fruit developmental stages. Specifically, the achene was dissected into ovary wall, ghost (seed coat and endosperm), and embryo; the receptacle was dissected into pith and cortex. FveTARs and FveYUCs were found more abundantly expressed in the ghost than in the embryo, suggesting that auxin biosynthesis occurred mainly in the ghost. However, exactly where auxin is produced inside the achene (seed coat or endosperm, or specific regions of the seedcoat or endosperm) is not known. Given the multiple family members in the genome, understanding which FveTARs and FveYUCs are responsible for the fertilization-induced auxin production within the achene or specific tissues of the root will be necessary. Local auxin production is highly relevant to auxin functions (Robert et al., 2013; Chen et al., 2014). Identification of the tissue and timing of auxin biosynthesis will provide crucial insights into auxin-mediated developmental processes and aid in future manipulation of auxin for increasing fruit yield.
Our study was conducted in wild strawberry, an emerging model for the cultivated strawberry due to its diploid genome and abundant genomic and molecular resources (Slovin et al., 2009; Kang et al., 2013; Edger et al., 2018). We first utilized an auxin-responsive synthetic promoter, DR5ver2 (Liao et al., 2015), to visualize dynamic auxin distributions in the F. vesca fruit before and after fertilization and in the F. vesca root. DR5ver2 is a new and improved synthetic promoter containing nine copies of the Aux cis-element fused to the β-glucuronidase (GUS) reporter. This new DR5ver2 reporter was shown to be more sensitive to auxin than the earlier version, DR5 (Liao et al., 2015). In addition, we generated transgenic F. vesca plants containing promoter::GUS reporters of two FveTAR and four FveYUC genes. Analyses of these reporter gene expression patterns revealed distinct spatial and temporal patterns, suggesting the unique roles each gene may play during fruit and root development. To investigate the biological significance of auxin in strawberry fruit development, CRISPR/Cas9 was used to knock out FveYUC10. The resulting fveyuc10 mutants did not exhibit obvious morphological defects, but showed a significant reduction of free auxin. Taken together, this work revealed the sites of auxin biosynthesis within achenes and the major auxin biosynthesis genes responsible in fruit and root with a resolution at the tissue and cell level; it indicates the endosperm is an important tissue for fertilization-induced auxin biosynthesis.
Materials and methods
Plant materials and growth conditions
Fragaria vesca strain Hawaii 4 (PI551572, National Clonal Germplasm Repository, USA, white-fruited) was used for transformation and served as the wild type control for qRT-PCR. The Arabidopsis Columbia accession was used as the wild type. These strawberry and Arabidopsis plants were cultivated in a growth room under a light intensity of 100 μmol m−2 s−1 with a 16/8 h light/dark photoperiod at 22 °C.
Plasmid construction
For DR5ver2::GUS, DR5ver2 with nine repeats of a higher affinity auxin response element (AuxREs, TGTCGG) was synthesized as previously described (Liao et al., 2015) and inserted into the binary vector pMDC162 (Curtis and Grossniklaus, 2003) before the GUS gene by gateway cloning (Invitrogen). For promoter::GUS constructs, the following promoters were used: 2005 bp promoter of FveTAR1 (gene37056 (ver2.0.a2); gene31791 (ver1.1); FvH4_5g05900 (ver4.0)), FveTAR2 (gene31790/FvH4_5g05880, 1907 bp), FveYUC4 (gene11728/FvH4_2g29930, 1958 bp), FveYUC5 (gene32686/FvH4_2g14550.1, 1971 bp), FveYUC10 (gene27796/FvH4_2g24750, 1411 bp), and FveYUC11 (gene06886/FvH4_4g17980, 2000 bp). The promoter fragments were amplified from the genomic DNA of YW5AF7, a seventh generation inbred line of F. vesca (Slovin et al., 2009), and inserted into the binary vector pHGWFS7 before the GFP-GUS dual reporter by gateway cloning. To construct the single guide RNA (sgRNA)-Cas9 vector for FveYUC10, two sgRNAs respectively targeting FveYUC10 at 8 bp (sgRNA1: AAGCGGCGGTGATAATAGT) and 728 bp (sgRNA2: ATGGAGACCTGGCCAAGTA) downstream of the translation initiation codon were designed using the web server CRISPR-P (http://cbi.hzau.edu.cn/cgi-bin/CRISPR). Two AtU6 promoter-sgRNA-AtU6 terminator cassettes were amplified by PCR using pCBC-DT1T2 as the template, and then the PCR fragments were inserted into pHSE401 (Xing et al., 2014) by Golden Gate Assembly and confirmed by Sanger sequencing. For overexpressing FveYUC10, the coding sequence was PCR amplified and cloned into the EcoRI- and NdeI-digested binary vector pRI101 to fuse with green fluorescent protein (GFP) by Gibson assembly. These constructs were transformed into Agrobacterium tumefaciens strain GV3101 for plant transformation. The primers used for making these constructs are shown in Supplementary Table S1 at JXB online.
Strawberry transformation
Strawberry transformation was carried out as previously described with minor modifications (Kang et al., 2013). Briefly, callus was induced on 5++ medium (1×Murashige and Skoog (MS), 2% sucrose, 3.4 mg l−1 benzyladenine, 0.3 mg l−1 indole-3-butyric acid (IBA), and 0.7% phytoagar, pH 5.8) from the leaf strips of Hawaii 4 for 2 weeks in the dark; co-cultivated with Agrobacterium GV3101 harboring each construct for 1 h in co-cultivation buffer (1×MS, 2% sucrose, 0.4 mg l−1 acetosyringone) with gentle shaking; and then transferred to the 5++ medium, and kept for 3 d in the dark. The leaf strips were washed with sterile water to remove Agrobacterium and transferred to 5++ medium with 250 mg l−1 timentin and 250 mg l−1 carbnicillin for 2 weeks in the dark. The calli were transferred to new medium every 14 d on the 5++ medium with 250 mg l−1 timentin, 250 mg l−1 carbnicillin and up to 4 mg l−1 hygromycin until shoots appeared. The shoots were transferred to the rooting medium (0.5×MS, 0.01 mg l−1 IBA, 2% glucose, 250 mg l−1 timentin, 250 mg l−1 carbnicillin, 4 mg l−1 hygromycin, and 0.7% phytoagar, pH5.8). After 1–2 months, the regenerated plants were transferred to the soil and cultivated in the growth room for future analysis. The transformation efficiency (based on number of viable shoots among the total number of leaf strips) is about 5%.
Stable transformation in Arabidopsis
Arabidopsis transformation was carried out by the floral-dip method (Clough and Bent, 1998). T1 transgenic seeds were screened on half-strength MS medium (M5524, Sigma-Aldrich) with 100 mg l−1 kanamycin. T2 plants were used for characterization.
GUS staining
The ovules or seeds were dissected out of the achenes under a stereomicroscope from T0 transgenic plants. More than 10 samples per tissue and developmental stage were examined. Representative images are shown in the figures. Receptacles and roots were also collected from T0 plants. The tissues were stored in cold 90% acetone during dissection and then kept at room temperature for 20 min. Next, acetone was removed and GUS staining solution (1 mM X-glucuronic acid, 0.1 mM EDTA, 0.1% Triton X-100, and 10 mM (for seed) or 2mM (for root) potassium ferri/ferrocyanide in 100 mM phosphate buffer, pH 7.0) was added to submerge all the material. After 30 min of vacuum, tissues were incubated overnight at 37 °C. The samples were then mounted on clearing solution (chloral hydrate: glycerol: H2O: 8:1:1) for 5 h and observed under differential interference contrast (DIC) optics using a Zeiss Axioscope A1 microscope with a ×0.5 optical adapter. The images were captured, analysed, and exported using ZEN2.3.
RNA extraction and qRT-PCR
Total RNA was extracted using a Plant Total RNA Isolation Kit (Sangon Biotech, Shanghai, China, no. SK8631) following the manufacturer’s instructions. RNA quality was examined by both gel electrophoresis for sharp rRNA bands and the NanoDrop 2000 Spectrophotometer to reach an OD260/280 of 1.8. Approximately 1 μg of total RNA was used for cDNA synthesis using a PrimeScriptTM RT reagent kit (TaKaRa, Shiga, Japan, cat. no. RR047A), which removes genomic contamination with gDNA eraser digestion before reverse transcription. A total volume of 10 μl qPCR reaction mixture contained 5 μl of 2×SYBR Green PCR master mix (cat. no. 172-5124, Bio-Rad), 1 μl of 5× diluted cDNA, 0.25 μl of each primer (Supplementary Table S1), and 3.5 μl ddH2O. Amplification was performed using a QuantStudio 7 Flex system (Applied Biosystems, USA). The amplification program consisted of one cycle of 50 °C for 2 min and 95 °C for 10 min, followed by 50 cycles of 95 °C for 15 s, 60 °C for 20 s, and 72 °C for 20 s. The fluorescent product was detected at the third step of each cycle. The expression level of each gene was calculated using the method (Livak and Schmittgen, 2001). Primers were designed to span exon to exon junctions if allowed. Gene11892 and actin (At3g18780) were used as the control genes. Three biological replicates were used.
Phylogenetic analysis
The protein sequences were obtained from the F. vesca genome annotation ver2.0.a2 (Li et al., 2018) or downloaded from The Arabidopsis Information Resource (TAIR) (http://www.arabidopsis.org/). The sequence alignment was performed using Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo). An unrooted phylogenetic tree was constructed using MEGA 7 (http://www.megasoftware.net/) with the neighbor-joining statistical method and bootstrap analysis (1000 replicates).
Heatmap
Sequence read counts of the FveTARs and FveYUCs were quantified with the reads per kilobase per million mapped reads (RPKM) unit based on the F. vesca RNA-seq dataset (Li et al., 2018). A heatmap was made by using MultiExperiment Viewer 4.8 (MeV4.8; http://www.tm4.org/mev/). In the heatmap, the average RPKM value among the replicates was used to represent the expression pattern of each gene (Supplementary Table S2).
Hormone treatment
Newly opened flowers were emasculated to prevent self-fertilization and then sprayed with 500 μM 1-naphthylacetic acid (NAA; Sigma-Aldrich) every 8 h. The ovules and fruit receptacles were collected for GUS staining at 2 and 5 d post-treatment, respectively. Roots obtained from adult plants grown in the substrate were immersed in 100 μM NAA for 3 h and then subjected to GUS staining.
Auxin quantification
Entire fruit (achenes plus receptacle) at stage 4 were collected, weighed, and then immediately frozen in liquid N2. The wild type and fveyuc10 (L2 and L3 pooled together) samples with four biological replicates were analysed by liquid chromatography–electrospray ionization–quadrupole time-of-flight tandem mass spectrometry as previously described (Ng et al., 2015).
Statistical analyses
Statistical analyses were performed with SPSS v22.0 (IBM Corp., Armonk, NY, USA). Pairwise comparisons were determined using Student’s t-test (**P<0.01). The comparison between multiple samples was determined using Tukey’s test, and significant differences at the P<0.05 level are indicated by different letters.
Accession numbers
Gene accessions for FveTARs and FveYUCs in the ver2.0.a2 annotation (Li et al., 2018) and the ver4.0 annotation (Edger et al., 2018) are as follows: gene03586/FvH4_4g25850 (FveTAA1), gene37056/FvH4_5g05900 (FveTAR1), gene31790/FvH4_5g05880 (FveTAR2), gene14327/FvH4_ 7g02760(FveTAR3), gene27578/FvH4_2g13010 (FveTAR4), gene01005/FvH4_2g15120.1 (FveYUC1), gene30882/FvH4_1g04800 (FveYUC2), gene08377/FvH4_2g20150.1 (FveYUC3), gene11728/FvH4_2g29930 (FveYUC4), gene32686/FvH4_2g14550.1 (FveYUC5), gene08779/FvH4_2g36220.1 (FveYUC6), gene00528/FvH4_4g05280 (FveYUC7), gene27796/FvH4_2g24750 (FveYUC10), and gene06886/FvH4_4g17980 (FveYUC11). Gene accessions for AtTARs and AtYUCs in Arabidopsis are as follows: At1g70560 (AtTAA1), At1g23320 (AtTAR1), At4g24670 (AtTAR2), At1g34040 (AtTAR3), At1g34060 (AtTAR4), At4g32540 (AtYUC1), At4g13260 (AtYUC2), At1g04610 (AtYUC3), At5g11320 (AtYUC4), At5g43890 (AtYUC5), At5g25620 (AtYUC6), At2g33230 (AtYUC7), At4g28720 (AtYUC8), At1g04180 (AtYUC9), At1g48910 (AtYUC10), and At1g21430 (AtYUC11).
Results
Auxin distribution and dynamics in the achene and receptacle fruit of F. vesca
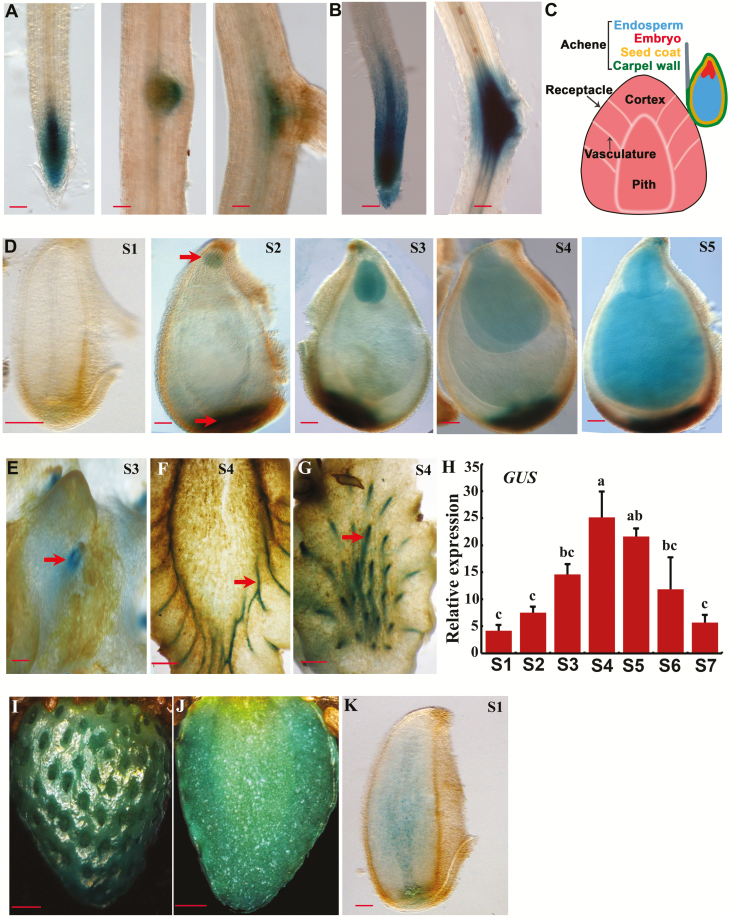
As strawberry is a unique fruit, revealing auxin distribution in its achenes and receptacle during early-stage fruit development would help understand the developmental process underlying fruit set. To do this, DR5ver2 was used to drive the GUS gene, and the reporter construct was stably transformed into the F. vesca accession Hawaii 4. A total of 12 independent transgenic lines were obtained and validated by PCR amplification of the DR5ver2 fragment (Supplementary Fig. S1A). As auxin distribution in roots was well documented in Arabidopsis, GUS staining was first carried out in F. vesca roots to assess whether DR5ver2::GUS acts correctly in these F. vesca transgenic lines in the T0 generation. GUS was expressed in the entire root tip with a higher level in the stele of the root meristematic zone (Fig. 1A). Compared with the expression pattern of DR5::GUS in Arabidopsis and garden strawberry (Ulmasov et al., 1997; Ottenschläger et al., 2003; Estrada-Johnson et al., 2017), GUS was more highly expressed, probably owing to the use of an enhanced DR5 (Liao et al., 2015). In addition, DR5ver2::GUS was expressed at the tip of a lateral root primordium before emergence and at the base of an emerged young lateral root (Fig. 1A). Moreover, GUS staining became stronger both at the root tip and at the lateral root primordium in response to NAA treatment (Fig. 1B), suggesting that the DR5ver2::GUS reporter behaves properly and can be used to monitor auxin distribution in F. vesca.
Fig. 1.
DR5ver2::GUS expression in roots and fruit of F. vesca. (A) GUS expression of the DR5ver2::GUS reporter lines in root. (B) GUS expression in root after NAA (100 μM) treatment. (C) Diagram of strawberry fruit showing the receptacle and an achene. The achene consists of embryo, ghost (endosperm and seed coat), and the ovary wall. (D) GUS expression of DR5ver2::GUS in ovule (stage 1) and seeds at stages 2–5. Arrows indicate embryo (top) and chalazal seed coat (bottom). (E–G) GUS expression of DR5ver2::GUS in receptacle. (E) Surface of the receptacle at stage 3. Arrow indicates the region where achene is attached to the receptacle. (F, G) Different sections of the receptacle at stage 4. Arrows indicate the vasculatures in sectioned receptacle fruit. (H) Expression level of GUS obtained by qRT-PCR in the receptacle of the DR5ver2::GUS reporter line at stages 1–7. Gene11892 was used as the internal control. Data are means ±SD obtained from three biological replicates. Significant differences at the P<0.05 level are indicated by different letters, tested by Tukey’s test. (I–K) GUS staining in receptacle (I, J) and ovule (K) at 5 and 2 d post-NAA (500 μM) treatment. Scale bars: 10 µm (A, B, D, K); 100 µm (E); 2 mm (F, G, I, J).
In strawberry, the achenes, i.e. the true fruit, sit on the surface of the receptacle (Fig. 1C); each achene consists of the ovary wall encasing a single seed. Each seed consists of seedcoat, endosperm, and embryo (Fig. 1C). The developmental stages of early fruit development were previously defined (Hollender et al., 2012; Kang et al., 2013) with embryo morphologies as temporal markers. Specifically, stage 1 is defined as flower anthesis, a pre-fertilization stage; stage 2 is only 2–3 d post-fertilization with a globular shaped embryo; stage 3 is defined as having fruits with a heart-shaped embryo; stage 4 fruit has a torpedo-shaped embryo; stage 5 has a fully grown embryo. The DR5ver2::GUS F. vesca transgenic lines were first examined in ovules (stage 1) and seeds at stages 2–5. There was no staining in ovules at anthesis (stage 1), indicating a very low level of free auxin (Fig. 1D). Starting from stage 2, which correlates post-fertilization, GUS was observed in the chalazal seed coat and embryo (Fig. 1D). GUS expression was increased from stage 2 to stage 5 in embryo and chalazal seed coat but less so in the endosperm (Fig. 1D).
Previous work suggested that post-fertilization-induced auxin in the achenes was transported to the receptacle to stimulate fleshy fruit growth (Nitch 1950; Kang et al., 2013). To reveal dynamic auxin accumulation in the receptacle, we examined GUS reporter expression in stage 3 and 4 receptacles of the DR5ver2::GUS transgenic lines. Interestingly, GUS was specifically expressed in the receptacle vasculature strands that connect to the achene, consistent with previous work (Estrada-Johnson, et al., 2017) and suggesting a likely auxin transport route from the chalazal seed coat to the vasculature veins of the receptacle (Fig. 1E, F, G). To quantitatively determine the auxin reporter expression, qRT-PCR was used to examine GUS transcripts driven by DR5ver2 in fruit receptacles at stages 1–5 plus later stages (stages 6 and 7). Stage 6 is equivalent to the white stage, while stage 7 is equivalent to the ripe/red stage. The GUS transcript level gradually increased from stage 1 to stage 4 and then slowly reduced from stage 4 to stage 7 (Fig. 1H). The stage 1–4 expression trend positively correlates with the early-stage receptacle fruit enlargement. Moreover, GUS expression level was greatly enhanced in the developing receptacle as well as ovule at anthesis in response to NAA treatment (Fig. 1I, J, K). Collectively, auxin distribution in seeds, roots, and fruit receptacles was revealed by the DR5ver2::GUS reporter gene expression in transgenic F. vesca plants, which not only corroborates prior studies suggestive of fertilization-induced auxin synthesis and transport from achene to the receptacle, but also reveals the likely transport route from chalazal seed coat to the receptacle via the vasculature strands connecting the two.
Phylogenetic and expression analyses of FveTARs and FveYUCs
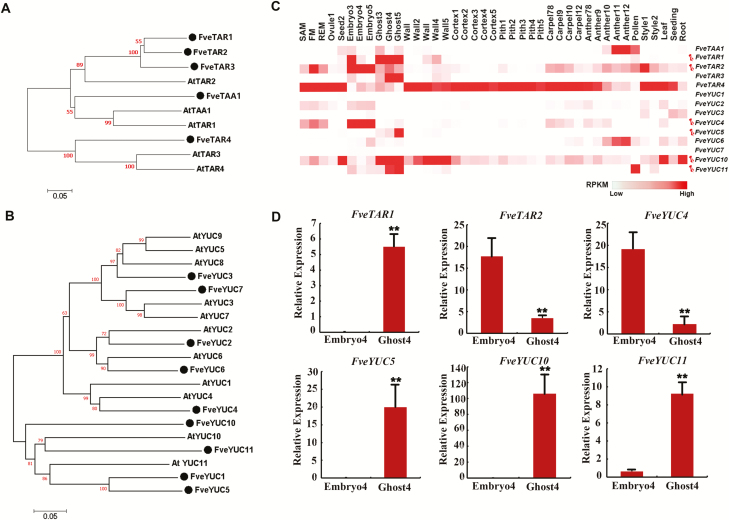
TARs and YUCs encode the two enzymes catalysing the main auxin biosynthetic pathway (Mashiguchi et al., 2011; Stepanova et al., 2011; Won et al., 2011). Although the phylogenetic analyses of these genes have been performed in both wild and cultivated strawberry (Kang et al., 2013; Liu et al., 2014; Estrada-Johnson et al., 2017), the recently published high quality genome and annotation of F. vesca (Edger et al., 2018; Li et al., 2018) necessitates an updated phylogenetic tree (Fig. 2A, B). Gene expression data of FveTARs and FveYUCs are included in the same figure to facilitate comparisons of gene expression patterns among family members (Fig. 2). Specifically, five FveTARs (FveTAA1 and FveTAR1–4) and nine FveYUCs (FveYUC1–7 and 10–11) were identified in F. vesca. FveTAR1, FveTAR2, and FveTAR3 shared a high level of similarity to AtTAR2; FveTAA1 was grouped with AtTAA1 and AtTAR1; FveTAR4 was grouped with AtTAR3 and AtTAR4 (Fig. 2A). FveYUCs were divided into two large groups. The first group contained FveYUC1, 5, 10, and 11, which were clustered with AtYUC10 and AtYUC11, while the second group contained the other five FveYUCs (2, 3, 4, 6, and 7) (Fig. 2B).
Fig. 2.
Phylogenetic analysis and expression pattern of FveTARs and FveYUCs in F. vesca. (A, B) The phylogenetic tree of TARs (A) and YUCs (B) in F. vesca and Arabidopsis. (C) Heatmap showing expression patterns of FveTARs and FveYUCs represented by normalized RPKM derived from the RNA-seq dataset (Li et al., 2018). Six genes selected for promoter activity analysis are marked by pins. (D) Expression level of the six selected genes obtained by qRT-PCR in embryo and ghost at stage 4. Gene11892 was used as the internal control. Data are means ±SD obtained from three biological replicates. **P<0.01, Student’s t-test.
The RNA expression of each of the FveTARs and FveYUCs was examined by mining the comprehensive RNA-seq data representing 41 different tissues, mostly floral organs and fruit tissues of F. vesca (Kang et al., 2013; Hollender et al., 2014; Toljamo et al., 2016; Li et al., 2018). Different members of FveTARs and FveYUCs exhibit distinct expression patterns (Fig. 2C; Supplementary Table S2). FveTAA1 is highly expressed in anthers at late developmental stages and mature pollens. FveTAR2 is more abundant in embryos. FveTAR1 and FveTAR3 are highly expressed in ghosts. FveTAR4 is highly and broadly expressed. In the YUC gene family, FveYUC1, 2, 3, and 7 are barely expressed. FveYUC6 is specifically expressed in anthers. FveYUC4 is more abundant in embryos, while FveYUC5, 10, and 11 are highly expressed in ghosts. Of note, none of the FveTARs or FveYUCs was significantly expressed in fruit receptacles, including cortex and pith, with the exception of FveTAR4, which appears to be a ubiquitously expressed gene. Finally, FveTAR1, FveTAR2, and four FveYUCs (4, 5, 10, and 11) were selected to investigate their expression further, as they are likely more relevant to auxin-mediated fruit set due to their abundant and specific expression in the seed. The expression of these six genes was confirmed by qRT-PCR in embryos and ghosts at stage 4, which was consistent with the transcriptome data (Fig. 2C, D).
Detailed analysis of GUS reporter expression of FveTAR2 and FveYUCs in seeds
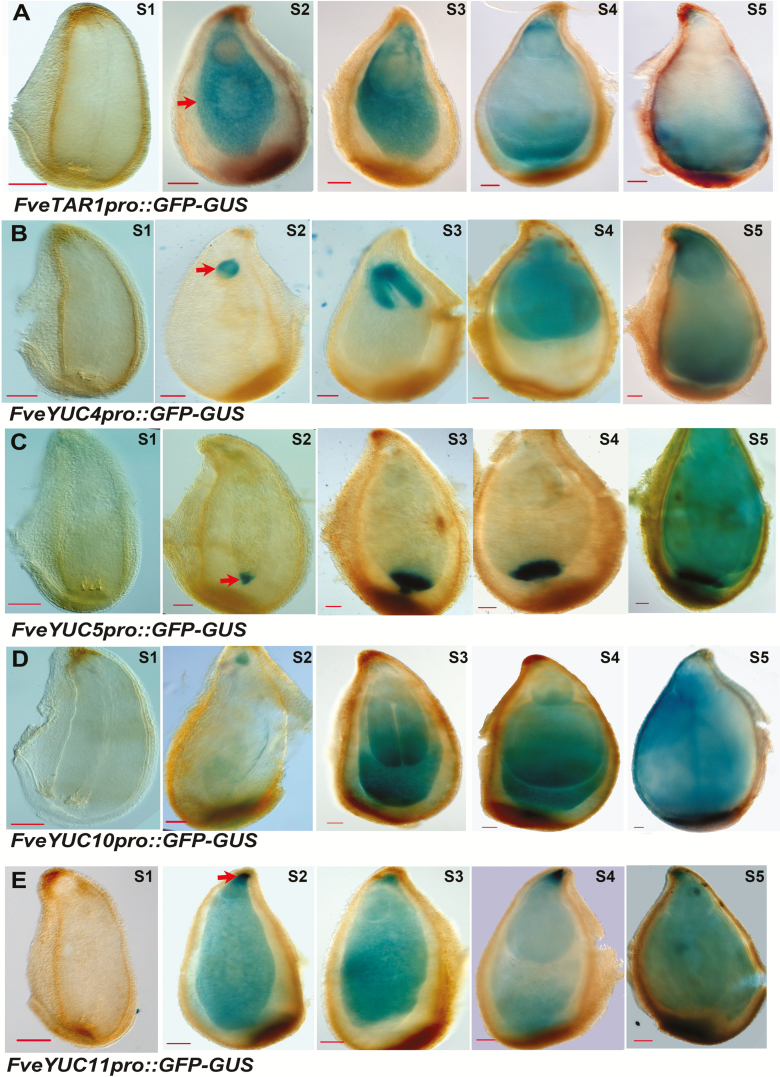
To gain finer tissue resolution of expression for each of the six chosen auxin biosynthetic genes, their promoters (~2 kb upstream of the translational start codon; Supplementary Fig. S2) were respectively isolated and used to drive the GFP-GUS dual reporter gene. The constructs were stably transformed into the F. vesca variety Hawaii 4. With the exception of FveTAR2, fertile transgenic plants for the other five genes were successfully obtained. Two to eight independent transgenic lines for each construct were confirmed by PCR (Supplementary Fig. S1) and then characterized. The ovules and seeds at stages 2–5 of other reporter lines were respectively stained with X-Gluc. For FveTAR1pro::GFP-GUS, no blue color was detected in ovules (stage 1, pre-fertilization), indicating low or no expression of TAR1; however, at stage 2 (soon after fertilization) to stage 5, the FveTAR1 reporter was almost exclusively expressed in the endosperms and at a high level (Fig. 3A). None of the four FveYUCs showed reporter expression in stage 1 ovules; however, their promoters were all induced to a high level post-fertilization (Fig. 3B–E). This strongly suggests that fertilization plays a key role in inducing the biosynthesis of auxin in the seed. Strikingly, these four FveYUCs genes displayed a great divergence in their post-fertilization expression patterns in seeds (Fig. 3B–E). FveYUC4 was predominantly expressed in the embryos at stages 2–5 (Fig. 3B), indicating that FveYUC4 may catalyse auxin biosynthesis primarily for regulating embryogenesis. Among the other three FveYUC genes, FveYUC5 was exclusively expressed in the chalazal endosperms from stage 2 to stage 5 (Fig. 3C) and FveYUC10 and FveYUC11 were more evenly expressed in the entire endosperms post-fertilization (Fig. 3D, E). YUC10’s GUS expression is not as robust as one would expect based on its high RNA-seq reads and qRT-PCR result (Fig. 2C, D). One explanation might be that the 1.4 kb promoter used in the construct lacks some important regulatory elements. FveYUC11 was also highly expressed at the micropylar pole of the seed next to the base of the embryo (Fig. 3E). Interestingly, no GUS staining was observed in fruit receptacles of all of the TARs and YUCs reporter lines (Supplementary Fig. S3), suggesting that auxin synthesis may not occur in the receptacle at early-stage fruit development. Taken together, GUS reporter expression revealed specific and diverse expression patterns of different FveTAR and FveYUC genes within the seeds during early-stage fruit development, highlighting the endosperm as the main site of post-fertilization auxin biosynthesis.
Fig. 3.
Expression pattern of FveTARs and FveYUCs in seeds. (A–E) GUS expression of the FveTAR1pro::GFP-GUS (A), FveYUC4pro::GFP-GUS (B), FveYUC5pro::GFP-GUS (C), FveYUC10pro::GFP-GUS (D), and FveYUC11pro::GFP-GUS (E) reporter lines in stage 1 ovule and stages 2–5 seed, respectively. Arrows indicate endosperm (A), embryo (B), chalazal endosperm (C), and the micropylar pole of the seed (E). Scale bars: 10 µm (A–E).
Expression pattern of FveTARs and FveYUCs in roots revealed by the GUS reporters
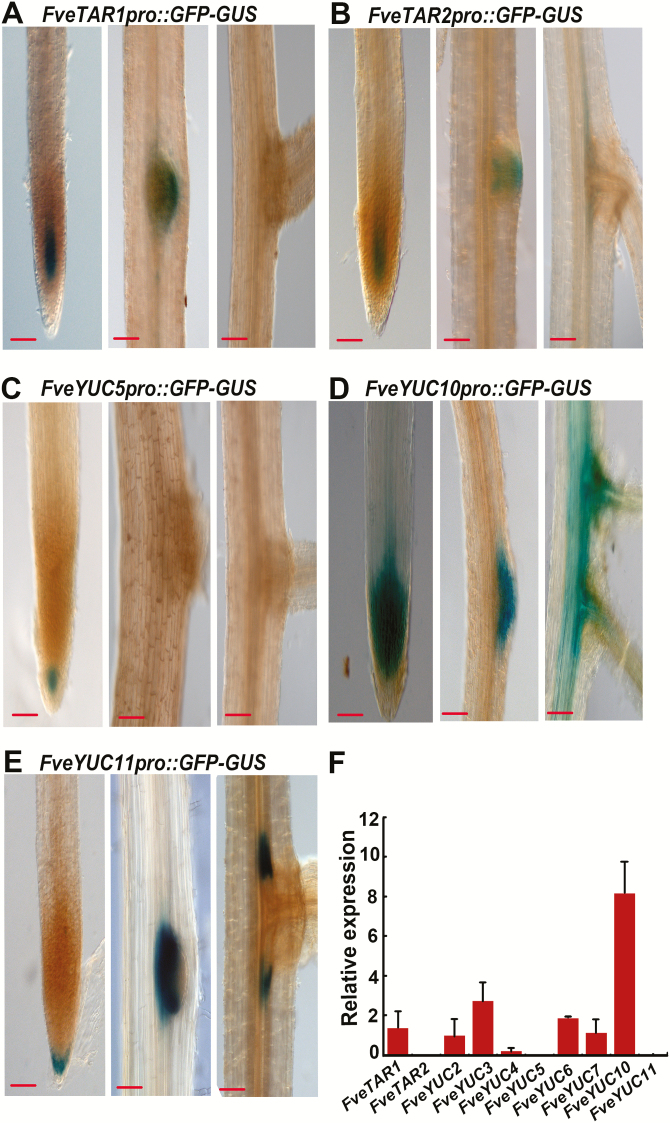
Onsite auxin biosynthesis is also crucial for root patterning (Benková et al., 2003). Hence, we examined the expression pattern in the roots of these reporter lines. Roots of the T0 transgenic plants were collected from soil and stained for GUS. The results showed that FveTAR1 and FveTAR2 were similarly expressed in the stele of the meristematic zone and in the emerging lateral root primordia (Fig. 4A, B). No staining was observed in roots of the FveYUC4pro::GFP-GUS reporter lines. FveYUC5 was only expressed around the quiescent center at a low level (Fig. 4C). By contrast, FveYUC10 was highly expressed in the root meristematic zone, in the entire lateral root primordia, and in the vasculature of subtending roots (Fig. 4D). FveYUC11 was expressed in the columella cells and at the margins of lateral roots (Fig. 4E). Among the six genes examined, FveYUC10 was the most abundantly expressed in roots, which was independently confirmed by qRT-PCR (Fig. 4F). The results revealed distinct expression patterns of these auxin biosynthetic genes in roots, demonstrating that the promoters used here possess distinct regulatory elements endowing these genes with specific expression patterns.
Fig. 4.
Expression pattern of FveTARs and FveYUCs in roots. (A–E) GUS expression of the FveTAR1pro::GFP-GUS (A), FveTAR2pro::GFP-GUS (B), FveYUC5pro::GFP-GUS (C), FveYUC10pro::GFP-GUS (D), and FveYUC11pro::GFP-GUS (E) reporter lines in root. Scale bars: 10 µm (A–E). (F) Expression level of FveTARs and FveYUCs in the roots of wild type obtained by qRT-PCR. Gene11892 was used as the internal control. Data are means ±SD obtained from three biological replicates.
Characterization of the fveyuc10 mutants created by CRISPR/Cas9 in F. vesca
Since FveYUC10 is most abundantly expressed in seeds and roots among the FveYUCs (Figs 2C, D, 4D), we hypothesized that genetic manipulation of FveYUC10 will likely give rise to severe morphological phenotypes and hence reveal the biological significance of auxin biosynthesis in strawberry fruit development. To test this possibility, FveYUC10 (Supplementary Fig. S4) driven by the constitutive 35S promoter was first transformed into wild type Arabidopsis ecotype Col-0. Expression of FveYUC10 was verified by qRT-PCR (Supplementary Fig. S5A). As overexpression of AtYUCs usually resulted in elongated hypocotyls (Hentrich et al., 2013; Zhao et al., 2001), the 35S::FveYUC10 transgenic plants in the T2 generation were grown together with the wild type control under white light. Consequently, the average length of 35S::FveYUC10 hypocotyls was 3.94 mm, significantly longer than the control (1.99 mm, P<0.01) (Supplementary Fig. S5B, C). This result indicates that FveYUC10 should be functional in auxin biosynthesis.
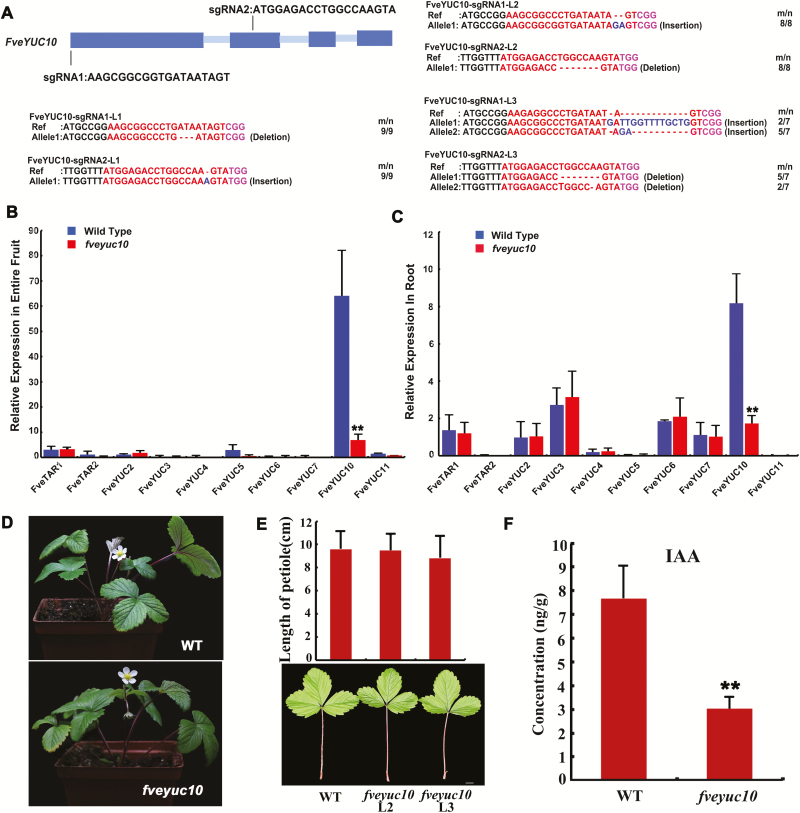
To investigate the function of FveYUC10 in strawberry, CRISPR/Cas9-mediated genome editing was applied to creating fveyuc10 mutants in F. vesca. Briefly, two sgRNAs respectively driven by Arabidopsis U6-26 and U6-29 promoters were designed for FveYUC10 (Fig. 5A). Three independent transgenic lines (L1–3) were obtained and analysed. CRISPR/Cas9-induced mutations were detected by Sanger sequencing of multiple clones of PCR-amplified fragments. The L1 and L2 lines each harbored homozygous mutations, while L3 contained bi-allelic mutations caused by both sgRNAs (Fig. 5A). In L1, the seventh amino acid was lost due to a 3 bp deletion at the sgRNA1 target site, and one nucleotide was inserted at the sgRNA2 site resulting in a truncated protein. In L2, the 2 bp insertion at the sgRNA1 site caused a premature stop codon. L3 had two alleles at the sgRNA1 and sgRNA2 sites. At the sgRNA1 site, one allele had a 13 bp insertion causing a premature stop codon; the other allele was the same as the L2 allele. At the sgRNA2 site, one of the two alleles had a seven-nucleotide deletion and the other a one-nucleotide deletion. Since all the mutations resulted in truncation of FveYUC10, the transcript level was determined by qRT-PCR in fveyuc10 mutants. The transcript level was greatly reduced in fruit of fveyuc10 (Fig. 5B), indicating potential nonsense-mediated decay. To examine whether other FveYUCs would be induced to compensate for the loss of FveYUC10 in fveyuc10 mutants, the expression levels of other FveYUCs were analysed by qRT-PCR in fruit, but their expression remained the same (Fig. 5B). Similar results were also obtained in roots (Fig. 5C). To summarize, the transgene of CRISPR-Cas9 induced a 100% mutation frequency at the FveYUC10 locus, and all the mutations should result in a complete loss of gene functions.
Fig. 5.
Characterization of the fveyuc10 mutants created by CRISPR/Cas9. (A) Genotyping of the three FveYUC10-CRISPR lines (L1–3) in the T0 generation. The protospacer adjacent motif sequence is indicated with magenta. The sgRNA is indicated with red. The mutation sites are indicated with blue. For the term m/n, n indicates the number of clones examined, and m indicates the number of clones showing the indicated genotype. (B) Expression level of FveTARs and FveYUCs in fruit (stage 4) of wild type and fveyuc10 (L3) obtained by qRT-PCR. (C) Expression level of FveTARs and FveYUCs in roots of wild type and fveyuc10 (L3) obtained by qRT-PCR. Gene11892 was used as the internal control. Data are means ±SD obtained from three biological replicates. (D) Morphology of wild type and the fveyuc10 (L3) mutant at the same age. (E) Morphology and length of leaves of wild type and fveyuc10 mutants (L2 and L3). Scale bar: 1 cm. No significant difference was found by Student’s t-test (n=21, 39, and 45 respectively). (F) Free auxin content in fruit of wild type and fveyuc10 (L2 and L3 pooled together) at stage 4. **P<0.01, Student’s t-test, n=4.
Next, the three fveyuc10 mutants were grown together with wild type to observe the phenotypes. There was no significant difference in the length of leaf petioles and rachises (Fig. 5D, E), and the time from fruit set to ripening and the fruit size also remained the same. Furthermore, we measured free auxin contents in the entire fruit at stage 4. The average free auxin content in wild type fruit was 7.65 ng g−1 fresh weight. By contrast, the fveyuc10 fruit contained free auxin of 3.04 ng g−1 fresh weight, accounting for roughly 40% of the wild type control (Fig. 5F, P<0.01). Taken together, loss of FveYUC10 alone was able to reduce free auxin content, but not sufficiently to cause significant morphological defects.
Discussion
Auxin plays critical roles in fruit initiation and fruit development in strawberry (Nitsch, 1950). Previously, we used RNA-seq to examine expression levels of auxin synthesis, transport, and signaling genes in wild strawberry (F. vesca) flower and fruit. An interesting finding was that fertilization-induced auxin biosynthesis genes were mainly expressed in the ghost (seed with embryo removed, consisting of seed coat and endosperm) rather than in the embryo within the seed (Kang et al., 2013). However, we do not know if the auxin biosynthesis genes are expressed in the endosperm or seed coat. Higher tissue resolution of gene expression can be achieved by laser capture microdissection, single-cell sequencing, spatially resolved transcriptome profiling, or reporter gene expression (Brady et al., 2007; Giacomello et al., 2017). In the current study, we generated promoter::reporters in transgenic F. vesca to directly visualize auxin distribution and specific cell types of auxin biosynthesis gene expression. Specifically, DR5ver2 reporter lines revealed that auxin accumulated in the chalazal seed coat and embryo within a seed as well as vasculatures in fruit receptacle (Fig. 1). In contrast, with the exception of FveYUC4, which is embryo-expressed and possibly functions in embryogenesis, FveTAR1 and FveYUC5, 10, and 11 reporters showed endosperm expression (Fig. 3). The above observed endosperm expression of auxin biosynthesis genes combined with DR2ver2::GUS reporter expression in the chalazal seed coat suggests that the endosperm-synthesized auxin likely pools at the chalazal seed coat due to specific transport activities. Indeed, auxin efflux transporter genes FvePIN1 and FvePIN5 were previously shown, via proFvePIN1::GUS and proFvePIN5::GUS reporter genes, to be highly expressed in the chalazal end of the seed coat (Kang et al., 2013), supporting this hypothesis.
DR5ver2::GUS revealed auxin distribution and dynamics in strawberry fruit
Thus far, three versions of DR5, the synthetic auxin-responsive reporter, have been published, namely DR5 (Ulmasov et al., 1997), DR5rev (Friml et al., 2002), and DR5ver2 (Liao et al., 2015). All three reporters report auxin signaling activity and hence are indirect indicators of auxin levels. Compared with DR5 and DR5rev, DR5ver2 displayed an expanded expression region with enhanced expression levels (Liao et al., 2015) and was chosen for this study.
We showed that DR5ver2::GUS was specifically expressed in root tips and responded to auxin application in the root and fruit of F. vesca (Fig. 1), indicating that it is a good indicator of auxin activity in strawberry. The strong expression of DR5ver2::GUS in the receptacle vasculature is consistent with previous results using DR5 in the cultivated strawberry (Estrada-Johnson et al., 2017) and indicates a possible traffic route of auxin from the endosperm, to the chalazal seed coat, and then to the receptacle via the vasculature connecting them. Expression of DR5ver2::GUS in the receptacle displayed an initial increase and then a reduction, a trend similar to the free auxin level change in the achenes at early fruit developmental stages (Symons et al., 2012). This suggests that the receptacle at the receiving end of the auxin transport experiences a similar dynamic change in auxin levels, albeit at a much lower level based on prior measurement of free auxin (Nitsch, 1950, 1955; Dreher and Poovaiah, 1982; Symons et al., 2012; Estrada-Johnson et al., 2017). Nevertheless, the increase in auxin in both achenes and receptacle coincides with and may be responsible for achene and receptacle enlargement during early-stage fruit development.
Auxin is mainly produced in the endosperms with potential roles for seed coat and fruit development
The formation of auxin gradients in embryo has drawn much attention as it is a good model system to study organogenesis (Benková et al., 2003), but where auxin is synthesized in other seed tissues other than embryo is not known. We showed that certain auxin biosynthesis genes were highly expressed in the endosperm (Fig. 3). For example, FveYUC5 was specifically expressed in the chalazal endosperm. Moreover, three of the five genes studied, namely FveTAR1, FveYUC10, and FveYUC11, are more highly expressed in the endosperm, highlighting endosperm as the primary auxin biosynthesis sites within the seeds. In fact, some YUCs in other plant species have been discovered to be expressed in the endosperm, such as AtYUC10 in Arabidopsis and ZmYUC1 in maize (Bernardi et al., 2012; Figueiredo et al., 2015). Therefore, auxin production in the endosperm seems to be a universal phenomenon in plants. The biological function of the endosperm-derived auxin remains to be determined. One hypothesis is that endosperm-derived auxin is primarily targeted for endosperm, seed coat, and fruit development with no or little role in embryogenesis. In support of this hypothesis, auxin was found essential for endosperm development in Arabidopsis and maize (Bernardi et al., 2012; Figueiredo et al., 2015). In addition, endosperm-derived auxin was also important for seed coat growth (Figueiredo et al., 2016). It is possible that endosperm-derived auxin is first transported to the seed coat (as shown by DR5ver2 in Fig. 1) and then to the fruit to stimulate their respective growth.
Auxin production and function in roots
TARs and YUCs play important roles in root development. Loss of their functions can cause moderate to very severe root defects in Arabidopsis (Cheng et al., 2007; Stepanova et al., 2008). This study provided a first look at the expression patterns of several FveTARs and FveYUCs in strawberry roots (Fig. 4). Auxin was considered to be produced in roots at significant levels with multiple sources in Arabidopsis (Ljung et al., 2005). Consistent with this report, FveTARs and FveYUCs exhibited diverse and distinct expression patterns in roots, including vascular tissues, root tips, lateral root primordia, and cells at the base surrounding lateral root primordia (Fig. 4). According to RNA-seq and qRT-PCR, FveYUC10, FveYUC3, and FveYUC6 were more abundantly expressed in roots, and may play important roles in root development. However, FveYUC11, which is expressed at an extremely low level in root based on RNA-seq (Fig. 2C) and qRT-PCR (Fig. 4F), showed a highly specific and unique tissue expression pattern at the base of the lateral primordium, indicating that FveYUC11 likely facilitates auxin biosynthesis for lateral root primordial emergence and growth. Our data demonstrate that GUS reporters can reveal extremely useful gene expression information not provided by RNA-seq or qRT-PCR.
Loss-of-function mutations in FveYUC10 did not cause morphological phenotypes
We found that FveYUC10 is the most abundantly expressed YUC gene in both seeds and roots in F. vesca (Kang et al., 2013), which was also verified in an independent study (Liu et al., 2014). In this study, fveyuc10 mutants, created by the CRISPR/Cas9-mediated genome editing (Yang et al., 2017), had a reduced level of auxin content in fruit (Fig. 5); however, no obvious morphological phenotypes were observed. In Arabidopsis, YUC single mutants usually do not exhibit obvious developmental defects due to functional redundancy (Cheng et al., 2006; Chen et al., 2014). However, other plant species may not have as many copies of YUC/TAA genes as Arabidopsis and single mutants could lead to significant defects, such as FveYUC6 RNAi in wild strawberry, de18 (defective endosperm18, ZmYUC1) and spi1 (sparse inflorescence1) in maize, and floozy in petunia (Tobeña-Santamaria et al., 2002; Gallavotti et al., 2008; Bernardi et al., 2012; Liu et al., 2014). The lack of phenotype in fveyuc10 in F. vesca could be explained by functional redundancy among multiple YUC genes in the F. vesca genome, or the existence of a feedback loop, or alternatively biosynthesis pathways (Tivendale et al., 2014) that could compensate for the loss of FveYUC10.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Validation of the positive transgenic plants examined by RT-PCR.
Fig. S2. Promoter sequences of FveTARs and FveYUCs used for plasmid construction.
Fig. S3. GUS staining of receptacles.
Fig. S4. Protein sequence of FveYUC10.
Fig. S5. Phenotypes of the 35S::FveYUC10 transgenic plants in Arabidopsis.
Table S1. The list of primers used in this study.
Table S2. Expression patterns of FveTARs and FveYUCs in the transcriptome database represented by RPKM.
Author contributions
CK, JF, and ZL conceived and designed the experiments. JF, CD, HL, and YH performed the experiments. CK, ZL, and JF wrote the paper. All authors have read and approved the manuscript.
Supplementary Material
Acknowledgements
The authors would like to thank Dr Xu Chen (Haixia Institute of Science and Technology) for helpful discussions. This work was supported by the National Natural Science Foundation of China (31572098) and Huazhong Agricultural University Scientific & Technological Self-innovation Foundation Grants (2014RC005) and (2014RC017). The authors declare no conflicts of interest associated with this manuscript.
References
- Benková E, Michniewicz M, Sauer M, Teichmann T, Seifertová D, Jürgens G, Friml J. 2003. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115, 591–602. [DOI] [PubMed] [Google Scholar]
- Bernardi J, Lanubile A, Li QB, Kumar D, Kladnik A, Cook SD, Ross JJ, Marocco A, Chourey PS. 2012. Impaired auxin biosynthesis in the defective endosperm18 mutant is due to mutational loss of expression in the ZmYuc1 gene encoding endosperm-specific YUCCA1 protein in maize. Plant Physiology 160, 1318–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady SM, Orlando DA, Lee JY, Wang JY, Koch J, Dinneny JR, Mace D, Ohler U, Benfey PN. 2007. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318, 801–806. [DOI] [PubMed] [Google Scholar]
- Chai YM, Jia HF, Li CL, Dong QH, Shen YY. 2011. FaPYR1 is involved in strawberry fruit ripening. Journal of Experimental Botany 62, 5079–5089. [DOI] [PubMed] [Google Scholar]
- Chen Q, Dai X, De-Paoli H, Cheng Y, Takebayashi Y, Kasahara H, Kamiya Y, Zhao Y. 2014. Auxin overproduction in shoots cannot rescue auxin deficiencies in Arabidopsis roots. Plant & Cell Physiology 55, 1072–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Dai X, Zhao Y. 2006. Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes & Development 20, 1790–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Dai X, Zhao Y. 2007. Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. The Plant Cell 19, 2430–2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U. 2003. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiology 133, 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darrow GM. 1966. The strawberry: history, breeding and physiology. New York: Holt, Rinehart & Winston. [Google Scholar]
- Dreher TW, Poovaiah BW. 1982. Changes in auxin content during development in strawberry fruits. Journal of Plant Growth Regulation 1, 267–276. [Google Scholar]
- Edger PP, VanBuren R, Colle M, et al.. 2018. Single-molecule sequencing and optical mapping yields an improved genome of woodland strawberry (Fragaria vesca) with chromosome-scale contiguity. GigaScience 7, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada-Johnson E, Csukasi F, Pizarro CM, et al.. 2017. Transcriptomic analysis in strawberry fruits reveals active auxin biosynthesis and signaling in the ripe receptacle. Frontiers in Plant Science 8, 889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiredo DD, Batista RA, Roszak PJ, Hennig L, Kohler C. 2016. Auxin production in the endosperm drives seed coat development in Arabidopsis. Elife 5, e20542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiredo DD, Batista RA, Roszak PJ, Köhler C. 2015. Auxin production couples endosperm development to fertilization. Nature Plants 1, 15184. [DOI] [PubMed] [Google Scholar]
- Friml J, Benková E, Blilou I, et al.. 2002. AtPIN4 mediates sink-driven auxin gradients and root patterning in Arabidopsis. Cell 108, 661–673. [DOI] [PubMed] [Google Scholar]
- Gallavotti A, Barazesh S, Malcomber S, Hall D, Jackson D, Schmidt RJ, McSteen P. 2008. Sparse inflorescence1 encodes a monocot-specific YUCCA-like gene required for vegetative and reproductive development in maize. Proceedings of the National Academy of Sciences, USA 105, 15196–15201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacomello S, Salmén F, Terebieniec BK, et al.. 2017. Spatially resolved transcriptome profiling in model plant species. Nature Plants 3, 17061. [DOI] [PubMed] [Google Scholar]
- Given NK, Venis MA, Gierson D. 1988. Hormonal regulation of ripening in the strawberry, a non-climacteric fruit. Planta 174, 402–406. [DOI] [PubMed] [Google Scholar]
- Hentrich M, Böttcher C, Düchting P, Cheng Y, Zhao Y, Berkowitz O, Masle J, Medina J, Pollmann S. 2013. The jasmonic acid signaling pathway is linked to auxin homeostasis through the modulation of YUCCA8 and YUCCA9 gene expression. The Plant Journal 74, 626–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollender CA, Geretz AC, Slovin JP, Liu Z. 2012. Flower and early fruit development in a diploid strawberry, Fragaria vesca. Planta 235, 1123–1139. [DOI] [PubMed] [Google Scholar]
- Hollender CA, Kang C, Darwish O, Geretz A, Matthews BF, Slovin J, Alkharouf N, Liu Z. 2014. Floral transcriptomes in woodland strawberry uncover developing receptacle and anther gene networks. Plant Physiology 165, 1062–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia HF, Chai YM, Li CL, Lu D, Luo JJ, Qin L, Shen YY. 2011. Abscisic acid plays an important role in the regulation of strawberry fruit ripening. Plant Physiology 157, 188–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang C, Darwish O, Geretz A, Shahan R, Alkharouf N, Liu Z. 2013. Genome-scale transcriptomic insights into early-stage fruit development in woodland strawberry Fragaria vesca. The Plant Cell 25, 1960–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Jia H, Chai Y, Shen Y. 2011. Abscisic acid perception and signaling transduction in strawberry: a model for non-climacteric fruit ripening. Plant Signaling & Behavior 6, 1950–1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Wei W, Feng J, Luo H, Pi M, Liu Z, Kang C. 2018. Genome re-annotation of the wild strawberry Fragaria vesca using extensive illumina- and SMRT-based RNA-seq datasets. DNA Research 25, 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao CY, Smet W, Brunoud G, Yoshida S, Vernoux T, Weijers D. 2015. Reporters for sensitive and quantitative measurement of auxin response. Nature Methods 12, 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Xie WF, Zhang L, Valpuesta V, Ye ZW, Gao QH, Duan K. 2014. Auxin biosynthesis by the YUCCA6 flavin monooxygenase gene in woodland strawberry. Journal of Integrative Plant Biology 56, 350–363. [DOI] [PubMed] [Google Scholar]
- Liu H, Ying YY, Zhang L, Gao QH, Li J, Zhang Z, Fang JG, Duan K. 2012. Isolation and characterization of two YUCCA flavin monooxygenase genes from cultivated strawberry (Fragaria×ananassa Duch.). Plant Cell Reports 31, 1425–1435. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2 −ΔΔC(T) method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Ljung K, Hull AK, Celenza J, Yamada M, Estelle M, Normanly J, Sandberg G. 2005. Sites and regulation of auxin biosynthesis in Arabidopsis roots. The Plant Cell 17, 1090–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashiguchi K, Tanaka K, Sakai T, et al. 2011. The main auxin biosynthesis pathway in Arabidopsis. Proceedings of the National Academy of Sciences, USA 108, 18512–18517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng JL, Hassan S, Truong TT, Hocart CH, Laffont C, Frugier F, Mathesius U. 2015. Flavonoids and auxin transport inhibitors rescue symbiotic nodulation in the Medicago truncatula cytokinin perception mutant cre1. The Plant Cell 27, 2210–2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitsch JP. 1950. Growth and morphogenesis of the strawberry as related to auxin. American Journal of Botany 37, 211–215. [Google Scholar]
- Nitsch JP. 1955. Free auxins and free tryptophane in the strawberry. Plant Physiology 30, 33–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottenschläger I, Wolff P, Wolverton C, Bhalerao RP, Sandberg G, Ishikawa H, Evans M, Palme K. 2003. Gravity-regulated differential auxin transport from columella to lateral root cap cells. Proceedings of the National Academy of Sciences, USA 100, 2987–2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert HS, Grones P, Stepanova AN, Robles LM, Lokerse AS, Alonso JM, Weijers D, Friml J. 2013. Local auxin sources orient the apical-basal axis in Arabidopsis embryos. Current Biology 23, 2506–2512. [DOI] [PubMed] [Google Scholar]
- Slovin JP, Schmitt K, Folta KM. 2009. An inbred line of the diploid strawberry Fragaria vesca f. semperflorens for genomic and molecular genetic studies in the Rosaceae. Plant Methods 5, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova AN, Robertson-Hoyt J, Yun J, Benavente LM, Xie DY, Dolezal K, Schlereth A, Jürgens G, Alonso JM. 2008. TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133, 177–191. [DOI] [PubMed] [Google Scholar]
- Stepanova AN, Yun J, Robles LM, Novak O, He W, Guo H, Ljung K, Alonso JM. 2011. The Arabidopsis YUCCA1 flavin monooxygenase functions in the indole-3-pyruvic acid branch of auxin biosynthesis. The Plant Cell 23, 3961–3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symons GM, Chua YJ, Ross JJ, Quittenden LJ, Davies NW, Reid JB. 2012. Hormonal changes during non-climacteric ripening in strawberry. Journal of Experimental Botany 63, 4741–4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Ferrer JL, Ljung K, et al.. 2008. Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133, 164–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tivendale ND, Ross JJ, Cohen JD. 2014. The shifting paradigms of auxin biosynthesis. Trends in Plant Science 19, 44–51. [DOI] [PubMed] [Google Scholar]
- Tobeña-Santamaria R, Bliek M, Ljung K, Sandberg G, Mol JN, Souer E, Koes R. 2002. FLOOZY of petunia is a flavin mono-oxygenase-like protein required for the specification of leaf and flower architecture. Genes & Development 16, 753–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toljamo A, Blande D, Kärenlampi S, Kokko H. 2016. Reprogramming of strawberry (Fragaria vesca) root transcriptome in response to Phytophthora cactorum. PLoS One 11, e0161078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. 1997. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. The Plant Cell 9, 1963–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won C, Shen X, Mashiguchi K, Zheng Z, Dai X, Cheng Y, Kasahara H, Kamiya Y, Chory J, Zhao Y. 2011. Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF ARABIDOPSIS and YUCCAs in Arabidopsis. Proceedings of the National Academy of Sciences, USA 108, 18518–18523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing HL, Dong L, Wang ZP, Zhang HY, Han CY, Liu B, Wang XC, Chen QJ. 2014. A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biology 14, 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada M, Greenham K, Prigge MJ, Jensen PJ, Estelle M. 2009. The TRANSPORT INHIBITOR RESPONSE2 gene is required for auxin synthesis and diverse aspects of plant development. Plant Physiology 151, 168–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Wu JJ, Tang T, Liu KD, Dai C. 2017. CRISPR/Cas9-mediated genome editing efficiently creates specific mutations at multiple loci using one sgRNA in Brassica napus. Scientific Reports 7, 7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Christensen SK, Fankhauser C, Cashman JR, Cohen JD, Weigel D, Chory J. 2001. A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291, 306–309. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.