Abstract
This article comments on:
Jiang J, Xi H, Dai Z, Lecourieux F, Yuan L, Liu X, Patra B, Wei Y, Li S, Wang L. 2019. VvWRKY8 negatively regulates VvSTS through direct interaction with VvMYB14 to balance resveratrol biosynthesis in grapevine. Journal of Experimental Botany 70, 715–729.
Keywords: Grapevine, phenylpropanoid biosynthesis, protein–protein interactions, regulation, resveratrol, secondary metabolism, VvMYB14, VvWRKY8
Transcription factors are key components in the regulation of metabolic pathways underlying numerous plant functions. Jiang et al. (2019) showed that the WRKY8 transcription factor fine-tunes biosynthesis of the phytoalexin resveratrol in grapevine through negative regulation of the stilbene synthase gene. This paves the way for new approaches in our understanding of the regulation of phytoalexin biosynthesis in plants and, through this, improved phytoalexin production in engineered disease resistance.
During their life cycle, plants face a large number of abiotic and biotic stresses including salt, cold, heat, drought, wounding, osmotic pressure, UV and pathogenic microorganisms. In order to cope, an intricate network has developed comprising stress-responsive signal transduction pathways and adaptive mechanisms involving both primary and secondary metabolism. Regulation of the plant transcriptome involved in these responses requires the action of an impressive predicted number of transcription factors (TFs), varying from 1500 to 1600 according to genome-wide identification analyses carried out in rice and Arabidopsis (Iida et al., 2005; Xiong et al., 2005; Agarwal et al., 2011).
WRKY proteins have been recognized as one of the ten largest families of TFs in higher plants, though they are absent in animals (Llorca et al., 2014). They are so called because of their characteristic DNA-binding domain of around 60 amino acids which contains, either once or twice, the quasi-invariant WRKYGQK amino acid sequence at the N-terminus and a zinc-finger structure at the C-terminus constituting the WRKY domain (Llorca et al., 2014). Both the WRKYGQK motif and the zinc finger are necessary for the DNA binding of TFs. Specifically, the WRKY domain binds the W-box (TTGACC/T) motifs of the promoters of target genes to modulate their expression (Llorca et al., 2014; Schluttenhofer and Yuan, 2015). In addition, the DNA sequences directly adjacent to the W-boxes have been shown to be important for the specific DNA-binding activity of the WRKY TFs (Ciolkowski et al., 2008).
Most WRKY TFs are located in the nucleus though some studies have reported the cytosolic localization of the Arabidopsis AtWRKY40, which inhibits the expression of ABA-responsive genes (Shang et al., 2010). TFs play a role as positive and negative regulators of major plant functions such as growth, development and senescence, defence, abiotic stresses and hormone signalling (Agarwal et al., 2011). For example, AtWRKY6 positively influences the promoter activity of the senescence SIRK gene while AtWRKY44 (TTG2) controls trichome and seed coat development in Arabidopsis (Johnson et al., 2002; Robatzek and Somssich, 2002). Importantly, WRKY TFs were also shown to be key regulators of plant secondary metabolism (Schluttenhofer and Yuan, 2015), including phytoalexin biosynthesis (Jiang et al., 2019).
WRKY TFs are key regulators of plant secondary metabolism
WRKY TFs intervene in numerous pathways of secondary metabolism relating to a wide array of biological functions in plants (Schluttenhofer and Yuan, 2015). They are also implicated in the biosynthesis of many metabolites of pharmaceutical significance. Artemisia annua AaWRKY1 positively regulates a cytochrome P450 monooxygenase in the biosynthetic route to the antimalarial drug artemisinin (Chen et al., 2017), whereas the Taxus chinensis TcWRKY1 regulates a 10‐deacetylbaccatin III‐10 β‐O‐acetyl transferase, a critical enzyme in the biosynthesis of the anticancer drug taxol (Li et al., 2013). TF expression is also linked to the biosynthesis of some phytoalexins, which are secondary metabolites of low molecular weight synthesized by plants as a response to both abiotic and biotic stresses and displaying antimicrobial activity (Jeandet et al., 2014). For example, AtWRKY40/18, Oryza sativa OsWRKY45 and Vitis vinifera VvWRKY8 TFs regulate the production of, respectively, camalexin in the Cruciferae (Pandey et al., 2010), momilactone, oryzalexin and phytocassane phytoalexins in the Poaceae (Akagi et al., 2014) and the resveratrol stilbene phytoalexin in the Vitaceae (Jiang et al., 2019). Although resveratrol has been the subject of a plethora of research (Jeandet et al., 2018), the mechanisms regulating the biosynthesis of this compound are still poorly understood. In their continuing efforts to decipher the regulatory mechanisms underlying the biosynthesis of resveratrol in Vitaceae, Jiang et al. (2019) report on the characterization of a WRKY transcription factor, VvWRKY8; this negatively regulates stilbene synthase, which catalyses the final committed step in the resveratrol pathway.
Regulation mechanisms of stilbene synthase in Vitaceae
Resveratrol is obtained from the universal phenylpropanoid pathway starting either from phenylalanine or tyrosine and leading through two or three steps to para-coumaroyl-CoA. Para-coumaroyl-CoA is then linked to three malonyl-CoA units to form resveratrol under the action of the plant polyketide synthase III, stilbene synthase (STS) (Jeandet et al., 2013, 2014, 2018) (Box 1). A few studies have reported on the regulatory mechanisms of the phenylpropanoid pathway and downstream pathways (lignins and flavonoids) in grapevine (Vitis vinifera). For example, the VvWRKY2 and VvMYB5a TFs were shown to activate transcription of the cinnamate-4-hydroxylase gene (VvC4H), suggesting their possible role in lignification processes in connection or not with plant disease resistance (Guillaumie et al., 2010) or in anthocyanin biosynthesis (Deluc et al., 2006). VvWRKY26 positively regulates chalcone synthase, the key enzyme of the flavonoid pathway as well as flavonoid hydroxylases (F3’H and F3’, 5’H) acting as decorating enzymes on the flavonoid core structure in grapevine (Amato et al., 2016).
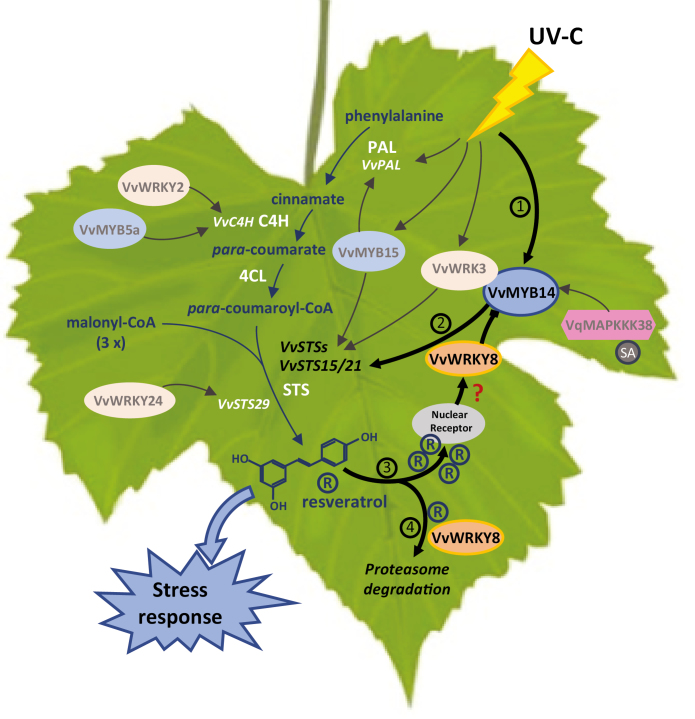
Box 1. Resveratrol biosynthesis in grapevine leaves: model of the transcriptional regulatory loop
Resveratrol is produced through the action of STS by condensation of para-coumaroyl-CoA and three malonyl CoA units. UV-C stress leads to induction of the VvMYB14 gene (1). Accumulation of VvMYB14, with the combinatorial effect of VvWRKY3, results in the up-regulation of the VvSTS29 gene (Vannozzi et al., 2018) (2). At high concentration, resveratrol (R) induces VvWRKY8 expression (possibly through a putative nuclear receptor), negatively regulating VvMYB14 and thereby stopping VsSTS expression and decreasing resveratrol production (3). At low resveratrol concentration, VvWRKY8 is channelled towards degradation through the ubiquitin ligase proteasome pathway, allowing the end of VvMYB14 repression and a new induction of VvSTS genes thereby increasing resveratrol production (4). Molecules of the resveratrol biosynthesis pathway are shown in blue; enzymes involved are white (PAL, phenylalanine ammonia lyase; C4H, cinnamate-4-hydroxylase; 4CL, p-coumarate-CoA ligase; STS, stilbene synthase) (Jeandet et al., 2018). The MYB TFs are shown as blue ellipses; WRKY TFs are orange ellipses. The darker colours and bold arrows highlight the different actors, with some involved in the regulatory loop described in Jiang et al. (2019).
The first study implicating TFs in the regulation of STS in response to stress conditions identified two R2R3-MYB TFs, VvMYB14 and VvMYB15, for which the corresponding genes were strongly co-expressed with two STS genes, VvSTS41/29 (Höll et al., 2013). A further study from Corso et al. (2015), based on transcriptome comparison of two grapevine genotypes with different drought susceptibility, led to the identification of five additional WRKY TFs (VvWRKY24/28/29/37/41) that were co-expressed simultaneously with eight VvSTS transcripts (VvSTS12/13/16/17/18/24/27/29) in roots and leaves. Recently, the integrated Gene Coexpression Network (GCN) analysis of STS and TFs suggested that a great number of TFs belonging to various families such as WRKYs, MYBs and even ERFs can putatively contribute to STS regulation (Wong et al., 2016; Vannozzi et al., 2018). Indeed, three MYB TFs (VvMYB13, VvMYB14 and VvMYB15) and four WRKY TFs (VvWRKY3, VvWRKY24, VvWRKY43 and VvWRKY53) are co-expressed with VvSTS genes following biotic or abiotic stresses, raising the question of a possible role of these TFs in the regulation of VvSTS gene expression in Vitis spp. Moreover, VvWRKY24 induces the expression of the VvSTS29 gene independently of VvMYB14 or VvMYB15 whereas VvWRKY3 and VvMYB14 have a combinatorial effect on the transcription of the VvSTS29 gene (Vannozzi et al., 2018) (Box 1). Genes of the MAPK pathway were also found to be involved in the activation of STS transcription in V. quinquangularis (Jiao et al., 2017). A Raf-like MAPKKK gene, VqMAPKKK38, was indeed shown to positively regulate STS in grapevine leaves in combination with the salicylic acid hormone (SA), likely by induction of VvMYB14 transcription. Overall, this highlights the intricate network underlying the regulation mechanisms of STS in grapevine. VvSTS genes can thus be regulated by the combinatorial action of MYB and WRKY transcription factors possibly in association with MAPKs (Box 1).
The research reported by Jiang et al. (2019) characterizes a negative regulator of resveratrol biosynthesis for the first time. It demonstrates a dosage-dependent inhibition of a VvSTS-inducing VvMYB14 TF by an N-terminus-mediated interaction with VvWRKY8 lacking transcriptional activity (Box 1). After UV-C exposure, the expression of VvSTSs, VvWRKY8 and VvMYB14 genes rose sharply in grapevine leaves. However, even if VvWRKY8 does not display any transcriptional activity in yeast, the transient VvWRKY8 overexpression in grapevine leaves led to a down-regulation of VvSTS15/21 and VvMYB14 gene expression along with a significant reduction of the resveratrol content. Further analysis showed that VvWRKY8 neither binds nor activates the promoter of VvSTS15/21 and VvMYB14 genes, rather it interacts directly with VvMYB14 at the N-terminus, thereby inhibiting the binding of VvMYB14 to the VvSTS15/21 promoter. Moreover, exogenous application of resveratrol in cell suspension cultures significantly increased VvWRKY8 expression, whereas VvSTS15/21 and VvMYB14 expression decreased. However, VvWRKY8 overexpressing cells display a higher VvWRKY8 accumulation when they are treated with a proteasome inhibitor, suggesting a possible role of the ubiquitin ligase system in regulating the activity of VvWRKY8. This in turn allows the fine tuning of resveratrol biosynthesis. Together these results indicate the existence of a negative regulatory loop involving the VvMYB14 activator TF and its negative regulator VvWRKY8, the key enzymes VvSTS15/21 and the final resveratrol product that allows fine regulation of the resveratrol biosynthetic pathway. This study provides the description of a key step that deepens our understanding of the regulatory mechanisms of resveratrol biosynthesis and more generally the biosynthesis of phytoalexins in plants.
Perspectives
New ways of investigation arise from the findings of Jiang et al. (2019). The fact that resveratrol production may result in the up-regulation of VvWRKY8, which in turn negatively regulates VvSTS genes to fine-tune the accumulation of this phytoalexin in grapevine tissues, triggers the question of the mechanisms underlying resveratrol interaction with VvWRKY8. As already noted, TFs are mainly located in the nucleus though some with cytosolic localization have been reported (Shang et al., 2010). It could then be hypothesized that a putative nuclear resveratrol-binding receptor is activated at a high resveratrol content level and then induces VvWRKY8 expression to negatively regulate the expression of STS through VvMYB14 (Box 1). Resveratrol has indeed already been found to bind to and activate a vitamin D nuclear receptor in various human tissues (Dampf Stone et al., 2015). In that sense, further research is needed to explore a possible potentiation of VvWRKY8 through nuclear receptor signalling.
The existence of a negative feedback in resveratrol biosynthesis including the regulatory loop resveratrol–putative nuclear receptor–VvWRKY8–VvMYB14–VvSTS is of great importance and may have interesting applications in the molecular engineering of phytoalexin pathways in plants. In fact, the main problems encountered in this research area stem from the impossibility of obtaining ectopic production of a given phytoalexin in engineered plants (Delaunois et al., 2009; Jeandet et al., 2017). The study of Jiang et al. (2019) suggests for the first time the existence of TF negatively-regulated phytoalexin production in grapevine. It may partly explain why the overexpression of phytoalexin genes in plants does not always result in high phytoalexin production levels. The down-regulation of VvWRKY8 expression by CRISPR-Cas technology combined with STS overexpression might represent an appropriate approach to improve resveratrol production and plant disease resistance. Strengthening VvWRKY8 channelling to the ubiquitin ligase proteasome pathway (Box 1) to decrease VvMYB14 repression would also constitute an interesting option for increasing resveratrol phytoalexin production in plants.
References
References
- Agarwal P, Reddy MP, Chikara J.. 2011. WRKY: its structure, evolutionary relationship, DNA-binding selectivity, role in stress tolerance and development of plants. Molecular Biology Reports 38, 3883–3896. [DOI] [PubMed] [Google Scholar]
- Akagi A, Fukushima S, Okada K, Jiang CJ, Yoshida R, Nakayama A, Shimono M, Sugano S, Yamane H, Takatsuji H.. 2014. WRKY45-dependent priming of diterpenoid phytoalexin biosynthesis in rice and the role of cytokinin in triggering the reaction. Plant Molecular Biology 86, 171–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amato A, Cavallini E, Zenoni S, Finezzo L, Begheldo M, Ruperti B, Tornielli GB.. 2016. A Grapevine TTG2-Like WRKY Transcription Factor Is Involved in Regulating Vacuolar Transport and Flavonoid Biosynthesis. Frontiers in Plant Science 7, 1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Yan T, Shen Q, et al. 2017. GLANDULAR TRICHOME-SPECIFIC WRKY 1 promotes artemisinin biosynthesis in Artemisia annua. The New phytologist 214, 304–316. [DOI] [PubMed] [Google Scholar]
- Ciolkowski I, Wanke D, Birkenbihl RP, Somssich IE.. 2008. Studies on DNA-binding selectivity of WRKY transcription factors lend structural clues into WRKY-domain function. Plant Molecular Biology 68, 81–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corso M, Vannozzi A, Maza E, et al. 2015. Comprehensive transcript profiling of two grapevine rootstock genotypes contrasting in drought susceptibility links the phenylpropanoid pathway to enhanced tolerance. Journal of Experimental Botany 66, 5739–5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dampf Stone A, Batie SF, Sabir MS, Jacobs ET, Lee JH, Whitfield GK, Haussler MR, Jurutka PW.. 2015. Resveratrol potentiates vitamin D and nuclear receptor signaling. Journal of Cellular Biochemistry 116, 1130–1143. [DOI] [PubMed] [Google Scholar]
- Delaunois B, Cordelier S, Conreux A, Clément C, Jeandet P.. 2009. Molecular engineering of resveratrol in plants. Plant Biotechnology Journal 7, 2–12. [DOI] [PubMed] [Google Scholar]
- Deluc L, Barrieu F, Marchive C, Lauvergeat V, Decendit A, Richard T, Carde JP, Mérillon JM, Hamdi S.. 2006. Characterization of a grapevine R2R3-MYB transcription factor that regulates the phenylpropanoid pathway. Plant Physiology 140, 499–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillaumie S, Mzid R, Méchin V, Léon C, Hichri I, Destrac-Irvine A, Trossat-Magnin C, Delrot S, Lauvergeat V.. 2010. The grapevine transcription factor WRKY2 influences the lignin pathway and xylem development in tobacco. Plant Molecular Biology 72, 215–234. [DOI] [PubMed] [Google Scholar]
- Höll J, Vannozzi A, Czemmel S, D’Onofrio C, Walker AR, Rausch T, Lucchin M, Boss PK, Dry IB, Bogs J.. 2013. The R2R3-MYB transcription factors MYB14 and MYB15 regulate stilbene biosynthesis in Vitis vinifera. The Plant Cell 25, 4135–4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida K, Seki M, Sakurai T, Satou M, Akiyama K, Toyoda T, Konagaya A, Shinozaki K.. 2005. RARTF: database and tools for complete sets of Arabidopsis transcription factors. DNA Research 12, 247–256. [DOI] [PubMed] [Google Scholar]
- Jeandet P, Clément C, Courot E, Cordelier S.. 2013. Modulation of phytoalexin biosynthesis in engineered plants for disease resistance. International Journal of Molecular Sciences 14, 14136–14170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeandet P, Courot E, Clément C, Ricord S, Crouzet J, Aziz A, Cordelier S.. 2017. Molecular Engineering of Phytoalexins in Plants: Benefits and Limitations for Food and Agriculture. Journal of Agricultural and Food Chemistry 65, 2643–2644. [DOI] [PubMed] [Google Scholar]
- Jeandet P, Hébrard C, Deville MA, Cordelier S, Dorey S, Aziz A, Crouzet J.. 2014. Deciphering the role of phytoalexins in plant-microorganism interactions and human health. Molecules 19, 18033–18056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeandet P, Sobarzo-Sánchez E, Clément C, Nabavi SF, Habtemariam S, Nabavi SM, Cordelier S.. 2018. Engineering stilbene metabolic pathways in microbial cells. Biotechnology Advances 36, 2264–2283. [DOI] [PubMed] [Google Scholar]
- Jiang J, Xi H, Dai Z, Lecourieux F, Yuan L, Liu X, Patra B, Wei Y, Li S, Wang L.. 2019. VvWRKY8 negatively regulates VvSTS through direct interaction with VvMYB14 to balance resveratrol biosynthesis in grapevine. Journal of Experimental Botany 70, 715–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Wang D, Wang L, Jiang C, Wang Y.. 2017. VqMAPKKK38 is essential for stilbene accumulation in grapevine. Horticulture Research 4, 17058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CS, Kolevski B, Smyth DR.. 2002. TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. The Plant Cell 14, 1359–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Zhang P, Zhang M, Fu C, Yu L.. 2013. Functional analysis of a WRKY transcription factor involved in transcriptional activation of the DBAT gene in Taxus chinensis. Plant Biology 15, 19–26. [DOI] [PubMed] [Google Scholar]
- Llorca CM, Potschin M, Zentgraf U.. 2014. bZIPs and WRKYs: two large transcription factor families executing two different functional strategies. Frontiers in Plant Science 5, 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey SP, Roccaro M, Schön M, Logemann E, Somssich IE.. 2010. Transcriptional reprogramming regulated by WRKY18 and WRKY40 facilitates powdery mildew infection of Arabidopsis. The Plant Journal 64, 912–923. [DOI] [PubMed] [Google Scholar]
- Robatzek S, Somssich IE.. 2002. Targets of AtWRKY6 regulation during plant senescence and pathogen defense. Genes & development 16, 1139–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluttenhofer C, Yuan L.. 2015. Regulation of specialized metabolism by WRKY transcription factors. Plant Physiology 167, 295–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y, Yan L, Liu ZQ, et al. 2010. The Mg-chelatase H subunit of Arabidopsis antagonizes a group of WRKY transcription repressors to relieve ABA-responsive genes of inhibition. The Plant Cell 22, 1909–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannozzi A, Wong DCJ, Höll J, Hmmam I, Matus JT, Bogs J, Ziegler T, Dry I, Barcaccia G, Lucchin M.. 2018. Combinatorial Regulation of Stilbene Synthase Genes by WRKY and MYB Transcription Factors in Grapevine (Vitis vinifera L.). Plant & cell physiology 59, 1043–1059. [DOI] [PubMed] [Google Scholar]
- Wong DCJ, Schlechter R, Vannozzi A, Höll J, Hmmam I, Bogs J, Tornielli GB, Castellarin SD, Matus JT.. 2016. A systems-oriented analysis of the grapevine R2R3-MYB transcription factor family uncovers new insights into the regulation of stilbene accumulation. DNA Research 23, 451–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Liu T, Tian C, Sun S, Li J, Chen M.. 2005. Transcription factors in rice: a genome-wide comparative analysis between monocots and eudicots. Plant Molecular Biology 59, 191–203. [DOI] [PubMed] [Google Scholar]