CARK1 preferentially interacts with and phosphorylates ABA receptors of subfamily III; the phosphorylation site RCAR11T78 plays a substantial role in the activation of the ABA response pathway.
Keywords: ABA signaling, abscisic acid, abiotic stress, Arabidopsis thaliana, drought, phosphorylation, seed germination, transient luciferase assay
Abstract
Abscisic acid (ABA) plays a vital role in responses to abiotic stresses that allow plants to cope with environmental challenges. In this study, we analyzed ABA receptors of subfamily III as the potential targets of Cytosolic ABA Receptor Kinase 1 (CARK1). We previously found that CARK1 phosphorylated the subfamily III member RCAR11 at a distinct threonine residue (T78). Our study now shows the physical interaction of CARK1 with the receptors RCAR12/13/14 in vitro and in vivo. The catalytically inactive form CARK1-N204A did not interact with the receptors. Phosphorylation of these ABA receptors in vitro occurred at a serine/threonine amino acid residue corresponding to T78 in RCAR11, which is located in the loop of β3 within a conserved site. Further analysis revealed that the phosphorylation of RCAR11T78 could increase the sensitivity of the pyr1pyl1pyl2pyl4 quadruple mutant (1124) to ABA, including the inhibition of root elongation and increasing drought tolerance. The analysis of CARK1:1124 complementation and the expression of ABA-related genes indicated that CARK1 could rescue the insensitivity of 1124 to ABA. Our results indicate that CARK1 tends to phosphorylate subfamily III ABA receptors, and the phosphosites RCAR11T78, RCAR12T105, RCAR13T101, and RCAR14S81 are the major sites involved in the activation of the ABA response pathway.
Introduction
The phytohormone abscisic acid (ABA) plays an important role in plant growth and developmental processes as well as in plant responses to environmental stresses, including salinity and drought (Finkelstein et al., 2002; Yoshida et al., 2002; Cutler et al., 2010; Fujita et al., 2011). Core ABA signaling components found in Arabidopsis are conserved, such as ABA receptors (Hauser et al., 2011). When binding to ABA, pyrabactin resistance 1 (PYR1)/PYR1-like (PYL)/regulatory components of ABA receptors (RCAR) (hereafter referred to as RCARs) interact with and inhibit clade A protein phosphatase 2C (PP2C). This releases SNF1-related protein kinase 2 (SnRK2)-type protein kinases from inhibition, subsequently allowing SnRK2 to phosphorylate and activate downstream components, such as transcription factors and ion channels (Furihata et al., 2006; Fujita et al., 2009; Lee et al., 2009; Zhou et al., 2015).
The Arabidopsis genome encodes 14 RCARs, which are divided into three subfamilies according to their sequence homology, namely subfamily I members (RCAR1–RCAR4), subfamily II (RCAR5–RCAR10) and subfamily III members (RCAR11–RCAR14) (Ma et al., 2009; Park et al., 2009; Nishimura et al., 2010). All of them show overlapping patterns of gene expressions (Winter et al., 2007; Gonzalez-Guzman et al., 2012; Antoni et al., 2013), and functional redundancy among them can be expected. For instance, RCAR11 and RCAR14 are found to be ubiquitylated by RSL1 and degraded by the 26S proteasome in Arabidopsis thaliana (Eduardo et al., 2015). At basal ABA levels, subfamily I members efficiently regulated ABA signaling, whereas those of subfamily II are moderately inhibited by PP2C activities (Zhao et al., 2013; Tischer et al., 2017). From a biochemical point of view, the receptors of subfamily I and II are characterized by a monomeric state, while those in subfamily III are in a dimeric state (Nishimura et al., 2009; Dupeux et al., 2011; Hao et al., 2011). The dimeric receptors show a lower affinity for ABA than the monomeric receptors (Ma et al., 2009; Santiago et al., 2009). Previous studies also found that subfamily III receptors exhibited ABA-dependent inhibition of PP2Cs, whereas other members inhibited the activities of PP2Cs even in the absence of ABA (Fujii et al., 2009; Miyazono et al., 2009; Tischer et al., 2017). The phenotypic analysis of ABA receptor mutants revealed that single mutants exhibited normal ABA sensitivity, except for pyl8, the lateral root growth of which was more sensitive to inhibition by ABA (Zhao et al., 2014). The quadruple mutant pyr1pyl1pyl2pyl4 (abbreviated as 1124) which is deficient in the three dimeric receptors (RCAR11, RCAR12, and RCAR14) and a monomeric receptor (RCAR10) shows a strong ABA insensitivity, including ABA-mediated inhibition of germination, root growth, and ABA-induced stomatal closure, providing evidence for the importance of the dimeric receptors in Arabidopsis (Park et al., 2009; Nishimura et al., 2010; Gonzalez-Guzman et al., 2012). However, the quadruple mutant still responds to ABA to some extent, indicating that all of the ABA receptors function similarly in ABA signaling (Gonzalez-Guzman et al., 2012).
The diversity of responses regulated by ABA and its dynamics in stress development associated with different ABA levels might require a large number of different ABA receptors to adjust signal transduction and metabolism in plants. Phosphorylation, a type of post-translational modification, has been found to play an important role in ABA signaling (Yoshida et al., 2002; Fujita et al., 2009; Zhou et al., 2015). The phosphorylation of RCAR11 or RCAR12 by target of rapamycin 1 (TOR1) would suppress ABA binding, and EL1-like kinases phosphorylate RCAR12 and then promote its degradation by 26S protease (Chen et al., 2018; Wang et al., 2018), suggesting that these kinases are negatively involved in ABA signaling. A recent study elaborates that the phosphorylation of RCAR3 and RCAR11 by CARK1 (Cytosolic ABA Receptor Kinase 1) positively promotes ABA signaling, and the N204 of CARK1, the critical active site, accounts for the interactions between CARK1 and RCAR3/RCAR11 (Zhang et al., 2018). CARK1 belongs to a putative Ser/Thr protein kinase RLCK VIII subfamily in Arabidopsis. Here, we report that CARK1 interacts with and phosphorylates RCAR12/13/14, and find the major phosphosites T105 of RCAR12, T101 of RCAR13, and S81 of RCAR14. Moreover, we reveal that a major phosphorylation site (T78) in RCAR11 is closely linked to the regulation of ABA-induced germination, root length, and drought tolerance, and affects the expression of ABA-related genes.
Materials and methods
Plant materials
Arabidopsis plants used in this study were in the Columbia (Col-0, WT), 1124 quadruple mutant (Gonzalez-Guzman et al., 2012), or Landsberg erecta ecotype (Ler) background. The CARK1 T-DNA insertion mutant cark1 (SALK_113377) was obtained from the Arabidopsis Biological Resources Center. Plants were grown in soil–vermiculite mixtures at 22 °C under 60% relative humidity with a photoperiod of 16 h light/8 h dark and 120 μmol m–2 s–1. For plate culture, seeds were first soaked in distilled water for 3 d at 4 °C. After stratification, seeds were surface sterilized and germinated on solid Murashige and Skoog (MS) medium containing 2% sucrose and 0.8% agar, pH 5.8.
In vitro GST pull-down assay
RCAR genes were cloned into the pGEX-6p-1 expression vector (Novagen, WI, USA) with a glutathione S-transferase (GST) tag fused at the N-terminal end. The primers are listed in Supplementary Table S1 at JXB online. These plasmids were transfected into Escherichia coli strain Rosetta (DE3), and protein expression and purification were performed for CARK1-KD (CARK1 kinase domain, residues 50–353) as previously described (Zhang et al., 2018). GST–RCARs were added to 25 μl of glutathione S–Sepharose 4B (GS4B, GE Healthcare, PA, USA) resin. His-CARK1-KDN204A or His-CARK1-KD was co-incubated with the GST–RCAR-bound resin for 1 h at room temperature. The resin was then extensively rinsed with buffer to remove unbound proteins. The resins were finally resuspended in 100 μl of buffer, and 10 μl of the suspension was subjected to 10% SDS–PAGE for analysis. A 0.2 μg aliquot of inputs was used in the assay, and the anti-His antibody (mouse derived Mab, TransGen Biotech; Beijing, China) was used in the western blot analysis.
Bimolecular fluorescence complementation (BiFC) assay
The full-length coding sequence (CDS) of RCAR9/12/13/14 was individually cloned into the binary nYFP (yellow fluorescent protein) vector via BamHI and SalI sites to generate the RCAR9/RCAR12/13/14–YFPN fusion protein. YFPC–CARK1 and YFPC–CARK1N204A constructions were as described by Zhang et al. (2018). Primer pairs for the construction of the vectors are listed in Supplementary Table S1. The BiFC assay was performed as previously described (Song et al., 2011). Briefly, various nYFP and cYFP fusion vectors were introduced into Agrobacterium tumefaciens strain GV3101. After incubation, Agrobacterium cells were harvested and resuspended in infiltration buffer (0.2 mM acetosyringone, 10 mM MgCl2, and 10 mM MES) to identical concentrations (OD600=0.5), and then transferred into the leaf cells of Nicotiana benthamiana by a needleless syringe. After 2 d, cells with YFP fluorescence were observed and imaged with a confocal laser-scanning microscope. The experiment was repeated three times, each time with three or four biological replicates.
Co-immunoprecipitation (Co-IP) assay
The full-length CDS of RCAR12/13/14 was cloned into the pHB vector via BamHI and SalI sites to generate pHB-3*flag-RCAR12/13/14, respectively. The full-length CDS of CARK1 and CARK1N204A were cloned into the pHB vector via SalI and XbaI sites to generate pHB-3*HA-CARK1 and pHB-3*HA-CARK1N204A. The recombined plasmids were transiently expressed in N. benthamiana by Agrobacterium infiltration as described above. Proteins were extracted and resuspended in IP buffer [50 mM HEPES, pH 7.5, 150 mM NaCl, 1% polyvinyloly pyrrolidone (PVPP), 10% glycerol, 1% Triton X-100, 2 mM DTT, 2 mM phenylmethylsulfonate fluoride (PMSF), and 1× protease inhibitor cocktail (Roche, Basel, Switzerland)]. After extraction in IP buffer, crude protein extracts (Input) were used for immunoprecipitation with Anti-Flag® M2 Magnetic Beads (Sigma-Aldrich). The beads were resuspended in 2× sample loading buffer and boiled at 98 °C for 10 min. The supernatant of the crude extracts was used as the input. Anti-HA and anti-Flag antibodies (Bioworld, Minneapolis, USA) were used in the immunoblot. Co-IP) was performed as previously described (Fiil et al., 2008).
In vitro kinase assay
RCAR12, RCAR12T105A, RCAR13, RCAR13T101A, RCAR14, and RCAR14S81A were cloned into the pET28a expression vector with a 6× His tag fused at the C-terminal side. The plasmids were then transformed into E. coli strain Rosetta (DE3). After the OD600 reached 0.5, the culture was cooled to 16 °C and supplemented with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG). The cells were harvested by centrifugation and the pellets were resuspended in lysis buffer containing 20 mM Tris–HCl (pH 8.0), 150 mM NaCl, and 5 mM MgCl2. The fusion proteins were purified by Ni-NTA affinity chromatography (Thermo Fisher Scientific, Rockford, IL, USA).
For autophosphorylation and transphosphorylation assays, 1 μM CARK1-KD or CARK1-KD mutant was diluted to 25 μl using reaction buffer (20 mM Tris, pH 7.5, 100 mM NaCl, 10 mM MgCl2, 2 mM DTT, and 10 mM ATP). The reaction mixture was incubated at 30 °C for 1 h and terminated by adding an equal volume of 2× SDS loading buffer. Horseradish peroxidase (HRP)-conjugated Phosphor-Threonine Antibody (Cell Signaling Technology, Beverly, MA, USA) was used. Western blots were developed with the ECL chemilluminesence detection system (Bio-Rad, Hercules, CA, USA).
Genotyping analysis of the CARK1 mutant and generation of various CARK1 transgenic plants
The construct for the complemented lines of Flag-tagged RCAR11/RCAR11T78A/E or HA-tagged CARK1 in the 1124 background (abbreviated as R11/R11A/R11E:1124) were generated as previously described (Zhang et al., 2018). The resulting constructs were transformed into N. benthamiana strain GV3101, which was subsequently infiltrated into the WT plants using the ‘floral dip’ method (Clough and Bent, 1998). All transgenic plants were screened on MS medium supplemented with hygromycin, and mRNA levels were verified with real-time PCR (RT-PCR) assays.
Physiological analysis
The germination assay was carried out as described by Lee et al. (2015) and the root length assay was as described by Cui et al. (2012). Approximately 100 seeds were placed on solid MS agar medium with different concentrations of ABA. Germination rates were scored 4 d after incubation. For root growth assay, >50 seeds from each line were first germinated vertically on MS medium for 3 d. Then, 20 seedlings of each line with similar root lengths were transferred in a vertical position to 1/2 MS medium supplemented with or without 40 μM or 30 μM ABA. The root length was determined 1 week after transfer. For the drought tolerance test, 2-week-old seedlings grown under the same conditions were subjected to drought stress treatment by withholding water for 10 d. After rehydration for 2 d, the morphological changes of plants were recorded.
Statistical analysis
Data are represented as the means ±SD. Statistical analysis was performed using Student’s t-test. Values of P<0.05 were considered significant, and values of P<0.01 were considered more significant.
RNA isolation and qRT-PCR analysis
Two-week-old seedlings were incubated in liquid MS medium with or without 50 μM ABA for 3 h. Total RNAs was extracted using the RNAiso Plus reagent from Takara (Otsu, Japan). The cDNAs were synthesized from 1 μg of total RNAs using the PrimeScript RT-PCR reagent Kit of Takara. qRT-PCR was performed using the Bio-Rad CFX96 real-time PCR detection system and SYBR Premix Ex Taq II from Takara. ACTIN2/8 was used as an internal control. RT-PCR was conducted with the gene-specific primers described by Zhang et al. (2018).
Transient luciferase (LUC) assay
Transcription activity of the interaction between CARK1/CARK1N204A (CARK1m) and RCARs against promoters of ABA signaling-responsive marker genes was determined using the dual luciferase assay of transiently transformed Arabidopsis protoplasts. Marker gene promoters (RAB18 and RD29B) were subcloned into the transient expression reporter vector pGreenII0800-LUC that contained the Cauliflower mosaic virus (CaMV) 35S promoter–REN cassette and the promoterless LUC cassette. RCAR12/13/14 were subcloned into the vector pBI221 via XbaI and BamHI sites. The internal control construct p35S::GUS and pBI221-CARK1/CARK1m vectors were used as effector constructs as previously described (Zhang et al., 2018). Primers used in this study are listed in Supplementary Table S1.
Preparation of Arabidopsis protoplasts of the cark1 T-DNA mutant plants and subsequent transfection of protoplasts were performed as described (Yoo et al., 2007). A no-effector construct was used as control. When indicated, 10 μM ABA was added to the incubation buffer immediately after transfection. Luciferase activity was measured by an LMax II384 luminometer (Molecular Devices, Bad Wildbad, Germany) using the Dual-Luciferase Reporter Assay System from Promega (Madison, WI, USA). Data are reported as the LUC/REN ratio. Three independent experiments were performed, and for each sample three technical replicates were analyzed.
Results
CARK1 interacts directly with RCAR12, RCAR13, and RCAR14
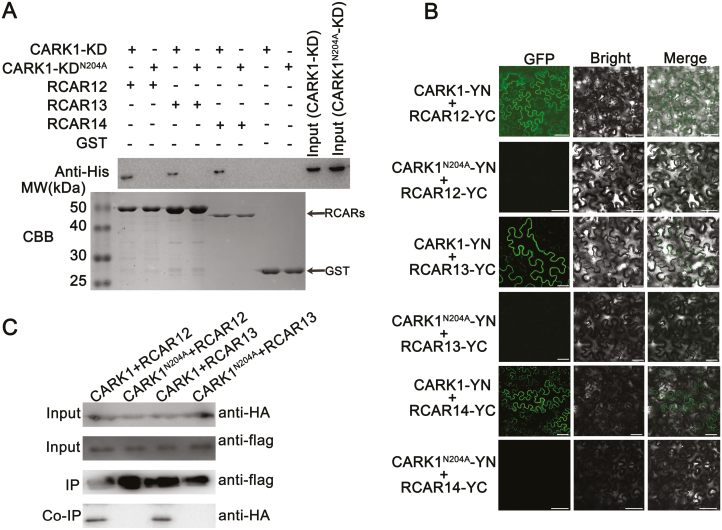
In a previous study, the results from the yeast two-hybrid assay demonstrated that CARK1 interacted with RCARs in vitro (Zhang et al., 2018). Here, a GST pull-down assay revealed that CARK1-KD interacted with RCAR12, RCAR13, and RCAR14 (Fig. 1A). However, these interactions were completely impaired by a single amino acid substitution in the kinase activity center of CARK1 (CARK1-KDN204A) (Fig. 1A). The BiFC assay showed that co-expression of CARK1–nYFP/cYFP–RCAR12, CARK1–nYFP/cYFP–RCAR13, or CARK1–nYFP/cYFP–RCAR14 in the leaves of N. benthamiana yielded YFP signals in the cytosol (Fig. 1B), while co-expression of a series of negative controls did not generate fluorescence (Supplementary Fig. S1A). The result of western blot showed that CARK1N204A was ectopically expressed in N. benthamiana (Supplementary Fig. S1B). However, no YFP signal was detected in leaves after co-expression of CARK1N204A–nYFP/cYFP–RCAR12, CARK1N204A–nYFP/cYFP–RCAR13, or CARK1N204A–nYFP/cYFP–RCAR14 (Fig. 1B). Similarly, the Co-IP assay revealed that the full-length CARK1, but not CARK1N204A, could interact with RCAR12 and RCAR13 in N. benthamiana (Fig. 1C). The previous study showed that CARK1 could interact with the subfamily I member, RCAR3; therefore, we also tested the interaction between CARK1 and RCAR9 (belonging to subfamily II of ABA receptors) (Zhang et al., 2018). The result showed that there was no fluorescence when CARK1 and RCAR9 were co-expressed (Supplementary Fig. S1A). Our investigations confirm that CARK1 indeed physically interacts with RCAR12, RCAR13, or RCAR14 in plant cells and the N204 of CARK1 is critical for CARK1’s kinase activity and for its interactions with RCAR12, RCAR13, and RCAR14.
Fig. 1.
CARK1 interacts with RCAR12, RCAR13, and RCAR14 in vitro and in vivo. (A) In vitro GST pull-down assay of CARK1-KD and CARK1-KDN204A interaction with RCAR12, RCAR13, or RCAR14. A 50 μg aliquot of 6× His-fused CARK1-KD or CARK1-KDN204A was incubated with 50 μg of GST-tagged RCAR12, RCAR13, or RCAR14. The immunoprecipitated proteins were analyzed by SDS–PAGE (bottom) and immunoblotting with antibodies against the His-tag (top). A 0.1 μg aliquot of both input proteins CARK1-KD and CARK1-KDN204A was used. (B) BiFC assay validates the interactions of RCAR12, RCAR13, and RCAR14 with CARK1, but not with mutant CARK1N204A, in the leaves of N. benthamiana. nYFP was fused to the full-length CARK1 and mutant CARK1N204A to form CARK1–nYFP and CARK1N204A–nYFP, and cYFP was fused to RCAR12, RCAR13, and RCAR14 to form cYFP–RCAR12, cYFP–RCAR13, and cYFP–RCAR14, respectively. Pictures were taken using a confocal laser-scanning microscope (LSM 710, Carl Zeiss). Scale bars=50 µm. (C) Co-immunoprecipitation (Co-IP) experiment of 3*HA-tagged CARK1/CARK1N204A and 3*Flag-tagged RCAR12 or RCAR13 was performed in N. benthamiana. Protein extracts were immunoprecipitated using an anti-Flag antibody and subjected to immunoblot analysis with an anti-HA antibody. All the experiments were conducted with at least three biological replicates.
CARK1 phosphorylates RCAR12/13/14 in vitro
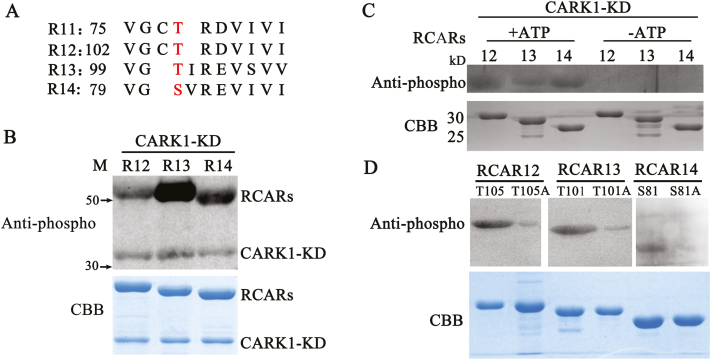
Based on the finding that CARK1 interacted with RCAR12/13/14, we next tested whether they could be phosphorylated by CARK1 in vitro. Alignment analysis revealed that ABA receptors are highly conserved (Fig. 2A) (Yin et al., 2009), suggesting their possible phosphorylation by CARK1. Therefore, we examined the ability of CARK1 to phosphorylate RCAR12/13/14 using a kinase assay in vitro. CARK1-KD was cloned and expressed with a His-tag, and RCARs as the substrates were purified as GST-tag proteins in an E. coli system. After separating the protein by SDS–PAGE, we detected the phosphorylated proteins on the gel using an anti-phosphothreonine antibody which could recognize the Ser and Thr phosphorylation sites and visualized total proteins with Coomassie Brilliant Blue (CBB) staining. As shown in Fig. 2B, an autophosphorylation band of ~38 kDa corresponding to His-CARK1-KD was observed in all samples, and the band of ~55 kDa showed that RCAR12, RCAR13, and RCAR14 were phosphorylated by CARK1. Similarly, CARK1 could also phosphorylate His-RCAR12, RCAR13, and RCAR14 (Fig. 2C). However, RCAR12, RCAR13, and RCAR14 were not phosphorylated by CARK1-KD without ATP (Fig. 2C). In a previous study, CARK1 was shown to phosphorylate the T78 site of RCAR11 in vitro and in vivo (Zhang et al., 2018). The T78 site of RCAR11 is located in the loop of β3 (Santiago et al., 2009). Protein kinases usually phosphorylated Ser and Thr residues on S/T-P motifs; the possible phosphorylation sites can be predicted in RCAR12/13/14 by an analysis of their homologous sequences (Fig. 2A), namely on RCAR12T105, RCAR13T101, and RCAR14S81. The analysis of the kinase assay in vitro showed that mutations of these sites to alanine (preventing phosphorylation) resulted in significantly suppressed phosphorylation by CARK1 (Fig. 2D). Therefore, our results further confirm the roles of these sites in phosphorylation (T78 of RCAR11, T105 of RCAR12, T101 of RCAR13, or S81 of RCAR14) and indicate a conserved CARK1-mediated phosphorylation of RCARs.
Fig. 2.
CARK1 phosphorylates RCAR12, RCAR13, and RCAR14. (A) Alignment of the phosphorylation sites with subfamily III receptor RCARs (R11–R14). (B–D) In vitro kinase assay. Phosphorylation of GST–RCAR12/13/14 (abbreviated as R12/13/14) by CARK1-KD (B). Phosphorylation of His-RCAR12/13/14 by CARK1-KD (C). Determination of the potential phosphorylation site of RCAR12/13/14 (D). Anti-phosphothreoinine antibody was used for the analysis of the western blot. The loading amounts of proteins were visualized on the Coomassie Brilliant Blue- (CBB) stained gel (bottom panel). Experiments were repeated at least three times.
The phosphorylation of RCAR11 affects the responses of plants to ABA
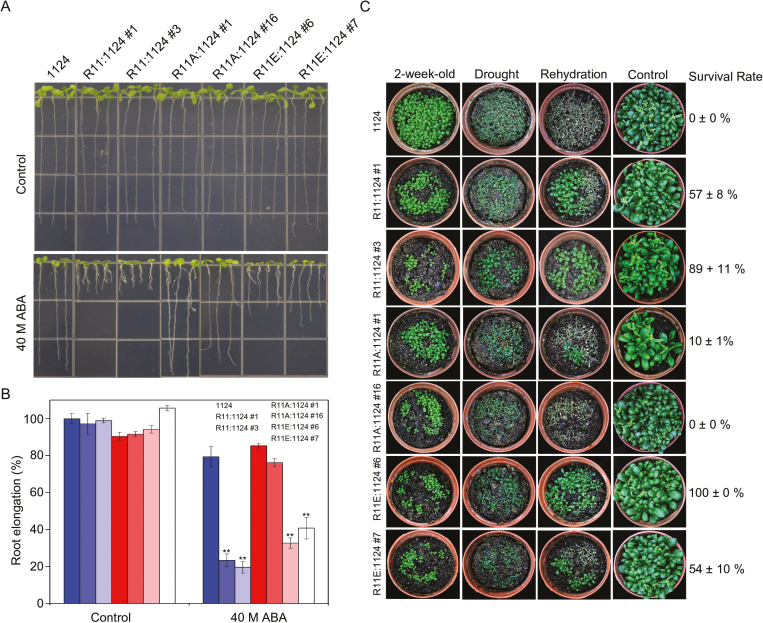
ABA is a key factor in controlling seedling growth including root length (Park et al., 2009; Nishimura et al., 2010; Gonzalez-Guzman et al., 2012). To elucidate whether the phosphorylation of ABA receptors by CARK1 has a corresponding function in ABA signaling, we generated various RCAR11 (R11) complemented transgenic lines in the 1124 mutant including the WT (R11:1124), phosphor-defective and phosphor-mimic forms on T78 (R11A:1124 and R11E:1124), respectively. The analysis of root elongation revealed that R11:1124 and R11E:1124 had shorter roots than those of 1124 in the presence of ABA, while the relative root length of 1124 was similar to that of R11A:1124 (Fig. 3A, B). However, the root length of the plants exhibited no difference in the absence of ABA (Fig. 3A, B). These results indicate that the phosphorylation of RCAR11 is a positive regulator of root growth inhibition in response to ABA.
Fig. 3.
Phosphorylation state of RCAR11 affects the response of plants to ABA. (A) The root architecture of plant seedlings was documented at 7 d post-transfer to medium (1/2 MS, 1% sucrose) with or without ABA. Seedlings were 3 d old at the time of transfer and had equal root lengths at that time. (B) Statistical analysis of the ABA-inhibited root growth in (A). Data are the means ±SD (n=20). (C) Drought tolerance assay. Two-week old plants before drought (top), after drought (second line), 2 d after rehydration (third line), and control without drought stress (bottom line). 1124, ABA receptor pyr1pyl1pyl2pyl4 quadruple mutant; R11:1124, RCAR11 overexpression in the 1124 quadruple mutant plants; R11A:1124, the phosphor-defective forms of RCAR11T78A overexpressed in the 1124 quadruple mutant plants; R11E:1124, a phosphor-mimic form of RCAR11T78E overexpressed in the 1124 quadruple mutant plants. *P<0.05, **P<0.01, Student’s t-test. All physiological analyses were conducted in triplicate.
To confirm our observations further, we next exposed the 1124 and the complemented plants to a dehydration condition by withholding water. There was no significant difference among them when plants grew under well-watered conditions, but all of them displayed wilting phenotypes after drought treatment for 10 d (Fig. 3C). However, after rewatering for 2 d, >50% of R11:1124 and R11E:1124 transgenic plants recovered, while R11A:1124 and 1124 plants did not recover, with the exception that R11A:1124 #1 was ~10% recovered (Fig. 3C). These results indicated that transgenic plants bearing phosphor-defective forms of RCAR11 (R11A:1124) almost completely lost ABA sensitivity and drought tolerance compared with the WT form of RCAR11 (R11:1124), while the phosphor-mimic form of RCAR11 (R11E:1124) could notably complement the phenotypes (Fig. 3).
CARK1 rescues the sensitivity of 1124 quadruple mutant plants to ABA
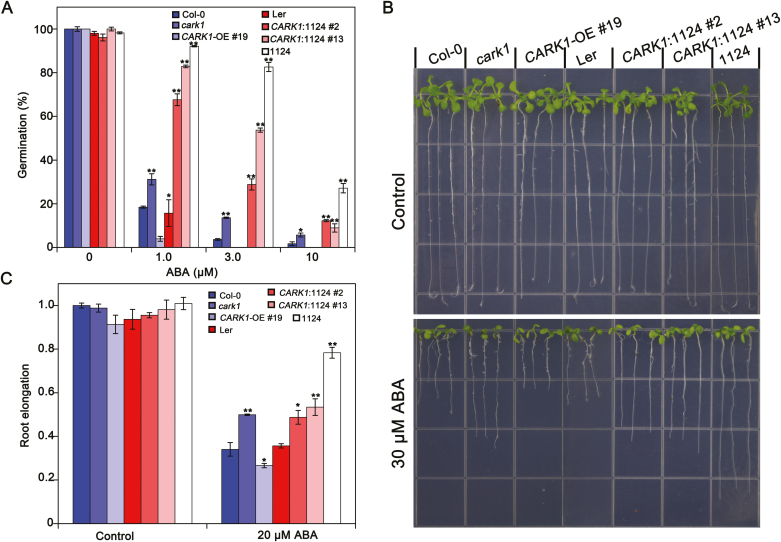
Subsequently, a complementation test was conducted. CARK1 was ectopically expressed in the 1124 mutant. Expression of the CARK1 transgene was detected in CARK1:1124 (lines #2 and #13) T3 complementation lines by RT-PCR and protein levels (Supplementary Fig. S2). These complementation progeny were used in the phenotypic analysis of ABA responses to determine whether the mutant phenotypes were rescued. As described previously (Park et al., 2009; Gonzalez-Guzman et al., 2012; Zhang et al., 2018), cark1 and1124 mutants were insensitive to ABA compared with WT plants during the germination and post-germination stages. The germination of CARK1:1124 transgenic seeds was higher than that of CARK1-OE #19 but lower than that of 1124 under ABA treatment (Fig. 4A). As shown in Fig. 4B and C, compared with the WT, CARK1-OE #19 showed increased sensitivity to ABA-inhibited primary root growth. The insensitivity of 1124 to ABA-mediated inhibition of root growth was notably diminished when CARK1 was overexpressed in 1124 (CARK1:1124) (Fig. 4B, C). These results indicate that the ABA-sensitive phenotypes of CARK1:1124 were partly rescued by reciprocal overexpression of CARK1.
Fig. 4.
RCARs act downstream of CARK1 in the ABA-induced inhibition of seed germination and root length. (A) Seed germination assay. Seeds (>100) were scored 3 d after stratification on MS medium supplemented with different concentrations of ABA. Values are the means ±SD (n=3). (B) The root architecture of seedlings was documented at 7 d post-transfer to media (1/2 MS, 1% sucrose) with or without ABA. Seedlings were 3 d old at the time of transfer and had equal root lengths at that time. (C) Statistical analysis of ABA-inhibited root growth in (B). Data are the means ±SD (n=20). Col-0, Columbia ecotype; cark1, CARK1 T-DNA mutant; CARK1-OE 19#, plants overexpressing CARK1 in Col-0; Ler (Landsberg erecta ecotype); CARK1:1124, overexpression of CARK1 in the 1124 quadruple mutant plants; 1124, ABA receptor pyr1pyl1pyl2pyl4 quadruple mutant. *P<0.05, **P<0.01, Student’s t-test. All physiological analyses were conducted in triplicate.
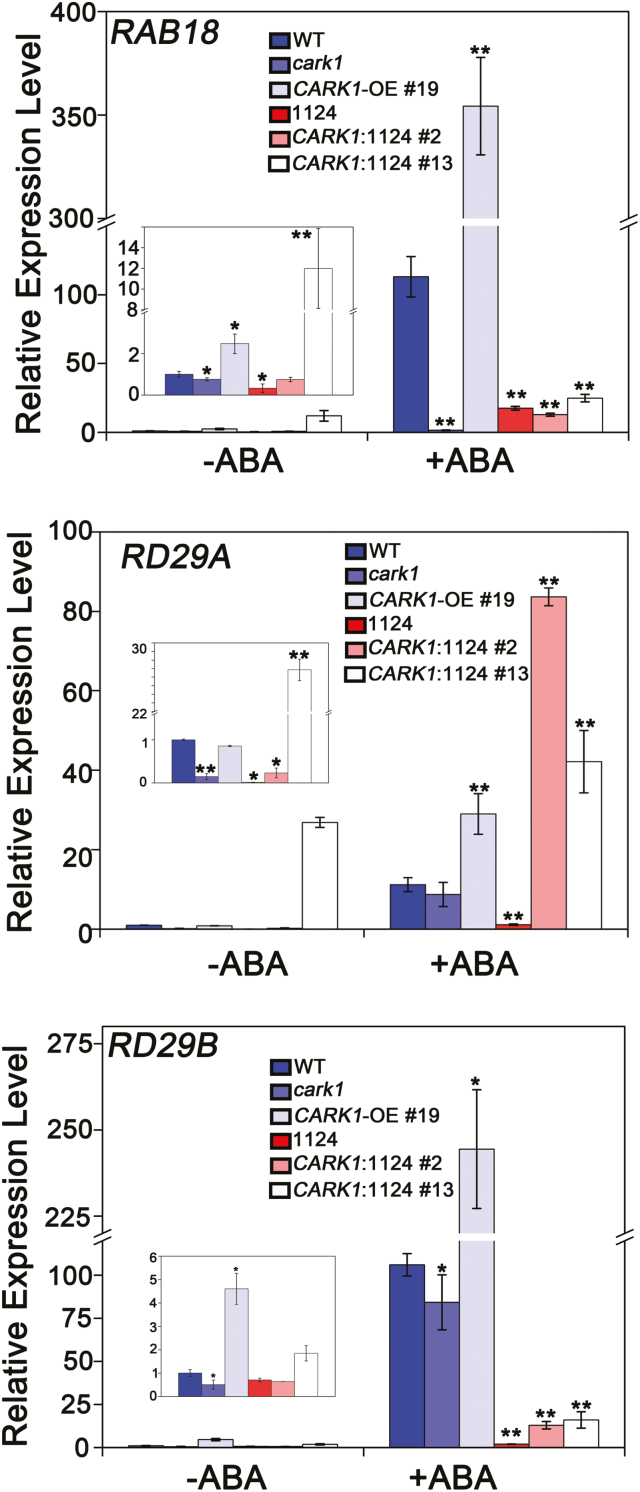
Furthermore, we examined the mechanism whereby CARK1 mediates the ABA signaling pathway in reciprocal transgenic plants. The expression of several ABA-related genes (RAB18, RD29A, and RD29B) was measured by qRT-PCR. The results showed that the expression levels of RAB18, RD29A, and RD29B in CARK1:1124 #13 were higher than those in 1124 without ABA treatment (Fig. 5). After the treatment with exogenous ABA for 3 h, the expression of these ABA-related genes in CARK1:1124 (lines #2 and #13) was significantly increased and induced compared with that in the WT. Taken together, the transgenic plants, overexpressing CARK1 in the 1124 background, promoted the expressions of ABA-related genes after treatment with exogenous ABA.
Fig. 5.
CARK1 attenuates the expression of several ABA-responsive genes. Relative expression levels of the ABA-responsive genes RAB18 (A), RD29A (B), and RD29B (C) in plants. Two-week-old seedlings were incubated in MS liquid medium with or without 50 μM ABA for 3 h. The transcriptional levels were determined by qRT-PCR analysis. Values are the means ±SD (n=3). ACTIN2/8 was used as an internal control. WT, Columbia ecotype; cark1, CARK1 T-DNA mutant; CARK1-OE 19#, plants overexpressing CARK1 in Col-0; 1124, ABA receptor pyr1pyl1pyl2pyl4 quadruple mutant; CARK1:1124, overexpression of CARK1 in the 1124 quadruple mutant plants.*P<0.05, **P<0.01, Student’s t-test. The experiment was conducted by triplicate.
From these results, we further revealed the significance of phosphorylation of ABA receptors in fine-tuning ABA signaling. On the other hand, CARK1 promotes ABA-mediated germination, root growth, and gene expression. Collectively, these studies support the hypothesis that kinase activity is critical for the function of CARK1 in ABA signaling.
CARK1 and RCARs synergistically promotes ABA-responsive gene expression in Arabidopsis protoplasts.
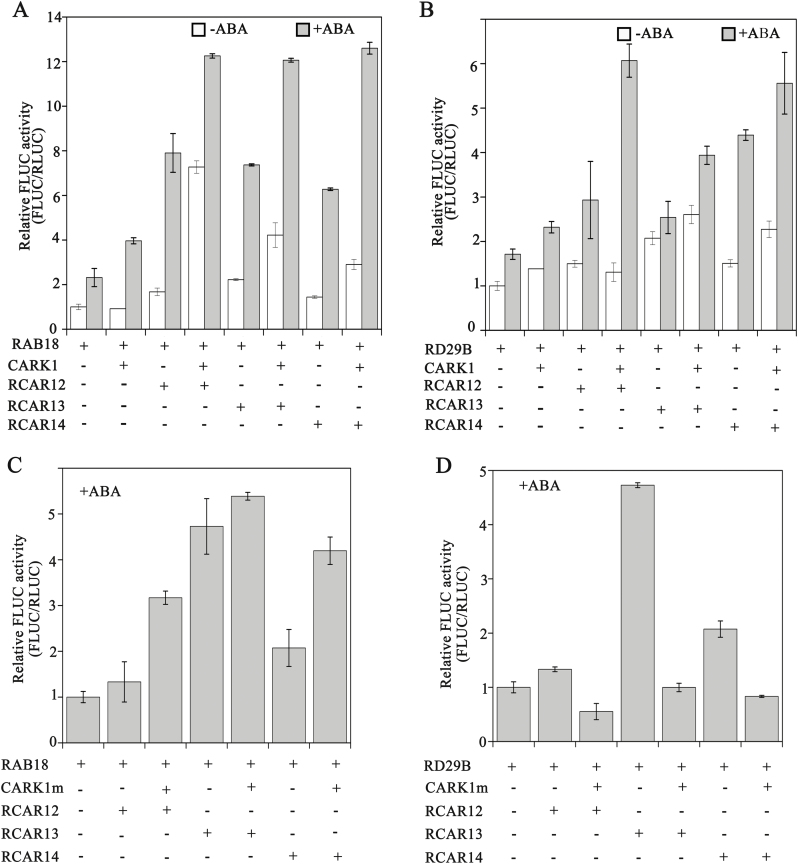
Transient LUC assays in cark1 mutant protoplasts were used to determine the effect of the interaction of CARK1 with ABA receptors on the expressions of the LUC gene driven by the RAB18 or RD29B promoter (Fig. 6). In the absence of exogenous ABA, the single transfection of CARK1 or RCAR12/13/14 had a minimal effect on the expression of RAB18-FLuc (1.2- to 2.3 fold) (Fig. 6A). On the other hand, the relative luciferase activity (LUC/REN) showed a significant enhancement (2.5- to 8-fold) when CARK1 and RCARs were co-transfected in cark1 protoplasts (Fig. 6A). Co-expression of CARK1 and RCARs in the presence of ABA noticeably increased the ABA response, on average >11-fold for the expression of RAB18-FLuc (Fig. 6A).
Fig. 6.
CARK1 and subfamily III ABA receptors synergistically regulate the expression of the reporter gene driven by the RAB18 or RAB29B promoter. Protoplasts of cark1 were transfected with 10 μg of effector DNA CARK1 (A and B) or CARK1N204A (abbreviated as CARK1m; C and D) for expression of the different RCARs (RCAR12/13/14) in the absence and presence of 10 μM exogenous ABA. As a negative control, empty effector vectors were used, taking the place of the effector plasmid. The relative activity of the vector control was set at 1. Experiments were repeated at least three times. Vertical bars indicate ±SD of three replicates.
Similarly, in the absence of exogenous ABA, the single transfection of CARK1 or RCAR13/14 had a minimal effect on the expression of RD29B-FLuc (2.5- to 4-fold) (Fig. 6B). On the other hand, the relative luciferase activity (LUC/REN) showed a significant enhancement (4- to 6-fold) when CARK1 and RCARs were co-transfected in cark1 protoplasts in the presence of ABA (Fig. 6B).
However, there were no obviously increased activities of luciferase when CARK1m and RCAR12/13/14 were co-expressed in cark1 protoplasts (Fig. 6C, D). Therefore, these findings support the notion that the phosphorylation of RCAR12/13/14 by CARK1 induces the expression of ABA-responsive genes and synergistically regulates ABA signaling.
Discussion
Unlike the monomeric state of ABA receptor subfamilies I and II (RCAR1–RCAR10), the members of subfamily III are dimeric receptors, except for RCAR13 that exists in a state of monomer–dimer shift (Nishimura et al., 2009; Dupeux et al., 2011; Hao et al., 2011). The dimeric receptors inhibit the activity of PP2Cs in an ABA-dependent manner and possess less sensitivity to ABA than do the monomeric receptors (Ma et al., 2009; Tischer et al., 2017). Despite this lower sensitivity, genetic evidence suggests that the dimeric receptors are important in all ABA responses measured to date (Okamoto et al., 2013; Ye et al., 2017).
A recent study revealed that CARK1 interacts with and phosphorylates RCAR11 at the site of T78 (Zhang et al., 2018). On the basis of the similarity of subfamily III receptors (Ma et al., 2009; Park et al., 2009), it is reasonable to hypothesize that an interaction might exist between the other members of subfamily III receptors and CARK1. In this study, we confirmed this hypothesis by GST pull-down, BiFC, and Co-IP assays (Fig. 1), suggesting that a kinase could target several receptors and regulate ABA signaling in plants.
The kinase assay in vitro revealed that recombinant CARK1 protein could phosphorylate RCAR12/13/14 (Fig. 2B–D), indicating a complex regulation of RCARs through phosphorylation. A recent study showed that the TOR kinase phosphorylates ABA receptors (RCAR12/PYL1S119 and RCAR10/PYL4S114) at a conserved serine residue to inhibit the binding of RCARs to ABA and then disrupts the ABA signaling pathway (Wang et al., 2018). Moreover, Arabidopsis EL1-like (AEL) casein kinases phosphorylate RCARs and promote their ubiquitination and degradation, ultimately leading to suppression of the ABA response (Chen et al., 2018). However, our lab previously found that CARK1 phosphorylates RCAR3 or RCAR11 to activate the ABA pathway (Zhang et al., 2018). The phosphorylated RCARs could alter their structure, sheltering from ABA binding (Wang et al., 2018), or exposing ubiquitination sites to be degraded by the 26S protease (Chen et al., 2018). Here, we confirmed that the phosphorylation of RCAR11 by CARK1 contributed to ABA signaling and stress responses. Therefore, we could propose that the phosphorylation of RCARs might play different roles in ABA signaling.
RCARs are highly conserved in plants, and phosphorylation of RCARs by CARK1 confers the possibility to modulate ABA signaling. Our results showed that the interaction of CARK1 with RCAR12, RCAR13, or RCAR14 would activate the ABA signaling pathway by inducing the expression of the ABA-responsive genes. Interestingly, GST pull-down assay demonstrated that CARK1 could physically interact with RCARs in vitro (Supplementary Fig. S3), but not every ABA receptor was found to interact with CARK1 in vivo (Supplementary Fig. S1). This is due to the fact that the structures of protein in an E. coli system are not identical to those in Arabidopsis. In this study, we demonstrate that subfamily III receptors (RCAR12, RCAR13, and RCAR14) physically interact with CARK1 and are phosphorylated by CARK1 at T105, T101, and S81, respectively, indicating that phosphorylation of RCAR12, RCAR13, or RCAR14 positively regulates ABA signaling. The phosphor-defective form of RCAR12/13/14 proteins presented a weak phosphorylated signal, which may due to other possible phosphosites phosphorylated by CARK1. Therefore, we propose that CARK1 tends to interact with and phosphorylate subfamily III receptors, and T105 of RCAR12, T101 of RCAR13, and S81 of RCAR14 are the major phosphosites of CARK1.
Further analysis confirmed that the RCAR11T78E (a phosphor-mimic form) transgenic plants in the 1124 background were more sensitive to ABA than the 1124 mutant plants. In contrast, the RCAR11T78A (a phosphor-defective form) plants were less sensitive. Hence, the phosphorylation of RCAR11 could modulate ABA signaling. The complementation of CARK1 in the 1124 mutant could rescue the sensitivity of plants to ABA, indicating that CARK1 phosphorylates ABA receptors and is involved in ABA signaling.
The analysis of ABA-mediated physiological responses revealed that CARK1:1124 transgenic plants were more sensitive to ABA than 1124 plants but less sensitive than CARK1-OE plants (Fig. 4). Therefore, CARK1 may phosphorylate other ABA receptors, such as RCAR13, which is consistent with the fact that CARK1 phosphorylates ABA receptors to modulate fine-tuning of ABA signaling. Interestingly, the R11:1124 transgenic plants were sensitive to ABA, being similar to R11E:1124 transgenic plants (Fig. 3), or even more sensitive (Fig. 3). We propose that one or more unknown kinases may also phosphorylate RCAR11 and then the ABA signal pathway is activated under stresses. In conclusion, the phosphorylation of ABA receptors plays an important role in ABA signal transduction, and CARK1 together with RCAR11/12/13/14 synergistically regulates ABA signaling, although the detailed mechanism is still unknown.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1 Controls of the BiFC assay.
Fig. S2 Construct used for plant and identification of transgenic lines by qRT-PCR and protein levels.
Fig. S3 In vitro GST pull-down assay of CARK1-KD interaction with RCARs.
Table S1. Primers used in this study.
Supplementary Material
Acknowledgements
This study was funded by the National transgenic project 2016ZX08009003-002 (to X.L), NSFC31671455 (to Y.Y.), and 973 Projects 2015CB755702 (to Y.Y.)
References
- Antoni R, Gonzalez-Guzman M, Rodriguez L, et al. . 2013. PYRABACTIN RESISTANCE1-LIKE8 plays an important role for the regulation of abscisic acid signaling in root. Plant Physiology 161, 931–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HH, Qu L, Xu ZH, Zhu JK, Xue HW. 2018. EL1-like casein kinases suppress ABA signaling and responses by phosphorylating and destabilizing the ABA receptors PYR/PYLs in Arabidopsis. Molecular Plant 11, 706–719. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Cui F, Liu L, Zhao Q, Zhang Z, Li Q, Lin B, Wu Y, Tang S, Xie Q. 2012. Arabidopsis ubiquitin conjugase UBC32 is an ERAD component that functions in brassinosteroid-mediated salt stress tolerance. The Plant Cell 24, 233–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. 2010. Abscisic acid: emergence of a core signaling network. Annual Review of Plant Biology 61, 651–679. [DOI] [PubMed] [Google Scholar]
- Dupeux F, Santiago J, Betz K, et al. . 2011. A thermodynamic switch modulates abscisic acid receptor sensitivity. EMBO Journal 30, 4171–4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eduardo B, Lesia R, Laura L-O, Miguel G-G, Enric S, Jesus M-B, Carla I, Ramon S, Pedro R. 2015. The single-subunit RING-type E3 ubiquitin ligase RSL1 targets PYL4 and PYR1 ABA receptors in plasma membrane to modulate abscisic acid signaling. The Plant Journal 80, 1057–1071. [DOI] [PubMed] [Google Scholar]
- Fiil BK, Qiu JL, Petersen K, Petersen M, Mundy J. 2008. Coimmunoprecipitation (co-IP) of nuclear proteins and chromatin immunoprecipitation (ChIP) from Arabidopsis. CSH Protocols 2008, pdb.prot5049. [DOI] [PubMed] [Google Scholar]
- Finkelstein RR, Gampala SS, Rock CD. 2002. Abscisic acid signaling in seeds and seedlings. The Plant Cell 14(Suppl), S15–S45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Chinnusamy V, Rodrigues A, Rubio S, Antoni R, Park SY, Cutler SR, Sheen J, Rodriguez PL, Zhu JK. 2009. In vitro reconstitution of an abscisic acid signalling pathway. Nature 462, 660–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y, Fujita M, Shinozaki K, Yamaguchi-Shinozaki K. 2011. ABA-mediated transcriptional regulation in response to osmotic stress in plants. Journal of Plant Research 124, 509–525. [DOI] [PubMed] [Google Scholar]
- Fujita Y, Nakashima K, Yoshida T, et al. . 2009. Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in Arabidopsis. Plant & Cell Physiology 50, 2123–2132. [DOI] [PubMed] [Google Scholar]
- Furihata T, Maruyama K, Fujita Y, Umezawa T, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K. 2006. Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proceedings of the National Academy of Sciences, USA 103, 1988–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Guzman M, Pizzio GA, Antoni R, et al. . 2012. Arabidopsis PYR/PYL/RCAR receptors play a major role in quantitative regulation of stomatal aperture and transcriptional response to abscisic acid. The Plant Cell 24, 2483–2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Q, Yin P, Li W, Wang L, Yan C, Lin Z, Wu JZ, Wang J, Yan SF, Yan N. 2011. The molecular basis of ABA-independent inhibition of PP2Cs by a subclass of PYL proteins. Molecular Cell 42, 662–672. [DOI] [PubMed] [Google Scholar]
- Hauser F, Waadt R, Schroeder JI. 2011. Evolution of abscisic acid synthesis and signaling mechanisms. Current Biology 21, R346–R355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SC, Lan W, Buchanan BB, Luan S. 2009. A protein kinase–phosphatase pair interacts with an ion channel to regulate ABA signaling in plant guard cells. Proceedings of the National Academy of Sciences, USA 106, 21419–21424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SJ, Lee MH, Kim JI, Kim SY. 2015. Arabidopsis putative MAP kinase kinase kinases Raf10 and Raf11 are positive regulators of seed dormancy and ABA response. Plant & Cell Physiology 56, 84–97. [DOI] [PubMed] [Google Scholar]
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E. 2009. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324, 1064–1068. [DOI] [PubMed] [Google Scholar]
- Miyazono K, Miyakawa T, Sawano Y, et al. . 2009. Structural basis of abscisic acid signalling. Nature 462, 609–614. [DOI] [PubMed] [Google Scholar]
- Nishimura N, Hitomi K, Arvai AS, Rambo RP, Hitomi C, Cutler SR, Schroeder JI, Getzoff ED. 2009. Structural mechanism of abscisic acid binding and signaling by dimeric PYR1. Science 326, 1373–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura N, Sarkeshik A, Nito K, et al. . 2010. PYR/PYL/RCAR family members are major in-vivo ABI1 protein phosphatase 2C-interacting proteins in Arabidopsis. The Plant Journal 61, 290–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto M, Peterson FC, Defries A, Park SY, Endo A, Nambara E, Volkman BF, Cutler SR. 2013. Activation of dimeric ABA receptors elicits guard cell closure, ABA-regulated gene expression, and drought tolerance. Proceedings of the National Academy of Sciences, USA 110, 12132–12137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Fung P, Nishimura N, et al. . 2009. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324, 1068–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago J, Dupeux F, Round A, Antoni R, Park SY, Jamin M, Cutler SR, Rodriguez PL, Márquez JA. 2009. The abscisic acid receptor PYR1 in complex with abscisic acid. Nature 462, 665–668. [DOI] [PubMed] [Google Scholar]
- Song S, Qi T, Huang H, Ren Q, Wu D, Chang C, Peng W, Liu Y, Peng J, Xie D. 2011. The Jasmonate-ZIM domain proteins interact with the R2R3-MYB transcription factors MYB21 and MYB24 to affect Jasmonate-regulated stamen development in Arabidopsis. The Plant Cell 23, 1000–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischer SV, Wunschel C, Papacek M, Kleigrewe K, Hofmann T, Christmann A, Grill E. 2017. Combinatorial interaction network of abscisic acid receptors and coreceptors from Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA 114, 10280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Zhao Y, Li Z, et al. . 2018. Reciprocal regulation of the TOR kinase and ABA receptor balances plant growth and stress response. Molecular Cell 69, 100–112.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. 2007. An ‘Electronic Fluorescent Pictograph’ browser for exploring and analyzing large-scale biological data sets. PLoS One 2, e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Zhou L, Liu X, Liu H, Li D, Cao M, Chen H, Xu L, Zhu JK, Zhao Y. 2017. A novel chemical inhibitor of ABA signaling targets all ABA receptors. Plant Physiology 173, 2356–2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin P, Fan H, Hao Q, Yuan X, Wu D, Pang Y, Yan C, Li W, Wang J, Yan N. 2009. Structural insights into the mechanism of abscisic acid signaling by PYL proteins. Nature Structural & Molecular Biology 16, 1230–1236. [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J. 2007. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nature Protocols 2, 1565–1572. [DOI] [PubMed] [Google Scholar]
- Yoshida R, Hobo T, Ichimura K, Mizoguchi T, Takahashi F, Aronso J, Ecker JR, Shinozaki K. 2002. ABA-activated SnRK2 protein kinase is required for dehydration stress signaling in Arabidopsis. Plant & Cell Physiology 43, 1473–1483. [DOI] [PubMed] [Google Scholar]
- Zhang L, Li X, Li D, et al. . 2018. CARK1 mediates ABA signaling by phosphorylation of ABA receptors. Cell Discovery 4, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Chan Z, Xing L, Liu X, Hou YJ, Chinnusamy V, Wang P, Duan C, Zhu JK. 2013. The unique mode of action of a divergent member of the ABA-receptor protein family in ABA and stress signaling. Cell Research 23, 1380–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Xing L, Wang X, Hou YJ, Gao J, Wang P, Duan CG, Zhu X, Zhu JK. 2014. The ABA receptor PYL8 promotes lateral root growth by enhancing MYB77-dependent transcription of auxin-responsive genes. Science Signaling 7, ra53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Hao H, Zhang Y, et al. . 2015. SOS2-LIKE PROTEIN KINASE5, an SNF1-RELATED PROTEIN KINASE3-type protein kinase, is important for abscisic acid responses in Arabidopsis through phosphorylation of ABSCISIC ACID-INSENSITIVE5. Plant Physiology 168, 659–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.