General base levels of chloroplast protein glycation were independent of rosette age or stress status. Yet under different abiotic stress treatments, a few highly specific changes in glycation of potential regulatory proteins were detected.
Keywords: Abiotic stress, advanced glycation end products (AGEs), chloroplast, diurnal, drought, photosynthesis, protein glycation
Abstract
Non-enzymatic post-translational modifications of proteins can occur when the nucleophilic amino acid side chains of lysine and arginine encounter a reactive metabolite to form advanced glycation end products (AGEs). Glycation arises predominantly from the degradation of reducing sugars, and glycation has been observed during metabolic stress from glucose metabolism in both animals and plants. The implications of glycating proteins on plant proteins and biology has received little attention, and here we describe a robust assessment of global glycation profiles. We identified 112 glycated proteins that were common under a range of growth conditions and abiotic stress treatments, but also showed rosette age, diurnal, and drought stress-specific targets. Among 18 drought stress-specific glycation targets included several thioredoxin and thioredoxin-like proteins. In vitro glycation of two carbohydrate metabolism enzymes led either to a reduction or to a complete inhibition of activity, demonstrating the impact of glycation on protein function. Taken together, our results suggest that stress-specific glycation patterns of a small number of regulatory proteins may have a much broader impact on downstream target proteins that are, for example, associated with primary metabolism.
Introduction
In higher organisms, post-translational modifications (PTMs) modulate proteomic diversity by creating chemical changes to proteins following translation. In plants, >300 different types of PTMs have been identified that regulate enzyme activity, localization, stability, and signal transduction (Jensen, 2004; Rocks et al., 2005; Stulemeijer and Joosten, 2008; Hunter, 2009; Deribe et al., 2010; Maeda et al., 2010; Duan and Walther, 2015). Attachment of carbohydrates to proteins to create glycoproteins is the most structurally diverse type of PTM that can significantly alter protein conformation (Parodi, 2000; Slawson et al., 2006). Glycoproteins are complex, and are widely believed to transfer biological information as part of complex signalling pathways (Gabius et al., 2011; Solís et al., 2015). Glycoproteins are either formed in a controlled manner via enzymatic (N- and O-glycosylation) reactions or in a spontaneous random manner by non-enzymatic (glycation) reactions. Both glycosylation and glycation are biologically important but differ significantly in their roles and reactions.
Glycation is a complex network of parallel and sequential reactions which is initiated by the carbonyl group of a reducing sugar (e.g. glucose, fructose, or galactose) reacting with a free amino group, followed by rearrangements to form early glycation products such as fructosyl-lysine (FL) and other ketoamines (McCance et al., 1993; Thornalley et al., 1999; Zhang et al., 2009; Younus and Anwar, 2016). Further rearrangements and oxidation reactions subsequently lead to a heterogeneous group called advanced glycation end products (AGEs) (Neglia et al., 1983; Baynes et al., 1989). Additionally, AGEs can also be formed directly and relatively rapidly via reactive carbonyl compounds such as glyoxal and methylglyoxal (MG) (Thornalley et al., 1999).
Due to their stable nature, AGEs can diffuse within an organism and propagate free radical reactions far from the site of formation, causing further damage to proteins, lipids, or DNA, acting as cytotoxic messengers (Hulbert et al., 2007). Thus, AGEs are the primary cause for diabetic-, ageing-, chronic inflammation-, neurodegenerative-, and cancer-associated complications (Cerami, 1985; Brownlee et al., 1988; McCance et al., 1993; Tessier, 2010; Younus and Anwar, 2016). Despite their toxic nature, there is evidence that breakdown products of AGEs also have essential roles in signalling cascades by switching on defence pathways through the binding of AGE-containing peptides to receptors of AGEs (RAGEs) (Park et al., 2010; Ott et al., 2014). Binding of AGE-containing peptides initiates pro-inflammatory responses, activating NADPH oxidase, the production of reactive oxygen species (ROS), such as hydrogen peroxide (H2O2), and mitogen-activated protein kinase signalling pathways, inducing gene expression (Lander et al., 1997; Dower and Qwarnstrom, 2003; Wautier and Schmidt, 2004; Alikhani et al., 2007a, b).
MG is an important glycating agent and, despite there being detailed analyses on its production and detoxification via the glyoxalase pathway, analysis with respect to its damaging effects to proteins in plants has received little attention (Norton et al., 1990; Maiti et al., 1997; Thornalley et al., 1999; Bechtold et al., 2009). However, the development of proteomic tools and methods for simultaneous detection of glycation products has now made the analysis of products possible (Ahmed et al., 2002; Thornalley et al., 2003). For example, early stage FL and nine AGE-modified amino acids were identified in Arabidopsis thaliana (Arabidopsis) leaves under controlled conditions (Bechtold et al., 2009). The physiological relevance of the occurrence of these products in Arabidopsis remains to be validated especially in relation to stress signalling. Nevertheless, it is well known that glucose levels in plants undergo diurnal variation, with the production of starch occurring during the light phase and starch degradation and release of glucose for respiration occurring during the dark phase. Elevations in mainly lysine-associated glycation products were observed during the dark period (Bechtold et al., 2009), which coincides with an increase in H2O2 production (Bechtold et al., 2004). Recent studies in Brassica napus and Arabidopsis identified 772 and 502 AGE-modified proteins in non-stressed leaf extracts, respectively (Bilova et al., 2016), and identified osmotic stress-specific (Paudel et al., 2016) and plant age-specific protein targets (Bilova et al., 2017) in Arabidopsis. The general purpose of these studies was to identify the pathways and mechanisms of glycation in plants, identifying likely glycation substrates and specific glycation hotspots in plants. However, methodologies used did not clearly distinguish between glycosylated and glycated proteins, which may have caused an overestimation of protein targets. In addition, Bilova et al. (2016, 2017) do not provide an exhaustive list of all identified protein targets, nor do selected protein targets match across the different studies, making it difficult to judge the general impact of glycation in plants.
In the present study, we compared glycation patterns in Arabidopsis in response to short-term stresses such as high light or heat, a slow developing drought stress (Ferguson et al., 2018), and changes in glycation targets during a diurnal period (Bechtold et al., 2009) on soil-grown plants. The aim was to determine baseline levels of glycation, and to identify stress- and diurnal-specific protein targets, which would allow us to link glycation and protein function to underlying physiological effects, especially under specific stress treatments.
Materials and methods
Plant material, plant growth, and stress conditions
Arabidopsis ecotype Col-0 was obtained from the European Arabidopsis Stock Centre and seeds were planted in soil (Scotts Levington’s F2+S) as described in Ferguson et al. (2018). Following germination, seedlings were grown under a 8 h to 16 h light to dark cycle at 23 °C, 60% relative humidity, and light intensity of 150 µmol m−2 s−1 before being transferred into separate pots after 2 weeks. For heat and light stress experiments, plants were kept under constant well-watered conditions for 4 weeks before transferring plants into either high light [IsoLight at 850 µmol m−2 s−1 (Technologica)] or heat [37 °C, 78% humidity, vapour pressure deficit 1 kPa (Fitotron)] conditions for 4 h.
For drought conditions, all pots were filled with an identical weight of soil mix in order to determine relative soil water content (rSWC) as described in Ferguson et al. (2018). Drought stress was performed on 5-week-old plants, and material was harvested at ~40% and ~20% rSWC including well-watered controls. Plant materials were obtained from two independent experiments with six replicates for each stress treatment experiment.
Chlorophyll fluorescence imaging
Plants were confirmed as responding to the heat or light stress conditions by dark adapting the plants before measuring the Chl a fluorescence parameter Fv/Fm using a fluorescence imager (Fluorimager, Technologica, Colchester UK).
Determination of glyoxalase II activity
Protein isolation and determination of glyoxalase II activity were carried out according to Bilova et al. (2016). For glyoxalase II activity, 50 μl of protein extracts were added to a mixture of 0.2 mM 5,5'-dithio-bis(2-nitrobenzoic acid (DTNB) and 1 mM S-d-lactoylglutathione in 20 mM NaH2PO4/Na2HPO4, pH 7.0 (450 μl). The absorbance at 412 nm was measured over 3–5 min (until no further change) with 30 s intervals using a spectrophotometer, and the activity was calculated using ε412 nm DTNB of 13 600 M–1 cm–1 (TNB2– ion has a yellow colour and absorbance at 412 nm) (Ellman, 1959).
Lipid hydroperoxides and H2O2 quantification
Lipid hydroperoxides (LHPOs) were quantified as described by Griffiths et al. (1997, 2000). Concentrations of LHPOs were determined using a 13(S)-hydroperoxy-9,11 (cis, trans)-octadecadienoic acid (13S-HPODE) calibration curve as standard. HPODE was prepared as described by Reeder and Wilson (1998) and calibration curves were constructed ranging from 1 μM to 150 μM.
H2O2 concentrations were determined by homogenizing 0.1 g (FW) of leaf tissue in 1 ml of 0.1 M HCl. Samples were centrifuged at 13000 rpm, 4 °C for 10 min, and the supernatant was cleared using activated charcoal. A 400 µl aliquot was then incubated with 2.6 ml of reaction mixture of 2.54 ml of 50 mM HEPES pH 7.5, 60 µl of 50 mM homovanillic acid, and 60 µl of 4 µM horseradish peroxidase (HRP; Sigma). Fluorescence was then measured at 425 nm emission (excitation of 315 nm) using an LS 50B fluorimeter (Perkin Elmer). H2O2 concentrations were determined using a H2O2 standard curve from 0.1 nmol to 10 nmol.
Carbohydrate analysis
Soluble carbohydrates (glucose, fructose, and sucrose) and starch were extracted from 20 mg FW of leaf material. Extracts were incubated in 80% (v/v) ethanol for 30 min at 80 °C, then washed four times with 80% (v/v) ethanol. Glucose, fructose, and sucrose were measured from the ethanol extract using an enzyme-based protocol according to Stitt and Quick (1989). The starch content was estimated from the ethanol-insoluble pellet according to Stitt et al. (1978).
Protein glycation
BSA, glyceraldehyde-3-phosphate dehydrogenase (GAPC1), and triosephosphate isomerase (TPI) were glycated according to Schmidt et al. (1992). BSA, GAPC1, and TPI were prepared with and without 0.4 M glucose and incubated at 37 °C in the dark for 3 weeks. Solutions were made using purified water, filtered prior to incubation, and contained various protease inhibitors to ensure minimal protein degradation and sterility during glycation. Solutions were then concentrated at 4 °C using a 5 kDa cut-off Centricon (Vivaspin) against 0.1 M sodium phosphate pH 7 buffer with two changes to remove any unspecific binding.
Protein isolation, Rubisco fractionation, and alkaline hydrolysis
Proteins were isolated from three biological replicates. Approximately 1 g (FW) of plant material was homogenized and extracted in 6 ml of extraction buffer [20 mM MgCl2, 2% β-mercaptoethanol, 0.1% protease inhibitor cocktail, 1 mM phenylmethylsulphonyl fluoride (PMSF), 2% NP-40, 500 mM Tris-HCl, pH 8.3]. Extracts were cleared by filtration through a 0.2 µm Minisart filter (Sartorius) and centrifuged at 3000 g, 4 °C for 20 min. Rubisco was isolated using a 50% polyethylene glycol (PEG) 6000 solution adapted from Widjaja et al. (2009), producing an almost Rubisco-free supernatant and a Rubisco PEG fraction.
Proteins were solubilized in 250 μl of solubilization buffer (SB; 7 M urea, 2 M thiourea, 50 mM DTT, 4% CHAPS, 0.4% SDS, 5 mM K2CO3). For alkaline hydrolysis, an equal volume (250 μl) of 0.1 M NaOH (pH 13) was added and samples were incubated at 45 °C for 6 h. Reactions were then neutralized by addition of 250 μl of 0.1 M HCl.
Boronate affinity chromatography (BAC)
Samples were adjusted to a volume of 2 ml with buffer A (250 mM ammonium acetate, 50 mM MgCl2, pH 8.1). Supernatants were loaded onto a 2 ml or 10 ml m-aminophenylboronic acid–agarose high performance Tricorn 5/100 column (GE healthcare) equilibrated with buffer A. Unbound non-glycated proteins were eluted in a 10 ml wash step with buffer A, before glycated proteins were eluted in 10 ml of 100% 0.1 M acetic acid using an ÄKTA pure purification system (GE Healthcare). Glycated fractions were collected and proteins were precipitated with acetone (4:1 acetone:water) overnight at –20 °C.
Mass spectrometry
Precipitated proteins were resuspended in ~50 μl of SB and 10 μl of SDS loading dye, and heated at 95 °C for 3 min. Samples were then loaded onto a 12.5% SDS gel. Samples were allowed to enter the resolving portion of the gel before cutting out the gel slices and placing them in low bind siliconized tubes (Greiner). Gel slices were subjected to in-gel trypsin digestion as previously described (Metodieva et al., 2013), and evaporated peptides were kept at –80 °C until required. The constituent tryptic peptides were concentrated and analysed by electrospray-ionization tandem MS on a hybrid high resolution LTQ/Orbitrap Velos instrument (Thermo Scientific, Hemel Hempstead, UK) interfaced to a split-less nanoscale HPLC (Ultimate 3000; Dionex, Loughborough, UK) as described previously (Greenwood et al., 2012; Metodieva et al., 2013; Croner et al., 2014).
MaxQuant was used to analyse the MS/MS data (Cox et al., 2014). Searches for the glycation sites were performed using MaxQuant Andromeda with masses of +58 Da for N-carboxymethyllysine/N-carboxymethylarginine (CML/CMA), +162 Da for FL, +39.99 Da for N-(5-hydro-4-imidazolon-2-yl)ornithine (G-H1), and +54 Da for N-(5-hydro-5-methyl-4-imidazolon-2-yl)ornithine (MG-H1).
Data analysis
All statistical analysis was performed using R 3.4.3. The data from the 10%, SN, and 20% fractions were combined to cover the total glycated proteome for each condition. The DEP 1.1.4 package (Zhang et al., 2018) was used to determine differential enrichment of glycated proteins between treatments. This package provides data filtering, variance normalization, imputation of missing values, and statistical testing. Proteins were filtered according to the presence in the biological replicates of, (i) a minimum of two out of three biological replicates (high stringency) or (ii) a minimum of one out of three of the biological replicates (low stringency) in at least one of the conditions tested.
One-way ANOVA and a post-hoc Tukey test was used to determine significance for the measurement of carbohydrates and stress biomarkers. Statistical significance for glycation targets was evaluated within the DEP 1.1.4 package (Zhang et al., 2018). Values on the bar charts represent the mean ±SE (n=6) with the asterisk indicating significant differences at P<0.05.
Western blotting
Protein solutions were quantified using the Bradford assay then diluted accordingly using SDS loading buffer. Samples were loaded on a 12.5% SDS–PAGE gel (BioRad) followed by transfer before incubation with the primary antibody (anti-CML at 1/1000 dilution, or anti-OGlcNAc at 1/5000). Goat anti-rabbit HRP secondary antibody added to anti-CML (1/2500, Promega), and goat anti-mouse HRP (1/5000, Thermo Fisher) added to anti-OGlcNAc were used to visualize the modifications using the Pierce ECL Western Blotting substrate (Thermo Scientific).
Cloning, overexpression, and purification of GAPC1 and TPI
Genes encoding GAPC1 (accession no. P25858 encoding 338 amino acids) and TPI (accession no. Q9SKP6 encoding 254 amino acids) were cloned into pET28a expression vectors (BaseClear) and overexpressed in Escherichia coli BL21(DE3) cells. Recombinant proteins were purified once expressed at 37 °C in 1 litre of LB cultures after induction with 0.4 M isopropyl β-d-1-thiogalactopyranoside (IPTG; Melford). Cells were harvested 16 h after induction at 3501 g for 20 min at 4 °C. Pellets were resuspended in buffer C (50 mM Tris–HCl, 500 mM NaCl, 20 mM imidazole, pH 8) then lysed using an EmulsiFlex-C5 cell disrupter (Avestin) followed by centrifugation at 38724 g for 20 min at 4 °C. The clarified supernatant was then loaded onto a 5 ml Ni-NTA Sepharose column (GE Healthcare) and protein was eluted with a linear imidazole gradient using buffer D (buffer C with 500 mM imidazole) using an ÄKTA purification system. Single peaks were collected and concentrated at 4 °C using a Centricon (Vivaspin) with a 10 kDa cut-off. Concentrated protein was purified further using a G75 or S200 Superdex size exclusion column (GE Healthcare) equilibrated with buffer E (10 mM NaH2PO4/Na2HPO4, pH 7, 100 mM NaCl). Concentrations of purified protein were determined spectrophotometrically [Cary 60 UV-Visible spectrophotometer (Agilent)] using extinction coefficients (ε) of 40910 M–1 cm-1 and 38960 M–1 cm–1 for GAPC1 and TPI, respectively as determined using the ExPASy server (Gasteiger et al., 2003).
Steady-state enzyme activity
Steady-state enzyme kinetics were carried out with purified, non-glycated and glycated GAPC1 and TPI enzymes. The assays were measured using a Hewlett-Packard 8453 diode-array spectrophotometer scanning between 190 nm and 1100 nm at 25 °C. The GAPC1 assay was carried out in a 1 ml cuvette in 10 mM NaH2PO4/Na2HPO4, pH 8.5, 100 mM NaCl. Each reaction mixture contained 10 mM NAD+, 3 mM DTT, and 1 μM GAPC1 enzyme which was base-lined before the reaction was started by addition of 0–5 mM dl-glyeraldehyde-3-phosphate (G3P; Sigma). The TPI assay was also carried out in 1 ml cuvettes in 10 mM NaH2PO4/Na2HPO4, pH 7.0, 100 mM NaCl. Each reaction contained 0.1 mM NADH, 5 U of α-glycerophosphate dehydrogenase, and 0–5 mM G3P, and reactions were started by addition of 1 μM TPI enzyme. Plots of the turnover rate constant (k, s–1) versus substrate concentration were then constructed, whereby k is the initial rate normalized to the enzyme concentration calculated from {(ΔA/ε/t)/[enzyme]} where ΔA is the absorbance change at 340 nm (decrease for TPI with oxidation of NADH and increase for GAPC1 assay with reduction of NAD+), ε is the extinction coefficient of the substrate oxidation product which is 6.22 mM–1 cm–1, t is the time in seconds, and [enzyme] is the total concentration of the enzyme in the assay. The Michaelis constant, turnover number, and catalytic efficiency were estimated by non-linear least squares fitting to the Michaelis–Menten model.
Results
Specific enrichment for non-enzymatically glycated peptides
To distinguish between N-/O-glycosylation and non-enzymatic glycation (see the Introduction), glycated BSA was used to ensure that the enrichment was specific to non-enzymatic glycation modifications. Glycated BSA and the total leaf extract were separated via BAC into non-glycated/non-glycosylated and glycated/glycosylated peptide fractions (see Supplementary Fig. S1 at JXB online). Glycated BSA and leaf extracts were subsequently subjected to mild alkaline hydrolysis, neutralized, and separated again using a BAC column. Glycated BSA did not show a reduction in peak area (Supplementary Fig.S1A), owing to the fact that BSA only contained non-enzymatically glycated residues. However, in total leaf extracts the glycated/glycosylated peak was reduced by ~75% following alkaline hydrolysis (Supplementary Fig. S1B).
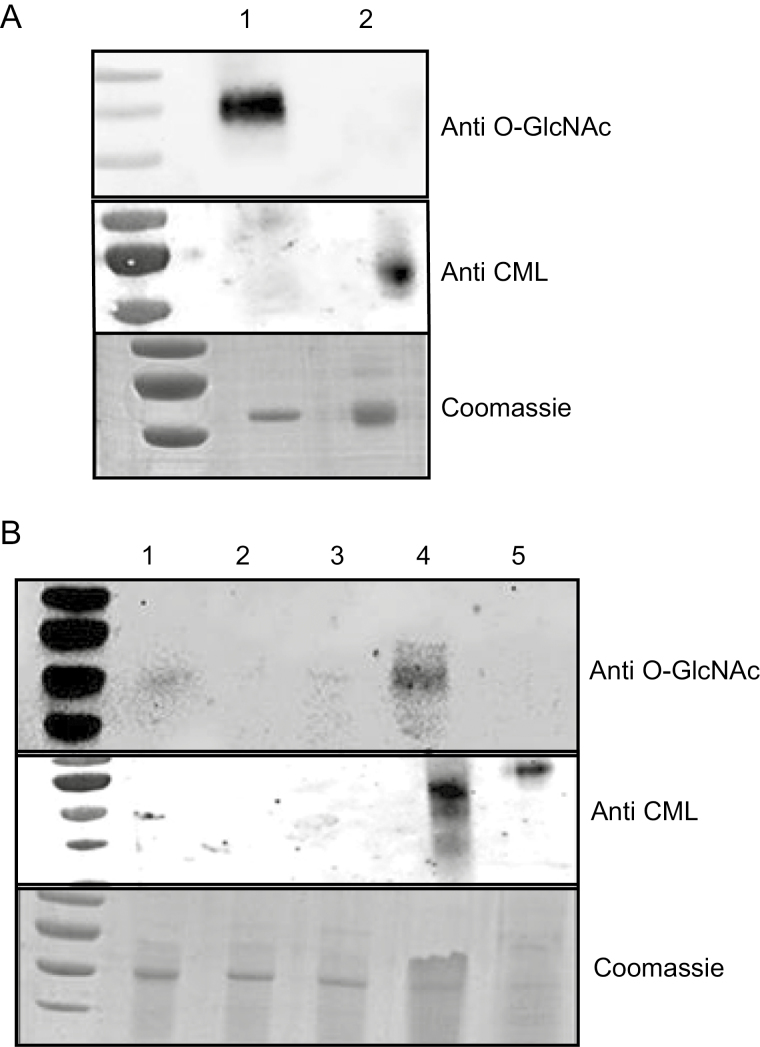
The separated glycated and non-glycated leaf fractions were subsequently analysed by western blot using glycation-specific anti-CML and O-glycosylation-specific anti-OGlcNAc antibodies. Specificity of the antibodies was verified using glycated and glycosylated BSA, respectively. The anti-CML antibody detected glycated BSA with minimal cross-reaction with O-glycosylated BSA (Fig. 1A), while anti-OGlcNAc exclusively identified O-glycosylated BSA (Fig. 1A). Importantly, anti-OGlcNAc and anti-CML both reacted with the leaf extract prior to alkaline hydrolysis (Fig. 1B), but following alkaline hydrolysis only the anti-CML reacted with the protein sample (Fig. 1B). Therefore, alkaline hydrolysis followed by BAC was successful in enriching for non-enzymatically glycated proteins, and illustrates that alkaline hydrolysis is essential in order to remove N- and O-glycosylated proteins.
Fig. 1.
Enrichment for protein glycation after alkaline hydrolysis. (A) Specificity of anti-CML and anti-O-GlcNAc antibodies using modified BSA. Lane 1, O-glycosylated BSA (Thermo Fisher, UK); lane 2, non-enzymatically glycated BSA (see the Materials and methods). Top: a western blot hybridized with anti-O-GlcNAc. Middle: a western blot hybridized with anti-CML. Bottom: a Coomassie stain of an SDS–PAGE gel transferred onto a nitrocellulose membrane before western blot analysis. (B) Enrichment for glycated proteins before and after alkaline hydrolysis of total leaf extracts. Lane 1, total protein extract; lanes 2 and 3, non glycated peptide peaks; lane 4, glycated peak prior to alkaline hydrolysis; 5, glycated peptide peak post-alkaline hydrolysis. Top: a western blot hybridized with anti-O-GlcNAc. Middle: a western blot hybridized with anti-CML antibody. Bottom: a Coomassie stain of an SDS–PAGE gel before transfer to a nitrocellulose membrane for western blot analysis.
Stress treatments lead to increases in sugar levels and other stress biomarkers
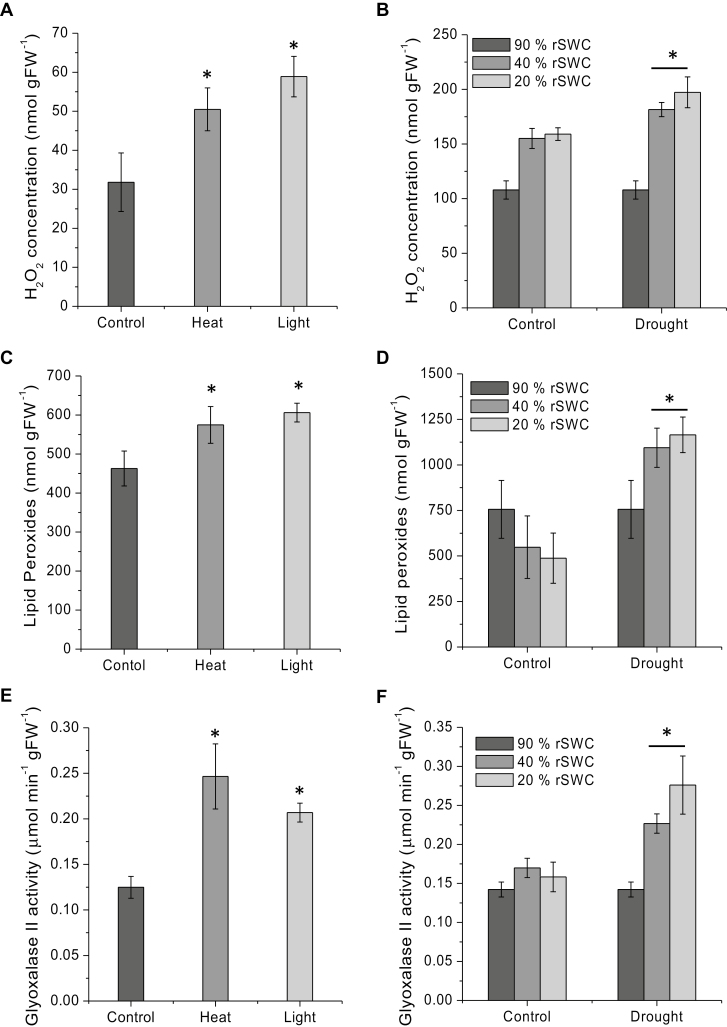
Arabidopsis rosettes were subjected to heat, light, and drought stress as described in Bechtold et al. (2009, 2013, 2016). To assess the physiological impact of heat or light stress, Chl a fluorescence imaging was used to measure the maximum quantum efficiency of PSII photochemistry (Fv/Fm). There was no significant difference between control and heat-stressed plants, with Fv/Fm values of 0.8 ± 0.006 and 0.806 ± 0.007 for control and heat, respectively. Only in the high light treatment was a significant decrease in Fv/Fm to 0.723 ± 0.006 observed (Supplementary Fig. S2). Heat, light, and drought stress treatments resulted in a significant increase in the production of H2O2 (Fig. 2A, B) and LHPOs (Fig. 2C, D). Glyoxalase II activity, a biomarker for increased glyoxal and methylglyoxal production, was also shown to increase significantly in either high light, heat, or drought stress treatments (Thornalley, 2003) (Fig. 2E, F). This suggests that the applied stress treatments were effective in inducing oxidative stress.
Fig. 2.
Analysis of stress response in Arabidopsis Col-0 plants under high light (IsoLight 850 µmol m−2 s−1), heat (37 °C, 78% humidity), and drought conditions. (A and B) H2O2 concentration (nmol gFW–1) measurements for (A) control, high light, and heat, and (B) drought stress treatments. H2O2 measurements were taken using homovanillic acid fluorescence by emission at 425 nm (excitation 315 nm) using an LS 50B fluorimeter. (C and D) Determination of lipid hydroperoxide content (nmol gFW–1) in (C) control, high light, and heat treatment, and (D) drought stress treatment. The concentrations were measured by ferrous oxidation-xylenol orange assay with absorption measurement at 560 nm using a Cary 60 UV-visible spectrophotometer. (Eand F) Glyoxalase II enzyme activity measurements of (E) control, high light, and heat, and (F) drought experiments. Measurements were taken with S-d-lactoylglutathione as substrate and the absorbance at 412 nm of DTNB was measured. * indicates a significant difference at P<0.05.
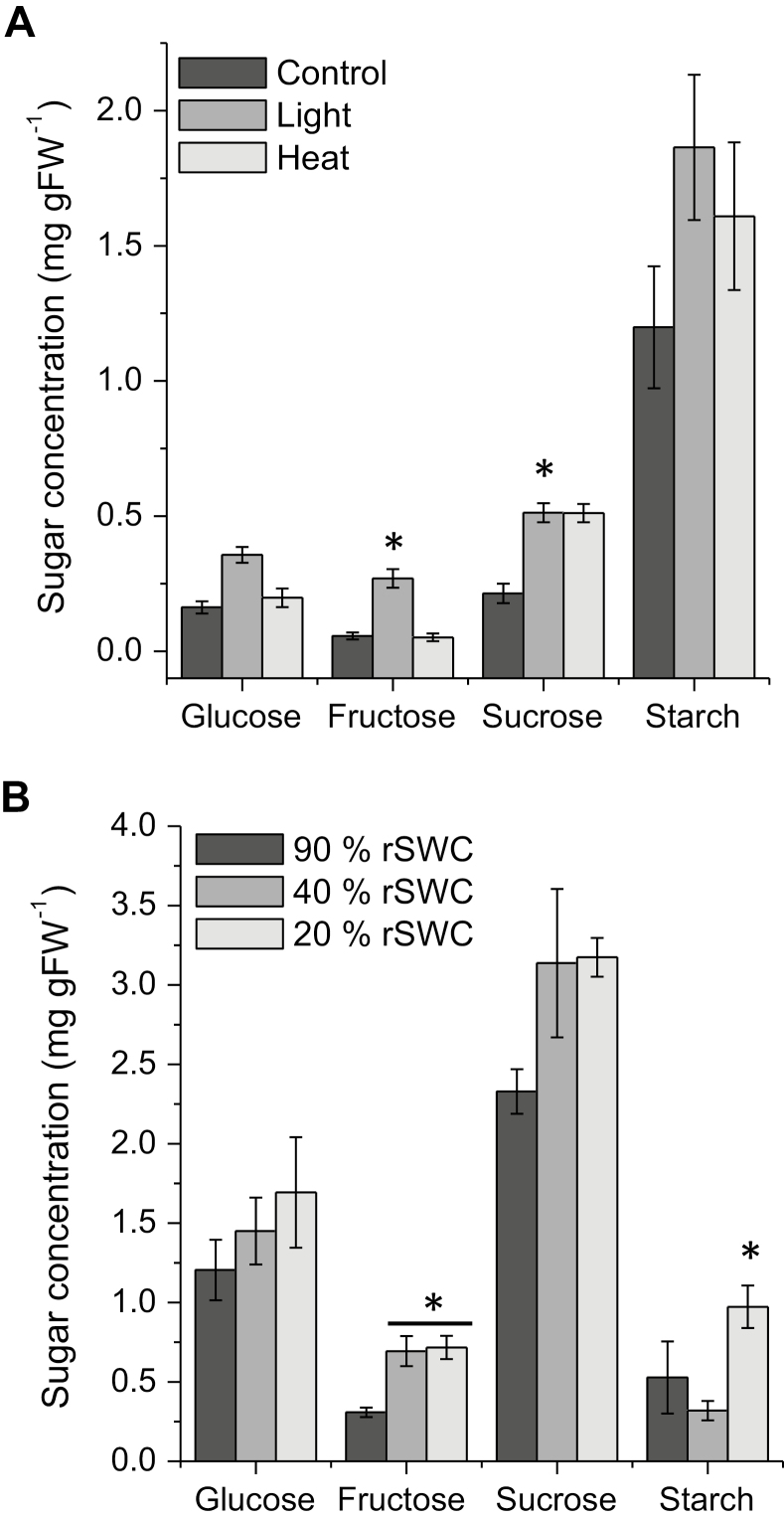
We also analysed glucose, fructose, sucrose, and starch content during stress treatments. High light or heat treatments led to significant increases in starch and sucrose content (Fig. 3A), while glucose and fructose only significantly increased during high light (Fig. 3A). Drought stress also significantly increased soluble carbohydrate levels but not the starch content (Fig. 3B). In addition, the expected diurnal changes in glucose, fructose, and sucrose were observed peaking during the light period, and decreasing during the night (Supplementary Fig. S3A).
Fig. 3.
Carbohydrate analysis of high light, heat, and drought experiments in Arabidopsis. (A) Control, high light, and heat, and (B) drought analysis of soluble (glucose, fructose, and sucrose) and insoluble (starch) carbohydrates. * indicates a significant difference at P<0.05.
A core set of 112 proteins are targeted for glycation
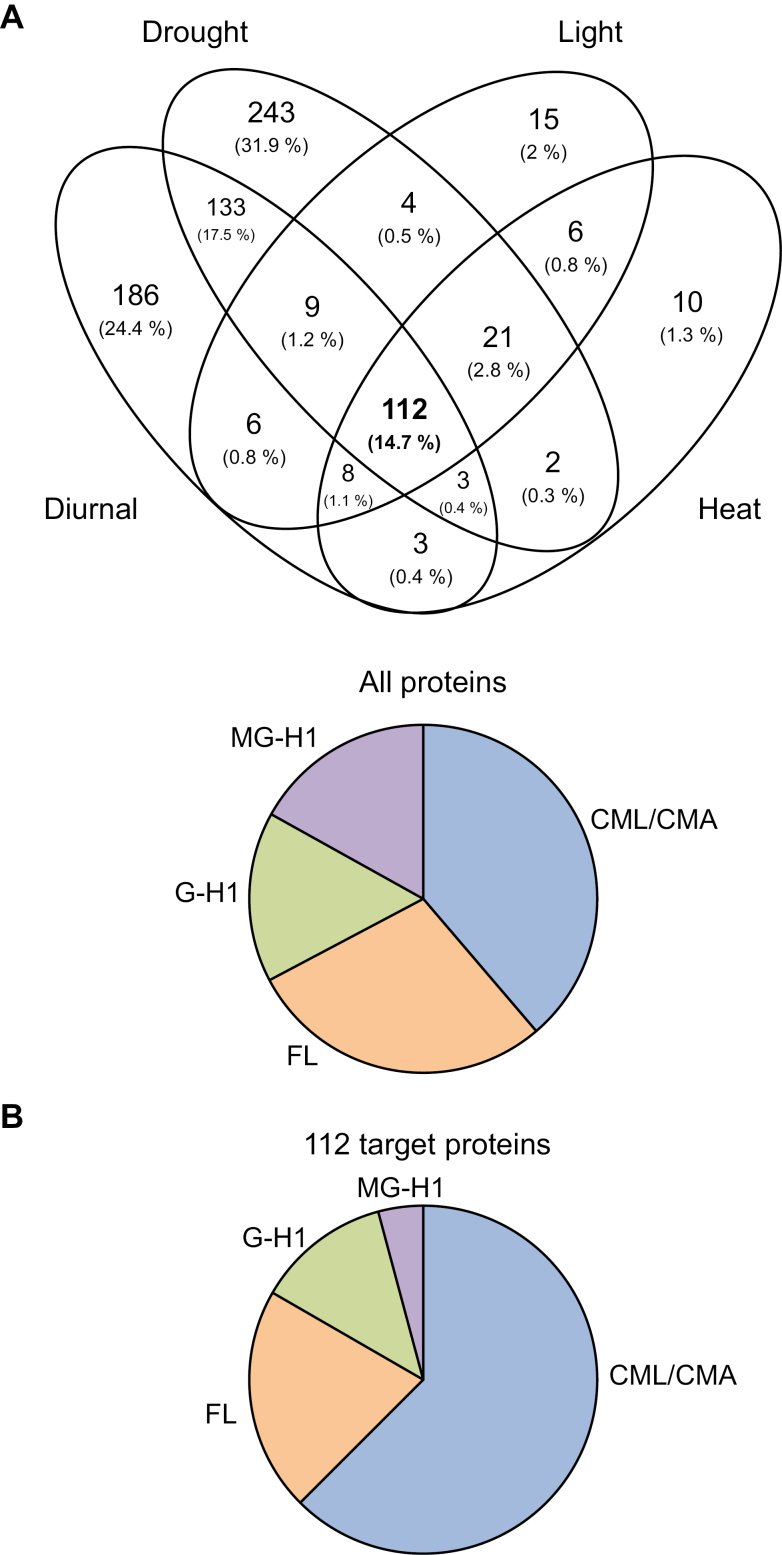
Previous studies of glycation profiles have reported >500 protein targets in Arabidopsis leaf extracts; however, methodologies were unclear regarding the removal of glycosylated proteins, and may have resulted in an overestimation of targets (Bilova et al., 2016). As described above, the use of alkaline hydrolysis removes the labile glycosylation and ribosylation moieties (Fig. 1; Supplementary Fig. S1), while leaving the stable covalently glycated peptides intact (Varki and Diaz, 1984). We identified 544, 182, 196, and 735 proteins during a diurnal period, heat, light, and drought experiments, respectively (see Supplementary Tables S1 and S2 for raw proteomics data). Generally, there were fewer targets in the heat and light experiments, which is probably due to plant age as high light and heat stress experiments were carried out on 4-week-old rosettes (Supplementray Table S3). These proteins contained thousands of peptides and 522, 502, 560, and 1863 glycation sites (modifications CML/CMA, FL, G-H1, and MG-H1) in the diurnal, heat, light, and drought experiments, respectively (Supplementary Table S3). An example mass spectrumof a CML glycated peptide belonging to the chloroplast-located PSBP1 oxygen-evolving enhancer protein 2-1 can be seen in Supplementary Fig. S4. All proteins were then subjected to a high stringency analysis, and any proteins with <2 unique peptides were removed, resulting in 459, 181, 165, and 527 proteins for a diurnal period, heat, light, and drought experiments, respectively. Comparison of all the proteins identified 112 common targets (Fig. 4A; Table 1). These 112 common targets were subsequently analysed using the DEP package (see the Materials and methods) (Zhang et al., 2018). Some of the most abundant proteins amongst these targets include the β-subunit of the chloroplastic ATP synthase and chloroplastic phosphoglycerate kinase. Across all experiments, four main glycation modifications were searched for: CML/CMA, FL, G-H1, and MG-H1. The percentage weightings of the glycan modifications remained similar for all experiments. The glycation percentage proportions for heat, light, and drought experiments were similar (Fig. 4B). However, when the glycation sites were analysed in the 112 targets, significant differences in the proportions of glycation modification were evident. In all the proteins, 38.7% CMA/CML and 4.2% MG-H1 were identified, which both increased, to 62.5% and 16.9%, respectively, in the 112 common targets (Fig. 4B). Interestingly, very few of the 112 core targets (Table 1) significantly changed during the heat, light, and drought stress treatments (Table 2). The diurnal (08.00, 16.00, 24 h) and rosette age experiments (4 weeks, controls of the high light and heat experiment; 5 weeks, 90% rSWC control at the start of the drought experiment; and 6 weeks, 90% rSWC control at the end of the drought experiment), showed the majority of significant changes in the abundance of glycation targets. There were eight significant protein targets between 08.00 h and 24.00 h in the diurnal experiment, and 17 significant protein targets between 4- and 6-week-old rosettes (Table 2; Supplementary Fig. S5A).
Fig. 4.
AGE-modified protein targets in Arabidopsis leaves. (A) Venn diagram showing the core 112 common proteins targeted for glycation identified in high light, heat, drought, and time-series experiments in Arabidopsis. The Venn diagram was constructed using Venny 2.1. (B) AGE-modified tryptic peptides obtained from Arabidopsis distributed by glycation modification in all the proteins in the high light, heat, and drought stress experiments, and in the 112 common target proteins. Searches for the glycation sites were performed using MaxQuant Andromeda with masses of +58 Da for N-carboxymethyllysine/N-carboxymethylarginine, +162 Da for fructosyl-lysine, +39.99 Da for N-(5-hydro-4-imidazolon-2-yl)ornithine, and +54 Da for N-(5-hydro-5-methyl-4-imidazolon-2-yl)ornithine.
Table 1.
The112 common AGE-modified proteins identified in high light, heat, drought, and time-series experiments in Arabidopsis plants
| Protein annotation | Genome identifier | Unique peptides | |
|---|---|---|---|
| Protein name | Accession no. | ||
| PROTOCHLOROPHYLLIDE OXIDOREDUCTASE C | O48741 | AT1G03630 | 4 |
| COPPER PROTEIN | Q9ZUX4 | AT2G27730 | 2 |
| SELENIUM-BINDING PROTEIN 1 | O23264 | AT4G14030 | 3 |
| LIPID TRANSFER PROTEIN 1 | Q42589 | AT2G38540 | 3 |
| ACTIN 8/ACTIN 2 | Q96293/Q96292 | AT1G49240/AT3G18780 | 4 - 5 |
| RIBOSOMAL PROTEIN L11 | Q9LFH5/Q9FF52/ P50883 | AT3G53430/AT5G60670/ AT2G37190 | 4 - 5 |
| UDP-GLUCOSE PYROPHOSPHORYLASE 1 | Q9M9P3 | AT3G03250 | 3 - 7 |
| ARABIDOPSIS THALIANA CYCLOPHILIN 1 | Q42406 | AT4G34870 | 4 |
| EMBRYO SAC DEVELOPMENT ARREST 38/SELENIUM-BINDING PROTEIN 2 | Q93WN0 | AT4G14040 | 4 |
| GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASE A SUBUNIT 2 | Q9LPW0 | AT1G12900 | 4 |
| LESION INITIATION 2 | Q9LR75 | AT1G03475 | 8 |
| GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE C SUBUNIT | P25858 | AT3G04120 | 2 |
| PSA E1 KNOCKOUT | Q9S831 | AT4G28750 | 2 - 4 |
| GLUTATHIONE S-TRANSFERASE PHI 9 | O80852 | AT2G30860 | 5 - 9 |
| RUBISCO SMALL SUBUNIT 3B/RUBISCO SMALL SUBUNIT 2B | P10798/P10797 | AT5G38410/AT5G38420 | 4 - 5 |
| HEAT SHOCK COGNATE PROTEIN 70-1 | P22953 | AT5G02500 | 2 - 5 |
| CYCLOPHILIN 19, ROTAMASE CYP 3 | Q38900 | AT2G16600 | 3 - 5 |
| PHOTOSYSTEM II SUBUNIT Q | Q41932 | AT4G05180 | 5 - 6 |
| GLUTAMATE-1-SEMIALDEHYDE 2,1-AMINOMUTASE 2 | Q42522 | AT3G48730 | 4 - 5 |
| RAN NUCLEAR PROTEIN GTPASE 3/2/1 | Q8H156/P41917/ P41916 | AT5G55190/AT5G20020/ AT5G20010 | 5 - 7 |
| 2-CYS PEROXIREDOXIN A | Q96291 | AT3G11630 | 5 |
| METHIONINE ADENOSYLTRANSFERASE 4 | Q9LUT2 | AT3G17390 | 5 |
| THYLAKOID FORMATION1 | Q9SKT0 | AT2G20890 | 5 - 8 |
| METHIONINE SYNTHASE 2 | Q9SRV5 | AT3G03780 | 5 - 6 |
| CATALASE 2 | P25819 | AT4G35090 | 6 |
| METHIONINE SULFOXIDE REDUCTASE A4 | P54150 | AT4G25130 | 5 - 6 |
| FIBRILLIN 4 | Q9LW57 | AT3G23400 | 6 - 7 |
| CHLOROPLAST STEM-LOOP BINDING PROTEIN OF 41 KDA | Q9LYA9 | AT3G63140 | 6 - 9 |
| GLUTATHIONE S-TRANSFERASE TAU 19 | Q9ZRW8 | AT1G78380 | 6 - 10 |
| GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASE A SUBUNIT 1 | P25856 | AT3G26650 | 6 - 7 |
| CYCLOPHILIN 20–3 | P34791 | AT3G62030 | 7 - 8 |
| ASPARTATE AMINOTRANSFERASE 5 | P46248 | AT4G31990 | 7 - 14 |
| O-ACETYLSERINE (THIOL) LYASE (OAS-TL) ISOFORM A1 | P47998 | AT4G14880 | 7 - 13 |
| PHOSPHOGLYCERATE KINASE FAMILY PROTEIN 2 | P50318 | AT1G56190 | 7 |
| GLUTATHIONE S-TRANSFERASE TAU 20 | Q8L7C9 | AT1G78370 | 7 - 10 |
| PEROXIREDOXIN Q | Q9LU86 | AT3G26060 | 5 - 7 |
| NONPHOTOCHEMICAL QUENCHING 4, PHOTOSYSTEM II SUBUNIT S | Q9XF91 | AT1G44575 | 5 - 7 |
| PHOTOSYSTEM II REACTION CENTER PROTEIN A | P83755 | ATCG00020 | 2 - 8 |
| EMBRYO DEFECTIVE 1395,HOMOLOGY-DEPENDENT GENE SILENCING 1, MATERNAL EFFECT EMBRYO ARREST 58, S-ADENOSYL-L-HOMOCYSTEIN HYDROLASE 1, | O23255 | AT4G13940 | 8 - 9 |
| TRANSLATIONALLY CONTROLLED TUMOR PROTEIN | P31265 | AT3G16640 | 5 - 8 |
| BETA CARBONIC ANHYDRASE 2 | P42737 | AT5G14740 | 8 |
| ASCORBATE PEROXIDASE 1 | Q05431 | AT1G07890 | 8 - 12 |
| THIAMINE4 | Q38814 | AT5G54770 | 4 - 8 |
| OXYGEN EVOLVING COMPLEX SUBUNIT 23 KDA | Q42029 | AT1G06680 | 8 |
| CHLOROPHYLL A/B BINDING PROTEIN 3/2/1 | Q8VZ87/P0CJ48/ P04778 | AT1G29910/AT1G29920/ AT1G29930 | 6 - 8 |
| GLYCERATE KINASE | Q944I4 | AT1G80380 | 8 - 10 |
| GLYCOLATE OXIDASE 1 | Q9LRR9 | AT3G14420 | 8 |
| PHOTOSYSTEM II SUBUNIT Q | Q9XFT3 | AT4G21280 | 8 - 9 |
| RIBOSOMAL L5P PROTEIN | O04603 | AT4G01310 | 7 - 9 |
| NUCLEOSIDE DIPHOSPHATE KINASE 1 | P39207 | AT4G09320 | 8 - 9 |
| MONODEHYDROASCORBATE REDUCTASE 6 | P92947 | AT1G63940 | 9 - 11 |
| NITRITE REDUCTASE 1 | Q39161 | AT2G15620 | 6 - 9 |
| DEHYDROASCORBATE REDUCTASE 5 | Q9FWR4 | AT1G19570 | 9 - 14 |
| ADP-RIBOSYLATION FACTOR 1/BFA-VISUALIZED EXOCYTIC TRAFFICKING DEFECTIVE 1/ATARFA1D | Q9LQC8/P36397/ P0DH91 | AT1G23490/AT2G47170/ AT1G70490 | 8 - 9 |
| CHLOROPLAST HEAT SHOCK PROTEIN 70–2 | Q9LTX9 | AT5G49910 | 6 - 9 |
| RIBOSE 5-PHOSPHATE ISOMERASE,TYPE A PROTEIN | Q9S726 | AT3G04790 | 6 - 9 |
| PEROXIDASE 34, PEROXIDASE CB | Q9SMU8 | AT3G49120 | 9 - 10 |
| MITOCHONDRIAL MALATE DEHYDROGENASE 1/2 | Q9ZP06/Q9LKA3 | AT1G53240/AT3G15020 | 9 - 10 |
| RIBULOSE BISPHOSPHATE CARBOXYLASE SMALL CHAIN 1A | P10795 | AT1G67090 | 6 - 10 |
| PROTOCHLOROPHYLLIDE OXIDOREDUCTASE B | P21218 | AT4G27440 | 10 - 12 |
| OXYGEN EVOLVING POLYPEPTIDE 1, MANGANESE-STABILIZING PROTEIN 1 | P23321 | AT5G66570 | 5 - 10 |
| GLUTATHIONE S-TRANSFERASE PHI 2/GLUTATHIONE S-TRANSFERASE 16, GLUTATHIONE S-TRANSFERASE F3 | P46422/Q9SLM6 | AT4G02520/AT2G02930 | 9 - 10 |
| ALDEHYDE DEHYDROGENASE 11A3 | Q1WIQ6 | AT2G24270 | 5 - 10 |
| GLYCOLATE OXIDASE 2 | Q9LRS0 | AT3G14415 | 6 - 11 |
| CYTOSOLIC NADP+-DEPENDENT ISOCITRATE DEHYDROGENASE | Q9SRZ6 | AT1G65930 | 11 - 13 |
| LIGHT HARVESTING COMPLEX OF PHOTOSYSTEM II 5 | Q9XF89 | AT4G10340 | 7 - 11 |
| PEROXISOMAL NAD-MALATE DEHYDROGENASE 2 | Q9ZP05 | AT5G09660 | 11 |
| ARGININE AMIDOHYDROLASE 2 | Q9ZPF5 | AT4G08870 | 10 - 11 |
| GLYCINE CLEAVAGE T-PROTEIN FAMILY | O65396 | AT1G11860 | 12 - 17 |
| EPITHIOSPECIFIER MODIFIER 1 | Q9LJG3 | AT3G14210 | 12 |
| PECTIN METHYLESTERASE 3 | O49006 | AT3G14310 | 8 - 13 |
| NAD(P)-BINDING ROSSMANN-FOLD PROTEIN | O80934 | AT2G37660 | 7 - 13 |
| PHOSPHORIBULOKINASE | P25697 | AT1G32060 | 12 - 13 |
| HIGH CYCLIC ELECTRON FLOW 1 | P25851 | AT3G54050 | 11 - 13 |
| CYTOSOLIC ISOFORM TRIOSE PHOSPHATE ISOMERASE | P48491 | AT3G55440 | 10 - 13 |
| ASCORBATE PEROXIDASE 4, THYLAKOID LUMEN 29, | P82281 | AT4G09010 | 12 - 13 |
| ATP SYNTHASE SUBUNIT 1 | P92549 | ATMG01190 | 11 - 13 |
| HYDROXYMETHYLBILANE SYNTHASE | Q43316 | AT5G08280 | 10 - 13 |
| MONODEHYDROASCORBATE REDUCTASE 1 | Q9LFA3 | AT3G52880 | 13 - 18 |
| FRUCTOSE-BISPHOSPHATE ALDOLASE 1 | Q9SJU4 | AT2G21330 | 8 - 13 |
| KETOL-ACID REDUCTOISOMERASE | Q05758 | AT3G58610 | 11 - 14 |
| ALANINE:GLYOXYLATE AMINOTRANSFERASE | Q56YA5 | AT2G13360 | 8 - 14 |
| CHLOROPLAST RNA BINDING | Q9SA52 | AT1G09340 | 13 - 14 |
| PLASTID ISOFORM TRIOSE PHOSPHATE ISOMERASE | Q9SKP6 | AT2G21170 | 11 - 14 |
| LOW EXPRESSION OF OSMOTICALLY RESPONSIVE GENES 2 | P25696 | AT2G36530 | 15 - 17 |
| CYSTEINE SYNTHASE 1 | P47999 | AT2G43750 | 13 - 15 |
| FRUCTOSE-BISPHOSPHATE ALDOLASE 2 | Q944G9 | AT4G38970 | 15 |
| GLYCINE DECARBOXYLASE P-PROTEIN 1 | Q94B78 | AT4G33010 | 8 - 15 |
| PHOSPHOGLYCERATE KINASE 1 | Q9LD57 | AT3G12780 | 13 - 15 |
| ALANINE-2-OXOGLUTARATE AMINOTRANSFERASE 1 | Q9LR30 | AT1G23310 | 15 - 20 |
| SEDOHEPTULOSE-BISPHOSPHATASE | P46283 | AT3G55800 | 16 - 17 |
| CATALASE 3 | Q42547 | AT1G20620 | 13 - 16 |
| HYDROXYPYRUVATE REDUCTASE | Q9C9W5 | AT1G68010 | 15 - 16 |
| GTP BINDING ELONGATION FACTOR TU FAMILY PROTEIN | Q8W4H7/Q8GTY0/ Q0WL56/P0DH99 | AT1G07930/AT5G60390/ AT1G07920/AT1G07940 | 15 - 17 |
| SERINE HYDROXYMETHYLTRANSFERASE 1 | Q9SZJ5 | AT4G37930 | 17 - 19 |
| ATP SYNTHASE C1 | Q01908 | AT4G04640 | 10 - 18 |
| COBALAMIN-INDEPENDENT METHIONINE SYNTHASE | O50008 | AT5G17920 | 19 - 20 |
| VARIEGATED 2/FTSH PROTEASE 8 | O80860/Q8W585 | AT2G30950/AT1G06430 | 10 - 19 |
| RAB GTPASE HOMOLOG E1B | P17745 | AT4G20360 | 17 - 19 |
| GLUTAMINE SYNTHETASE 2 | Q43127 | AT5G35630 | 14 - 19 |
| GLUCOSIDE GLUCOHYDROLASE 2 | Q9C5C2 | AT5G25980 | 12 - 19 |
| SNOWY COTYLEDON 1 | Q9SI75 | AT1G62750 | 5 - 19 |
| GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE B SUBUNIT | P25857 | AT1G42970 | 20 - 21 |
| ALLENE OXIDE SYNTHASE | Q96242 | AT5G42650 | 15 - 20 |
| CORONATINE INDUCED 1 | Q9SUR6 | AT4G23600 | 11 - 20 |
| MITOCHONDRIAL ATP SYNTHASE BETA-SUBUNIT | P83484/P83483/ Q9C5A9 | AT5G08690/AT5G08670/ AT5G08680 | 20 - 23 |
| TRANSKETOLASE 1 | Q8RWV0 | AT3G60750 | 26 - 30 |
| CHAPERONIN-60ALPHA | P21238 | AT2G28000 | 18 - 28 |
| ATP SYNTHASE SUBUNIT ALPHA | P56757 | ATCG00120 | 21 - 30 |
| FERREDOXIN-DEPENDENT GLUTAMATE SYNTHASE | Q9ZNZ7 | AT5G04140 | 29 - 31 |
| LIPOXYGENASE 2 | P38418 | AT3G45140 | 31 - 39 |
| ATP SYNTHASE SUBUNIT BETA | P19366 | ATCG00480 | 35 - 40 |
Table 2.
Significant protein glycation changes in the 112 common targets during high light, heat, diurnal time-series, and rosette ageing
| Light | |||
|---|---|---|---|
| Name | ID | TAIR ID | Control/high light |
| RPI3 | Q9S726 | AT3G04790 | –2.44 |
| TPI | Q9SKP6 | AT2G21170 | –∞ |
| Heat | |||
| Name | ID | TAIR ID | Control/heat |
| TPI | Q9SKP6 | AT2G21170 | –∞ |
| Time-series | |||
| Name | ID | TAIR ID | 08.00 h/24.00 h |
| AOC2 | Q9LS02 | AT3G25770 | –1.7 |
| ASP5 | P46248 | AT4G31990 | 2.9 |
| BAS1 | Q96291 | AT3G11630 | –1.62 |
| CORI3 | Q9SUR6 | AT4G23600 | –1.37 |
| FTSH2 | O80860 | AT2G30950 | 1.68 |
| OASB | P47999 | AT2G43750 | –1.59 |
| PRK | P25697 | AT1G32060 | –0.937 |
| PRXQ | Q9LU86 | AT3G26060 | –1.77 |
| PURA | Q96529 | AT3G57610 | –1.24 |
| RAN3 | Q8H156 | AT5G55190 | 2.27 |
| Rosette age | |||
| Name | ID | TAIR ID | 4 weeks/6 weeks |
| A2 | Q8W4H7 | AT1G07930 | 1.92 |
| CAT2 | P25819 | AT4G35090 | –2.47 |
| CICDH | Q9SRZ6 | AT1G65930 | –1.8 |
| CTIMC | P48491 | AT3G55440 | –2.36 |
| CYP18-4 | Q42406 | AT4G34870 | –2.78 |
| FBP | P25851 | AT3G54050 | –2.26 |
| GGAT1 | Q9LR30 | AT1G23310 | –2.99 |
| GLU1 | Q9ZNZ7 | AT5G04140 | –2.03 |
| GSA2 | Q42522 | AT3G48730 | 1.77 |
| LOX2 | P38418 | AT3G45140 | –1.44 |
| P25697 | P25697 | AT1G32060 | –1.46 |
| P83484 | P83484 | AT5G08690 | 1.2 |
| PER34 | Q9SMU8 | AT3G49120 | –1.93 |
| RBCS-1A | P10795 | AT1G67090 | –3.74 |
| RBCS-3B | P10798 | AT5G38410 | –2.38 |
| TGG2 | Q9C5C2 | AT5G25980 | –3.99 |
| TL29 | P82281 | AT4G09010 | –1.49 |
Values represent the label-free quantification log2 ratios for the different experiments at a significance of P<0.05. TPI was present in high light- and heat-stressed samples but absent in the controls
Gene Ontology (GO) analysis of the 112 target proteins revealed that the majority of these targets were classified as carrying out catalytic activity (82.5%) and thus may be considered as having an enzymatic function (Table 3). Furthermore, the majority of the proteins were located within the chloroplast (89.7%; Table 3). The few significant perturbations in the spectral counts and label-free quantification intensities in these 112 targets even under quite severe stress conditions (e.g. heat or light; Table 2) suggests that these proteins represent a base level of continually glycated proteins in plant cells and reveal that a high proportion of chloroplast enzymes are frequently glycated.
Table 3.
Gene Ontology analysis of the core 112 proteins targeted for glycation in Arabidopsis
| Term | Count | % | Fold enrichment | P-value |
|---|---|---|---|---|
| Bonferroni | ||||
| Molecular function | ||||
| Oxidoreductase activity | 35 | 36.1 | 4.1 | 1.41E-10 |
| Cofactor binding | 19 | 19.6 | 7.2 | 2.94E-08 |
| Catalytic activity | 80 | 82.5 | 1.6 | 1.73E-07 |
| Copper ion binding | 12 | 12.4 | 9.3 | 1.76E-05 |
| Peroxidase activity | 9 | 9.3 | 12.7 | 1.59E-04 |
| Ion binding | 44 | 45.4 | 2 | 1.81E-04 |
| Oxidoreductase activity | 35 | 36.1 | 4.1 | 1.41E-10 |
| Cofactor binding | 19 | 19.6 | 7.2 | 2.94E-08 |
| Cellular compartment | ||||
| Apoplast | 64 | 66 | 33.6 | 1.18E-82 |
| Chloroplast stroma | 59 | 60.8 | 23.4 | 1.94E-65 |
| Chloroplast | 87 | 89.7 | 5.9 | 6.22E-57 |
| Chloroplast envelope | 49 | 50.5 | 23.4 | 2.37E-52 |
| Thylakoid | 30 | 30.9 | 37.8 | 5.29E-36 |
| Chloroplast thylakoid | 22 | 22.7 | 27.7 | 1.67E-22 |
| Stromule | 14 | 14.4 | 100.8 | 2.35E-21 |
| Biological process | ||||
| Reductive pentose-phosphate cycle | 12 | 12.4 | 114.4 | 1.67E-18 |
| Oxidative photosynthetic carbon pathway | 4 | 4.1 | 84.8 | 0.002832 |
| Gluconeogenesis | 5 | 5.2 | 68.1 | 1.67E-04 |
| Photorespiration | 8 | 8.2 | 30.5 | 1.72E-06 |
| Glycolytic process | 11 | 11.3 | 29.5 | 7.95E-10 |
| Chlorophyll biosynthetic process | 5 | 5.2 | 22.7 | 0.016572 |
| Reductive pentose-phosphate cycle | 12 | 12.4 | 114.4 | 1.67E-18 |
The top eight terms (fold enrichment) for biological process, cellular compartment, and biological process are shown. Analysis was carried out using The Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.8 (Huang et al., 2009a, b).
Treatment-specific targets of glycation
We had initially hypothesized that stress treatments leading to oxidative stress and increased sugar levels would either significantly change specific protein targets or substantially increase levels of glycation within proteins. Yet, the 112 common target proteins remained stable during the stress treatments, with very few exceptions (see above; Table 3). However, comparisons also revealed unique glycation targets for all treatments (Fig. 4A), with few significant differences in the diurnal and drought experiments compared with their respective controls (Tables 4, 5). For the high light- and heat-specific glycation targets, however, no significant differences were observed.
Table 4.
Specific proteins targeted for glycation unique to the diurnal experiment in Arabidopsis
| Name | ID | TAIR ID | 16.00 h versus 24.00 h | 08.00 h versus 24.00 h | 08.00 h versus 16.00 h |
|---|---|---|---|---|---|
| ADK1 | Q9SF85-2 | AT3G09820 | 4.11 | 2.16 | –1.95 |
| RPL23AB | Q9M3C3 | AT3G55280 | 2.63 | 2.21 | –0.418 |
| ALDH5F1 | Q9SAK4 | AT1G79440 | –0.492 | –1.46 | –0.973 |
| MPPA2 | O04308 | AT3G16480 | –1.3 | 1.63 | 2.93 |
| Q0WW26 | Q0WW26 | AT4G34450 | –2.41 | –0.551 | 1.86 |
| GLN1-3 | Q9LVI8 | AT3G17820 | –3.03 | –0.948 | 2.08 |
| XYL1 | Q9S7Y7 | AT1G68560 | –1.98 | 1.18 | 3.16 |
| AGP31 | Q9FZA2-2 | AT1G28290 | –1.49 | 2.32 | 3.8 |
| P59259 | P59259 | AT5G59970 AT1G07660/ AT1G07820/ AT2G28740/ AT3G45930/ AT3G46320/ AT3G53730/ AT5G59690/ | –6.16 | –4.96 | 1.2 |
Label-free quantification log2 intensities, with those that are significant (P<0.05) shown in bold.
Table 5.
Specific proteins targeted for glycation unique to the drought experiments in Arabidopsis
| Name | ID | TAIR ID | Control (90% rSWC) versus drought (20% rSWC) |
|---|---|---|---|
| THM1 | O48737 | AT1G03680 | –2.31 |
| PDX11 | O80448 | AT2G38230 | 1.36 |
| PsbB | P56777 | ATCG00680 | 2.30 |
| AIR12 | Q94BT2 | AT3G07390 | 1.69 |
| CSY4 | P20115-2 | AT2G44350 | –1.13 |
| DHAR3 | Q8LE52 | AT5G16710 | –2.00 |
| EMB1144 | P57720 | AT1G48850 | 1.67 |
| FDH1 | Q9S7E4 | AT5G14780 | –2.18 |
| FQR1 | Q9LSQ5 | AT5G54500 | –1.78 |
| GRF10 | P48347 | AT1G22300 | –2.07 |
| At14a | P0DI79 | AT3G28300 | –1.54 |
| PetA | A4QKU4 | ATCG00540 | 1.77 |
| PPA6 | Q9LXC9 | AT5G09650 | –2.19 |
| Q9C7N5 | Q9C7N5 | AT1G29660 | –1.57 |
| TRXF1 | Q9XFH8 | AT3G02730 | –1.68 |
| RPL1 | Q9LY66 | AT3G63490 | 1.47 |
| TRX5 | Q39241 | AT1G45145 | –1.59 |
| VHA-B1 | P11574 | AT1G76030 | 2.06 |
Values represent the label-free quantification log2 ratios (control/drought) at a P<0.05.
Diurnal changes in glycation targets shows a spike in glycation levels at the end of the light period
Specific AGEs have previously been shown to change diurnally and in response to abiotic stress (Bechtold et al., 2009; Paudel et al., 2016), Overall, there were few significant changes in the112 common target proteins (see above; Table 2) or in time-specific target proteins (Table 4). Interestingly, while there were few significant differences in proteins targeted for glycation in a time-specific manner, the percentage of glycated peptides did increase, with an ~31% increase from 08.00 h to 16.00 h (Supplementary Fig. S3B). This suggests that the proteins targeted for glycation remain constant but the level of glycation within these proteins accumulates during the day, coinciding with an increase in sugar and starch levels (Supplementary Fig. S3A).
Drought stress increases glycation of thioredoxin proteins associated with the regulation of Calvin–Benson cycle enzymes
In contrast to heat and high light stresses, there were 18 unique protein targets that significantly differed in abundance between well-watered (90% rSWC) and severely drought-stressed plants (20% rSWC; Table 5; Supplementary Fig. S5B; Bechtold et al., 2018). Among these drought-specific targets were three thioredoxins, two present in the chloroplast (thioredoxin M1 and thioredoxin F1) and one present in the cytosol (thioredoxin H5; Table 5), and a glutathione S-transferase containing a thioredoxin-like fold (Table 5). In animals, thioredoxin activity is regulated by different post-translational modifications including glycation (Yuan et al., 2010). This suggests that glycation of plant thioredoxins may potentially impact key functions of thioredoxin and thioredoxin-like proteins in regulating the structure and function of diverse target proteins.
Glycation of recombinant GAPC1 and TPI enzymes leads to significant reduction in catalytic efficiency
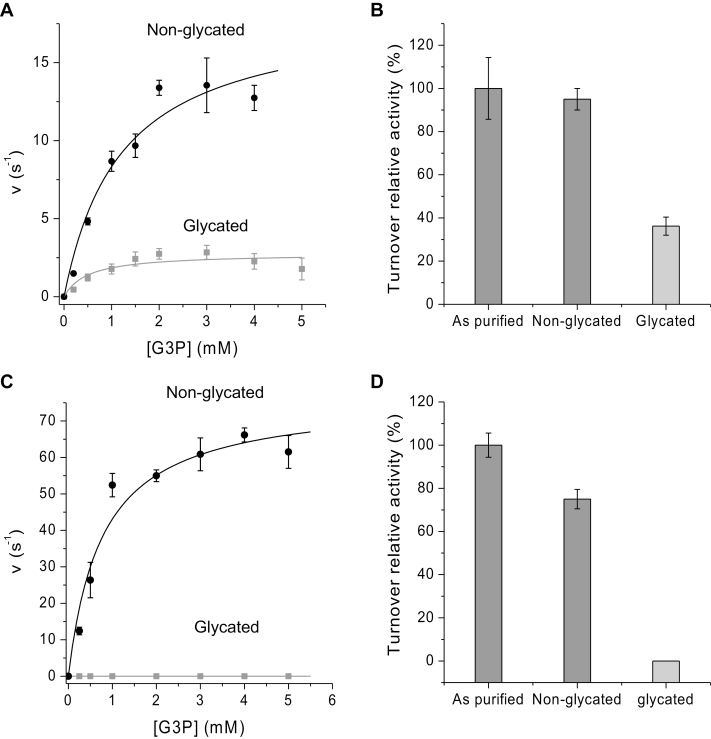
The above GO analysis suggested that ~82.5% of the 112 common target proteins were associated with catalytic function (Tables 1, 3). Among these, cytosolic and chloroplastic GAPC1 and GAPA2 (glyceraldehyde-3-phosphate dehydrogenase C1 and A2, respectively) and CTPI and TPI (cytosolic and chloroplastic triosephosphate isomerase, respectively) were identified. To investigate whether glycation affects enzyme activity and whether there is a difference between cellular compartments, cytosolic GAPC1 (accession no. P25858) and chloroplastic TPI (accession no. Q9SKP6) were chosen from the 112 protein targets for further testing in vitro. Purified recombinant enzymes migrated as single bands on a denaturing polyacrylamide gel consistent with their molecular mass (~39 kDa for GAPC1 and ~29 kDa for TPI) (Supplementary Fig. S6A). GAPC1 and TPI were glycated with glucose at 37 °C (see the Material and methods). Following a 3 week incubation with and without glucose, glycated proteins were enriched using BAC. Importantly, control samples incubated without glucose also displayed a small glycated peak, suggesting that a basal level glycation also occurred in E. coli, and that glycated residues were stable during a 3 week incubation period at 37 °C (Supplementary Fig. S6B). Incubation with glucose led to a substantial increase in the glycation peak (Supplementary Fig. S6B). Activity assays for GAPC1 and TPI (see the Materials and methods) were conducted under steady-state conditions using (i) purified enzyme; (ii) buffer only-treated enzyme; and (iii) glycated enzyme. From steady-state kinetics, TPI displayed Michaelis–Menten kinetics for G3P with kcat, Km, and kcat/Km (Table 6) in a similar range to previously observed Km values for TPI and other plant triosephosphate isomerases (Turner et al., 1965; Tang et al., 1999; Chen and Thelen, 2010; Enriquez-Flores et al., 2011; Zaffagnini et al., 2014; Dumont et al., 2016; Lopez-Castillo et al., 2016). Following incubation at 37 °C with buffer only, TPI displayed similar kinetics with 5% lower turnover than the relative turnover (kcat/Km value) of the ‘as-purified’ enzyme sample (Fig. 5A). However, once incubated with glucose at 37 °C for 3 weeks, the catalytic efficiency of TPI decreased by 63.1% (Fig. 5A, B; Table 6). The activity of GAPC1 displayed Michaelis–Menten kinetics (Table 6) but, unlike for TPI, kcat, Km, and kcat/Km have not been previously reported. When GAPC1 was incubated at 37 °C with buffer only, the activity decreased by 25%. However, incubation of GAPC1with glucose for 3 weeks resulted in a completely inactive enzyme (Fig. 5C, D). Therefore, the catalytic efficiencies of both GAPC1 and TPI were inhibited when non-enzymatically glycated, with GAPC1 showing a much more dramatic reduction.
Table 6.
Steady-state kinetic parameters for Arabidopsis enzymes glyceraldehyde-3-phosphate dehydrogenase (GAPC1 at pH 8.5) and triosephosphate isomerase (TPI at pH 7.0)
| Enzyme | pH | K m (mM) | k cat (s–1) | k cat/Km (M–1 s–1) |
|---|---|---|---|---|
| TPI non-glycated | 7.0 | 1.21 (±0.39) | 18.37 (±2.25) | 1.5 × 104 |
| TPI glycated | 7.0 | 0.5 (±0.30) | 2.77 (±0.39) | 5.54 × 103 |
| GAPC1 non-glycated | 8.5 | 0.77 (±0.21) | 76.4 (±5.91) | 9.9 × 104 |
| GAPC1 glycated | 8.5 | 0 | 0 | 0 |
K m, kcat, and kcat/Km were determined from Michaelis–Menten plots.
Fig. 5.
Steady-state enzyme kinetics of Arabidopsis glyceraldehyde-3-phosphate dehydrogenase (GAPC1) and triosephosphate isomerase (TPI). (A) Plots of turnover rate constants of TPI versus dl-glyceraldehyde-3-phosphate (G3P) substrate concentration. Non-glycated TPI shown in black and glycated TPI after 3 weeks in grey, with the data points fitted to the Michaelis–Menten equation to yield a Km value and turnover rate (kcat). (B) The percentage relative turnover activity of purified TPI, non-glycated and glycated after 3 weeks. (C) Plots of turnover rate constants of GAPC1 versus G3P substrate concentration in 10 mM NaH2PO4/Na2HPO4, pH 8.5, 100 mM NaCl. Non-glycated GAPC1 shown in black and glycated GAPC1 after 3 weeks in grey, with the data points fitted to the Michaelis–Menten equation to yield a Km value and turnover rate (kcat). (D) The percentage relative turnover activity of purified GAPC1, non-glycated and glycated after 3 weeks. Km, kcat, and kcat/Km values are given in Table 6. All experiments were performed at 25 °C using a Hewlett-Packard 8453 diode-array spectrophotometer.
MS reveals different glycation patterns of GAPC1 and TPI
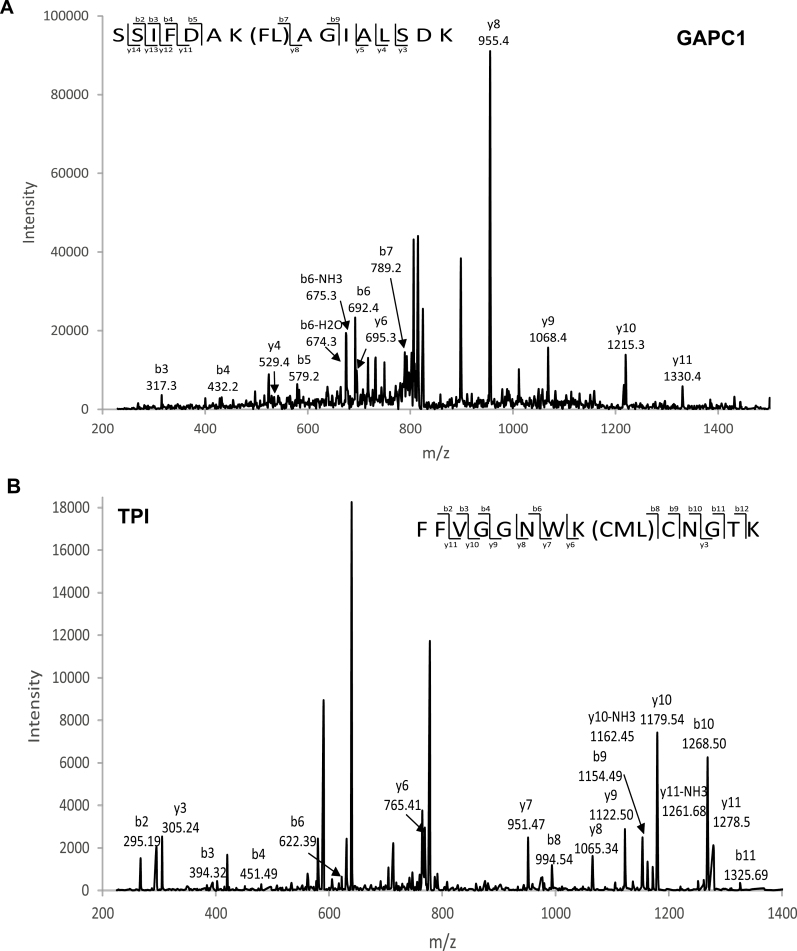
To assess the changes in glycation sites and to gain an insight into why the catalytic activity of GAPC1 was completely abolished while TPI was able to maintain nearly 40% of its activity, we analysed specific glycation sites using MS. The acquired MS/MS spectrum positively identified GAPC1 and TPI (GAPC1 measured mass of 39077 Da, and TPI measured mass of 29273 Da). The GAPC1 protein contains 338 amino acids with 3.8% Arg and 9.8% Lys residues. We identified 78 peptides corresponding to GAPC1 of which 40 contained at least one of the modifications we searched for (CEL, CML, FL, G-H1, and MG-H1). The number of glycation sites increased from 6 in the non-glycated protein sample to 20 in the glycated sample. Furthermore, the number of glycated peptides increased from 10 to 69 (3% to 19% of the total amino acid count) from the non-glycated to the glycated GAPC1 protein sample. The glycation sites were mapped onto a single monomer of GAPC1 from the biologically relevant tetramer in solution (Fig. 6A) [PDB 4Z0H (Zaffagnini et al., 2016)] by determining the probability with which the modification was present (Fig. 6). The modification patterns identified few glycation sites near the active site before incubation with glucose (Fig. 6B). In contrast, after glucose treatment, a significantly higher prevalence of seemingly random glycation sites was observed surrounding the active site of GAPC1 (Fig. 6C). This increase in glycation sites around the active site could explain the complete reduction in catalytic efficiency. An example of an FL-modified glycated peptide SSIFDAK(FL)AGIALSDK, from GAPC1 is shown in Fig. 7A. This peptide was not present in the non-glycated protein but increased to 11 peptides identified after glucose incubation.
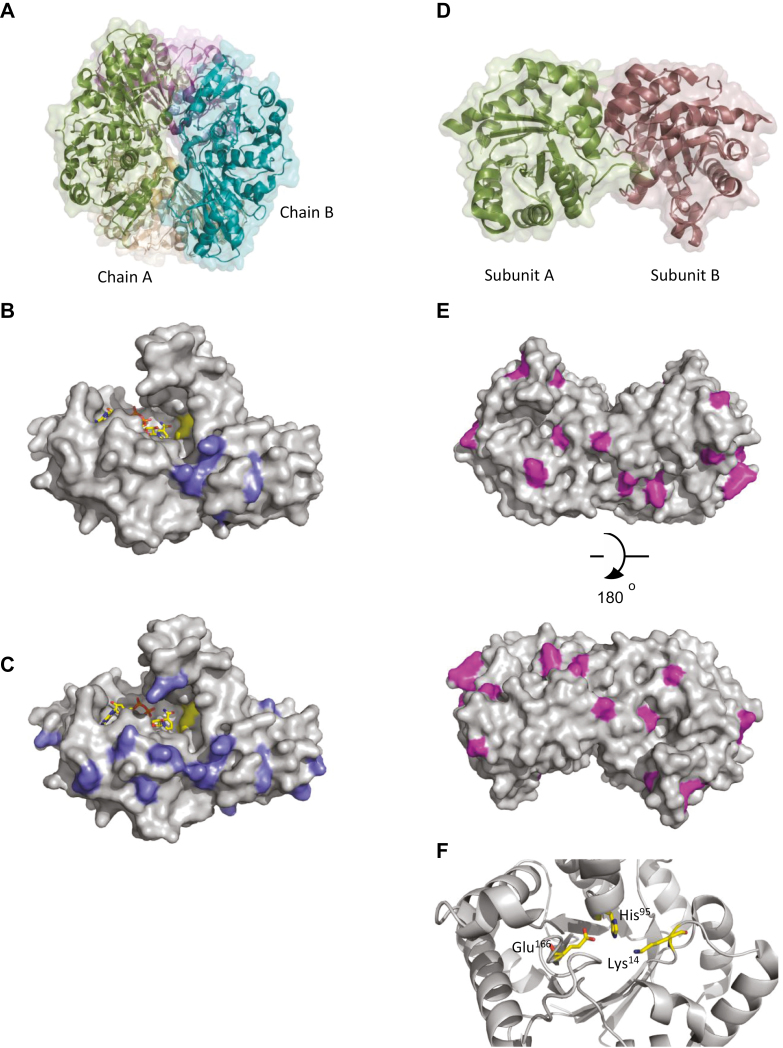
Fig. 6.
Structure of Arabidopsis enzymes glyceraldehyde-3-phosphate dehydrogenase C1 (GAPC1) and triosephosphate isomerase (TPI). (A) Biological tetramer assembly of GAPC1; each molecule is coloured differently (Chain A, white; Chain B, teal; symmetry-related molecules, green and purple). (B) Monomer of GAPC1 as purified from E. coli with the surface represented as grey, NAD+ shown in yellow sticks, and glycation sites illustrated on the surface in purple. (C) GAPC1 after 3 weeks glycation with 400 mM glucose; NAD+ is shown as yellow sticks and glycation sites as purple on a grey surface representation [PDB 4Z0H (Zaffagnini et al., 2016)]. (D) Biological homodimeric assembly of TPI: subunit A in green and subunit B in dark red. (E) Surface representation of TPI with glycation sites indicated in pink in both purified and glycated TPI. (F) Catalytic active site of TPI with the main chain shown in grey cartoon and the active site residues (Lys14, Glu166, and His96) shown as yellow sticks [PDB entry 4OHQ (Lopez-Castillo et al., 2016)].
Fig. 7.
MS/MS data confirming protein glycation sites. (A) Tandem mass spectra of m/z 699.57 corresponding to the y122+ ion of the tryptic AGE-modified peptide SSIFDAK(FL)AGIALSDK, representing glyceraldehyde-3-phosphate dehydrogenase C1 (GAPC1), and (B) tandem mass spectra of the tryptic AGE-modified peptide FFVGGNWK(CML)CNGTK, representing triosephosphate isomerase (TPI).
The chloroplast-localized TPI functions as a homodimer and displayed a different glycation pattern from GAPC1 (Fig. 6D) [PDB entry 4OHQ (Lopez-Castillo et al., 2016)]. TPI contains 254 amino acids with 3.8% Arg and 7.3% Lys. We identified 57 peptides of which 28 contained a glycation site. Thirteen glycation sites were identified for both non-glycated and glycated samples, with the non-glycated protein containing one extra site at residue Lys139 (Fig. 6E). Interestingly, TPI displayed a much higher proportion of glycation directly purified from E. coli (8% compared with 3% in GAPC1) and, once glycated, the percentage increased from 8% to 13% (Fig. 6E). In total, 12/13 Lys and 1 Arg glycation site were identified within the protein, with these sites remaining the same before and after glycation (with the exception of Lys139). Instead, the abundance of glycated peptides at these sites increased following glycation (Fig. 6E). This suggests that the chloroplast enzyme experiences a higher level of glycation under the same conditions as the cytosolic enzyme, when expressed in E. coli. Lys14 within the active site of TPI was glycated, containing a CML when directly purified from E. coli (Fig. 6F). An example MS/MS spectrum of the peptide containing the CML modification (FFVGGNWK(CML)CNGTK) on Lys14 of TPI can be seen in Fig. 7B. Importantly, the number of this glycated peptide remained constant after glycation, which suggests that the lowered activity for TPI was not due to increased glycation of the active site Lys residue.
Discussion
General glycation levels during abiotic stress and rosette ageing
Previous research revealed that up to 502 proteins are targets for glycation in Arabidopsis leaves under various conditions (Bilova et al., 2016). We reasoned that perturbations of primary metabolic pathways through stress treatments that lead to increases in sugar levels and ROS (i.e. H2O2) formation may also lead to altered glycation profiles in plants, as has been previously shown for specific AGE modifications (Bechtold et al., 2009). Furthermore, in yeast and mammalian cells, high levels of glucose are associated with toxicity and pathogenesis through increased ROS production via glucose auto-oxidation and metabolism (Bonnefont-Rousselot, 2002; Russell et al., 2002), and consequently experiments were chosen that would alter accumulation of sugar levels and oxidative stress production. We detected 460, 165, 181 and 527 glycated proteins for diurnal, heat, light, and drought responses, respectively, of which 112 proteins were common between the different treatments (Table 1). Interestingly, the short-term stress treatments, such as heat and high light, despite leading to significant oxidative stress (Fig. 2A–C) and increases in soluble sugars (Fig. 3A), did not result in a broad significant increase in the level of glycation within the 112 core targets, or in the emergence of many stress-specific glycation targets (Tables 1, 4). Most of the significant changes were observed during rosette ageing, drought, and the diurnal time-series, which were associated with less severe but probably more sustained changes in sugar, ROS, and glyoxalase I activity levels (Figs 2B, D–F; Supplementary Fig. S5A). This aligns with the role of protein glycation in the loss of seed viability during long-term seed storage (Wettlaufer and Leopold, 1991; Murthy and Sun, 2000; Murthy et al., 2003), which is associated with sugar hydrolysis, accumulation of glucose, and lipid peroxidation in both accelerated and naturally aged seeds (Murthy and Sun, 2000; Murthy et al., 2003). Seed ageing has also been closely connected to the formation of free radicals, oxidative stress, and a reduction in antioxidant capacities (Kranner et al., 2006, 2010; Roach et al., 2010), and suggests that the release of sugars and the accumulation of early and late stage glycation products may propagate free radical production. More recently, the age-related changes in the Arabidopsis glycation profile of proteins have been investigated, and only minimal changes in protein abundance were found. It was therefore suggested that glycation targets are conserved, with only a limited number of proteins being glycated during ageing (Bilova et al., 2017). Minimal changes in protein abundance corroborate the findings from our diverse experiments, including a range of abiotic stress treatments (Paudel et al., 2016; Bilova et al., 2017). Yet direct comparison of protein targets, where available, indicated that there was overall little overlap between osmotic stress- and age-specific targets (Paudel et al., 2016; Bilova et al., 2017) with our 112 common, drought- and development-specific targets (Supplementary Fig. S7).
The identification of a number of significant AGE sites during plant ageing led to the proposal of ‘glycation hotspotsֹ within the plant proteome (Bilova et al., 2017), which were in parts of the proteins essential for enzyme catalysis, activity, and interaction with other molecules (Bilova et al., 2017). Glycation hotspots have also been shown for some mammalian proteins, such as serum albumin (Barnaby et al., 2011) and recombinant antibodies (Zhang et al., 2008), but whether hotspots serve a specific function is unclear. Intriguingly, the number of glycated proteins identified under low stringency conditions was higher when compared with applying a more stringent filter (Supplementary Table S3). However, the number of identified peptides and modification sites was reduced significantly when applying a more stringent cut-off (see the Results). The dramatic decrease in protein numbers and modification sites with increased filter stringency highlights the somewhat random nature of glycation within plant cells. In addition, due to the great variety of glycation products (Baynes et al., 1989), it was not possible to search all of the glycation products present in our samples fully. However, we are confident that all identified proteins were specifically glycated due to their fractionation on the BAC and exclusive hybridization with glycation-specific antibodies (Fig. 1). More importantly, the 112 identified common targets were subject to more stringent filtering criteria, indicating a much greater consistency of glycation amongst this core set of proteins across the many different treatments.
Within the 112 common targets, we did observe increases in the number of glycated peptides during diurnal changes, with the highest number of glycated peptides occurring at the end of the light period (Supplementary Fig. S3), coinciding with the highest carbohydrate concentrations (Supplementary Fig. S3). To establish whether this could be related to particular glycation hotspots in proteins, we subsequently analysed two well-known recombinant plant enzymes for glycation sites. The results indicated that TPI may contain glycation hotspots, maintaining identical glycation between control and severely glycating conditions (Fig. 6D–F), while in GAPC1 the glycation pattern appeared to be much more random, with many more sites emerging under severely glycating conditions (Fig. 6A–C, discussed below).
Impact of glycation on enzyme activity in different subcellular compartments
In animals, glycation of GAPDH has been shown to reduce catalytic efficiency dramatically (Muronetz et al., 2016). We therefore selected chloroplastic TPI and cytosolic GAPC1 enzymes as part of the 112 core glycation targets (Table 1), and demonstrated that glycation affected enzyme activities, although with different sensitivities (Fig. 5). This could suggest that the subcellular localization of both enzymes impacts on the resilience to glycation, where the enzyme present in the cytosol is more susceptible to glycation than the chloroplast-located enzyme (Fig. 5). Alternatively, the existence of glycation hotspots versus random glycation may be due purely to the different three-dimensional protein structures and neighbouring amino acids (Sáenz-Suárez et al., 2016; Bilova et al., 2017) leading to site-specific increases of glycation levels, independent of subcellular localization and functionality. While it has previously been shown that glycation leads to a 70% decrease in Rubisco activity and increased protease susceptibility when incubated with the potent glycation agent ascorbate (Yamauchi et al., 2002), it is not clear whether specific glycation hotspots emerge under those conditions. Recently, reduced chloroplast TPI activity was shown to lead to stunted and chlorotic seedlings that accumulated dihydroxyacetone phosphate (DHAP) and MG (Chen and Thelen, 2010), a highly reactive glycating agent. An increased resilience of this enzyme towards glycation would therefore be paramount to maintain cellular function.
Intriguingly, the substrates responsible for glycation, notably MG, hexoses, ascorbate (Yamauchi et al., 2002), and sugar-phosphates (Fortpied et al., 2005), are all produced and/or present at high concentrations within the chloroplast. In addition, the only known protective enzyme, fructosamine 3-kinase (FN3K), capable of reversing early glycation products (Szwergold et al., 2001) is also located within the chloroplast exhibiting a 700 times higher activity than the equivalent enzyme in human erythrocytes (Fortpied et al., 2005). Together with the majority of targets being chloroplast located (89.7%; Table 3), this suggests that chloroplast enzymes may not only be extensive targets for glycation, but may also have evolved to withstand increased glycation pressures. From the limited data available on glycation of plant proteins in vivo, it appears that chloroplast enzymes are able to maintain some degree of activity (Fig. 5A, C; Yamauchi et al., 2002), while the cytosolic enzyme GAPC1 does not (Fig. 5B, D). Interestingly, TPI is absent in control samples and only emerges as a glycation target under high light and heat stress treatments, indicating minimal glycation under control conditions (Table 2). In comparison, GAPC1 is present both in the controls and in high light or heat treatments, and even though the degree of protein glycation cannot be assessed in vivo we have shown in vitro that glycation severely affects catalytic turnover. Further analysis of chloroplast and cytosolic enzymes is required to test this hypothesis.
Drought-specific targets are linked to redox regulation of the Calvin cycle and abiotic stress responses
Among the drought-specific glycation targets, few are directly linked to abiotic stress responses and/or signalling. However, we identified some interesting functions and regulatory capacities that may impact on wider signalling pathways. Among the proteins with increased glycation levels under drought conditions were several enzymes with known regulatory functions (Table 5) For example, the plastidial pyrophosphatase is a glycation target (AtPPa6, At5g09650, Table 5), and it has previously been shown that altering a PPa6 homologue affects primary metabolism in tobacco. Furthermore, PPa6 gene silencing resulted in drought sensitivity due to reduced abscisic acid content (George et al., 2010). Consequently, the removal of PPi has been suggested to play an important role in maintaining plastidial metabolism (George et al., 2010). Increased glycation of PPa6 under drought stress and the potential reduction of enzyme activity could therefore contribute to physiological drought responses. Similarly, At14a, a small transmembrane protein, showed increased glycation levels during drought stress (Table 5, At3g28300), and the close homologue At14a-Like1 protein was linked to plant growth and proline accumulation under drought conditions involving protein disulphide isomerase 5 and NAI2 (Kumar et al., 2015). The increase of glycation in three thioredoxins under drought conditions could indirectly have a much wider impact through their specific target proteins, especially since thioredoxin activity is regulated by different post-translational modifications and reduction of thioredoxin activity contributes to organ injury in many diseases (Sano et al., 2002; Tsutsui et al., 2003; Kondo et al., 2004). Recently, the reduction of thioredoxin 1 activity through glycation was demonstrated to play a critical role in lipo-polysaccharide-related cell death (Yuan et al., 2010), and thioredoxin-1 glycation was shown to be irreversible, resulting in permanent inactivation of this important signalling protein (Yuan et al., 2010).
In plants, thioredoxins participate in the regulation of protein targets involved in plant development and in response to (a)biotic stresses, carry out redox sensing and signal transduction functions (Rouhier and Jacquot, 2005), and participate in the repair of oxidized proteins during environmental stress (Vieira Dos Santos and Rey, 2006).
For example, thioredoxin f1 is essential for the light-dependent reduction of specific Calvin–Benson enzymes such as fructose bisphosphatase (FBPase), sedoheptulose-1,7-biphosphatase (SBPase), phosphoribulokinase (PRK), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and Rubisco activase (Michelet et al., 2013; Nikkanen et al., 2017). Importantly, many of the thioredoxin f downstream targets are among the 112 conserved continually glycated proteins, but did not significantly increase during drought stress (Table 1), suggesting that specific glycation of a small number of key regulatory enzymes under stress may affect a greater number of downstream proteins and physiological processes, such as the down-regulation of photosynthesis during drought stress (Bechtold et al., 2016).
Conclusions
In this study, we demonstrated that glycation proceeds spontaneously in plants already under relatively benign growth conditions, resulting in many common glycation targets under different growth conditions. Glycation targets are largely located within the chloroplast, the site of production of high levels of sugars during photosynthesis, daily sugar fluctuations, and increased sugar levels during stressful conditions, particularly drought. This is analogous to animal systems where glycation is also associated with energy-producing metabolic processes, and non-enzymatic glycation together with free radical oxidation are considered to be the cause of many diabetes-related complications (see the Introduction). Intriguingly, glycated peptides have been associated with signalling processes leading to transcriptional changes in animal systems (see the Introduction). Whether glycated proteins and/or their breakdown products play a role in signalling responses in plants remains to be investigated. However, with the knowledge of specific targets, we can begin to unravel physiological functions with regard to specific plant responses.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Enrichment for glycated peptides using boronate affinity chromatography before and after alkaline hydrolysis.
Fig. S2. The effect of high light and heat on the Chl a efficiency.
Fig. S3. Carbohydrate analysis of diurnal period experiments in Arabidopsis.
Fig. S4. Mass spectrum of a glycated peptide from PSBP1 oxygen-evolving enhancer protein 2-1.
Fig. S5. Volcano plots from DEP R analysis of glycated proteins from Arabidopsis.
Fig. S6. SDS–PAGE gel analysis of GAPC1 and TPI, and boronate affinity chromatography of TPI from Arabidopsis.
Fig. S7. Comparison of glycated protein targets with published data sets (Bilova et al., 2016, 2017; Paudel et al., 2016).
Table S1. The raw mass spectrum data for the protein groups identified from the time-series experiment in Arabidopsis.
Table S2. The raw mass spectrum data for the protein groups identified from the high light, heat, and drought experiments in Arabidopsis.
Table S3. Summary of glycated proteins, peptides, and modification sites for CML/CMA, M-GH1, GH1, and FL
Supplementary Material
Acknowledgements
AC was supported by The Leverhulme Trust project grant RPG-2015-176 awarded to UB. The authors would like to thank G. Metodieva and M. Metodiev for running the mass spectrometry equipment at Essex University.
References
- Ahmed N, Argirov OK, Minhas HS, Cordeiro CA, Thornalley PJ. 2002. Assay of advanced glycation endproducts (AGEs): surveying AGEs by chromatographic assay with derivatization by 6-aminoquinolyl-N-hydroxysuccinimidyl-carbamate and application to Nε-carboxymethyl-lysine- and Nε-(1-carboxyethyl)lysine-modified albumin. Biochemical Journal 364, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alikhani M, Alikhani Z, Boyd C, MacLellan CM, Raptis M, Liu R, Pischon N, Trackman PC, Gerstenfeld L, Graves DT. 2007a. Advanced glycation end products stimulate osteoblast apoptosis via the MAP kinase and cytosolic apoptotic pathways. Bone 40, 345–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alikhani M, Maclellan CM, Raptis M, Vora S, Trackman PC, Graves DT. 2007b. Advanced glycation end products induce apoptosis in fibroblasts through activation of ROS, MAP kinases, and the FOXO1 transcription factor. American Journal of Physiology. Cell Physiology 292, C850–C856. [DOI] [PubMed] [Google Scholar]
- Barnaby OS, Cerny RL, Clarke W, Hage DS. 2011. Comparison of modification sites formed on human serum albumin at various stages of glycation. Clinica Chimica Acta 412, 277–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baynes JW, Watkins NG, Fisher CI, Hull CJ, Patrick JS, Ahmed MU, Dunn JA, Thorpe SR. 1989. The Amadori product on protein: structure and reactions. Progress in Clinical and Biological Research 304, 43–67. [PubMed] [Google Scholar]
- Bechtold U, Albihlal WS, Lawson T, et al.. 2013. Arabidopsis HEAT SHOCK TRANSCRIPTION FACTORA1b overexpression enhances water productivity, resistance to drought, and infection. Journal of Experimental Botany 64, 3467–3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold U, Ferguson JN, Mullineaux PM. 2018. To defend or to grow: lessons from Arabidopsis C24. Journal of Experimental Botany 69: 2809–2821. [DOI] [PubMed] [Google Scholar]
- Bechtold U, Murphy DJ, Mullineaux PM. 2004. Arabidopsis peptide methionine sulfoxide reductase2 prevents cellular oxidative damage in long nights. The Plant Cell 16, 908–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold U, Penfold CA, Jenkins DJ, et al.. 2016. Time-series transcriptomics reveals that AGAMOUS-LIKE22 affects primary metabolism and developmental processes in drought-stressed Arabidopsis. The Plant Cell 28, 345–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold U, Rabbani N, Mullineaux PM, Thornalley PJ. 2009. Quantitative measurement of specific biomarkers for protein oxidation, nitration and glycation in Arabidopsis leaves. The Plant Journal 59, 661–671. [DOI] [PubMed] [Google Scholar]
- Bilova T, Lukasheva E, Brauch D, et al.. 2016. A snapshot of the plant glycated proteome: structural, functional, and mechanistic aspects. Journal of Biological Chemistry 291, 7621–7636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilova T, Paudel G, Shilyaev N, et al.. 2017. Global proteomic analysis of advanced glycation end products in the Arabidopsis proteome provides evidence for age-related glycation hot spots. Journal of Biological Chemistry 292, 15758–15776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnefont-Rousselot D. 2002. Glucose and reactive oxygen species. Current Opinion in Clinical Nutrition and Metabolic Care 5, 561–568. [DOI] [PubMed] [Google Scholar]
- Brownlee M, Cerami A, Vlassara H. 1988. Advanced glycosylation end products in tissue and the biochemical basis of diabetic complications. New England Journal of Medicine 318, 1315–1321. [DOI] [PubMed] [Google Scholar]
- Cerami A. 1985. Hypothesis. Glucose as a mediator of aging. Journal of the American Geriatrics Society 33, 626–634. [DOI] [PubMed] [Google Scholar]
- Chen M, Thelen JJ. 2010. The plastid isoform of triose phosphate isomerase is required for the postgerminative transition from heterotrophic to autotrophic growth in Arabidopsis. The Plant Cell 22, 77–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, Mann M. 2014. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Molecular & Cellular Proteomics 13, 2513–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croner RS, Stürzl M, Rau TT, Metodieva G, Geppert CI, Naschberger E, Lausen B, Metodiev MV. 2014. Quantitative proteome profiling of lymph node-positive vs. -negative colorectal carcinomas pinpoints MX1 as a marker for lymph node metastasis. International Journal of Cancer 135, 2878–2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deribe YL, Pawson T, Dikic I. 2010. Post-translational modifications in signal integration. Nature Structural & Molecular Biology 17, 666–672. [DOI] [PubMed] [Google Scholar]
- Dower SK, Qwarnstrom EE. 2003. Signalling networks, inflammation and innate immunity. Biochemical Society Transactions 31, 1462–1471. [DOI] [PubMed] [Google Scholar]
- Duan G, Walther D. 2015. The roles of post-translational modifications in the context of protein interaction networks. PLoS Computational Biology 11, e1004049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont S, Bykova NV, Pelletier G, Dorion S, Rivoal J. 2016. Cytosolic triosephosphate isomerase from Arabidopsis thaliana is reversibly modified by glutathione on cysteines 127 and 218. Frontiers in Plant Science 7, 1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellman GL. 1959. Tissue sulfhydryl groups. Archives of Biochemistry and Biophysics 82, 70–77. [DOI] [PubMed] [Google Scholar]
- Enríquez-Flores S, Rodríguez-Romero A, Hernández-Alcántara G, et al.. 2011. Determining the molecular mechanism of inactivation by chemical modification of triosephosphate isomerase from the human parasite Giardia lamblia: a study for antiparasitic drug design. Proteins 79, 2711–2724. [DOI] [PubMed] [Google Scholar]
- Ferguson JN, Humphry M, Lawson T, Brendel O, Bechtold U. 2018. Natural variation of life-history traits, water use, and drought responses in Arabidopsis. Plant Direct 2, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortpied J, Gemayel R, Stroobant V, van Schaftingen E. 2005. Plant ribulosamine/erythrulosamine 3-kinase, a putative protein-repair enzyme. Biochemical Journal 388, 795–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabius HJ, André S, Jiménez-Barbero J, Romero A, Solís D. 2011. From lectin structure to functional glycomics: principles of the sugar code. Trends in Biochemical Sciences 36, 298–313. [DOI] [PubMed] [Google Scholar]
- Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. 2003. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Research 31, 3784–3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George GM, van der Merwe MJ, Nunes-Nesi A, Bauer R, Fernie AR, Kossmann J, Lloyd JR. 2010. Virus-induced gene silencing of plastidial soluble inorganic pyrophosphatase impairs essential leaf anabolic pathways and reduces drought stress tolerance in Nicotiana benthamiana. Plant Physiology 154, 55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood C, Metodieva G, Al-Janabi K, Lausen B, Alldridge L, Leng L, Bucala R, Fernandez N, Metodiev MV. 2012. Stat1 and CD74 overexpression is co-dependent and linked to increased invasion and lymph node metastasis in triple-negative breast cancer. Journal of Proteomics 75, 3031–3040. [DOI] [PubMed] [Google Scholar]
- Griffiths G, Jones HE, Eaton CL, Stobart AK. 1997. Effect of n-6 polyunsaturated fatty acids on growth and lipid composition of neoplastic and non-neoplastic canine prostate epithelial cell cultures. The Prostate 31, 29–36. [DOI] [PubMed] [Google Scholar]
- Griffiths G, Leverentz M, Silkowski H, Gill N, Sánchez-Serrano JJ. 2000. Lipid hydroperoxide levels in plant tissues. Journal of Experimental Botany 51, 1363–1370. [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. 2009a. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Research 37, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. 2009b. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols 4, 44–57. [DOI] [PubMed] [Google Scholar]
- Hulbert AJ, Pamplona R, Buffenstein R, Buttemer WA. 2007. Life and death: metabolic rate, membrane composition, and life span of animals. Physiological Reviews 87, 1175–1213. [DOI] [PubMed] [Google Scholar]
- Hunter T. 2009. Tyrosine phosphorylation: thirty years and counting. Current Opinion in Cell Biology 21, 140–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen ON. 2004. Modification-specific proteomics: characterization of post-translational modifications by mass spectrometry. Current Opinion in Chemical Biology 8, 33–41. [DOI] [PubMed] [Google Scholar]
- Kondo N, Ishii Y, Kwon YW, Tanito M, Horita H, Nishinaka Y, Nakamura H, Yodoi J. 2004. Redox-sensing release of human thioredoxin from T lymphocytes with negative feedback loops. Journal of Immunology 172, 442–448. [DOI] [PubMed] [Google Scholar]
- Kranner I, Birtić S, Anderson KM, Pritchard HW. 2006. Glutathione half-cell reduction potential: a universal stress marker and modulator of programmed cell death?Free Radical Biology & Medicine 40, 2155–2165. [DOI] [PubMed] [Google Scholar]
- Kranner I, Roach T, Beckett RP, Whitaker C, Minibayeva FV. 2010. Extracellular production of reactive oxygen species during seed germination and early seedling growth in Pisum sativum. Journal of Plant Physiology 167, 805–811. [DOI] [PubMed] [Google Scholar]
- Kumar MN, Hsieh YF, Verslues PE. 2015. At14a-Like1 participates in membrane-associated mechanisms promoting growth during drought in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA 112, 10545–10550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander HM, Tauras JM, Ogiste JS, Hori O, Moss RA, Schmidt AM. 1997. Activation of the receptor for advanced glycation end products triggers a p21(ras)-dependent mitogen-activated protein kinase pathway regulated by oxidant stress. Journal of Biological Chemistry 272, 17810–17814. [DOI] [PubMed] [Google Scholar]
- Lopez-Castillo LM, Jiménez-Sandoval P, Baruch-Torres N, Trasviña-Arenas CH, Díaz-Quezada C, Lara-González S, Winkler R, Brieba LG. 2016. Structural basis for redox regulation of cytoplasmic and chloroplastic triosephosphate isomerases from Arabidopsis thaliana. Frontiers in Plant Science 7, 1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda A, Okano K, Park PS, Lem J, Crouch RK, Maeda T, Palczewski K. 2010. Palmitoylation stabilizes unliganded rod opsin. Proceedings of the National Academy of Sciences, USA 107, 8428–8433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiti MK, Krishnasamy S, Owen HA, Makaroff CA. 1997. Molecular characterization of glyoxalase II from Arabidopsis thaliana. Plant Molecular Biology 35, 471–481. [DOI] [PubMed] [Google Scholar]
- McCance DR, Dyer DG, Dunn JA, Bailie KE, Thorpe SR, Baynes JW, Lyons TJ. 1993. Maillard reaction products and their relation to complications in insulin-dependent diabetes mellitus. Journal of Clinical Investigation 91, 2470–2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metodieva G, Nogueira-de-Souza NC, Greenwood C, Al-Janabi K, Leng L, Bucala R, Metodiev MV. 2013. CD74-dependent deregulation of the tumor suppressor scribble in human epithelial and breast cancer cells. Neoplasia 15, 660–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelet L, Zaffagnini M, Morisse S, et al.. 2013. Redox regulation of the Calvin–Benson cycle: something old, something new. Frontiers in Plant Science 4, 470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muronetz V, Barinova K, Schmalhausen E. 2016. Glycation of glyceraldehyde-3-phosphate dehydrogenase in the presence of glucose and glyceraldehyde-3-phosphate. Journal of the International Society of Antioxidants 3, 1–4. [Google Scholar]
- Murthy UM, Kumar PP, Sun WQ. 2003. Mechanisms of seed ageing under different storage conditions for Vigna radiata (L.) Wilczek: lipid peroxidation, sugar hydrolysis, Maillard reactions and their relationship to glass state transition. Journal of Experimental Botany 54, 1057–1067. [DOI] [PubMed] [Google Scholar]
- Murthy UM, Sun WQ. 2000. Protein modification by Amadori and Maillard reactions during seed storage: roles of sugar hydrolysis and lipid peroxidation. Journal of Experimental Botany 51, 1221–1228. [DOI] [PubMed] [Google Scholar]
- Neglia CI, Cohen HJ, Garber AR, Ellis PD, Thorpe SR, Baynes JW. 1983. 13C NMR investigation of nonenzymatic glucosylation of protein. Model studies using RNase A. Journal of Biological Chemistry 258, 14279–14283. [PubMed] [Google Scholar]
- Nikkanen L, Toivola J, Diaz MG, Rintamaki E. 2017. Chloroplast thioredoxin systems: prospects for improving photosynthesis. Philosophical Transactions of the Royal Society B: Biological Science 372, pii: 20160474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton SJ, Talesa V, Yuan WJ, Principato GB. 1990. Glyoxalase I and glyoxalase II from Aloe vera: purification, characterization and comparison with animal glyoxalases. Biochemistry International 22, 411–418. [PubMed] [Google Scholar]
- Ott C, Jacobs K, Haucke E, Navarrete Santos A, Grune T, Simm A. 2014. Role of advanced glycation end products in cellular signaling. Redox Biology 2, 411–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H, Adsit FG, Boyington JC. 2010. The 1.5 Å crystal structure of human receptor for advanced glycation endproducts (RAGE) ectodomains reveals unique features determining ligand binding. Journal of Biological Chemistry 285, 40762–40770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parodi AJ. 2000. Protein glucosylation and its role in protein folding. Annual Review of Biochemistry 69, 69–93. [DOI] [PubMed] [Google Scholar]
- Paudel G, Bilova T, Schmidt R, et al.. 2016. Osmotic stress is accompanied by protein glycation in Arabidopsis thaliana. Journal of Experimental Botany 67, 6283–6295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeder BJ, Wilson MT. 1998. Mechanism of reaction of myoglobin with the lipid hydroperoxide hydroperoxyoctadecadienoic acid. Biochemical Journal 330, 1317–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roach T, Beckett RP, Minibayeva FV, Colville L, Whitaker C, Chen H, Bailly C, Kranner I. 2010. Extracellular superoxide production, viability and redox poise in response to desiccation in recalcitrant Castanea sativa seeds. Plant, Cell & Environment 33, 59–75. [DOI] [PubMed] [Google Scholar]
- Rocks O, Peyker A, Kahms M, Verveer PJ, Koerner C, Lumbierres M, Kuhlmann J, Waldmann H, Wittinghofer A, Bastiaens PI. 2005. An acylation cycle regulates localization and activity of palmitoylated Ras isoforms. Science 307, 1746–1752. [DOI] [PubMed] [Google Scholar]
- Rouhier N, Jacquot JP. 2005. The plant multigenic family of thiol peroxidases. Free Radical Biology & Medicine 38, 1413–1421. [DOI] [PubMed] [Google Scholar]
- Russell JW, Golovoy D, Vincent AM, Mahendru P, Olzmann JA, Mentzer A, Feldman EL. 2002. High glucose-induced oxidative stress and mitochondrial dysfunction in neurons. FASEB Journal 16, 1738–1748. [DOI] [PubMed] [Google Scholar]
- Sáenz-Suárez H, Poutou-Pinales RA, González-Santos J, Barreto GE, Rieto-Navarrera LP, Sáenz-Moreno JA, Landázuri P, Barrera-Avellaneda LA. 2016. Prediction of glycation sites: new insights from protein structural analysis. Turkish Journal of Biology 40, 12–25. [Google Scholar]
- Sano H, Sata T, Nanri H, Ikeda M, Shigematsu A. 2002. Thioredoxin is associated with endotoxin tolerance in mice. Critical Care Medicine 30, 190–194. [DOI] [PubMed] [Google Scholar]
- Schmidt AM, Vianna M, Gerlach M, Brett J, Ryan J, Kao J, Esposito C, Hegarty H, Hurley W, Clauss M. 1992. Isolation and characterization of two binding proteins for advanced glycosylation end products from bovine lung which are present on the endothelial cell surface. Journal of Biological Chemistry 267, 14987–14997. [PubMed] [Google Scholar]
- Slawson C, Housley MP, Hart GW. 2006. O-GlcNAc cycling: how a single sugar post-translational modification is changing the way we think about signaling networks. Journal of Cellular Biochemistry 97, 71–83. [DOI] [PubMed] [Google Scholar]
- Solís D, Bovin NV, Davis AP, Jiménez-Barbero J, Romero A, Roy R, Smetana K Jr, Gabius HJ. 2015. A guide into glycosciences: how chemistry, biochemistry and biology cooperate to crack the sugar code. Biochimica et Biophysica Acta 1850, 186–235. [DOI] [PubMed] [Google Scholar]
- Stitt M, Bulpin PV, ap Rees T. 1978. Pathway of starch breakdown in photosynthetic tissues of Pisum sativum. Biochimica et Biophysica Acta 544, 200–214. [DOI] [PubMed] [Google Scholar]
- Stitt M, Quick P. 1989. Photosynthetic carbon partitioning—its regulation and possibilities for manipulation. Physiologia Plantarum 77, 633–641. [Google Scholar]
- Stulemeijer IJ, Joosten MH. 2008. Post-translational modification of host proteins in pathogen-triggered defence signalling in plants. Molecular Plant Pathology 9, 545–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szwergold BS, Howell S, Beisswenger PJ. 2001. Human fructosamine-3-kinase: purification, sequencing, substrate specificity, and evidence of activity in vivo. Diabetes 50, 2139–2147. [DOI] [PubMed] [Google Scholar]
- Tang GL, Wang YF, Bao JS, Chen HB. 1999. Overexpression in Escherichia coli and characterization of the chloroplast triosephosphate isomerase from spinach. Protein Expression and Purification 16, 432–439. [DOI] [PubMed] [Google Scholar]
- Tessier FJ. 2010. The Maillard reaction in the human body. The main discoveries and factors that affect glycation. Pathologie-Biologie 58, 214–219. [DOI] [PubMed] [Google Scholar]
- Thornalley PJ. 2003. Glyoxalase I—structure, function and a critical role in the enzymatic defence against glycation. Biochemical Society Transactions 31, 1343–1348. [DOI] [PubMed] [Google Scholar]
- Thornalley PJ, Battah S, Ahmed N, Karachalias N, Agalou S, Babaei-Jadidi R, Dawnay A. 2003. Quantitative screening of advanced glycation endproducts in cellular and extracellular proteins by tandem mass spectrometry. iochemical Journal 375, 581–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornalley PJ, Langborg A, Minhas HS. 1999. Formation of glyoxal, methylglyoxal and 3-deoxyglucosone in the glycation of proteins by glucose. Biochemical Journal 344, 109–116. [PMC free article] [PubMed] [Google Scholar]
- Tsutsui T, Koide H, Fukahori H, et al.. 2003. Adenoviral transfection of hepatocytes with the thioredoxin gene confers protection against apoptosis and necrosis. Biochemical and Biophysical Research Communications 307, 765–770. [DOI] [PubMed] [Google Scholar]
- Turner DH, Blanch ES, Gibbs M, Turner JF. 1965. Triosephosphate isomerase of pea seeds. Plant Physiology 40, 1146–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A, Diaz S. 1984. The release and purification of sialic acids from glycoconjugates: methods to minimize the loss and migration of O-acetyl groups. Analytical Biochemistry 137, 236–247. [DOI] [PubMed] [Google Scholar]
- Vieira Dos Santos C, Rey P. 2006. Plant thioredoxins are key actors in the oxidative stress response. Trends in Plant Science 11, 329–334. [DOI] [PubMed] [Google Scholar]
- Wautier JL, Schmidt AM. 2004. Protein glycation: a firm link to endothelial cell dysfunction. Circulation Research 95, 233–238. [DOI] [PubMed] [Google Scholar]
- Wettlaufer SH, Leopold AC. 1991. Relevance of Amadori and Maillard products to seed deterioration. Plant Physiology 97, 165–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widjaja I, Naumann K, Roth U, Wolf N, Mackey D, Dangl JL, Scheel D, Lee J. 2009. Combining subproteome enrichment and Rubisco depletion enables identification of low abundance proteins differentially regulated during plant defense. Proteomics 9, 138–147. [DOI] [PubMed] [Google Scholar]
- Yamauchi Y, Ejiri Y, Tanaka K. 2002. Glycation by ascorbic acid causes loss of activity of ribulose-1,5-bisphosphate carboxylase/oxygenase and its increased susceptibility to proteases. Plant & Cell Physiology 43, 1334–1341. [DOI] [PubMed] [Google Scholar]
- Younus H, Anwar S. 2016. Prevention of non-enzymatic glycosylation (glycation): implication in the treatment of diabetic complication. International Journal of Health Sciences 10, 261–277. [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, Jiao X, Lau WB, Wang Y, Christopher TA, Lopez BL, Ramachandrarao SP, Tao L, Ma XL. 2010. Thioredoxin glycation: a novel posttranslational modification that inhibits its antioxidant and organ protective actions. Free Radical Biology & Medicine 49, 332–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaffagnini M, Fermani S, Calvaresi M, Orrù R, Iommarini L, Sparla F, Falini G, Bottoni A, Trost P. 2016. Tuning cysteine reactivity and sulfenic acid stability by protein microenvironment in glyceraldehyde-3-phosphate dehydrogenases of Arabidopsis thaliana. Antioxidants & Redox Signaling 24, 502–517. [DOI] [PubMed] [Google Scholar]
- Zaffagnini M, Michelet L, Sciabolini C, Di Giacinto N, Morisse S, Marchand CH, Trost P, Fermani S, Lemaire SD. 2014. High-resolution crystal structure and redox properties of chloroplastic triosephosphate isomerase from Chlamydomonas reinhardtii. Molecular Plant 7, 101–120. [DOI] [PubMed] [Google Scholar]
- Zhang B, Yang Y, Yuk I, Pai R, McKay P, Eigenbrot C, Dennis M, Katta V, Francissen KC. 2008. Unveiling a glycation hot spot in a recombinant humanized monoclonal antibody. Analytical Chemistry 80, 2379–2390. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Ames JM, Smith RD, Baynes JW, Metz TO. 2009. A perspective on the Maillard reaction and the analysis of protein glycation by mass spectrometry: probing the pathogenesis of chronic disease. Journal of Proteome Research 8, 754–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Smits AH, van Tilburg GB, Ovaa H, Huber W, Vermeulen M. 2018. Proteome-wide identification of ubiquitin interactions using UbIA-MS. Nature Protocols 13, 530–550. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.