Abstract
Making cancer treatment more effective is one of the grand challenges in our health care system. However, many drugs have entered clinical trials but so far showed limited efficacy or induced rapid development of resistance. We urgently need multi-targeted drug combinations, which shall selectively inhibit the cancer cells and block the emergence of drug resistance. The book chapter focuses on mathematical and computational tools to facilitate the discovery of the most promising drug combinations to improve efficacy and prevent resistance. Data integration approaches that leverage drug-target interactions, cancer molecular features, and signaling pathways for predicting, understanding, and testing drug combinations are critically reviewed.
Keywords: Drug combinations, Informatics approaches, Data integration, Mathematical modeling
1. Introduction
1.1. The State of the Art in Cancer Drug Discovery
Aberrant alteration of protein kinases plays fundamental signaling transduction roles in human cancer progression. Over the last decade, the overall efforts in cancer drug discovery have made a clear shift to focus on targeted drugs directed toward those deregulated kinases in cancers but not in normal tissue. Drugs that aim at selectively inhibiting deregulated kinases have shown unprecedented promise for effective cancer treatment with minimal toxicity. However, we are seeing that hundreds of such targeted drugs have entered clinical trials but have most often had disappointing efficacy due to varying treatment responses. This is most likely because we do not have sufficient understanding on which patient subpopulations are the expected responders and what the predictive biomarkers for treatment efficacy are.
To achieve the ultimate objective for precision medicine, large numbers of patient-derived samples with a wide range of molecular and clinical data are critical. The emergence of next-generation sequencing has enabled us to read the whole genome or the exome of cancer cells, sparking great expectation to identify novel protein targets for more effective and selective treatment opportunities. An example of such large-scale efforts is The Cancer Genome Atlas (TCGA), where genome sequencing data has been made available for more than 10,000 patient samples of 33 major types and subtypes of cancer [1]. These studies have revealed not only a remarkable degree of genetic heterogeneity between and within tumors but also enormous loads of passenger alterations confounded with often less obvious driver alterations [2]. To make matters worse, even when driver alterations can be found, they do not necessarily link to their expected treatment responses due to the rewiring of signaling pathways, such as the unresponsiveness to B-Raf proto-oncogene (BRAF) inhibition in many cancers including melanoma [3]. Furthermore, many known driver genes are not pharmacologically druggable as there is a lack of available drugs in the clinics [4]. How to best utilize the existing chemical probes to design a more potent drug has thus become a pressing task in medicinal chemistry. Taken together, it is clear that we still have a major gap between our understanding of the cancer genome and the in vivo behavior of cancers. As more and more cancer genome sequence data from tumors is generated, we will discover novel links, but to effectively make the links between cancer genomes, epigenomes, and cancer therapeutics, we will need powerful functional screening and profiling methodologies. The functional profiling techniques include high-throughput drug screening, RNA-based interference (RNAi), and more recently CRISPR-Cas9 genome-editing approaches. By the analysis of how tumor cells respond to the genetic or pharmacological perturbations, one may identify the therapeutic targets which are functionally related to the driver mutations while avoiding targets associated with unwanted side effects. The availability of functional screening and molecular profiling for the same patient samples, together with a comprehensive understanding of drug-target interactions and cancer signaling pathways, should dramatically improve our ability to develop data integration and modeling approaches to fill the knowledge gap between molecular biology and pharmacological response of a cancer, to achieve the ultimate goal being the rational design of targeted therapies given the disease profiles of individual patients [5, 6].
1.2. Need for Rational Designs of Drug Combinations
However, multiple clinical studies have shown that even when there is a dramatic initial treatment response, cancer cells with high mutational potential and functional redundancy can easily develop drug resistance by emerging activation of compensating or bypassing pathways [7, 8]. In fact, acquired resistance to therapy is not only common but also expected to be responsible for the poor prognosis of most cancers. Compared to standard chemotherapy, the progression-free survival rates remain poor for targeted therapies despite the improvements in the initial responses; for lung cancer patients receiving gefitinib, an epidermal growth factor receptor (EGFR) inhibitor; and for melanoma patients receiving vemurafenib, a BRAF (V600E) inhibitor [9, 10]. It is now widely acknowledged that effective cancer treatments need to go beyond the traditional “one disease, one drug, one target” paradigm, which is often too simplistic for the understanding and treatment of many complex diseases [11]. Polypharmacology, on the contrary, focuses on developing multi-targeted drugs or drug combinations, which has recently been introduced as an alternative paradigm to drug discovery showing great promises to reach effective and sustained clinical responses [12–14]. A combination of targeted drugs can potentially reduce the chances of resistance by inhibiting redundant pathways. Effective drug combinations allow for reduced dosages and therefore also minimize the toxicity and other side effects associated with high doses of single drugs. On the other hand, systematic exploration of targeted-drug combinations has also revealed functional links between drug efficacies and cancer genetic dependencies and thus supports the feasibility of using chemical probes to identify novel synthetic lethal and synergistic interactions as biomarkers for cancer diagnosis and treatment [15, 16]. However, despite the emerging possibilities for perturbing gene functions with a wide spectrum of RNAi/CRISPR libraries or using diverse compound collections, synergistic interactions between genes and/or drugs have remained extremely rare, posing challenges for developing more efficient search strategies for the discovery of drug combinations [17].
Similar to single drugs, the identification of drug combinations is largely driven by serendipity or a trial-and-error process using high-throughput screening [18]. However, such a brute-force search strategy becomes quickly infeasible, as the number of possible drug combinations may go up exponentially beyond what any automated experimentation can handle. Furthermore, the heterogeneity of cancer genomics makes drug combination discovery a rather daunting task as there are more than 200 subtypes, each of which is characterized by a unique profile of molecular alterations that may requires very specific treatment options to achieve sustainable efficacy [19]. Therefore, a rational design that could make the best use of the existing knowledge to prioritize the most promising drug combinations has the potential to greatly speed up the currently rather suboptimal drug screening efforts. Despite the pressing needs for such rational design strategies, there is a lack of systematic experimental-computational approaches that offer the possibility to predict and prioritize the most potential drug combinations warranting further experimental and clinical validations for specific cancer types. Missing such system-level drug combination design strategies has increasingly become the major bottleneck hindering the future development of cancer drug discovery.
Due to the complexity in cancer biology as well as in drug-target interactions, it remains a highly challenging endeavor for a drug combination to reach the clinic successfully. I would argue that a paradigm shift in both experimental and computational frameworks is needed to achieve such an ambitious goal. Novel experimental designs that enable the profiling of pharmacological and molecular biology features from the same cancer patients are essential for us to understand the mechanisms of drug combinations in a specific pathological context. Furthermore, there has always been a critical need for more efficient and robust informatics tools which can maximize the knowledge discovery from the ever-increasing massive medicinal data [20]. The data integration at the systems pharmacology level may eventually help us identify the drug response biomarkers with which we can predict the treatment outcomes for patient subpopulations or even individual patients [21].
The remaining sections of the chapter follow closely the recent development of experimental and computational methods for rational design of drug combinations. Informatics and modeling approaches that have shown potential to facilitate our understanding of drug combination effects are highlighted from the perspective of drug-target identification and modeling of cancer signaling pathways. To understand the mechanisms of action of compounds, identifying the most disease-relevant drug-target interactions is essential. There exist quite a few major databases that host the experimental bioactivity data. We provide an overview of the current database resources and also highlight the challenge of knowledge discovery from the heterogeneous public and literature content. To predict the combination of drugs, many mathematical and machine learning models have been proposed. Special focus will be given to the latest development of network pharmacology models to link the drug perturbations and molecular biology. Upon a valid informatics approach to predict the most potential drug combinations, experimental validation will be needed. Formal statistical testing methods to characterize the degree of interaction in the drug combination experimental data are briefly reviewed. Finally, personalize medicine strategies to apply these computational-experimental pipelines to integrate the drug screen data and molecular profiles for individual patient-derived cancer samples will be summarized.
2. Informatics Approaches to Make Sense of Drug-Target Interactions
Most drug molecules, despite initially designed to be very specific, often produce therapeutic and adverse effects by modulating multiple cellular targets [22]. Kinase inhibitors, in particular, are often competing with the high concentrations of adenosine triphosphate (ATP) for binding of the targets and therefore result in promiscuous interactions with many targets that share similar ATP-binding sites. The efficacy and toxicity of such a compound is usually arising from the interplay between the multiplexed drug-target interactions and the dynamic responses of the biological systems. Improved understanding of such polypharmacological effects is thus crucial for the development of more effective and safe drug treatments. To predict a drug or drug combination response, there is a clear need to obtain more comprehensive and context-dependent drug-target interaction data on a proteome-wide scale. Public drug-target interaction databases, such as ChEMBL [23] and PubChem [24], contain the vast majority of experimental bioactivity data curated from the literature. However, these deposited bioactivity values often lack sufficient annotations on the underlying experimental conditions, making it difficult to evaluate the reliability of the derived drug-target relationships when comparing multiple studies done usually in different assay formats [25]. On the other hand, computational methods have been utilized to infer novel drug-target interactions from existing experimental data. The predictive features that were identified from chemical or proteomic fingerprints have provided useful information to guide the design of more specific and potent compounds. However, how to leverage these predictive features derived from single drug-target interactions for the optimization of multi-targeted drugs remains largely unknown.
2.1. Information Retrieval from the Literature and Databases Resources
Targeted drugs such as kinase inhibitors are promiscuous, with multiple on-target and off-target binding contributing to both the drugs’ efficacy and side effects. Therefore, it is of ultimate importance to confirm the therapeutic significance of a drug-target interaction by measuring its binding affinity, the change of enzymatic activity of the target protein, and its downstream effectors in the signaling pathways. Recent technological advances have led to an explosion of rapid and cost-effective bioactivity assays to probe the drug-target interactions for kinase inhibitors. These bioassays can be generally classified into three major categories: binding assays, functional assays, and cellular assays [26]. Binding assays and functional assays often rely on recombinant or purified enzymes, which make it suitable to screen a large number of kinase inhibitors against a wide coverage of human kinome in vitro. For example, the KINOMEscan bioassay platform has been developed to generate binding affinity data for over 80 kinase inhibitors across more than 400 kinases including their disease-relevant mutants [27–30]. Metz and colleagues have applied functional assays to collect the enzymatic inhibition profiles across 172 kinases for 1493 publically available compounds [31]. In the same year, another major large-scale functional profiling was reported for 178 kinase inhibitors and 300 wild-type kinases, where the enzymatic activities were measured at 500 nM of the compounds in the presence of 10 μM ATP [32]. A more recent functional screen using the same assay format has been focused on 76 clinically important mutant kinases and 183 compounds [33]. Despite the differences in the binding and functional assays, the overall consistency of these studies seems reasonably good, indicating that the binding affinity of a drug-target interaction can predict its inhibition of catalytic activity [25, 34]. On the other hand, cellular assays measure the drug-target interactions in the living cells and thus allow for the confirmation of inhibition activity in the native biological systems. The trade-off, however, is the low-throughput compared to biochemistry-based binding and functional assays. One recent breakthrough of this kind is the use of heat shock protein 90 (HSP90) chaperone to effectively detect ligand binding to kinases in living cells, which led to the target profiling of 30 kinase inhibitors against more than 300 kinases [35]. Compared to binding and inhibition assays, cellular assays often utilize native kinases in a particular cellular context and thus are expected to reflect more the actual kinase function and regulation in the biological systems [36].
These kinase profiling data have revealed a much higher level of target promiscuity for many compounds previously thought to be very specific. The unexpected drug-target interactions may potentially lead to the discovery of new therapeutic indications for many existing compounds, especially if the drugs are Food and Drug Administration (FDA) approved for which the safety profiles have already been established. For example, a compound axitinib was originally approved as a specific vascular endothelial growth factor receptor (VEGFR) inhibitor for renal cancer but recently has been shown in the KINOMEscan platform to bind strongly to the T315I mutant of breakpoint cluster region (BCR)-Abelson murine leukemia viral oncogene homolog 1 (ABL1) fusion gene, which confers acquired imatinib resistance in leukemia. Treating of a chronic myeloid leukemia patient with axitinib resulted in a rapid clearance of BCR-ABL1(T315I)-positive cells from the bone marrow, providing further clinical evidence for axitinib’s potential to be “repositioned” as an effective drug for leukemia [37]. In the more recent screening of 183 kinase inhibitors against 76 mutant kinases, an FDA-approved EGFR inhibitor erlotinib has been shown to inhibit the T674I mutant of platelet-derived growth factor receptor alpha (PDGFRα) which also induces the imatinib resistance in many cancers [33].
With drug-target profiling at the kinome scale being increasingly reported, there have been extensive efforts to deposit those bioactivity data from the literature into public repositories to facilitate the knowledge sharing. PubChem Bioassay and ChEMBL are currently the two major databases, where a vast amounts of experimental drug-target interaction data are curated and updated regularly. In PubChem Bioassay, the majority of the data is uploaded by a list of data providers including various research organizations and other databases including ChEMBL. In contrast, ChEMBL is utilizing a top-down strategy by manually curating the published data from the main medicinal chemistry journals, many of which have been annotated with detailed experimental conditions and source information. However, caution should be taken when one needs to extract relevant data for drug-target interactions, as a target by the definition in ChEMBL could mean a single protein or protein complex, or in some cases, it may refer to a cell, a tissue, or a whole organism. Both PubChem Bioassay and ChEMBL provide a number of web tools to facilitate the queries. For example, PubChem Bioassay provides an identifier exchange program to mapping compound IDs between different systems. ChEMBL provides a similar web service called UniChem for cross-reference [38]. These ID mapping systems will greatly expand the search space of drug interactions into many related database resources to facilitate the integration of molecular biology and pharmacology data. Other major databases such as BindingDB [39] and GtoPdb [40] contain a less number of data points, but with a higher level of expert curation and annotation.
Utilizing the bioactivity data from these major databases, more focused data curation efforts for kinase drug-target interactions have also been made. For example, by applying a model-based method called KiBA, Tang and colleagues have compiled a drug-target interaction matrix of 52,598 compounds with 467 kinases by integration of multiple bioassay types extracted from ChEMBL [25]. The DrugKiNET database has manually curated the bioactivity data for over 800 compounds against 413 kinases from multiple sources, based on which it provides the in silico predictions of 200,000 drug-target interactions. Table 1 provides a brief summary of the aforementioned bioactivity databases and, as a comparison, also lists several less quantitative databases such as DrugBank, TTD, and MATADOR, where the names of commonly known primary targets are given but often missing their corresponding experimental bioactivity values.
Table 1. Major database resources for extracting drug-target interactions.
| Category | Database (URL) | Data statistics | Essential features |
|---|---|---|---|
| Quantitative | PubChem Bioassay (https://pubchem.ncbi.nlm.nih.gov/) | >2.8 M compounds >8 K protein targets |
Data are uploaded by >50 organizations worldwide |
| Versatile web services facilitate ID mapping, information retrieval, and structural-based data analysis | |||
| ChEMBL (https://www.ebi.ac.uk/chembl/) | >1.5 M compounds >110 K targets |
Data are extracted from major medicinal publications with a mix of automated and manual curation | |
| Web interfaces enable the queries of compounds, targets, and documents as well as an easier download of the result data | |||
| The UniChem ID mapping tool produces cross-references between >20 different databases | |||
| BindingDB (https://www.bindingdb.Org/) | >0.5 M compounds >6 K protein targets |
Recently a major update has been made for the data content and the web interface | |
| Journals that are not covered by ChEMBL are further curated | |||
| Quantitative bioactivity data are extracted from recent US patents | |||
| GtoPdb (http://www.guidetopharmacology.org) | >6 K compounds >1.3 K protein targets |
Data is produced by a group committed researchers with a deep level of expert curation and annotation | |
| Kinase targets might be underrepresented compared to the other target classes such as GPCRs and ion channels | |||
| DrugKiNET (http://www.drugkinet.ca/) | >800 compounds >400 kinase targets |
A company data curation effort focusing on kinase inhibitors and their targets | |
| Links to the publication sources are available | |||
| KiBA (http://pubs.acs.org/doi/suppl/10.1021/ci400709d) | >52 K compounds >400 kinase targets |
An informatics approach to integrate replicates from multiple assay formats as a summary bioactivity score | |
| Data is freely available as the supplementary material accompanying the methodology publication | |||
| K-Map (http://tanlab.ucdenver.edu/kMap/) | 250 kinase inhibitor >400 kinase targets |
A web-based visualization program for investigating kinase inhibitor target promiscuity | |
| Descriptive | DrugBank (http://www.drugbank.ca/) | >10 K compounds >4 K protein targets |
Focusing more on the existing knowledge on the pharmacology, ADMET, and primary targets |
| TTD (http://database.idrb.cqu.edu.cn/TTD/TTD.asp) | >30 K compounds >2 K protein targets |
Focusing on the disease pathways of the drug-target interactions; More than 100 drug combinations of known mechanisms of action are listed | |
| MATADOR (http://matador.embl.de/) | >800 compounds >2.9 K protein targets |
Direct and indirect drug-target interactions are included | |
2.2. Computational Methods for Predicting Drug-Target Interactions
Despite the increasing availability of high-throughput kinase profiling, the number of drug targets that are confirmed in such an experiment is still very limited compared to the vast majority of unknowns. In light of high-quality experimental data, in silico prediction methods aim to capture important chemical or molecular features that may predict the likelihood of an active compoundtarget interaction. These predictive features are particularly needed for our understanding of kinase inhibitors, as the target promiscuity has been observed for many seemingly unrelated kinases. Machine learning methods such as RandomForest, support vector machines (SVMs), and ElasticNet have been frequently applied in many large-scale predictions for various classes of drug targets including kinases [41], ion channels [42], G-protein-coupled receptors (GPCRs) [43], and nuclear receptors [44]. Table 2 lists a few popular machine learning methods, their implementation in R (https://cran.r-project.org/), and a few recent applications for drug-target predictions. The readers are referred to, e.g., Ding et al. for a more comprehensive review on this topic [45]. A common principle of these machine learning approaches is to predict novel drug-target interactions based on the features that are inferred from similar compounds and targets in a training data. Compounds are often represented by a list of chemical descriptors which can be either atom-type based or structure based. Utilizing a distance metric such as the Tanimoto coefficient, a similarity matrix for the compounds can be derived. On the other hand, target similarity is often determined by the amino acid sequences or the fingerprints derived from the 3D structures of the proteins. A machine learning model then utilizes the training data to determine the importance of these compound and target features. Depending on the areas of applications, a prediction can be made either as a classification of active versus inactive interactions or as a regression on the actual bioactivity values for novel compounds on untested targets.
Table 2. Representative examples of similarity-based machine learning methods for drug-target predictions.
| Type | Method | Description | Software packages in R | Applications |
|---|---|---|---|---|
| Classification | Naive Bayes | A Bayesian classification algorithm which learns model parameters for each feature independently | bnclassify klaR |
A multi-label Naive Bayes classifier trained on compound fingerprint data to predict primary targets for >156 K compounds [46] |
| Predicting potential compounds for 2507 protein targets utilizing the bioactivity data from ChEMBL. The web service is available [47] | ||||
| Regression | Partial least squares | A multivariate statistical algorithm identifies predictive features for a drug-target interaction | pls gpls |
A comprehensive review on the use of PLS for relating compound structure and bioactivity [48] |
| Dual | Random Forest | An ensemble method to make collective predictions by the majority voting from multiple random decision trees | RandomForest RRF |
5858 drugs and 14,490 drug-target interactions were retrieved from DrugBank and molecular descriptors for a drug pair was constructed. RandomForest was used to predict positive drug pairs that share the same targets [49] |
| SVM | A kernel-based supervised learning method to project the high-dimensional data into a lower-dimension hyperplane to separate the data into two classes | e1071 Kernlab |
Identification of potential binding pockets based on the three-dimensional protein structures, with which the potential drug targets can be predicted. A public web server called D3TPredictor is also available [50] | |
3. Mathematical Modeling for the Prediction of Drug Combinations
Since many cancers rely on the hyperactivation of specific signaling pathways, it is no surprise that protein kinases have been considered as one of the most druggable classes of anticancer targets. Despite their fundamental roles in cancer biology, only a few kinases are fully functionally annotated. With the improved understanding of the up/downstream effects of the kinase targets in the signaling pathways, the mechanisms of action of kinase inhibitors can be elucidated more systematically in the context of specific cellular environments. More importantly, the use of signaling pathways may help also link the drug responses with the genetic alterations and thus facilitate the translation of the vast genomic information into disease diagnosis and treatment strategies. The knowledge of cancer signaling pathways, when incorporated into the context-dependent drug-target interaction data and molecular feature data, should provide a rich set of information to construct a network pharmacology model. Such a network modeling should provide the functional links between disease biomarkers and drug targets, by which the cellular response of a multi-targeted drug or drug combination can be predicted and tested in follow-up experiments.
Given that the drug-target interaction data of high confidence can be obtained from both experimental and computational methods, the next important question is how to utilize specific chemistry and molecular biology information to explain the observed interactions between the compounds and furthermore predict the drug combination effects for a new cellular environment? The prediction of drug-drug interactions is largely relying on our understanding of drug targets as well as their mechanistic links in the context of molecular biology. There exist multiple methods where different modeling techniques adopted. Based on the input data that is needed, I make a rough classification of the existing drug combination prediction methods into gene expression based, signaling network based, and drug-target based.
3.1. Gene Expression-Based Methods
Gene expression-based methods infer the drug combination effects from the cellular responses of drug perturbations such as the transcriptomics changes before and after drug treatments. One popular source of such data has been provided by the Connectivity Map (CMap) study, where 1309 compounds have been screened of gene expression signatures at the genome scale against a panel of five cell lines using the microarray techniques [51]. The rational of relating drugs with transcriptomics is that the mechanisms of actions of drugs often result in the biological processes or pathways that may be enriched in the gene expression profiles. The gene expression-based methods have been shown great potential in the DREAM7 drug combination challenge, where the participants were asked to predict the degrees of synergy on 91 drug pairs on a B-cell lymphoma cancer cell line [52]. The winning method DIGRE utilized the gene expression signatures between paired drugs to derive a linear regression model where the residual effect can be attributed to a drug synergy score [53]. Using a similar concept, the CMap data has also been applied in a Combinatorial Drug Assembler model where the drug pairs with a higher overlap in their gene expression patterns are predicted to be more synergistic, with a certain level of experimental validations done for non-small cell lung cancer and triple-negative breast cancer [54].
3.2. Signaling Network-Based Methods
Signaling network-based methods annotate the cancer signaling pathways with a set of mass action and enzyme kinetics equations, by which one can derive the quantitative prediction of the dynamic changes of the cancer cells responding to drug perturbations. Therefore, accurate prior knowledge on the drug targets and their related proteins in the signaling pathways is needed for the prediction of drug combinations. Ideally, the topology of the signaling network and its associated kinetic parameters should be translated into a series of ordinary or partial differential equations (ODEs or PDEs) to capture the behaviors of the cellular system. The knockdown effects of a particular drug combination can be inferred by solving the differential equations, linking the target perturbation with the cell response phenotypes such as viability or toxicity. Compared to the gene expression-based methods, the differential equation-based methods link the drug-target interactions with the signaling pathways, which can provide mechanistic explanations on the observed drug interactions. For example, an ODE-based modeling has been applied to infer the effects of a combined inhibition of cyclin-dependent kinase 4 (CDK4) and insulin-like growth factor 1 receptor (IGF1R) on the AKT signaling pathway [55]. A limitation of such a method, however, is that the detailed kinetic parameters for a particular cellular context are often difficult to obtain due to experimental complexity. For cases where the quantitate prediction is impossible, an alternative approach has been proposed to utilize the logical rules that are derived from the literature to build a Boolean network [56]. Despite the apparent oversimplification of the cellular dynamic systems, such a logic-based method may still allow for a binary prediction on the drug perturbations. Furthermore, solving the differential equations often requires extensive computer simulations that practically hinder the applications for large signaling networks. A recent methodological improvement proposed by Molinelli et al. involved a probabilistic algorithm called belief propagation which can efficiently estimate the output of the ordinary equations, so that the effects of hundreds of proteins can be modeled simultaneously [15].
3.3. Drug-Target-Based Methods
Signaling network-based models rely on empirical cellular dynamic models, which may not be directly applicable for individualized drug combination prediction since accurate kinetic parameters under a particular cancer cellular environment are largely unknown. Gene expression profiles for drug responses, on the other hand, are not yet routinely profiled in a typical high-throughput drug screening setup and thus may provide limited translational potential in clinical settings. Further, many existing computational methods often rely on the primary targets (i.e., intended on-targets) of a cancer drug to infer the mechanism of action. However, it has been increasingly recognized that targeted drugs especially kinase inhibitors elicit their therapeutic efficacy through not only on-targets but also unintended off-targets. Application of these methods on kinase inhibitors without considering the full spectrum of drug-target interactions might lead to insufficient understanding of the drug combination effects.
Recently, there have been initial efforts to infer the drug combination effects by exploiting the similarity of drugs in terms of their proteome-level drug-target profiles [57, 58]. The main strategy is to consider a drug combination as a combination of targets, by which the sensitivity of a drug combination can be estimated by checking whether its targets are essential for cancer survival. Such a target-based approach requires two sources of input data, the drug-target interaction profiles, and the monotherapy drug sensitivities. Drug-target interaction data, as mentioned in the previous section, can be either obtained from public databases or from the prediction of computational methods. The monotherapy drug sensitivity data is then utilized to determine the essentiality of the targets. Due to the technical advances in high-throughput drug screening, such monotherapy drug sensitivity data has been increasingly available in public databases such as CTRP, CCLE, GDSC, and NCI-60, where a large number of compounds have been tested on a vast majority of cancer cell lines (Table 3). Given that the monotherapy drug sensitivities are often cell line specific, the target-based methods may accordingly capture the cell-specific essential targets with which the potential drug combinations can be predicted also at the individual cell level. This may potentially lead to a personalized medicine design that allows further experimental validation of the most promising drug combinations for a specific cancer type [59, 60].
Table 3. Monotherapy data resources for target-based drug combination predictions.
| Database | Description | URL |
|---|---|---|
| The Cancer Therapeutics Response Portal (CTRP) | 481 Compounds have been tested against 860 cancer cell lines for which the molecular profiles can be accessed from the CCLE database | http://www.broadinstitute.org/ctrp/ |
| The Cancer Cell Line Encyclopedia (CCLE) | http://www.broadinstitute.org/ccle | |
| The Genomics of Drug Sensitivity in Cancer (GDSC) | 140 Compounds have been tested against a maximal of 672 cancer cell lines for which the molecular profiles are available in the COSMIC database | http://www.cancerrxgene.org/ |
| http://cancer.sanger.ac.uk/cosmic | ||
| NCI-60 | 232 Compounds tested against 60 cancer cell lines | https://dtp.cancer.gov/ |
| http://discover.nci.nih.gov/cellminer/ | ||
4. Statistical Analyses for Assessing the Synergy in Experimental Drug Combination Data
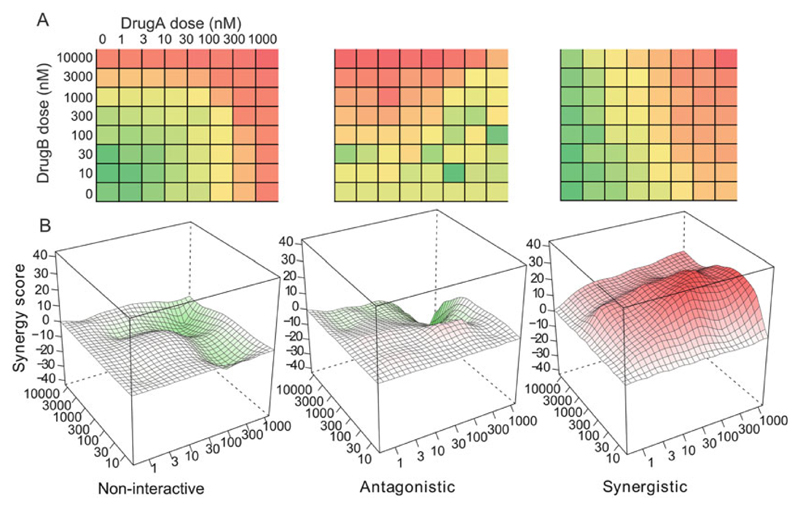
To be able to access the clinical significance of a drug combination, high-throughput screening that probes the cellular responses has become the standard approach. The current high-throughput drug combination screening typically enables a pair of drugs tested in a dose-response matrix. Based on the degree of interactions, a drug combination is commonly classified as synergistic, antagonistic, and noninteractive (Fig. 1). A synergistic drug combination is expected to boost up the effects more than what a single drug can achieve and thus has been extensively pursued in the clinics. However, despite the increasing popularity of this topic, there is currently a lack of consensus on the definitions of synergy, which often leads to significant confusion on the interpretation of experimental data [61]. Further, a generally accepted guideline for the choice of appropriate experimental designs and formal statistical testing are largely missing. With the rapid emergence of automated screening platforms, there is a critical need to develop standardized statistical methods for evaluating the significance of the most promising combination therapies.
Fig. 1. The quantitative scoring of drug combination screen data.
(a) A typical high-throughput drug combination screen utilizes a dose-response matrix design where all possible dose combinations for a drug pair can be tested. Colors in the dose-response matrices indicate different levels of phenotypic responses of the cancer cell. (b) Depending on the interaction patterns from the dose-response matrices, the drug combinations need to be quantified by a synergy score and depending on the distribution of synergy scores over the dose matrix as noninteractive, antagonistic, or synergistic
4.1. Reference Models for Noninteraction
To be able to assess the degree of synergy, the null hypothesis of no synergy needs to be clearly defined. There are three common reference models for the characterization of no synergy: highest single-agency model [62], Loewe additivity model [63], and Bliss independence model [64]. The highest single-agency (HSA) model assumes that the expected combination effect is the maximal effect that any individual drug can achieve, representing the common sense that a drug combination should produce additional efficacy compared to monotherapy. The synergy according to the HSA model, due to its simplicity in the calculation and easy interpretation as clear clinical benefits, has become the criteria of FDA for the approval of drug combinations. However, the use of HSA model may produce a high level of false discovery rate in preclinical drug screen studies as it does not correct for the dose-additive effect of a drug combined with itself [65]. To make the classification criteria for synergy more stringent, the Loewe additivity and Bliss independence models are preferred in many of the drug combination screen studies. The Loewe additivity model has been favored in studies where individual drug dose-response curves can be fitted using similar parameters [66–68]. The synergy over the Loewe model might reveal a drug combination with common mechanisms of action (e.g., acting on the same targets or different targets in the same signaling pathway), which produce similar dose-response patterns. The Bliss independence model, on the other hand, can be justified in cases where two drugs are targeting different pathways and thus elicit their effects independent of each other [69]. The Bliss independence model may identify synergy when the dose responses for the two drugs are expected to be different [70, 71]. However, these case studies also concluded that the Loewe and Bliss models seldom agreed completely with each other, and there are examples where a drug combination is synergistic according to one model but antagonistic according to the other [72]. Recently, there have been efforts to combine the advantages of Loewe and Bliss models into a synergy interaction landscape model, which showed promises to identify more informative interaction patterns over the tested dose-response matrices [73]. However, whether there exists a standardized guideline for choosing the optimal reference model is still a topics under considerable debate [61, 74–76]. As a summary, Table 4 listed the main assumptions of these commonly used models and the relevant software implementation tools.
Table 4. Comparing the different reference models for characterizing drug synergy.
| Model | Expected effect of no synergy | Related concepts and implementation tools |
|---|---|---|
| HSA | The highest single-agent effect | FDA approval guideline |
| Loewe | The effect as if a drug is combined with itself | Combination index [75] Isobologram [77] URSA [74] |
| Bliss | The effect as if two drugs are acting independently | Synergy landscape [78] |
| ZIP | The effect as if two drugs do not change the potency of each other | Synergy landscape [73] |
| SANE | The effect if two drugs are conditionally independent | Combenefit http://www.cruk.cam.ac.uk/research-groups/jodrell-group/combenefit |
4.2. Experimental Design and Statistical Testing
Despite the many ways of defining synergy, there is lack of common agreement on what the synergy is and how to statistically test it using what experiments. In line with previous synergy models, one may think of using the Bliss model due to its simplicity in the experimental design and interpretations. In a typical highthroughput drug screening setting, the drug combination is usually tested in a dose-response matrix format where multiple doses have been tested in combination, and the phenotypic readouts have been given for each cell in the matrix. The advantage of using doseresponse matrix is that higher resolution can be obtained about where and how much synergy effects can be detected. One would propose the use of dose-response matrix experimental data for better data analysis and more confident experimentation. For reliable estimation, replicates are always needed, and the clinical significance, for example, the dose ranges and the effect size, is always kept in mind for the decision-making [79].
5. Conclusions
A pressing challenge in the development of personalized cancer medicine is to understand how to make the most out of genomic information from a patient when evaluating treatment options. Compared to the current cytotoxic drugs, which typically affect both normal and cancerous cells, targeted-drug combinations address the fundamental challenges of drug resistance and clinical safety. This book chapter presents an overview on the innovative informatics strategies to suggest effective treatments that can lead to improved efficacy and reduce the number of expensive but inefficient treatments. Furthermore, the herein described approaches ranging from drug-target predictions to mathematical modeling of target-disease signaling networks pave the way to move beyond the current trial-and-error clinical assessment of drug combinations toward more systematic prediction and evaluation of the most effective drug-target combinations for each patient. The informatics approaches for analyzing drug combination data will also contribute significantly to the standardization of the experiment protocols and in the long run should facilitate the replicability and interpretation of experimental results.
As a critical component in rational design of drug combinations, computational methods will enable us to effectively reduce the search space for determining the most promising combinations and prioritizing their experimental evaluation. However, to achieve its eventual clinical translation, identifying cancer patients who will respond to the combination therapy is crucially important but to date remains an unresolved issue. The state-of-the-art patient stratification is often done via genomic characterization of the cancer samples, but the genomic similarity does not necessarily predict the drug response similarity. In contrast, the ex vivo drug screening using patient-derived samples has shown tremendous translation potential, as those drug sensitivity profiles often provide more clinically relevant information on the possible treatment options for individual patients. Recently there have been a few intensive drug screening and molecular profiling campaigns for patientderived samples (e.g., [80]). Data integration approaches to combine the molecular profiling of tumors with comprehensive testing of their drug sensitivity and resistance would make it possible to identify novel personalized combinatorial therapies in particular for chemotherapy-resistant patients. By efficiently integrating the informatics approaches with the patient-derived drug testing and molecular profiling platforms, the cost-effective and widely applicable computational modeling strategies have the potential to speed up the experimental work and promote translational breakthrough in personalized medicine.
Acknowledgments
This work was supported by the European Research Council Starting Grant project DrugComb (grant number: 716063 to J.T.).
References
- 1.Hoadley KA. Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell. 2014;158:929–944. doi: 10.1016/j.cell.2014.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vogelstein B. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prahallad A, Sun C, Huang S, Di Nicolantonio F, Salazar R, Zecchin D, Beijersbergen RL, Bardelli A, Bernards R. Unresponsiveness of colon cancer to BRAF (V600E) inhibition through feedback activation of EGFR. Nature. 2012;483:100–103. doi: 10.1038/nature10868. [DOI] [PubMed] [Google Scholar]
- 4.Cox AD, Fesik SW, Kimmelman AC, Luo J, Der CJ. Drugging the undruggable RAS: mission possible? Nat Rev Drug Discov. 2014;13:828–851. doi: 10.1038/nrd4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sanchez-Garcia F, Villagrasa P, Matsui J, Kotliar D, Castro V, Akavia UD, Chen BJ, Saucedo-Cuevas L, Rodriguez Barrueco R, Llobet-Navas D, Silva JM, et al. Integration of genomic data enables selective discovery of breast cancer drivers. Cell. 2014;159:1461–1475. doi: 10.1016/j.cell.2014.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pagliarini R, Shao W, Sellers WR. Oncogene addiction: pathways of therapeutic response, resistance, and road maps toward a cure. EMBO Rep. 2015;16:280–296. doi: 10.15252/embr.201439949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wagle N, Emery C, Berger MF, Davis MJ, Sawyer A, Pochanard P, Kehoe SM, Johannessen CM, Macconaill LE, Hahn WC, Meyerson M, et al. Dissecting therapeutic resistance to RAF inhibition in melanoma by tumor genomic profiling. J Clin Oncol. 2011;29:3085–3096. doi: 10.1200/JCO.2010.33.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Knight ZA, Lin H, Shokat KM. Targeting the cancer kinome through polypharmacology. Nat Rev Cancer. 2010;10:130–137. doi: 10.1038/nrc2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maemondo M, Inoue A, Kobayashi K, Sugawara S, Oizumi S, Isobe H, Gemma A, Harada M, Yoshizawa H, Kinoshita I, Fujita Y, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. 2010;362:2380–2388. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- 10.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, Dummer R, Garbe C, Testori A, Maio M, Hogg D, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Imming P, Sinning C, Meyer A. Drugs, their targets and the nature and number of drug targets. Nat Rev Drug Discov. 2006;5:821–834. doi: 10.1038/nrd2132. [DOI] [PubMed] [Google Scholar]
- 12.Hopkins AL. Network pharmacology: the next paradigm in drug discovery. Nat Chem Biol. 2008;4:682–690. doi: 10.1038/nchembio.118. [DOI] [PubMed] [Google Scholar]
- 13.Al-Lazikani B, Banerji U, Workman P. Combinatorial drug therapy for cancer in the post-genomic era. Nat Biotechnol. 2012;30:679–692. doi: 10.1038/nbt.2284. [DOI] [PubMed] [Google Scholar]
- 14.Tang J, Aittokallio T. Network pharmacology strategies toward multi-target anticancer therapies: from computational models to experimental design principles. Curr Pharm Des. 2014;20:20–36. doi: 10.2174/13816128113199990470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Molinelli EJ, Korkut A, Wang W, Miller ML, Gauthier NP, Jing X, Kaushik P, He Q, Mills G, Solit DB, Pratilas CA, et al. Perturbation biology: inferring signaling networks in cellular systems. PLoS Comput Biol. 2013;9 doi: 10.1371/journal.pcbi.1003290. e1003290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goltsov A, Langdon SP, Goltsov G, Harrison DJ, Bown J. Customizing the therapeutic response of signaling networks to promote antitumor responses by drug combinations. Front Oncol. 2014;4:13. doi: 10.3389/fonc.2014.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brough R, Frankum JR, Costa-Cabral S, Lord CJ, Ashworth A. Searching for synthetic lethality in cancer. Curr Opin Genet Dev. 2011;21:34–41. doi: 10.1016/j.gde.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 18.Latosińska JN, Latosińska M. Anticancer drug discovery—from serendipity to rational design. In: El-Shemy H, editor. Drug discovery. InTech; 2013. ISBN: 978-953-51-0906-8. [Google Scholar]
- 19.Forbes SA, Beare D, Gunasekaran P, Leung K, Bindal N, Boutselakis H, Ding M, Bamford S, Cole C, Ward S, Kok CY, et al. COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015;43:D805–D811. doi: 10.1093/nar/gku1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tan AC, Ryall KA, Huang PH. Expanding the computational toolbox for interrogating cancer kinomes. Pharmacogenomics. 2016;17:95–97. doi: 10.2217/pgs.15.154. [DOI] [PubMed] [Google Scholar]
- 21.Ryall KA, Tan AC. Systems biology approaches for advancing the discovery of effective drug combinations. J Cheminform. 2015;7:7. doi: 10.1186/s13321-015-0055-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xie L, Xie L, Kinnings SL, Bourne PE. Novel computational approaches to polypharmacology as a means to define responses to individual drugs. Annu Rev Pharmacol Toxicol. 2012;52:361–379. doi: 10.1146/annurev-pharmtox-010611-134630. [DOI] [PubMed] [Google Scholar]
- 23.Bento AP, Gaulton A, Hersey A, Bellis LJ, Chambers J, Davies M, Krüger FA, Light Y, Mak L, McGlinchey S, Nowotka M, et al. The ChEMBL bioactivity database: an update. Nucleic Acids Res. 2014;42:D1083–D1090. doi: 10.1093/nar/gkt1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y, Xiao J, Suzek TO, Zhang J, Wang J, Zhou Z, Han L, Karapetyan K, Dracheva S, Shoemaker BA, Bolton E, et al. PubChem’s bioassay database. Nucleic Acids Res. 2012;40:D400–D412. doi: 10.1093/nar/gkr1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang J, Szwajda A, Shakyawar S, Xu T, Hintsanen P, Wennerberg K, Aittokallio T. Making sense of large-scale kinase inhibitor bioactivity data sets: a comparative and integrative analysis. J Chem Inf Model. 2014;54:735–743. doi: 10.1021/ci400709d. [DOI] [PubMed] [Google Scholar]
- 26.Wang Y, Ma H. Protein kinase profiling assays: a technology review. Drug Discov Today Technol. 2015;18:1–8. doi: 10.1016/j.ddtec.2015.10.007. [DOI] [PubMed] [Google Scholar]
- 27.Fabian MA, Biggs WH, 3rd, Treiber DK, Atteridge CE, Azimioara MD, Benedetti MG, Carter TA, Ciceri P, Edeen PT, Floyd M, Ford JM, et al. A small molecule-kinase interaction map for clinical kinase inhibitors. Nat Biotechnol. 2005;23:329–336. doi: 10.1038/nbt1068. [DOI] [PubMed] [Google Scholar]
- 28.Karaman MW, Herrgard S, Treiber DK, Gallant P, Atteridge CE, Campbell BT, Chan KW, Ciceri P, Davis MI, Edeen PT, Faraoni R, et al. A quantitative analysis of kinase inhibitor selectivity. Nat Biotechnol. 2008;26:127–132. doi: 10.1038/nbt1358. [DOI] [PubMed] [Google Scholar]
- 29.Zarrinkar PP, Gunawardane RN, Cramer MD, Gardner MF, Brigham D, Belli B, Karaman MW, Pratz KW, Pallares G, Chao Q, Sprankle KG, et al. AC220 is a uniquely potent and selective inhibitor of FLT3 for the treatment of acute myeloid leukemia (AML) Blood. 2009;114:2984–2992. doi: 10.1182/blood-2009-05-222034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davis MI, Hunt JP, Herrgard S, Ciceri P, Wodicka LM, Pallares G, Hocker M, Treiber DK, Zarrinkar PP. Comprehensive analysis of kinase inhibitor selectivity. Nat Biotechnol. 2011;29:1046–1051. doi: 10.1038/nbt.1990. [DOI] [PubMed] [Google Scholar]
- 31.Metz JT, Johnson EF, Soni NB, Merta PJ, Kifle L, Hajduk PJ. Navigating the kinome. Nat Chem Biol. 2011;7:200–202. doi: 10.1038/nchembio.530. [DOI] [PubMed] [Google Scholar]
- 32.Anastassiadis T, Deacon SW, Devarajan K, Ma H, Peterson JR. Comprehensive assay of kinase catalytic activity reveals features of kinase inhibitor selectivity. Nat Biotechnol. 2011;29:1039–1045. doi: 10.1038/nbt.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Duong-Ly KC, Devarajan K, Liang S, Horiuchi KY, Wang Y, Ma H, Peterson JR. Kinase inhibitor profiling reveals unexpected opportunities to inhibit disease-associated mutant kinases. Cell Rep. 2016;14:772–781. doi: 10.1016/j.celrep.2015.12.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rudolf AF, Skovgaard T, Knapp S, Jensen LJ, Berthelsen J. A comparison of protein kinases inhibitor screening methods using both enzymatic activity and binding affinity determination. PLoS One. 2014;9 doi: 10.1371/journal.pone.0098800. e98800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taipale M, Krykbaeva I, Whitesell L, Santagata S, Zhang J, Liu Q, Gray NS, Lindquist S. Chaperones as thermodynamic sensors of drug-target interactions reveal kinase inhibitor specificities in living cells. Nat Biotechnol. 2013;31:630–637. doi: 10.1038/nbt.2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patricelli MP, Nomanbhoy TK, Wu J, Brown H, Zhou D, Zhang J, Jagannathan S, Aban A, Okerberg E, Herring C, Nordin B, et al. In situ kinase profiling reveals functionally relevant properties of native kinases. Chem Biol. 2011;18:699–710. doi: 10.1016/j.chembiol.2011.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pemovska T, Johnson E, Kontro M, Repasky GA, Chen J, Wells P, Cronin CN, McTigue M, Kallioniemi O, Porkka K, Murray BW, et al. Axitinib effectively inhibits BCR-ABL1 (T315I) with a distinct binding conformation. Nature. 2015;519:102–105. doi: 10.1038/nature14119. [DOI] [PubMed] [Google Scholar]
- 38.Chambers J, Davies M, Gaulton A, Hersey A, Velankar S, Petryszak R, Hastings J, Bellis L, McGlinchey S, Overington JP. UniChem: a unified chemical structure cross-referencing and identifier tracking system. J Cheminform. 2013;5:3. doi: 10.1186/1758-2946-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gilson MK, Liu T, Baitaluk M, Nicola G, Hwang L, Chong J. BindingDB in 2015: a public database for medicinal chemistry, computational chemistry and systems pharmacology. Nucleic Acids Res. 2016;44:D1045–D1053. doi: 10.1093/nar/gkv1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Southan C, Sharman JL, Benson HE, Faccenda E, Pawson AJ, Alexander SP, Buneman OP, Davenport AP, McGrath JC, Peters JA, Spedding M, et al. The IUPHAR/BPS guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. Nucleic Acids Res. 2016;44:D1054–D1068. doi: 10.1093/nar/gkv1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cao DS, Zhou GH, Liu S, Zhang LX, Xu QS, He M, Liang YZ. Large-scale prediction of human kinase-inhibitor interactions using protein sequences and molecular topological structures. Anal Chim Acta. 2013;792:10–18. doi: 10.1016/j.aca.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 42.Huang C, Zhang R, Chen Z, Jiang Y, Shang Z, Sun P, Zhang X, Li X. Predict potential drug targets from the ion channel proteins based on SVM. J Theor Biol. 2010;262:750–756. doi: 10.1016/j.jtbi.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 43.Hu J, Li Y, Yang JY, Shen HB, Yu DJ. GPCR-drug interactions prediction using random forest with drug-association-matrix-based post-processing procedure. Comput Biol Chem. 2016;60:59–71. doi: 10.1016/j.compbiolchem.2015.11.007. [DOI] [PubMed] [Google Scholar]
- 44.Oak N, Jayaraman VK. Identification of ligand binding pockets on nuclear receptors by machine learning methods. Protein Pept Lett. 2014;21:808–814. doi: 10.2174/09298665113209990061. [DOI] [PubMed] [Google Scholar]
- 45.Ding H, Takigawa I, Mamitsuka H, Zhu S. Similarity-based machine learning methods for predicting drug-target interactions: a brief review. Brief Bioinform. 2014;15:734–747. doi: 10.1093/bib/bbt056. [DOI] [PubMed] [Google Scholar]
- 46.Nidhi GM, Davies JW, Jenkins JL. Prediction of biological targets for compounds using multiple-category Bayesian models trained on chemogenomics databases. J Chem Inf Model. 2006;46:1124–1133. doi: 10.1021/ci060003g. [DOI] [PubMed] [Google Scholar]
- 47.Pogodin PV, Lagunin AA, Filimonov DA, Poroikov VV. PASS targets: ligand-based multi-target computational system based on a public data and naive Bayes approach. SAR QSAR Environ Res. 2015;26:783–793. doi: 10.1080/1062936X.2015.1078407. [DOI] [PubMed] [Google Scholar]
- 48.Stanton DT. QSAR and QSPR model interpretation using partial least squares (PLS) analysis. Curr Comput Aided Drug Des. 2012;8:107–127. doi: 10.2174/157340912800492357. [DOI] [PubMed] [Google Scholar]
- 49.Park K, Kim D. Drug-drug relationship based on target information: application to drug target identification. BMC Syst Biol. 2011;5:S12. doi: 10.1186/1752-0509-5-S2-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu YT, Li Y, Huang ZF, Xu ZJ, Yang Z, Chen ZX, Chen KX, Shi JY, Zhu WL. Multi-algorithm and multi-model based drug target prediction and web server. Acta Pharmacol Sin. 2014;35:419–431. doi: 10.1038/aps.2013.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qu XA, Rajpal DK. Applications of connectivity map in drug discovery and development. Drug Discov Today. 2014;17:23–24. doi: 10.1016/j.drudis.2012.07.017. [DOI] [PubMed] [Google Scholar]
- 52.Bansal M, Yang J, Karan C, Menden MP, Costello JC, Tang H, Xiao G, Li Y, Allen J, Zhong R, Chen B, et al. A community computational challenge to predict the activity of pairs of compounds. Nat Biotechnol. 2014;32:1213–1222. doi: 10.1038/nbt.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang J, Tang H, Li Y, Zhong R, Wang T, Wong S, Xiao G, Xie Y. DIGRE: drug-induced genomic residual effect model successful prediction of multidrug effects. CPT Pharmacometrics Syst Pharmacol. 2015;4:e1. doi: 10.1002/psp4.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee JH, Kim DG, Bae TJ, Rho K, Kim JT, Lee JJ, Jang Y, Kim BC, Park KM, Kim S. CDA: combinatorial drug discovery using transcriptional response modules. PLoS One. 2012;7(8) doi: 10.1371/journal.pone.0042573. e42573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miller ML, Molinelli EJ, Nair JS, Sheikh T, Samy R, Jing X, He Q, Korkut A, Crago AM, Singer S, Schwartz GK, et al. Drug synergy screen and network modeling in dedifferentiated liposarcoma identifies CDK4 and IGF1R as synergistic drug targets. Sci Signal. 2013;6:ra85. doi: 10.1126/scisignal.2004014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Morris MK, Saez-Rodriguez J, Sorger PK, Lauffenburger DA. Logic-based models for the analysis of cell signaling networks. Biochemistry. 2010;49:3216–3224. doi: 10.1021/bi902202q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pal R, Berlow N, et al. A kinase inhibition map approach for tumor sensitivity prediction and combination therapy design for targeted drugs. Pac Symp Biocomput. 2012:351–362. [PubMed] [Google Scholar]
- 58.Tang J, Karhinen L, Xu T, Szwajda A, Yadav B, Wennerberg K, Aittokallio T. Target inhibition networks: predicting selective combinations of druggable targets to block cancer survival pathways. PLoS Comput Biol. 2013;9 doi: 10.1371/journal.pcbi.1003226. e1003226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pemovska T, Kontro M, Yadav B, Edgren H, Eldfors S, Szwajda A, Almusa H, Bespalov MM, Ellonen P, Elonen E, Gjertsen BT, et al. Individualized systems medicine strategy to tailor treatments for patients with chemorefractory acute myeloid leukemia. Cancer Discov. 2013;3:1416–1429. doi: 10.1158/2159-8290.CD-13-0350. [DOI] [PubMed] [Google Scholar]
- 60.He L, Wennerberg K, Aittokallio T, Tang J. TIMMA-R: an R package for predicting synergistic multi-targeted drug combinations in cancer cell lines or patient-derived sample. Bioinformatics. 2015;31:1866–1868. doi: 10.1093/bioinformatics/btv067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tang J, Wennerberg K, Aittokallio T. What is synergy? The Saariselkä agreement revisited. Front Pharmacol. 2015;6:181. doi: 10.3389/fphar.2015.00181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Berenbaum MC. What is synergy? Pharmacol Rev. 1989;41:93–141. [PubMed] [Google Scholar]
- 63.Loewe S. The problem of synergism and antagonism of combined drugs. Arzneimittelforschung. 1953;3:285–290. [PubMed] [Google Scholar]
- 64.Bliss CI. The toxicity of poisons applied jointly. Ann Appl Biol. 1939;26:585–615. [Google Scholar]
- 65.Borisy AA, Elliott PJ, Hurst NW, Lee MS, Lehar J, Price ER, Serbedzija G, Zimmermann GR, Foley MA, Stockwell BR, Keith CT. Systematic discovery of multicomponent therapeutics. Proc Natl Acad Sci U S A. 2003;100:7977–7982. doi: 10.1073/pnas.1337088100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lehár J, Zimmermann GR, Krueger AS, Molnar RA, Ledell JT, Heilbut AM, Short GF, 3rd, Giusti LC, Nolan GP, Magid OA, Lee MS, et al. Chemical combination effects predict connectivity in biological systems. Mol Syst Biol. 2007;3:80. doi: 10.1038/msb4100116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lehár J, Krueger AS, Avery W, Heilbut AM, Johansen LM, Price ER, Rickles RJ, Short GF, 3rd, Staunton JE, Jin X, Lee MS, et al. Synergistic drug combinations tend to improve therapeutically relevant selectivity. Nat Biotechnol. 2009;27:659–666. doi: 10.1038/nbt.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cokol M, Chua HN, Tasan M, Mutlu B, Weinstein ZB, Suzuki Y, Nergiz ME, Costanzo M, Baryshnikova A, Giaever G, Nislow C, et al. Systematic exploration of synergistic drug pairs. Mol Syst Biol. 2011;7:544. doi: 10.1038/msb.2011.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fitzgerald JB, Schoeberl B, Nielsen UB, Sorger PK. Systems biology and combination therapy in the quest for clinical efficacy. Nat Chem Biol. 2006;2:458–466. doi: 10.1038/nchembio817. [DOI] [PubMed] [Google Scholar]
- 70.Tan X, Hu L, Luquette LJ, 3rd, Gao G, Liu Y, Qu H, Xi R, Lu ZJ, Park PJ, Elledge SJ. Systematic identification of synergistic drug pairs targeting HIV. Nat Biotechnol. 2012;30:1125–1130. doi: 10.1038/nbt.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mathews Griner LA, Guha R, Shinn P, Young RM, Keller JM, Liu D, Goldlust IS, Yasgar A, McKnight C, Boxer MB, Duveau DY, et al. High-throughput combinatorial screening identifies drugs that cooperate with ibrutinib to kill activated B-cell–like diffuse large B-cell lymphoma cells. Proc Natl Acad Sci U S A. 2014;111:2349–2354. doi: 10.1073/pnas.1311846111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jansen G, Lee AY, Epp E, Fredette A, Surprenant J, Harcus D, Scott M, Tan E, Nishimura T, Whiteway M, Hallett M, et al. Chemogenomic profiling predicts antifungal synergies. Mol Syst Biol. 2009;5:338. doi: 10.1038/msb.2009.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yadav B, Wennerberg K, Aittokallio T, Tang J. Searching for drug synergy in complex dose-response landscapes using an interaction potency model. Comput Struct Biotechnol J. 2015;13:504–513. doi: 10.1016/j.csbj.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Greco WR, Bravo G, Parsons JC. The search for synergy: a critical review from a response surface perspective. Pharmacol Rev. 1995;47:331–385. [PubMed] [Google Scholar]
- 75.Chou TC. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 2010;70:440–446. doi: 10.1158/0008-5472.CAN-09-1947. [DOI] [PubMed] [Google Scholar]
- 76.Lee JJ, Kong M, Ayers GD, Lotan R. Interaction index and different methods for determining drug interaction in combination therapy. J Biopharm Stat. 2007;17:461–480. doi: 10.1080/10543400701199593. [DOI] [PubMed] [Google Scholar]
- 77.Tallarida RJ. An overview of drug combination analysis with isobolograms. J Pharmacol Exp Ther. 2006;319:1–7. doi: 10.1124/jpet.106.104117. [DOI] [PubMed] [Google Scholar]
- 78.Zhao W, Sachsenmeier K, Zhang L, Sult E, Hollingsworth RE, Yang H. A new bliss independence model to analyze drug dombination data. J Biomol Screen. 2014;19:817–821. doi: 10.1177/1087057114521867. [DOI] [PubMed] [Google Scholar]
- 79.Almohaimeed B, Donev AN. Experimental designs for drug combination studies. Comput Stat Data Anal. 2014;71:1077–1087. [Google Scholar]
- 80.Crystal AS, Shaw AT, Sequist LV, Friboulet L, Niederst MJ, Lockerman EL, Frias RL, Gainor JF, Amzallag A, Greninger P, Lee D, et al. Patient-derived models of acquired resistance can identify effective drug combinations for cancer. Science. 2014;19:1480–1486. doi: 10.1126/science.1254721. [DOI] [PMC free article] [PubMed] [Google Scholar]