Abstract
Background:
Semaphorin (Sema)/Plexin (Plxn) signaling is important for many aspects of neuronal development, however, the transcriptional regulation imposed by this signaling pathway is unknown. Previously, we identified an essential role for Sema6A/PlxnA2 signaling in regulating proliferation and cohesion of retinal precursor cells (RPCs) during early eye development. This study used RNA isolated from control, Sema6A-deficient and PlxnA2-deficient zebrafish embryos in a microarray analysis to identify genes that were differentially expressed when this signaling pathway was disrupted.
Results:
We uncovered a set of 58 transcripts, and all but 1 were up-regulated in both sema6A and plxnA2 morphants. We validated gene expression changes in subset of candidates that are suggested to be involved in proliferation, migration or neuronal positioning. We further functionally evaluated one gene, rasl11b, as contributing to disrupted proliferation in sema6A and plxna2 morphants. Our results suggest rasl11b negatively regulates proliferation of RPCs in the developing zebrafish eye.
Conclusions:
Microarray analysis has generated a resource of target genes downstream of Sema6A/PlxnA2 signaling, which can be further investigated to elucidate the downstream effects of this well-studied neuronal and vascular guidance signaling pathway.
Keywords: retina, eye development, rasl11b, microarray, proliferation, rx3:GFP
Introduction
Neuronal migration and positioning are important aspects of nervous system development. During the early stages of neuronal development, guidance cues are essential to lead neurons and their processes to correct target tissues. One well-studied group of guidance cues are the Semaphorins (Semas). Semas are a family of transmembrane and secreted proteins that typically signal through Plexin (Plxn) receptors. There are eight subclasses of Semas. Semas 1 and 2 are found in invertebrates, 3–7 are found in vertebrates, and Sema V is found in the genome of nonneuro-tropic DNA viruses (Neufeld and Kessler, 2008). The Plexin family is composed of two invertebrate Plxns, A and B, and four classes of vertebrate Plxns, A1–4, B1–3, C1, and D1 (Tamagnone et al., 1999). Semas and Plxns have tissue-specific expression patterns, and many Semas can signal through multiple Plxn family members. Semas were initially discovered with respect to their role as repulsive guidance cues for migrating axons (Luo et al., 1993), although it is now appreciated that they have much broader roles in development. They have been implicated in vasculogenesis (Serini et al., 2003), tumorigenesis (Neufeld and Kessler, 2008), immunity (Shi et al., 2000), and bone development (Behar et al., 1996). Here, we focus specifically on the interaction and downstream effects of Sema6A and PlxnA2 in early eye development.
During eye development, retinal precursor cells (RPCs) are directed from the optic stalk into the correct domain of the developing retina (Rembold et al., 2006). We previously demonstrated that Sema6A and its receptor, PlxnA2, are necessary for the proliferation and cohesion of RPCs within the early embryonic eye vesicles (Ebert et al., 2014). More recently, Sema6A and PlxnA2 were shown to be required for interkinetic nuclear migration of RPCs within optic vesicles (Belle et al., 2016). In addition, Sema6/PlxnA signaling is required for neuronal positioning in the cerebellum (Renaud et al., 2008; Renaud and Chedotal, 2014), lami-nation of the hippocampus (Tawarayama et al., 2010), and guidance of the corticospinal tract (Rünker et al., 2008), among other roles. Several studies have begun to uncover the intracellular signaling mechanisms downstream of activated PlxnA receptors (Oinuma et al., 2004; Toyofuku et al., 2005; He et al., 2009); however, before this study, it was unknown what transcriptional regulation occurred downstream of Sema6A/PlxnA2 signaling.
Microarray analysis using RNA extracted from 18 somite zebrafish embryos deficient in either Sema6A or PlxnA2 has enabled the identification of several downstream transcriptional targets of Sema6A/PlxnA2 signaling during early stages of neuronal development. Further characterization of one of these genes, RAS-like, family 11, member B (rasl11b), revealed its role in regulating RPC proliferation.
Results and Discussion
Microarray Analysis Identifies Transcriptional Changes Downstream of Sema6A/PlxnA2 Signaling
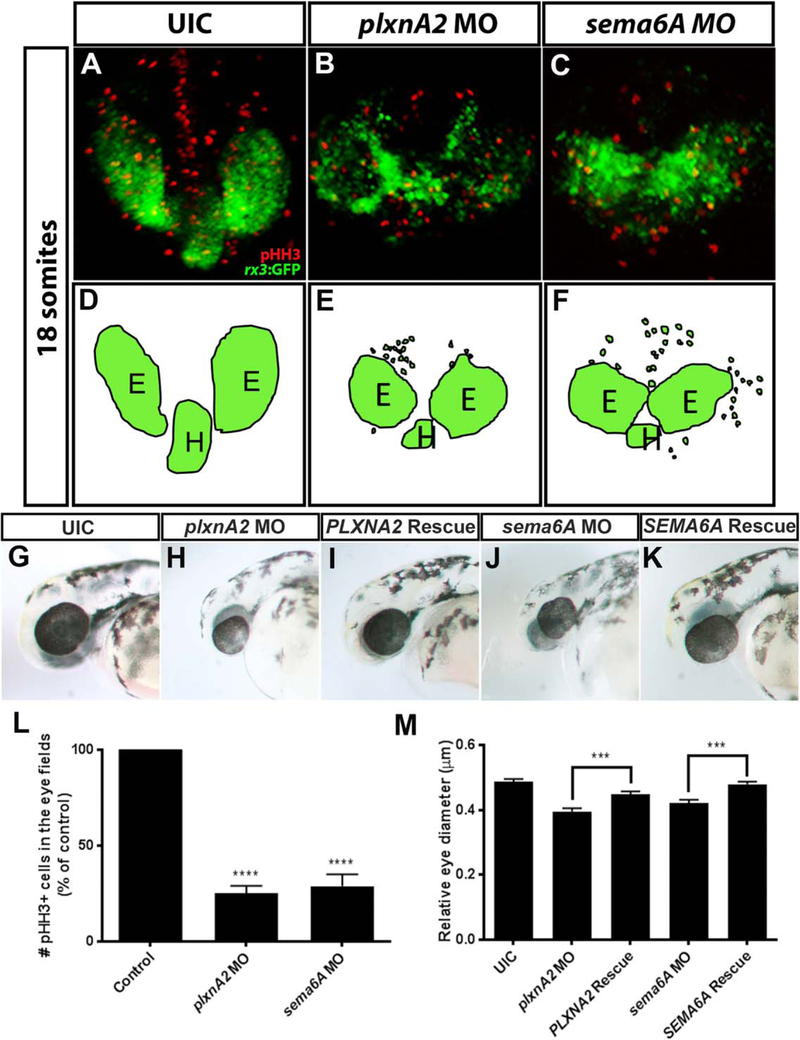
Confirming our previous results (Ebert et al., 2014), morpholino antisense oligonucleotides (MOs) targeted to either sema6A or plxnA2 resulted in decreased proliferation and cohesion of RPCs within migrating optic vesicles compared with control embryos. To observe these phenotypes, we used the rx3:GFP zebrafish transgenic line that expresses green fluorescent protein (GFP) in RPCs and the hypothalamus (Rembold et al., 2006). Using confocal microscopy, we identified GFP-positive RPCs outside of the eye field and optic vesicles that were closer together in morphant embryos (Fig. 1A–F). Using a mitotic marker, phospho-Histone H3 (pHH3), we observed less proliferation within the GFP-positive optic vesicles (Fig. 1A–C,L; control 100%, N = 3, n = 30; plxnA2 MO 25.11% ± 0.72 N = 3, n = 30, P < 0.0001; sema6A MO 28.86% ± 1.12, N = 3, n = 30; P < 0.0001, one-way analysis of variance [ANOVA], multiple comparisons test).
Fig. 1.
Sema6A and plxnA2 knockdown leads to loss of cohesion and decreased proliferation in eye vesicles. A–C: Confocal dorsal views of 18 somite stage rx3:GFP embryos co-labeled with pHH3. L: Both sema6A and plxnA2 morphants display ectopic retinal precursor cells outside of the eye field as well as significantly reduced pHH3 positive cells in the developing eyes compared with controls, as quantified (control 100%, N = 3, n = 30; plxnA2 MO 25.11% ± 0.72%, N = 3, n = 30, ****P < 0.0001; sema6A MO 28.86% ± 1.12%, N = 3, n = 30; ****P < 0.0001, one-way ANOVA, multiple comparisons post-hoc test). D–F: Schematics of rx3:GFP dorsal view. G,H,J,M: Decreased proliferation is later observed as smaller eyes at 48 hpf (control 0.49 μm ± 0.008 μm, N = 3, n = 30; plxnA2 MO 0.40 μm ± 0.01 μm, N = 3, n = 29, ****P < 0.0001; sema6A MO 0.42 μm ± 0.009 μm, N = 3, n = 31; ****P < 0.0001, one-way ANOVA, multiple comparisons test). G,I,K,M: Full length human PLXNA2 and SEMA6A mRNA partially rescues smaller eye phenotypes (control 0.49 μm ± 0.008 μm, N = 3, n = 30; PLXNA2 rescue 0.45 μm ± 0.009 μm, N = 3, n = 32, ***P < 0.001; SEMA6A rescue 0.48 μm ± 0.008 μm, N = 3, n = 28; ***P < 0.001, one-way ANOVA, multiple comparisons test. Error bars indicate SEM). E, eye; H, hypothalamus.
As these embryos develop further, the decreased proliferation results in smaller eyes at 48 hr post fertilization (hpf) (Fig. 1G,H,J,M; eye diameter relative to head length; Control 0.49 μm ± 0.008 μm, N = 3, n = 30; plxnA2 MO 0.40 μm ± 0.01 μm, N = 3, n = 29, P < 0.0001; sema6A MO 0.42 μm ± 0.009 μm, N = 3, n = 31, P < 0.0001, one-way ANOVA, Multiple comparisons test). Off-target effects and specificity of both sema6A and plxnA2 morpholinos have been addressed with secondary non-overlapping morpholino constructs, partial rescue of phenotypes by means of full-length mRNA injections, and penetrance verified by the absence of wildtype gene products by reverse transcriptase-polymerase chain reaction (RT-PCR) (Ebert et al., 2014). To re-address morpholino specificity in this study, co-injections of morpholino with full-length human SEMA6A or PLXNA2 mRNA partially rescued eye size (Fig. 1I,K,M; eye diameter relative to head length; control 0.49 μm ± 0.008 μm, N 3, n = 30; PLXNA2 rescue 0.45 μm ± 0.009 μm, N =3, n = 32, P < 0.001; SEMA6A rescue 0.48 μm ± 0.008 μm, N= 3, n= 28, P < 0.001; one-way ANOVA, multiple comparisons test).
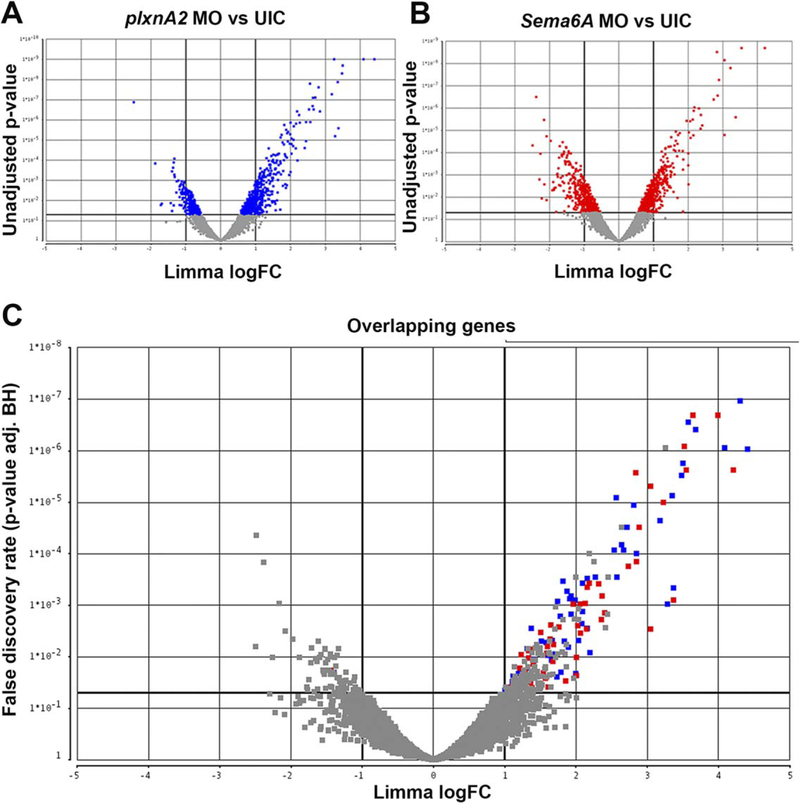
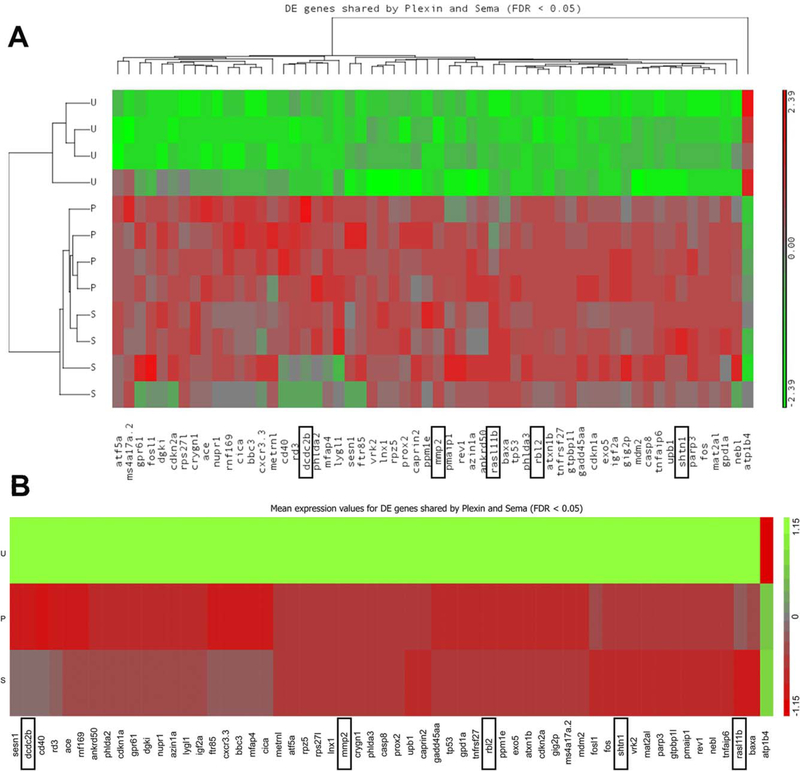
Overexpression of SEMA6A or PLXNA2 human mRNA alone exhibited no significant developmental phenotypes (data not shown). Considering that eye development takes several hours, we wanted to investigate what transcriptional changes were occurring downstream of Sema6A/PlxnA2 signaling that could be controlling this process. We compared gene expression levels of 18 somite plxnA2 morphants compared with uninjected control embryos (Fig. 2A) and 18 somite sema6A morphants compared with uninjected control embryos (Fig. 2B). In a micro-array of 4885 zebrafish genes, we identified 58 significantly (greater than two-fold) differentially regulated genes that were in common to both morphants (Table 1, 2C, GEO accession# GSE86246). Strikingly, of the 58 genes, 57 were up-regulated and only 1 was down-regulated, suggesting that Sema6A/PlxnA2 signaling is largely repressive to downstream transcriptional targets (Figs. 2C, 3). Using gene ontology information, we prioritized genes predicted to be involved in cell migration and proliferation for validation and further functional studies due to the pheno-types observed in morphant embryos.
Fig. 2.
Volcano plots denoting differential gene expression. Each point represents a gene in the microarray. A: Gene expression changes between plxnA2 MO and uninjected controls (UIC). B: sema6A MO compared with UIC. Gray are nonsignificant. C: Overlapping genes in both plxnA2 and sema6A plots. Blue squares indicate significantly different expression levels in plxnA2 MO-UIC, red squares in sema6A MOUIC. Gray are nonsignificant in at least one condition, with under a two-fold change difference in expression or P > 0.05.
TABLE 1.
List of 58 genes that are differentially regulated downstream of Sema6a/PlxnA2 signaling at 18 somites, common to both morphants compared to uninjected controls. GEO accession # GSE86246
| zfin symbol | ensembl gene ID | logFC Sema-UIC |
logFC Plexin-UIC |
|---|---|---|---|
| ace | ENSDARG00000079166 | 2.02 | 2.67 |
| ankrd50 | ENSDARG00000007077 | 1.59 | 1.99 |
| Atf5a | ENSDARG00000068096 | 2 | 2 |
| Atp1b4 | ENSDARG00000053262 | −1.44 | −1.4 |
| Atxn1b | ENSDARG00000060862 | 3.99 | 4.3 |
| Azin1a | ENSDARG00000052789 | 1.36 | 1.6 |
| baxa | ENSDARG00000020623 | 1.35 | 1 |
| bbc3 | ENSDARG00000069282 | 2.37 | 3.35 |
| caprin2 | ENSDARG00000020749 | 1.36 | 1.29 |
| casp8 | ENSDARG00000058325 | 3.55 | 3.5 |
| cd40 | ENSDARG00000054968 | 1.57 | 2.84 |
| Cdkn1a | ENSDARG00000076554 | 2.13 | 2.53 |
| cdkn2a | ENSDARG00000037262 | 3.05 | 3.28 |
| cica | ENSDARG00000071150 | 1.28 | 1.88 |
| Crygn1 | ENSDARG00000087437 | 2.31 | 2.27 |
| cxcr3.3 | ENSDARG00000070669 | 1.09 | 1.51 |
| dcdc2b | ENSDARG00000053744 | 1.25 | 2.16 |
| Dgki | ENSDARG00000063578 | 1.55 | 1.55 |
| exo5 | ENSDARG00000042727 | 3.22 | 3.48 |
| Fos | ENSDARG00000031683 | 3.05 | 2.81 |
| fosl1 | ENSDARG00000015355 | 2.4 | 2.03 |
| ftr85 | ENSDARG00000034707 | 1.5 | 2.09 |
| gadd45aa | ENSDARG00000043581 | 2.85 | 3.18 |
| gig2p | ENSDARG00000088260 | 1.23 | 1.32 |
| gpd1a | ENSDARG00000043701 | 1.64 | 1.74 |
| gpr61 | ENSDARG00000003900 | 1.85 | 2.2 |
| Gtpbp1l | ENSDARG00000042900 | 2.84 | 2.56 |
| igf2a | ENSDARG00000018643 | 1.51 | 1.82 |
| Inx1 | ENSDARG00000043323 | 1.4 | 1.41 |
| Lygl1 | ENSDARG00000056874 | 1.38 | 1.66 |
| mat2al | ENSDARG00000063665 | 2.89 | 2.64 |
| mdm2 | ENSDARG00000033443 | 1.76 | 1.91 |
| metrnl | ENSDARG00000007289 | 1.67 | 1.71 |
| mfap4 | ENSDARG00000090557 | 2.73 | 4.09 |
| mmp2 | ENSDARG00000017676 | 1.33 | 1.31 |
| ms4al7a.2 | ENSDARG00000093546 | 1.61 | 1.73 |
| nebl | ENSDARG00000021200 | 2 | 1.79 |
| nupr1 | ENSDARG00000094300 | 1.64 | 1.93 |
| parp3 | ENSDARG00000003961 | 1.96 | 1.77 |
| phlda2 | ENSDARG00000042874 | 1.28 | 1.63 |
| Pmaip1 | ENSDARG00000089307 | 2.15 | 1.89 |
| phlda3 | ENSDARG00000037804 | 3.64 | 3.57 |
| ppm1e | ENSDARG00000026499 | 1.06 | 1.1 |
| prox2 | ENSDARG00000041952 | 1.69 | 1.67 |
| rasl11b | ENSDARG00000015611 | 2.18 | 1.58 |
| rb12 | ENSDARG00000045636 | 3.52 | 3.68 |
| rd3 | ENSDARG00000031600 | 1.21 | 1.93 |
| rev1 | ENSDARG00000018296 | 1.6 | 1.41 |
| mf169 | ENSDARG00000042825 | 2.05 | 2.71 |
| rps27l | ENSDARG00000090186 | 2.06 | 2.08 |
| rpz5 | ENSDARG00000075718 | 3.37 | 3.37 |
| sesn1 | ENSDARG00000020693 | 1.49 | 2.57 |
| shtn1 | ENSDARG00000039697 | 1.21 | 1.12 |
| tnfaip6 | ENSDARG00000093440 | 1.33 | 1.19 |
| tnfrsf27 | ENSDARG00000079403 | 4.21 | 4.4 |
| tp53 | ENSDARG00000035559 | 1.23 | 1.37 |
| upb1 | ENSDARG00000011521 | 2.16 | 2.09 |
TABLE 2.
Primer Sequences and Accession Numbers Used for Antisense Riboprobe Generation and RT-PCR
| Name | Forward primer | Reverse primer | Accession # |
|---|---|---|---|
| rasl11b | ATGCGTCTGATCCAGAACATG | TAATACGACTCACTATAGGGGTCACACTGAAGTGACGGTGC | NM_200140.1 |
| dcdc2b | TGCTTCGTACTGGAGCTGTG | TGCTTCGTACTGGAGCTGTG | NM_001037689.2 |
| shtn-1 | TTTAGGTGACACTATAGAAGAGAGGCCTTACGGAAGCTGAAT | TAATACGACTCACTATAGGGGAGATGCCCAAGAGCTTCTTTTGT | NM_001198761.1 |
| rbl2 | TTTAGGTGACACTATAGAAGGGAGATCAAACCCTTCGCTCTG | TAATACGACTCACTATAGGGGAGAAAGAAGCCGTTGATCAGCAT | XM_002666954.5 |
| mmp2 | TTTAGGTGACACTATAGAAGGGCCCTTCAAATTCTTGGGTGA | TAATACGACTCACTATAGGGGAGACAATTTTCTCCGGAAGCTCA | NM_198067.1 |
Fig. 3.
Microarray heat map of 58 differentially regulated genes in common to both sema6A MO-UIC and plxnA2 MO-UIC. Differential expression is represented on a green to red scale with expression values standardized to a mean of zero and standard deviation of one. A: Results from each of the four replicates are shown. B: Average expression change of four replicates. Boxes indicate validated genes. U, uninjected control; S, sema6A MO; P, plxnA2 MO.
In Situ Hybridization and RT-PCR Validation of Microarray Results
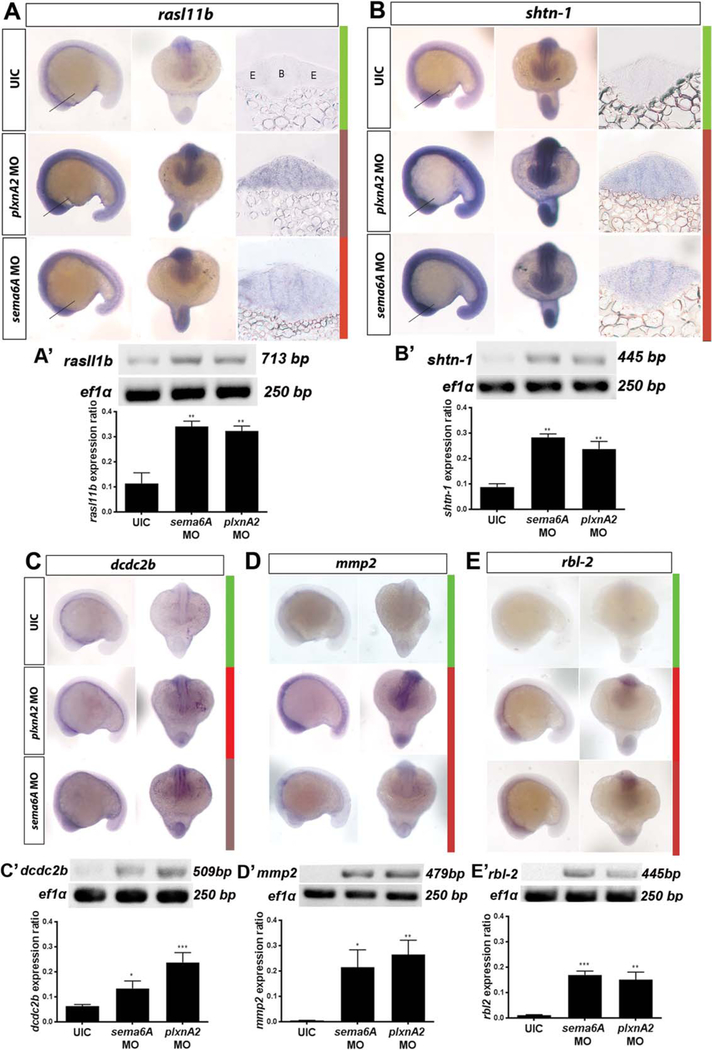
Zebrafish embryos were injected with either sema6A or plxnA2 morpholinos at the one cell stage, and fixed at the 18 somite stage, dehydrated, and processed for in situ hybridization. Digoxigenin (DIG)-labeled antisense riboprobes were generated for RAS-like, family 11, member B (rasl11b), shootin-1 (shtn-1), doublecortin domain-containing 2b (dcdc2b), matrix metalloproteinase 2 (mmp2), and retinoblastoma-like protein 2 (rbl2), and in situ hybridization was performed on control and morphant embryos. Probe intensity for all five candidate genes was consistent with microarray results and offered additional spatial information (Fig. 4A–E). Transverse sections of rasl11b and shtn-1 labeled embryos show neuronal expression, including in the developing eye, at this early stage (Fig. 4A,B, right columns).
Fig. 4.
Microarray validation. In situ hybridization and RT-PCR validation of differential gene expression in plxnA2 and sema6A morphants at 18 somites. A–E: Top, middle, and bottom panels show uninjected control, plxnA2 MO injected, and sema6A MO injected embryos, respectively. Left and right panels show lateral and dorsal views, respectively, for each probe under each condition. Colored bars show corresponding heat map expression changes for each gene. UIC, uninjected control. A,B: Additional right panels show transverse 20 μm sections through 18 somite stage brain and eye vesicles processed for in situ hybridization with rasl11b and shtn-1 probes. Lines in whole-mounts indicate corresponding section locations. A′–E′: RT-PCR gels and quantification of gene expression changes for each gene as a ratio to ef1α control levels (rasl11b control 0.1137 ± 0.04, N = 3, rasl11b sema6A MO 0.34 ± 0.02, N = 3, **P < 0.01; rasl11b plxnA2 MO 0.32 ± 0.01, N = 3, **P < 0.01; shtn-1 control 0.08 ± 0.01, N = 3, shtn-1 sema6A MO 0.28 ± 0.01, N = 3, **P < 0.01; shtn-1 plxnA2 MO 0.24 ± 0.03, N = 3, **P < 0.01; dcdc2b control 0.06 ± 0.004, N = 3, dcdc2b sema6A MO 0.13 ± 0.02 N = 3, *P < 0.05; dcdc2b plxnA2 MO 0.24 ± 0.02, N = 3, ***P < 0.001; mmp2 control 0.004 ± 0.001, N = 5, mmp2 sema6A MO 0.21 ± 0.07, N = 5, *P < 0.05; mmp2 plxnA2 MO 0.27 ± 0.06, N= 5, **P < 0.01; rbl2 control 0.01 ± 0.002, N = 4, rbl2 sema6A 0.17 ± 0.02, N = 4, ***P < 0.001; rbl2 plxnA2 MO 0.15 ± 0.03, N = 4, **P < 0.01; one-way ANOVA, multiple comparisons test. Error bars indicate SEM). B, brain. E, eye.
We observed overlap between previously reported plxnA2 and sema6A expression (Ebert et al., 2014) and the candidate genes, supporting a relationship between Sema6A/PlxnA2 signaling and these genes. We generated cDNA from uninjected control, sema6A morphant, and plxnA2 morphant embryos at 18 somites and performed RT-PCR (Fig. 4A′–E′). The results are consistent with the microarray and in situ expression data, showing increases in transcriptional levels of the candidate genes in both morphants compared with uninjected control (Fig. 4A′–E′; rasl11b control 0.1137 ± 0.04, N = 3; sema6A MO 0.34 ± 0.02, N = 3, P < 0.01; plxnA2 MO 0.32 ± 0.01, N = 3, P < 0.01; shtn-1 control 0.08 ± 0.01, N = 3; sema6A MO 0.28 ± 0.01, N = 3, P < 0.01; plxnA2 MO 0.24 ± 0.03, N = 3, P < 0.01; dcdc2b control 0.06 ± 0.004, N = 3; sema6A MO 0.13 ± 0.02, N = 3, P < 0.05; plxnA2 MO 0.24 ± 0.02, N = 3, P < 0.001; mmp2 control 0.004 ± 0.001, N = 5; sema6A MO 0.21 ± 0.07, N = 5, P < 0.05; plxnA2 MO 0.27 ± 0.06, N = 5, P < 0.01; rbl2 control 0.01 ± 0.002, N = 4; sema6A MO 0.17 ± 0.02, N = 4, P < 0.001; plxnA2 MO 0.15 ± 0.03, N = 4, P < 0.01; one-way ANOVA, multiple comparisons test).
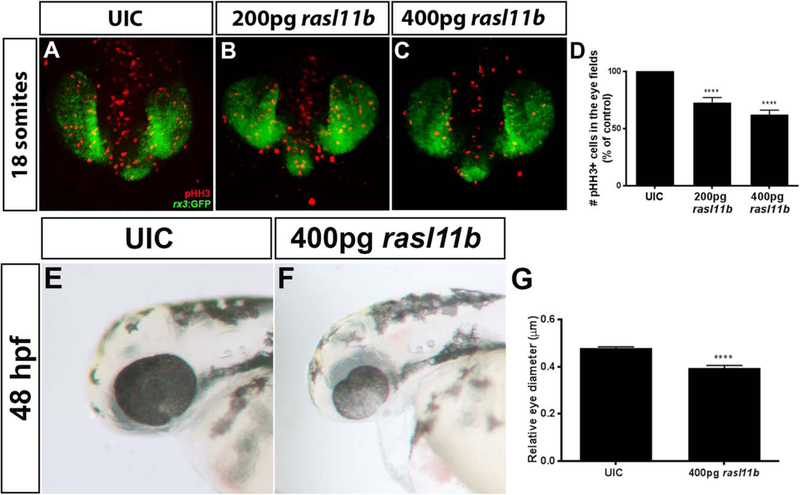
Overexpression of rasl11b Leads to Decreased Proliferation in Optic Vesicles
Microarray results indicated that rasl11b has a 2.18 log-fold change (logFC) in sema6A morphants and a 1.58 logFC in plxnA2 morphants (Table 1). We chose to further investigate this gene because of its predicted role in cell proliferation and expression in the developing eye field (Fig. 4A, right column). To determine if increased rasl11b expression contributes to decreased proliferation of RPCs, we injected full-length zebrafish rasl11b mRNA into single-cell embryos. We visualized proliferation (pHH3) in the early eye field (rx3:GFP) of 18 somite embryos injected with 200 pg or 400 pg rasl11b mRNA. Overexpression of rasl11b resulted in decreased RPC proliferation in a dose-dependent manner (Fig. 5A–D; control 100%, N = 3, n = 30; 200 pg rasl11b 72.87% ± 4.25%, N = 3, n = 38, P < 0.0001; 400 pg rasl11b 62.19% ± 4.01%, N = 3, n = 31, P < 0.0001; one-way ANOVA, multiple comparisons test). At 48 hpf, overexpression of rasl11b resulted in smaller eyes (Fig. 5E–G; eye diameter relative to head length; control 0.48 μm ± 0.005 μm, N = 3, n = 30; 400 pg rasl11b 0.39 μm ± 0.01 μm, N = 3, n = 36, P < 0.0001; one-way ANOVA, multiple comparisons test). We=show that rasl11b is negatively regulated downstream of Sema6A/PlxnA2 signaling and when overexpressed, decreases RPC proliferation and eye size.
Fig. 5.
Overexpression of rasl11b decreases proliferation of RPCs. A–C: Confocal dorsal views of 18 somite rx3:GFP embryos co-labeled with pHH3. D: Increasing doses of rasl11b resulted in decreased proliferation within the GFP positive eye field (control 100%, N = 3, n = 30; 200 pg rasl11b 72.87% ± 4.25% N = 3, n = 38, ****P < 0.0001; 400 pg rasl11b 62.19% ± 4.01%, N = 3, n = 31, ****P < 0.0001; one-way ANOVA, multiple comparisons test; error bars indicate SEM). E–G: Overexpression of rasl11b results in smaller eyes at 48 hpf (eye diameter relative to head length; control 0.48 μm ± 0.005 mm N = 3, n = 30; 400 pg rasl11b 0.39 μm ± 0.01 μm, N = 3, n = 36; ****P < 0.0001; one-way ANOVA, multiple comparisons test; error bars indicate SEM).
Proposed Signaling Mechanism of rasl11b Impinging on Ras Signaling
Rasl11b is a small atypical GTPase that has been implicated in prostate cancer (Louro et al., 2004). In zebrafish, Rasl11b inhibits mesendodermal and prechordal plate formation during embryo-genesis, and interacts with the one-eyed pinhead signaling pathway (Pezeron et al., 2008). Rasl11b is highly conserved in vertebrates and is atypical to most Ras-like family members in two ways. First, it is cytosolic and lacks carboxy terminal lipid modification sites which allow for membrane anchoring (Pezeron et al., 2008). Second, it has a lower GTPase activity than Ras, and is more often in its active GTP-bound state (Colicelli, 2004). Ras proteins are well known to be involved in the mitogen-activated protein kinase (MAPK) pathway, therefore, we hypothesize that Rasl11b acts as a negative regulator of MAPK by outcompeting Ras for its effectors such as Raf, leading to decreases in RPC proliferation seen in morphant embryos.
It is known that Plexins have an intracellular split GAP (GTPase activating protein) domain that can regulate Ras-family small GTPases (Negishi et al., 2005; Pasterkamp, 2005). Small GTPases act as molecular switches: “on” when GTP-bound, and “off” when GDP-bound (Bos et al., 2007). GAPs increase GTP hydrolysis and thereby increase the “off,” GDP-bound form of the protein. Plxn intracellular GAP domains are inactive when Plxns are in inactive, open conformations. Upon Sema binding, PlxnAs undergo a conformational change, which forms an active GAP domain, in addition to activating downstream effector proteins (He et al., 2009). When Sema6A/PlxnA2 signaling is disrupted, rasl11b expression increased (Table 1; Fig. 4A). We propose that when nonmembrane bound Rasl11b is present, it inhibits membrane-tethered Ras signaling through its lower GTPase activity and its ability to recruit either Raf or the small Ras activating guanine nucleotide exchange factor (GEF), son of sevenless (SOS), away from the membrane. This could sequester Raf or SOS away from Ras, therefore, Ras is predominantly found in its GDP-bound state, reducing signal propagation, and resulting in decreased pro-proliferative effects of the MAPK signaling cascade.
Proposed Contributions of shtn-1, dcdc2b, rbl2, and mmp2 to Morphant Phenotypes
Additional validated genes may also contribute to morphant phenotypes. Microarray results demonstrated that shtn-1 has a 1.21 logFC in sema6A morphants and a 1.12 logFC in plxnA2 morphants. Shtn-1 is involved in the formation of cellular asymmetric polarization signals, which direct the cell during migration, as seen in the orientation of neurite outgrowth of cultured hippo-campal neurons (Toriyama et al., 2006). To polarize, cells need to have carefully regulated amounts of Shtn-1. Overexpression of Shtn-1 in rat hippocampal neurons leads to its impaired asymmetric distribution and induces the formation of surplus axons (Toriyama et al., 2006). Alternatively, Shtn-1 may control the actin cytoskeleton in migrating cells, in that Shtn-1 couples factin retrograde flow to L1-CAM aiding with “clutch engagement” at the leading edge of the extending axons of rat hippocampal neurons in culture (Shimada et al., 2008). Thus, we hypothesize that an increase in Shtn-1 could impair the coordinated migration of RPCs, as individual cells fail to either receive the correct polarization signals and/or regulate their actin cytoskeleton.
Dcdc2b has a 1.25 logFC in sema6a and a 2.16 logFC in plxnaA2 morphants (Table 1). It is one of a family of 11 DCX domain-containing proteins. DCX domain-containing proteins are atypical microtubule-associated proteins. They bind to tubulin by means of their DCX domains to regulate microtubule stabilization and also couple microtubule-actin binding through various accessory proteins (Dijkmans et al., 2010). In zebrafish, mutations in dcdc2b have uncovered a role in the function of microtubules within cilia of hair cells, correlating with deficits that lead to deafness in humans (Grati et al., 2015). It is known that cytoskeletal microtubule dynamics at the trailing edge of migratory cells are critical for cellular movement, in addition to generating cellular polarity, both of which guide the direction of migration (Ganguly et al., 2012). We hypothesize that the increase in expression of dcdc2b in sema6A and plxnA2 morphants is partially responsible for the abnormal RPC migration seen within optic vesicles. Increases in dcdc2b could stabilize microtubules, inhibiting microtubule depolymerization at the trailing edge of migrating RPCs. Stabilization would potentially not only slow normal cellular migration, but disrupt the normal direction of migration, and explain why RPCs within morphant optic vesicles do not migrate as a tightly unified group.
Rbl2, sometimes referred to as p130, is a member of the pocket protein family, along with p107 and retinoblastoma (Rb) (Cobrinik, 2005). Mutations to the Rb family of genes can lead to a rare form of retinal cancer, retinoblastoma, in which cell cycle regulation is lost (Murphree and Benedict, 1984; Friend et al., 1986). Rbl2 has a 3.52 logFC in sema6a and a 3.68 logFC in plxnaA2 morphants (Table 1). These proteins are known to bind to members of the E2F family of transcription factors to regulate the cell cycle. Rbl2 binds to E2F4 to form a repressor complex in mitotic cells to block cell cycle progression (Dingar et al., 2012). The ability of Rbl2 to repress transcription is in part because it can recruit methyltransferases to histones, causing chromatin condensation, and, therefore, decreasing the accessibility of transcriptional machinery to the promoters of genes that promote the cell cycle (Lai et al., 2001). We, therefore, hypothesize that increases in rbl2 levels in morphant embryos contributes to impaired proliferation of RPCs by blocking the cell cycle.
Matrix metalloproteinases (MMP) are well known for their role in central nervous system development and amphibian regeneration through their ability to degrade extracellular matrix (ECM) proteins (Brinckerhoff and Matrisian, 2002). mmp2 has a 1.33 logFC in sema6a and a 1.31 logFC in plxnaA2 morphants (Table 1). Recently, mmp2 was shown to be important for degrading nonpermissive ECM to allow for zebrafish retinal ganglion cell axon outgrowth during recovery from injury (Lemmens et al., 2016). Increases in MMP2 could lead to increased degradation of the ECM surrounding RPCs, and removal of key proteins required for cell migration. Alternatively, increases in MMP2 at the leading edge of migrating RPCs could allow for the ease of pseudopodia through surrounding tissue and cause RPCs to over-migrate.
Conclusions
We confirm through morpholino knockdown, that Sema6A/PlxnA2 signaling regulates proliferation and cohesive migration of RPCs in developing optic vesicles in zebrafish. Here, we provide a resource of genes that are transcriptionally regulated downstream of Sema6A/PlxnA2 signaling. Initial characterization of one gene, rasl11b, uncovered its contributing role to the proliferation of RPCs, providing insight into the mechanisms that control this key developmental process. The broadness of Sema/Plxn signaling is only just becoming widely appreciated, and further research into downstream targets and signal propagation is necessary to fully understand the diverse roles of this signaling pair.
Experimental Procedures
Zebrafish Husbandry
rx3:GFP zebrafish embryos were generously provided by Jochen Wittbrodt, University of Heidelberg, Germany and developmentally staged as previously described (Kimmel et al., 1995). All procedures were approved by the University of Vermont Institutional Animal Care and Use Committee (IACUC).
Injections and Rescue Constructs
MOs (Gene Tools, Philomath, OR), or mRNA were injected at the one-cell stage (Kimmel et al., 1995). Morpholino concentrations and sequences were as follows: plxnA2 e2i2, 2 ng (AAAAGCGATGTCTTTCTCACCTTCC); sema6A, 4 ng (TGCTGATATCCTGCACTCACCTCAC). Both sema6A and plxnA2 morpholino constructs have been previously validated using RT-PCR to show successful knockdown (Ebert et al., 2014). Off target effects and specificity of morpholinos was addressed by rescue by full length human mRNA and co-injection of sema6A or plxnA2 and p53 morpholino. Second nonoverlapping morpholino constructs reproduced the same phenotypes, see Ebert et al. (2014). Capped and tailed full-length mRNA, (mMESSAGE mMACHINE kit, Ambion, Austin, TX, cat# AM1344; Poly(A) tailing kit, Invitrogen Cat# AM1350) was generated from plasmids containing human plxnA2 (Origene, Cat# RC221024) or sema6A (Invitrogen, Cat# MDR1734–202803771) and injected into embryos at the one cell stage for a final concentration of 200pg or 400pg/embryo. Full-length, zebrafish rasl11b mRNA was generated from pCS2 -DRRasl11b generously provided by Dr. Philippe Mourrain þ(Paris, France).
Microarray Assay
Uninjected control, sema6A morphant, and plxnA2 morphant embryos were staged at 18 somites and total RNA was isolated from 100 embryos using TRIzol (Invitrogen) followed by ethanol precipitation. Each treatment had four replicates generated from separate injection experiments. Data collection and analyses performed by the IBEST Genomics Resources CORE at the University of Idaho was supported by grants from the National Center for Research Resources (P30GM103324) and the National Institute of General Medical Sciences (8 P20 GM103397–10) from the National Institutes of Health. RNA from 18 somite stage control, sema6A and plxnA2 morphant embryos was analyzed using the NimbleGen Zebrafish 12 × 135 k Array based on Zv9 [090818_Zv7_EXPR], following the NimbleGen arrays user’s guide version 5.1.
Microarray Data Analysis
NimbleScan software (NimbleGen. Madison, WI) was used to align a chip-specific grid to control features and extract raw intensity data for each probe and each array. Chip images were then visually checked for each array and verified not to contain any significant spatial artifacts. Raw intensity data was then read into the R statistical computing environment and checked for quality. Chip intensity distributions, boxplots, and hierarchical clusters were compared and checked for any unusual global patterns. Each array was then background corrected and normalized using the quantile normalization procedure and finally each pro-beset was summarized using the median polish procedure as described with the robust multichip average (RMA) procedure (Irizarry et al., 2003a,b; Bolstad et al., 2003). The median polish procedure is a robust method for summarizing all probes contained within each probeset to a single expression value for each gene, taking into account individual probe effects. Probesets with low levels of expression variation across all samples (interquartile range < 0.5) were removed from further analysis, reducing the overall number of statistical tests to be performed. Differential expression was then assessed using a linear model with an empirical Bayesian adjustment to the variances (Smyth, 2005) and comparisons of interest were extracted using contrasts. The Benjamini and Hochberg (BH) method was used to control for the expected false discovery rate given multiple tests (Benjamini and Hochberg, 1995). Probesets were considered statistically differentially expressed with a BH adjusted P-value of < 0.05. Both raw intensity data (.pair files) and RMA-normalized expression values are available in the National Center for Biotechnology Information Gene Expression Omnibus database (NCBI GEO accession no. GSE86246).
Whole-Mount In Situ Hybridization
Zebrafish embryos were staged at 18 somites, fixed in 4% paraformaldehyde overnight at 48C, and stored at −20°C in 100% methanol until use. In situ hybridization was performed as described previously (Thisse and Thisse, 2008). DIG-labeled anti-sense RNA probes were generated using primers listed in Table 2 (IDT, Coralville, IA). PCR products were cloned into pCR 2.1 TOPO vector (Invitrogen, Carlsbad, CA) and sequencing at the UVM Cancer Center Advanced Genome Technologies Core, Burlington, VT. Embryos were oriented in 4% methyl cellulose on depression slides and imaged using a Nikon SMZ800 dissecting light microscope at 50 × magnification.
RT-PCR
cDNA was synthesized using SuperScript II reverse transcriptase (Invitrogen) from isolated control, plxnA2 morphant or sema6A morphant total RNA (described above). Gene specific primers (Table 2) were used to amplify approximately 500 base pair amplicons in 10 μl PCR reactions from 1 μg cDNA using PCR Mastermix (Thermo). ef1α primers were used as parallel control reactions. PCRs were run on a 1% agarose gel (Invitrogen). Gels were imaged using a Syngene INGENIUS3 system and GENEsys V1.5.5.0 imaging software. Densitometry was performed using Adobe Photoshop CS6. Band intensities were normalized to background, and a ratio of gene band intensity to ef1α control levels was obtained for each sample. At least three independent replicates were averaged. Graphs and statistical analyses were obtained using Prism 6.
Sectioning, Imaging, Cell Counts, and Eye Measurements
Embryos were dehydrated in 100% ETOH for 30 min before embedding using a JB4 Embedding Kit (Polysciences, Inc., Warrington, PA) and sectioned on a Leica RM2265 microtome at 20 μm. Brightfield images were obtained using an Olympus iX71 inverted light microscope and figures were assembled using Adobe Photoshop CS6, in which image brightness and contrast were optimized. Eye size measurements were obtained using SPOT imaging software version 5.2. To control for any changes in overall embryo size, eye size was calculated as a ratio of eye diameter divided by the distance from the anterior tip of the head to the posterior otic placode for each embryo. Proliferating cell counts were recorded when pHH3 positive cells were seen within GFP positive optic vesicles only. For confocal imaging, embryos were mounted in 0.5% low-melt agarose on glass bottom dishes (CLS-1811 ChemGlass, NJ) and imaged at 20 × using 2-μm steps on a Nikon Eclipse Ti inverted microscope. Z Stacks were subjected to a Kalman stack filter in Image J and were presented as maximum intensity projections. RGB images were generated in Adobe Photoshop CS6.
Whole-Mount Immunohistochemistry
Eighteen somite stage embryos were fixed in 4% paraformalde-hyde overnight at 4°C, rehydrated in PBST (phosphate buffered saline + 1% Triton), incubated in immuno block (PBST + 10% normal goat serum) for 1 hr at room temperature, primaryþ antibody in immuno block overnight at 4°C, secondary antibody for 2 hr at room temperature and stored in PBST until imaged. Antibodies used: phospho-Histone H3, pHH3 (1:1,000, Cell Signaling, Cat# 3377, Danvers, MA), anti-rabbit Alexa Flour 555 (1:1,000, Cell Signaling, Cat# 4413S).
Acknowledgments
VGN Bioinformatics Core services were supported by an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under grant number P20GM103449. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of NIGMS or NIH. Thank you to Dr. Paula Deming, Department of Nuclear Medicine and Radiological Sciences, University of Vermont for guidance and insight. Authors have no conflict of interest to declare.
Grant sponsor: U.S. National Science Foundation; Grant number: IOS1456846; Grant sponsor: Foundation Fighting Blindness Canada.
ABBREVIATIONS:
- ANOVA
analysis of variance
- BH
Benjamini and Hochberg
- dcdc2b
doublecortin domain-containing 2b
- DIG
digoxigenin
- ECM
extracellular matrix
- GAP
GTPase activating protein
- GEF
guanine nucleotide exchange factor
- GFP
green fluorescent protein
- hpf
hours post fertilization
- logFC
log-fold change
- MAPK
mitogen-activated protein kinase
- mmp
matrix metalloproteinase
- MO
morpholino
- PBST
(phosphate buffered saline + 1% Triton
- pHH3
phospho-Histone H3
- Plxn
plexin
- rasl11b
RAS-like, family 11, member B
- rbl2
retinoblastoma-like protein 2
- RMA
robust multichip average
- RPCs
retinal precursor cells
- RT-PCR
reverse transcriptase-polymerase chain reaction
- Sema
semaphorin
- shtn-1
Shootin-1
References
- Behar O, Golden JA, Mashimo H, Schoen FJ, Fishman MC. 1996. Semaphorin III is needed for normal patterning and growth of nerves, bones and heart. Nature 383:525–528. [DOI] [PubMed] [Google Scholar]
- Belle M, Parray A, Belle M, Chedotal A, Nguyen-Ba-Charvet KT. 2016. PlexinA2 and Sema6A are required for retinal progenitor cell migration. Dev Growth Differ 58:492–502. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol 57:289–300. [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. 2003. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19:185–193. [DOI] [PubMed] [Google Scholar]
- Bos JL, Rehmann H, Wittinghofer A. 2007. GEFs and GAPs: Critical elements in the control of small G proteins. Cell 129:865–877. [DOI] [PubMed] [Google Scholar]
- Brinckerhoff CE, Matrisian LM. 2002. Matrix metalloproteinases: a tail of a frog that became a prince. Nat Rev Mol Cell Biol 3:207–214. [DOI] [PubMed] [Google Scholar]
- Cobrinik D 2005. Pocket proteins and cell cycle control. Oncogene 24:2796–2809. [DOI] [PubMed] [Google Scholar]
- Colicelli J 2004. Human RAS superfamily proteins and relatedGTPases. Science’s STKE 2004:RE13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkmans TF, van Hooijdonk LW, Fitzsimons CP, Vreugdenhil E. 2010. The doublecortin gene family and disorders of neuronal structure. Cent Nerv Syst Agents Med Chem 10:32–46. [DOI] [PubMed] [Google Scholar]
- Dingar D, Konecny F, Zou J, Sun X, von Harsdorf R. 2012. Anti-apoptotic function of the E2F transcription factor 4 (E2F4)/p130, a member of retinoblastoma gene family in cardiac myocytes. J Mol Cell Cardiol 53:820–828. [DOI] [PubMed] [Google Scholar]
- Ebert AM, Childs SJ, Hehr CL, Cechmanek PB, McFarlane S. 2014. Sema6a and Plxna2 mediate spatially regulated repulsion within the developing eye to promote eye vesicle cohesion. Development 141:2473–2482. [DOI] [PubMed] [Google Scholar]
- Friend SH, Bernards R, Rogelj S, Weinberg RA, Rapaport JM, Albert DM, Dryja TP. 1986. A human DNA segment with properties of the gene that predisposes to retinoblastoma and osteosarcoma. Nature 323:643–646. [DOI] [PubMed] [Google Scholar]
- Ganguly A, Yang H, Sharma R, Patel KD, Cabral F. 2012. The role of microtubules and their dynamics in cell migration. J Biol Chem 287:43359–43369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mh Grati, Chakchouk, Ma Q, Bensaid M, Desmidt A, Turki N, Yan D, Baanannou A, Mittal R, Driss N, Blanton S, Farooq A, Lu Z, Liu XZ, Masmoudi S. 2015. A missense mutation in DCDC2 causes human recessive deafness DFNB66, likely by interfering with sensory hair cell and supporting cell cilia length regulation. Hum Mol Genet 24:2482–2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H, Yang T, Terman JR, Zhang X. 2009. Crystal structure of the plexin A3 intracellular region reveals an autoinhibited conformation through active site sequestration. Proc Natl Acad Sci U S A 106:15610–15615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. 2003a. Summaries of affymetrix GeneChip probe level data. Nucleic Acids Res 31:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. 2003b. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4:249–264. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. 1995. Stages of embryonic development of the zebrafish. Dev Dyn 203:253–310. [DOI] [PubMed] [Google Scholar]
- Lai A, Kennedy BK, Barbie DA, Bertos NR, Yang XJ, Theberge MC, Tsai S-C, Seto E, Zhang Y, Kuzmichev A, Lane WS, Reinberg D, Harlow E, Branton PE. 2001. RBP1 recruits the mSIN3-histone deacetylase complex to the pocket of retinoblastoma tumor suppressor family proteins found in limited discrete regions of the nucleus at growth arrest. Mol Cell Biol 21:2918–2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmens K, Bollaerts I, Bhumika S, De Groef L, Van Houcke J, Darras VM, Van Hove I, Moons L. 2016. Matrix metalloproteinases as promising regulators of axonal regrowth in the injured adult zebrafish retinotectal system. J Comp Neurol 524:1472–1493. [DOI] [PubMed] [Google Scholar]
- Louro R, Nakaya HI, Paquola AC, Martins EA, da Silva AM, Verjovski-Almeida S, Reis EM. 2004. RASL11A, member of a novel small monomeric GTPase gene family, is down-regulated in prostate tumors. Biochem Biophys Res Commun 316:618–627. [DOI] [PubMed] [Google Scholar]
- Luo Y, Raible D, Raper JA. 1993. Collapsin: A protein in brain that induces the collapse and paralysis of neuronal growth cones. Cell 75:217–227. [DOI] [PubMed] [Google Scholar]
- Murphree AL, Benedict WF. 1984. Retinoblastoma: clues to human oncogenesis. Science 223:1028. [DOI] [PubMed] [Google Scholar]
- Negishi M, Oinuma I, Katoh H. 2005. Plexins: axon guidance and signal transduction. Cell Mol Life Sci 62:1363–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld G, Kessler O. 2008. The semaphorins: versatile regulators of tumour progression and tumour angiogenesis. Nat Rev Cancer 8:632–645. [DOI] [PubMed] [Google Scholar]
- Oinuma I, Ishikawa Y, Katoh H, Negishi M. 2004. The semaphorin 4D receptor plexin-B1 is a GTPase activating protein for R-Ras. Science 305:862–865. [DOI] [PubMed] [Google Scholar]
- Pasterkamp RJ. 2005. R-Ras fills another GAP in semaphorin signalling. Trends Cell Biol 15:61–64. [DOI] [PubMed] [Google Scholar]
- Pezeron G, Lambert G, Dickmeis T, Strahle U, Rosa FM, Mourrain P. 2008. Rasl11b knock down in zebrafish suppresses one-eyed-pinhead mutant phenotype. PLoS One 3:e1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rembold M, Loosli F, Adams RJ, Wittbrodt J. 2006. Individual cell migration serves as the driving force for optic vesicle evagination. Science 313:1130–1134. [DOI] [PubMed] [Google Scholar]
- Renaud J, Chedotal A. 2014. Time-lapse analysis of tangential migration in Sema6A and PlexinA2 knockouts. Mol Cell Neurosci 63:49–59. [DOI] [PubMed] [Google Scholar]
- Renaud J, Kerjan G, Sumita I, Zagar Y, Georget V, Kim D, Fouquet C, Suda K, Sanbo M, Suto F, Ackerman SL, Mitchell KJ, Fujisawa H, Chedotal A. 2008. Plexin-A2 and its ligand, Sema6A, control nucleus-centrosome coupling in migrating granule cells. Nat Neurosci 11:440–449. [DOI] [PubMed] [Google Scholar]
- Rünker AE, Little GE, Suto F, Fujisawa H, Mitchell KJ. 2008. Semaphorin-6A controls guidance of corticospinal tract axons at multiple choice points. Neural Dev 3:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serini G, Valdembri D, Zanivan S, Morterra G, Burkhardt C, Caccavari F, Zammataro L, Primo L, Tamagnone L, Logan M, Tessier-Lavigne M, Taniguchi M, Puschel AW, Bussolino F. 2003. Class 3 semaphorins control vascular morphogenesis by inhibiting integrin function. Nature 424:391–397. [DOI] [PubMed] [Google Scholar]
- Shi W, Kumanogoh A, Watanabe C, Uchida J, Wang X, Yasui T, Yukawa K, Ikawa M, Okabe M, Parnes JR, Yoshida K, Kikutani H. 2000. The class IV semaphorin CD100 plays nonredundant roles in the immune system: defective B and T cell activation in CD100-deficient mice. Immunity 13:633–642. [DOI] [PubMed] [Google Scholar]
- Shimada T, Toriyama M, Uemura K, Kamiguchi H, Sugiura T, Watanabe N, Inagaki N. 2008. Shootin1 interacts with actin retrograde flow and L1-CAM to promote axon outgrowth. J Cell Biol 181:817–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. 2005. limma: linear models for microarray data In: Gentleman R, Carey VJ, Huber W, Irizarry RA, Dudoit S, editors. Bioinformatics and computational biology solutions using R and bioconductor. New York, NY: Springer New York; p 397–420. [Google Scholar]
- Tamagnone L, Artigiani S, Chen H, He Z, Ming GI, Song H, Chedotal A, Winberg ML, Goodman CS, Poo M, Tessier-Lavigne M, Comoglio PM. 1999. Plexins are a large family of receptors for transmembrane, secreted, and GPI-anchored semaphorins in vertebrates. Cell 99:71–80. [DOI] [PubMed] [Google Scholar]
- Tawarayama H, Yoshida Y, Suto F, Mitchell KJ, Fujisawa H. 2010. Roles of semaphorin-6B and plexin-A2 in lamina-restricted projection of hippocampal mossy fibers. J Neurosci 30:7049–7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse C, Thisse B. 2008. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat Protoc 3:59–69. [DOI] [PubMed] [Google Scholar]
- Toriyama M, Shimada T, Kim KB, Mitsuba M, Nomura E, Katsuta K, Sakumura Y, Roepstorff P, Inagaki N. 2006. Shootin1: a protein involved in the organization of an asymmetric signal for neuronal polarization. J Cell Biol 175:147–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyofuku T, Yoshida J, Sugimoto T, Zhang H, Kumanogoh A, Hori M, Kikutani H. 2005. FARP2 triggers signals for Sema3A-mediated axonal repulsion. Nat Neurosci 8:1712–1719. [DOI] [PubMed] [Google Scholar]