Abstract
The goals of the KIR component of the 17th International HLA and Immunogenetics Workshop (IHIW) were to encourage and educate researchers to begin analyzing KIR at allelic resolution, and to survey the nature and extent of KIR allelic diversity across human populations. To represent worldwide diversity, we analyzed 1269 individuals from ten populations, focusing on the most polymorphic KIR genes, which express receptors having three immunoglobulin (Ig)-like domains (KIR3DL1/S1, KIR3DL2 and KIR3DL3). We identified 13 novel alleles of KIR3DL1/S1, 13 of KIR3DL2 and 18 of KIR3DL3. Previously identified alleles, corresponding to 33 alleles of KIR3DL1/S1, 38 of KIR3DL2, and 43 of KIR3DL3, represented over 90% of the observed allele frequencies for these genes. In total we observed 37 KIR3DL1/S1 allotypes, 40 for KIR3DL2 and 44 for KIR3DL3. As KIR allotype diversity can affect NK cell function, this demonstrates potential for high functional diversity worldwide. Allelic variation further diversifies KIR haplotypes. We determined KIR3DL3~KIR3DL1/S1~KIR3DL2 haplotypes from five of the studied populations, and observed multiple population-specific haplotypes in each. This included 234 distinct haplotypes in European Americans, 191 in Ugandans, 35 in Papuans, 95 in Egyptians and 86 in Spanish populations. For another 35 populations, encompassing 642,105 individuals we focused on KIR3DL2 and identified another 375 novel alleles, with approximately half of them observed in more than one individual. The KIR allelic level data gathered from this project represents the most comprehensive summary of global KIR allelic diversity to date, and continued analysis will improve understanding of KIR allelic polymorphism in global populations. Further, the wealth of new data gathered in the course of this workshop component highlights the value of collaborative, community-based efforts in immunogenetics research, exemplified by the IHIW.
Keywords: KIR3DL1/S1, KIR3DL2, KIR3DL3
Introduction
The Killer-cell Immunoglobulin-like Receptor (KIR) region is located on human chromosome 19q13.4 [1–4]. KIR molecules are primarily expressed on natural killer (NK) cells [5] and a small percentage of T-cells [6]. KIR interact with specific amino acid motifs expressed by some human leukocyte antigen (HLA) class I molecules [5], and function to modulate the cytolysis of infected and/or otherwise altered cells, such as neoplastic cells. The KIR gene complex is characterized by structural variation that creates multiple gene-content haplotypes. In addition, each of the KIR genes exhibits allelic variability [7], which generates considerable intra- and inter-population diversity [8]. This diversity can influence immune responses against pathogens, which has the potential to alter the fitness of individuals [9, 10]. Specific combinations of KIR with their cognate HLA ligands are associated with autoimmunity [3, 11, 12], infectious diseases [13, 14], cancer [15, 16], pregnancy outcomes [17, 18], are crucial in determining clinical outcomes of hematopoietic stem cell transplantation (HCT), and solid organ transplants [19–22].
The allelefrequencies.net database (AFND) has collected KIR datasets from 245 populations across the globe [23]. A similar resource was recently developed called the KIR and Disease Database (KDDB), which gathered KIR associations from 204 published articles, and indicates a growing interest in KIR in epidemiological studies. These associations consisted of 32 autoimmune diseases, 19 infectious diseases, 16 cancer, eight chronic inflammatory diseases, three related to pregnancy, and one psychiatric disease. [24]. The complex polymorphism observed in this gene family, when combined with the high sequence similarity among KIR genes [25, 26], imposes technical difficulties for sequencing and genotyping to full allelic resolution. Thus, despite the fact that KIR gene content polymorphism has been extensively studied, KIR allelic diversity has been characterized in only a handful of well-defined populations [27–32].
KIR gene content variation was examined during previous International HLA and Immunogenetics Workshop (IHIW) studies. In the 15th and 16th IHIW, the KIR anthropology component (Population Global Distribution of KIR and Ligand) aimed to accumulate and examine the KIR and HLA frequencies in individuals recruited from distinct populations worldwide [33, 34], in order to replicate the earlier findings of coevolution of KIR and HLA [30, 33, 35, 36]. The preliminary studies conducted by Hiby et al. (2004) while investigating the role of maternal KIR and fetal HLA-C in preeclampsia, first raised the question whether KIR and HLA class I coevolution is related to reproductive fitness [17]. Single et al. (2007) demonstrated evidence of KIR-HLA coevolution, by showing a negative correlation of the frequency of KIR3DS1 with HLA-Bw4 [35], followed by several other studies corroborating the coevolution of KIR with HLA [30, 33, 36]. Further evidence of KIR-HLA coevolution was demonstrated in the 16th IHIW, in which 105 populations were examined and a strong positive correlation of KIR2DL3 and its ligand HLA-C1 was observed [34].
The goal of the 17th IHIW KIR component was to collect KIR allelic data to characterize the nature and extent of allelic diversity across human populations using primarily next generation sequencing (NGS) technology. As NGS for KIR has not yet been implemented in several laboratories that study KIR, Sanger sequencing was also welcomed [30, 37]. All the participants performing KIR genotyping were required to validate their method by genotyping a control panel, however, the reference laboratories performed most genotyping. Many investigators participated in the KIR component by providing DNA specimens sequenced by one of the reference laboratories. Here, we present a summary of the KIR component of the 17th IHIW working group meeting, and the KIR allelic data generated from the 45 worldwide populations that were analyzed. Our preliminary analysis focused on the KIR genes that encode three Ig domain receptors because they have been most extensively characterized to the allelic level and their diversity has been shaped by natural selection [38].
Materials and methods
Participants from eleven laboratories submitted KIR allelic genotyping data from a total of 45 populations. Five populations were analyzed through the entire coding sequence for KIR3DL1/S1, KIR3DL2 and KIR3DL3 polymorphism, four for KIR3DL2 and one for KIR3DL1/S1. Exons 4 and 5 from KIR3DL2 were analyzed in the remaining 35 populations. The participants either used NGS platforms or Sanger sequencing to generate KIR allelic data locally, or contributed DNA samples to be sequenced at the workshop reference laboratory at Stanford University. The list of all populations, including sample size, KIR genes, sequencing method, sample contributor and the location where sequencing was performed is given in Table 1. Additionally, Single molecule real-time (SMRT) KIR gene sequencing was performed for 19 IHIW cell lines from populations including European, black southern African, Warao Amerindian and Chinese.
Table 1:
Details of KIR allele-level sequencing of workshop populations, including sample size, KIR genes, KIR typing method, sample contributor and sequencing location
| Population | N | Genes | Method | Sample contributor | $Sequencing |
|---|---|---|---|---|---|
| Uganda | 174 | KIR3DL1/S1, KIR3DL2 and KIR3DL3 | NGS | Traherne/Moffett | Local |
| Egypt | 136 | KIR3DL1/S1, KIR3DL2 and KIR3DL3 | NGS | Elfishawi | Stanford |
| European American | 378 | KIR3DL1/S1, KIR3DL2 and KIR3DL3 | NGS | Hollenbach/Oksenberg | Stanford |
| Papua New Guinea | 185 | KIR3DL1/S1, KIR3DL2 and KIR3DL3 | NGS | Mentzer/Oppenheimer | Stanford |
| Spain | 153 | KIR3DL1/S1, KIR3DL2 and KIR3DL3 | NGS | GETHIT# | Stanford |
| Curitiba | 42 | KIR3DL2 | Sanger | Augusto/Petzl-Erler | Local |
| Kaingang | 30 | KIR3DL2 | Sanger | Augusto/Petzl-Erler | Local |
| Guarani | 49 | KIR3DL2 | Sanger | Augusto/Petzl-Erler | Local |
| Japanese-Brazilian | 22 | KIR3DL2 | Sanger | Augusto/Petzl-Erler | Local |
| Mexican Mestizos | 100 | KIR3DL1S1 | Sanger | Gorodezky | Local |
| Germany | 564253 | KIR3DL2 exons 4 and 5 | NGS | DKMS | Local |
| Poland | 6509 | KIR3DL2 exons 4 and 5 | NGS | DKMS | Local |
| Kosovo | 649 | KIR3DL2 exons 4 and 5 | NGS | DKMS | Local |
| Serbia | 857 | KIR3DL2 exons 4 and 5 | NGS | DKMS | Local |
| Croatia | 1947 | KIR3DL2 exons 4 and 5 | NGS | DKMS | Local |
| Brazil | 381 | KIR3DL2 exons 4 and 5 | NGS | DKMS | Local |
| Syria | 554 | KIR3DL2 exons 4 and 5 | NGS | DKMS | Local |
| Bosnia-Herzegovina | 992 | KIR3DL2 exons 4 and 5 | NGS | DKMS | Local |
| Sri Lanka | 1809 | KIR3DL2 exons 4 and 5 | NGS | DKMS | Local |
| Austria | 1374 | KIR3DL2 exons 4 and 5 | NGS | DKMS | Local |
| Czech Republic | 620 | KIR3DL2 exons 4 and 5 | NGS | DKMS | Local |
| Kazakhstan | 1701 | KIR3DL2 exons 4 and 5 | NGS | DKMS | Local |
| Spain | 1053 | KIR3DL2 exons 4 and 5 | NGS | DKMS | Local |
| France | 865 | KIR3DL2 exons 4 and 5 | NGS | DKMS | Local |
| India | 393 | KIR3DL2 exons 4 and 5 | NGS | DKMS | Local |
| USA | 903 | KIR3DL2 exons 4 and 5 | NGS | DKMS | Local |
| Vietnam | 546 | KIR3DL2 exons 4 and 5 | NGS | DKMS | Local |
| Greece | 2695 | KIR3DL2 exons 4 and 5 | NGS | DKMS | Local |
| Hungary | 833 | KIR3DL2 exons 4 and 5 | NGS | DKMS | Local |
| Romania | 1425 | KIR3DL2 exons 4 and 5 | NGS | DKMS | Local |
| Afghanistan | 541 | KIR3DL2 exons 4 and 5 | NGS | DKMS | Local |
| Great Britain | 755 | KIR3DL2 exons 4 and 5 | NGS | DKMS | Local |
| Burundi | 398 | KIR3DL2 exons 4 and 5 | NGS | DKMS | Local |
| Albania | 469 | KIR3DL2 exons 4 and 5 | NGS | DKMS | Local |
| Lebanon | 437 | KIR3DL2 exons 4 and 5 | NGS | DKMS | Local |
| Other | 8326 | KIR3DL2 exons 4 and 5 | NGS | DKMS | Local |
| Russia | 5288 | KIR3DL2 exons 4 and 5 | NGS | DKMS | Local |
| Switzerland | 405 | KIR3DL2 exons 4 and 5 | NGS | DKMS | Local |
| Portugal | 1450 | KIR3DL2 exons 4 and 5 | NGS | DKMS | Local |
| Turkey | 26119 | KIR3DL2 exons 4 and 5 | NGS | DKMS | Local |
| Netherlands | 981 | KIR3DL2 exons 4 and 5 | NGS | DKMS | Local |
| Iran | 1059 | KIR3DL2 exons 4 and 5 | NGS | DKMS | Local |
| Italy | 4416 | KIR3DL2 exons 4 and 5 | NGS | DKMS | Local |
| Morocco | 449 | KIR3DL2 exons 4 and 5 | NGS | DKMS | Local |
| Ukraine | 653 | KIR3DL2 exons 4 and 5 | NGS | DKMS | Local |
GETHIT means for Spanish Working Group in Histocompatibility and Transplant Immunology study;
Local sequencing means KIR genotyping was performed by the participant’s lab either using a NGS exome capture method [38] for KIR genes containing three Ig receptors (Traherne/Moffett lab) or Sanger sequencing for KIR3DL1S1 (Gorodezky lab) and KIR3DL2 (Augusto/Petzl-Erler) or an in-house developed NGS short amplicon approach for KIR3DL2 (DKMS lab). For Stanford Sequencing,KIR genotyping was performed at Stanford using a NGS exome capture method [38].
NGS genotyping of KIR genes containing three Ig-like domains
To determine the sequences of KIR genes containing three Ig-like domains, a previously described capture/enrichment method, followed by NGS [39] was applied. DNA isolated from healthy unrelated blood donors from the following populations was used: Ugandan (n = 174); Egyptian (n = 136); European American (USA) (n = 376); Papuan (n = 185); and Spanish (n = 153). The Ugandan, Egyptian and Spanish populations have been previously examined for KIR gene content [40–42]. Similarly, the European American sample was described in a recent HLA study [43]. The Papuan sample consists of individuals from both the highland and lowland regions, as described [44].
Sanger sequencing for genotyping KIR3DL1/S1 and KIR3DL2
KIR3DL2 was genotyped using sequence-based typing in samples from Brazil, which included Euro-descendants from Curitiba (n = 42), non-mixed Brazilians with Japanese ancestry (n = 22) and Amerindians from the Kaingang (n = 30) and Guarani (n = 49) populations. The Brazilian populations have been previously described for KIR gene-content [45–47]. Exons 3, 4, 5, 7–9 were amplified with gene-specific primers and the products were sequenced with Big Dye terminator kit (Applied Biosystems) according to the manufacturer’s instructions. Specific PCR-SSP primers were designed to resolve two common ambiguities; where it was otherwise not possible to distinguish the genotype KIR3DL2*002+*010 from KIR3DL2*010+*015, and the genotype KIR3DL2*001+*007 from KIR3DL2*006+*010. Primer sequences are available upon request. KIR3DL1/S1 was genotyped using sequence-based typing as reported earlier [30] in unrelated healthy Mexican Mestizos (n = 59). The Mexican Mestizos population KIR gene-content variation was examined in an earlier report [37].
Large scale KIR3DL2 sequencing
Sequence data for exons 4 and 5 of KIR3DL2 was generated from a total of 642,105 individuals from 35 populations (Table 1). PCR amplicons were generated from these exons individually, and then sequenced using Illumina paired-end technology (HiSeq or MiSeq). Alleles were called using the neXtype algorithm [48] and IPD-KIR library version 2.7.0 (Release, 14th July 2017) as the reference [7].
SMRT KIR gene sequencing for IHIW cell lines
In addition to the populations described above, KIR allele sequences were also generated for a small panel of IHIW cell lines. Briefly, samples underwent PCR targeting individual KIR genes to amplify full-length alleles (5´ UTR to 3´ UTR). Amplicons of the same locus were pooled together and sequenced on Pacific Biosciences’ RSII platform using a movie time of six hours to obtain maximum read depth. A combination of Pacific Biosciences’ SMRTAnalysis and Anthony Nolan’s AlleleTeaSet software (Anthony Nolan Research Institute, London, UK) were used to demultiplex and analyze the sequences. For the purposes of this study, the coding domain sequences were extracted from the phased, full-length sequence for further analysis.
Data analysis
All data analysis including allele counts, and frequency estimations were performed in the R environment for statistical computing and visualization [49]. The haplotype analysis was carried out using the R ‘haplo.stats’ package [50].
The KIR Component Meeting
The KIR component meeting of the 17th IHIW was held during two breakout sessions. Each participant presented the results of the population data submitted by their group. Additionally, updates on the state of KIR haplotype reference sequences, KIR in Allelefrequencies.net database, KIR nomenclature, and the IPD-KIR database were presented. Finally, there was an overview of PING (Pushing Immunogenetics to the Next Generation) software package [39], which is a bioinformatics pipeline for the analysis of next-generation sequencing KIR data. A supplementary file describes the schedule of the KIR component meeting, titles of the presentation and details of the presenters (Supplementary File S1).
Results
Allelic diversity of KIR3DL1/S1, KIR3DL2 and KIR3DL3
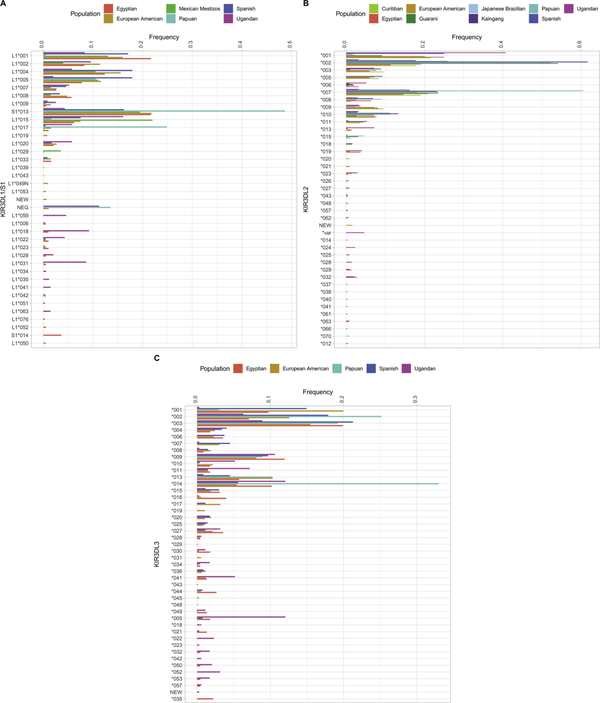
We analyzed KIR3DL1/S1, KIR3DL2 and KIR3DL3, which encode receptors having three Ig domains. These genes have been the most extensively characterized to date, and their allelic diversity has been shown to be shaped by natural selection [38]. We observed 33 previously identified alleles of KIR3DL1/S1, 38 of KIR3DL2 and 43 of KIR3DL3. We also identified 13 novel alleles for KIR3DL1/S1, 13 for KIR3DL2 and 18 for KIR3DL3 genes. The validation of these novel alleles is underway. Thus, the total numbers of alleles identified in the workshop samples were 46 for KIR3DL1/S1, 51 for KIR3DL2 and 61 for KIR3DL3 (Table 2), and these encode 37, 40 and 44 distinct KIR allotypes respectively (Table 2). Considering the modest sample sizes analyzed compared with HLA (more than 30 million to date [51]), this suggests that there are many more alleles remaining to be discovered and that the extent of KIR polymorphism identified in human populations could ultimately equal or exceed the extent of HLA polymorphism.
Table 2:
Allelic variations of one centromeric (KIR3DL3) and two telomeric (KIR3DL1/S1 and KIR3DL2) genes diversifies KIR haplotypes in European American, Ugandan, Papuan, Egyptian and Spanish populations
| Centromeric | Telomeric | |||
|---|---|---|---|---|
| Population | KIR3DL3 | KIR3DL1/S1 | KIR3DL2 | Frequency |
| European American¥(Total observed 234) | *003 | S1*013 | *007 | 0.051 |
| *001 | S1*013 | *007 | 0.043 | |
| *001 | L1*004 | *005 | 0.035 | |
| *001 | L1*002 | *002 | 0.028 | |
| *002 | L1*001 | *001 | 0.028 | |
| *002 | L1*005 | *001 | 0.027 | |
| *003 | L1*002 | *002 | 0.026 | |
| *013 | L1*001 | *001 | 0.020 | |
| *013 | S1*013 | *007 | 0.020 | |
| *001 | L1*001 | *001 | 0.018 | |
| Ugandan¥(Total observed 191) | *005 | L1*001 | *001 | 0.049 |
| *002 | L1*015 | *001 | 0.031 | |
| *014 | L1*031 | *001 | 0.028 | |
| *010 | L1*017 | *023 | 0.026 | |
| *014 | L1*018 | *001 | 0.023 | |
| *011 | L1*018 | *001 | 0.020 | |
| *022 | NEG | *006 | 0.020 | |
| *009 | L1*022 | *001 | 0.019 | |
| *004 | L1*059a | - | 0.017 | |
| *003 | L1*007 | *008 | 0.017 | |
| Papuan¥(Total observed 35) | *002 | S1*013 | *007 | 0.165 |
| *014 | S1*013 | *007 | 0.142 | |
| *014 | NEG | *007 | 0.100 | |
| *003 | S1*013 | *007 | 0.100 | |
| *014 | L1*017 | *002 | 0.075 | |
| *002 | L1*017 | *002 | 0.066 | |
| *003 | L1*017 | *002 | 0.060 | |
| *009 | S1*013 | *007 | 0.051 | |
| *013 | L1*005 | *010 | 0.047 | |
| *013 | L1*017 | *002 | 0.028 | |
| Egyptian¥(Total observed 95) | *003 | L1*001 | *001 | 0.045 |
| *009 | L1*001 | *001 | 0.034 | |
| *003 | S1*013 | *007 | 0.034 | |
| *003 | L1*002 | *002 | 0.027 | |
| *003 | L1*007 | *008 | 0.027 | |
| *001 | L1*020 | *009 | 0.027 | |
| *014 | S1*013 | *007 | 0.027 | |
| *013 | L1*001 | *001 | 0.020 | |
| *027 | L1*001 | *001 | 0.020 | |
| *015 | L1*002 | *002 | 0.020 | |
| Spanish¥(Total observed 86) | *003 | L1*002 | *002 | 0.055 |
| *002 | L1*005 | *001 | 0.050 | |
| *001 | L1*001 | *001 | 0.047 | |
| *001 | L1*004 | *005 | 0.047 | |
| *003 | S1*013 | *007 | 0.046 | |
| *009 | S1*013 | *007 | 0.040 | |
| *002 | S1*013 | *007 | 0.040 | |
| *002 | L1*004 | *003 | 0.035 | |
| *002 | L1*001 | *001 | 0.031 | |
| *001 | L1*015 | *002 | 0.026 | |
| #Total Alleles | 61 | 46 | 51 | |
| $Total Allotypes | 44 | 37 | 40 | |
The number of distinct haplotypes identified by analyzing one centromeric (KIR3DL3) and two telomeric (KIR3DL1/S1 and KIR3DL2) genes;
Number of distinct alleles (including novel alleles);
Number of distinct allotypes (including novel allotypes);
L1*059 is an allele of KIR3DL1/2v, a fusion gene derived from KIR3DL1 and KIR3DL2.
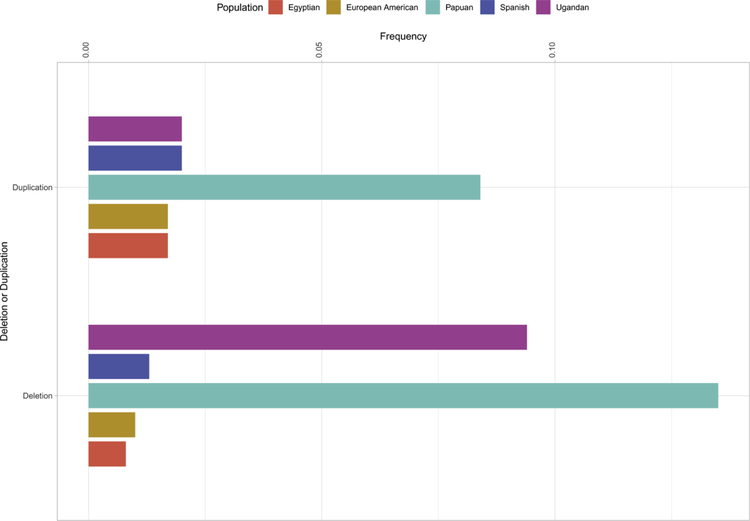
The allele frequencies of KIR receptors having three immunoglobulin (Ig)-like domains namely; KIR3DL1/S1, KIR3DL2, and KIR3DL3 as well as the duplication/deletion polymorphism of KIR3DL1/S1 detected in the 10 populations analyzed are given in Figures 1 and 2, respectively. These frequencies are deposited in the allele frequency net database (AFND) database (http://www.allelefrequencies.net/default.asp). Data were examined at the polypeptide sequence resolution, which is equivalent to the first three digits in the allele name, as described in IPD/KIR Database (https://www.ebi.ac.uk/ipd/kir/). The frequencies range from 0.1% to 48.7% for the various alleles of KIR3DL1/S1 (Figure 1A), 0.1% to 61.7% for KIR3DL2 (Figure 1B) and 0.1% to 33% for KIR3DL3 (Figure 1C) in total across all populations. The number of those alleles classified as rare (those with a frequency of <1% in any given population) was 31 for KIR3DL1/S1 (67.4%), 38 for KIR3DL2 (74.5%), and 38 for KIR3DL3 (62.3%). Thus, both common and rare alleles contributed substantially to the rich worldwide diversity of KIR. In addition to allelic variation, deletions and duplications of the entire KIR3DL1/S1 gene were also observed (Figure 2). The highest frequency of deletions and duplications were observed in the Papuan population (13.5% and 8.4%, respectively). Meanwhile, no deletions and/or duplications were observed for KIR3DL2 (except for KIR3DL1/2v, a fusion gene derived from KIR3DL1 and KIR3DL2) [52].
Figure 1: Allele frequency distribution in workshop populations for KIR receptors having three immunoglobulin (Ig)-like domains.
Figure 1A: KIR3DL1/S1 allele frequency distribution in workshop populations. Figure 1B: KIR3DL2 allele frequency distribution in workshop populations. Figure 1C: KIR3DL3 allele frequency distribution in workshop populations.
Figure 2:
KIR3DL1/S1 structural variations in workshop populations.
Haplotypic diversity of KIR3DL1/S1, KIR3DL2 and KIR3DL3 genes
Specific KIR alleles and haplotypes are associated with better education of NK cells and/or control of specific pathogens [14, 53]. Diversity in KIR haplotypes may therefore contribute to improved population survival. To estimate the extent of haplotype diversity we analyzed the five populations that were genotyped for KIR3DL3, KIR3DL1/S1 and KIR3DL2; European American, Ugandan, Papuan, Egyptian and Spanish (Table 2).ld> KIR3DL3 is located in the segment of the KIR region oriented towards the centromere of chromosome 19, and KIR3DL1/S1 and KIR3DL2 in the telomere oriented segment [4]. Since, the centromeric and telomeric KIR genes are separated by a region that contain a recombination hotspot [54, 55], we analyzed both full and telomeric-only haplotypes. We observed 503 distinct population-specific KIR3DL3~KIR3DL1/S1~KIR3DL2 haplotypes and 158 distinct population-specific KIR3DL1/S1~KIR3DL2 haplotypes. Additionally, we found six shared haplotypes, five of which, 3DL1*001~3DL2*001, 3DL1*005~3DL2*010, 3DS1*013~3DL2*007, 3DL1*015~3DL2*002, and 3DL3*003~3DS1*013~3DL2*007 were present in all five populations (Table 3), and one (3DL3*002~3DS1*013~3DL2*007) was present in all except the Egyptian population (Table 3).
Table 3:
A summary of six haplotypes shared across five populations analyzed
| A. Haplotypes of KIR3DL3, KIR3DL1/S1 and KIR3DL2 | |||||||
| Centromeric | Telomeric | Haplotype Frequency | |||||
| KIR3DL3 | KIR3DL1/S1 | KIR3DL2 | European American | Spanish | Egyptian | Ugandan | Papuan |
| *003 | S1*013 | *007 | 0.051 | 0.046 | 0.034 | 0.003 | 0.100 |
| *002 | S1*013 | *007 | 0.016 | 0.040 | - | 0.003 | 0.165 |
| B. Haplotypes of KIR3DL1/S1 and KIR3DL2 | |||||||
| Telomeric | Haplotype Frequency | ||||||
| KIR3DL1/S1 | KIR3DL2 | European American | Spanish | Egyptian | Ugandan | Papuan | |
| L1*001 | *001 | 0.115 | 0.124 | 0.174 | 0.072 | 0.005 | |
| L1*005 | *010 | 0.017 | 0.019 | 0.050 | 0.011 | 0.100 | |
| S1*013 | *007 | 0.189 | 0.138 | 0.116 | 0.020 | 0.481 | |
| L1*015 | *002 | 0.071 | 0.071 | 0.012 | 0.011 | 0.014 | |
Our analysis of allelic variation in KIR3DL3~KIR3DL1/S1~KIR3DL2 haplotypes revealed 234 distinct haplotypes in European Americans, 191 in Ugandans, 35 in Papua New Guineans, 95 in Egyptians, and 86 in the Spanish population (Table 2). The top ten most frequent KIR3DL3~KIR3DL1/S1~KIR3DL2 haplotypes are listed in Table 2. Limiting to KIR3DL1/S1~KIR3DL2 haplotypes, we identified 66 distinct haplotypes in European Americans, 81 in Ugandans, 16 in Papuans, 40 in Egyptians, and 24 in the Spanish population (Table 4). The top 10 most frequent KIR3DL1/S1~KIR3DL2 haplotypes in each population are listed in Table 4.
Table 4:
The 10 most frequent KIR3DL1/S1 and KIR3DL2 haplotypes in European American, Ugandan, Papuan, Egyptian and Spanish populations
| Telomeric | |||
|---|---|---|---|
| Population | KIR3DL1/S1 | KIR3DL2 | Frequency |
| European American¥(Total observed 66) | S1*013 | *007 | 0.189 |
| L1*001 | *001 | 0.115 | |
| L1*002 | *002 | 0.112 | |
| L1*005 | *001 | 0.079 | |
| L1*015 | *002 | 0.071 | |
| L1*004 | *003 | 0.071 | |
| L1*004 | *005 | 0.063 | |
| L1*008 | *009 | 0.040 | |
| L1*001 | *011 | 0.028 | |
| L1*020 | *009 | 0.027 | |
| Ugandan¥(Total observed 81) | L1*015 | *001 | 0.092 |
| L1*031 | *001 | 0.077 | |
| L1*001 | *001 | 0.072 | |
| L1*004 | *003 | 0.052 | |
| L1*059a | - | 0.046 | |
| L1*018 | *001 | 0.041 | |
| L1 NEG | *019 | 0.040 | |
| L1*022 | *001 | 0.036 | |
| L1*015 | *013 | 0.033 | |
| L1 NEG | *006 | 0.032 | |
| Papuan¥(Total observed 16) | S1*013 | *007 | 0.481 |
| L1*017 | *002 | 0.243 | |
| L1 NEG | *007 | 0.124 | |
| L1*005 | *010 | 0.100 | |
| L1*015 | *002 | 0.014 | |
| L1*001 | *001 | 0.005 | |
| L1*005 | *001 | 0.005 | |
| S1*013 | *010 | 0.005 | |
| L1 NEG | *010 | 0.005 | |
| L1 NEG | *070 | 0.005 | |
| Egyptian¥(Total observed 40) | L1*001 | *001 | 0.174 |
| S1*013 | *007 | 0.116 | |
| L1*002 | *002 | 0.093 | |
| L1*004 | *005 | 0.070 | |
| L1*004 | *003 | 0.052 | |
| L1*005 | *010 | 0.050 | |
| L1*008 | *009 | 0.047 | |
| S1*013 | *006 | 0.041 | |
| L1*014 | *032 | 0.035 | |
| L1*005 | *001 | 0.031 | |
| Spanish¥(Total observed 24) | L1*005 | *001 | 0.138 |
| S1*013 | *007 | 0.138 | |
| L1*001 | *001 | 0.124 | |
| L1*002 | *002 | 0.100 | |
| L1*004 | *005 | 0.100 | |
| L1*004 | *003 | 0.095 | |
| L1*015 | *002 | 0.071 | |
| L1*007 | *008 | 0.048 | |
| L1*001 | *011 | 0.038 | |
| L1*009 | *011 | 0.024 | |
The number of distinct haplotypes identified by analyzing two telomeric genes (KIR3DL1/S1 and KIR3DL2);
L1*059 is an allele of KIR3DL1/2v, a fusion gene derived from KIR3DL1 and KIR3DL2.
KIR3DL2 single nucleotide variations in exons 4 and 5
To achieve a high-depth analysis in an extremely large sample size, we focused on exons 4 and 5 from KIR3DL2, which encode for the extracellular D1 and D2 domains of the KIR3DL2 molecule, which are most likely to contact the HLA ligand directly [56]. We targeted 642,105 individuals from 35 populations and examined single nucleotide variations. We observed SNP variation in 78.5% (467 of 595) of all nucleotides that comprise these two exons. Among the observed single nucleotide substitutions, 67.4% (315 of 467) are nonsynonymous, and the reminders encode for either synonymous (31.3%) or premature stop codons (1.3%) (Table 5). Almost half of these nucleotide variations were observed in more than one individual, and the remainder in a single individual each (singletons). As expected, the number of these singletons increases with sample size (Supplementary Figure S1). Out of 375 KIR3DL2 allelic variants identified in this study, 275 were population-specific and 221 were found in the German population, which is the population with the largest sample size.
Table 5:
KIR3DL2 variation in exons 4 and 5 (D1 and D2 domains) among 642,105 individuals from 35 populations
| KIR3DL2 variation | Count |
|---|---|
| Allelic variation | |
| 1 nucleotide change | 288 |
| 2 nucleotide changes | 82 |
| 3 nucleotide changes | 5 |
| Sum of all allelic variants | 375 |
| Variant description | |
| Single nucleotide polymorphism | 467 |
| Synonymous substitutions | 146 |
| Non-synonymous substitutions | 315 |
| Premature stop codon | 6 |
| Total number of nucleotide sites | 595 |
| Number of Amino Acid changes | Codon |
| 0 | 23 |
| 1 | 77 |
| 2 | 66 |
| 3 | 24 |
| 4 | 7 |
| 5 | 1 |
| Polypeptide sites | 199 |
KIR diversity in IHIW cell lines
Data from a total of 19 IHIW cell lines from populations including European, black southern African, Warao Amerindian and Chinese were submitted for analysis. Different subsets of genes were investigated for each sample, resulting in the definition of 105 allele types in total, including 45 distinct alleles. The use of long read sequencing allowed the resolution of previous phase ambiguity over the large intron 5/6 (>5 Kbp) in KIR3DL3. In addition, novel KIR3DL3 and KIR2DL1 alleles were characterized in the cell lines AKIBA and SPO010, respectively, correcting previous allele typing [57]. Further characterization of a broader panel of IHIW cell lines using SMRT DNA sequencing is ongoing, helping to maintain the functionality of this valuable resource.
Future directions
The 17th IHIW KIR component has effectively applied the IHIW paradigm as a model for studying global KIR allelic diversity. Collaboration and multi-centric efforts were essential both to encourage the adoption of high-resolution KIR genotyping, and to generate KIR allelic data in an unprecedented scale from diverse ancestries. These data will be the basis of a more thorough examination of the KIR diversity in order to improve our understanding of KIR in human health and disease, as well as to provide a resource for immunogenetic databases for future research. The KIR allelic data gathered in this project represents the most comprehensive summary of global KIR3DL1/S1, KIR3DL3 and KIR3DL2 allelic diversity to date and provides an increased understanding of KIR allelic polymorphism and KIR evolution. The intention of the organizers is to continue this work during the 18th IHIW that will be held in Amsterdam in 2021, with the hope that more laboratories will adopt KIR allelic genotyping approaches and that a greater number of populations will be analyzed for all KIR genes.
Supplementary Material
Supplementary Figure S1: Singleton SNPs increase with sample size in the large-scale analysis of KIR3DL2 exons 4 and 5. The number of single nucleotide polymorphisms (SNPs) observed in a single sample for each population examined are shown on the y-axis, while the sample size for the population is given on the x-axis.
Acknowledgements
We wish to thank all participants in this project, those who contributed to discussions during the project meeting in Asilomar, and the organizers of the 17th IHIW. Thanks to Illumina for supplying the sequencing and capture reagents, and Kapa Biosystems for supplying some of the library preparation reagents. National Institutes of Health (NIH) grant U19NS095774 (JAH, PJN, JRO, MKM), NIH RO1 AI17892 (PP) and NIH P01 CA111412 (SGEM) supported this work. This project has received funding for KIR genotyping of the Ugandan samples from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement No. 695551); and Wellcome programme grants and enhancement awards related to the collection and study of Ugandan cohort (MR/L020041/1, 094073/Z/10/Z, 094073/Z/10/B). We are grateful to the participants in the Papua New Guinea Highlands for taking part in this study and for Dr. Willie Pomat and Dr. George Koki from the PNG Institute for Medical Research for their assistance in collecting samples associated with the Papua New Guinea cohort, and AJM was supported by a Wellcome Trust Fellowship with reference 106289/Z/14/Z. We also thank Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Science Without Borders program award, Fundação Araucária, Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) that supported the genotyping of the Brazilian samples.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Liu WR, Kim J, Nwankwo C, Ashworth LK, Arm JP: Genomic organization of the human leukocyte immunoglobulin-like receptors within the leukocyte receptor complex on chromosome 19q13.4. Immunogenetics 2000;51:659. [DOI] [PubMed] [Google Scholar]
- [2].Wende H, Colonna M, Ziegler A, Volz A: Organization of the leukocyte receptor cluster (LRC) on human chromosome 19q13.4. Mamm Genome 1999;10:154. [DOI] [PubMed] [Google Scholar]
- [3].Misra MK, Damotte V, Hollenbach JA: The immunogenetics of neurological disease. Immunology 2018;153:399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Wilson MJ, Torkar M, Haude A, Milne S, Jones T, Sheer D et al. : Plasticity in the organization and sequences of human KIR/ILT gene families. Proc Natl Acad Sci U S A 2000;97:4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Colonna M, Moretta A, Vely F, Vivier E: A high-resolution view of NK-cell receptors: structure and function. Immunol Today 2000;21:428. [DOI] [PubMed] [Google Scholar]
- [6].Bjorkstrom NK, Beziat V, Cichocki F, Liu LL, Levine J, Larsson S et al. : CD8 T cells express randomly selected KIRs with distinct specificities compared with NK cells. Blood 2012;120:3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Robinson J, Mistry K, McWilliam H, Lopez R, Marsh SG: IPD--the Immuno Polymorphism Database. Nucleic Acids Res 2010;38:D863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Parham P, Norman PJ, Abi-Rached L, Guethlein LA: Human-specific evolution of killer cell immunoglobulin-like receptor recognition of major histocompatibility complex class I molecules. Philos Trans R Soc Lond B Biol Sci 2012;367:800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Knapp S, Warshow U, Hegazy D, Brackenbury L, Guha IN, Fowell A et al. : Consistent beneficial effects of killer cell immunoglobulin-like receptor 2DL3 and group 1 human leukocyte antigen-C following exposure to hepatitis C virus. Hepatology 2010;51:1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Carrillo-Bustamante P, Kesmir C, de Boer RJ: Virus encoded MHC-like decoys diversify the inhibitory KIR repertoire. PLoS Comput Biol 2013;9:e1003264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Hollenbach JA, Ladner MB, Saeteurn K, Taylor KD, Mei L, Haritunians T et al. : Susceptibility to Crohn’s disease is mediated by KIR2DL2/KIR2DL3 heterozygosity and the HLA-C ligand. Immunogenetics 2009;61:663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Augusto DG, Lobo-Alves SC, Melo MF, Pereira NF, Petzl-Erler ML: Activating KIR and HLA Bw4 ligands are associated to decreased susceptibility to pemphigus foliaceus, an autoimmune blistering skin disease. PLoS One 2012;7:e39991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Martin MP, Qi Y, Gao X, Yamada E, Martin JN, Pereyra F et al. : Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat Genet 2007;39:733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Khakoo SI, Thio CL, Martin MP, Brooks CR, Gao X, Astemborski J et al. : HLA and NK cell inhibitory receptor genes in resolving hepatitis C virus infection. Science 2004;305:872. [DOI] [PubMed] [Google Scholar]
- [15].Misra MK, Prakash S, Moulik NR, Kumar A, Agrawal S: Genetic associations of killer immunoglobulin like receptors and class I human leukocyte antigens on childhood acute lymphoblastic leukemia among north Indians. Hum Immunol 2016;77:41. [DOI] [PubMed] [Google Scholar]
- [16].Boudreau JE, Giglio F, Gooley TA, Stevenson PA, Le Luduec JB, Shaffer BC et al. : KIR3DL1/ HL A-B Subtypes Govern Acute Myelogenous Leukemia Relapse After Hematopoietic Cell Transplantation. J Clin Oncol 2017;35:2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Hiby SE, Walker JJ, O’Shaughnessy KM, Redman CW, Carrington M, Trowsdale J et al. : Combinations of maternal KIR and fetal HLA-C genes influence the risk of preeclampsia and reproductive success. J Exp Med 2004;200:957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Nakimuli A, Chazara O, Hiby SE, Farrell L, Tukwasibwe S, Jayaraman J et al. : A KIR B centromeric region present in Africans but not Europeans protects pregnant women from pre-eclampsia. Proc Natl Acad Sci U S A 2015;112:845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Cooley S, Trachtenberg E, Bergemann TL, Saeteurn K, Klein J, Le CT et al. : Donors with group B KIR haplotypes improve relapse-free survival after unrelated hematopoietic cell transplantation for acute myelogenous leukemia. Blood 2009;113:726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Cooley S, Weisdorf DJ, Guethlein LA, Klein JP, Wang T, Marsh SG et al. : Donor killer cell Ig-like receptor B haplotypes, recipient HLA-C1, and HLA-C mismatch enhance the clinical benefit of unrelated transplantation for acute myelogenous leukemia. J Immunol 2014;192:4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ruggeri L, Capanni M, Urbani E, Perruccio K, Shlomchik WD, Tosti A et al. : Effectiveness of donor natural killer cell alloreactivity in mismatched hematopoietic transplants. Science 2002;295:2097. [DOI] [PubMed] [Google Scholar]
- [22].Kunert K, Seiler M, Mashreghi MF, Klippert K, Schonemann C, Neumann K et al. : KIR/HLA ligand incompatibility in kidney transplantation. Transplantation 2007;84:1527. [DOI] [PubMed] [Google Scholar]
- [23].Gonzalez-Galarza FF, Takeshita LY, Santos EJ, Kempson F, Maia MH, da Silva AL et al. : Allele frequency net 2015 update: new features for HLA epitopes, KIR and disease and HLA adverse drug reaction associations. Nucleic Acids Res 2015;43:D784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Takeshita LY, Gonzalez-Galarza FF, dos Santos EJ, Maia MH, Rahman MM, Zain SM et al. : A database for curating the associations between killer cell immunoglobulin-like receptors and diseases in worldwide populations. Database (Oxford) 2013;2013:bat021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Middleton D, Gonzelez F: The extensive polymorphism of KIR genes. Immunology 2010;129:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Barten R, Torkar M, Haude A, Trowsdale J, Wilson MJ: Divergent and convergent evolution of NK-cell receptors. Trends Immunol 2001;22:52. [DOI] [PubMed] [Google Scholar]
- [27].Middleton D, Meenagh A, Gourraud PA: KIR haplotype content at the allele level in 77 Northern Irish families. Immunogenetics 2007;59:145. [DOI] [PubMed] [Google Scholar]
- [28].Gendzekhadze K, Norman PJ, Abi-Rached L, Graef T, Moesta AK, Layrisse Z et al. : Co-evolution of KIR2DL3 with HLA-C in a human population retaining minimal essential diversity of KIR and HLA class I ligands. Proc Natl Acad Sci U S A 2009;106:18692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Vierra-Green C, Roe D, Hou L, Hurley CK, Rajalingam R, Reed E et al. : Allele-level haplotype frequencies and pairwise linkage disequilibrium for 14 KIR loci in 506 European-American individuals. PLoS One 2012;7:e47491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Norman PJ, Hollenbach JA, Nemat-Gorgani N, Guethlein LA, Hilton HG, Pando MJ et al. : Co-evolution of human leukocyte antigen (HLA) class I ligands with killer-cell immunoglobulin-like receptors (KIR) in a genetically diverse population of sub-Saharan Africans. PLoS Genet 2013;9:e1003938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Nemat-Gorgani N, Edinur HA, Hollenbach JA, Traherne JA, Dunn PP, Chambers GK et al. : KIR diversity in Maori and Polynesians: populations in which HLA-B is not a significant KIR ligand. Immunogenetics 2014;66:597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Nemat-Gorgani N, Hilton HG, Henn BM, Lin M, Gignoux CR, Myrick JW et al. : Different Selected Mechanisms Attenuated the Inhibitory Interaction of KIR2DL1 with C2(+) HLA-C in Two Indigenous Human Populations in Southern Africa. J Immunol 2018;200:2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Hollenbach JA, Meenagh A, Sleator C, Alaez C, Bengoche M, Canossi A et al. : Report from the killer immunoglobulin-like receptor (KIR) anthropology component of the 15th International Histocompatibility Workshop: worldwide variation in the KIR loci and further evidence for the coevolution of KIR and HLA. Tissue Antigens 2010;76:9. [DOI] [PubMed] [Google Scholar]
- [34].Hollenbach JA, Augusto DG, Alaez C, Bubnova L, Fae I, Fischer G et al. : 16(th) IHIW: population global distribution of killer immunoglobulin-like receptor (KIR) and ligands. Int J Immunogenet 2013;40:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Single RM, Martin MP, Gao X, Meyer D, Yeager M, Kidd JR et al. : Global diversity and evidence for coevolution of KIR and HLA. Nat Genet 2007;39:1114. [DOI] [PubMed] [Google Scholar]
- [36].Augusto DG, Petzl-Erler ML: KIR and HLA under pressure: evidences of coevolution across worldwide populations. Hum Genet 2015;134:929. [DOI] [PubMed] [Google Scholar]
- [37].Contreras G, Alaez C, Murguia A, Garcia D, Flores H, Gorodezky C: Distribution of the killer cell immunoglobulin-like receptors in Mexican Mestizos. Tissue Antigens 2007;69 Suppl 1:125. [DOI] [PubMed] [Google Scholar]
- [38].Parham P, Norman PJ, Abi-Rached L, Guethlein LA: Variable NK cell receptors exemplified by human KIR3DL1/S1. J Immunol 2011;187:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Norman PJ, Hollenbach JA, Nemat-Gorgani N, Marin WM, Norberg SJ, Ashouri E et al. : Defining KIR and HLA Class I Genotypes at Highest Resolution via High-Throughput Sequencing. Am J Hum Genet 2016;99:375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Nakimuli A, Chazara O, Farrell L, Hiby SE, Tukwasibwe S, Knee O et al. : Killer cell immunoglobulin-like receptor (KIR) genes and their HLA-C ligands in a Ugandan population. Immunogenetics 2013;65:765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Elfishawi SM, Mossallam GI, El-Fattah RA, El-Haddad A, Kamel AM: The effect of killer cell immunoglobulin-like receptor genotype on outcome of hematopoietic stem cell transplantation from matched sibling. Hum Immunol 2017;78:684. [DOI] [PubMed] [Google Scholar]
- [42].Vilches C, Castano J, Gomez-Lozano N, Estefania E: Facilitation of KIR genotyping by a PCR-SSP method that amplifies short DNA fragments. Tissue Antigens 2007;70:415. [DOI] [PubMed] [Google Scholar]
- [43].Creary LE, Mallempati KC, Gangavarapu S, Caillier SJ, Oksenberg JR, Fernandez-Vina MA: Deconstruction of HLA-DRB1*04:01:01 and HLA-DRB1*15:01:01 class II haplotypes using next-generation sequencing in European-Americans with multiple sclerosis. Mult Scler 2018:1352458518770019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Bergstrom A, Oppenheimer SJ, Mentzer AJ, Auckland K, Robson K, Attenborough R et al. : A Neolithic expansion, but strong genetic structure, in the independent history of New Guinea. Science 2017;357:1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Augusto DG, Amorim LM, Farias TD, Petzl-Erler ML: KIR and HLA genotyping of Japanese descendants from Curitiba, a city of predominantly European ancestry from Southern Brazil. Hum Immunol 2016;77:336. [DOI] [PubMed] [Google Scholar]
- [46].Augusto DG, Piovezan BZ, Tsuneto LT, Callegari-Jacques SM, Petzl-Erler ML: KIR gene content in amerindians indicates influence of demographic factors. PLoS One 2013;8:e56755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Augusto DG, Zehnder-Alves L, Pincerati MR, Martin MP, Carrington M, Petzl-Erler ML: Diversity of the KIR gene cluster in an urban Brazilian population. Immunogenetics 2012;64:143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Lange V, Bohme I, Hofmann J, Lang K, Sauter J, Schone B et al. : Cost-efficient high-throughput HLA typing by MiSeq amplicon sequencing. BMC Genomics 2014;15:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Ihaka R, Gentleman R: R: A language for data analysis and graphics. Journal of Computational and Graphical Statistics, 1996;5:299. [Google Scholar]
- [50].Sinnwell J, Schaid D: haplo.stats: Statistical Analysis of Haplotypes with Traits and Covariates when Linkage Phase is Ambiguous. R package version 1.7.1. 2015. [Google Scholar]
- [51].Robinson J, Guethlein LA, Cereb N, Yang SY, Norman PJ, Marsh SGE et al. : Distinguishing functional polymorphism from random variation in the sequences of >10,000 HLA-A, -B and -C alleles. PLoS Genet 2017;13:e1006862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Norman PJ, Abi-Rached L, Gendzekhadze K, Hammond JA, Moesta AK, Sharma D et al. : Meiotic recombination generates rich diversity in NK cell receptor genes, alleles, and haplotypes. Genome Res 2009;19:757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Boudreau JE, Hsu KC: Natural Killer Cell Education and the Response to Infection and Cancer Therapy: Stay Tuned. Trends Immunol 2018;39:222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Vendelbosch S, de Boer M, van Leeuwen K, Pourfarzad F, Geissler J, van den Berg TK et al. : Novel insights in the genomic organization and hotspots of recombination in the human KIR locus through analysis of intergenic regions. Genes Immun 2015;16:103. [DOI] [PubMed] [Google Scholar]
- [55].Norman PJ, Cook MA, Carey BS, Carrington CV, Verity DH, Hameed K et al. : SNP haplotypes and allele frequencies show evidence for disruptive and balancing selection in the human leukocyte receptor complex. Immunogenetics 2004;56:225. [DOI] [PubMed] [Google Scholar]
- [56].Vivian JP, Duncan RC, Berry R, O’Connor GM, Reid HH, Beddoe T et al. : Killer cell immunoglobulin-like receptor 3DL1-mediated recognition of human leukocyte antigen B. Nature 2011;479:401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Bultitude WP, Gymer AW, Robinson J, Mayor NP, Marsh SGE: The novel KIR2DL1 allele, KIR2DL1*037, defined in the cell line SPO010 (IHW9036). HLA 2018;91:547. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1: Singleton SNPs increase with sample size in the large-scale analysis of KIR3DL2 exons 4 and 5. The number of single nucleotide polymorphisms (SNPs) observed in a single sample for each population examined are shown on the y-axis, while the sample size for the population is given on the x-axis.