Abstract
Mitochondria are found in a variety of shapes, from small round punctate structures to a highly interconnected web. This morphological diversity is important for function, but complicates quantification. Consequently, early quantification efforts relied on various qualitative descriptors that understandably reduce the complexity of the network leading to challenges in consistency across the field. Recent application of state-of-the-art computational tools have resulted in more quantitative approaches. This prospective highlights the implementation of MitoGraph, an open-source image analysis platform for measuring mitochondrial morphology initially optimized for use with Saccharomyces cerevisiae. Here Mitograph was assessed on five different mammalian cells types, all of which were accurately segmented by MitoGraph analysis. MitoGraph also successfully differentiated between distinct mitochondrial morphologies that ranged from entirely fragmented to hyper-elongated. General recommendations are also provided for confocal imaging of labeled mitochondria (using mito-YFP, MitoTracker dyes and immunostaining parameters). Widespread adoption of MitoGraph will help achieve a long-sought goal of consistent and reproducible quantification of mitochondrial morphology.
Keywords: Mitochondrial morphology, Mitochondrial fission, Mitochondrial fusion, Confocal microscopy, Image analysis
Graphical abstract
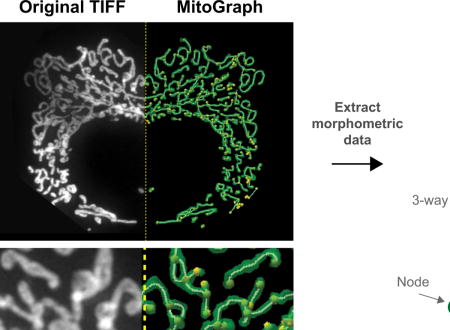
1.1 Introduction
The relationship between biological form and function is well appreciated [1–4]. One example in cell biology is illustrated by mitochondria, where alterations in mitochondrial function result in dramatic modifications to its structure and vice-versa [5–7]. The gross morphology of mitochondria is organized into an intricate network that can dynamically interconvert through frequent fission and fusion events that appear tightly regulated to control overall morphology [6,8–12]. The dynamic nature of mitochondria reflects organelle function since morphology varies with developmental stage, tissue type, and metabolic need as first noted over one hundred years ago [13] and illustrated throughout the past century [14,15], including beautiful examples of microcellular cinematography [16,17].
More recently, the genes responsible for mitochondrial fusion and fission have been identified and many belong to the superfamily of dynamin mechanoenzymes [18–23]. These are large GTPases that mediate many membrane remodeling events throughout the cell, including the mitochondria [24,25]. Mutations in both fusion and fission genes cause rare neurometabolic disorders and even neonatal lethality suggesting that mitochondrial form may impact mitochondrial function [9,26,27]. In addition, changes in mitochondrial morphology occur in many human diseases including Alzheimer’s disease [28], amyotrophic lateral sclerosis [29], Parkinson’s disease [30–34], hypertrophic cardiomyopathy [35], and diabetes [36], however in many of these cases it is unclear whether the alterations in morphology are correlative, rather than causative. These observations highlight the importance of mitochondrial morphology.
Quantification of mitochondrial morphology is challenging given the tremendous heterogeneity in mitochondrial length and degree of branching. This heterogeneity has plagued the field with diverse nomenclature and classification. For example, the plethora of terms have been used to define the mitochondrial network (fragmented, short, elongated, tubular, long, reticular, interconnected, fused, hyperfused, netted, netlike, collapsed, aggregated, etc) make it challenging to thoroughly understand mitochondrial phenotypes associated with various experimental parameters. This issue is compounded by morphological phenotypes being routinely classified by the predominant morphology within a given cell despite the heterogeneity observed. For example, it is commonplace to classify cells as containing either elongated, fragmented, or collapsed mitochondria when in reality a single cell’s mitochondrial network rarely consists of a single type of morphology. Such classifications have been useful, but belie the underlying complexity of the mitochondrial network within a single cell, making the results highly subjective and difficult to translate between labs. Recently, this problem has been addressed with the development of methods applying various image processing algorithms to more robustly quantify mitochondrial morphology (Table 1). One early method for quantification involved manually measuring mitochondrial length using image analysis programs, however, difficulty arose when measuring highly interconnected mitochondria [37]. FRAP analysis of a matrix-targeted fluorescent protein helped to report on mitochondrial connectivity [37–40], but gave little insight into the actual structural differences in a mitochondrial network. The first attempts at applying morphometric image analysis tools to mitochondria used ImageJ or Image Pro Plus 5.1 (Media Cybernetics) to dissect the mitochondrial network into individual objects and ultimately reported on the length of the mitochondria (aspect ratio) and the network characteristics (form factor) [41–43]. During the last 10 years, many other approaches have been published and can mainly be classified into two categories: 1) “Morphological” programs that automate the classification of mitochondria into individually defined categories that are often identified through machine learning strategies and 2) “Morphometric” programs that automate the measurement of individual mitochondrial parameters such as volume, length, number of nodes, etc. Many of these approaches use 2D image projections, while only a subset use 3D image stacks. A comparison of 2D to 3D image analysis found it imperative to use 3D images when examining mitochondrial fragmentation in addition to mitochondrial biomass [44].
Table 1.
Mitochondrial morphology image analysis programs
| (Input parameters | Measurements | Analysis Platform | Validated on | Ref | |
|---|---|---|---|---|---|
| Computational classification of mitochondrial shape | |||||
| mito-GFP label; acquired with Leica DM16000 or Leica SP5 confocal microscope | Used random forest algorithm to classify mitochondria into blob, donut and elongated | MATLAB; Gemident | BEAS-2B, human primary bronchial epithelial cells, A549 | [46] | |
| Computational imaging to use mitochondria as biomarker | |||||
| TOMM-20 immunostain; acquired with BX-63 upright automated fluorescence microscope equipped with 100x oil objective; z-stack 0.5μm; 2560 × 2160 pixel resolution. Used 2D image for analysis. | Used random forest algorithm to classify mitochondria into punctate, intermediate and filamentous | MATLAB | A2780, OVCA-429, A549, Caco-2, Panc- 1, H146, Jurkat, Daudi, patient derived xenograft samples | [47] | |
| Quantitative analysis of mitochondrial morphology using high content imaging, machine learning and morphological binning | |||||
| MitoTracker Deep Red label; acquired with wide-field fluorescence with offline deconvolution using GE InCell 2000 Analyzer; 40x 0.6 NA objective; pixel size 0.185μm; z-stack 1.55μm; used 3D image for analysis | Mitochondria defined in 4 subclasses (networked, rod-like, punctate, and large/round). Measured total mitochondrial area & object count, individual subclass area, average fiber length | GE INCel Developer Toolbox | 661 w photoreceptor cells | [48] | |
| Multi-parametric analysis of mitochondria | |||||
| mito-GFP label; acquired with wide-field DeltaVision RT deconvolution microscope; 63X 1.40 NA oil objective, z-stack 0.22μm; used 2D image for analysis | Random Forest classifier using supervised classification model (separate into 3 classes: networked, fragmented, swollen) | Cell-Profiler software | MCF-7 | [49] | |
| MicroP: automatic morphological subtyping | |||||
| MitoTracker Orange label; acquired with an Olympus fluorescence microscope (IX-71); 60× 1.45 NA objective; pixel (size 165nm, used 2D image for analysis | Total number/area of mitochondria. Classify mitochondria into subtypes: small globules, swollen globules, loops, straight tubules, twisted tubules, branched tubules. | MATLAB | CHO-K1 | [50] | |
| Mitochondrial shape analysis with ImageJ | |||||
| mito-dsRED label; acquired with Fluoview 300 or 500 confocal 100x (1.3 NA)/60x objective (1.4 NA). | Area, major/minor axis of best fitting ellipse, aspect ratio, number ofmitochondria | FIJI | CV1 | [43] | |
| TMRM labeled/Cytochrome c oxidase immunostain; acquired with LSM 510 LSCM or Leica epifluorescence microscope; used 2D image for analysis | Mitochondrial area, perimeter and major/minor axis of the bounding ellipse. Morphometric parameters: aspect ratio & form factor | FIJI | HeLa, PC12, hippocampal neurons | [42,51,52] | |
| Momito: novel algorithm to identify stress-induced alterations in mitochondrial connectivity | |||||
| TOM20 or mtHSP70 immunostain; acquired with Leica TSC SP8 confocal microscope; 63x/1.4 NA oil objective; used 2D image for analysis | Identifies mitochondrial tubules, junctions and endpoints. Measures mitochondrial length. | ImageJ & Momito algorithm | U2OS, WT and OPA1 KO | [53] | |
| MiNa: simple ImageJ macro to analyzing mitochondrial morphology | |||||
| mito-mEFP label; acquired on Carl Zeiss Axio Observer.Z1 inverted epifluorescence microscope; 63 × 1.4 NA oil objective; z-stack 0.25-0.3μm; used 2D image for analysis | Mitochondrial footprint, branch length (mean, median, standard deviation), number of branches (individual, networks, mean, median, standard deviation) | FIJI | SH-SY5Y, C2C12, MEF | [54] | |
| Mytoe: automatic analysis of mitochondrial dynamics | |||||
| mito-DsRED2 label; acquired on a Nikon Eclipse Ti with a Wallac-Perkin Ember Ultraview spinning disk confocal system 100x objective; used 3D image for analysis | Length, thickness, turtuosity, intensity, speed, direction, wiggle ratio, distance to centroid/nuclear membrane/cell membrane, orientation relative to x-axis/centroid, number of branches, total area, degree clustering, colocalization | PC stand alone application or MATLAB | U2OS | [55] | |
| MitoGraph: quantifying mitochondrial content | |||||
| mito-GFP/mitodsRed2 label; acquired on a Nikon Eclipse Ti chassis with a Yokogawa CSUX spinning-disk head; 100x 1.49 NA TIRF objective; 0.056 μm pixel size; Z-stack 0.2 μm; used 3D image for analysis | Total cell measurements (volume, total length, average width), individual mitochondrial measurements (nodes, edges, length), network characteristics | C++ program Visualization Toolkit (VTK)) | S. cerevisae | [45,56] | |
| MitoMap: quantification of the mitochondrial network | |||||
| mito-LOC label; acquired with Deltavision 3D-SIM OMX system; 100× 1.4 NA oil objective; z-stack 0.125μm; used 3D image for analysis | Mean, sum, volume, surface area, compactness, distribution isotropy, isoperimetric quotient, sphericity, SA:V, radius variance | FIJI | S. cerevisae | [57] | |
| Mitochondrial morphological features associated with fission and fusion events | |||||
| mito-EYFP label; acquired with Nikon Ti Eclipse fluorescent microscope; 60x oil objective | Area, extent, solidity, perimeter, eccentricity and Euler number | MATLAB | U2OS | [58] | |
| Simultaneous quantitative measurement and automated analysis of mitochondrial morphology | |||||
| Rhodamine 123 label; acquired with a Nikon Diaphot inverted microscope; 40× 1.4 NA oil objective; image size 512×480 pixels | Area, perimeter, max/min radius, radius ration, max/min diameter, length, width, aspect ratio, form factor, number | Image Pro Plus 5.1 | Healthy fibroblasts | [41] | |
| Multi-plexed high-content analysis of mitochondrial morphofunction | |||||
| TMRM labeled; acquired with a BD Pathway 855 system for automated wide field image acquisition equipped; 40× objective pixel size 210.5 μm × 160.4 μm | Aspect ratio, area, density, axis minor, diameter, radius, perimeter, roundness, length. width, count, box, feret, density and margination | Image Pro Plus & MATLAB | patient human skin fibroblasts | [59] | |
| Automated quantification and integrative analysis of 2D/3D mitochondria | |||||
| Mito-GFP label; acquired with LSM 510; 63x oil objective (1.40 NA); pixel size 0.0845 μm; z-step 0.364 μm; used 2D & 3D images for analysis | 2D/3D shape analysis (count; area/volume; perimeter/surface area; formfactor/sphericity factor), 2D/3D network analysis (branch: count, points, length, diameter & volume); 2D/3D Network - Shape analysis | Image-Pro Plu software | HUVEC | [44] | |
In this prospective, we detail one such method, MitoGraph, which is among the programs that uses 3D images to measure individual mitochondrial morphometric characteristics in addition to the overall volume of the total mitochondrial network. MitoGraph was initially developed for quantifying mitochondrial amount in budding yeast cells [45]. More recently, MitoGraph has been validated to perform graph theory-based quantitative analysis of mitochondrial networks, which provides information about mitochondrial volume, total length, degree of branching, and other network characteristics (paper under revision). Here, we first provide recommendations for best practices for imaging mitochondria in both live and fixed cells and offer detailed instructions on the application of MitoGraph to fixed mammalian cells. MitoGraph successfully quantified changes in mitochondrial morphology including enhanced mitochondrial fragmentation due to treatment with a chemical uncoupler and mitochondrial hyper-elongation due to genetic ablation of a key fission protein. Furthermore, we demonstrate the utility of this software by applying MitoGraph analysis to four additional cell types. We anticipate that the application of MitoGraph (or other quantitative methods) to a wide variety of cell types will ultimately lead to a deeper understanding of the relationship between mitochondrial form and function.
1.2 Method
Cell culture
HMEC-1 cells (human microvascular endothelial cells; ATCC) were cultured in MCDB-131 medium supplemented with 10 ng/mL EGF (Gibco; Thermo Fisher Scientific), 1 μg/mL Hydrocortisone (Sigma), 10 nM Glutamine (Gibco; Thermo Fisher Scientific), 10% FBS (Gemini) and 1mM HEPES(Thermo Fisher Scientific). C2C12 (mouse myoblast; ATCC) cells were cultured in DMEM (Gibco; Thermo Fisher Scientific) supplemented with 10% FBS (Gemini). HEK293 and 293T cells (generous gift from Dr. J. Park; MCW) were cultured in high glucose DMEM (Gibco; Thermo Fisher Scientific) supplemented with 10% bovine growth serum (Hyclone; GE Healthcare). C6 (rat glial cells; generous gift from Dr. A. Beyer; MCW) were cultured in DMEM medium supplemented with 10% FBS (Gibco; Thermo Fisher Scientific). H1299 cells (human non-small cell lung carcinoma cell line derived from the lymph node; generous gift from Dr. A. Beyer; MCW) were cultured in RPMI-1640 supplemented with 10% FBS (Gibco; Thermo Fisher Scientific).
Transfection
The manufacturer’s protocol for Avalanche-Omni (EZ Biosystems) was followed. HMEC-1 cells (175,000) were plated in a 35-mm imaging dish using medium lacking antibiotics. 24 hrs post plating the cells were prepared for transfection. 2.5μg of total DNA (0.3μg - 2.5μg Mito-YFP and 2.2μg - 0μg pcDNA3.1(-)) was added to 250μL of Opti-MEM and mixed by briefly vortexing. The vial of Avalanche-Omni was briefly vortexed and 3.5μL added to the DNA:Opti-MEM mixture and immediately vortexed for 5 sec. The complexes were incubated at RT for 15 min and added dropwise into each well. After 2 hrs, the transfection medium was replaced with warm cell culture medium and the cells were incubated overnight for 14-24 hrs. The cells were then imaged live or fixed in 4% PFA. Images of the mitochondria were then collected.
Immunofluorescence
Cells were plated on either an imaging dish (MatTek; P35G-1.5-14-C)) or a clean and sterilized No. 1.5 cover glass placed in a 6-well tissue culture dish. Once the cells achieved 70-80% confluency, the medium was aspirated and replaced with pre-warmed to 37°C PBS/4% paraformaldehyde (4% PFA: Electron Microscopy Sciences). The 4% PFA fixative was added quickly to the side of the culture dish to minimize dislodging of cells, which were incubated with gently shaking at room temperature for 20-25 minutes. Fixative was removed and replaced with PBS. Cells were stored at 4°C until ready for immunostaining. Following fixation, the cells were: washed once in PBS, permeabilized by incubating with PBS/0.15% Triton X-100 for 15 min, washed twice in PBS, incubated with 5% normal goat serum/primary antibody (1:500 Rb anti-Tom20; Santa Cruz) for 2 hr at RT, washed three times in PBS/0.05% TWEEN 20, incubated with PBS/secondary antibody for 1 hr at RT (1:500 anti-Rb Alexa 568; Invitrogen), washed 2X in PBS/0.05% TWEEN 20, once with PBS and rinsed in water prior to mounting. The coverslips were then inverted and mounted on glass slides in mounting medium (50 mM Tris pH 9.0, 45% glycerol (v/v) containing 2 mg/ml of the anti-fade reagent p-phenylenediamine (Sigma)).
MitoTracker staining
Using DMSO, the MitoTracker probes were diluted to 1mM. Cells were incubated with 25-200 nM of either MitoTracker Red CMXRos (Ex 579nm; Em 599nm), Orange CMTMRos (Ex 554nm; Em 576nm) or Deep Red FM (Ex 644nm; Em 665nm) for 15 min. Following incubation, cells were washed 3 times in pre-warmed, equilibrated medium (note the manufacture protocol does not mention these extra washes, but if you skip this step you will end up with more unsavory background), and either imaged live or fixed in 4% PFA . Note, all three MitoTracker probes assessed were retained to some degree post fixation (see section 1.3.1.3). If the end goal is to compare membrane potential levels between samples, it is imperative to ensure consistent image settings between samples. If using an oil objective, use fresh oil each time a slide is switched to avoid any loss in intensity due to lack of oil. We recommend comparing any results against live cells TMRM staining to ensure accuracy of the measurements.
Microscopy
The images were acquired using a spinning disk confocal (Olympus Ix83 DSU) microscope (DSU in place unless otherwise noted), equipped with a manual magnification changer deck module and a Hamamatsu ImagEM X2 EM-CCD Digital Camera (C9100-23B). Z-slices were set to 0.2 micron unless otherwise noted. Cells were imaged using either a 100x oil objective (UPlanSApo 1.4 NA oil; 0.17 mm; FN26.5) with the magnification changer set to 1x zoom (pixel size = 0.1600 μm) or a 60x silicon objective (UPlanSApo 1.30 NA sil; 0.15-0.19 mm, FN22) with the magnification changer set to 1.6x zoom (pixel size = 0.1667 μm) or 1x zoom (pixel size = 0.2667 μm).
Post-image processing
Prior to MitoGraph analysis, it is often useful to view the full mitochondrial network, which typically span multiple z-planes and require the creation of maximum intensity z-projections (using FIJI ((https://fiji.sc); Image → Stacks → Z Project)). Note all images in the figures presented here (unless otherwise noted) are z-projections. If imaging multiple colors, avoid red/green combinations that can be challenging for individuals with color blindness to discern. FIJI now offers an easy way to adjust your images: starting with an RGB image replace the red with magenta using the following FIJI menu commands: Image→ Color → Replace red with magenta. FIJI also has the capability to simulate color blindness to ensure your audience can visualize the data properly. To facilitate image to image comparison, we typically combine individual maximum intensity z-projection images into a single stack using FIJI (Image → Stacks → Images to Stack), which facilitates the adjustment of brightness and contrast ensuring all images in the stack are equally adjusted. See section 1.3.2.1 below for a helpful custom FIJI macro that can be used to create the stacked images in bulk. For the MitoGraph analysis we used stacks of z-projection images to assist in outlining the cells, but used the raw 3D (newly cropped) stacks for subsequent analysis.
MitoGraph node validation
MitoGraph first segments the input image into a binary file containing mitochondria in white and the background in black. Mitograph then creates a skeletonized representation of the mitochondrial network that is subdivided into discrete components, referred to here as edges, that are joined at nodes (see Supplemental Figure 3 for a visual representation). Nodes can occur at termini of the mitochondrial reticulum and branch points between adjoining mitochondria. Nodes generated by MitoGraph can be improperly assigned due to under- or over-segmentation of the initial input image. Under-segmentation of continuous mitochondrial tubules generates fragmented pieces of mitochondria and false nodes at the extremities of these ill-defined fragments. Over-segmentation creates spurious connections (‘internal’ branching nodes) between mitochondrial tubules that are close to each other. While under-segmentation is related to areas of dim signal, problems of over-segmentation are, in general, related to mitochondrial tubules that are challenging to clearly resolve given the optical resolution of the acquired images. For validation of nodes, of particular interest are the false nodes along the z-direction (an overlap error), which typically has the lowest optical resolution. This overlap error mainly happens when two mitochondrial tubules lying along xy-plane are separated by a distance smaller than the resolution limit in the z direction and appear to overlap each other and may, or may not, represent a real branch point. This specific type of overlap error does not occur in wild-type budding yeast because these mitochondria are constrained to the cell cortex [60], preventing tubule overlap. Therefore MitoGraph 3.0 was not optimized nor validated for this type of error (paper under revision). For this reason, we decided to quantify the number of false nodes in the experiments presented here and specifically included the fraction of these false nodes caused by this type of overlap error. In order to calculate this error rate, fifteen 5μm × 5μm regions were randomly selected from each cell stack and analyzed. For each region, we counted: 1) the total number of nodes, 2) how many of these nodes appeared false based on visual inspection of the original confocal image, and 3) the fraction of false nodes that are caused by overlapping mitochondrial tubules. Only nodes of visually discernible regions of mitochondria were taken into account. Results shown in Table 2 show the total error rate of ~7% in node detection in mammalian cells (compared to ~5% in yeast cells) is mostly due to overlapping mitochondrial tubules.
Table 2.
Error rate of MitoGraph nodes detection in different types of mammalian cells.
1.3 Results
1.3.1 Imaging the Mitochondrial Network
The mitochondrial network is in constant flux, frequently undergoing fission and fusion events as it responds to cellular demands. During cellular stress, such as inner membrane depolarization, apoptosis or mitophagy, the mitochondria undergo dramatic fragmentation [6,61–64]. Given this tight coupling to cellular stress, imaging mitochondria requires minimizing cellular stress to avoid introduction of unwanted artifacts that may confound analyses. Here we describe recommendations to reduce artifacts when visualizing mitochondria by: immunostaining for a mitochondrial protein, expression of mitochondrial targeted fluorescent probes, and treatment with potentiometric, fluorescent dyes. We recommend supplementing the information presented here with previous thorough methods on mitochondrial imaging [38,65–67] as well as general recommendations for fluorescent imaging [68,69].
1.3.1.1 Immunostaining
Cells should first be plated on a surface compatible with the microscope objectives, typically 60x or 100x. Many microscope objectives for biological specimens require a No. 1.5 cover glass and deviation from that may induce spherical aberrations, although some objectives have a correction collar allowing other cover glass thickness to be used. If the cell type permits it, the cell density should be optimized to avoid crowding. Readily detectable single cells with clear boundaries between adjacent mitochondria facilitates image cropping and subsequent individual cell measurements. Once 50-70% confluency is reached, the cells are fixed and immunostained for a mitochondrial protein ensuring use of a robust antibody. Image the cells keeping in mind the higher the resolution, the easier it will be to resolve subtle features of the mitochondrial network. The use of a confocal microscope helps to reduce out of focus light (see Figure 1 DSU out (widefield) versus DSU in (spinning disk confocal mode)). If a confocal is not available, then deconvolution methods can enhance the xy resolution and reduce the hazy appearance from out of focus light [70] (see supplemental Figure 1A and 1D; original versus deconvolved).
Figure 1. Mitochondrial imaging parameters.
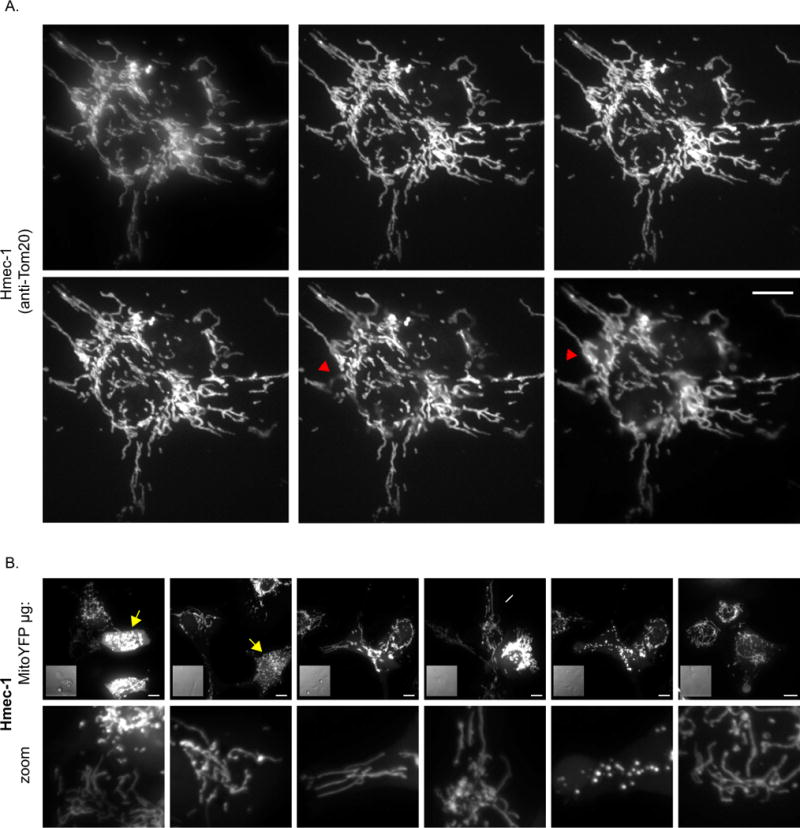
A) An HMEC-1 cell immunostained for Tom20 was imaged using the indicated z-spacing parameters, keeping all other settings constant. The top left cell was imaged with the DSU out (widefield mode), while the remainder of the cells were imaged with the DSU in place (confocal mode). Red arrowhead denotes out of focus mitochondria. B) HMEC-1 cells transiently expressing Mito-YFP. The cells were transfected using a constant total amount of DNA, but varying the amount Mito-YFP and filler DNA (pcDNA 3.1(-)). The first five images were collected using live cells, while the far-right image derives from cells fixed in 4% PFA, following the live cell image session. The 0.6μg condition was treated with 10μM CCCP and the same cells were imaged 15-20 min after the addition of CCCP. Note each of the control conditions contain normal elongated mitochondria and the fraction of fragmented mitochondria decrease as Mito-YFP is decreased. Yellow arrow denotes fragmented/clumped mitochondria; white arrow denotes normal elongated mitochondria. Scale bar represents 10μm.
Mitochondria often span the entire thickness of the cell and therefore sufficient z-stack spacing is needed to visualize the full depth and breadth of the mitochondrial network. In comparison to small round cells, the mitochondrial network of large flat cells are easier to visualize and require less time to image due to fewer z-stacks. Here we use HMEC-1 cells which are relatively flat, immortalized human microvascular endothelial cells grown on 2D cover glass. Many microscope systems recommend an optimal z-spacing that is objective specific, for example the 60x objective used here has an optimal z-spacing of 0.2μm. For HMEC-1 cells, imaging at 0.2, 0.5, 1, and 2 μm z-spacing illustrate that 0.2 - 1μm spacing is sufficient to image the entire network, while 2μm spacing – or opting to image without z-stacks – results in substantial mitochondrial regions out of focus (Figure 1). Therefore, we recommend when routinely imaging mitochondria with a 60x objective to use a minimum z-spacing 1μm, and if acquiring publication quality images, 0.2 - 0.5 μm z-spacing is ideal. If the downstream analysis will involve deconvolution (to either confocal or widefield images), then ensure the use of fine z-space spacing (0.1 - 0.2μm) and also collect a few slices above and below the last detectable fluorescence to capture out of focus light, which is helpful for proper deconvolution.
1.3.1.2 Transfection of mitochondrial markers
The past several decades have been an exciting time for microscopists with the ever-growing toolbox of fluorescent probes for examining cellular events in real time. Thanks to the use of these probes, the mitochondrial network has been captured as a function of time to highlight their dynamic nature [9,64,71]. Probes such as mito-YFP have facilitated the observation of mitochondrial fission and fusion events, as well as the connectivity of the mitochondrial network through FRAP analysis [37–40]. Photoactivatable probes, such as mito-PAGFP, have been used extensively to quantify mitochondrial fusion rates and assess the mitochondrial network [72–76]. Other fluorescent proteins boast unique properties that allow them to act as biosensors for various mitochondrial functions (see [77,78] for a recent reviews). Thanks to the generous donation by the late Mike Davidson, a champion microscopist, Addgene has a vast array of fluorescent proteins that are targeted to the mitochondria. Table 3 displays the diverse array of mitochondrial probes available, including those that are photoactivatable, photoconvertible as well as those that can be used as tools to understanding physiological processes such as mitophagy. Most of these probes target a fluorescent protein to the mitochondria by expressing a mitochondrial targeting sequence from the cytochrome c oxidase subunit 8a [79–81]. Plasmids encoding these proteins typically use the CMV promoter and can result in high expression levels leading to spurious fragmentation and clumping of the mitochondria and, even at low expression levels, cause swelling of the mitochondria (Fig 1B). Accordingly, transfection conditions need to be optimized. Lowering the total amount of mito-YFP plasmid while maintaining a constant total DNA concentration for transfection reduces unwanted fragmentation and clumping of mitochondria (Figure 1B). For some transfection reagents, reducing the amount mito-YFP, and not using so-called “filler” DNA may reduce overall transfection efficiency.
Table 3.
Mitochondrial Targeted Fluorescent Proteins
| Mitochondrial T argeted Fluorescent Protein | Excitation | Emission | Addgene # | Ref | |
|---|---|---|---|---|---|
| Fluorescent probes used to visualize mitochondria | |||||
| Blue fluorescent proteins | |||||
| mito-eBFP2 | 383 | 448 | 55248 | [82] | |
| mito-tagBFP | 402 | 457 | 49151 | [83] | |
| Cyan fluorescent proteins | |||||
| mito-eCFP | 433 | 475 | 55345 | * | |
| mito-mCerulean | 433 | 475 | 55384 | * | |
| mito-mCerulean3 | 433 | 475 | 55433 | [84] | |
| Green fluorescent proteins | |||||
| mito-mEmerald | 487 | 509 | 54160 | [85] | |
| mito-Clover | 505 | 515 | 56529 | [86] | |
| TOMM20-mEGFP | 488 | 507 | 87423 | [87] | |
| Yellow fluorescent proteins | |||||
| mito-YFP | 513 | 527 | 56596 | [88] | |
| mito-YPet | 513 | 527 | 56637 | * | |
| mito-mV enus | 515 | 528 | 56619 | * | |
| mito-mCitrine | 516 | 529 | 56563 | * | |
| Orange fluorescent proteins | |||||
| mito-mKO | 548 | 561 | 57848 | [88] | |
| mito-mKOet | 548 | 561 | 57931 | ||
| mito- mOrange2 | 549 | 565 | 57969 | [89] | |
| mito-mKO2 | 551 | 565 | 57886 | ||
| Red fluorescent proteins | |||||
| mito-tdTurboRFP | 553 | 576 | 58062 | * | |
| mito-tdTomato | 554 | 581 | 58115 | [90] | |
| mito-mT agRFP-T | 555 | 584 | 58023 | [91] | |
| mito-mT agRFP-T2 | 555 | 584 | 58041 | * | |
| mito-DsRed2 | 558 | 583 | 55838 | * | |
| mito-mRuby | 558 | 605 | 55874 | * | |
| mito-mRuby2 | 559 | 600 | 55905 | [86] | |
| mito-mStrawberry | 574 | 596 | 55941 | * | |
| mito-mCherry | 587 | 610 | 55102 | [92] | |
| Far-red fluorescent proteins | |||||
| mito-HcRed1 | 588 | 618 | 55842 | * | |
| mito-mPlum | 590 | 649 | 55988 | [91] | |
| mito- mRaspberry | 598 | 625 | 55931 | ||
| Infrared fluorescent protein | |||||
| mito-mIFP | 683 | 704 | 56231 | [93] | |
| Photo-reversible fluorescent proteins | |||||
| mito-iLOV ~ Photo-reversible fluorescent protein that outperforms eGFP | 450 | 495 | 56519 | * | |
| mito-Dronpa ~ Reversibly switchable photoactivatable fluorescent protein; switched off by 488nm; reversed by weak 405 nm light | 503 | 518 | 57288 | [94] | |
| Photo-convertible or photo-activatable tools for monitoring dynamics events | |||||
| mito-PAGFP ~ Fluorescence activated after illumination with 405 nm light. Used extensively to monitor mitochondrial fusion. | 504 | 517 | 23348 | [73] | |
| mito-PA-mCherry1 ~ Fluorescence activated after illumination with 405 nm light. | 564 | 595 | 57189 | [95] | |
| mito-PS-CFP2 ~ Irreversible photo-conversion from a cyan to a green in response to 405 nm light | 400 490 | 468/511 | 57240 | ||
| mito-mini-SOG ~ Versatile label for correlated light and electron microscopy | 448 473 | 500/528 | 57773 | [96] | |
| mito-PA-mCherry-mini-SOG ~ Versatile label for correlated light and electron microscopy | 448 473 | 500/528 | 57795 | [96] | |
| mito-dendra2 | Photo-conversion from green to red in response to 405 nm light | 490 553 | 507/573 | 55796 | [76] |
| mito-mKikGR | 507 583 | 517/593 | 57327 | * | |
| mito-mEos2 | 505 569 | 516/581 | 57401 | [97] | |
| mito-Eos | 505 569 | 516/581 | 57566 | * | |
| mito-tdEos | 505 569 | 516/581 | 57644 | [98] | |
| Mitochondrial sensors & tools | |||||
| mito-Timer ~ Fluorescence shifts over time from green to red as the protein matures. Used to measure mitochondrial content, structure, oxidative stress and damage | 483 558 | 500/583 | 52659 | [99,100] | |
| pTRE-Tight-mito-Timer ~ Timer protein targeted to the mitochondria under a Tet-inducible promoter | 483/558 | 500/583 | 50547 | [101] | |
| mito-mKeima-Red ~ pH sensor: excitation spectrum sensitive to pH. 440 nm = neutral environment; 586 nm = acidic environment. Used to measure mitophagy. | 440 | 620 | 56018 | [100] * | |
| mito-QC (mCherry-GFP-Fis1101-152) ~ pH sensor: low pH in the lysosome quenches GFP, mCherry remains stable. Used to monitor mitophagy. Unlike mKeima, probe is fixable. | 488/587 | 507/610 | n/a | [100,102,103] | |
| pLPCX mito Grx1 -roGFP2 ~ redox sensor: Redox sensitive probe that reports on cellular glutathione redox potential | 408/488 | 500-530 | 64977 | [104] | |
| mito-SypHer ~ pH sensitive circularly permuted YFP | 488 | 500 - 550 | 48251 | [105,106] | |
| dMitoHyper ~ hydrogen peroxide sensitive circularly permuted YFP | 420/500 | 516 | Evrogen | [107] | |
| mito-CEPIA2 ~ mitochondrial calcium indicator (Kd = 0.16 μM) | 488 | 500-550 | 58218 | [108] | |
| mito-CEPIA3 ~ mitochondrial calcium indicator (Kd = 11 μM) | 58219 | ||||
| mito-CEPIA4 ~ mitochondrial calcium indicator (Kd = 59 μM) | 58220 | ||||
| pKillerRed-dMito ~ damage inducing: Light-induced production of ROS due to exposure to 540-590 nm light | 585 | 610 | Evrogen | [109] | |
Davidson lab unpublished
Further optimization is achieved by modifying the exposure time to the transfection reagent and altering overall transfection time; these modifications can help minimize transfection induced toxicity and also help minimize over expression artifacts. While optimizing transient transfection using mito-YFP is helpful at reducing unwanted side-effects, creating a stable cell line using lentiviral or retroviral methods, although time consuming, is the best option to reduce the possibility of the mitochondrial fluorescent protein interfering with mitochondrial morphology [38]. Another method to ensure low expression uses the CRISPR/Cas9 technology to insert fluorescent protein tags into endogenous loci [87]. The Allen Institute deposited a variety of these constructs at Addgene, including the mitochondrial protein Tom20 tagged with mEGFP (Table 3).
One benefit from working with cells expressing mitochondrial targeted fluorescent proteins is the versatility to use either fixed or live cells. Note that mitochondrial structure is visible in both control and depolarized (CCCP treated) cells (yellow arrow; Figure 1B), unlike many potentiometric dyes used to visualize the mitochondrial structure in live cells (see section below). As mentioned above, the mitochondrial network is exquisitely sensitive to changes in the cell environment and, as such, care needs to be taken in maintaining the cell environment, especially regarding the pH and temperature of the medium. If possible use a heated stage equipped with 5% CO2. If CO2 isn’t available including 10-25 mM HEPES in the medium can extend cell survival time outside a CO2 controlled environment (for thorough review on Live Cell Imaging see [110]). Alternatively, use a medium that does not require CO2 for buffering such as CO2 independent or Leibovitz L-15 medium (ThermoFisher Scientific), although it is essential to test cells to ensure the alternative medium is tolerated. Make sure to use phenol-red free medium for all live cell imaging to reduce background fluorescence, especially when imaging cyan fluorescent proteins (CFPs) as phenol red fluoresces at excitation wavelengths used for CFPs (~440 nm) (note that phenol-red can also impact imaging of green fluorescent protein (GFP) cell lines as well) [110]. If planning an extended live cell imaging session, pre-warm all microscope components for 30 min - 1 hr prior to imaging to help minimize focus drift that can happen during imaging. When adding drugs to live cells, pre-equilibrate medium used for imaging in the incubator for at least 30 min - 1 hr prior to use and add the drugs in this medium at 2X their final concentration; this ultimately can help with consistency between samples. Minimize exposure time, z-slices and excessive time points to avoid oversampling and reduce phototoxicity. Mitochondria are sensitive to imaging parameters and can lose membrane potential and fragment depending on a number of factors including light intensity, duration, and the photophysical properties of the probe [38,74,109,111,112]. In some cases, excessive imaging can cause cell retraction and even rupture. If problems arise using traditional laser scanning confocal methods, consider alternative imaging approaches such as spinning disc confocal, wide-field coupled with deconvolution, two-photon, total internal reflection or light sheet microscopy which may help reduce cellular stress and the mitochondrial fragmentation that may occur when imaging live cells [113]. Especially promising is the newest technology used to image mitochondria, lattice light-sheet microscopy, which allows faster image collection and reduces photodamage [114].
1.3.1.3 Potentiometric Dyes
A large variety of dyes are used to illuminate mitochondria, many of which are cationic and taken up into respiring organelles based on the electrical potential across the inner membrane. Among these dyes are compounds such as TMRM, TMRE, Rho123, JC-1 and DiO6 that can report on membrane potential. A thorough description of each of these probes is beyond the scope of this paper (see ref [115–124]), but limitations of JC-1 are well documented and include issues with the fluorescence deriving from sources (i.e. H2O2, pH, nuclei [125–127]) other than mitochondrial membrane potential [124,128]. Aside from monitoring membrane potential, dyes have been designed to report on mitochondrial-derived superoxide [106,129]. One such dye, MitoSox, is a frequently used superoxide indicator, but at higher concentrations leads to minor off target nuclear staining [130,131] and inhibition of mitochondrial function [131–133]. Furthermore, MitoSox fluorescence is not a reliable indicator of the amount of mitochondrial superoxide for two reasons. First, MitoSox is metabolized into two fluorescent compounds, only one of which reports on mitochondrial superoxide. Second, MitoSox can intercalate mtDNA resulting in an ~20-fold higher fluorescence that is independent of redox status [134]. Fortunately, newer mitochondrial superoxide probes, including MitoB [135,136] and MitoNeoD [137], have been developed to overcome these shortcomings. Another mitochondrial dye, which monitors temperature gradients (mito thermo yellow) has been used to suggest the provocative idea that mitochondria may be physiologically maintained around 50°C [138,139].
TMRE or TMRM (~15-30 nM for most cell types) is recommended for accurate measurement of the mitochondrial membrane potential, however, only in live cells as the dyes are released upon fixation [124]. Since not all investigators have ready access to live cell imaging instrumentation, the focus of this section is on mitochondrial morphology of fixed cells using a subset of the MitoTracker family of dyes designed to be well retained post-fixation: MitoTracker Orange CMTMRos, MitoTracker Red CMXRos, and MitoTracker Deep Red FM (Thermo Fisher Scientific) [140]. These probes contain a mildly thiol reactive chloromethyl moiety that reacts with free thiols on proteins to form aldehyde-fixable conjugates. The recommended concentration range is 25 - 200 nM and a comparison under normal live cell conditions shows that 25 nM of each probe produces mitochondrial images of high quality (Figure 2A). By contrast, treatment with 200 nM of each probe increases the population of HMEC-1 cells with swollen and fragmented mitochondria (see yellow and red arrows; Figure 2A). 200nM of MitoTracker Orange CMTMRos also gave rise to non-mitochondrial fluorescence that appeared localized within the nucleus (orange arrow; Figure 2A). The lower concentrations of MitoTracker probes were also more sensitive at revealing the drastic loss in membrane potential upon the exposure to the chemical uncoupler FCCP, which causes depolarization across the inner membrane (compare panels ii to iii in Figure 2A).
Figure 2. Mitochondrial morphology as assessed by Mitotracker Red CMXRos, Orange CMTMRos and Deep Red FM staining.
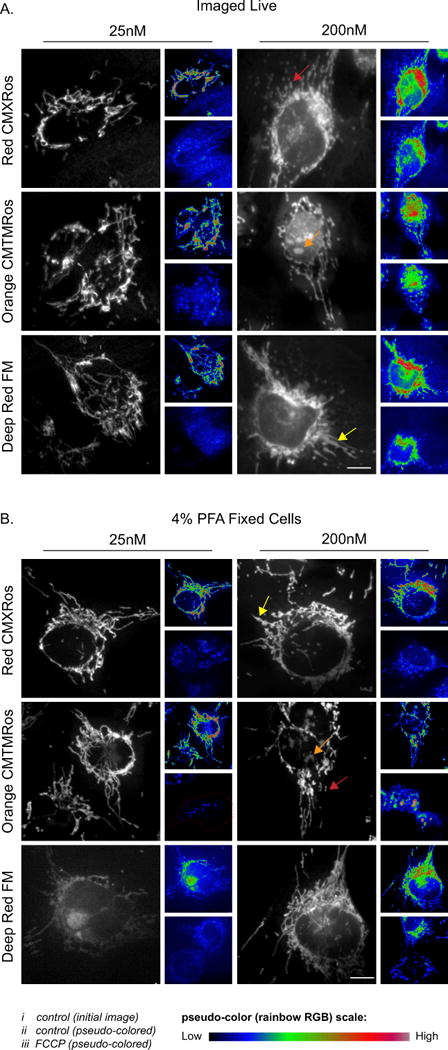
A-B) HMEC-1 cells were incubated with varying concentrations of Mitotracker probes and either imaged live (A) or fixed (B) using 4% PFA. i (initial image); ii & iii (initial image (i) or 10 μM FCCP treated cell both pseudo colored using FIJI Rainbow RGB lookup table). Note in A (iii) is the same cell 15-20min post addition of FCCP, while in (B) the images are of different cells. The arrows denote: red (fragmented mitochondria), yellow (swollen mitochondria) and orange (off target nuclear staining). Scale bar represents 10μm.
To address retention of MitoTracker probes post fixation, samples were pretreated with 10 μM FCCP or vehicle, then incubated with the MitoTracker probes, followed by three washes and fixation. For vehicle treated cells, MitoTracker Deep Red FM was only partially retained at 25 nM causing insufficient mitochondrial signal. By contrast, 200 nM was more fully retained giving sufficient signal. MitoTracker Red CMXRos and Orange CMTMRos were adequately retained in vehicle treated cells at both concentrations, however 200 nM treatment lead to a minor increase in the amount of swollen, fragmented mitochondria (see yellow and red arrows; Figure 2B). Prior disruption of the membrane potential reduced the mitochondrial signal of all 3 probes at both concentrations; however, with 200 nM treatment the mitochondrial signal persisted, especially with MitoTracker Orange CMTMRos and Deep Red FM. Similar to live cells, 200 nM MitoTracker Orange CMTMRos also had problematic non-mitochondrial signal. In summary, either 25 nM MitoTracker Red CMXRos or MitoTracker Orange CMTMRos are well retained post-fixation with minimal artifacts and are recommended for routine mitochondrial imaging when immunostaining is not an option.
1.3.2 Quantifying the mitochondrial network
Image analysis methods have recently made it possible to quantitatively measure the mitochondrial network (see Table 1 for examples). The following section focuses on the implementation of one of these programs, MitoGraph [45,56]. MitoGraph is an open-source, C++ program that is a fully automated image processing software for analysis of three-dimensional (3D) mitochondrial structures. MitoGraph was initially applied and validated to measure the mitochondrial volume in the budding yeast Saccharomyces cerevisiae and then applied to measurements of network topology (paper in revision). Aside from mitochondrial volume a large amount of information can be extracted from the data generated with MitoGraph (detailed below). Here we demonstrate MitoGraph’s versatility and applicability to mammalian cells by quantifying the mitochondrial network from five different mammalian cell types, ranging from entirely fragmented to elongated. We also provide recommendations for successful application of the software. One advantage of MitoGraph is fewer parameters to tune, ultimately allowing more consistency between data sets. For a thorough description on validation, methodology and software functionality see the original MitoGraph publications [45,56]. MitoGraph 3.0 can be downloaded for free at https://github.com/vianamp/MitoGraph. It is also recommended the user download the two FIJI macros listed on the MitoGraph download page for batch processing of z-stacks and cropping cell images.
1.3.2.1 MitoGraph Protocol (for visual depiction of workflow see Figure 3 and Supplemental Movie 1)
Figure 3.
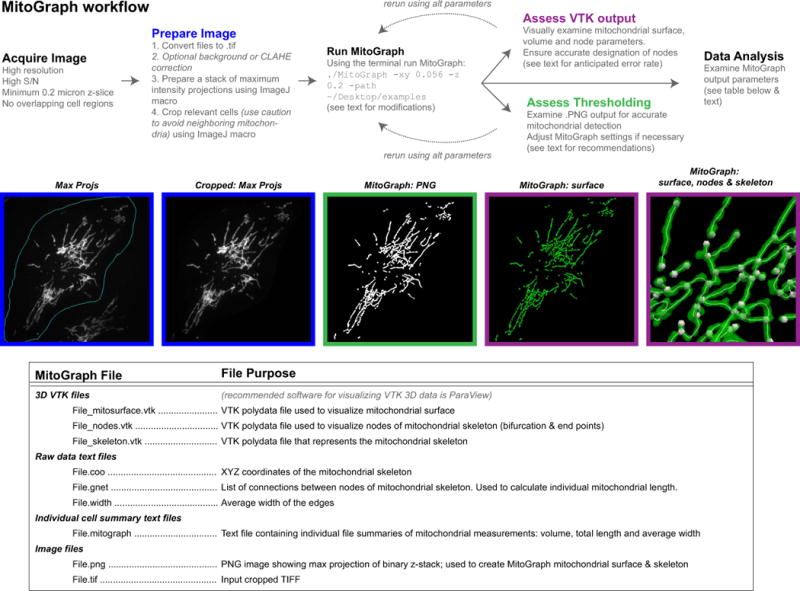
MitoGraph method (see text for detailed description).
Obtain high quality images. MitoGraph performs optimally on images with a high S/N ratio, a small z-spacing (0.2μm used throughout the data presented here) and sufficient xy resolution to depict mitochondrial tubules within 3 to 6 pixels. For accurate downstream analyses ensure the entire cell is imaged, setting the top and bottom of the z-stack position to above or below the last in focus remnant of mitochondria. If individual cell data is desired, ensure when imaging cells that are adjacent to others that the mitochondrial network of neighboring cells be readily excluded via subsequent image cropping. Avoid cells that grow on top of each other, or in such tight colonies that it is difficult to successfully separate the adjacent mitochondrial networks (for examples see: Supplemental Figure 2; problem 5 and [45].
-
Prepare single cell 3D images for MitoGraph analysis. Although MitoGraph does not require confocal images of individual cells to function, it is highly recommended the user isolate individual cells in order to extract cellular trends from subsequent data analysis of the MitoGraph output. In addition to the MitoGraph software, we recommend downloading the two custom FIJI macros that simplify bulk image processing and creation of 3D image files containing a single cell. The first macro, GenMaxProjsFrom.ijm, creates a stack of maximum intensity projections of your entire dataset and allows for manual selection of the all cells to be analyzed by outlining multiple regions of interests (ROIs). The other macro, CropCells.ijm, then uses these designated ROIs and crops the raw original image stack and creates a new 3D image file containing a single cell, which serves as the input into the MitoGraph software. Here we outline recommendations for usage of those macros, the troubleshooting guide in Supplemental Figure 2 provides examples.
-
○
The GenMaxProjsFrom.ijm macro requires single channel .tif files. First convert your image files to .tif, then ensure that all, and only, the files you want to measure with MitoGraph are in one folder and direct the macro to run on that folder. Close all images before you attempt to run the macro. The program we use for imaging (Metamorph) generates .TIF files for the raw image files; these can be easily renamed in bulk using the terminal command ( find . -depth 1 -name ‘*.TIF’ -exec rename -f ‘s/\.TIF/\.tif/’ ‘{}’ \;). Ensure the resulting file is saved as MaxProjs.tif in the same folder as the original files.
-
○
Two modifications are recommended for the CropCells.ijm macro,:
Since the macro was created for yeast cells, we modified the macro to accommodate the larger ROI needed to crop a mammalian cell (Supplemental Figure 2; problem 1). Redefine the final size to accommodate the larger mammalian cells (in the script for CropCells.ijm find “//Defining the size of the singl cell images:_xy = 200” and replace 200 with 512).
Although the addition of Gaussian noise [45] outside the cropped region of interest is helpful in reducing the spurious segmentation at the boundary of the cropped ROI, we found that in certain instances this correction adds excessive speckling outside the cropped ROI or causes the MitoGraph analysis to stall on specific cells (see Supplemental Figure 2; problem 2). Therefore, we recommend removing the added noise feature for the first pass through the data analysis (in the CropCells.ijm macro comment out the added noise component by adding a “//” before “run(“Add Specified Noise...”, “slice standard=” + 0.5*std);”. Any images (usually less than 10% of the cells) that have the spurious segmentation (see Supplemental Figure 2: problem 4) can then be re-processed by enabling added noise.
-
○
Before running the CropCells.ijm macro, outline cells that can be easily segmented from neighboring cells. Use caution when drawing the ROI, if individual cells are not appropriately outlined, artifacts will persist on the subsequent MitoGraph analysis (for examples see: Supplemental Figure 2; problem 5 and [45]. Using the MaxProjs.tif stack of image projections outline all the cells you want to analyze. Apply the FIJI shortcut to access the ROI manager (press ‘t’ to open ROI manager, select ‘show all’ as well as ‘labels’ to keep track of all the cells you mark) and draw regions around the cell perimeter and save the ROIs as RoiSet.zip, ensuring the file is in the same folder as the .tif files and MaxProjs.tif. Close all image windows and direct the CropCells.ijm macro to run on the folder containing the initial images, the MaxProjs.tif file and the RoiSet.zip file. A new folder ‘cells’ is created and contains all of the cropped cells; use these for input to MitoGraph.
-
○
Run MitoGraph. MitoGraph is run from the terminal. First change the directory to the location of MitoGraph ( cd ~/Desktop/MitoGraph) and make the file executable ( chmod +xxx MitoGraph). Then run MitoGraph with the following command line using default settings ( ./MitoGraph -xy 0.1667 -z 0.2 -path ~/Desktop/cells) where the flag –xy 0.1667 refers to the xy pixel size in μm, –z 0.2 refers to z spacing also in μm and –path designates where the file is located. Make sure to correctly annotate the pixel size of your images as using the incorrect pixel dimensions will distort the MitoGraph output. Additional flags can be added to further fine tune the MitoGraph analysis. These include –rad (tubule radius; default 0.15μm), –scales (scale range to fine tune low S/N images; default 1.0 1.5 6), –threshold (post-divergence threshold value between 0.0 and 1.0 that can be adjusted for low S/N images; default 0.1667), –adaptive 10 that forces MitoGraph to split the image in 10x10 equally sized blocks and use a block-dependent value of global threshold. A thorough description of these parameters is provided in the original publication [45]. Most of the images here were processed using the following MitoGraph command: ./MitoGraph -xy 0.1667 -z 0.2 -scales 1.0 1.3 4 -adaptive 10 -path ~/Desktop/cells/.
Assess MitoGraph Thresholding. Examine all PNG images produced by the MitoGraph program and ensure accurate representation of the entire network - some optimization of parameters may be necessary. Look for any lack of detection of mitochondrial structures (Supplemental Figure 1E; in which case additional flags such as scales and adaptive may be useful) or false signal assignment where there are no mitochondria present in the original image (Supplemental Figure 1C arrow; here the scales and adaptive need further fine tuning or removal). Our initial attempts with default MitoGraph settings (as above) on mammalian cells did not adequately detect all the mitochondrial network, especially regarding the long, thin tubules with low S/N observed upon genetic ablation of Drp1 (Supplemental Figure 1E and Supplemental Figure 2, problem 2). However, after adjusting different MitoGraph parameters (we added in the flags –scales 1.0 1.3 4 and –adaptive 10), using MitoGraph v3.0 and modifying our image acquisition settings (used the 60x objective at 1.6x zoom (instead of 1x zoom) and a higher exposure setting to increase the S/N ratio (increased to 1 sec exposure from a 500 msec exposure with the DSU in place) we obtained a nearly complete binary representation of the original image (Figure 5). Near the nucleus in the highly-networked region is often where the MitoGraph segmentation is subpar, likely due to saturated pixels and especially dense tubules preventing accurate discrimination of mitochondrial tubules, given the resolution limits of light microscopy. A brief pilot test of MitoGraph v3.0 on deconvolved widefield and confocal images demonstrated that deconvolution may be beneficial as it improves segmentation, but we recommend further troubleshooting and fine tuning of MitoGraph settings before combining deconvolution with MitoGraph analysis (Supplemental Figure 1 C, E).
Assess VTK output. MitoGraph produces three 3D files that can be viewed using the freely available ParaView software (https://www.paraview.org/). See Supplemental Movie 1 for a visual depiction of the 3D data generated from the MitoGraph output files. First open the mitosurface file (_mitosurface.vtk) to reveal the surface rendering view (once the file is opened, you must click apply to view the file). Scale down the opacity to allow viewing of the subsequent components. Open the skeleton file (_skeleton.vtk), click apply to view the outline within the mitochondrial surface, adjust the color and line thickness to allow easier visualization of the skeleton. Next, open the nodes file (_nodes.vtk) and click apply to view in context of the mitochondrial surface and skeleton. Nodes represent free mitochondrial ends and junction points that are derived from the mitochondrial skeleton. Node assignments are key to the successful downstream data analysis portion of MitoGraph. Visually inspect if end points were properly assigned and junctions points were properly detected. In the experiments presented here the current version of MitoGraph has a node assignment error rate of ~7%, which primarily derived from overlapping tubules (see Methods and Table 2). While exploring MitoGraph parameters, we also tried the Enhanced Local Contrast (CLAHE) FIJI plugin, which enhanced detection of low S/N tubules, but at a cost of increased misassigned nodes thereby rendering the data analysis not reliable (not shown).
-
Data Analysis. Originally designed to report on mitochondrial volume, MitoGraph reports on total mitochondrial length, mitochondrial volume (using two different approaches: one based on the skeleton and mitochondrial radius and the other is based on voxels) and relative mitochondrial width (note the width parameter has not been fully validated yet). Aside from these global measurements, MitoGraph measurements allow extraction of a large number of network connectivity parameters by skeletonizing the segmented mitochondrial network into a set of edges (representing the mitochondrial tubules), and nodes (representing endpoints and branch points). Separate individual mitochondria within a network can thus also be analyzed. The connection between these nodes is referred to as an edge. Supplemental Figure 3 schematically illustrates this segmentation and the downstream mitochondrial network metrics that are calculated. Supplemental Figure 4 has single cell examples of a wide range of mitochondrial phenotypes, from highly fused to highly fragmented and the corresponding metrics that were extracted from the MitoGraph analysis. For the sake of simplicity, we now will refer to an individual mitochondrion within the network as a “connected component” to help differentiate individual components versus the entire mitochondrial network.
-
○
First, a list of the total length, total edge number and total node number for each connected component must be generated. The length of each edge present in the network is displayed in the gnet text file. The first column represents the node number at the beginning of the segment, the second column represents the node number at the end of the segment, and the third column represents the distance between these two nodes in μm. From this information, we used an R-script to extract the following information for each connected component: total length, edges and nodes. These are calculated from the sum of the distance between the connected nodes. For example, in the branched example in Supplemental Figure 3, three individual edges are connected by one central node; thereby forming a connected component that is 20 μm in total length with 4 total nodes and 3 total edges.
-
○
Once the list of connected components is generated for each cell assessed, the total mitochondrial length, total edge number, total node number are calculated by summing each corresponding metric of the connected component; the total number of connected components is also tallied. PHI represents the relative size of the largest connected component to the total mitochondrial size (note PHI increases in scenarios where the mitochondrial network is highly interconnected; Supplemental Figure 4). The average edge length is calculated by taking the total mitochondrial length and dividing by the total edge number. Each metric that is not a ratio or an average needs to be normalized to some metric of cell size given the heterogeneity of cell sizes in mammalian tissue culture cells. We chose to normalize total nodes, edges and connected components to the total mitochondrial length for each cell. Lastly, an additional measurement of the mitochondrial network connectivity is extracted from the node data by assessing the degree distribution, Pk, which indicates the fraction of nodes with k-neighbors. We assessed the ratio found in free ends (k=1), three-way junctions (k=3) and four-way junctions (k=4). The AVG degree measurement is the sum of each individual ratio multiplied by its corresponding k factor.
-
○
Each of the metrics reports on unique network properties. PHI is highest in scenarios where the entire mitochondrial network is connected and lowest in scenarios where the entire network is uniformly fragmented. Average edge length is highest when the node to node distance is greatest, typically with very long, not highly interconnected mitochondria like those present in Drp1 KO cells (Supplemental Figure 4B-C). Total nodes, edges and connected components increase in highly fragmented mitochondria. In a network that is entirely fragmented up to 90% of the nodes are found in free ends (only one neighbor), while in a scenario where fission is inhibited and the network is hyperfused around 50% of the nodes are found in free ends (Supplemental Figure 4B-C). The AVG degree is highest in highly interconnected examples such as H1299 and to a lesser extent in Drp1 KO cells (Supplemental Figure 4B-C). Based on these differences we sought to capture the diverse nature of the mitochondrial network into a single score, the MitoGraph Connectivity Score. We calculated this score by taking the sum of the parameters elevated in highly fused networks (PHI, AVG edge length and AVG degree) and divided by parameters elevated in highly fragmented networks (nodes; edges and connected components (all normalized to total length)). Although MitoGraph Connectivity Score has not been fully validated, it does mirror the morphology trend as would be predicted by eye (Supplemental Figure 4C). It can be used in addition to reporting all MitoGraph parameters, but should not be considered a substitute as it does not capture the complexity of the network.
-
○
-
Using the R-scripts. Here we supply two R-scripts that should assist data analysis efforts (https://github.com/Hill-Lab/MitoGraph-Contrib-RScripts.git). First download R and Rstudio (www.rstudio.com) [141]. To run the scripts: 1) place the R-scripts in the folder containing all of the files generated by MitoGraph, 2) open with R-studio, set the working directory to the source file location (session → set working directory → to source file location), paste in the path for the folder and run the script using command+option+r.
-
○
The first R-script (CreateSummary.R) produces two files: output.csv and output-summary.csv. Output.csv contains data from individual connected components for all of the .gnet files within that directory and lists the file name, number of nodes and edges as well as total length for each connected component within the dataset. Output-summary.csv calculates the following for each image acquired: 1) total nodes, edges and length for all connected components, 2) the number of connected components, 3) PHI, 4) average edge length, 5) total edges, nodes and connected components all normalized to total length. The output-summary.csv also includes a summary from the degree distribution analysis that contains the fraction of free ends (k=1), 3-way (k=3) and 4-way junctions (k=4) and the average degree. The output-summary.csv also contains a compilation of the .mitograph text file data including: 1) volume from voxels (μm3), 2) average width (μm), 3) standard deviation width (μm), 4) total length (μm) and 5) volume from length (μm3). Finally, the output-summary.csv also contains the MitoGraph Connectivity Score for each image.
-
○
The second R-script (CreatePlots.R) creates a variety of plots to help visualize any alterations in the metrics assessed through the MitoGraph analysis and contains statistical measurements of the differences using ANOVA followed by TUKEY analysis. Since file naming schemes likely will vary, first manually edit the output-summary.csv file and add a column titled “Condition” and populate with the relevant condition or treatment information in the order you want it displayed on the graphs. See Supplemental Figure 5 for an example of a report generated using this script on one of the datasets presented here.
-
○
Figure 5. MitoGraph analysis under conditions favoring mitochondrial fusion.
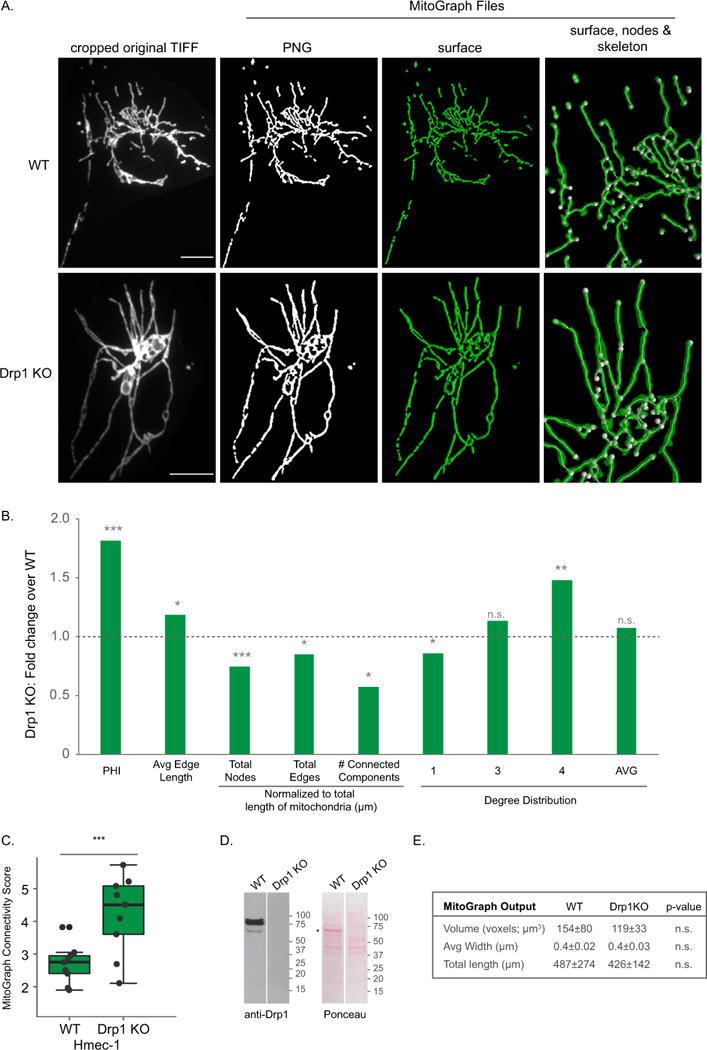
A-E) HMEC-1 cells (WT or Drp1 KO) were fixed in 4% PFA, immunostained for Tom20, imaged using the 60x objective (zoom 1.6) and processed by MitoGraph v3.0 using the following command (identical to Figures 4–6): ./MitoGraph -xy 0.1667 -z 0.2 -scales 1.0 1.3 4 -adaptive 10 -path ~/Desktop/cells/. A) The top row represents the original cropped TIFF file, the bottom 3 rows are output files of MitoGraph (the binary PNG, the 3D surface representation and a zoom in on the surface, skeleton and nodes of the mitochondrial network. B) The .GNET files were processed using an R-script to compile the following metrics (see Supplemental Figures 3 and 4 for detailed descriptions of how these metrics were calculated): PHI (ratio of the largest connected component); average edge length; total nodes, edges and connected components (normalized to total length of mitochondria); the degree distribution of the nodes (1 (free ends), 3 (3-way junction), 4 (4-way junction) and Avg Degree). Raw data is shown in Supplemental Figure 6, while the fold change between averages of at least 10 cells are presented here. C) The MitoGraph connectivity score which is calculated from the sum of metrics elevated in a fused state (PHI, Avg Edge Length and Avg Degree) divided by sum of metrics which are elevated in a fragmented state (total nodes, edges and connected components (normalized)). D) Western blot analysis of lysates from WT and Drp1KO HMEC-1 cells. *Extra band likely due to FBS in the medium. E) The AVG±STDEV of the indicated parameters, p-value was calculated using an unpaired Students t-test. Scale bar represents 10μm.
1.3.2.2 Validating MitoGraph on mammalian cells
MitoGraph was shown in yeast to help elucidate mitochondrial phenotypes in mitochondrial fission and fusion mutants [45]. Based on this success, we sought to test MitoGraph in a few key scenarios to evaluate its utility. First, we focused on determining if MitoGraph analysis can distinguish a highly fragmented mitochondrial network from a normal healthy mitochondrial network. We used mito-YFP transfected HMEC-1 cells (generated above) treated with CCCP to uncouple and fragment the mitochondria, and compared them against untreated cells. MitoGraph analysis in these cells resulted in a binary PNG image that almost entirely mirrored the original TIFF image (Fig 4A) and was validated for correct assignment of nodes (only 7.2% of total nodes were misassigned, with 3.6% due to overlapping tubules - see Methods and Table 2). Based on these positive attributes, we processed the MitoGraph data output. A plot of the raw data and all the metrics assessed is presented in Supplemental Figure 6, while Figure 4B represents the fold change between the averages of at least 10 of CCCP treated HMEC-1 cells over untreated WT HMEC-1 cells. As shown in Fig 4B, metrics associated with enhanced fission are increased over WT (total nodes, edges and connected components) while metrics associated with enhanced fusion are decreased (PHI, Avg edge length and Avg degree). The largest difference between control and CCCP treated cells is within the total number of connected components, with CCCP treatment leading to a 2.3 fold increase in the total number of connected components. The average MitoGraph Connectivity Score significantly decreased from 2.11 to 1.04 with CCCP treatment (Figure 4C). Overall, mitochondria in CCCP treated cells are shorter, less interconnected and form more connected components than in WT control HMEC-1 cells. Furthermore, the total length and volume of the mitochondria were also impacted by the CCCP treatment (Figure 4D). Overall, these qualities are in line with the expected results for short fragmented mitochondria found in CCCP treated cells.
Figure 4. MitoGraph analysis under conditions favoring mitochondrial fission.
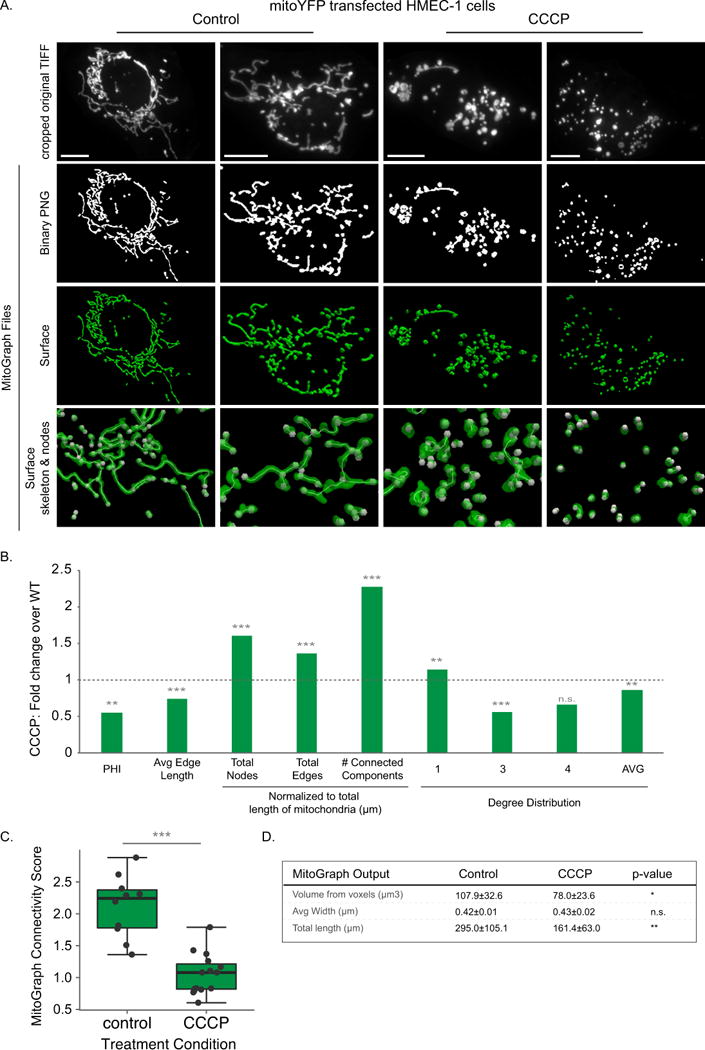
A-D) HMEC-1 cells transiently expressing Mito-YFP were treated with 10 μM CCCP for 15-20 min to fragment the mitochondria, fixed in 4% PFA, imaged using the 60x objective (zoom 1.6) and processed by MitoGraph v3.0 using the following command (identical to Figures 5–6): ./MitoGraph -xy 0.1667 –z 0.2 -scales 1.0 1.3 4 -adaptive 10 -path ~/Desktop/cells/. A) The top row represents the original cropped TIFF file, the bottom 3 rows are output files of MitoGraph (the binary PNG, the 3D surface representation and a zoom in on the surface with the skeleton and nodes of the mitochondrial network also depicted. B) The .GNET files were processed using an R-script to compile the following metrics (see Supplemental Figures 3 and 4 for detailed descriptions of how these metrics were calculated): PHI (ratio of the largest connected component); average edge length; total nodes, edges and connected components (normalized to total length of mitochondria); the degree distribution of the nodes (1 (free ends), 3 (3-way junction), 4 (4-way junction) and Avg Degree). Raw data is shown in Supplemental Figure 6, while the fold change between averages of at least 10 cells are presented here. C) The MitoGraph connectivity score, which is calculated from the sum of metrics elevated in a fused state (PHI, Avg Edge Length and Avg Degree) divided by sum of metrics which are elevated in a fragmented state (total nodes, edges and connected components (normalized)). D) The AVG±STDEV of the indicated parameters, p-value was calculated using an unpaired Students t-test. Scale bar represents 10μm.
Next, we validated the other end of morphology spectrum and compared WT morphology to a hyper-elongated phenotype. Using a Drp1 null HMEC-1 clone generated using CRISPR/Cas9 (Harwig & Hill unpublished data), we compared these hyper-elongated mitochondria to WT HMEC-1 mitochondria in Tom20 immunostained cells. Like the mito-YFP experiments shown in Figure 4, Tom20 immunostained mitochondria, when imaged at a high resolution and exposure settings enabling a high S/N ratio, performed well on MitoGraph analysis. The binary PNG image again mirrored the cropped TIFF signal and when the node assignments were validated we found that the total error rate was 7.7%, while 6.3% were due to overlapping tubules (see Methods and Table 2). A plot of the raw data and all the metrics assessed is presented in Supplemental Figure 6B, while Figure 5B represents the fold change between the averages of at least 10 of Drp1KO HMEC-1 cells over WT HMEC-1 cells. Relative to WT mitochondria, Drp1KO mitochondria have elevated metrics associated with a highly fused state (PHI, Avg Edge Length and to a lesser extent AVG degree) while metrics associated with highly fragmented networks are decreased (total nodes, edges and connected components). Drp1 KO mitochondria have an average MitoGraph Connectivity Score that is significantly higher than WT mitochondria (4.2 versus 2.9) (Figure 5C). PHI, the measurement of fraction of mitochondria found in the largest component, resulted in the largest fold difference between WT and Drp1 KO HMEC-1 cells. The volume, total length and average width of mitochondria in WT versus Drp1 KO were not significantly different. Overall, the enhanced length and the more intricate network found in Drp1KO cells matches what is visually observed, thereby enhancing the confidence in the MitoGraph analysis.
The HMEC-1 cells used for the MitoGraph analysis in Fig 4 and 5 were very flat cells with easily discernible highly elongated mitochondria. Next, we examined how well MitoGraph performed on a variety of other cells types: C6 (rat brain glial cells), C2C12 (mouse myoblasts), HEK293 (human embryonic kidney cells) and H1299 (human non-small cell lung carcinoma). As shown in Figure 6, the mitochondrial morphology was vastly different in these 4 cells lines. The C6 cells were very flat with an easily discernable mitochondria that were relatively elongated, but not highly interconnected network (note: these cells frequently overlap each other requiring caution in image acquisition). C2C12 cells were also very flat and had shorter, but more interconnected mitochondria than the C6 cells. HEK293 cells had a heterogeneous relatively elongated mitochondrial network and were very thick and required nearly twice as many z-slices as the other 3 cells lines tested. H1299 cells had highly interconnected, elongated mitochondria that were frequently found in striking loop-like structures. Node validation revealed that MitoGraph performed well on all 4 cells lines assessed and nodes were misassigned less than 10% of the time. Closer examination of the total error rates and fraction due to overlapping tubules revealed that H1299 had the lowest total error rate, while C6, C2C12 and HEK293 had very similar error rates (see Methods and Table 2). As shown in Figure 6, H1299 cells contained the most highly connected mitochondrial network (highest PHI and AVG degree and lowest number of connected components), while C6 cells had the longest edge length, but was the least connected network of the cells examined (lowest PHI and AVG degree). C6 cells scored surprisingly high with the MitoGraph connectivity score, likely due to the contribution of the increased average edge length. H1299 cells, which have the most striking unique mitochondrial network scored highest on the MitoGraph connectivity score, with statistically significant differences from all 3 other cells types assessed in this panel (Figure 6C). Overall, the MitoGraph analysis added quantitative data that agreed with the confocal images shown in Figure 6A.
Figure 6. MitoGraph analysis of a variety of cell types.
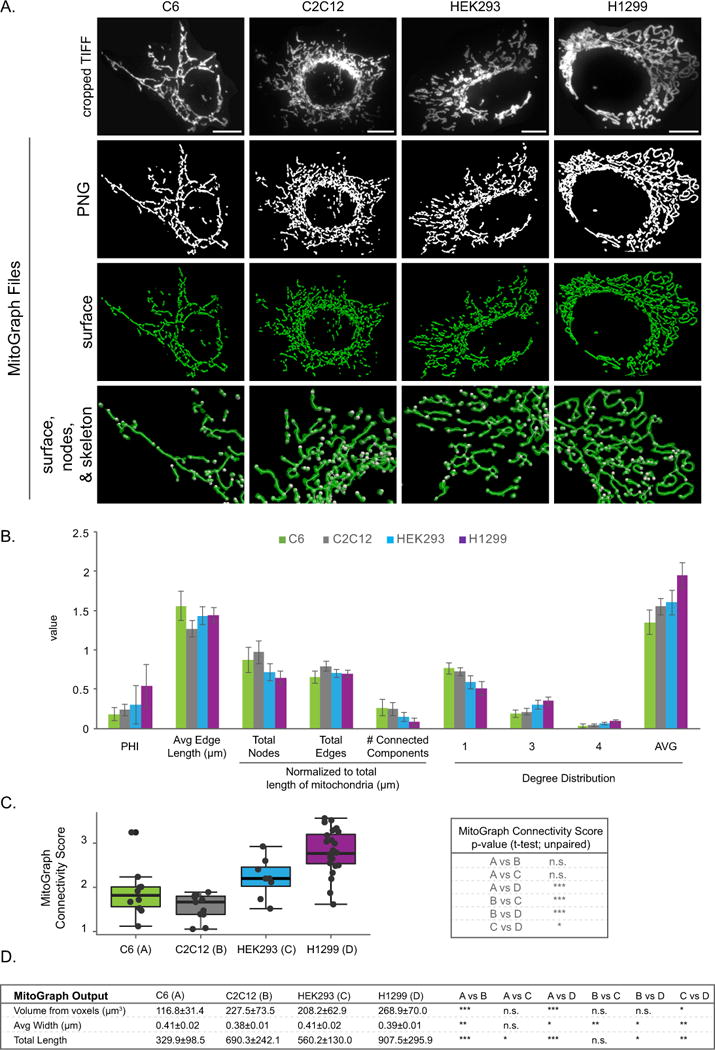
A-C) C6, C2C12, HEK293 and H1299 cells were fixed in 4% PFA, immunostained for Tom20, imaged using the 100x objective (zoom 1) and processed by MitoGraph v3.0 using the following command (identical to Figures 4–5): ./MitoGraph -xy 0.1600 -z 0.2 -scales 1.0 1.3 4 -adaptive 10 -path ~/Desktop/cells/. A) The top row represents the original cropped TIFF file, the bottom 3 rows are output files of MitoGraph (the binary PNG, the 3D surface representation and a zoom in on the surface, skeleton and nodes of the mitochondrial network. B) The .GNET files were processed using an R-script to compile the following metrics (see Supplemental Figures 3 and 4 for detailed descriptions of how these metrics were calculated): PHI (ratio of the largest connected component); average edge length; total nodes, edges and connected components (normalized to total length of mitochondria); the degree distribution of the nodes (1 (free ends), 3 (3-way junction), 4 (4-way junction) and Avg Degree). C) The MitoGraph connectivity score which is calculated from the sum of metrics elevated in a fused state (PHI, Avg Edge Length and Avg Degree) divided by sum of metrics which are elevated in a fragmented state (total nodes, edges and connected components (normalized)). D) The AVG±STDEV of the indicated parameters, p-value was calculated using an unpaired Students t-test. Scale bar represents 10μm.
1.4 Discussion
As dynamic organelles, mitochondria are exquisitely sensitive to cellular stresses and as such must be handled with care. Here we outlined necessary optimization strategies for imaging mitochondria including recommendations to 1) completely image the entire network using z-stack spacing of at least 1μm for routine imaging and 0.2 - 0.5 μm spacing if performing MitoGraph analysis or acquiring publication quality images, 2) optimize DNA amount for mitochondrial-targeted fluorescent proteins transfections to minimize fragmentation, 3) use of 25nM MitoTracker probes to avoid cytotoxic effects during live cell imaging and 4) use 25nM of MitoTracker Red CMXRos and MitoTracker Orange CMTMRos to minimize off target fluorescence, morphological side effects and to visualize the network post-fixation. Finally, we supplied a detailed protocol and validation of the mitochondrial network analysis program MitoGraph using five mammalian cell types.
The experimental conditions and results obtained here using mito-YFP and MitoTracker dyes agree with previous findings. Specifically, Mitra et al. mentioned that a 48hr transient transfection of mito-DsRred can cause mitochondrial fragmentation and ultimately recommended short transient transfections with low expression of the markers [38]. Similarly, treating cells with too high of a concentration of MitoTracker can lead to deleterious effect on the cells. In rat brain mitochondria, exposure to high concentrations of MitoTracker dyes (~3 μM range) altered mitochondrial respiration by increasing resting respiration and decreasing maximal respiration, however low concentrations (50nM) had little to no effect [142]. Several reviews provide more detailed discussions on the appropriate use of MitoTracker and potentiometric dyes[115–117,121–123].
Given the interest in quantifying mitochondrial morphology, a number of methods and software have been recently developed (Table 1). Throughout the years we have installed a subset of the freeware programs and applied them to our existing images. The ideal software would be free, easy-to-use, and accurately reflect the entire mitochondrial network. The purpose of this prospective is not to compare and contrast each method, but rather to highlight how MitoGraph stood out from a subset we tested. MitoGraph is free, works on 3D images, has minimal tunable parameters and most notably, once optimized, works on a variety of datasets using identical settings, making batch processing straightforward. Our very first attempt using MitoGraph on S. cerevisiae expressing mito-RFP was successful and resulted in proper segmentation of yeast mitochondria (once the xy pixel width was set to reflect the microscope’s settings). Transitioning from yeast to mammalian cells required more troubleshooting, especially for the problematic long thin low S/N tubules found in Drp1 KO HMEC-1 cells (Supplemental Figure 2; Problem 3). After several iterations, we discovered that we needed to increase our resolution, increase our S/N ratio and modify the input parameters for the MitoGraph analysis. We achieved this by using a magnification changer to boost the pixel size from 0.2667μm to 0.1667μm, doubling the exposure time (500 msec to 1sec), adding in the flag –scales 1.0 1.3 4 and the flag –adaptive 10 so that MitoGraph calculates a global threshold for each slice instead of using the same value for the entire image.
We also switched to MitoGraph 3.0, the newest version, which implements a region based-threshold and outputs mitochondrial tubules width. This region based thresholding in addition to the adaptive flag modality allows for fine tuning the threshold between slices and within regions of a single slice, thereby accurately segmenting more regions of the mitochondrial network. This adaptive thresholding is especially important in mammalian cells where the perinuclear region containing mitochondria is often substantially brighter than lower density regions closer to the cell periphery. Once a dataset was successfully imaged and analyzed with MitoGraph, the same settings were applied to additional experimental rounds and found to properly segment the resulting images. Importantly, thanks to the adaptive thresholding, MitoGraph performed equally well on diverse cell types that had considerable differences in the level of Tom20 staining (Supplemental Figure 7). MitoGraph does take a significant amount of CPU time to complete the segmentation and analysis, but is amenable to implementing on a GPU cluster. Furthermore, MitoGraph has the following limitations: 1) an average of 7.2±1% total nodes are misassigned, with the majority of the misassignments due to overlapping tubules and 2) the current parameters do not always properly segment the highly interconnected, often-saturated perinuclear mitochondria. Despite these limitations, MitoGraph analysis statistically differentiated between fragmented and normal mitochondria; normal mitochondria and hyper-elongated mitochondria and differentiated between four unique cell types that varied in cell thickness with a range of mitochondrial morphologies in tissue culture cells. Depending on the resolution, MitoGraph may also be useful for analyzing images using intravital microscopy on live animals [143,144]. Such intravital imaging has been beneficial in cancer studies [145–149] and imaging of mitochondria in intact animal disease models [150–152]. Future application of MitoGraph will undoubtedly lead to a much deeper understanding of mitochondrial form and function.
Supplementary Material
A,D) Original widefield (A; DSU out) versus confocal (D; DSU IN) images from Figure 1 (original), previously acquired with Metamorph software, were input into the cellSens software (Olympus) and were processed with the blind iterative deconvolution module (deconvolved). Note the improvement deconvolution added to both widefield and confocal images. B-C, E-F) Either the original or deconvolved images from (A, D) were cropped and processed with MitoGraph v3.0 using the default MitoGraph settings (B, E; ./MitoGraph -xy 0.1667 -z 0.2 -path ~/Desktop/cells/) or the settings optimized for this study (C, F; ./MitoGraph -xy 0.1667 -z 0.2 -scales 1.0 1.3 4 -adaptive 10 -path ~/Desktop/cells/). Note the TIFF images output from the cellSens deconvolution algorithm were incompatible with MitoGraph v3.0 and were first processed with a the FIJI 3D median filter (set to 1,1,1) which was then MitoGraph v3.0 compatible. Arrows denote examples of non-mitochondrial areas detected when the settings are not properly optimized. The circles denote examples of mitochondrial regions that are absent in the segmented MitoGraph image indicating poor thresholding by the default MitoGraph settings, likely due to poor S/N of the original image. The images outlined in green designate the optimal settings used throughout the manuscript. Scale bar represents 10μm.
Supplemental Figure 2: Troubleshooting guide. See figure for description.
The top panel indicates network measurements generated for each cell, while the bottom portion focuses on the metrics gathered for an individual mitochondrion or the connected component that result for connections between multiple nodes. Central to understanding MitoGraph measurements is understanding the definition of nodes (mitochondrion end points or junction points between branches of the network) and edges (cylindrical connection between nodes, essentially each branch is an edge). The total length, nodes and edges are calculated for each connected component found within the cells From this information the overall network size or total mitochondrial length is calculated by taking the sum of the length of each connected component. The number of connected components is tallied and the ratio of the largest connected component to the total network size is calculated and is designated as PHI. The average edge length is calculated by taking the total mitochondrial length and dividing it by the total edge number. The total nodes and edges are tallied and finally the nodes are assessed for the fraction found in free ends (k=1) versus those found in 3-way and 4-way junctions (k = 3/4) and the average degree calculated from those measurements.
A) Six distinct mitochondrial phenotypes arranged from highly fused to highly fragmented (see figure for the indicated cell type; see Figures 3–5 for original images). B) Individual cell metrics copied as is from the .mitograph text files. Total length, nodes and connected components generated from the R-script CreateSummary.R that extracts information from the .GNET file (contains node assignments and edge length for each edge in the network). From this information the normalized metrics are calculated as described on the figure. Note that PHI is generated by first determining the largest connected component found in a given cell and then dividing that by the total length of the entire network. C) Graphical depiction of the data presented in the table in (B). Note that some parameters increase upon enhanced fragmentation, while others decrease upon enhanced fragmentation. The MitoGraph Connectivity Score represents this relationship by summing the factors elevated in a highly fused scenario (PHI, Avg edge length and Avg Degree) and dividing by those factors elevated in a highly fragmented scenario (Total nodes, edges and connected components (all normalized to length)).
Output from R Studio of the dataset from Figure 5 processed with following R-scripts: CreateSummary.R followed by CreatePlots.R.
A, B) 10 images were collected for HMEC-1 WT or CCCP treated cells (A; see Figure 4 legend for details) or Hmec-1 WT of Drp1 KO (B; see Figure 5 for details). From those 10 images, cells were individually cropped and processed as depicted by Supplemental Figures 3 and 4.
Cropped images of 4 individual cells per cell type (293T, C2C12, C6, H1299 and HEK293) of Tom20 immunostained mitochondria (left) and the resulting MitoGraph PNG binary image (right). Using FIJI, the cropped images were combined into a single stack to permit applying the same brightness/contrast to all the images and the stack was then displayed as a 4 × 5 montage using a 1.00 scale. Note that the cells were stained and imaged using identical settings and despite the differences in Tom20 staining, MitoGraph effectively segmented the mitochondria in all cell types. 293T cells were problematic showing rogue lines or regions from neighboring cells in all the cells assessed and therefore was not processed for downstream analysis (see Supplemental Figure 2 Problem 5).
Acknowledgments
We thank Dr. A. Marchese for the generous use of his laboratory’s confocal microscope, Dr. J. Park for sharing HEK293 and 293T cells and Dr. A. Beyer for sharing C6 and H1299 cells. We also thank Dr. A. Posey for the generous use of his artistic representation of mitochondria and W. Hausner for microscopy discussions and training sessions.
Funding. This was work was supported by the National Institutes of Health grants: R01-GM067180 (RBH), R01-HL128240(MEW) and R41-AG056253 (RBH).
Abbreviations
- CCCP
Carbonyl cyanide m-chlorophenyl hydrazine
- DiO6
3,3′-Dihexyloxacarbocyanine Iodide
- DSU
Disk-scanning unit (spinning disk)
- FCCP
carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone
- FRAP
Fluorescence Recovery After Photobleaching
- JC-1
tetraethylbenzimidazolylcarbocyanine iodide
- Mito-YFP
mitochondrial targeted yellow fluorescent protein
- PFA
paraformaldehyde
- ROI
Region of interest RT (room temperature)
- Rho123
Rhodamine 123
- S/N
Signal/Noise ratio
- TMRE
Tetramethylrhodamine, ethyl ester
- TMRM
Tetramethylrhodamine, methyl ester
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions. M.C.H. and R.B.H. conceived the project and wrote the manuscript. S.M.R. and M.P.V. contributed to the design of manuscript and validation experiments. M.C.H. collected all images and analyzed all the data. M.C.H., M.P.V., J.J.H. and J.M.E. wrote R and other scripts for analyzing the data generated by MitoGraph. M.P.V. validated the node assignments. S.M.R. and M.P.V. provided critical comments on the manuscript. All co-authors read and approved the manuscript.
References
- 1.Dohrn A. On the Morphology of the Arthropoda. J Anat Physiol. 1868;2:80–86. [PMC free article] [PubMed] [Google Scholar]
- 2.Keith A, Flack M. The Form and Nature of the Muscular Connections between the Primary Divisions of the Vertebrate Heart. J Anat Physiol. 1907;41:172–189. [PMC free article] [PubMed] [Google Scholar]
- 3.Banavar JR, Cooke TJ, Rinaldo A, Maritan A. Form, function, and evolution of living organisms. Proc Natl Acad Sci U S A. 2014;111:3332–3337. doi: 10.1073/pnas.1401336111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ball P. Shapes: Nature’s patterns: a tapestry in three parts. OUP; Oxford: 2009. [Google Scholar]
- 5.Wang C, Youle R. Cell biology: Form follows function for mitochondria. Nature. 2016;530:288–289. doi: 10.1038/530288a. [DOI] [PubMed] [Google Scholar]
- 6.Friedman JR, Nunnari J. Mitochondrial form and function. Nature. 2014;505:335–343. doi: 10.1038/nature12985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cogliati S, Enriquez JA, Scorrano L. Mitochondrial Cristae: Where Beauty Meets Functionality. Trends Biochem Sci. 2016;41:261–273. doi: 10.1016/j.tibs.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 8.Detmer SA, Chan DC. Functions and dysfunctions of mitochondrial dynamics. Nat Rev Mol Cell Biol. 2007;8:870–879. doi: 10.1038/nrm2275. [DOI] [PubMed] [Google Scholar]
- 9.Archer SL. Mitochondrial dynamics–mitochondrial fission and fusion in human diseases. N Engl J Med. 2013;369:2236–2251. doi: 10.1056/NEJMra1215233. [DOI] [PubMed] [Google Scholar]
- 10.van der Bliek AM, Shen Q, Kawajiri S. Mechanisms of mitochondrial fission and fusion. Cold Spring Harb Perspect Biol. 2013;5 doi: 10.1101/cshperspect.a011072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang HC, Simon V, Swayne TC, Pon L. Methods in Cell Biology. Academic Press; 2001. Chapter 19 Visualization of mitochondrial movement in yeast; pp. 333–351. [DOI] [PubMed] [Google Scholar]
- 12.Egner A, Jakobs S, Hell SW. Fast 100-nm resolution three-dimensional microscope reveals structural plasticity of mitochondria in live yeast. Proc Natl Acad Sci U S A. 2002;99:3370–3375. doi: 10.1073/pnas.052545099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lewis MR, Lewis WH. Mitochondria in Tissue Culture. Science. 1914;39:330–333. doi: 10.1126/science.39.1000.330. [DOI] [PubMed] [Google Scholar]
- 14.Cowdry EVI. Surface Film Theory of the Function of Mitochondria. Am Nat. 1926;60:157–165. [Google Scholar]
- 15.Bourne GH. XX Recent discoveries concerning mitochondria and Golgi apparatus and their significance in cellular physiology. J R Microsc Soc. 1950;70:367–380. doi: 10.1111/j.1365-2818.1950.tb04444.x. [DOI] [PubMed] [Google Scholar]
- 16.Frederic J, Chevremont M. Recherches sur les chondriosomes de cellules vivantes par la microscopie et la microcinématographie en contraste de phase. Arch Biol. 1952;63:109–131. [PubMed] [Google Scholar]
- 17.Frederic J. Cytological research on the normal chondriome or the chondriome subjected to experimentation in living cells cultivated in vitro. Arch Biol. 1958;69:167–349. [PubMed] [Google Scholar]
- 18.Smirnova E, Shurland DL, Ryazantsev SN, van der Bliek AM. A human dynamin-related protein controls the distribution of mitochondria. J Cell Biol. 1998;143:351–358. doi: 10.1083/jcb.143.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Otsuga D, Keegan BR, Brisch E, Thatcher JW, Hermann GJ, Bleazard W, Shaw JM. The dynamin-related GTPase, Dnm1p, controls mitochondrial morphology in yeast. J Cell Biol. 1998;143:333–349. doi: 10.1083/jcb.143.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Santel A, Frank S, Gaume B, Herrler M, Youle RJ, Fuller MT. Mitofusin-1 protein is a generally expressed mediator of mitochondrial fusion in mammalian cells. J Cell Sci. 2003;116:2763–2774. doi: 10.1242/jcs.00479. [DOI] [PubMed] [Google Scholar]
- 21.Santel A, Fuller MT. Control of mitochondrial morphology by a human mitofusin. J Cell Sci. 2001;114:867–874. doi: 10.1242/jcs.114.5.867. [DOI] [PubMed] [Google Scholar]
- 22.Hermann GJ, Thatcher JW, Mills JP, Hales KG, Fuller MT, Nunnari J, Shaw JM. Mitochondrial fusion in yeast requires the transmembrane GTPase Fzo1p. J Cell Biol. 1998;143:359–373. doi: 10.1083/jcb.143.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hales KG, Fuller MT. Developmentally regulated mitochondrial fusion mediated by a conserved, novel, predicted GTPase. Cell. 1997;90:121–129. doi: 10.1016/s0092-8674(00)80319-0. [DOI] [PubMed] [Google Scholar]
- 24.Faelber K, Gao S, Held M, Posor Y, Haucke V, Noe F, Daumke O. Oligomerization of dynamin superfamily proteins in health and disease. Prog Mol Biol Transl Sci. 2013;117:411–443. doi: 10.1016/B978-0-12-386931-9.00015-5. [DOI] [PubMed] [Google Scholar]
- 25.Antonny B, Burd C, De Camilli P, Chen E, Daumke O, Faelber K, Ford M, Frolov VA, Frost A, Hinshaw JE, Kirchhausen T, Kozlov MM, Lenz M, Low HH, McMahon H, Merrifield C, Pollard TD, Robinson PJ, Roux A, Schmid S. Membrane fission by dynamin: what we know and what we need to know. EMBO J. 2016;35:2270–2284. doi: 10.15252/embj.201694613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen H, Chan DC. Mitochondrial dynamics–fusion, fission, movement, and mitophagy–in neurodegenerative diseases. Hum Mol Genet. 2009;18:R169–76. doi: 10.1093/hmg/ddp326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chan DC. Mitochondrial dynamics in disease. N Engl J Med. 2007;356:1707. doi: 10.1056/NEJMp078040. [DOI] [PubMed] [Google Scholar]
- 28.Baloyannis SJ. Mitochondrial alterations in Alzheimer’s disease. J Alzheimers Dis. 2006;9:119–126. doi: 10.3233/jad-2006-9204. [DOI] [PubMed] [Google Scholar]
- 29.Chung MJ, Suh YL. Ultrastructural changes of mitochondria in the skeletal muscle of patients with amyotrophic lateral sclerosis, Ultrastruct. Pathol. 2002;26:3–7. doi: 10.1080/01913120252934260. [DOI] [PubMed] [Google Scholar]
- 30.Exner N, Treske B, Paquet D, Holmström K, Schiesling C, Gispert S, Carballo-Carbajal I, Berg D, Hoepken HH, Gasser T, Krüger R, Winklhofer KF, Vogel F, Reichert AS, Auburger G, Kahle PJ, Schmid B, Haass C. Loss-of-function of human PINK1 results in mitochondrial pathology and can be rescued by parkin. J Neurosci. 2007;27:12413–12418. doi: 10.1523/JNEUROSCI.0719-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trimmer PA, Swerdlow RH, Parks JK, Keeney P, Bennett JP, Jr, Miller SW, Davis RE, Parker WD., Jr Abnormal mitochondrial morphology in sporadic Parkinson’s and Alzheimer’s disease cybrid cell lines. Exp Neurol. 2000;162:37–50. doi: 10.1006/exnr.2000.7333. [DOI] [PubMed] [Google Scholar]
- 32.Clark IE, Dodson MW, Jiang C, Cao JH, Huh JR, Seol JH, Yoo SJ, Hay BA, Guo M. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature. 2006;441:1162–1166. doi: 10.1038/nature04779. [DOI] [PubMed] [Google Scholar]
- 33.Stichel CC, Zhu XR, Bader V, Linnartz B, Schmidt S, Lübbert H. Mono- and double-mutant mouse models of Parkinson’s disease display severe mitochondrial damage. Hum Mol Genet. 2007;16:2377–2393. doi: 10.1093/hmg/ddm083. [DOI] [PubMed] [Google Scholar]
- 34.Beal MF. Mitochondrial Disorders. Springer; Paris: 2002. Mitochondria in neurodegeneration; pp. 17–35. [Google Scholar]
- 35.Ekeren GJ, Stadhouders AM, Egberink GJM, Sengers RCA, Danils O, Kubat K. Hereditary mitochondrial hypertrophic cardiomyopathy with mitochondrial myopathy of skeletal muscle, congenital cataract and lactic acidosis. Vichows Archiv A Pathol Anat. 1987;412:47–52. doi: 10.1007/BF00750730. [DOI] [PubMed] [Google Scholar]
- 36.Shenouda SM, Widlansky ME, Chen K, Xu G, Holbrook M, Tabit CE, Hamburg NM, Frame AA, Caiano TL, Kluge MA, Duess MA, Levit A, Kim B, Hartman ML, Joseph L, Shirihai OS, Vita JA. Altered mitochondrial dynamics contributes to endothelial dysfunction in diabetes mellitus. Circulation. 2011;124:444–453. doi: 10.1161/CIRCULATIONAHA.110.014506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karbowski M, Norris KL, Cleland MM, Jeong SY, Youle RJ. Role of Bax and Bak in mitochondrial morphogenesis. Nature. 2006;443:658–662. doi: 10.1038/nature05111. [DOI] [PubMed] [Google Scholar]
- 38.Mitra K, Lippincott-Schwartz J. Analysis of mitochondrial dynamics and functions using imaging approaches. Curr Protoc Cell Biol Chapter. 2010;4 doi: 10.1002/0471143030.cb0425s46. Unit 4.25.1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cleland MM, Norris KL, Karbowski M, Wang C, Suen DF, Jiao S, George NM, Luo X, Li Z, Youle RJ. Bcl-2 family interaction with the mitochondrial morphogenesis machinery. Cell Death Differ. 2011;18:235–247. doi: 10.1038/cdd.2010.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Szabadkai G, Simoni AM, Chami M, Wieckowski MR, Youle RJ, Rizzuto R. Drp-1-dependent division of the mitochondrial network blocks intraorganellar Ca2+ waves and protects against Ca2+-mediated apoptosis. Mol Cell. 2004;16:59–68. doi: 10.1016/j.molcel.2004.09.026. [DOI] [PubMed] [Google Scholar]
- 41.Koopman WJH, Visch HJ, Smeitink JAM, Willems PHGM. Simultaneous quantitative measurement and automated analysis of mitochondrial morphology, mass, potential, and motility in living human skin fibroblasts. Cytometry A. 2006;69:1–12. doi: 10.1002/cyto.a.20198. [DOI] [PubMed] [Google Scholar]
- 42.Cribbs JT, Strack S. Functional characterization of phosphorylation sites in dynamin-related protein 1. Methods Enzymol. 2009;457:231–253. doi: 10.1016/S0076-6879(09)05013-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.De Vos KJ, Allan VJ, Grierson AJ, Sheetz MP. Mitochondrial function and actin regulate dynamin-related protein 1-dependent mitochondrial fission. Curr Biol. 2005;15:678–683. doi: 10.1016/j.cub.2005.02.064. [DOI] [PubMed] [Google Scholar]
- 44.Nikolaisen J, Nilsson LIH, Pettersen IKN, Willems PHGM, Lorens JB, Koopman WJH, Tronstad KJ. Automated quantification and integrative analysis of 2D and 3D mitochondrial shape and network properties. PLoS One. 2014;9:e101365. doi: 10.1371/journal.pone.0101365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Viana MP, Lim S, Rafelski SM. Quantifying mitochondrial content in living cells. Methods Cell Biol. 2015;125:77–93. doi: 10.1016/bs.mcb.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 46.Ahmad T, Aggarwal K, Pattnaik B, Mukherjee S, Sethi T, Tiwari BK, Kumar M, Micheal A, Mabalirajan U, Ghosh B, Sinha Roy S, Agrawal A. Computational classification of mitochondrial shapes reflects stress and redox state. Cell Death Dis. 2013;4:e461. doi: 10.1038/cddis.2012.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Giedt RJ, Fumene Feruglio P, Pathania D, Yang KS, Kilcoyne A, Vinegoni C, Mitchison TJ, Weissleder R. Computational imaging reveals mitochondrial morphology as a biomarker of cancer phenotype and drug response. Sci Rep. 2016;6:32985. doi: 10.1038/srep32985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leonard AP, Cameron RB, Speiser JL, Wolf BJ, Peterson YK, Schnellmann RG, Beeson CC, Rohrer B. Quantitative analysis of mitochondrial morphology and membrane potential in living cells using high-content imaging, machine learning, and morphological binning. Biochim Biophys Acta. 2015;1853:348–360. doi: 10.1016/j.bbamcr.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reis Y, Bernardo-Faura M, Richter D, Wolf T, Brors B, Hamacher-Brady A, Eils R, Brady NR. Multi-parametric analysis and modeling of relationships between mitochondrial morphology and apoptosis. PLoS One. 2012;7:e28694. doi: 10.1371/journal.pone.0028694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peng J-Y, Lin C-C, Chen Y-J, Kao L-S, Liu Y-C, Chou C-C, Huang Y-H, Chang F-R, Wu Y-C, Tsai Y-S, Hsu C-N. Automatic morphological subtyping reveals new roles of caspases in mitochondrial dynamics. PLoS Comput Biol. 2011;7:e1002212. doi: 10.1371/journal.pcbi.1002212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Merrill RA, Dagda RK, Dickey AS, Cribbs JT, Green SH, Usachev YM, Strack S. Mechanism of neuroprotective mitochondrial remodeling by PKA/AKAP1. PLoS Biol. 2011;9:e1000612. doi: 10.1371/journal.pbio.1000612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Merrill RA, Flippo KH, Strack S. Measuring Mitochondrial Shape with ImageJ. In: Strack S, Usachev YM, editors. Techniques to Investigate Mitochondrial Function in Neurons. Springer New York; New York, NY: 2017. pp. 31–48. [Google Scholar]
- 53.Ouellet M, Guillebaud G, Gervais V, Lupien St-Pierre D, Germain M. A novel algorithm identifies stress-induced alterations in mitochondrial connectivity and inner membrane structure from confocal images. PLoS Comput Biol. 2017;13:e1005612. doi: 10.1371/journal.pcbi.1005612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Valente AJ, Maddalena LA, Robb EL, Moradi F, Stuart JA. A simple ImageJ macro tool for analyzing mitochondrial network morphology in mammalian cell culture. Acta Histochem. 2017;119:315–326. doi: 10.1016/j.acthis.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 55.Lihavainen E, Mäkelä J, Spelbrink JN, Ribeiro AS. Mytoe: automatic analysis of mitochondrial dynamics. Bioinformatics. 2012;28:1050–1051. doi: 10.1093/bioinformatics/bts073. [DOI] [PubMed] [Google Scholar]
- 56.Rafelski SM, Viana MP, Zhang Y, Chan Y-HM, Thorn KS, Yam P, Fung JC, Li H, da F Costa L, Marshall WF. Mitochondrial network size scaling in budding yeast. Science. 2012;338:822–824. doi: 10.1126/science.1225720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vowinckel J, Hartl J, Butler R, Ralser M. MitoLoc: A method for the simultaneous quantification of mitochondrial network morphology and membrane potential in single cells. Mitochondrion. 2015;24:77–86. doi: 10.1016/j.mito.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Westrate LM, Drocco JA, Martin KR, Hlavacek WS, MacKeigan JP. Mitochondrial morphological features are associated with fission and fusion events. PLoS One. 2014;9:e95265. doi: 10.1371/journal.pone.0095265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Iannetti EF, Smeitink JAM, Beyrath J, Willems PHGM, Koopman WJH. Multiplexed high-content analysis of mitochondrial morphofunction using live-cell microscopy. Nat Protoc. 2016;11:1693–1710. doi: 10.1038/nprot.2016.094. [DOI] [PubMed] [Google Scholar]
- 60.Lackner LL, Ping H, Graef M, Murley A, Nunnari J. Endoplasmic reticulum-associated mitochondria-cortex tether functions in the distribution and inheritance of mitochondria. Proc Natl Acad Sci U S A. 2013;110:E458–67. doi: 10.1073/pnas.1215232110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Youle RJ, van der Bliek AM. Mitochondrial fission, fusion, and stress. Science. 2012;337:1062–1065. doi: 10.1126/science.1219855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol. 2011;12:9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Martinou JC, Youle RJ. Mitochondria in apoptosis: Bcl-2 family members and mitochondrial dynamics. Dev Cell. 2011;21:92–101. doi: 10.1016/j.devcel.2011.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Youle RJ, Karbowski M. Mitochondrial fission in apoptosis. Nat Rev Mol Cell Biol. 2005;6:657–663. doi: 10.1038/nrm1697. [DOI] [PubMed] [Google Scholar]
- 65.Goffart S, Martinsson P, Malka F, Rojo M, Spelbrink JN. The mitochondria of cultured mammalian cells: II. Expression and visualization of exogenous proteins in fixed and live cells. Methods Mol Biol. 2007;372:17–32. doi: 10.1007/978-1-59745-365-3_2. [DOI] [PubMed] [Google Scholar]
- 66.Mariotti FR, Corrado M, Campello S. Following mitochondria dynamism: confocal analysis of the organelle morphology. Methods Mol Biol. 2015;1241:153–161. doi: 10.1007/978-1-4939-1875-1_13. [DOI] [PubMed] [Google Scholar]
- 67.Marchi S, Bonora M, Patergnani S, Giorgi C, Pinton P. Methods to Assess Mitochondrial Morphology in Mammalian Cells Mounting Autophagic or Mitophagic Responses. Methods Enzymol. 2017;588:171–186. doi: 10.1016/bs.mie.2016.09.080. [DOI] [PubMed] [Google Scholar]
- 68.Pawley J. The 39 steps: a cautionary tale of quantitative 3-D fluorescence microscopy. Biotechniques. 2000;28:884–6,888. doi: 10.2144/00285bt01. [DOI] [PubMed] [Google Scholar]
- 69.North AJ. Seeing is believing? A beginners’ guide to practical pitfalls in image acquisition. J Cell Biol. 2006;172:9–18. doi: 10.1083/jcb.200507103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Verdaasdonk JS, Stephens AD, Haase J, Bloom K. Bending the rules: widefield microscopy and the Abbe limit of resolution. J Cell Physiol. 2014;229:132–138. doi: 10.1002/jcp.24439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Berman SB, Chen Y-B, Qi B, McCaffery JM, Rucker EB, 3rd, Goebbels S, Nave K-A, Arnold BA, Jonas EA, Pineda FJ, Hardwick JM. Bcl-x L increases mitochondrial fission, fusion, and biomass in neurons. J Cell Biol. 2009;184:707–719. doi: 10.1083/jcb.200809060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Karbowski M, Cleland MM, Roelofs BA. Photoactivatable green fluorescent protein-based visualization and quantification of mitochondrial fusion and mitochondrial network complexity in living cells. Methods Enzymol. 2014;547:57–73. doi: 10.1016/B978-0-12-801415-8.00004-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Karbowski M, Arnoult D, Chen H, Chan DC, Smith CL, Youle RJ. Quantitation of mitochondrial dynamics by photolabeling of individual organelles shows that mitochondrial fusion is blocked during the Bax activation phase of apoptosis. J Cell Biol. 2004;164:493–499. doi: 10.1083/jcb.200309082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Twig G, Graf SA, Wikstrom JD, Mohamed H, Haigh SE, Elorza A, Deutsch M, Zurgil N, Reynolds N, Shirihai OS. Tagging and tracking individual networks within a complex mitochondrial web with photoactivatable GFP. Am J Physiol Cell Physiol. 2006;291:C176–84. doi: 10.1152/ajpcell.00348.2005. [DOI] [PubMed] [Google Scholar]
- 75.Rehman J, Zhang HJ, Toth PT, Zhang Y, Marsboom G, Hong Z, Salgia R, Husain AN, Wietholt C, Archer SL. Inhibition of mitochondrial fission prevents cell cycle progression in lung cancer. FASEB J. 2012;26:2175–2186. doi: 10.1096/fj.11-196543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pham AH, McCaffery JM, Chan DC. Mouse lines with photo-activatable mitochondria to study mitochondrial dynamics. Genesis. 2012;50:833–843. doi: 10.1002/dvg.22050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang Y, Avalos JL. Traditional and novel tools to probe the mitochondrial metabolism in health and disease, Wiley Interdiscip. Rev Syst Biol Med. 2017;9:e1373. doi: 10.1002/wsbm.1373. [DOI] [PubMed] [Google Scholar]
- 78.De Michele R, Carimi F, Frommer WB. Mitochondrial biosensors. Int J Biochem Cell Biol. 2014;48:39–44. doi: 10.1016/j.biocel.2013.12.014. [DOI] [PubMed] [Google Scholar]
- 79.Rizzuto R, Nakase H, Darras B, Francke U, Fabrizi GM, Mengel T, Walsh F, Kadenbach B, DiMauro S, Schon EA. A gene specifying subunit VIII of human cytochrome c oxidase is localized to chromosome 11 and is expressed in both muscle and non-muscle tissues. J Biol Chem. 1989;264:10595–10600. [PubMed] [Google Scholar]
- 80.Rizzuto R, Simpson AW, Brini M, Pozzan T. Rapid changes of mitochondrial Ca2+ revealed by specifically targeted recombinant aequorin. Nature. 1992;358:325–327. doi: 10.1038/358325a0. [DOI] [PubMed] [Google Scholar]
- 81.Rizzuto R, Brini M, Pizzo P, Murgia M, Pozzan T. Chimeric green fluorescent protein as a tool for visualizing subcellular organelles in living cells. Curr Biol. 1995;5:635–642. doi: 10.1016/s0960-9822(95)00128-x. [DOI] [PubMed] [Google Scholar]
- 82.Shaner NC, Patterson GH, Davidson MW. Advances in fluorescent protein technology. J Cell Sci. 2007;120:4247–4260. doi: 10.1242/jcs.005801. [DOI] [PubMed] [Google Scholar]
- 83.Friedman JR, Lackner LL, West M, DiBenedetto JR, Nunnari J, Voeltz GK. ER tubules mark sites of mitochondrial division. Science. 2011;334:358–362. doi: 10.1126/science.1207385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Markwardt ML, Kremers GJ, Kraft CA, Ray K, Cranfill PJC, Wilson KA, Day RN, Wachter RM, Davidson MW, Rizzo MA. An improved cerulean fluorescent protein with enhanced brightness and reduced reversible photoswitching. PLoS One. 2011;6:e17896. doi: 10.1371/journal.pone.0017896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Planchon TA, Gao L, Milkie DE, Davidson MW, Galbraith JA, Galbraith CG, Betzig E. Rapid three-dimensional isotropic imaging of living cells using Bessel beam plane illumination. Nat Methods. 2011;8:417–423. doi: 10.1038/nmeth.1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lam AJ, St-Pierre F, Gong Y, Marshall JD, Cranfill PJ, Baird MA, McKeown MR, Wiedenmann J, Davidson MW, Schnitzer MJ, Tsien RY, Lin MZ. Improving FRET dynamic range with bright green and red fluorescent proteins. Nat Methods. 2012;9:1005–1012. doi: 10.1038/nmeth.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Roberts B, Haupt A, Tucker A, Grancharova T, Arakaki J, Fuqua MA, Nelson A, Hookway C, Ludmann SA, Mueller IA, Yang R, Horwitz R, Rafelski SM, Gunawardane RN. Systematic gene tagging using CRISPR/Cas9 in human stem cells to illuminate cell organization. Mol Biol Cell. 2017;28:2854–2874. doi: 10.1091/mbc.E17-03-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rizzo MA, Davidson MW, Piston DW. Fluorescent protein tracking and detection: fluorescent protein structure and color variants. Cold Spring Harb Protoc. 2009;2009 doi: 10.1101/pdb.top63. db.top63. [DOI] [PubMed] [Google Scholar]
- 89.Shaner NC, Lin MZ, McKeown MR, Steinbach PA, Hazelwood KL, Davidson MW, Tsien RY. Improving the photostability of bright monomeric orange and red fluorescent proteins. Nat Methods. 2008;5:545–551. doi: 10.1038/nmeth.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ai HW, Hazelwood KL, Davidson MW, Campbell RE. Fluorescent protein FRET pairs for ratiometric imaging of dual biosensors. Nat Methods. 2008;5:401–403. doi: 10.1038/nmeth.1207. [DOI] [PubMed] [Google Scholar]
- 91.Day RN, Davidson MW. The fluorescent protein palette: tools for cellular imaging. Chem Soc Rev. 2009;38:2887–2921. doi: 10.1039/b901966a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Olenych SG, Claxton NS, Ottenberg GK, Davidson MW. The fluorescent protein color palette. Curr Protoc Cell Biol. 2007 doi: 10.1002/0471143030.cb2105s36. Chapter 21. Unit 21.5. [DOI] [PubMed] [Google Scholar]
- 93.Yu D, Baird MA, Allen JR, Howe ES, Klassen MP, Reade A, Makhijani K, Song Y, Liu S, Murthy Z, Zhang SQ, Weiner OD, Kornberg TB, Jan YN, Davidson MW, Shu X. A naturally monomeric infrared fluorescent protein for protein labeling in vivo. Nat Methods. 2015;12:763–765. doi: 10.1038/nmeth.3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kremers GJ, Hazelwood KL, Murphy CS, Davidson MW, Piston DW. Photoconversion in orange and red fluorescent proteins. Nat Methods. 2009;6:355–358. doi: 10.1038/nmeth.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.York AG, Ghitani A, Vaziri A, Davidson MW, Shroff H. Confined activation and subdiffractive localization enables whole-cell PALM with genetically expressed probes. Nat Methods. 2011;8:327–333. doi: 10.1038/nmeth.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shu X, Lev-Ram V, Deerinck TJ, Qi Y, Ramko EB, Davidson MW, Jin Y, Ellisman MH, Tsien RY. A genetically encoded tag for correlated light and electron microscopy of intact cells, tissues, and organisms. PLoS Biol. 2011;9:e1001041. doi: 10.1371/journal.pbio.1001041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.McKinney SA, Murphy CS, Hazelwood KL, Davidson MW, Looger LL. A bright and photostable photoconvertible fluorescent protein. Nat Methods. 2009;6:131–133. doi: 10.1038/nmeth.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Shtengel G, Galbraith JA, Galbraith CG, Lippincott-Schwartz J, Gillette JM, Manley S, Sougrat R, Waterman CM, Kanchanawong P, Davidson MW, Fetter RD, Hess HF. Interferometric fluorescent super-resolution microscopy resolves 3D cellular ultrastructure. Proc Natl Acad Sci U S A. 2009;106:3125–3130. doi: 10.1073/pnas.0813131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Laker RC, Xu P, Ryall KA, Sujkowski A, Kenwood BM, Chain KH, Zhang M, Royal MA, Hoehn KL, Driscoll M, Adler PN, Wessells RJ, Saucerman JJ, Yan Z. A novel MitoTimer reporter gene for mitochondrial content, structure, stress, and damage in vivo. J Biol Chem. 2014;289:12005–12015. doi: 10.1074/jbc.M113.530527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Williams JA, Zhao K, Jin S, Ding WX. New methods for monitoring mitochondrial biogenesis and mitophagy in vitro and in vivo. Exp Biol Med. 2017;242:781–787. doi: 10.1177/1535370216688802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hernandez G, Thornton C, Stotland A, Lui D, Sin J, Ramil J, Magee N, Andres A, Quarato G, Carreira RS, Sayen MR, Wolkowicz R, Gottlieb RA. MitoTimer: a novel tool for monitoring mitochondrial turnover. Autophagy. 2013;9:1852–1861. doi: 10.4161/auto.26501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Allen GFG, Toth R, James J, Ganley IG. Loss of iron triggers PINK1/Parkin-independent mitophagy. EMBO Rep. 2013;14:1127–1135. doi: 10.1038/embor.2013.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.McWilliams TG, Prescott AR, Allen GFG, Tamjar J, Munson MJ, Thomson C, Muqit MMK, Ganley IG. Mito-QC illuminates mitophagy and mitochondrial architecture in vivo. J Cell Biol. 2016;214:333–345. doi: 10.1083/jcb.201603039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gutscher M, Pauleau AL, Marty L, Brach T, Wabnitz GH, Samstag Y, Meyer AJ, Dick TP. Real-time imaging of the intracellular glutathione redox potential. Nat Methods. 2008;5:553–559. doi: 10.1038/nmeth.1212. [DOI] [PubMed] [Google Scholar]
- 105.Poburko D, Santo-Domingo J, Demaurex N. Dynamic regulation of the mitochondrial proton gradient during cytosolic calcium elevations. J Biol Chem. 2011;286:11672–11684. doi: 10.1074/jbc.M110.159962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wang W, Gong G, Wang X, Wei-LaPierre L, Cheng H, Dirksen R, Sheu S-S. Mitochondrial Flash: Integrative Reactive Oxygen Species and pH Signals in Cell and Organelle Biology. Antioxid Redox Signal. 2016;25:534–549. doi: 10.1089/ars.2016.6739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Belousov VV, Fradkov AF, Lukyanov KA, Staroverov DB, Shakhbazov KS, Terskikh AV, Lukyanov S. Genetically encoded fluorescent indicator for intracellular hydrogen peroxide. Nat Methods. 2006;3:281–286. doi: 10.1038/nmeth866. [DOI] [PubMed] [Google Scholar]
- 108.Suzuki J, Kanemaru K, Ishii K, Ohkura M, Okubo Y, Iino M. Imaging intraorganellar Ca2+ at subcellular resolution using CEPIA. Nat Commun. 2014;5:4153. doi: 10.1038/ncomms5153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bulina ME, Chudakov DM, Britanova OV, Yanushevich YG, Staroverov DB, Chepurnykh TV, Merzlyak EM, Shkrob MA, Lukyanov S, Lukyanov KA. A genetically encoded photosensitizer. Nat Biotechnol. 2006;24:95–99. doi: 10.1038/nbt1175. [DOI] [PubMed] [Google Scholar]
- 110.Ettinger A, Wittmann T. Fluorescence live cell imaging Methods. Cell Biol. 2014;123:77–94. doi: 10.1016/B978-0-12-420138-5.00005-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Khodjakov A, Rieder C, Mannella CA, Kinnally KW. Laser micro-irradiation of mitochondria: is there an amplified mitochondrial death signal in neural cells? Mitochondrion. 2004;3:217–227. doi: 10.1016/j.mito.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 112.Amchenkova AA, Bakeeva LE, Chentsov YS, Skulachev VP, Zorov DB. Coupling membranes as energy-transmitting cables. I. Filamentous mitochondria in fibroblasts and mitochondrial clusters in cardiomyocytes. J Cell Biol. 1988;107:481–495. doi: 10.1083/jcb.107.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Icha J, Weber M, Waters JC, Norden C. Phototoxicity in live fluorescence microscopy, and how to avoid it. Bioessays. 2017;39 doi: 10.1002/bies.201700003. [DOI] [PubMed] [Google Scholar]
- 114.Chen B-C, Legant WR, Wang K, Shao L, Milkie DE, Davidson MW, Janetopoulos C, Wu XS, Hammer JA, 3rd, Liu Z, English BP, Mimori-Kiyosue Y, Romero DP, Ritter AT, Lippincott-Schwartz J, Fritz-Laylin L, Mullins RD, Mitchell DM, Bembenek JN, Reymann A-C, Böhme R, Grill SW, Wang JT, Seydoux G, Tulu US, Kiehart DP, Betzig E. Lattice light-sheet microscopy: imaging molecules to embryos at high spatiotemporal resolution. Science. 2014;346:1257998. doi: 10.1126/science.1257998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Emaus RK, Grunwald R, Lemasters JJ. Rhodamine 123 as a probe of transmembrane potential in isolated rat-liver mitochondria: spectral and metabolic properties. Biochim Biophys Acta. 1986;850:436–448. doi: 10.1016/0005-2728(86)90112-x. [DOI] [PubMed] [Google Scholar]
- 116.Chacon E, Reece JM, Nieminen AL, Zahrebelski G, Herman B, Lemasters JJ. Distribution of electrical potential, pH, free Ca2+, and volume inside cultured adult rabbit cardiac myocytes during chemical hypoxia: a multiparameter digitized confocal microscopic study. Biophys J. 1994;66:942–952. doi: 10.1016/S0006-3495(94)80904-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Scaduto RC, Jr, Grotyohann LW. Measurement of mitochondrial membrane potential using fluorescent rhodamine derivatives. Biophys J. 1999;76:469–477. doi: 10.1016/S0006-3495(99)77214-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Porcelli AM, Pinton P, Ainscow EK, Chiesa A, Rugolo M, Rutter GA, Rizzuto R. Methods in Cell Biology. Academic Press; 2001. Targeting of reporter molecules to mitochondria to measure calcium, ATP, and pH; pp. 353–380. [DOI] [PubMed] [Google Scholar]
- 119.Tanji K, Bonilla E. Optical imaging techniques (histochemical, immunohistochemical, and in situ hybridization staining methods) to visualize mitochondria. Methods Cell Biol. 2007;80:135–154. doi: 10.1016/S0091-679X(06)80006-3. [DOI] [PubMed] [Google Scholar]
- 120.Lemasters JJ, Ramshesh VK. Imaging of mitochondrial polarization and depolarization with cationic fluorophores. Methods Cell Biol. 2007;80:283–295. doi: 10.1016/S0091-679X(06)80014-2. [DOI] [PubMed] [Google Scholar]
- 121.Bouchier-Hayes L, Muñoz-Pinedo C, Connell S, Green DR. Measuring apoptosis at the single cell level. Methods. 2008;44:222–228. doi: 10.1016/j.ymeth.2007.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Perry SW, Norman JP, Barbieri J, Brown EB, Gelbard HA. Mitochondrial membrane potential probes and the proton gradient: a practical usage guide. Biotechniques. 2011;50:98–115. doi: 10.2144/000113610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Cottet-Rousselle C, Ronot X, Leverve X, Mayol JF. Cytometric assessment of mitochondria using fluorescent probes. Cytometry A. 2011;79:405–425. doi: 10.1002/cyto.a.21061. [DOI] [PubMed] [Google Scholar]
- 124.Duchen MR, Surin A, Jacobson J. Methods in Enzymology. Academic Press; 2003. [17] Imaging mitochondrial function in intact cells; pp. 353–389. [DOI] [PubMed] [Google Scholar]
- 125.Reers M, Smith TW, Chen LB. J-aggregate formation of a carbocyanine as a quantitative fluorescent indicator of membrane potential. Biochemistry. 1991;30:4480–4486. doi: 10.1021/bi00232a015. [DOI] [PubMed] [Google Scholar]
- 126.Duchen MR, McGuinness O, Brown LA, Crompton M. On the involvement of a cyclosporin A sensitive mitochondrial pore in myocardial reperfusion injury. Cardiovasc Res. 1993;27:1790–1794. doi: 10.1093/cvr/27.10.1790. [DOI] [PubMed] [Google Scholar]
- 127.Chinopoulos C, Tretter L, Adam-Vizi V. Depolarization of In Situ Mitochondria Due to Hydrogen Peroxide-Induced Oxidative Stress in Nerve Terminals. J Neurochem. 1999;73:220–228. doi: 10.1046/j.1471-4159.1999.0730220.x. [DOI] [PubMed] [Google Scholar]
- 128.Nicholls DG, Ward MW. Mitochondrial membrane potential and neuronal glutamate excitotoxicity: mortality and millivolts. Trends Neurosci. 2000;23:166–174. doi: 10.1016/s0166-2236(99)01534-9. [DOI] [PubMed] [Google Scholar]
- 129.Kalyanaraman B, Hardy M, Podsiadly R, Cheng G, Zielonka J. Recent developments in detection of superoxide radical anion and hydrogen peroxide: Opportunities, challenges, and implications in redox signaling. Arch Biochem Biophys. 2017;617:38–47. doi: 10.1016/j.abb.2016.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Liu Y, Schubert DR. The specificity of neuroprotection by antioxidants. J Biomed Sci. 2009;16:98. doi: 10.1186/1423-0127-16-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Polster BM, Nicholls DG, Ge SX, Roelofs BA. Use of potentiometric fluorophores in the measurement of mitochondrial reactive oxygen species. Methods Enzymol. 2014;547:225–250. doi: 10.1016/B978-0-12-801415-8.00013-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Cheng G, Zielonka J, Kalyanaraman B. Modulatory effects of MitoSOX on cellular bioenergetics: a cautionary note. Free Radical Biology and Medicine. 2011;51:S37–S38. [Google Scholar]
- 133.Roelofs BA, Ge SX, Studlack PE, Polster BM. Low micromolar concentrations of the superoxide probe MitoSOX uncouple neural mitochondria and inhibit complex IV. Free Radic Biol Med. 2015;86:250–258. doi: 10.1016/j.freeradbiomed.2015.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zielonka J, Kalyanaraman B. Hydroethidine- and MitoSOX-derived red fluorescence is not a reliable indicator of intracellular superoxide formation: another inconvenient truth. Free Radic Biol Med. 2010;48:983–1001. doi: 10.1016/j.freeradbiomed.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Cochemé HM, Quin C, McQuaker SJ, Cabreiro F, Logan A, Prime TA, Abakumova I, Patel JV, Fearnley IM, James AM, Porteous CM, Smith RAJ, Saeed S, Carré JE, Singer M, Gems D, Hartley RC, Partridge L, Murphy MP. Measurement of H2O2 within living Drosophila during aging using a ratiometric mass spectrometry probe targeted to the mitochondrial matrix. Cell Metab. 2011;13:340–350. doi: 10.1016/j.cmet.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Cochemé HM, Logan A, Prime TA, Abakumova I, Quin C, McQuaker SJ, Patel JV, Fearnley IM, James AM, Porteous CM, Smith RAJ, Hartley RC, Partridge L, Murphy MP. Using the mitochondria-targeted ratiometric mass spectrometry probe MitoB to measure H2O2 in living Drosophila. Nat Protoc. 2012;7:946–958. doi: 10.1038/nprot.2012.035. [DOI] [PubMed] [Google Scholar]
- 137.Shchepinova MM, Cairns AG, Prime TA, Logan A, James AM, Hall AR, Vidoni S, Arndt S, Caldwell ST, Prag HA, Pell VR, Krieg T, Mulvey JF, Yadav P, Cobley JN, Bright TP, Senn HM, Anderson RF, Murphy MP, Hartley RC. MitoNeoD: A Mitochondria-Targeted Superoxide Probe. Cell Chem Biol. 2017;24:1285–1298.e12. doi: 10.1016/j.chembiol.2017.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Arai S, Suzuki M, Park SJ, Yoo JS, Wang L, Kang NY, Ha HH, Chang YT. Mitochondria-targeted fluorescent thermometer monitors intracellular temperature gradient. Chem Commun. 2015;51:8044–8047. doi: 10.1039/c5cc01088h. [DOI] [PubMed] [Google Scholar]
- 139.Chretien D, Benit P, Ha HH, Keipert S, El-Khoury R, Chang YT, Jastroch M, Jacobs H, Rustin P, Rak M. Mitochondria Are Physiologically Maintained At Close To 50. C, bioRxiv. 2017:133223. doi: 10.1101/133223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Poot M, Zhang YZ, Krämer JA, Wells KS, Jones LJ, Hanzel DK, Lugade AG, Singer VL, Haugland RP. Analysis of mitochondrial morphology and function with novel fixable fluorescent stains. J Histochem Cytochem. 1996;44:1363–1372. doi: 10.1177/44.12.8985128. [DOI] [PubMed] [Google Scholar]
- 141.R.C. Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2016. p. 2017. [Google Scholar]
- 142.Buckman JF, Hernández H, Kress GJ, Votyakova TV, Pal S, Reynolds IJ. MitoTracker labeling in primary neuronal and astrocytic cultures: influence of mitochondrial membrane potential and oxidants. J Neurosci Methods. 2001;104:165–176. doi: 10.1016/s0165-0270(00)00340-x. [DOI] [PubMed] [Google Scholar]
- 143.Weigert R, Sramkova M, Parente L, Amornphimoltham P, Masedunskas A. Intravital microscopy: a novel tool to study cell biology in living animals. Histochem Cell Biol. 2010;133:481–491. doi: 10.1007/s00418-010-0692-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Masedunskas A, Porat-Shliom N, Tora M, Milberg O, Weigert R. Intravital microscopy for imaging subcellular structures in live mice expressing fluorescent proteins. J Vis Exp. 2013 doi: 10.3791/50558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Hoffman RM. The multiple uses of fluorescent proteins to visualize cancer in vivo. Nat Rev Cancer. 2005;5:796–806. doi: 10.1038/nrc1717. [DOI] [PubMed] [Google Scholar]
- 146.Hoffman RM, Yang M. Subcellular imaging in the live mouse. Nat Protoc. 2006;1:775–782. doi: 10.1038/nprot.2006.109. [DOI] [PubMed] [Google Scholar]
- 147.Hoffman RM, Yang M. Color-coded fluorescence imaging of tumor-host interactions. Nat Protoc. 2006;1:928–935. doi: 10.1038/nprot.2006.119. [DOI] [PubMed] [Google Scholar]
- 148.Hoffman RM, Yang M. Whole-body imaging with fluorescent proteins. Nat Protoc. 2006;1:1429–1438. doi: 10.1038/nprot.2006.223. [DOI] [PubMed] [Google Scholar]
- 149.Hoffman RM. Application of GFP imaging in cancer. Lab Invest. 2015;95:432–452. doi: 10.1038/labinvest.2014.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Hall AM, Rhodes GJ, Sandoval RM, Corridon PR, Molitoris BA. In vivo multiphoton imaging of mitochondrial structure and function during acute kidney injury. Kidney Int. 2013;83:72–83. doi: 10.1038/ki.2012.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Porat-Shliom N, Chen Y, Tora M, Shitara A, Masedunskas A, Weigert R. In vivo tissue-wide synchronization of mitochondrial metabolic oscillations. Cell Rep. 2014;9:514–521. doi: 10.1016/j.celrep.2014.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Takihara Y, Inatani M, Eto K, Inoue T, Kreymerman A, Miyake S, Ueno S, Nagaya M, Nakanishi A, Iwao K, Takamura Y, Sakamoto H, Satoh K, Kondo M, Sakamoto T, Goldberg JL, Nabekura J, Tanihara H. In vivo imaging of axonal transport of mitochondria in the diseased and aged mammalian CNS. Proc Natl Acad Sci U S A. 2015;112:10515–10520. doi: 10.1073/pnas.1509879112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A,D) Original widefield (A; DSU out) versus confocal (D; DSU IN) images from Figure 1 (original), previously acquired with Metamorph software, were input into the cellSens software (Olympus) and were processed with the blind iterative deconvolution module (deconvolved). Note the improvement deconvolution added to both widefield and confocal images. B-C, E-F) Either the original or deconvolved images from (A, D) were cropped and processed with MitoGraph v3.0 using the default MitoGraph settings (B, E; ./MitoGraph -xy 0.1667 -z 0.2 -path ~/Desktop/cells/) or the settings optimized for this study (C, F; ./MitoGraph -xy 0.1667 -z 0.2 -scales 1.0 1.3 4 -adaptive 10 -path ~/Desktop/cells/). Note the TIFF images output from the cellSens deconvolution algorithm were incompatible with MitoGraph v3.0 and were first processed with a the FIJI 3D median filter (set to 1,1,1) which was then MitoGraph v3.0 compatible. Arrows denote examples of non-mitochondrial areas detected when the settings are not properly optimized. The circles denote examples of mitochondrial regions that are absent in the segmented MitoGraph image indicating poor thresholding by the default MitoGraph settings, likely due to poor S/N of the original image. The images outlined in green designate the optimal settings used throughout the manuscript. Scale bar represents 10μm.
Supplemental Figure 2: Troubleshooting guide. See figure for description.
The top panel indicates network measurements generated for each cell, while the bottom portion focuses on the metrics gathered for an individual mitochondrion or the connected component that result for connections between multiple nodes. Central to understanding MitoGraph measurements is understanding the definition of nodes (mitochondrion end points or junction points between branches of the network) and edges (cylindrical connection between nodes, essentially each branch is an edge). The total length, nodes and edges are calculated for each connected component found within the cells From this information the overall network size or total mitochondrial length is calculated by taking the sum of the length of each connected component. The number of connected components is tallied and the ratio of the largest connected component to the total network size is calculated and is designated as PHI. The average edge length is calculated by taking the total mitochondrial length and dividing it by the total edge number. The total nodes and edges are tallied and finally the nodes are assessed for the fraction found in free ends (k=1) versus those found in 3-way and 4-way junctions (k = 3/4) and the average degree calculated from those measurements.
A) Six distinct mitochondrial phenotypes arranged from highly fused to highly fragmented (see figure for the indicated cell type; see Figures 3–5 for original images). B) Individual cell metrics copied as is from the .mitograph text files. Total length, nodes and connected components generated from the R-script CreateSummary.R that extracts information from the .GNET file (contains node assignments and edge length for each edge in the network). From this information the normalized metrics are calculated as described on the figure. Note that PHI is generated by first determining the largest connected component found in a given cell and then dividing that by the total length of the entire network. C) Graphical depiction of the data presented in the table in (B). Note that some parameters increase upon enhanced fragmentation, while others decrease upon enhanced fragmentation. The MitoGraph Connectivity Score represents this relationship by summing the factors elevated in a highly fused scenario (PHI, Avg edge length and Avg Degree) and dividing by those factors elevated in a highly fragmented scenario (Total nodes, edges and connected components (all normalized to length)).
Output from R Studio of the dataset from Figure 5 processed with following R-scripts: CreateSummary.R followed by CreatePlots.R.
A, B) 10 images were collected for HMEC-1 WT or CCCP treated cells (A; see Figure 4 legend for details) or Hmec-1 WT of Drp1 KO (B; see Figure 5 for details). From those 10 images, cells were individually cropped and processed as depicted by Supplemental Figures 3 and 4.
Cropped images of 4 individual cells per cell type (293T, C2C12, C6, H1299 and HEK293) of Tom20 immunostained mitochondria (left) and the resulting MitoGraph PNG binary image (right). Using FIJI, the cropped images were combined into a single stack to permit applying the same brightness/contrast to all the images and the stack was then displayed as a 4 × 5 montage using a 1.00 scale. Note that the cells were stained and imaged using identical settings and despite the differences in Tom20 staining, MitoGraph effectively segmented the mitochondria in all cell types. 293T cells were problematic showing rogue lines or regions from neighboring cells in all the cells assessed and therefore was not processed for downstream analysis (see Supplemental Figure 2 Problem 5).


