Abstract
Genotyping-by-sequencing (GBS) provides high SNP coverage and has recently emerged as a popular technology for genetic and breeding applications in bread wheat (Triticum aestivum L.) and many other plant species. Although GBS can discover millions of SNPs, a high rate of missing data is a major concern for many applications. Accurate imputation of those missing data can significantly improve the utility of GBS data. This study compared imputation accuracies among four genome references including three wheat references (Chinese Spring survey sequence, W7984, and IWGSC RefSeq v1.0) and one barley reference genome by comparing imputed data derived from low-depth sequencing to actual data from high-depth sequencing. After imputation, the average number of imputed data points was the highest in the B genome (~48.99%). The D genome had the lowest imputed data points (~15.02%) but the highest imputation accuracy. Among the four reference genomes, IWGSC RefSeq v1.0 reference provided the most imputed data points, but the lowest imputation accuracy for the SNPs with < 10% minor allele frequency (MAF). The W7984 reference, however, provided the highest imputation accuracy for the SNPs with < 10% MAF.
Introduction
Wheat (Triticum aestivum L.) is a major staple food crop in the world. The fast-growing world population demands wheat production to be increased up to 70% by 2050 to feed estimated world population of approximately nine billion [1–3]. Application of advanced genomic technologies in breeding can speed up genetic improvement of new wheat varieties to meet the challenge [4]. Next-generation-sequencing (NGS) technologies have revolutionized throughput and greatly reduced DNA sequencing cost, which makes it feasible for routine screening of breeding materials [5]. Genotyping-by-sequencing (GBS) is one application that sequences a subset of a complex genome and also multiplexes various numbers of samples to lower the genotyping cost [6]. GBS can discover and genotype single nucleotide polymorphisms (SNPs) simultaneously and it is a valuable platform for crop breeding and genomic research [6].
SNPs are the most abundant type of sequence variations in plant genomes [7] and therefore suitable for studies that require a large number of markers to be assayed such as marker-trait association analysis, genetic map construction, quantitative trait locus (QTL) screening, genomic selection, and analysis of population structure and genetic variation [8]. High-throughput SNP genotyping platforms have been successfully used for diploid crops such as maize [9] and barley [10]. Wheat, however, is polyploid and has a huge genome (~17 Gb) with abundant repetitive DNA (> 80%), which present major challenges to direct sequencing the genome for developing high-density SNP maps [11]. Recently, a GBS protocol [5] has been optimized for cereal crops including wheat and can generate thousands of SNPs with reasonably low cost [12–14]. However, abundance of missing data due to low sequencing coverage significantly reduces number of usable SNPs and lowers marker density [5]. High marker density will improve accuracy of many downstream analyses such as QTL mapping, genome-wide association studies (GWAS) and genomic selection [15, 16]. Sequencing depth and library complexity and quality may all affect number of missing data. Increase in sequencing depth can lower missing data rate, but also increase sequencing cost. Marker imputation using available information from reference genomes can increase usable SNPs without increasing sequencing cost. Several imputation algorithms including IMPUTE [17], MaCH [18], fastPHASE [19], BEAGLE [20] have been developed to assign allelic status of missing values to genotypic data. Among those algorithms, IMPUTE and MaCH use hidden Markov model (HMM) and Markov chain Monte Carlo (MCMC) iterations to conduct subsampling, and the haplotypes in each iteration are considered as a sample from the haplotype pool. FastPHASE and BEAGLE, however, cluster haplotypes and collapsed total number of haplotypes into a smaller number of “ancestral” haplotypes [21]. Although both BEAGLE and fastPHASE use a hidden Markov model, BEAGLE is more parsimonious by allowing fewer possible transitions and emissions. In addition, fastPHASE fixes the number of clusters in the model, whereas BEAGLE allows dynamic change of number of clusters to fit localized linkage disequilibrium (LD) patterns [22]. Therefore, BEAGLE has been used to impute missing data in many studies [23–25]. Length of LD blocks greatly affects imputation accuracy because recombination breaks allelic associations. The markers that are common between samples and a reference panel serve as anchors to guide genotype imputation of any missing haplotypes within an LD block [26].
Imputation strategies may vary from species to species depending on availability of reference genomes and well-saturated reference linkage maps in a species. A more complete reference genome allows proper alignment and ordering of the sequenced tags and helps impute low coverage data [27]. To date, three wheat reference genomes and one barley reference genome have been reported [28]. Among the three wheat reference genomes, Chinese Spring survey sequence (CSSS) [29] has 10.2 Gb of sequences generated from Illumina NGS and W7984 reference has 9.1 Gb of sequences that were assembled using large-insert libraries and the three homoeologous genomes were assembled separately [30]. W7984 reference has lower genome coverage than CSSS, but higher assembly quality. Wheat IWGSC RefSeq v1.0 reference is the newest version of wheat reference genome with the best assembly quality, contains 14.5 Gb sequences with 94% genome coverage and was assembled using POPSEQ data and HiC map (chromosome conformation capture) [https://wheat-urgi.versailles.inra.fr/Seq-Repository/Assemblies]. Here we used the four reference genomes to compare imputation efficiencies and accuracies of wheat GBS data.
Materials and methods
Plant materials
A total of 384 accessions of Iranian wheat accessions [http://biogeo.ucdavis.edu/projects/iranwheat] were kindly provided by the United States Department of Agriculture (USDA) germplasm collection (https://npgsweb.ars-grin.gov/gringlobal/search.aspx), International Center for the Improvement of Maize and Wheat (CIMMYT), University of Tehran (UT), and Seed and Plant Improvement Institute (SPII), Karaj, Iran [31]. The wheat collection includes 276 Iranian landraces collected from different climates between 1937 and 1968 and 108 cultivars released in Iran between 1942 and 2014. Genomic DNA of the accessions were extracted from two-week-old seedling leaves using a modified cetyltrimethyl ammonium bromide (CTAB) method [32]. DNA concentration was quantified using the Quant-iT PicoGreen dsDNA Assay (Life Technologies Inc., NY) and normalized to 20 ng/μl.
GBS library preparation and sequencing
The GBS library was constructed following Poland et al. [13]. In brief, genomic DNA of each sample was double-digested with PstI (CTGCAG) and MspI (CCGG) restriction enzymes (New England BioLabs Inc., Ipswich, MA, USA), and ligated to barcoded adapters using T4 ligase (New England BioLabs Inc.). All the ligated products were pooled and cleaned up using the QIAquick PCR Purification Kit (Qiagen Inc., Valencia, CA, USA). Primers complementary to both adaptors were used for PCR. PCR amplification started at 95 °C for 5 min, followed by 16 cycles of 95 °C for 30 s, 62 °C for 20 s and 68 °C for 1 min and ended by a final extension step at 72 °C for 5 min. The PCR product was then cleaned up again using the QIAquick PCR Purification Kit, and quantified using Bioanalyzer 7500 Agilent DNA Chips (Agilent Technologies, Inc.). After size-selection for 250–300 bp fragments in an E-gel system (Life Technologies Inc.), concentration of the library was evaluated using a Qubit 2.0 fluorometer and Qubit dsDNA HS Assay Kit (Life Technologies Inc.). The size-selected library was sequenced on an Ion Proton system (Life Technologies Inc.).
Sequence reads were first trimmed to 64 bp, and identical reads were grouped into sequence tags. Unique sequence tags were aligned internally to identify SNPs within the tags allowing mismatches of up to 3 bp. SNPs were called using the Universal Network Enabled Analysis Kit (UNEAK) GBS pipeline [33] in TASSEL 3.0 bioinformatics analysis package [34]. Tags with low quality score (< 15) were removed. SNPs with heterozygotes or a minor allele frequency > 10% were discarded to reduce the false positive markers. Only SNPs with lower than 80% missing data were used for this study. BLASTn analysis was carried out to align sequence tags to the four genome references including one from the barley reference genome [28], and three from wheat reference genomes, the flow-sorted Chinese Spring survey sequence (CSSS) [29], the Popseq W7984 sequence reference [30] and IWGSC RefSeq v1.0 [https://wheat-urgi.versailles.inra.fr/Seq-Repository/Assemblies]. The purpose of using the barley reference genome is to show efficiency of using reference genomes of closely related species to impute missing data in cases where reference genome sequence is absent in some species. If a SNP could be mapped in multiple chromosome positions, the position with the lowest E-value was used to represent the SNP location.
In this study, imputation was performed using BEAGLE v3.3.2 [20] and the four genome reference genomes. BEAGLE used a phasing algorithm to determine haplotype phase for each individual and to impute the missing values based upon allele frequencies. This was done by constructing local haplotype clusters and then sampling a number of haplotypes for each individual from a special class of HMM. Probability of each possible haplotype was estimated using the genotypic information and a forward-backward algorithm [35]. Then, new haplotypes for the individuals were sampled according to the conditional probabilities to reconstruct the local haplotype cluster as input for next iteration. This process was repeated several times. To achieve a high level of phasing accuracy in the end, the most-likely haplotypes for each individuals were imputed using the Viterbi algorithm [35].
Imputation accuracy was calculated by comparing the imputed SNPs after five, six and seven sequencing runs to the actual SNPs called from eight sequencing runs. Two files including one file of the actual SNP data from five (359 million reads), six (421 million reads), seven (488 million reads) and eight (566 million reads) sequencing runs, and an imputed data file generated from the five, six and seven sequencing runs were compared to calculate the number of correctly imputed data points. The ratio between correctly imputed and total imputed data points was used to estimate imputation accuracy [36]. To evaluate the relationship between imputation accuracy and allele frequency, allele frequencies from the original data file were calculated for each SNP.
Results
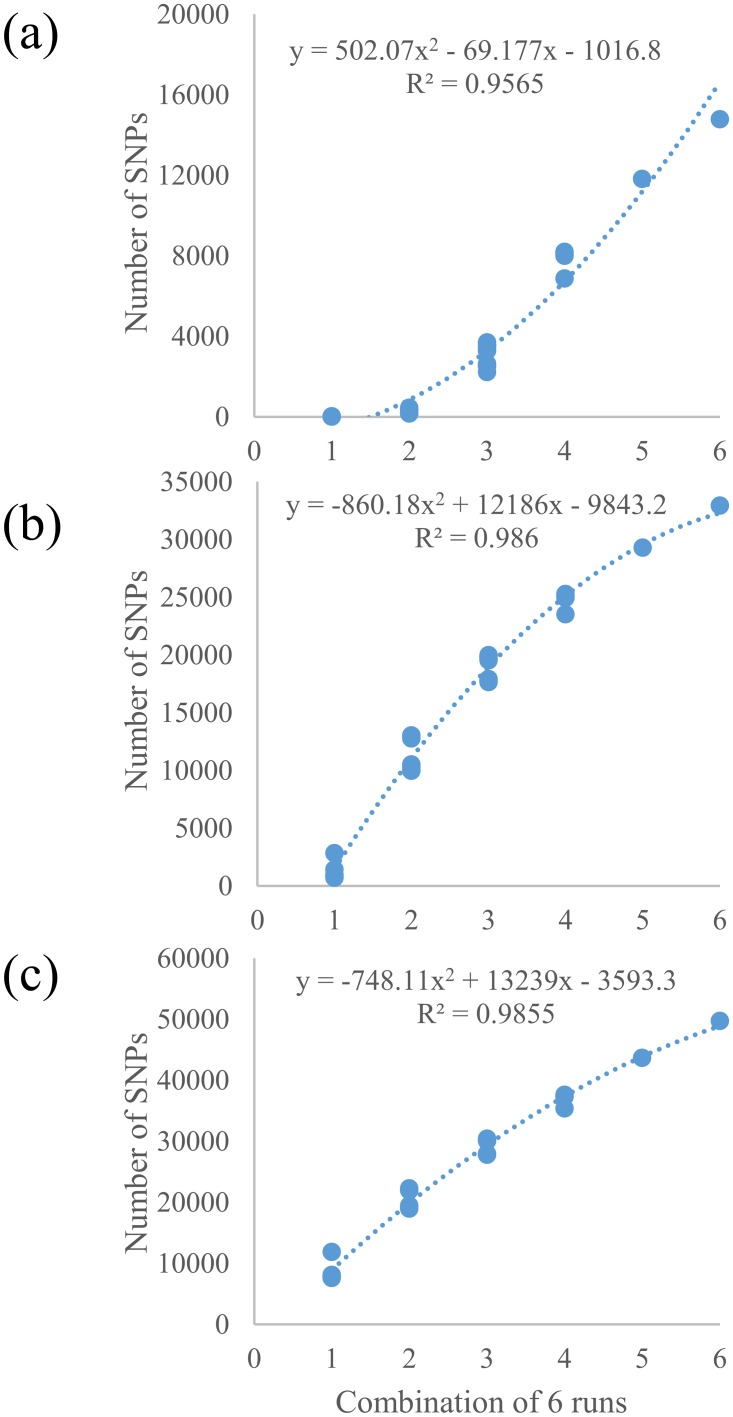
Two GBS libraries were generated for the 384 wheat accessions with 276 landraces and 12 cultivars in library 1 and 96 cultivars in library 2. To minimize missing data, an average of two sequencing runs was performed for each plate of 96 samples, therefore, library 1 with three plates of samples was run a total of six times and library 2 with one plate of samples was run twice. Eight sequencing runs generated a total of 566,439,207 reads from the two libraries with 81% (458,363,607) of high-quality barcoded reads, from which 133,039 unique SNPs were identified including 16,506, 38,642 and 65,560 SNPs with <20%, <50% and <80% missing data, respectively. To determine the relationship between number of GBS-SNPs and number of sequencing runs, numbers of SNPs were calculated for each increased run from first to six sequencing runs of the library 1 (Fig 1). The number of SNPs with <20% missing data was concave up as the run number increased (Fig 1a) but concave down for the numbers of SNPs with <50% and <80% missing data (Fig 1b and 1c).
Fig 1. Relationship between numbers of sequencing runs and the numbers of SNPs with (a) <20%, (b) <50%, and (c) <80% missing data for the first library of 288 Iranian wheat genotypes.
The average SNP density was 3.87 SNPs per Mbp when the SNPs with <80% missing data were counted (Table 1). The sequence tags containing the SNPs with <80% missing data were used to blast against each of the four references to map those SNPs to unique chromosome locations. The barley reference genome mapped the lowest percentage of sequencing tags (23.14%, Table 1), whereas IWGSC RefSeq v1.0 reference mapped the highest (94.94%, Table 2) among the four references. CSSS (55.34%, Table 3) and W7984 (85.61%, Table 4) were in between. Among the three wheat genomes, B genome had the most mapped SNPs and D genome the least across the four references, thus B chromosomes had much higher marker density than that in the D chromosomes. Among the four reference genomes, IWGSC RefSeq v1.0 reference provided the highest SNP density in almost all chromosomes. Among 21 chromosomes, chromosomes 2B and 3B had the highest SNP density, and chromosome 4D had the lowest (Tables 1–4).
Table 1. Summary of single nucleotide polymorphism types identified in three wheat genomes using the barley genome reference.
| Allele | 1A | 2A | 3A | 4A | 5A | 6A | 7A | A genome | 1B | 2B | 3B | 4B | 5B | 6B | 7B | B genome | 1D | 2D | 3D | 4D | 5D | 6D | 7D | D genome | NA | Total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chromosome_size(Mbp) | 798 | 899 | 828 | 856 | 827 | 705 | 814 | 5,727 | 849 | 928 | 993 | 821 | 870 | 913 | 900 | 6,274 | 605 | 729 | 771 | 649 | 750 | 713 | 728 | 4,945 | - | 16,946 |
| No. of SNP | 803 | 1,211 | 574 | 316 | 410 | 874 | 977 | 5,165 | 910 | 1,750 | 1,537 | 471 | 1,277 | 689 | 613 | 7,247 | 414 | 709 | 160 | 57 | 330 | 470 | 620 | 2,760 | 50,388 | 65,560 |
| Density (SNP/Mbp) | 1.01 | 1.35 | 0.69 | 0.37 | 0.50 | 1.24 | 1.20 | 0.90 | 1.07 | 1.89 | 1.55 | 0.57 | 1.47 | 0.75 | 0.68 | 1.16 | 0.68 | 0.97 | 0.21 | 0.09 | 0.44 | 0.66 | 0.85 | 0.56 | - | 3.87 |
| Transition | 537 | 812 | 378 | 196 | 280 | 603 | 639 | 3,445 | 623 | 1,161 | 1,035 | 298 | 836 | 454 | 417 | 4,824 | 247 | 444 | 98 | 43 | 200 | 288 | 394 | 1,714 | 35,012 | 44,995 |
| A/G | 257 | 413 | 160 | 83 | 135 | 287 | 307 | 1,642 | 300 | 512 | 477 | 131 | 390 | 215 | 196 | 2,221 | 108 | 213 | 42 | 24 | 86 | 132 | 193 | 798 | 16,495 | 21,156 |
| C/T | 234 | 322 | 182 | 86 | 126 | 242 | 269 | 1,461 | 261 | 491 | 419 | 134 | 363 | 191 | 173 | 2,032 | 104 | 172 | 43 | 16 | 87 | 128 | 161 | 711 | 14,387 | 18,591 |
| T/C | 23 | 39 | 14 | 17 | 9 | 34 | 38 | 174 | 29 | 68 | 67 | 15 | 31 | 29 | 26 | 265 | 16 | 32 | 6 | 2 | 14 | 13 | 21 | 104 | 1,957 | 2,500 |
| G/A | 23 | 38 | 22 | 10 | 10 | 40 | 25 | 168 | 33 | 90 | 72 | 18 | 52 | 19 | 22 | 306 | 19 | 27 | 7 | 1 | 13 | 15 | 19 | 101 | 2,173 | 2,748 |
| Transversion | 266 | 399 | 196 | 120 | 130 | 271 | 338 | 1,720 | 287 | 589 | 502 | 173 | 441 | 235 | 196 | 2,423 | 167 | 265 | 62 | 14 | 130 | 182 | 226 | 1,046 | 15,376 | 20,565 |
| A/T | 37 | 69 | 26 | 20 | 17 | 43 | 41 | 253 | 35 | 77 | 81 | 32 | 62 | 47 | 22 | 356 | 29 | 38 | 12 | 3 | 16 | 21 | 37 | 156 | 2,137 | 2,902 |
| A/C | 52 | 113 | 42 | 30 | 26 | 58 | 75 | 396 | 60 | 139 | 119 | 40 | 118 | 53 | 45 | 574 | 46 | 56 | 16 | 1 | 32 | 41 | 50 | 242 | 3,597 | 4,809 |
| T/A | 3 | 8 | 6 | 4 | 4 | 5 | 15 | 45 | 6 | 11 | 16 | 2 | 3 | 8 | 5 | 51 | 3 | 3 | 1 | 0 | 1 | 1 | 2 | 11 | 314 | 421 |
| T/G | 3 | 7 | 2 | 1 | 2 | 5 | 8 | 28 | 9 | 25 | 18 | 6 | 7 | 4 | 8 | 77 | 5 | 6 | 2 | 1 | 2 | 7 | 3 | 26 | 453 | 584 |
| C/A | 8 | 10 | 5 | 2 | 5 | 4 | 9 | 43 | 9 | 10 | 11 | 2 | 9 | 1 | 8 | 50 | 3 | 11 | 3 | 0 | 2 | 6 | 3 | 28 | 437 | 558 |
| C/G | 88 | 115 | 71 | 41 | 37 | 97 | 102 | 551 | 101 | 194 | 131 | 44 | 152 | 65 | 69 | 756 | 48 | 87 | 12 | 5 | 46 | 61 | 67 | 326 | 4,665 | 6,298 |
| G/T | 64 | 69 | 36 | 18 | 31 | 44 | 70 | 332 | 54 | 108 | 108 | 41 | 75 | 46 | 33 | 465 | 25 | 54 | 13 | 3 | 24 | 32 | 52 | 203 | 3,140 | 4,140 |
| G/C | 11 | 8 | 8 | 4 | 8 | 15 | 18 | 72 | 13 | 25 | 18 | 6 | 15 | 11 | 6 | 94 | 8 | 10 | 3 | 1 | 7 | 13 | 12 | 54 | 633 | 853 |
| Ts % | 66.87 | 67.05 | 65.85 | 62.03 | 68.29 | 68.99 | 65.40 | 66.70 | 68.46 | 66.34 | 67.34 | 63.27 | 65.47 | 65.89 | 68.03 | 66.57 | 59.66 | 62.62 | 61.25 | 75.44 | 60.61 | 61.28 | 63.55 | 62.10 | 69.48 | 68.63 |
| Tv % | 33.13 | 32.95 | 34.15 | 37.97 | 31.71 | 31.01 | 34.60 | 33.30 | 31.54 | 33.66 | 32.66 | 36.73 | 34.53 | 34.11 | 31.97 | 33.43 | 40.34 | 37.38 | 38.75 | 24.56 | 39.39 | 38.72 | 36.45 | 37.90 | 30.52 | 31.37 |
| Ts/Tv ratio | 2.02 | 2.04 | 1.93 | 1.63 | 2.15 | 2.23 | 1.89 | 2.00 | 2.17 | 1.97 | 2.06 | 1.72 | 1.90 | 1.93 | 2.13 | 1.99 | 1.48 | 1.68 | 1.58 | 3.07 | 1.54 | 1.58 | 1.74 | 1.64 | 2.28 | 2.19 |
Table 2. Summary of single nucleotide polymorphism types identified in three wheat genomes using the wheat the IWGSC RefSeq v1.0.
| Allele | 1A | 2A | 3A | 4A | 5A | 6A | 7A | A genome | 1B | 2B | 3B | 4B | 5B | 6B | 7B | B genome | 1D | 2D | 3D | 4D | 5D | 6D | 7D | D genome | NA |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of SNP | 3,399 | 4,001 | 2943 | 3,684 | 2,850 | 2,683 | 4,556 | 24,116 | 4,424 | 5,083 | 5,350 | 2,055 | 4,054 | 4,776 | 4,055 | 29,797 | 1,370 | 2,029 | 1,162 | 466 | 798 | 993 | 1,510 | 8,328 | 3,319 |
| Density (SNP/Mbp) | 4.26 | 4.45 | 3.55 | 4.3 | 3.45 | 3.81 | 5.6 | 4.21 | 5.21 | 5.48 | 5.39 | 2.5 | 4.66 | 5.23 | 4.51 | 4.75 | 2.26 | 2.78 | 1.51 | 0.72 | 1.06 | 1.39 | 2.07 | 1.68 | - |
| Transition | 2,390 | 2,873 | 2,023 | 2,523 | 1,946 | 1,833 | 3,183 | 16,771 | 3,072 | 3,502 | 3,729 | 1,389 | 2,774 | 3,337 | 2,872 | 20,675 | 863 | 1,310 | 754 | 321 | 537 | 623 | 958 | 5,366 | 2,183 |
| A/G | 1,121 | 1,397 | 975 | 1,185 | 886 | 847 | 1,452 | 7,863 | 1,482 | 1,643 | 1,721 | 650 | 1,273 | 1,587 | 1,344 | 9,700 | 412 | 604 | 340 | 160 | 251 | 305 | 466 | 2,538 | 1,055 |
| C/T | 1,004 | 1,172 | 812 | 1,051 | 855 | 766 | 1,347 | 7,007 | 1,263 | 1,384 | 1,529 | 596 | 1,190 | 1,380 | 1,196 | 8,538 | 353 | 526 | 315 | 129 | 217 | 253 | 392 | 2,185 | 861 |
| T/C | 131 | 157 | 112 | 130 | 85 | 105 | 186 | 906 | 145 | 221 | 215 | 67 | 144 | 181 | 162 | 1,135 | 45 | 94 | 48 | 12 | 31 | 32 | 55 | 317 | 142 |
| G/A | 134 | 147 | 124 | 157 | 120 | 115 | 198 | 995 | 182 | 254 | 264 | 76 | 167 | 189 | 170 | 1,302 | 53 | 86 | 51 | 20 | 38 | 33 | 45 | 326 | 125 |
| Transversion | 1,009 | 1,128 | 920 | 1,161 | 904 | 850 | 1,373 | 7,345 | 1,352 | 1,581 | 1,621 | 666 | 1,280 | 1,439 | 1,183 | 9,122 | 507 | 719 | 408 | 145 | 261 | 370 | 552 | 2,962 | 1,136 |
| A/T | 140 | 181 | 126 | 180 | 116 | 109 | 184 | 1,036 | 195 | 234 | 236 | 96 | 162 | 207 | 145 | 1,275 | 69 | 98 | 56 | 21 | 45 | 54 | 97 | 440 | 151 |
| A/C | 219 | 296 | 209 | 263 | 207 | 204 | 341 | 1,739 | 307 | 373 | 390 | 154 | 306 | 357 | 285 | 2,172 | 116 | 160 | 94 | 28 | 61 | 89 | 115 | 663 | 235 |
| T/A | 18 | 28 | 23 | 22 | 19 | 10 | 28 | 148 | 27 | 36 | 41 | 13 | 25 | 29 | 23 | 194 | 9 | 12 | 11 | 2 | 4 | 6 | 4 | 48 | 31 |
| T/G | 17 | 25 | 20 | 29 | 36 | 20 | 33 | 180 | 33 | 56 | 59 | 20 | 33 | 40 | 45 | 286 | 15 | 22 | 14 | 2 | 6 | 12 | 12 | 83 | 35 |
| C/A | 24 | 31 | 21 | 28 | 25 | 29 | 38 | 196 | 36 | 41 | 43 | 19 | 44 | 28 | 43 | 254 | 6 | 23 | 9 | 4 | 9 | 12 | 15 | 78 | 30 |
| C/G | 314 | 319 | 297 | 362 | 262 | 274 | 397 | 2,225 | 429 | 468 | 456 | 187 | 390 | 440 | 385 | 2,755 | 160 | 234 | 125 | 45 | 68 | 103 | 180 | 915 | 403 |
| G/T | 243 | 213 | 183 | 212 | 207 | 167 | 302 | 1,527 | 264 | 308 | 335 | 154 | 267 | 276 | 218 | 1,822 | 104 | 134 | 83 | 36 | 59 | 74 | 101 | 591 | 200 |
| G/C | 34 | 35 | 41 | 65 | 32 | 37 | 50 | 294 | 61 | 65 | 61 | 23 | 53 | 62 | 39 | 364 | 28 | 36 | 16 | 7 | 9 | 20 | 28 | 144 | 51 |
| Ts % | 70.31 | 71.81 | 68.74 | 68.49 | 68.28 | 68.32 | 69.86 | 69.54 | 69.44 | 68.90 | 69.70 | 67.59 | 68.43 | 69.87 | 70.83 | 69.39 | 62.99 | 64.56 | 64.89 | 68.88 | 67.29 | 62.74 | 63.44 | 64.43 | 65.77 |
| Tv % | 29.69 | 28.19 | 31.26 | 31.51 | 31.72 | 31.68 | 30.14 | 30.46 | 30.56 | 31.10 | 30.30 | 32.41 | 31.57 | 30.13 | 29.17 | 30.61 | 37.01 | 35.44 | 35.11 | 31.12 | 32.71 | 37.26 | 36.56 | 35.57 | 34.23 |
| Ts/Tv ratio | 2.37 | 2.55 | 2.20 | 2.17 | 2.15 | 2.16 | 2.32 | 2.28 | 2.27 | 2.22 | 2.30 | 2.09 | 2.17 | 2.32 | 2.43 | 2.27 | 1.70 | 1.82 | 1.85 | 2.21 | 2.06 | 1.68 | 1.74 | 1.81 | 1.92 |
Table 3. Summary of single nucleotide polymorphism types identified in three wheat genomes using the wheat Chinese Spring survey sequence (CSSS) assembly.
| Allele | 1A | 2A | 3A | 4A | 5A | 6A | 7A | A genome | 1B | 2B | 3B | 4B | 5B | 6B | 7B | B genome | 1D | 2D | 3D | 4D | 5D | 6D | 7D | D genome | NA |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of SNP | 2,151 | 2,578 | 1,188 | 1,647 | 898 | 1,926 | 2,057 | 12,445 | 2,464 | 3,782 | 4,307 | 1,519 | 3,408 | 1,661 | 1,517 | 18,658 | 906 | 1,019 | 348 | 114 | 657 | 911 | 1,220 | 5,175 | 29,282 |
| Density (SNP/Mbp) | 2.7 | 2.87 | 1.43 | 1.92 | 1.09 | 2.73 | 2.53 | 2.17 | 2.9 | 4.08 | 4.34 | 1.85 | 3.92 | 1.82 | 1.69 | 2.97 | 1.5 | 1.4 | 0.45 | 0.18 | 0.88 | 1.28 | 1.68 | 1.05 | - |
| Transition | 1,490 | 1,830 | 813 | 1,113 | 606 | 1,326 | 1,417 | 8,595 | 1,707 | 2,671 | 2,994 | 1,026 | 2,372 | 1,116 | 1,049 | 12,935 | 568 | 647 | 216 | 92 | 440 | 574 | 796 | 3,333 | 20,132 |
| A/G | 704 | 897 | 371 | 489 | 273 | 591 | 645 | 3,970 | 793 | 1,214 | 1,394 | 472 | 1,114 | 527 | 474 | 5,988 | 263 | 293 | 88 | 53 | 210 | 262 | 379 | 1,548 | 9,650 |
| C/T | 632 | 732 | 346 | 478 | 277 | 577 | 619 | 3,661 | 722 | 1,117 | 1,219 | 449 | 995 | 472 | 462 | 5,436 | 235 | 271 | 93 | 33 | 183 | 251 | 326 | 1,392 | 8,102 |
| T/C | 74 | 104 | 46 | 68 | 19 | 69 | 75 | 455 | 89 | 153 | 169 | 56 | 116 | 59 | 65 | 707 | 36 | 45 | 13 | 4 | 25 | 29 | 46 | 198 | 1,140 |
| G/A | 80 | 97 | 50 | 78 | 37 | 89 | 78 | 509 | 103 | 187 | 212 | 49 | 147 | 58 | 48 | 804 | 34 | 38 | 22 | 2 | 22 | 32 | 45 | 195 | 1,240 |
| Transversion | 661 | 748 | 375 | 534 | 292 | 600 | 640 | 3,850 | 757 | 1,111 | 1,313 | 493 | 1,036 | 545 | 468 | 5,723 | 338 | 372 | 132 | 22 | 217 | 337 | 424 | 1,842 | 9,150 |
| A/T | 87 | 136 | 50 | 71 | 39 | 90 | 91 | 564 | 106 | 145 | 195 | 76 | 125 | 93 | 58 | 798 | 45 | 57 | 19 | 4 | 33 | 50 | 61 | 269 | 1,271 |
| A/C | 146 | 199 | 83 | 130 | 56 | 125 | 159 | 898 | 155 | 267 | 313 | 96 | 253 | 131 | 120 | 1,335 | 77 | 83 | 33 | 4 | 51 | 77 | 106 | 431 | 2,145 |
| T/A | 9 | 17 | 9 | 8 | 13 | 8 | 17 | 81 | 15 | 18 | 29 | 10 | 17 | 21 | 9 | 119 | 7 | 7 | 1 | 0 | 1 | 3 | 4 | 23 | 198 |
| T/G | 8 | 13 | 8 | 8 | 12 | 17 | 17 | 83 | 20 | 36 | 46 | 19 | 28 | 9 | 18 | 176 | 13 | 9 | 4 | 1 | 6 | 14 | 8 | 55 | 270 |
| C/A | 17 | 22 | 8 | 11 | 7 | 21 | 15 | 101 | 21 | 39 | 32 | 10 | 40 | 6 | 16 | 164 | 8 | 12 | 3 | 0 | 8 | 11 | 9 | 51 | 242 |
| C/G | 203 | 197 | 127 | 174 | 91 | 193 | 181 | 1,166 | 240 | 335 | 368 | 147 | 316 | 151 | 156 | 1,713 | 106 | 121 | 38 | 8 | 66 | 98 | 138 | 575 | 2,844 |
| G/T | 167 | 142 | 76 | 103 | 62 | 126 | 138 | 814 | 165 | 238 | 282 | 118 | 215 | 105 | 74 | 1,197 | 68 | 68 | 24 | 5 | 43 | 63 | 80 | 351 | 1,778 |
| G/C | 24 | 22 | 14 | 29 | 12 | 20 | 22 | 143 | 35 | 33 | 48 | 17 | 42 | 29 | 17 | 221 | 14 | 15 | 10 | 0 | 9 | 21 | 18 | 87 | 402 |
| Ts % | 69.27 | 70.99 | 68.43 | 67.58 | 67.48 | 68.85 | 68.89 | 69.06 | 69.28 | 70.62 | 69.51 | 67.54 | 69.60 | 67.19 | 69.15 | 69.33 | 62.69 | 63.49 | 62.07 | 80.70 | 66.97 | 63.01 | 65.25 | 64.41 | 68.75 |
| Tv % | 30.73 | 29.01 | 31.57 | 32.42 | 32.52 | 31.15 | 31.11 | 30.94 | 30.72 | 29.38 | 30.49 | 32.46 | 30.40 | 32.81 | 30.85 | 30.67 | 37.31 | 36.51 | 37.93 | 19.30 | 33.03 | 36.99 | 34.75 | 35.59 | 31.25 |
| Ts/Tv ratio | 2.25 | 2.45 | 2.17 | 2.08 | 2.08 | 2.21 | 2.21 | 2.23 | 2.25 | 2.40 | 2.28 | 2.08 | 2.29 | 2.05 | 2.24 | 2.26 | 1.68 | 1.74 | 1.64 | 4.18 | 2.03 | 1.70 | 1.88 | 1.81 | 2.20 |
Table 4. Summary of single nucleotide polymorphism types identified in three wheat genomes using the wheat W7984 assembly.
| Allele | 1A | 2A | 3A | 4A | 5A | 6A | 7A | A genome | 1B | 2B | 3B | 4B | 5B | 6B | 7B | B genome | 1D | 2D | 3D | 4D | 5D | 6D | 7D | D genome | NA |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of SNP | 2,907 | 3,434 | 2,645 | 3,299 | 1,921 | 2,455 | 4,023 | 20,684 | 3,813 | 4,649 | 5,067 | 1,986 | 3,686 | 4,455 | 3,792 | 27,448 | 1,232 | 1,804 | 1,128 | 428 | 897 | 1,039 | 1,465 | 7,993 | 9,435 |
| Density (SNP/Mbp) | 3.64 | 3.82 | 3.19 | 3.85 | 2.32 | 3.48 | 4.94 | 3.61 | 4.49 | 5.01 | 5.10 | 2.42 | 4.24 | 4.88 | 4.21 | 4.37 | 2.04 | 2.47 | 1.46 | 0.66 | 1.20 | 1.46 | 2.01 | 1.62 | - |
| Transition | 2,061 | 2,454 | 1,819 | 2,291 | 1,296 | 1,689 | 2,803 | 14,413 | 2,615 | 3,172 | 3,541 | 1,348 | 2,521 | 3,103 | 2,671 | 18,971 | 776 | 1,149 | 717 | 296 | 588 | 662 | 934 | 5,122 | 6,489 |
| A/G | 957 | 1,190 | 858 | 1,053 | 597 | 787 | 1,294 | 6,736 | 1,250 | 1,465 | 1,635 | 609 | 1,160 | 1,468 | 1,240 | 8,827 | 368 | 518 | 325 | 160 | 276 | 330 | 434 | 2,411 | 3,182 |
| C/T | 881 | 1,000 | 741 | 975 | 570 | 703 | 1,184 | 6,054 | 1,081 | 1,274 | 1,455 | 591 | 1,083 | 1,287 | 1,123 | 7,894 | 322 | 469 | 302 | 108 | 243 | 270 | 395 | 2,109 | 2,534 |
| T/C | 108 | 139 | 103 | 112 | 46 | 92 | 158 | 758 | 136 | 205 | 203 | 75 | 126 | 165 | 149 | 1,059 | 32 | 86 | 46 | 16 | 29 | 34 | 53 | 296 | 387 |
| G/A | 115 | 125 | 117 | 151 | 83 | 107 | 167 | 865 | 148 | 228 | 248 | 73 | 152 | 183 | 159 | 1,191 | 54 | 76 | 44 | 12 | 40 | 28 | 52 | 306 | 386 |
| Transversion | 846 | 980 | 826 | 1,008 | 625 | 766 | 1,220 | 6,271 | 1,198 | 1,477 | 1,526 | 638 | 1,165 | 1,352 | 1,121 | 8,477 | 456 | 655 | 411 | 132 | 309 | 377 | 531 | 2,871 | 2,946 |
| A/T | 99 | 170 | 112 | 149 | 83 | 98 | 165 | 876 | 174 | 199 | 226 | 84 | 159 | 188 | 135 | 1,165 | 67 | 88 | 53 | 17 | 48 | 59 | 90 | 422 | 439 |
| A/C | 189 | 252 | 193 | 229 | 130 | 181 | 299 | 1,473 | 273 | 356 | 357 | 142 | 274 | 337 | 272 | 2,011 | 107 | 145 | 98 | 28 | 69 | 86 | 99 | 632 | 693 |
| T/A | 12 | 22 | 26 | 14 | 19 | 12 | 19 | 124 | 24 | 31 | 37 | 16 | 23 | 26 | 24 | 181 | 9 | 12 | 11 | 1 | 3 | 5 | 8 | 49 | 67 |
| T/G | 18 | 19 | 18 | 28 | 28 | 23 | 22 | 156 | 32 | 49 | 56 | 21 | 32 | 36 | 43 | 269 | 10 | 21 | 12 | 5 | 7 | 10 | 14 | 79 | 80 |
| C/A | 19 | 28 | 21 | 24 | 14 | 24 | 32 | 162 | 33 | 37 | 42 | 20 | 39 | 26 | 43 | 240 | 9 | 23 | 5 | 3 | 11 | 12 | 13 | 76 | 80 |
| C/G | 276 | 273 | 247 | 315 | 186 | 248 | 371 | 1,916 | 377 | 447 | 426 | 184 | 355 | 414 | 358 | 2,561 | 143 | 215 | 130 | 34 | 91 | 109 | 171 | 893 | 928 |
| G/T | 211 | 186 | 171 | 196 | 144 | 151 | 265 | 1,324 | 232 | 299 | 325 | 149 | 239 | 270 | 199 | 1,713 | 89 | 120 | 83 | 35 | 66 | 71 | 109 | 573 | 530 |
| G/C | 22 | 30 | 38 | 53 | 21 | 29 | 47 | 240 | 53 | 59 | 57 | 22 | 44 | 55 | 47 | 337 | 22 | 31 | 19 | 9 | 14 | 25 | 27 | 147 | 129 |
| Ts % | 70.90 | 71.46 | 68.77 | 69.45 | 67.46 | 68.80 | 69.67 | 69.68 | 68.58 | 68.23 | 69.88 | 67.88 | 68.39 | 69.65 | 70.44 | 69.12 | 62.99 | 63.69 | 63.56 | 69.16 | 65.55 | 63.72 | 63.75 | 64.08 | 68.78 |
| Tv % | 29.10 | 28.54 | 31.23 | 30.55 | 32.54 | 31.20 | 30.33 | 30.32 | 31.42 | 31.77 | 30.12 | 32.12 | 31.61 | 30.35 | 29.56 | 30.88 | 37.01 | 36.31 | 36.44 | 30.84 | 34.45 | 36.28 | 36.25 | 35.92 | 31.22 |
| Ts/Tv ratio | 2.44 | 2.50 | 2.20 | 2.27 | 2.07 | 2.20 | 2.30 | 2.30 | 2.18 | 2.15 | 2.32 | 2.11 | 2.16 | 2.30 | 2.38 | 2.24 | 1.70 | 1.75 | 1.74 | 2.24 | 1.90 | 1.76 | 1.76 | 1.78 | 2.20 |
Transitions were the most observed nucleotide variations (68.63%) including A/G (32.27%), C/T (28.36%), C/G (9.61%), A/C (7.34%), G/T (6.31%) and A/T (4.43%) transition types (Table 1). Using the barley reference genome, more transition-type SNPs were identified in A (3,445) and B (4,824) genomes than that in the D genome (1,714). The transition/transversion (Ts/Tv) SNP ratios from the A and B genomes (2.0) were relatively higher than that (1.64) from the D genome (Table 1). A similar trend in SNP types was observed for the CSSS assembly, but its transition/transversion SNP ratios were higher than those from the barley reference genome with 2.23 for the A genome, 2.26 for the B genome and 1.81 for the D genome (Table 3). The W7984 assembly and IWGSC RefSeq v1.0 had similar transition/transversion ratios to the CSSS assembly, but with slightly higher numbers of total SNPs (Tables 2 and 4).
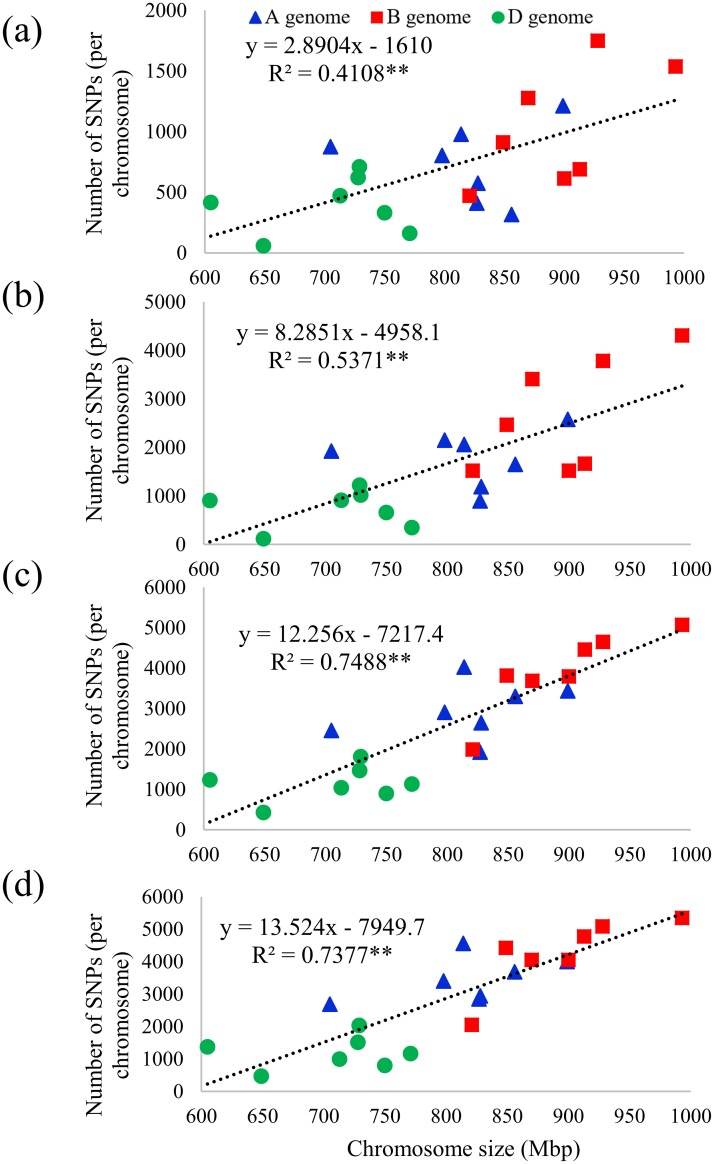
The numbers of SNPs per chromosome were significantly correlated to the chromosome sizes (Mbp) in all four references. Although they were all significant, the correlations were much lower for the barley reference genome (R2 ~ 0.41**, Fig 2a) and wheat CSSS assembly (R2 ~ 0.54**, Fig 2b) than those for W7984 assembly (R2 ~ 0.75**, Fig 2c) and IWGSC RefSeq v1.0 (R2 ~ 0.74**, Fig 2d).
Fig 2. Relationship between the number of SNPs per chromosome and chromosome sizes when (a) barley genome, (b) Chinese Spring survey sequence (CSSS), (c) W7984, and (d) IWGSC RefSeq v1.0 reference was used to call SNPs.
Five sequencing runs (three runs of the library 1 and two runs of the library 2) generated 355,197,375 total reads and 5,571 SNPs with less than 20% missing data. The number of SNPs was almost doubled (10,213) after adding one additional sequencing run of library 1 (total six runs) and tripled (16,506) after adding three sequencing runs of library 1 (total eight runs). For the number of SNPs with <80% missing data, increasing number of sequencing runs from five (45,624 SNPs) to eight (65,560 SNPs) only increased 44% SNPs. However, imputation reduced much more missing data than increasing sequencing runs from five to eight although imputation efficiency is different among the four references (Table 5). The numbers of SNPs with <20% missing data increased only three times when sequencing runs were increased from five to eight (Table 5), but increased 2.8 times (the barley reference genome), 5.1 times (CSSS), 7.0 times (W7984) and 7.8 times (IWGSC RefSeq v1.0) after imputation from the data of five runs. Imputation using IWGSC RefSeq v1.0 reference generated the most SNPs with only 4.9% missing data in the final imputed dataset; the barley reference genome imputed the least SNPs with 65.5% missing data after imputation; and wheat CSSS and W7984 were in between with 38.0% and 12.5% missing data after imputation (Table 5). These results indicate that although both imputation and increasing sequencing depth can quickly fill up missing data, imputation can reduce more missing data and therefore detect more SNPs than increasing sequencing depth with the highest increase for SNPs with <20% missing data.
Table 5. Numbers of SNPs called after five (3 runs of the library 1 and 2 runs of the library 2), six (4 runs of the library 1 and 2 runs of the library 2), seven (5 runs of the library 1 and 2 runs of the library 2) and eight (6 runs of the library 1 and 2 runs of the library 2) sequencing runs of the two GBS libraries with and without imputation using barley genome and Chinese Spring survey sequence (CSSS) and W7984 assembly and Wheat IWGSC RefSeq v1.0 reference.
| Imputation methods | Missing data | Number of SNPs | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Five sequencing runs (355,197,375 reads) | Six sequencing runs (419,373,662 reads) | Seven sequencing runs (486,646,481 reads) | Eight sequencing runs (566,439,207 reads) | ||||||||||
| <20% | <50% | <80% | <20% | <50% | <80% | <20% | <50% | <80% | <20% | <50% | <80% | ||
| Without imputation | 5,571 | 26,773 | 45,624 | 10,213 | 31,472 | 53,241 | 13,746 | 35,380 | 59,766 | 16,506 | 38,642 | 65,560 | |
| Barley | 15,738 | 30,442 | 45,624 | 20,142 | 35,320 | 53,241 | 23,526 | 39,269 | 59,766 | 26,284 | 42,706 | 65,560 | |
| Wheat CSSS | 28,285 | 36,811 | 45,624 | 34,135 | 42,782 | 53,241 | 38,783 | 47,831 | 59,766 | 42,877 | 52,357 | 65,560 | |
| Wheat W7984 | 39,906 | 42,532 | 45,624 | 46,819 | 49,634 | 53,241 | 52,529 | 55,605 | 59,766 | 57,703 | 60,900 | 65,560 | |
| Wheat IWGSC RefSeq v1.0 | 43,365 | 44,410 | 45,624 | 50,735 | 51,924 | 53,241 | 57,065 | 58,361 | 59,766 | 62,740 | 64,083 | 65,560 | |
Imputation accuracy was calculated for each reference by comparing the SNP data imputed from five, six or seven sequencing runs to the real SNP data from eight runs (Table 6). All four references provided high imputation accuracy. Among them, the IWGSC RefSeq v1.0 provided the lowest imputation accuracy (84.16%) although it imputed the most data points in all run combinations. The other three references had relatively higher accuracies from 87.31% for CSSS reference to 89.80% for W7984 reference (Table 6). The number of imputed data points was the highest using five sequencing runs, and the lowest using the data from eight sequencing runs. For all references and sequencing runs, even though the imputed data points per chromosome were much lower in the D genome than those in the A and B genomes in general, the D genome had the highest imputation accuracy and the B genome the lowest (Table 6).
Table 6. Numbers of data points per chromosome after imputation with barley genome, Chinese Spring survey sequence (CSSS), W7984 and IWGSC RefSeq v1.0 references and imputation accuracy calculated by comparing imputed data of five, six and seven runs with actual SNP data generated from eight sequencing runs.
| Run | Reference | Missing data points | A genome | B genome | D genome | Total |
|---|---|---|---|---|---|---|
| Five runs | Barley | Total data points | 157,689 | 221,204 | 85,281 | 464,174 |
| Correctly imputed | 141,111 | 195,450 | 77,230 | 413,791 | ||
| Accuracy | 89.49 | 88.36 | 90.56 | 89.15 | ||
| CSSS | Total data points | 374,452 | 448,904 | 125,864 | 949,220 | |
| Correctly imputed | 339,919 | 401,393 | 114,507 | 855,819 | ||
| Accuracy | 90.78 | 89.42 | 90.98 | 90.16 | ||
| W7984 | Total data points | 556,620 | 637,680 | 186,281 | 1,380,581 | |
| Correctly imputed | 502,417 | 568,400 | 169,075 | 1,239,892 | ||
| Accuracy | 90.26 | 89.14 | 90.76 | 89.81 | ||
| IWGSC RefSeq v1.0 | Total data points | 691,863 | 841,893 | 243,017 | 1,776,773 | |
| Correctly imputed | 584,917 | 694,667 | 209,385 | 1,488,969 | ||
| Accuracy | 84.54 | 82.51 | 86.16 | 83.80 | ||
| Six runs | Barley | Total data points | 23,138 | 22,979 | 11,552 | 57,669 |
| Correctly imputed | 20,356 | 20,016 | 10,294 | 50,666 | ||
| Accuracy | 87.98 | 87.11 | 89.11 | 87.86 | ||
| CSSS | Total data points | 74,520 | 103,888 | 29,095 | 207,503 | |
| Correctly imputed | 63,312 | 86,484 | 25,494 | 175,290 | ||
| Accuracy | 84.96 | 83.25 | 87.62 | 84.48 | ||
| W7984 | Total data points | 393,307 | 517,916 | 154,857 | 1,066,080 | |
| Correctly imputed | 363,152 | 473,970 | 143,550 | 980,672 | ||
| Accuracy | 92.33 | 91.51 | 92.70 | 91.99 | ||
| IWGSC RefSeq v1.0 | Total data points | 467,224 | 574,541 | 162,471 | 1,204,236 | |
| Correctly imputed | 397,638 | 475,980 | 140,790 | 1,014,408 | ||
| Accuracy | 85.11 | 82.85 | 86.66 | 84.24 | ||
| Seven runs | Barley | Total data points | 25,242 | 33,810 | 12,195 | 71,247 |
| Correctly imputed | 22,637 | 29,634 | 11,207 | 63,478 | ||
| Accuracy | 89.68 | 87.65 | 91.90 | 89.10 | ||
| CSSS | Total data points | 67,293 | 104,296 | 25,254 | 196,843 | |
| Correctly imputed | 59,247 | 89,933 | 22,647 | 171,827 | ||
| Accuracy | 88.04 | 86.23 | 89.68 | 87.29 | ||
| W7984 | Total data points | 154,672 | 214,222 | 59,594 | 428,488 | |
| Correctly imputed | 135,735 | 186,183 | 53,429 | 375,347 | ||
| Accuracy | 87.76 | 86.91 | 89.65 | 87.60 | ||
| IWGSC RefSeq v1.0 | Total data points | 233,321 | 289,654 | 80,048 | 603,023 | |
| Correctly imputed | 199,037 | 240,517 | 69,725 | 509,279 | ||
| Accuracy | 85.31 | 83.04 | 87.10 | 84.45 |
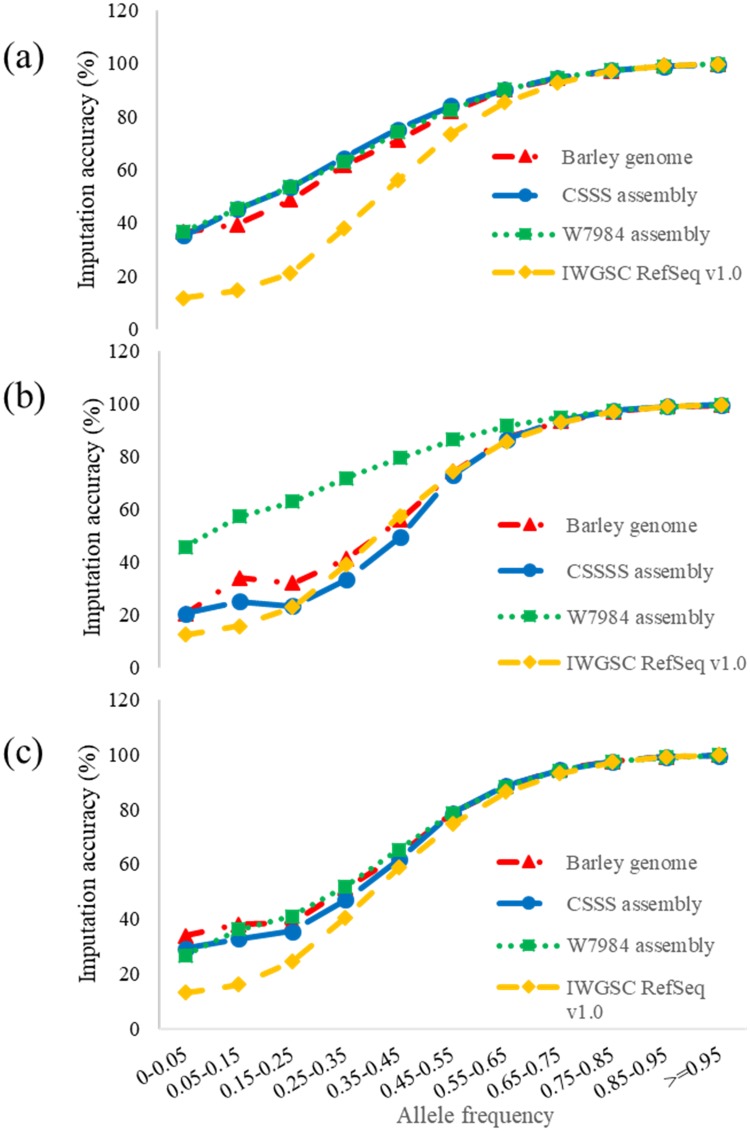
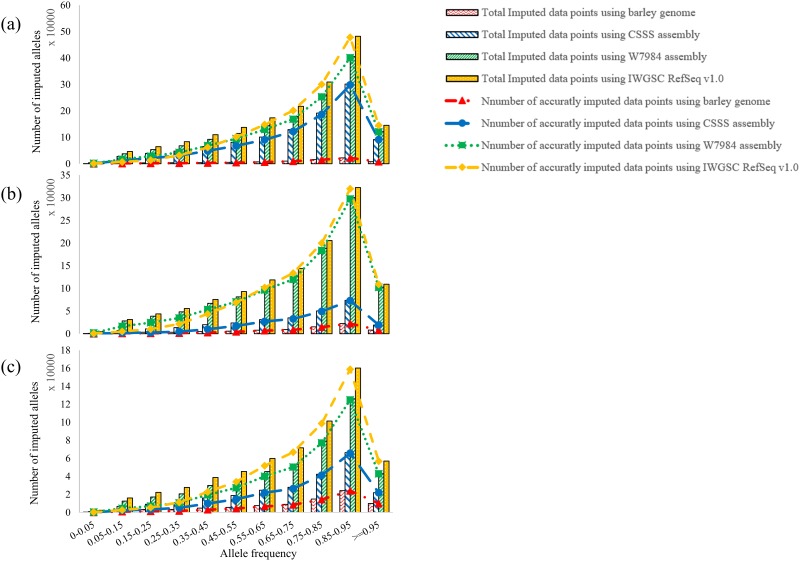
The imputation accuracy increased with the increase in allele frequency from 25.7% accuracy for allele frequency < 5% and 99.7% accuracy for allele frequency > 95% (Fig 3). The positive correlations were observed for all four references. The imputation accuracy reached 94% when allele frequency was >65%. Relatively lower imputation accuracy of IWGSC RefSeq v1.0 than other references mainly occurred at those alleles with frequency <35% (Fig 3b and 3c) where W7984 assembly provided more accurate imputation than other references. About 75% of imputed SNPs were distributed in allele frequencies between 0.55 and 0.95 (Fig 4), and had high mean imputation accuracies from 88.0% to 99.7%. The difference in imputation accuracy in this allele frequency range was negligible among the four references (Fig 4).
Fig 3. Relationship between imputation accuracy and allele frequency for (a) five, (b) six, (c) seven runs imputed with eight runs using barley genome, Chinese Spring survey sequence (CSSS), W7984 and IWGSC RefSeq 1.0 in Iranian wheat GBS data.
Fig 4. Number of total and correctly imputed alleles for different allele frequencies for (a) five, (b) six, (c) seven runs imputed with eight runs using barley genome, Chinese Spring survey sequence (CSSS), W7984 and IWGSC RefSeq 1.0 in Iranian wheat GBS data.
Discussion
Advancements in next-generation sequencing technology and high-throughput SNP genotyping can greatly accelerate crop breeding process if properly deployed [37]. GBS technology not only significantly improves throughput, but also greatly reduces SNP genotyping costs by reducing genome complexity and multiplexing samples [13]. Although GBS can generate a large number of SNP markers, its application in association mapping and genomics-assisted breeding can be limited by massive amount of missing data when low coverage sequencing is conducted [4, 38]. Biologically, missing SNP calls in GBS datasets can be due to presence-absence variation and/or differential methylation in restriction sites. Technically, genome complexity, low library quality, and sequence coverage [27] are among the major contributors. Library complexity can be reduced by digesting sample DNA with restriction enzymes such as PstI and MspI. A combination of PstI and MspI enzymes has been successfully used for high quality wheat library construction [13]. The sequencing coverage is a function of genome complexity, multiplexing level, and output of a NGS platform [39]. In the current study, we constructed two libraries, library 1 with three plates of samples and library 2 with one plate of samples. The library 1 had six sequencing runs and the library 2 had two, thus each sample in both libraries had the same sequence depth. However, the samples in the library 1 produced more SNPs (14,781, 32,926 and 49,717 SNPs at <20%, <50%, and <80% missing data, respectively) than those from the library 2 (10,630, 23,931 and 26,198 SNPs at <20%, <50%, and <80% missing data, respectively), suggesting that raising multiplexing level and sequencing multiple times can significantly increase SNP number and reduce missing data in comparison with a library at lower multiplex level with the same sequencing depth. The results in this study showed that increasing number of SNPs with <20% missing data was concave up by increasing run number (Fig 1a) while increasing numbers of SNPs with <50% and <80% missing data were concave down (Fig 1b and 1c), suggesting increasing run number can quickly reduce missing data, but slowly increase in total numbers of SNPs.
Typically, two strategies can be used to reduce missing data: increasing sequencing depth or imputing missing data using a reference genome. Increasing sequence depth can be achieved through lowering multiplexing level in a fixed run number or increasing sequencing runs of a highly multiplexed library. Both methods will result in an increase in per-sample cost. As indicated previously, lowering multiplex level may not increase SNP number as expected. Thus, increasing number of sequencing runs can be an option. In this study, the first library was run six times and the number of SNPs was significantly increased especially for the number of SNPs with <20% missing data (Fig 1), indicating that increasing number of runs significantly decreased the number of missing data and therefore increased number of usable SNPs. However, this also increased the per-sample cost significantly. Imputing missing data is an effective approach to minimize missing data without increasing sequencing costs. Imputed data can be very accurate if a high-quality genome reference is available and SNP markers can be accurately aligned on the physical map [27]. However, in the case that genome reference is absent or incomplete, such imputation is challenging. In this study, we compared imputation efficiency and accuracy among four genome references including the barley reference genome and three wheat references (CSSS, W7984 and IWGSC RefSeq v1.0) and found that all the references are useful for ordering GBS-SNPs and can significantly reduce missing data points and provide accurate imputation to leverage the application GBS markers in wheat [4] although imputation efficiency varied with completeness of genome references.
Before imputation, we were able to bioinformatically map 15,172 (~23.14% of total SNPs called), 36,278 (~55.34%), 56,125 (~85.61%) and 62,241 (94.94%) SNPs out of 65,560 SNPs with <80% missing data to the barley reference genome (Table 1), CSSS (Table 2), W7984 (Table 3) and IWGSC RefSeq v1.0 (Table 4) references, respectively. That the most SNPs were mapped to IWGSC RefSeq v1.0 among the four reference genomes may be due to that the newest reference has the best genome coverage. For all four references, the highest number of SNPs were mapped to the B genome with 47.77%, 51.43%, 48.91% and 47.87% of totally mapped SNPs to the barley reference genome, CSSS, W7984 and IWGSC RefSeq v1.0 references, respectively, and the lowest number of SNPs were mapped to the D genome with 18.19%, 14.26%, 14.24% and 13.38% of totally mapped SNPs, respectively (Tables 1, 2, 3 and 4). The number of SNPs mapped to the A and B genomes were 1.8 and 3.6 times higher than those mapped on the D genome, whereas the differences in the numbers of mapped SNPs between the D genome and the A and B genomes from previous reports were even higher (about two-fold higher) than observed in this study [40–42], reflecting the most recent polyploidy bottleneck of hexaploid wheat [2, 43]. During the evolution of modern bread wheat, there has been extensive gene flow between hexaploid T. aestivum and tetraploid emmer wheat (AABB), while gene flow between the hexaploid and Ae. tauschii (DD) might have not occurred [44–47], which might explain higher polymorphism on the A and B genomes than on the D genome [31, 48]. The greatest number of SNPs was mapped to the chromosome 3B and the least number of SNPs was mapped to the chromosome 4D, which agrees with Edae et al. [4] using W7984 and CSSS assemblies.
The number of transition-type SNPs (68.63%) with majority of A/G (32.27%) and C/T (28.36%) transition was much higher than transversion-type SNPs with an average Ts/Tv SNP ratio of 2.19 (Table 1), which agrees with several previous studies on hexaploid wheat [31, 49–53] and barley [54–56] where Ts/Tv SNP ratios was from 1.59 to 2.12. A/G and C/T types of mutations are usually due to methylation of cytosine that can be easily achieved from spontaneous deamination and transition to a thymine [57]. In this study, the Ts/Tv SNP ratios from the A and B genomes were significantly higher than that from the D genome, which most likely due to high methylation occurred in the A and B genomes during the two rounds of polyploidization [49], whereas D genome has been through only one round of such polyploidization during the hexaploid wheat evolution [58].
Among the four reference genomes, IWGSC RefSeq v1.0 has the best wheat genome sequence coverage and assembly quality therefore it is expected that the IWGSC RefSeq v1.0 generated the most imputed data points (1,776,773) from the five sequencing runs and the barley reference genome (464,174) generated the least (Table 6). However, W7984 assembly had the highest imputation accuracy. An obvious relationship was not observed between imputation accuracy and chromosome size and between imputation accuracy and number of missing data per chromosome. The percentages of imputed SNPs were much higher in the A (~38.69%) and B (~48.03%) genomes than that in the D genome (~13.28). The numbers of imputed data points in the A and B chromosomes were much higher than that from the D chromosomes, but imputation accuracy of the SNPs in the D chromosomes was the highest (Table 6). This could be owing to the low polymorphism level that resulted in a relatively low number of imputed SNPs in the D genome [59]. A high LD level on the D chromosomes may also contribute to its higher imputation accuracy than that in the A and B genomes [43, 60–63] as observed in several other studies [36, 64–66].
A high positive correlation was observed between imputation accuracy and allele frequency. Based on different references and runs, the greatest number of imputed data points was observed in allele frequencies of 0.55 to 0.95 with imputation accuracy from 88 to 99% (Fig 4a, 4b and 4c). Although the numbers of imputed data points using IWGSC RefSeq v1.0 were higher than those using other three references in different allele frequencies, especially after six runs, the imputation accuracy using IWGSC RefSeq v1.0 was much lower than other references in the lower allele frequency. However, imputation accuracy for 75% of missing data were similar among four genome references, and only the missing data with lower allele frequencies showed difference in imputation accuracy among references. SNPs with a low minor allele frequency (MAF < 10%) was reported to have significantly lower power to detect true trait-marker association [67], thus, they were removed in many GWAS [68]. Since IWGSC RefSeq v1.0 imputed the most missing data points, it can be used to impute missing data if SNPs with MAF < 10% was removed in a study. However, in the cases where rare variants with MAF <10% might play a more important role than common SNPs with a MAF >10% [69], imputation using W7984 may improve imputation accuracy.
Conclusions
Imputation using genome references is an effective tool to fill up massive missing genotypic data generated from GBS. Among the four references (the barley reference genome and wheat reference genomes of CSSS, W7984 and IWGSC RefSeq v1.0) used for imputation, IWGSC RefSeq v1.0 imputed the greatest number of missing data points with adequate imputation accuracy, especially for those alleles with high frequencies. For those alleles with low allele frequency, W7984 assembly showed the best imputation accuracy although imputed number of missing data points was slightly lower than the IWGSC RefSeq v1.0 reference. Therefore, they both can be used as reference genomes to impute missing GBS data in wheat breeding and genetic research.
Acknowledgments
Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the USDA. USDA is an equal opportunity provider and employer.
Data Availability
All relevant data are within the paper and SNP data files are deposited in Figshare. SNP data files for 5, 6, 7, and 8 runs can be each accessed at https://figshare.com/s/92cd9f23d9dddb224304; https://figshare.com/s/b402031d86d969a7cb0f; https://figshare.com/s/83b4abe205a8a5409017; https://figshare.com/s/253e39735d226b78b6bd. All the original GBS reads will be available upon request. Please send request to Guihua.Bai@ARS.USDA.GOV, Hard Winter Wheat Genetics Research, ARS-USDA, Manhattan, KS 66506, USA.
Funding Statement
This project is partly funded by US Wheat and Barley Scab Initiative and the National Research Initiative Competitive Grants 2017-67007-25939 to GB and 2017-67007-25929 to GB from the National Institute of Food and Agriculture, U.S. Department of Agriculture. Funders had no role in the study design, data collection and analysis, decision to publication, or manuscript preparation.
References
- 1.FAO. High Level Expert Forum—How to Feed the World in 2050. Food and Agricultural Organization of the United Nations Rome; 2009.
- 2.Marcussen T, Sandve SR, Heier L, Spannagl M, Pfeifer M, Jakobsen KS, et al. Ancient hybridizations among the ancestral genomes of bread wheat. Science. 2014;345(6194):1250092 10.1126/science.1250092 [DOI] [PubMed] [Google Scholar]
- 3.Ray DK, Mueller ND, West PC, Foley JA. Yield trends are insufficient to double global crop production by 2050. PloS One. 2013;8(6):e66428 10.1371/journal.pone.0066428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Edae EA, Bowden RL, Poland J. Application of Population Sequencing (POPSEQ) for Ordering and Imputing Genotyping-by-Sequencing Markers in Hexaploid Wheat. G3. 2015;5(12):2547–53. 10.1534/g3.115.020362 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler ES, et al. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PloS One. 2011;6(5):e19379 10.1371/journal.pone.0019379 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He J, Zhao X, Laroche A, Lu ZX, Liu H, Li Z. Genotyping-by-sequencing (GBS), an ultimate marker-assisted selection (MAS) tool to accelerate plant breeding. Front Plant Sci. 2014;5:484 10.3389/fpls.2014.00484 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Batley J, Edwards D. SNP applications in plants Association mapping in plants: Springer; 2007. p. 95–102. [Google Scholar]
- 8.Kumar S, Banks TW, Cloutier S. SNP Discovery through Next-Generation Sequencing and Its Applications. Int J Plant Genomics. 2012;2012:831460 10.1155/2012/831460 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yan J, Kandianis CB, Harjes CE, Bai L, Kim E-H, Yang X, et al. Rare genetic variation at Zea mays crtRB1 increases β-carotene in maize grain. Nature Genet. 2010;42(4):322 10.1038/ng.551 [DOI] [PubMed] [Google Scholar]
- 10.Sato K, Nankaku N, Takeda K. A high-density transcript linkage map of barley derived from a single population. Heredity. 2009;103(2):110 10.1038/hdy.2009.57 [DOI] [PubMed] [Google Scholar]
- 11.Bernardo A, Wang S, St Amand P, Bai G. Using Next Generation Sequencing for Multiplexed Trait-Linked Markers in Wheat. PloS One. 2015;10(12):e0143890 10.1371/journal.pone.0143890 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poland J, Endelman J, Dawson J, Rutkoski J, Wu S, Manes Y, et al. Genomic Selection in Wheat Breeding using Genotyping-by-Sequencing. Plant Genome. 2012;5(3):103 10.3835/plantgenome2012.06.0006 [DOI] [Google Scholar]
- 13.Poland JA, Brown PJ, Sorrells ME, Jannink J-L. Development of high-density genetic maps for barley and wheat using a novel two-enzyme genotyping-by-sequencing approach. PloS One. 2012;7(2):e32253 10.1371/journal.pone.0032253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang YF, Poland JA, Wight CP, Jackson EW, Tinker NA. Using genotyping-by-sequencing (GBS) for genomic discovery in cultivated oat. PloS One. 2014;9(7):e102448 10.1371/journal.pone.0102448 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meuwissen TH, Goddard ME. Accurate prediction of genetic values for complex traits by whole genome resequencing. Genetics. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Druet T, Macleod I, Hayes B. Toward genomic prediction from whole-genome sequence data: impact of sequencing design on genotype imputation and accuracy of predictions. Heredity. 2014;112(1):39 10.1038/hdy.2013.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nature Genet. 2007;39(7):906 10.1038/ng2088 [DOI] [PubMed] [Google Scholar]
- 18.Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34(8):816–34. 10.1002/gepi.20533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scheet P, Stephens M. A fast and flexible statistical model for large-scale population genotype data: applications to inferring missing genotypes and haplotypic phase. Am J Hum Genet. 2006;78(4):629–44. 10.1086/502802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Browning BL, Browning SR. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am J Hum Genet. 2009;84(2):210–23. 10.1016/j.ajhg.2009.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Whalen A, Gorjanc G, Ros-Freixedes R, Hickey JM. Assessment of the performance of different hidden Markov models for imputation in animal breeding. bioRxiv. 2017:227157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pei Y-F, Li J, Zhang L, Papasian CJ, Deng H-W. Analyses and comparison of accuracy of different genotype imputation methods. PloS One. 2008;3(10):e3551 10.1371/journal.pone.0003551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Verma S, Gupta S, Bandhiwal N, Kumar T, Bharadwaj C, Bhatia S. High-density linkage map construction and mapping of seed trait QTLs in chickpea (Cicer arietinum L.) using Genotyping-by-Sequencing (GBS). Sci Rep. 2015;5:17512 10.1038/srep17512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chan AW, Hamblin MT, Jannink J-L. Evaluating imputation algorithms for low-depth genotyping-by-sequencing (GBS) data. PloS One. 2016;11(8):e0160733 10.1371/journal.pone.0160733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hussain W, Baenziger PS, Belamkar V, Guttieri MJ, Venegas JP, Easterly A, et al. Genotyping-by-sequencing derived high-density linkage map and its application to QTL mapping of flag leaf traits in bread wheat. Sci Rep. 2017;7(1):16394 10.1038/s41598-017-16006-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y, Lin G, Li C, Stothard P. Genotype imputation methods and their effects on genomic predictions in cattle. Springer Science Reviews. 2016;4(2):79–98. [Google Scholar]
- 27.Poland JA, Rife TW. Genotyping-by-Sequencing for Plant Breeding and Genetics. Plant Genome. 2012;5(3):92 10.3835/plantgenome2012.05.0005 [DOI] [Google Scholar]
- 28.Mayer KF, Waugh R, Brown JW, Schulman A, Langridge P, Platzer M, et al. A physical, genetic and functional sequence assembly of the barley genome. Nature. 2012;491(7426):711–6. 10.1038/nature11543 . [DOI] [PubMed] [Google Scholar]
- 29.International Wheat Genome Sequencing C. A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum L.) genome. Science. 2014;345(6194):1251788 10.1126/science.1251788 . [DOI] [PubMed] [Google Scholar]
- 30.Chapman JA, Mascher M, Buluc A, Barry K, Georganas E, Session A, et al. A whole-genome shotgun approach for assembling and anchoring the hexaploid bread wheat genome. Genome Biol. 2015;16:26 10.1186/s13059-015-0582-8 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alipour H, Bihamta MR, Mohammadi V, Peyghambari SA, Bai G, Zhang G. Genotyping-by-Sequencing (GBS) Revealed Molecular Genetic Diversity of Iranian Wheat Landraces and Cultivars. Front Plant Sci. 2017;8:1293 10.3389/fpls.2017.01293 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard R. Ribosomal DNA spacer-length polymorphisms in barley: Mendelian inheritance, chromosomal location, and population dynamics. Proc Natl Acad Sci. 1984;81(24):8014–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu F, Lipka AE, Glaubitz J, Elshire R, Cherney JH, Casler MD, et al. Switchgrass genomic diversity, ploidy, and evolution: novel insights from a network-based SNP discovery protocol. PLoS Genet. 2013;9(1):e1003215 10.1371/journal.pgen.1003215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics. 2007;23(19):2633–5. 10.1093/bioinformatics/btm308 [DOI] [PubMed] [Google Scholar]
- 35.Rabiner LR. A tutorial on hidden Markov models and selected applications in speech recognition. Proc IEEE. 1989;77(2):257–86. [Google Scholar]
- 36.He S, Zhao Y, Mette MF, Bothe R, Ebmeyer E, Sharbel TF, et al. Prospects and limits of marker imputation in quantitative genetic studies in European elite wheat (Triticum aestivum L.). BMC Genomics. 2015;16(1):168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thomson MJ. High-Throughput SNP Genotyping to Accelerate Crop Improvement. Plant Breed Biotechnol. 2014;2(3):195–212. 10.9787/pbb.2014.2.3.195 [DOI] [Google Scholar]
- 38.Davey JW, Hohenlohe PA, Etter PD, Boone JQ, Catchen JM, Blaxter ML. Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nature Rev Genet. 2011;12(7):499–510. 10.1038/nrg3012 . [DOI] [PubMed] [Google Scholar]
- 39.Andolfatto P, Davison D, Erezyilmaz D, Hu TT, Mast J, Sunayama-Morita T, et al. Multiplexed shotgun genotyping for rapid and efficient genetic mapping. Genome Res. 2011;21(4):610–7. 10.1101/gr.115402.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Allen AM, Barker GL, Berry ST, Coghill JA, Gwilliam R, Kirby S, et al. Transcript‐specific, single‐nucleotide polymorphism discovery and linkage analysis in hexaploid bread wheat (Triticum aestivum L.). Plant Biotechnol J. 2011;9(9):1086–99. 10.1111/j.1467-7652.2011.00628.x [DOI] [PubMed] [Google Scholar]
- 41.Cavanagh CR, Chao S, Wang S, Huang BE, Stephen S, Kiani S, et al. Genome-wide comparative diversity uncovers multiple targets of selection for improvement in hexaploid wheat landraces and cultivars. Proc Natl Acad Sci. 2013;110(20):8057–62. 10.1073/pnas.1217133110 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Allen AM, Barker GL, Wilkinson P, Burridge A, Winfield M, Coghill J, et al. Discovery and development of exome‐based, co‐dominant single nucleotide polymorphism markers in hexaploid wheat (Triticum aestivum L.). Plant Biotechnol J. 2013;11(3):279–95. 10.1111/pbi.12009 [DOI] [PubMed] [Google Scholar]
- 43.Chao S, Dubcovsky J, Dvorak J, Luo M-C, Baenziger SP, Matnyazov R, et al. Population-and genome-specific patterns of linkage disequilibrium and SNP variation in spring and winter wheat (Triticum aestivum L.). BMC Genomics. 2010;11(1):727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Berkman PJ, Visendi P, Lee HC, Stiller J, Manoli S, Lorenc MT, et al. Dispersion and domestication shaped the genome of bread wheat. Plant Biotechnol J. 2013;11(5):564–71. 10.1111/pbi.12044 [DOI] [PubMed] [Google Scholar]
- 45.Caldwell KS, Dvorak J, Lagudah ES, Akhunov E, Luo MC, Wolters P, et al. Sequence polymorphism in polyploid wheat and their d-genome diploid ancestor. Genetics. 2004;167(2):941–7. 10.1534/genetics.103.016303 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dvorak J, Akhunov ED, Akhunov AR, Deal KR, Luo M-C. Molecular characterization of a diagnostic DNA marker for domesticated tetraploid wheat provides evidence for gene flow from wild tetraploid wheat to hexaploid wheat. Mol Biol Evol. 2006;23(7):1386–96. 10.1093/molbev/msl004 [DOI] [PubMed] [Google Scholar]
- 47.Talbert L, Smith L, Blake N. More than one origin of hexaploid wheat is indicated by sequence comparison of low-copy DNA. Genome. 1998;41(3):402–7. [Google Scholar]
- 48.Lai K, Lorenc MT, Lee HC, Berkman PJ, Bayer PE, Visendi P, et al. Identification and characterization of more than 4 million intervarietal SNPs across the group 7 chromosomes of bread wheat. Plant Biotechnol J. 2015;13(1):97–104. 10.1111/pbi.12240 . [DOI] [PubMed] [Google Scholar]
- 49.Lorenc MT, Hayashi S, Stiller J, Lee H, Manoli S, Ruperao P, et al. Discovery of Single Nucleotide Polymorphisms in Complex Genomes Using SGSautoSNP. Biology. 2012;1(2):370–82. 10.3390/biology1020370 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Winfield MO, Wilkinson PA, Allen AM, Barker GL, Coghill JA, Burridge A, et al. Targeted re‐sequencing of the allohexaploid wheat exome. Plant Biotechnol J. 2012;10(6):733–42. 10.1111/j.1467-7652.2012.00713.x [DOI] [PubMed] [Google Scholar]
- 51.Manickavelu A, Jighly A, Ban T. Molecular evaluation of orphan Afghan common wheat (Triticum aestivum L.) landraces collected by Dr. Kihara using single nucleotide polymorphic markers. BMC Plant Biol. 2014;14(1):320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cui F, Zhang N, Fan XL, Zhang W, Zhao CH, Yang LJ, et al. Utilization of a Wheat660K SNP array-derived high-density genetic map for high-resolution mapping of a major QTL for kernel number. Sci Rep. 2017;7(1):3788 10.1038/s41598-017-04028-6 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rimbert H, Darrier B, Navarro J, Kitt J, Choulet F, Leveugle M, et al. High throughput SNP discovery and genotyping in hexaploid wheat. PloS One. 2018;13(1):e0186329 10.1371/journal.pone.0186329 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Turuspekov Y, Ormanbekova D, Rsaliev A, Abugalieva S. Genome-wide association study on stem rust resistance in Kazakh spring barley lines. BMC Plant Biol. 2016;16 Suppl 1:6 10.1186/s12870-015-0686-z . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kono TJ, Fu F, Mohammadi M, Hoffman PJ, Liu C, Stupar RM, et al. The Role of Deleterious Substitutions in Crop Genomes. Mol Biol Evol. 2016;33(9):2307–17. 10.1093/molbev/msw102 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bayer MM, Rapazote-Flores P, Ganal M, Hedley PE, Macaulay M, Plieske J, et al. Development and Evaluation of a Barley 50k iSelect SNP Array. Front Plant Sci. 2017;8:1792 10.3389/fpls.2017.01792 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rosenberg MS, Subramanian S, Kumar S. Patterns of transitional mutation biases within and among mammalian genomes. Mol Biol Evol. 2003;20(6):988–93. 10.1093/molbev/msg113 [DOI] [PubMed] [Google Scholar]
- 58.Lai K. Genome diversity in Triticum aestivum. PhD thesis, The University of Queensland, St Lucia, QLD. 2015.
- 59.Jordan KW, Wang S, Lun Y, Gardiner LJ, MacLachlan R, Hucl P, et al. A haplotype map of allohexaploid wheat reveals distinct patterns of selection on homoeologous genomes. Genome Biol. 2015;16:48 10.1186/s13059-015-0606-4 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Akhunov ED, Akhunova AR, Anderson OD, Anderson JA, Blake N, Clegg MT, et al. Nucleotide diversity maps reveal variation in diversity among wheat genomes and chromosomes. BMC Genomics. 2010;11(1):702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chao S, Zhang W, Dubcovsky J, Sorrells M. Evaluation of genetic diversity and genome-wide linkage disequilibrium among US wheat (Triticum aestivum L.) germplasm representing different market classes. Crop Science. 2007;47(3):1018–30. [Google Scholar]
- 62.Chen X, Min D, Yasir TA, Hu Y-G. Genetic diversity, population structure and linkage disequilibrium in elite Chinese winter wheat investigated with SSR markers. PloS One. 2012;7(9):e44510 10.1371/journal.pone.0044510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang S, Wong D, Forrest K, Allen A, Chao S, Huang BE, et al. Characterization of polyploid wheat genomic diversity using a high-density 90,000 single nucleotide polymorphism array. Plant Biotechnol J. 2014;12(6):787–96. 10.1111/pbi.12183 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang Z, Druet T. Marker imputation with low-density marker panels in Dutch Holstein cattle. J Dairy Sci. 2010;93(11):5487–94. 10.3168/jds.2010-3501 [DOI] [PubMed] [Google Scholar]
- 65.Rutkoski JE, Poland J, Jannink J-L, Sorrells ME. Imputation of unordered markers and the impact on genomic selection accuracy. G3. 2013;3(3):427–39. 10.1534/g3.112.005363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Torkamaneh D, Belzile F. Scanning and Filling: Ultra-Dense SNP Genotyping Combining Genotyping-By-Sequencing, SNP Array and Whole-Genome Resequencing Data. PloS One. 2015;10(7):e0131533 10.1371/journal.pone.0131533 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ardlie KG, Lunetta KL, Seielstad M. Testing for population subdivision and association in four case-control studies. Am J Hum Genet. 2002;71(2):304–11. 10.1086/341719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cupples LA, Arruda HT, Benjamin EJ, D’Agostino RB, Demissie S, DeStefano AL, et al. The Framingham Heart Study 100K SNP genome-wide association study resource: overview of 17 phenotype working group reports. BioMed Central; 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McClellan J, King M-C. Genetic heterogeneity in human disease. Cell. 2010;141(2):210–7. 10.1016/j.cell.2010.03.032 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper and SNP data files are deposited in Figshare. SNP data files for 5, 6, 7, and 8 runs can be each accessed at https://figshare.com/s/92cd9f23d9dddb224304; https://figshare.com/s/b402031d86d969a7cb0f; https://figshare.com/s/83b4abe205a8a5409017; https://figshare.com/s/253e39735d226b78b6bd. All the original GBS reads will be available upon request. Please send request to Guihua.Bai@ARS.USDA.GOV, Hard Winter Wheat Genetics Research, ARS-USDA, Manhattan, KS 66506, USA.