Abstract
LATERAL ORGAN BOUNDARIES DOMAIN (LBD) proteins, a family of plant-specific transcription factors harboring a conserved Lateral Organ Boundaries (LOB) domain, are regulators of plant organ development. Recent studies have unraveled additional pivotal roles of the LBD protein family beyond defining lateral organ boundaries, such as pollen development and nitrogen metabolism. The structural basis for the molecular network of LBD-dependent processes remains to be deciphered. Here, we solved the first structure of the homodimeric LOB domain of Ramosa2 from wheat (TtRa2LD) to 1.9 Å resolution. Our crystal structure reveals structural features shared with other zinc-finger transcriptional factors, as well as some features unique to LBD proteins. Formation of the TtRa2LD homodimer relied on hydrophobic interactions of its coiled-coil motifs. Several specific motifs/domains of the LBD protein were also involved in maintaining its overall conformation. The intricate assembly within and between the monomers determined the precise spatial configuration of the two zinc fingers that recognize palindromic DNA sequences. Biochemical, molecular modeling, and small-angle X-ray scattering experiments indicated that dimerization is important for cooperative DNA binding and discrimination of palindromic DNA through a molecular calipers mechanism. Along with previously published data, this study enables us to establish an atomic-scale mechanistic model for LBD proteins as transcriptional regulators in plants.
Keywords: structural biology, transcription factor, plant molecular biology, small-angle X-ray scattering (SAXS), zinc finger, cooperative binding, LBD proteins, LOB domain, molecular model, TtRa2LD
Introduction
The plant-specific LATERAL ORGAN BOUNDARIES DOMAIN (LBD)4 proteins are expressed at the adaxial base of initiating lateral organs and regulate plant pattern formation by precisely coordinating temporal and spatial developmental programs. It has been well-documented that these proteins are involved in a large number of developments, such as meristem programming (1, 2), inflorescence morphogenesis (3, 4), leaf patterning (5, 6), and lateral root formation (7, 8). Recent studies have expanded their functional diversity beyond the definition of lateral organ boundaries. The LBD family proteins also play pivotal roles in pollen development (9, 10), vascular differentiation (11), nitrogen metabolism (12), plant regeneration (13, 14), photomorphogenesis (15), disease susceptibility (16, 17), and some specific developmental processes in wood and grass species (18, 19).
In the model plant Arabidopsis, the LBD gene family comprises 43 members (20). A genome survey of the recently published maize (Zea mays) and rice (Oryza sativum) genomes has revealed the presence of 44 and 35 LBD genes in the two plant species, respectively (21, 22). Ramosa2, a maize LBD protein, is involved in floral organ branching and architecture. The ra2 mutants display striking phenotypic heterogeneities in inflorescence developments, including additional tassel branching, spikelet multimers, and disorganized rows of ear kernels (3, 4).
LBD proteins are defined by a conserved Lateral Organ Boundaries (LOB) domain, which comprises a conserved CX2CX6CX3C motif (referred to as zinc finger). This domain is often followed by three conserved residues Gly–Ala–Ser (termed the GAS motif) harboring an invariant glycine residue and a C-terminal leucine zipper–like motif (LZLM). Although the zinc finger is assumed to be responsible for DNA binding, the LZLM is thought to be a protein dimerization domain because the C-terminal sequence includes five repetitions of hydrophobic amino acid residues, such as valine, isoleucine, and leucine, with six-residue intervals, and it has characteristics similar to a coiled-coil motif (20, 23). A DNA-binding study has shown that Arabidopsis LBD proteins, such as LOB, LBD4, and ASYMMETRIC LEAVES2 (AS2), specifically bind a 6-bp consensus LBD-response element (LBD-RE, GCGGCG), which is conserved in the four-nucleotide core sequence CGGC (24). It is well-documented that LBD proteins are nuclear proteins, and the LBD-RE sequence is present in promoters of several target genes of LBD proteins (25–30). Therefore, LBD proteins are DNA-binding transcription factors. To date, about 129,288 plant transcription factors from 83 species have been identified, which are classified into 58 families (31). Transcription factors are usually classified into different families according to their DNA-binding domains. Currently, three-dimensional structures of DNA-binding domains, including GRAS, ARF, NAC, SBP, WRKY, B3, and AP2/ERF, have been elucidated, which represent about 10 plant-specific transcription factor families (32–38). However, the structure and mechanistic basis of regulation of the LBD family of transcription factors are still unsolved.
In this study, we crystallized and determined the crystal structure of the LOB domain of an LBD protein from wheat. The structure mainly consists of a zinc finger, a GAS motif consisting of two α-helices, a highly conserved five-residue motif (Asp–Pro–Val–Tyr–Gly, referred to as DPVYG motif), and an amphipathic α-helix with the feature of leucine zipper–like coiled-coil element (Fig. 1). The intricate assembly within and between the monomers determines the exact spatial arrangement of the two zinc fingers through which the palindrome sequences are recognized and bound. These structural features were further confirmed by biochemical and biophysical studies. The structure, together with our molecular modeling and SAXS experiments, enables us to gain a deep insight into the structure-based mechanism for DNA-binding specificity in the LBD family of transcription factors.
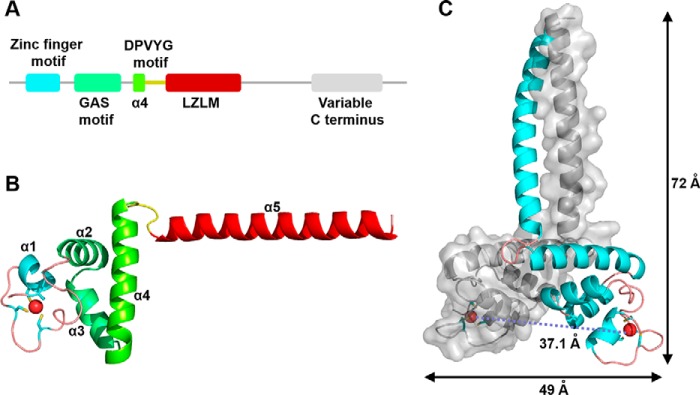
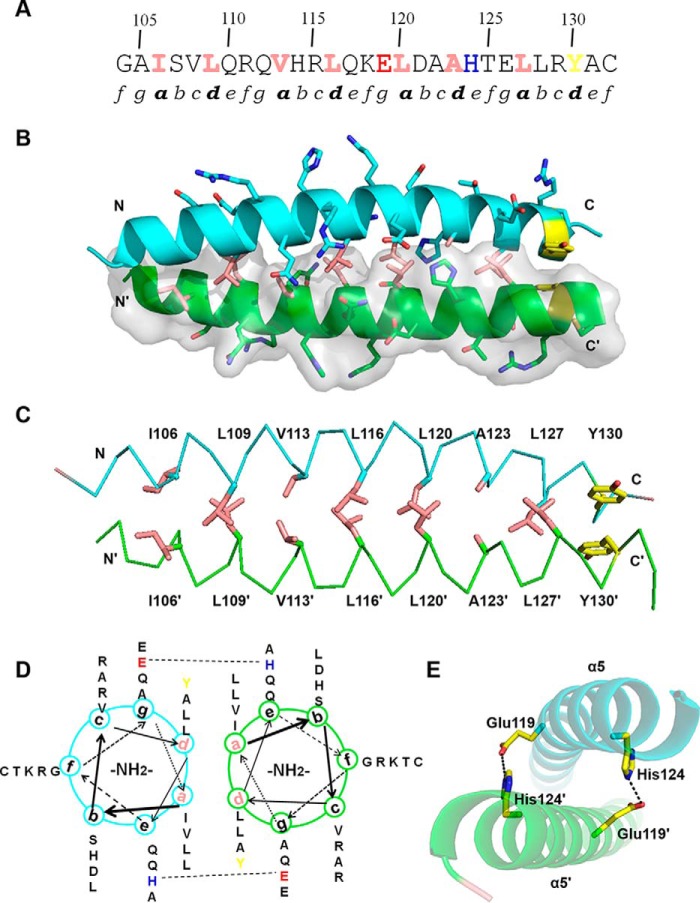
Figure 1.
Overall structure of TtRa2LD. A, motif organization with individual motifs labeled and colored: zinc-finger motif (cyan), GAS motif (green), α4 (chartreuse), DPVYG motif (dark yellow), LZLM (crimson), and the variable C terminus (gray). B, ribbon representation of TtRa2LD monomer. Motifs are colored as in A, and Zn2+ ion is shown as a red sphere. C, structure of TtRa2LD dimer, with one monomer shown in cartoon representation and the other in surface rendering.
Results
Overall crystal structure of the LOB domain of Ramosa2 from wheat
The LBD family proteins consist of an N-terminal LOB domain and a variable C-terminal region that regulates downstream gene expression (1). Our phylogenetic analysis and multiple sequence alignment revealed that the LOB domain is highly conserved throughout the plant kingdom (Fig. S1). To gain insight into the structural and mechanistic basis of the functions of LBD transcription factors, we screened several LBD proteins from different species such as Arabidopsis, maize, rice, and wheat. Finally, the LOB domain of Ramosa2 from wheat (Triticum turgidum) (termed as TtRa2LD) harboring residues 17–147 was successfully expressed in Escherichia coli, purified to homogeneity, and crystallized. The structure of TtRa2LD was solved to 1.9 Å resolution by single-wavelength anomalous dispersion (SAD) using the natively-bound zinc ions (Table S1).
The crystal structure shows that a dimeric TtRa2LD was crystallized in space group P21 21 21, which contains two subunits per asymmetric unit. The two identical subunits mainly fold into five α-helices, and each monomer is composed of five distinct modules as follows: a compact zinc-finger domain (residues 26–50); a GAS motif composed of two α-helices (α2 and α3, residues 51–76) that are perpendicular to each other; an α-helical coiled-coil leucine zipper element (α5, residues 102–132) that is connected to the GAS motif through α4 (residues 77–96); and the DPVYG motif (residues 97–101) (Fig. 1 and Fig. S1B). N- and C-terminal ends (residues 17–25 and 133–147, respectively) do not exhibit defined electron density, suggesting that they are disordered. Interestingly, the orientation of the α-helical dimerization element α5 is approximately perpendicular to α4, and thus the dimeric TtRa2LD globally takes a T-like form, which extends ∼72 Å in length and ∼49 Å in width (Fig. 1C). The two protein subunits of TtRa2LD dimer are related by a 2-fold noncrystallographic axis and are nearly identical with a root-mean-square deviation (r.m.s.d.) of 1.57 Å over all protein atoms (r.m.s.d. of 0.93 Å calculated over Cα atoms).
At first glance, the global structure of TtRa2LD is similar to some previously solved three-dimensional structures of dimeric DNA-binding proteins, such as TFIIIA-like “zinc finger” or the steroid receptor-like DNA-binding module, but structural similarity searches with DALI (39) and PDBeFold (40) on the entire protein show that there is no homologue in the Protein Data Bank (PDB). Search of homologues of the separated GAS motif and zinc finger did not give significant results, indicating that the GAS motif and zinc finger folds with unique structural features. However, the coiled-coil dimerization domains are conserved and can be superimposed on the leucine zipper domains of GAL4 with an average r.m.s.d. on Cα of 1.0 Å. Superposition of TtRa2LD on the structure of GAL4 in complex with DNA (PDB code 1D66) reveals that the rest of TtRa2LD occupies the DNA position (Fig. 7A). Therefore, the structure of GAL4–DNA complex does not give clues that allow us to predict how TtRa2LD binds DNA.
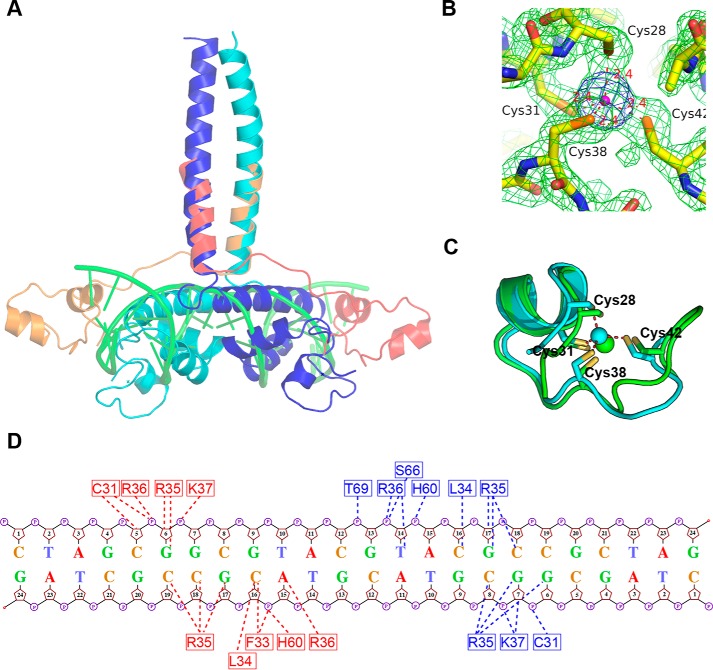
Figure 7.
Structural superposition of TtRa2LD and GAL4–DNA complex. A, superposition of TtRa2LD on GAL4–DNA complex. The dimeric TtRa2LD is colored in blue and cyan; GAL4 in red and orange; and DNA in green. B, 2Fo − Fc electron density of the zinc coordination environment contoured at 1.8σ (green) and anomalous electron density map contoured at 4σ (blue). Cys-28, Cys-31, Cys-38, and Cys-42 coordinate firmly with the central zinc ion (magenta). C, superposition of TtRa2LD Cys-4 zinc finger (cyan) on GAL4 partial Zn2/Cys-6 zinc finger (green), giving r.m.s.d. of 1.44 Å over 16 Cα. The TtRa2LD zinc finger is related to the Zn2/Cys-6-like finger in GAL4 with the first two Cys in an α-helix and the other two in a loop. D, schematic showing interactions of TtRa2LD with the palindromic LBD motifs based on the model from computational modeling.
Zinc-finger domain and its integral structure for DNA binding
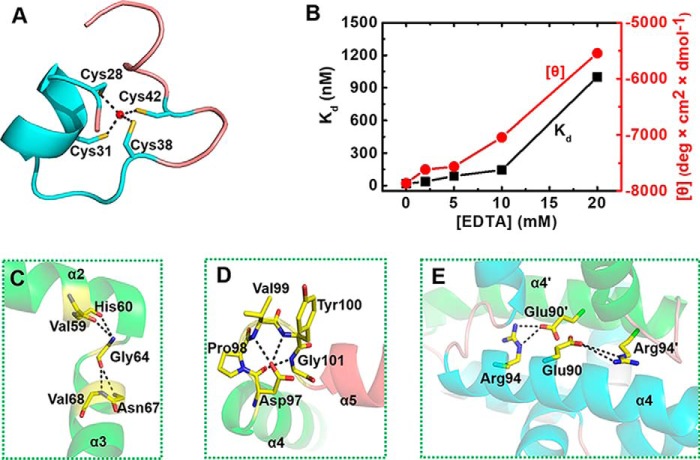
The zinc finger of TtRa2LD is composed of an α-helix (α1) with two extended random-coil segments at both ends (Fig. 1B). The conformation of the zinc finger is stabilized by tetrahedral coordination of a zinc ion with four conserved cysteine residues: Cys-28, Cys-31, Cys-38, and Cys-42 (Fig. 2A). As shown in the electron density map in Fig. 7B, the high resolution of 1.9 Å allows us to unambiguously identify the distances between the sulfhydryl atoms and the zinc ion: 2.41 Å (Zn-Sγ of Cys-28), 2.36 Å (Zn-Sγ of Cys-31), 2.40 Å (Zn-Sγ of Cys-38), and 2.36 Å (Zn-Sγ of Cys-42), with all the distances well within the ideal distances of zinc-binding sites in proteins (41). Although the zinc finger appears reminiscent of the Cys-4–type motif, it has no structural homology with previously solved Cys-4 zinc fingers such as those found in GATA (42), Sec23 (43), RecQ (44), RecR (45), or RecO (46). The zinc finger found in TtRa2LD is rather closer to the Cys-6 zinc finger found in GAL4 (PDB code 1D66) (47) and is related to the Zn2/Cys-6–like finger (fold group 5 according to the zinc finger classification) with the first two Cys in an α-helix and the other two in a loop (41). Superposition of TtRa2LD zinc finger on GAL4 zinc finger gives an r.m.s.d. of 1.44 Å over 16 Cα (Fig. 7C). The two zinc-finger motifs in the TtRa2LD dimer are congruent, and the distance between their centers is about 37.1 Å (Fig. 1C).
Figure 2.
Detailed structural interactions of residues in the conserved motifs. A, view of the Zn2+-binding motif (residues 26–50). Four highly conserved cysteine residues (Cys-28, Cys-31, Cys-38, and Cys-42) are arrayed around a Zn2+ ion (ball-and-stick). B, zinc-dependent structural formation of TtRa2LD. The DNA-binding activity (in terms of dissociation constant Kd) and the CD value [θ] (at 209 nm wavelength) were determined with increasing concentrations of EDTA. The detail experiments conditions are described under “Experimental procedures.” C, detailed view of the highly conserved glycine residue (Gly-64) with the residues in α2 and α3. D, relative conformation of α4 and α5 are determined by an interacting network of amino acid residues in the conserved DPVYG motif. E, Arg-94 and Glu-90 on α4 of one subunit form two salt bridges with Glu-90′ and Arg-94′ of α4′.
To confirm the structure of the zinc finger and better understand its intrinsic properties, we quantified the Zn2+ content using 4-(2-pyridylazo)resorcinol (PAR) and the thiol group of untreated or Zn2+-extracted TtRa2LD using DTNB (48). The results indicated that TtRa2LD was co-purified with a stoichiometric amount of Zn2+. In addition, dialysis of TtRa2LD against DTT and EDTA removed ∼93% of the bound Zn2+ and uncovered additionally ∼4 solvent-accessible thiol groups, supporting the notion that the Zn2+-binding site is composed of ∼4 cysteines near the surface of TtRa2LD (Table 1). We next asked whether the presence of Zn2+ ion is necessary for the DNA-binding activity of TtRa2LD. We pre-incubated the protein with increasing concentrations of EDTA, and we then measured its DNA-binding activity under equilibrium conditions. The results show that the DNA-binding activity was seriously compromised at high EDTA concentrations (Fig. 2B). Furthermore, analysis of circular dichroism (CD) spectra of TtRa2LD in the far-UV region further revealed that the reduction of DNA-binding activity was accompanied by an alteration of the secondary structures with increasing EDTA concentrations (Fig. 2B), confirming that the Zn2+ ion is essential for keeping the zinc-finger configuration for efficient DNA binding. These biochemical data correlate well with the determined crystal structure of TtRa2LD.
Table 1.
Bound Zn2+ and free thiol groups determined with TtRa2LD
All parameters were determined as described under “Experimental procedures.”
| Protein | [Protein] | [Zn2+] | Thiolfree/protein |
|---|---|---|---|
| nmol | nmol | ||
| TtRa2LD | 0.70 | 0.68 (0.97:1)a | 1.82 |
| Treated TtRa2LDb | 1.21 | 0.08 (0.07:1) | 5.90 |
a Ratio is between [Zn2+] and [protein].
b TtRa2LD protein was treated with EDTA/DTT.
Both GAS motif and α-helix 4 are implicated in protein stability, dimerization, and DNA binding
Our structural analysis revealed that the highly conserved GAS motif folds into two α-helices (α2 and α3) that are linked by an invariant glycine residue (Gly-64) (Fig. 1). The two α-helices are approximately perpendicular to each other and stabilized by a set of hydrophobic interactions implicated by Val-59 and His-60 of α2 and Asn-67 and Val-68 of α3, whereas Gly-64 is located at the central position of the hydrophobic core (Fig. 2C).
It was observed that a natural mutation in Arabidopsis AS2 (named as2–5) results in abnormal phenotype with the leaves having obvious humps at the base of the leaf lamina and an asymmetric lamina in addition to the prominent leaf lobes (5). Sequence analysis in the as2–5 mutant revealed that a single guanine nucleotide was replaced by an adenine nucleotide in the coding region for the N-terminal half of AS2, resulting in substitution of the glycine residue at position 64 by a glutamic acid residue (23). This glycine residue is conserved in all members of the LBD family and is located in the linker region between α2 and α3. The Gly-64 with φ/ψ value of 92.6/143.4° is in an allowed region of the Ramachandran plot only accessible for glycine (49). Therefore, this glycine residue is essential for the structure and/or function of LBD proteins. When such interaction was perturbed by replacing Gly-64 with Glu, the resulting mutant protein G64E misfolded and aggregated (results not shown), impeding further biochemical characterization, which suggests that Gly-64 plays an essential role in folding and/or protein conformational stability.
The α-helix 4 involved in the dimer formation is linked to α5 through a conserved loop (DPVYG motif). The perpendicular conformation between α4 and α5 is stabilized by a set of interactions of the residues on this loop (Fig. 2D). Asp-97 forms three hydrogen bonds with Val-99, Tyr-100, and Gly-101, respectively. The exceptional conformational rigidity of proline may affect the secondary structure of the protein near a proline residue, and the effects on interactions of the clustered residues are emphasized by a mutation of Pro-98 (P98L). The P98L mutant becomes insoluble and heavily aggregated. These results are consistent with previous studies on LBD16 and LBD18 in Arabidopsis, which showed that mutations of the conserved proline residues in LBD16 and LBD18 compromise seriously their DNA-binding activity and biological function in lateral root formation (50). More interestingly, Arg-94 and Glu-90 on α4 of one subunit form two salt bridges with Glu-90′ and Arg-94′ of α4′, stabilizing the TtRa2LD dimer (Fig. 2E). To probe the effects of these interactions on protein function, Glu-90 was replaced by Ala. Under equilibrium experimental conditions, quantitative determination and/or comparison of the DNA-binding activities between the WT and the modified TtR2LD proteins were assessed by fluorescence anisotropy-based binding assay (FABA). The WT TtRa2LD displays robust binding activities with slightly higher affinity for the palindrome sequence (AH3–ER6) than LBD-RE motif (AH3–12) (Fig. 3 and Table 2), as judged from the determined dissociation constants (Kd = 16.8 and 10.7 nm for AH3–12 and AH3–ER6, respectively). However, the dissociation constants of E90A determined with the two DNA substrates are increased by 4.5- and 3-fold, respectively (Fig. 3 and Table 2). Furthermore, the possibility that the reduced DNA-binding activity is due to abnormal mutant folding was ruled out by the identical CD spectra of both WT and mutant proteins (Fig. S3A, hereafter, all mutants were analyzed by CD with results shown in this figure). Altogether, these results show that the interactions between α4- and α4′-helices may precisely determine their relative position, enhance the dimerization, and consequently ensure the correct zinc-finger conformation for DNA binding.
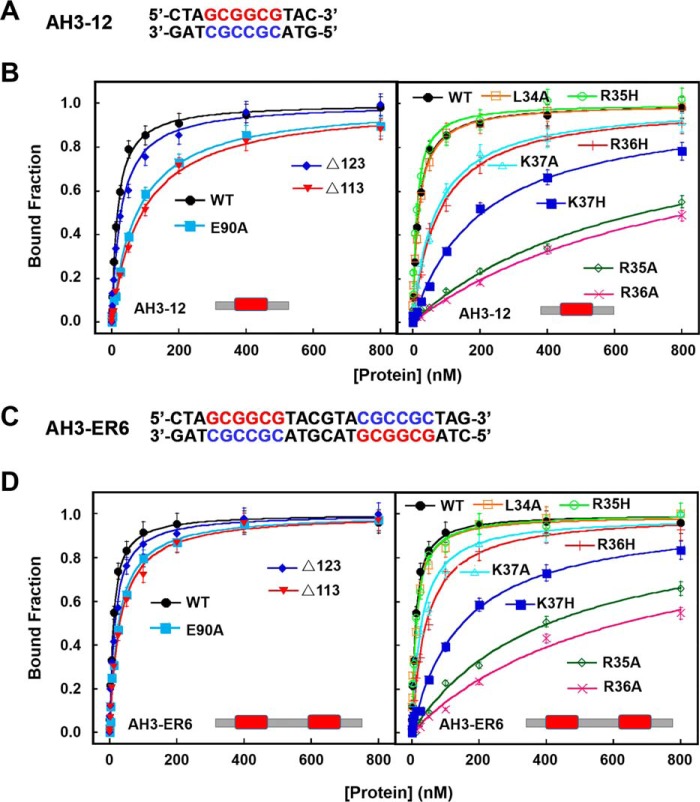
Figure 3.
DNA-binding activities of WT and mutated TtRa2LD determined with FABA. A, fluorescent-labeled DNA sequence of AH3–12. B, FABAs were performed with 2.5 nm AH3–12 and increasing concentrations of WT and mutant proteins as indicated. The determined dissociation constants are summarized in Table 2. C, fluorescent-labeled DNA sequence of AH3–ER6. D, same assays as in B, but AH3–ER6 was used. The determined dissociation constants are summarized in Table 2.
Table 2.
Dissociation constants (Kd) of wild-type and mutated TtRa2LD with two different DNAs
Values were determined with 2.5 nm DNA by steady-state FABA.
| Protein |
Kd |
|
|---|---|---|
| AH3–12 | AH3-ER6 | |
| nm | ||
| WT | 16.8 ± 1.0 | 10.7 ± 0.6 |
| E90A | 75.7 ± 4.3 | 26.7 ± 1.8 |
| L34A | 17.4 ± 1.2 | 12.3 ± 1.3 |
| R35A | 681 ± 118 | 407 ± 48 |
| R35H | 11.4 ± 1.0 | 12.9 ± 1.0 |
| R36A | 831 ± 111 | 623 ± 128 |
| R36H | 77.5 ± 6.7 | 43.3 ± 3.5 |
| K37A | 68.6 ± 5.2 | 28.3 ± 2.9 |
| K37H | 204 ± 19 | 151 ± 18 |
| TMa | NDb | ND |
| Δ123 | 27.8 ± 2.1 | 15.8 ± 1.2 |
| Δ113 | 90.8 ± 5.4 | 30.7 ± 2.1 |
| Δα5 | ND | ND |
a TM indicates triple mutant R35A/R36A/K37A.
b ND is not determined; values are too low to be determined unambiguously.
Homodimerization is crucial for DNA binding
Residues 102–131 fold as an amphipathic α-helix with the typical feature of leucine zipper–like coiled-coil element defined by heptad repeats (abcdefg) (51). About four heptad repeats lie continuously with 3.6 amino acids per helical turn, and each helix forms a smooth curvature (Figs. 1C and 4A). A detailed view of the side-chain packing and a helical wheel diagram shows that the hydrophobic amino acid residues such as leucine, isoleucine, and valine are predominantly at positions a and d, and the hydrophilic residues such as Lys, Arg, Glu, and Gln are at positions b, c, e, f, and g (Fig. 4, B and D). The two helices are intertwined with each other to form a left-handed coiled-coil. Fig. 4C shows the hydrophilic residues at positions b, c, e, f, and g that force the hydrophobic regions of the side chains at positions a and d to form an extensive hydrophobic core and participate in interhelical hydrophobic interactions. Consistent with previous observations that interhelical salt bridges are one of the essential features of coiled-coil formation and usually required for dimer stability, our structure reveals that Glu-119 at position g and His-124 at position e of α5 form two salt bridges with that of α5′ (Fig. 4E).
Figure 4.
Dimerization interface of LZLM. A, primary amino acid sequence of α5 contains two partial and three complete repetitive heptad repeats (labeled as a–g). Hydrophobic residues at positions a and d are colored in pink and the C-terminal Tyr residue in yellow. Glu-119 and His-124 are showed in red and blue, respectively. B, dimerization interface of α5. One monomer is shown in cartoon representation and the other in surface rendering. Side chains of hydrophobic residues at positions a and d are shown as pink sticks. C, hydrophobic interactions and π-stacking at the dimerization interface. Side chains of hydrophobic residues at positions a and d are shown as pink sticks and π-stacking side chains in yellow. D, helical wheel diagram of α5. Glu-119 and His-124, which are involved in an interhelical salt bridge, are highlighted. E, detailed view of salt bridges between α5 and α5′. Glu-119 and His-124 of α5 form interhelical salt bridges with His-124′ and Glu-119′ of α5′, respectively.
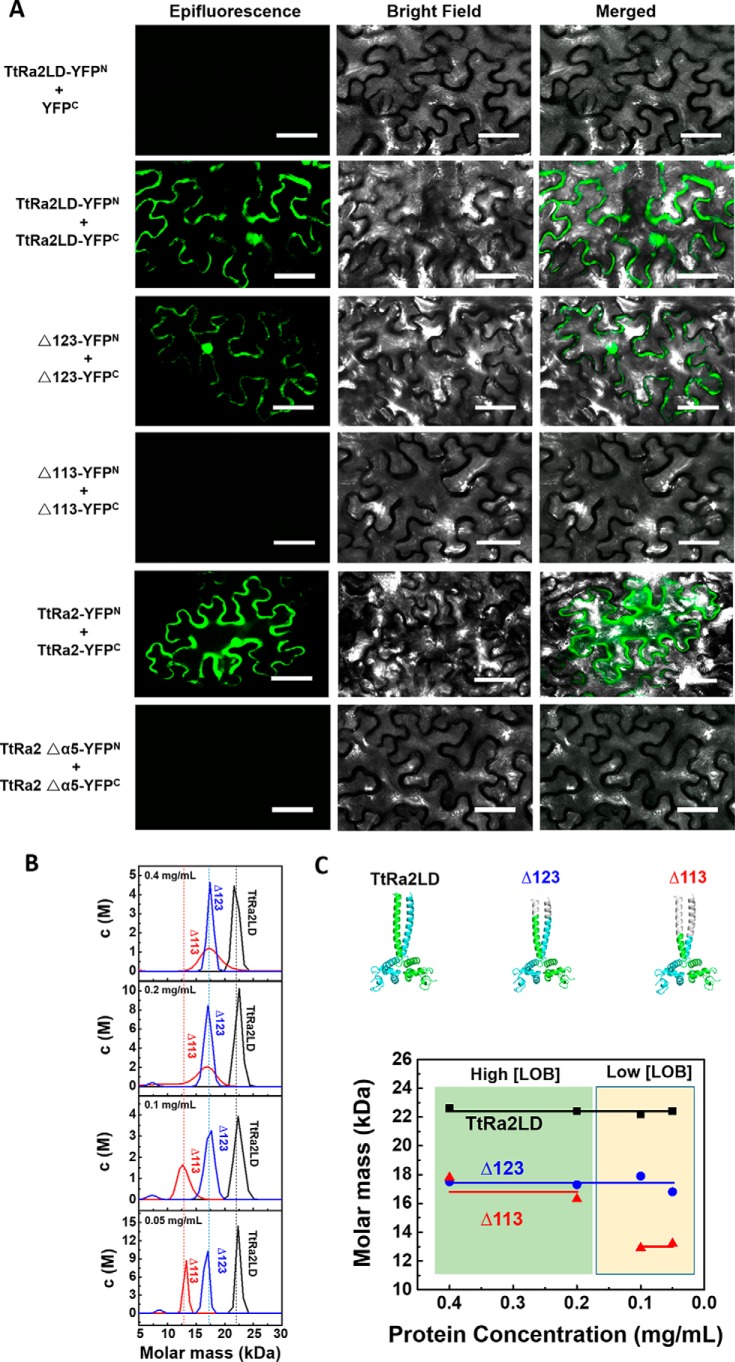
We used bimolecular fluorescence complementation (BiFC) assay to investigate the dimer formation under a normal physiological environment and to probe the coiled-coil structure–mediated dimerization. For this purpose, tobacco (Nicotiana benthamiana) leaves were transfected with vectors TtRa2LD-YFPN/YFPC (residues 17–147) as a negative control, TtRa2LD-YFPN/TtRa2LD-YFPC, TtRa2LDΔ123-YFPN/TtRa2LDΔ123-YFPC (residues 17–123, termed as Δ123), TtRa2LDΔ113-YFPN/TtRa2LDΔ113-YFPC (residues 17–113, termed as Δ113), TtRa2-YFPN/TtRa2-YFPC (residues 1–257), and TtRa2Δα5-YFPN/TtRa2Δα5-YFPC, allowing the cells to express TtRa2LD, two progressively truncated TtRa2LD (Δ123 and Δ113), the full-length TtRa2 and the α5 truncated TtRa2, respectively. As expected, no false-positive BiFC signal can be observed in leaf cells transfected with vectors TtRa2LD-YFPN/YFPC (Fig. 5).
Figure 5.
In vivo homodimerization of TtRa2LD determined by BiFC assay and analytical ultracentrifugation analyses of the WT and mutant TtRa2LD proteins. A, representative confocal images of tobacco leaf cells expressing WT and modified TtRa2 proteins. Epifluorescence and bright-field images of epidermal leaf cells were captured by Zeiss LSM510 system. Scale bar, 50 μm. B, curves for molar mass distribution (c(M)) of TtRa2LD and mutants Δ123 and Δ113 (protein contraction from 0.05 to 0.4 mg/ml). C, cartoon model of TtRa2LD and mutants Δ123 and Δ113 (upper panels) and the relationship between molar mass and protein concentration of TtRa2LD and mutants Δ123 and Δ113 from analytical ultracentrifugation results (lower panels).
The transfected lower epidermis leaf cells with TtRa2LD-YFPN/YFPC, Δ123-YFPN/YFPC, and TtRa2-YFPN/YFPC displayed green fluorescence, but those transfected with plasmids expressing the truncated proteins (Δ113 and TtRa2Δα5) did not (Fig. 5, left panel), demonstrating that deletion of 2/3 or the full-length coiled-coil element (Δα5) completely abrogates the dimerization of TtRa2LD or TtRa2. From the fact that the cells transfected with Δ123-YFPN/YFPC displayed subtle reduction in green signal, we suggest that the two salt bridges together with the last 10 amino acids of the C-terminal coiled-coil are dispensable for dimer stabilization. To understand the relationship between the dimerization and DNA-binding ability, TtRa2LD, Δ123, and Δ113 were purified, and their DNA-binding activities were determined with the two kinds of DNA substrates: AH3–12 and AH3–ER6. Compared with TtRa2LD, Δ123 displayed a comparable DNA-binding ability with the two DNA substrates (e.g. KdWT = 10.7 nm versus KdΔ123 = 15.8 nm for AH3–ER6) (Fig. 3 and Table 2). In the cases of Δα5 (complete truncation of the coiled-coil element) and Δ113 (2/3 deletion of the coiled-coil element), the dimerization signal in cells becomes invisible, and their DNA binding to the single LBD motif and the palindromic sequence are completely inactivated or severely reduced (Figs. 3 and 4 and Table 2). We further analyzed the oligomeric states of the two truncated mutants (Δ113 and Δ123) in parallel with TtRa2LD by ultracentrifugation assay. In accordance with the crystal structure and the observation in BiFC assay as described above, both TtRa2LD and Δ123 exist in solution as very stable dimer at the all protein concentrations used (Fig. 5B). In contrast, only the higher protein concentrations stabilize the mutant Δ113 as a dimer, and lower protein concentrations enhance the dissociation of Δ113 into a monomer (Fig. 5B). Taken together, the correlation between in vivo and in vitro structure–activity relationships allows us to conclude that the coiled-coil–mediated TtRa2LD homodimerization is crucial for DNA binding and recognition of potential target sites.
Molecular mechanism of DNA recognition and DNA specific binding
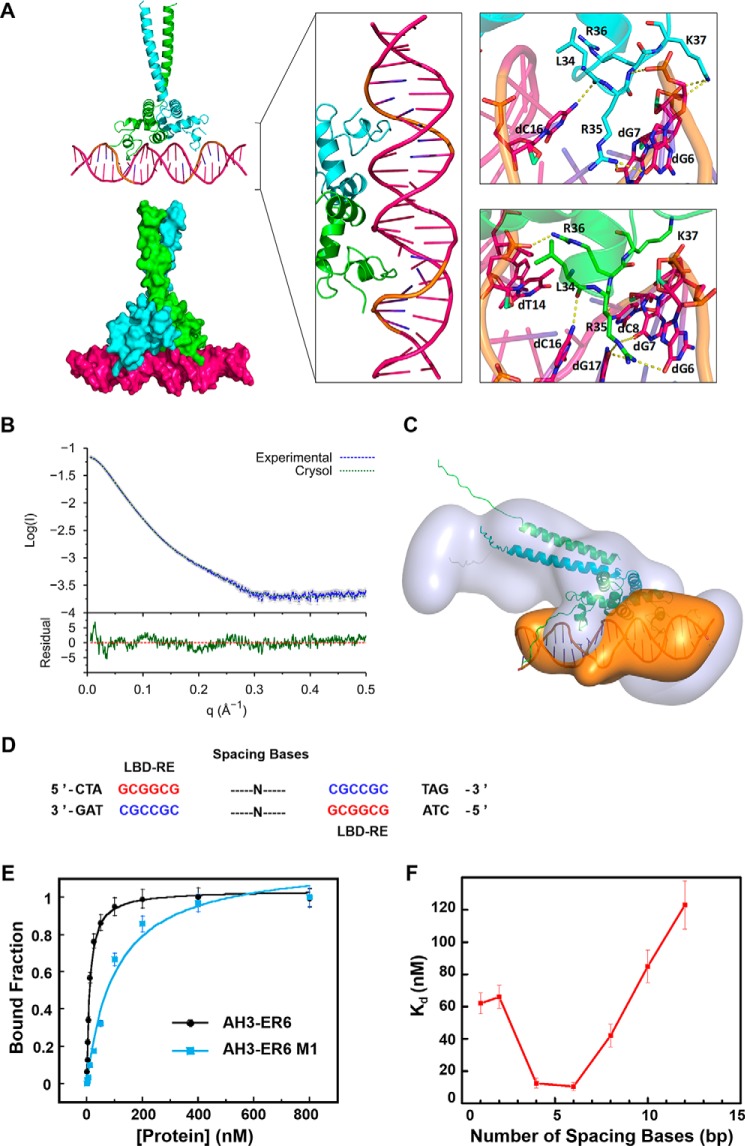
We were unable to get crystals of the protein–DNA complex. Only DNA AH3–ER6, from a protein–DNA complex mixture, was crystallized, and its structure was solved at 1.8 Å resolution, showing a typical B-DNA form. Based on the crystal structures of the protein and AH3–ER6, the structural model of the complex (TtRa2LD-AH3–ER6) was obtained by flexible docking. The stability of the modeled complex was then further submitted to 100-ns molecular dynamics with GROMACS. The interactions modeled by the flexible docking are maintained during the molecular dynamics simulation, and the overall complex becomes more symmetric with the same kind of interactions in the two inverted repeat binding sites (Fig. 6A).
Figure 6.
Molecular model of the TtRa2LD–AH3–ER6 complex. A, modeled TtRa2LD–DNA complex structure. In the left panel, the modeled complex structures are shown in cartoon and surface, respectively. The middle panel is a detailed view of the protein–DNA complex in which two zinc fingers contact with the major groove of the DNA. The right panel shows a closer view of the hydrogen-bond contacts between the highly conserved amino acids and the LBD motif (5′-GCGGCG-3′). Two TtRa2LD monomers were colored in green and cyan, respectively. DNA is colored in hot pink, and the LBD motif is colored in orange. B, SAXS profile of TtRa2LD–AH3–ER6 complex. Error bars are shown in gray. The fit and the residual of the atomic model calculated by CRYSOL is shown with its residual (Iexp − Imodel/σexp (χ2 = 4.24), together with the residual defined as (Iexp(q) − Icalc(q))/Sexp(q), corresponding to the difference between the experimental and the computed intensities weighed by experimental errors (standard deviations). C, ab initio model of TtRa2LD–AH3–ER6 complex. D, DNA sequences of AH3–ERN spaced by different spacing lengths (n = 1–12 bp). E, FABA analysis of TtRa2LD with fluorescently labeled AH3–ER6 sequence, and its derivative AH3–ER6 M1 in which one LBD–ER motif CGCCGC is replaced by AATTTAT. F, relationship between the DNA-binding affinity and the number of spacing bases. A series of fluorescently labeled AH3–ER6 derivatives in which the two LBD–ER motifs CGCCGC are spaced by different numbers of base pairs as indicated in Table S5. The dissociation constants (Kd) were determined by the FABA.
To confirm the model from computational simulation, three scattering curves of DNA, protein, and protein–DNA complex were respectively recorded from SEC–SAXS experiments. SAXS data of isolated components, DNA and protein, fit with the structures derived from X-ray crystallography (Fig. S5, A and B, respectively). A reliable model was constructed by multiphase analysis with the MONSA program, by taking the advantage that the different electron density contrasts of the protein and DNA allow us to locate them in the protein–DNA complex (Fig. 6, B and C; Figs. S6, S8, S9). Although symmetry was not imposed during calculation, the SAXS ab initio envelope reveals a dimeric structure. The protein envelope surrounds the nucleic acid on both sides, which may correspond to the zinc-finger domains. The longest part of the protein envelope corresponds to the coiled-coil helix bundle. The connection between α4 and the helix bundle may be flexible, and the LZLM domain is bent toward DNA, forming a 45° angle regarding its position in the crystal structure. The two zinc fingers have a distance of 34 Å, which represents a turn of double-stranded B-DNA. Both zinc fingers can thus be in the major groove simultaneously (Fig. 6C). The loop (residues 34–37) downstream α-helix 1 penetrates the major groove and contacts with both sugar–phosphate backbone and certain bases (Fig. 6, A and C, and Fig. 7). The residues involved in those interactions are Leu-34, Arg-35, Arg-36, and Lys-37. Leu-34 interacts with the phosphate of C3 (L34:NH-C3:O1P/O2P). The consecutive highly conserved and positively charged residues, including Arg-35, Arg-36, and Lys-37, are implicated in both base and backbone interactions (Fig. 7 and Fig. S2). Interestingly, it appears that Arg-35 is the key residue that recognizes the bases in the conserved core sequence (GCGGCG). Arg-35 side chain forms hydrogen bonds with G3 (R35:NH2-G3:O6 and R35:NH2-G3:N7), G4 (R35:NH2-G4:06 and R35:NE-G4:06), and C3 (R35:NH2-C3:N4) on the complementary strand. In addition to the zinc finger, the perpendicular α2 and α3 of the GAS motif interact with the sugar–phosphate backbone of the minor groove, where His-60 and Ser-66 interact with groups of phosphate of the spacer sequence (CGTA) (Fig. 7).
To further confirm the molecular modeling and SAXS-determined model, several mutants were prepared and assayed for DNA binding. Seven single mutants (L34A, R35A, R35H, R36A, R36H, K37A, and K37H) and one triple mutant (R35A/R36A/K37A, TM) were analyzed for their DNA-binding activities with AH3–12 and AH3–ER6 by the FABA assay. Consistent with our model, FABA confirmed that DNA bindings of R35A and R36A were reduced by about 40-and 60-fold, respectively (Fig. 3 and Table 2). When Arg was replaced by His, while R35H displays DNA-binding affinity that is comparable with WT protein, the binding ability of R36H was reduced ∼4-fold (Fig. 3 and Table 2). The DNA-binding activity of the triple mutant is too low to be determined precisely. Interestingly, the “conservative mutation” K37H severely compromises DNA binding more than K37A (KdK37A = 28.3 versus KdR37H = 150.6 nm). This is possibly due to a steric effect of the His side chain that may prohibit the flexibility of the lateral chain (Fig. 3 and Table 2). The reduced DNA binding of R36H may explain why a natural Ra2-mum4 mutant of maize corresponding to R36H shows abnormal phenotype with disorganized rows of kernels in the ear (3).
One striking crystal structural feature of TtRa2LD is that the spatial conformation of the dimeric form is well-defined not only by canonical coiled-coil element but also by two salt bridges between α4 and α4′ (Fig. 2E) and two pairs of hydrogen bonds between the two GAS motifs (Fig. S4). Although it appears that the spatial conformations of the two zinc fingers may be strictly positioned due to the series of interactions mentioned above, our SAXS data reveal that the zinc finger and the GAS motif may be more flexible in solution. Taken together, these observations suggest that TtRa2LD may be cooperative for DNA binding, although it is tolerant for palindromic DNAs bearing different spacing lengths.
Indeed, the reduction in DNA binding with the palindrome sequence mutated in one GCGGCG motif is more than 4-fold (Fig. 6E, KdWT = 10.7 nm for AH3–ER6 versus 52.3 nm for AH3–ER6 M1), indicating that the binding is cooperative but not additive. Although many LBD proteins recognize the canonical GCGGCG element, the physiological function of each member does not overlap (25–30). It is possible that the optimal DNA binding affinity of each LBD protein is also determined by spacer length, allowing a given LBD transcription factor to recognize a given target site with defined spacer length. To test this hypothesis, we performed FABA experiments using AH3–ERN oligonucleotides in which the two inverted GCGGCG sites are spaced by N bases (n = 1, 4, 6, 8, 10 and 12 bp, Fig. 6D). TtRa2LD displays optimal binding affinity toward DNA probes with a space between 4 and 6 bp as revealed by the determined dissociation constants. The DNA-binding affinity was seriously compromised when the space is more than 8 or less than 4 bp (Fig. 6F). The above phenomena and DNA-binding properties are resembling that of DNA-binding auxin response factors (33).
Discussion
Our results presented here provide a high-resolution description of the exquisite structural details of different modules in the LOB domain that was previously identified merely by sequence alignments. Our structure reveals that the previously mentioned C-block in fact forms a C4-type zinc finger. Together with GAS motif and the perpendicular conformation between α4 and α5, the zinc finger constitutes an interaction network that precisely determines the spatial configuration of DNA recognition and binding site. The perpendicular conformation between α4 and α5 is stabilized by a set of interactions of the residues on this loop and constitutes a stable coiled-coil–containing dimer. Until now, only three structures of the zinc-finger transcription factors in plants, including Arabidopsis thaliana SUPERMAN, WRKY4, and SBP-family transcription factors, are available, although the zinc finger in the WRKY4 protein does not directly participate in DNA binding (35, 37, 52). The plant TFIIIA-type zinc-finger proteins are characterized by long spacers of various lengths between adjacent fingers and a unique sequence, QALGGH, within a putative DNA-contacting surface of each finger (53, 54). However, the LBD protein harbors only one zinc-finger motif without the conserved sequence QALGGH. In accordance with these sequence characteristics, the conformation of the zinc finger determined from TtRa2LD does not match those determined in plants; rather, it resembles more the Cys-6 zinc finger found in GAL4 (Fig. 7), indicating that the zinc-finger motif of LBD proteins differs from the previously characterized zinc fingers of transcription factors in plants and possesses unique structural features.
A rigorous molecular model constructed from molecular dynamics simulations and SAXS ab initio envelope provides the molecular basis for DNA recognition and binding specificity. The zinc-finger motif recognizes the conserved LBD motif (GCGGCG) largely at the major groove. The zinc finger binds to the conserved LBD motif involving both base contacts and backbone interactions, and the specific contacts between the GCGGCG elements in major groove and a cluster of residues downstream from the α-helix1 (Leu-34, Arg-35, Arg-36, and Lys-37) play a particularly important role in the recognition of the conserved bases and DNA binding. The above interpretations are consistent with our molecular modeling data and have been confirmed by site-directed mutagenesis. Moreover, the structural information helps us to understand and interpret the previously reported phenotypes of naturally occurring and genetically produced mutants (3, 23, 50).
Homo- and heterodimerization are generally observed within LBD proteins (10, 50). However, it was not clear whether LBD proteins function as monomers in cells. According to our present crystal structure, and cell-based BiFC and high-speed ultracentrifugation analysis, dimerization is an intrinsic property of LBD proteins and is absolutely required for their cellular functions. Interestingly, our structure reveals that dimerization of LBD proteins is not merely ensured by the classical coiled-coil dimerization elements as mentioned previously in the literature, but it is also further strengthened by the additional dimerization α-helices (α4–α4′, α2′–α3, and α3–α2′) (Fig. 2E and Fig. S4) that lie between the zinc fingers and the coiled-coil module. Thus, the two monomeric LBD proteins are assembled into a stable dimer in which the distance between the two zinc-finger motifs and their stereochemical conformations are precisely determined. Interestingly, our SAXS experiments revealed that the relative positions of the zinc fingers may be more flexible in solution, and it is therefore not surprising that the dimeric TtRa2LD displays asymmetric DNA binding: the two zinc fingers recognize somewhat differently the palindromic sequences (Fig. 7D). In fact, it has been widely reported that a homodimeric transcription factor orients the zinc fingers to take an asymmetric configuration to recognize the target sequences. This may be a biologically resourceful and common mechanism for target DNA recognition (47).
Furthermore, the dimeric structural features of LOB domain indicate that the DNA binding of LBD proteins can involve either one or two binding sites; thus DNA binding may be cooperative, and the relatively constrained spacing between the two zinc fingers may determine its palindrome DNA target. Indeed, our binding experiments confirmed the cooperative property for DNA binding. With a series of DNA substrates harboring the palindromic sequence and varying in spacing lengths, we showed that the dimeric LOB domain acts as “molecular calipers” to measure the optimal spacing in potential DNA targets. In this regard, it is reminiscent of GAL4 family proteins in which the sequences linking the zinc-finger motifs and the coiled-coil elements are not structured, leaving a great flexibility of spacing for the two zinc fingers. Although the structures of heterodimers of LBD proteins are not yet available, the heterodimers formed in cells must possess structural divergences in the zinc-finger regions that might recognize different palindromic DNAs, localizing different transcriptional sites. Thus, these dimeric structural properties of LOB domain help us to understand how LBD family members perform diverse developmental programs through the cooperative actions of homo- and heterodimeric LBD-mediated molecular pathways.
Experimental procedures
Protein expression and purification
The LOB domain of T. turgidum L. Ramosa2 (residues 17–147) was amplified from cDNA clones using primers as listed in Table S3 and Prime STAR HS DNA polymerase (Takara), and it was expressed in E. coli strain 2566 (New England Biolabs) with a modified pET21a vector with a His tag, a maltose-binding protein (MBP) tag, and a TEV cleavage site fused at its N terminus. When the culture reached an A600 of 0.6, protein expression was induced by adding 0.3 mm isopropyl 1-thio-β-d-galactopyranoside for 12 h at 18 °C. After harvesting the cells, the pellets were resuspended in amylose lysis buffer (20 mm Tris-HCl, pH 7.4, 200 mm NaCl, 10% glycerol (v/v)). Cells were lysed by passing the suspension twice through a Low Temperature High Pressure Cell Disrupter at 1100 bar, and then the supernatant was generated by centrifugation at 12,000 rpm for 40 min. The sample was loaded onto a amylose column (New England Biolabs) and washed with 20 column volumes wash buffer (20 mm Tris-HCl, pH 7.4, 1 m NaCl, 10% glycerol (v/v)). Thereafter, target protein was eluted with elution buffer (20 mm Tris-HCl, pH 7.4, 200 mm NaCl, 0.5% maltose (w/v)). The eluted protein was incubated with TEV protease for 12 h at 4 °C and loaded on an equilibrated Ni2+ affinity column to remove the cleavaged N-terminal tag. The sample was dialyzed against buffer A (20 mm Tris-HCl, pH 7.4, 50 mm NaCl, 5% glycerol (v/v)) and loaded on a Hitrap SP column (GE Healthcare). The protein was then eluted by a NaCl gradient (50–1000 mm in buffer A). After a final step of Superdex 200 size-exclusion chromatography, the final purified protein was concentrated to 20 mg/ml and stored at −80 °C. The protein concentration was determined by UV absorbance at 280 nm, and the purity was checked by SDS-PAGE (typically >95%).
Crystallization, data collection, and structure determination
Crystals were grown from a 2.5 + 2.5-μl drop of 16 mg/ml protein solution and the crystallization buffer in a 24-well sitting-drop crystallization plate at 20 °C. Initial needle-shaped crystals were obtained in a solution containing 0.1 m sodium citrate tribasic dihydrate, pH 5.5, and 22% PEG1000. Optimized crystals for X-ray diffraction were acquired using a solution containing 0.1 m sodium citrate tribasic dihydrate, pH 6.4, 28% PEG1000, and 3 mm ZnCl2. Rod-shaped crystals were observed within 5 days and reached maximum sizes within 2 weeks. They were picked and immediately frozen in liquid nitrogen.
X-ray diffraction data were collected at 100 K on beamline BL19U1 at the Shanghai Synchrotron Radiation Facility using a Pilatus 6M detector (Dectris) and processed using XDS (version March 1, 2015) (55) through AutoPROC pipeline. The crystal structure of TtRa2LD was solved by single-wavelength anomalous dispersion (SAD) with AutoSHARP (56) and CCP4 suite (57) by using an anomalous signal of zinc at the wavelength of 0.9785 Å. In AutoSHARP pipeline, two anomalous scatterers were found in the asymmetric unit using ShelxC/D and then the anomalous scatterer parameters and phases were calculated and refined with SHARP to an anomalous phasing power of 0.63 and a Figure of Merit (FOM) of 0.23. Correct hand was chosen unambiguously after density modification with Solomon using a solvent content of 47.9%, improving the overall FOM to 0.65. Automatic building was done by Arp/Warp (version 7.5), and 200 residues were docked in the sequence. Final structure was manually rebuilt with Coot (58) and refined to 1.9 Å resolution with Phenix (59).
Site-directed mutagenesis and FABA
Deletions or site-directed point mutations of TtRa2LD related to DNA binding were introduced into DNA fragments corresponding to TtRa2LD through PCR using primers as listed in Table S3. Then the fragments were cloned into the modified pET21a vector containing an N-terminal His–MBP–TEV tag. DNA sequencing verified the introduction of the wanted mutations and confirmed that no undesired mutation was present. Expression and purification of the mutant TtRa2LD proteins are similar to that for the WT TtRa2LD.
The oligonucleotides used for FABA experiments (Table S5) were annealed from complementary single-stranded sequences to form the DNA duplexes. The resulting oligonucleotides were purified to remove the unwanted DNA by using a Hitrap Q column (GE Healthcare) after annealing. FABA for DNA binding was performed in reaction buffer containing 10 mm Tris-HCl, pH 7.5, 1 mm DTT, and 5% glycerol (v/v) by using an Infinite F200 instrument (TECAN). The wavelengths of fluorescence excitation and emission were 485 and 535 nm, respectively. The 6-carboxyfluorescein–labeled oligonucleotides were used for the DNA-binding assay. Twelve 2-fold dilution steps (800 to 0 nm) of the purified proteins with a final volume of 100 μl were incubated with 2.5 nm fluorescent-labeled probes in a 96-well plate at 30 °C for 10 min before measurements. Curves were fit individually using KaleidaGraph 4.03 software. The dissociation constant Kd was determined using Equation 1,
| (Eq. 1) |
where Δr is the measured anisotropy in millipolarization; Δrmax is the anisotropy at saturation, and P is the protein concentration.
Quantitation of bound Zn2+ and free thiol groups in TtRa2LD
The Zn2+ content of the TtRa2LD used for crystallization was measured by taking advantage of the light absorbance variation of 4-(2-pyridylazo)resorcinol (PAR) at 500 nm when bound to Zn2+ (60). To quantify more precisely the zinc content of the protein, all of the buffers were pre-treated with Chelex 100 resin. TtRa2LD was dialyzed against the EDTA-free Chelex-treated buffer passed over a 10-cm column of Chelex-100 and re-concentrated. To facilitate zinc release, the protein (1 nmol in a volume of 20 μl) was first denatured with Chelex-treated 7 m guanidine-HCl and then transferred to a 1-ml cuvette, and the volume was adjusted to 0.9 ml with a buffer containing 20 mm Tris-HCl, pH 8.0, and 150 mm NaCl. PAR was added into the cuvette for a final concentration of 100 μm. The absorbance at 500 nm was measured. The quantity of zinc ion was determined from a standard curve of ZnCl2 samples in a range of concentrations using the sample preparation procedure as described above but with TtRa2LD omitted.
The zinc-demetallated TtRa2LD was obtained by dialysis of purified TtRa2LD against buffer B (20 mm Tris-HCl, pH 8.0, 300 mm NaCl, 10 mm EDTA, 1.5 mm DTT, 10% glycerol (v/v)) overnight at 4 °C. The [thiolsfree] in TtRa2LD and the Zn2+-extracted TtRa2LD were determined by measuring the absorbance at 412 nm following reaction of DTNB with thiols in the protein as described in the literature (61). 10 μl of DTNB (10 mm in 0.1 m sodium phosphate, pH 8.0) was added to 5 ml of untreated or Zn2+-extracted TtRa2LD in a buffer containing 20 mm Tris-HCl, pH 8.0, 300 mm NaCl, and 10% glycerol (v/v), and diluted into 150 ml of 0.1 m sodium phosphate, pH 8.0. Samples were incubated at room temperature for 1 h, and their absorbances at 412 nm were measured. These values were converted to [thiolsfree] as described in the literature (61).
BiFC
TtRa2LD, Δ123, Δ113, TtRa2, and TtRa2 Δα5 DNA were cloned into BiFC binary vectors pSPYNE-35S and pSPYCE-35S containing the N- and C-terminal fragments of yellow fluorescence protein (62) via XbaI and XhoI as a translational fusion, yielding the pSPYNE-35S-TtRa2LD, pSPYCE-35S-TtRa2LD, pSPYNE-35S-Δ123, pSPYCE-35SΔ123, pSPYNE-35S-Δ113, pSPYCE-35SΔ113, pSPYNE-35S-TtRa2, pSPYCE-35S-TtRa2, pSPYNE-35S-TtRa2Δα5, and pSPYCE-35S-TtRa2Δα5 constructs. Oligonucleotides used for constructing BiFC vectors are listed in Table S4. All constructs were verified by sequencing and introduced into Agrobacterium strain GV3101 via electroporation method. Agrobacterium infiltration for transient expression of the constructs was accomplished in the leaves of 4–6-week-old N. benthamiana grown in soil under greenhouse conditions. The p19 protein of tomato bushy stunt virus was used to suppress gene silencing. Agrobacterium strains containing the BiFC-partner constructs and the p19 helper strain were mixed together at A600 of 0.9:0.9:0.3, incubated away from light for 2 h at room temperature, and then co-infiltrated into the tobacco leaves. Fluorescence of lower epidermal cell layers of tobacco leaves was visualized with a Zeiss LSM510 system about 2–3 days after infiltration. Argon laser (488 nm) was utilized for fluorescence excitation.
Analytical ultracentrifugation
Analytical ultracentrifugation experiment was performed using an XL-I analytical ultracentrifuge (Beckman Coulter, Fullerton, CA) equipped with a four-cell An-60 Ti rotor. The purified protein (0.4–0.05 mg/ml) in 50 mm Tris-HCl, pH 7.5, and 500 mm NaCl was centrifuged at 20 °C and 60,000 × g for 8 h, with the same buffer as control. To determine the molecular weight of the protein, the data were analyzed using the software Sedfit (63).
Overall structure analysis by CD spectra
All the CD spectra of the WT and mutant TtRa2LD proteins were collected on a MOS-450/AF-CD spectrometer (Bio-Logic, French) at room temperature. All proteins were diluted to 0.1 mg/ml in 0.2 m phosphate buffer, pH 7.5, and a quartz cell with a path length of 1 mm was used. Each spectrum was the average of three scans.
DNA segment docking
The molecular model of AH3–ER6 DNA duplex was generated using the 3D-dart web server. Modeling of the TtRa2LD-AH3–ER6 protein–DNA complex was first performed with rigid body approach using ZDOCK. 2000 models were generated and clustered using the Rosetta energy function (64). The top cluster was further used as an initial model for flexible docking using HADDOCK 2.2 web server (65). For HADDOCK calculations, active residues for TtRa2LD were chosen as the closest to the DNA according to the initial rigid body model. Passive residues were automatically defined around the active residues by HADDOCK. All of the bases of the DNA duplex sequence were considered active in the docking. The standard docking procedure was used: from the initial 1000 complex structures generated by rigid-body docking, the 200 lowest energy structures were further refined in explicit water after semi-flexible simulated annealing. A cluster analysis was performed on the finally docked structures corresponding to the 200 best solutions with lowest intermolecular energies based on a 7.5-Å r.m.s.d. cutoff criterion. The clusters were ranked according to the averaged Z-score of their top 10 structures.
The best solution was then submitted to 100 ns molecular dynamics simulation with GROMACS using AMBER99SB and AMBER94 force fields for protein and DNA, respectively. Before molecular dynamics, the system was equilibrated in an explicit dodecahedron TIP3 water box at 300 K and 1 bar. Trajectory was analyzed with the tools distributed with GROMACS.
SAXS
SAXS experiments were carried out at 20 °C with SEC–HPLC coupled to SAXS data collection at the SWING beamline (SOLEIL Synchrotron, Saint-Aubin, France). The samples of protein, DNA, and protein–DNA complexes were injected at a concentration of 10 mg/ml on BioSEC3 HPLC column (Agilent) at a flow rate of 0.2 ml/min in a buffer containing 10 mm Tris-HCl, pH 7.5, 30 mm NaCl, and 5% glycerol (v/v). Scattering data were collected on the PCCD170 detector (Aviex), and data reduction and processing of images were done with Foxtrot (66). Analysis of the HPLC–SAXS profiles was performed using Foxtrot and US–SOMO HPLC–SAXS module of Ultrascan2 software (67). Briefly, I(q,t) stack of data frames I(q) collected over time t was transformed into I(t,q) stack allowing visualization of peaks at I(t,qmin) for the lowest value of q recorded (qmin = 0.007 Å−1). I(0) and the radius of gyration (Rg) were calculated over the profile with Guinier approximation up to q limit satisfying qRg <1.3. Peaks that were not baseline-resolved were deconvoluted and modeled with Gaussian approximation tools included in US–SOMO. SAXS profiles I(t,q) for the extracted peaks were generated and then frames with constant Rg values were averaged to produce SAXS reference data I(q) for each peak. SEC–SAXS profiles with treatment of peaks are shown in Fig. S6. Pair-distance distribution function and the maximum particle dimension (Dmax) were calculated using the GNOM4 program from ATSAS 2.8 suite (Fig. S7) (68). Ab initio envelopes for isolated protein and DNA were determined using DAMMIF with experimental Rg and Dmax values as constraints. Ab initio modeling of TtRa2LD–AH3–ER6 protein–DNA complex was done with MONSA multiphase approach (69) using a combination of SAXS data of the protein, DNA, and complex and defining two phases with distinct electronic density contrasts for protein and DNA. The atomic model of the complex was adjusted by rigid body modeling with SASREF and local adjustments with MODELLER (70). Finally, the atomic model profiles were calculated and fitted to the experimental data using CRYSOL and aligned on ab initio bead models with SUPCOMB. All of the SAXS parameters are summarized in Table S2.
Author contributions
W.-F. C., L.-Y. H., and X.-B. W. purified, crystallized the proteins. X.-B. W. performed BiFC experiments. W.-F. C. performed analytical ultracentrifugation experiment. W.-F. C. collected crystal diffraction data. S. R. determined the crystal structures. S. R. and X.-G. X. performed SAXS experiments and analyzed SAXS data. W.-F. C. and X.-B. W. performed DNA binding. S. R. performed molecular dynamics simulations. W.-F. C., X.-B. W., S.-X. D., S. R., and X.-G. X. analyzed the data and wrote the paper. X.-G. X. conceived the project.
Supplementary Material
Acknowledgments
We thank the staff of BL19U1 beamline at Shanghai Synchrotron Radiation Facility for assistance with structure data collection. We thank Xiao-Xia Yu (Institute of Biophysics, Chinese Academy of Sciences) for performing analytical ultracentrifugation. We are grateful to Dr. Javier Perez and team for SAXS data collection at the SWING beamline (SOLEIL Synchrotron, France). We also acknowledge Professor Shang-Ting Zhao for much help in BiFC experiments.
This work was supported by National Natural Science Foundation of China Grants 31370798, 11304252, 11574252, and 31301632, Northwest A & F University Startup Funding Grant Z101021102, and the International Associated Laboratory (LIA, “G-quadruplex-HELI”). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S9, Tables S1–S5, and supporting Refs. 1–2.
The atomic coordinates and structure factors (code 5ly0) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- LBD
- LATERAL ORGAN BOUNDARIES DOMAIN
- LOB
- Lateral Organ Boundary
- SAXS
- small-angle X-ray scattering
- SEC
- size-exclusion chromatography
- LSLM
- leucine zipper–like motif
- PDB
- Protein Data Bank
- TEV
- tobacco etch virus
- r.m.s.d.
- root-mean-square deviation
- RE
- response element
- BiFC
- bimolecular fluorescence complementation
- FOM
- Figure of Merit
- FABA
- fluorescence anisotropy-based binding assay
- DTNB
- 5,5′-dithiobis(2-nitrobenzoic acid)
- PAR
- 4-(2-pyridylazo)resorcinol
- MBP
- maltose-binding protein.
References
- 1. Majer C., and Hochholdinger F. (2011) Defining the boundaries: structure and function of LOB domain proteins. Trends Plant Sci. 16, 47–52 10.1016/j.tplants.2010.09.009 [DOI] [PubMed] [Google Scholar]
- 2. Xu C., Luo F., and Hochholdinger F. (2016) LOB domain proteins: beyond lateral organ boundaries. Trends Plant Sci. 21, 159–167 10.1016/j.tplants.2015.10.010 [DOI] [PubMed] [Google Scholar]
- 3. Bortiri E., Chuck G., Vollbrecht E., Rocheford T., Martienssen R., and Hake S. (2006) ramosa2 encodes a LATERAL ORGAN BOUNDARY domain protein that determines the fate of stem cells in branch meristems of maize. Plant Cell 18, 574–585 10.1105/tpc.105.039032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vollbrecht E., Springer P. S., Goh L., Buckler E. S. 4th., and Martienssen R. (2005) Architecture of floral branch systems in maize and related grasses. Nature 436, 1119–1126 10.1038/nature03892 [DOI] [PubMed] [Google Scholar]
- 5. Semiarti E., Ueno Y., Tsukaya H., Iwakawa H., Machida C., and Machida Y. (2001) The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana regulates formation of a symmetric lamina, establishment of venation and repression of meristem-related homeobox genes in leaves. Development 128, 1771–1783 [DOI] [PubMed] [Google Scholar]
- 6. Jun J. H., Ha C. M., and Fletcher J. C. (2010) BLADE-ON-PETIOLE1 coordinates organ determinacy and axial polarity in arabidopsis by directly activating ASYMMETRIC LEAVES2. Plant Cell 22, 62–76 10.1105/tpc.109.070763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Okushima Y., Fukaki H., Onoda M., Theologis A., and Tasaka M. (2007) ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell 19, 118–130 10.1105/tpc.106.047761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lee H. W., Cho C., and Kim J. (2015) Lateral organ boundaries domain 16 and 18 act downstream of the AUXIN1 and LIKE-AUXIN3 auxin influx carriers to control lateral root development in Arabidopsis. Plant Physiol. 168, 1792–1806 10.1104/pp.15.00578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mangeon A., Lin W. C., and Springer P. S. (2012) Functional divergence in the Arabidopsis LOB-domain gene family. Plant Signal. Behav. 7, 1544–1547 10.4161/psb.22320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kim M. J., Kim M., Lee M. R., Park S. K., and Kim J. (2015) LATERAL ORGAN BOUNDARIES DOMAIN (LBD)10 interacts with SIDECAR POLLEN/LBD27 to control pollen development in Arabidopsis. Plant J. 81, 794–809 10.1111/tpj.12767 [DOI] [PubMed] [Google Scholar]
- 11. Soyano T., Thitamadee S., Machida Y., and Chua N. H. (2008) ASYMMETRIC LEAVES2-LIKE19/LATERAL ORGAN BOUNDARIES DOMAIN30 and ASL20/LBD18 regulate tracheary element differentiation in Arabidopsis. Plant Cell 20, 3359–3373 10.1105/tpc.108.061796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rubin G., Tohge T., Matsuda F., Saito K., and Scheible W. R. (2009) Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in Arabidopsis. Plant Cell 21, 3567–3584 10.1105/tpc.109.067041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sugimoto K., Jiao Y., and Meyerowitz E. M. (2010) Arabidopsis regeneration from multiple tissues occurs via a root development pathway. Dev. Cell 18, 463–471 10.1016/j.devcel.2010.02.004 [DOI] [PubMed] [Google Scholar]
- 14. Liu J., Sheng L., Xu Y., Li J., Yang Z., Huang H., and Xu L. (2014) WOX11 and 12 are involved in the first-step cell fate transition during de novo root organogenesis in Arabidopsis. Plant Cell 26, 1081–1093 10.1105/tpc.114.122887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mangeon A., Bell E. M., Lin W. C., Jablonska B., and Springer P. S. (2011) Misregulation of the LOB domain gene DDA1 suggests possible functions in auxin signalling and photomorphogenesis. J. Exp. Bot. 62, 221–233 10.1093/jxb/erq259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Thatcher L. F., Powell J. J., Aitken E. A., Kazan K., and Manners J. M. (2012) The lateral organ boundaries domain transcription factor LBD20 functions in Fusarium wilt susceptibility and jasmonate signaling in Arabidopsis. Plant Physiol. 160, 407–418 10.1104/pp.112.199067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hu Y., Zhang J., Jia H., Sosso D., Li T., Frommer W. B., Yang B., White F. F., Wang N., and Jones J. B. (2014) Lateral organ boundaries 1 is a disease susceptibility gene for citrus bacterial canker disease. Proc. Natl. Acad. Sci. U.S.A. 111, E521–E529 10.1073/pnas.1313271111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yordanov Y. S., Regan S., and Busov V. (2010) Members of the LATERAL ORGAN BOUNDARIES DOMAIN transcription factor family are involved in the regulation of secondary growth in Populus. Plant Cell 22, 3662–3677 10.1105/tpc.110.078634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ge L., Peng J., Berbel A., Madueño F., and Chen R. (2014) Regulation of compound leaf development by PHANTASTICA in Medicago truncatula. Plant Physiol. 164, 216–228 10.1104/pp.113.229914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shuai B., Reynaga-Peña C. G., and Springer P. S. (2002) The lateral organ boundaries gene defines a novel, plant-specific gene family. Plant Physiol. 129, 747–761 10.1104/pp.010926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang Y. M., Zhang S. Z., and Zheng C. C. (2014) Genomewide analysis of LATERAL ORGAN BOUNDARIES Domain gene family in Zea mays. J. Genet. 93, 79–91 10.1007/s12041-014-0342-7 [DOI] [PubMed] [Google Scholar]
- 22. Yang Y., Yu X., and Wu P. (2006) Comparison and evolution analysis of two rice subspecies LATERAL ORGAN BOUNDARIES domain gene family and their evolutionary characterization from Arabidopsis. Mol. Phylogenet. Evol. 39, 248–262 10.1016/j.ympev.2005.09.016 [DOI] [PubMed] [Google Scholar]
- 23. Iwakawa H., Ueno Y., Semiarti E., Onouchi H., Kojima S., Tsukaya H., Hasebe M., Soma T., Ikezaki M., Machida C., and Machida Y. (2002) The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana, required for formation of a symmetric flat leaf lamina, encodes a member of a novel family of proteins characterized by cysteine repeats and a leucine zipper. Plant Cell Physiol. 43, 467–478 10.1093/pcp/pcf077 [DOI] [PubMed] [Google Scholar]
- 24. Husbands A., Bell E. M., Shuai B., Smith H. M., and Springer P. S. (2007) LATERAL ORGAN BOUNDARIES defines a new family of DNA-binding transcription factors and can interact with specific bHLH proteins. Nucleic Acids Res. 35, 6663–6671 10.1093/nar/gkm775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Berckmans B., Vassileva V., Schmid S. P., Maes S., Parizot B., Naramoto S., Magyar Z., Alvim Kamei C. L., Koncz C., Bögre L., Persiau G., De Jaeger G., Friml J., Simon R., Beeckman T., and De Veylder L. (2011) Auxin-dependent cell cycle reactivation through transcriptional regulation of Arabidopsis E2Fa by lateral organ boundary proteins. Plant Cell 23, 3671–3683 10.1105/tpc.111.088377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bell E. M., Lin W. C., Husbands A. Y., Yu L., Jaganatha V., Jablonska B., Mangeon A., Neff M. M., Girke T., and Springer P. S. (2012) Arabidopsis lateral organ boundaries negatively regulates brassinosteroid accumulation to limit growth in organ boundaries. Proc. Natl. Acad. Sci. U.S.A. 109, 21146–21151 10.1073/pnas.1210789109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Majer C., Xu C., Berendzen K. W., and Hochholdinger F. (2012) Molecular interactions of ROOTLESS CONCERNING CROWN AND SEMINAL ROOTS, a LOB domain protein regulating shoot-borne root initiation in maize (Zea mays L.). Philos. Trans. R. Soc. Lond. B Biol. Sci. 367, 1542–1551 10.1098/rstb.2011.0238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Muthreich N., Majer C., Beatty M., Paschold A., Schützenmeister A., Fu Y., Malik W. A., Schnable P. S., Piepho H. P., Sakai H., and Hochholdinger F. (2013) Comparative transcriptome profiling of maize coleoptilar nodes during shoot-borne root initiation. Plant Physiol. 163, 419–430 10.1104/pp.113.221481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Xu C., Tai H., Saleem M., Ludwig Y., Majer C., Berendzen K. W., Nagel K. A., Wojciechowski T., Meeley R. B., Taramino G., and Hochholdinger F. (2015) Cooperative action of the paralogous maize lateral organ boundaries (LOB) domain proteins RTCS and RTCL in shoot-borne root formation. New Phytol. 207, 1123–1133 10.1111/nph.13420 [DOI] [PubMed] [Google Scholar]
- 30. Koppolu R., Anwar N., Sakuma S., Tagiri A., Lundqvist U., Pourkheirandish M., Rutten T., Seiler C., Himmelbach A., Ariyadasa R., Youssef H. M., Stein N., Sreenivasulu N., Komatsuda T., and Schnurbusch T. (2013) Six-rowed spike4 (Vrs4) controls spikelet determinacy and row-type in barley. Proc. Natl. Acad. Sci. U.S.A. 110, 13198–13203 10.1073/pnas.1221950110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jin J., Zhang H., Kong L., Gao G., and Luo J. (2014) PlantTFDB 3.0: a portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res. 42, D1182–D1187 10.1093/nar/gkt1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li S., Zhao Y., Zhao Z., Wu X., Sun L., Liu Q., and Wu Y. (2016) Crystal structure of the GRAS domain of SCARECROW-LIKE 7 in Oryza sativa. Plant Cell 28, 1025–1034 10.1105/tpc.16.00018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Boer D. R., Freire-Rios A., van den Berg W. A., Saaki T., Manfield I. W., Kepinski S., López-Vidrieo I., Franco-Zorrilla J. M., de Vries S. C., Solano R., Weijers D., and Coll M. (2014) Structural basis for DNA binding specificity by the auxin-dependent ARF transcription factors. Cell 156, 577–589 10.1016/j.cell.2013.12.027 [DOI] [PubMed] [Google Scholar]
- 34. Allen M. D., Yamasaki K., Ohme-Takagi M., Tateno M., and Suzuki M. (1998) A novel mode of DNA recognition by a β-sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA. EMBO J. 17, 5484–5496 10.1093/emboj/17.18.5484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yamasaki K., Kigawa T., Inoue M., Tateno M., Yamasaki T., Yabuki T., Aoki M., Seki E., Matsuda T., Nunokawa E., Ishizuka Y., Terada T., Shirouzu M., Osanai T., Tanaka A., et al. (2004) A novel zinc-binding motif revealed by solution structures of DNA-binding domains of Arabidopsis SBP-family transcription factors. J. Mol. Biol. 337, 49–63 10.1016/j.jmb.2004.01.015 [DOI] [PubMed] [Google Scholar]
- 36. Yamasaki K., Kigawa T., Inoue M., Tateno M., Yamasaki T., Yabuki T., Aoki M., Seki E., Matsuda T., Tomo Y., Hayami N., Terada T., Shirouzu M., Osanai T., Tanaka A., et al. (2004) Solution structure of the B3 DNA-binding domain of the Arabidopsis cold-responsive transcription factor RAV1. Plant Cell 16, 3448–3459 10.1105/tpc.104.026112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yamasaki K., Kigawa T., Inoue M., Tateno M., Yamasaki T., Yabuki T., Aoki M., Seki E., Matsuda T., Tomo Y., Hayami N., Terada T., Shirouzu M., Tanaka A., Seki M., Shinozaki K., and Yokoyama S. (2005) Solution structure of an Arabidopsis WRKY DNA-binding domain. Plant Cell 17, 944–956 10.1105/tpc.104.026435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ernst H. A., Olsen A. N., Larsen S., and Lo Leggio L. (2004) Structure of the conserved domain of ANAC, a member of the NAC family of transcription factors. EMBO Rep. 5, 297–303 10.1038/sj.embor.7400093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Holm L., and Rosenström P. (2010) Dali server: conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549 10.1093/nar/gkq366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Krissinel E., and Henrick K. (2004) Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 60, 2256–2268 10.1107/S0907444904026460 [DOI] [PubMed] [Google Scholar]
- 41. Krishna S. S., Majumdar I., and Grishin N. V. (2003) Structural classification of zinc fingers: survey and summary. Nucleic Acids Res. 31, 532–550 10.1093/nar/gkg161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Omichinski J. G., Clore G. M., Schaad O., Felsenfeld G., Trainor C., Appella E., Stahl S. J., and Gronenborn A. M. (1993) NMR structure of a specific DNA complex of Zn-containing DNA-binding domain of GATA-1. Science 261, 438–446 10.1126/science.8332909 [DOI] [PubMed] [Google Scholar]
- 43. Bi X., Corpina R. A., and Goldberg J. (2002) Structure of the Sec23/24-Sar1 pre-budding complex of the COPII vesicle coat. Nature 419, 271–277 10.1038/nature01040 [DOI] [PubMed] [Google Scholar]
- 44. Bernstein D. A., Zittel M. C., and Keck J. L. (2003) High-resolution structure of the E. coli RecQ helicase catalytic core. EMBO J. 22, 4910–4921 10.1093/emboj/cdg500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lee B. I., Kim K. H., Park S. J., Eom S. H., Song H. K., and Suh S. W. (2004) Ring-shaped architecture of RecR: implications for its role in homologous recombinational DNA repair. EMBO J. 23, 2029–2038 10.1038/sj.emboj.7600222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Leiros I., Timmins J., Hall D. R., and McSweeney S. (2005) Crystal structure and DNA-binding analysis of RecO from Deinococcus radiodurans. EMBO J. 24, 906–918 10.1038/sj.emboj.7600582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Marmorstein R., Carey M., Ptashne M., and Harrison S. C. (1992) DNA recognition by GAL4: structure of a protein–DNA complex. Nature 356, 408–414 10.1038/356408a0 [DOI] [PubMed] [Google Scholar]
- 48. Ren H., Dou S. X., Zhang X. D., Wang P. Y., Kanagaraj R., Liu J. L., Janscak P., Hu J. S., and Xi X. G. (2008) The zinc-binding motif of human RECQ5β suppresses the intrinsic strand-annealing activity of its DExH helicase domain and is essential for the helicase activity of the enzyme. Biochem. J. 412, 425–433 10.1042/BJ20071150 [DOI] [PubMed] [Google Scholar]
- 49. Lakshmi B., Sinduja C., Archunan G., and Srinivasan N. (2014) Ramachandran analysis of conserved glycyl residues in homologous proteins of known structure. Protein Sci. 23, 843–850 10.1002/pro.2468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lee H. W., Kim M. J., Park M. Y., Han K. H., and Kim J. (2013) The conserved proline residue in the LOB domain of LBD18 is critical for DNA-binding and biological function. Mol. Plant 6, 1722–1725 10.1093/mp/sst037 [DOI] [PubMed] [Google Scholar]
- 51. O'Shea E. K., Klemm J. D., Kim P. S., and Alber T. (1991) X-ray structure of the GCN4 leucine zipper, a two-stranded, parallel coiled coil. Science 254, 539–544 10.1126/science.1948029 [DOI] [PubMed] [Google Scholar]
- 52. Isernia C., Bucci E., Leone M., Zaccaro L., Di Lello P., Digilio G., Esposito S., Saviano M., Di Blasio B., Pedone C., Pedone P. V., and Fattorusso R. (2003) NMR structure of the single QALGGH zinc finger domain from the Arabidopsis thaliana SUPERMAN protein. Chembiochem 4, 171–180 10.1002/cbic.200390028 [DOI] [PubMed] [Google Scholar]
- 53. Takatsuji H. (1998) zinc-finger transcription factors in plants. Cell. Mol. Life Sci. 54, 582–596 10.1007/s000180050186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Fukushima S., Yoshida M., and Takatsuji H. (2012) Role of linkers between zinc fingers in spacing recognition by plant TFIIIA-type zinc-finger proteins. J. Amino Acids 2012, 848037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kabsch W. (2010). XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 10.1107/S0907444909047337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Vonrhein C., Blanc E., Roversi P., and Bricogne G. (2007) Automated structure solution with autoSHARP. Methods Mol. Biol. 364, 215–230 [DOI] [PubMed] [Google Scholar]
- 57. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., et al. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 10.1107/S0907444910045749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hunt I. F., Murphy N. J., Cleaver A. E., Faraji B., Swendseid M. E., Coulson A. H., Clark V. A., Browdy B. L., Cabalum T., and Smith J. C. Jr. (1984) zinc supplementation during pregnancy: effects on selected blood constituents and on progress and outcome of pregnancy in low-income women of Mexican descent. Am. J. Clin. Nutr. 40, 508–521 10.1093/ajcn/40.3.508 [DOI] [PubMed] [Google Scholar]
- 61. Riddles P. W., Blakeley R. L., and Zerner B. (1983) Reassessment of Ellman's reagent. Methods Enzymol. 91, 49–60 10.1016/S0076-6879(83)91010-8 [DOI] [PubMed] [Google Scholar]
- 62. Walter M., Chaban C., Schütze K., Batistic O., Weckermann K., Näke C., Blazevic D., Grefen C., Schumacher K., Oecking C., Harter K., and Kudla J. (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40, 428–438 10.1111/j.1365-313X.2004.02219.x [DOI] [PubMed] [Google Scholar]
- 63. Schuck P. (2000) Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 78, 1606–1619 10.1016/S0006-3495(00)76713-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ashworth J., and Baker D. (2009) Assessment of the optimization of affinity and specificity at protein–DNA interfaces. Nucleic Acids Res. 37, e73 10.1093/nar/gkp242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. van Zundert G. C. P., Rodrigues J. P. G. L. M., Trellet M., Schmitz C., Kastritis P. L., Karaca E., Melquiond A. S. J., van Dijk M., de Vries S. J., and Bonvin A. M. J. J. (2016) The HADDOCK2.2 Web Server: user-friendly integrative modeling of biomolecular complexes. J. Mol. Biol. 428, 720–725 10.1016/j.jmb.2015.09.014 [DOI] [PubMed] [Google Scholar]
- 66. David G., and Pérez J. (2009) Combined sampler robot and high-performance liquid chromatography: a fully automated system for biological small-angle X-ray scattering experiments at the Synchrotron SOLEIL SWING beamline. J. Appl. Crystallogr. 42, 892–900 10.1107/S0021889809029288 [DOI] [Google Scholar]
- 67. Brookes E., Vachette P., Rocco M., and Pérez J. (2016) US-SOMO HPLC-SAXS module: dealing with capillary fouling and extraction of pure component patterns from poorly resolved SEC–SAXS data. J. Appl. Crystallogr. 49, 1827–1841 10.1107/S1600576716011201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Petoukhov M. V., Franke D., Shkumatov A. V., Tria G., Kikhney A. G., Gajda M., Gorba C., Mertens H. D., Konarev P. V., and Svergun D. I. (2012) New developments in the ATSAS program package for small-angle scattering data analysis. J. Appl. Crystallogr. 45, 342–350 10.1107/S0021889812007662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Svergun D. I. (1999) Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys. J. 76, 2879–2886 10.1016/S0006-3495(99)77443-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Sali A., and Blundell T. L. (1993) Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779–815 10.1006/jmbi.1993.1626 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.