Abstract
Viral infections still threaten human health all over the world, and many people die from viral diseases every year. However, there are no effective vaccines or drugs for preventing or managing most viral diseases. Thus, the discovery and development of broad-spectrum antiviral agents remain urgent. Here, we expressed and purified a venom peptide, Ev37, from the scorpion Euscorpiops validus in a prokaryotic system. We found that rEv37 can inhibit dengue virus type 2 (DENV-2), hepatitis C virus (HCV), Zika virus (ZIKV), and herpes simplex virus type 1 (HSV-1) infections in a dose-dependent manner at noncytotoxic concentrations, but that it has no effect on Sendai virus (SeV) and adenovirus (AdV) infections in vitro. Furthermore, rEv37 alkalized acidic organelles to prevent low pH–dependent fusion of the viral membrane–endosomal membrane, which mainly blocks the release of the viral genome from the endosome to the cytoplasm and then restricts viral late entry. Taken together, our results indicate that the scorpion venom peptide Ev37 is a broad-spectrum antiviral agent with a specific molecular mechanism against viruses undergoing low pH–dependent fusion activation during entry into host cells. We conclude that Ev37 is a potential candidate for development as an antiviral drug.
Keywords: peptides, infection, dengue virus (DENV), hepatitis C virus (HCV), virus entry, antiviral agent, antiviral mechanism, antiviral peptide, dengue virus, scorpion venom peptide, Zika virus, herpes simplex virus, membrane fusion, endosome
Introduction
There are many viruses threatening human health throughout the world. They include Flaviviridae (dengue virus, HCV,3 Zika virus, West Nile virus, and Japanese encephalitis virus), Coronaviridae (severe acute respiratory syndrome), Retroviridae (HIV-1), and other viruses (1). Many people die every year due to viral infection. Dengue virus, including four serotypes (DENV 1–4), is an emerging threat to billions of people worldwide (2). Some disappointing therapeutic trials in dengue infection investigating both antivirals and adjunctive therapies have been carried out in recent years, such as balapiravir in Vietnam, and dengvaxia, the first clinically approved dengue vaccine, recently was recommended for use in areas where the disease is highly prevalent because of its side effects (3, 4). However, there are no effective vaccines or drugs for dengue virus. HCV accounts for 170 million chronic infections worldwide and is still a major health threat to people. Although there are some highly potent direct-acting antiviral drugs that have been approved, their high costs and limited effectiveness drive us to find more effective drugs (5). Zika virus (ZIKV) is also a member of the Flaviviridae family that is a large burden to people and can cause fetal and newborn microcephaly (6). There are no effective drugs and vaccines for ZIKV to date. Therefore, the discovery and development of broad-spectrum antiviral agents are still urgent.
Scorpion venom contains many kinds of peptides that have a wide range of biological properties. Some venom peptides that are classified as anti-microbial peptides exhibit anti-microbial activities. For example, Hp1404 can inhibit Staphylococcus aureus by penetrating the membrane (7), and BmKbpp can suppress Gram-negative bacteria (8). Some scorpion venom peptides also showed antiviral activities. For instance, Ctry2459 and its derived peptides can inhibit HCV infection by directly inactivating viral particles (9). Eval418 and its derived peptides can suppress HSV-1 infection by blocking the initial step (10). Mucroporin-M1 can inhibit HBV replication by activating the MAPK pathway and down-regulating HNF4α in vitro and in vivo (11). A recombinant peptide scorpine inhibited DENV-2 (NGC strain) replication in C6/36 cells (12). Scorpio maurus palmatus venom can inhibit HCV infection through virucidal effects (13). However, the molecular mechanisms of scorpion venom peptides against DENV-2 remain poorly understood.
Here, we found that a scorpion venom peptide, Ev37, derived from Euscorpiops validus can inhibit DENV-2 (TSV01 strain) infection in a dose-dependent manner at noncytotoxic concentrations. Previously, the cDNA sequence of the Ev37 mature peptide was inserted into the expression vector pGEX-6p-1 and expressed with a GST tag as a fusion protein by our group (14). The recombinant peptide Ev37 (rEv37) was found to exhibit no antibacterial activity against S. aureus and Escherichia coli (14). To acquire high production of rEv37, we optionally selected pET-32a as the expression vector and purified rEv37 by nickel column affinity chromatography. rEv37 was found to inhibit DENV-2, HCV, ZIKV, and HSV-1 in a concentration-dependent manner but had no effect on SeV and AdV. Moreover, rEv37 was shown to mainly inhibit the viral post-entry stage of viral membrane–endosomal membrane fusion with low pH dependence but had no effect on viral attachment and replication. Furthermore, its antiviral mechanism was revealed to be related to the alkalization of acidic organelles.
Results
Expression, purification, and characterization of the recombinant scorpion venom peptide Ev37
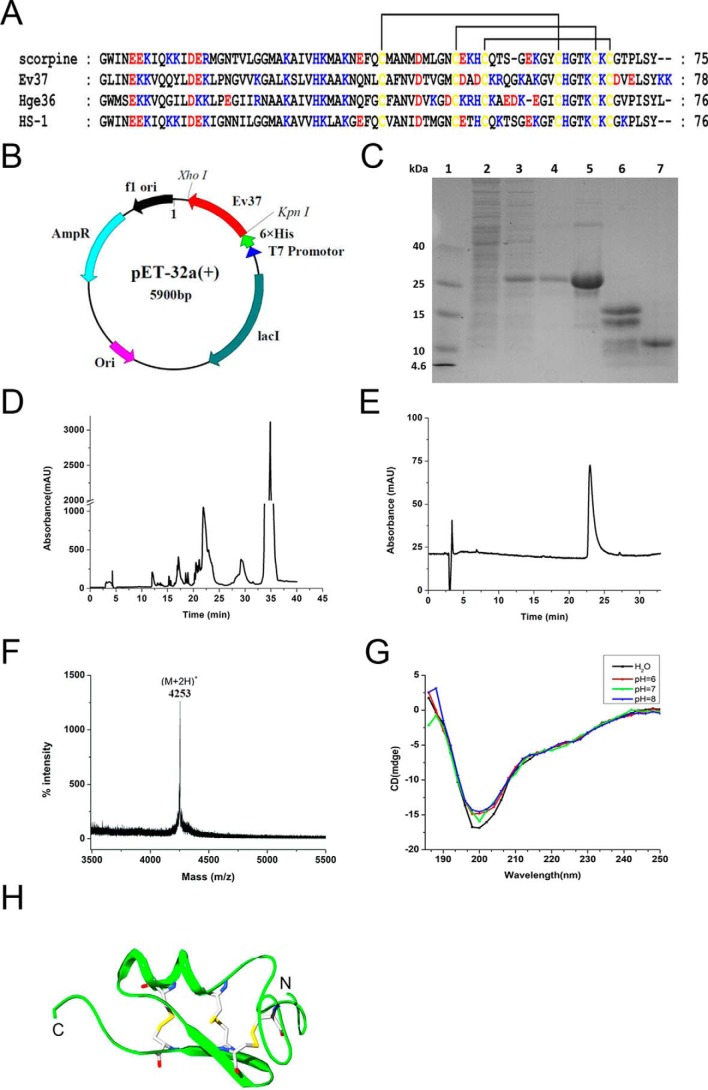
The amino acid sequence of the mature peptide Ev37 was obtained as described previously and then blasted in NCBI (http://blast.ncbi.nlm.nih.gov) to obtain three highly similar peptides, scorpine, Hge36, and HS-1, derived from the scorpion venoms of Pandinus imperator, Hadrurus gertschi, and Heterometrus laoticus, respectively. The amino acid sequence alignment indicates that Ev37 is a scorpine-like peptide, whose N terminus is likely an α-helix, and C terminus is a conserved CSαβ scaffold. Considering that the 3D structure of the peptide Hge36 with three pairs of disulfide bonds between six cysteines (C1–C4, C2–C5, and C3–C6) was determined, we established the structural model of the peptide Ev37 using Hge36 as a homology model (Protein Data Bank code 5IPO) with SWISS-MODEL (https://swissmodel.expasy.org/) (Fig. 1, A and H).
Figure 1.
Expression, purification, and identification of rEv37. A, amino acid sequence alignment of four scorpine-like peptides from scorpion venoms. Scorpine, Ev37, Hge36, and HS-1 were characterized from P. imperator, E. validus, H. gertschi, and H. laoticus, respectively. Highly conserved cysteine residues are highlighted in yellow, and they formed three pairs of disulfide bonds. Blue, basic residues; red, acidic residues; and yellow, cysteine residues. B, recombinant plasmid of Ev37. The cDNA coding sequence of the Ev37 mature peptide was cloned into the expression vector pET-32a with the restriction enzyme sites of KpnI–XhoI to express rEv37 in E. coli BL21(DE3). C, SDS-PAGE analysis of rEv37 expressed and purified in E. coli BL21(DE3). Lane 1, molecular mass markers; lane 2, noninduced cell-free extract of E. coli containing pET-32a-rEv37; lane 3, total cell-free extract induced with IPTG; lane 4, purified His fusion protein after affinity chromatography; lane 5, fusion protein after desalting by dialysis; lane 6, fusion protein after cleavage by enterokinase; and lane 7, rEv37 after HPLC purification. D, HPLC profile of rEv37 products after cleavage by enterokinase. E, purity analysis of rEv37 with the analytical column. F, mass spectra for rEv37 as measured by MALDI-TOF MS. G, CD spectrum analysis of rEv37. The CD spectrum was recorded in different pH solutions. H, 3D structure model of Ev37. The 3D structure of the peptide Ev37 was established using Hge36 as a homology model.
To acquire rEv37, the cDNA sequence of its mature peptide was inserted into the expression vector pET-32a with restriction enzyme sites KpnI and XhoI (Fig. 1B). The peptide rEv37 was expressed and purified as described under “Materials and methods.” The rEv37 was eluted at ∼22 min and was collected (Fig. 1D). The products in each process were detected by SDS-polyacrylamide gel (Fig. 1C). The results showed that there was an obvious band nearly at 25 kDa after being induced, which was a fusion protein, including Ev37 and the His tag portion of the pET-32a vector (Fig. 1C, lane 5). The purity of the peptide rEv37 was analyzed using an analytical XDB-C18 reversed-phase HPLC (RP-HPLC) column (Welch Materials, 5 μm, 4.6 × 150 mm) (Fig. 1E). The collected peptide in Fig. 1D was determined by MALDI-TOF–MS to measure its molecular mass. The experimental bivalent peak (M + 2H+) was 4253 (m/z), which was closely equal to its theoretical molecular mass of 8503.96 Da for the peptide containing three pairs of disulfide bonds (Fig. 1F). To further detect the secondary structure of rEv37, the peptide was determined by circular dichroism (CD) spectroscopy in different pH solutions. As expected, rEv37 not only appeared as an α-helix and β-sheet in different pH solutions but also had no conformational change of secondary structure in different pH solutions (Fig. 1G).
rEv37 inhibits DENV-2, HCV, and ZIKV in vitro at noncytotoxic concentrations
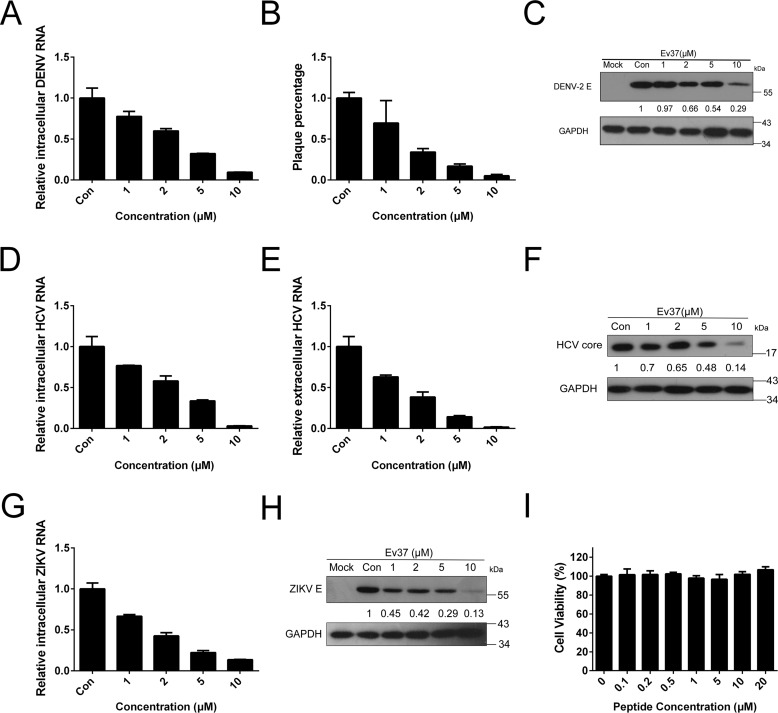
With the rapid increase in the number of characterized scorpion venom compounds, many new drug candidates have been identified as potential medicines to inhibit viral infection (15). Scorpine-like peptides, such as scorpine (16), can inhibit DENV-2 infection as demonstrated earlier. Here, we studied whether the recombinant scorpion venom peptide Ev37 can inhibit DENV-2 (TSV01 strain) in a concentration-dependent manner in the hepatoma cell line Huh7. The inhibitory ability of the peptide rEv37 against DENV-2 infection was determined via real-time fluorescent quantitative PCR (qRT-PCR) for mRNA in the infected cells and was then found to reduce 23, 41, 68, and 91% of DENV-2 at the concentrations of 1, 2, 5, and 10 μm, respectively (Fig. 2A). A plaque-forming assay was also used to determine virus particle numbers in the supernatant, and the inhibition rates were 31, 66, 84, and 95% at concentrations of 1, 2, 5, and 10 μm, respectively (Fig. 2B). rEv37 could also inhibit DENV-2 at the protein level in a concentration-dependent manner as shown by Western blotting using anti-E protein antibody (1:1000) (Fig. 2C). The cytotoxic effect of rEv37 was measured in Huh7 cells using the MTT cytotoxicity assay as described previously (17). rEv37 had no cytotoxicity on Huh7 cells at 20 μm (CC50 = 116.3 μm). So, rEv37 can inhibit DENV-2 infection under noncytotoxic concentrations (Fig. 2I).
Figure 2.
Antiviral activities of rEv37 against DENV, HCV, and ZIKV in a dose-dependent manner at noncytotoxic concentrations. A–C, concentration-dependent inhibitory activity of rEv37 against DENV-2 (TSV01) infection in vitro. Different concentrations of the peptide rEv37 reduced DENV-2 (TSV01) infection. Intracellular DENV-2 RNA (A), extracellular DENV-2 particles (B), and intracellular DENV-2 protein levels (C) were analyzed by qRT-PCR, plaque formation, and Western blotting, respectively. D–F, concentration-dependent inhibitory activity of rEv37 against HCV (JFH-1) infection in vitro. Increasing concentrations of the peptide rEv37 reduced intracellular HCV RNA (D), extracellular HCV RNA (E), and intracellular HCV core protein (F) as shown by qRT-PCR and Western blotting, respectively. G and H, concentration-dependent antiviral activity of rEv37 against ZIKV in vitro. The peptide rEv37 inhibited intracellular ZIKV RNA (G) and intracellular ZIKV E protein (H) as shown by qRT-PCR and Western blotting, respectively. I, cytotoxicity of rEv37 on Huh7 cells. The cytotoxic activity of rEv37 on Huh7 cells was measured by an MTT cytotoxicity assay. rEv37 inhibited viral infection under noncytotoxic concentrations.
To detect the activity of rEv37 against other viruses, we first examined the effect of rEv37 on HCV (JFH-1 strain) and ZIKV (PRVABC59 strain), which enter the host cells by a similar process as DENV-2. The viral entry process includes attachment, binding, endocytosis, fusion, and uncoating (18). The intracellular RNA and protein levels were measured by qRT-PCR and Western blotting, respectively (Fig. 2, D–H). Similar to DENV-2, rEv37 could reduce HCV (97%) and ZIKV (87%) infection at the mRNA level at a concentration of 10 μm.
rEv37 affects the early stages of the DENV-2 and HCV life cycles
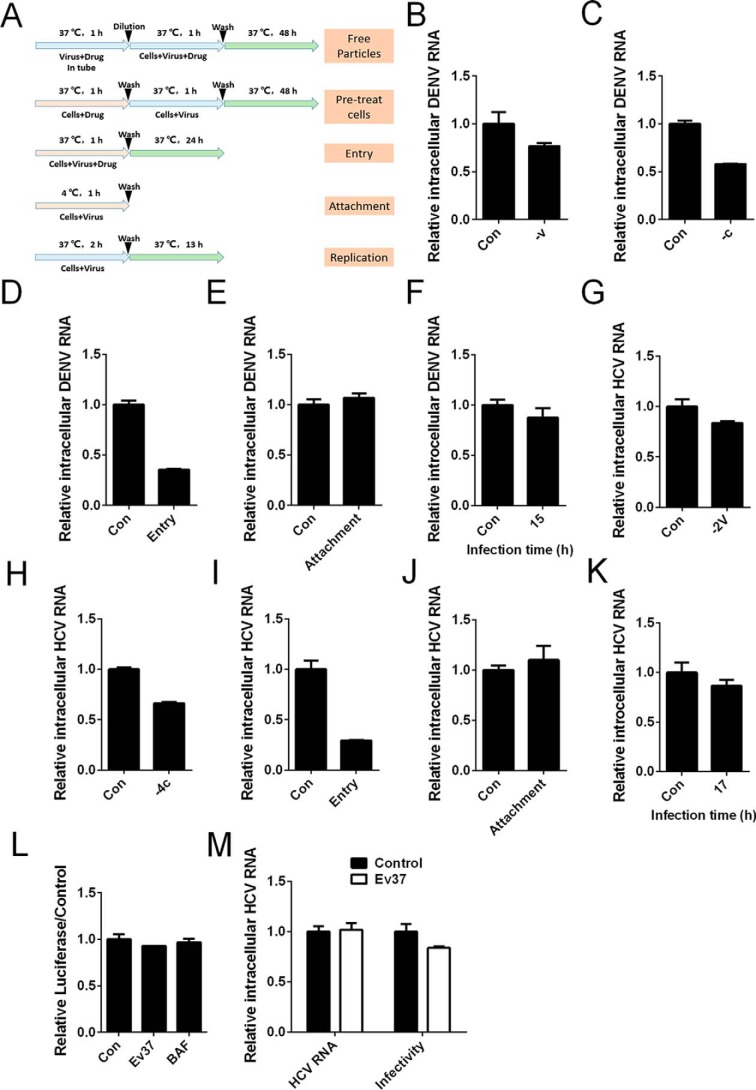
DENV-2 enters the host cells through multiple steps, as described above. To determine which step in the viral life cycle rEv37 utilizes to inhibit DENV-2, we performed some experiments following treatment (Fig. 3A). To detect whether rEv37 was directly virucidal for DENV-2 particles, virus was incubated with rEv37 for 1 h at 37 °C before adding to cells. As the results showed, rEv37 was slightly virucidal for DENV-2 particles (23%) (Fig. 3B) and had a similar effect on HCV particles (16%) (Fig. 3G). Meanwhile, rEv37 reduced DENV-2 infection (42%) and HCV infection (34%) when cells were pretreated with rEv37 for 1 h (DENV-2) or 4 h (HCV) (Fig. 3, C and H). These results suggest that rEv37 had an antiviral effect by acting on the host cells, but the mechanism was unknown.
Figure 3.
Restriction of rEv37 in the early stages of the DENV-2 and HCV life cycles. A, treatment schematic diagram of rEv37 to cells and/or viruses as described under “Materials and methods.” B and G, measurement of virion infectivity after pretreatment with rEv37. rEv37 was incubated with viral particles for the indicated time at 37 °C, and then the mixture was diluted and the cells infected for 1 h. C and H, measurements of the effect on cells pretreated with rEv37. D and I, effect of rEv37 on viral entry. Cells, virus, and rEv37 were incubated at 37 °C for 1 h. Then, we washed the unabsorbed virus, followed by culturing for 24 h. E and J, effect of rEv37 on viral attachment. Cells, virus particles, and 10 μm rEv37 were cultured at 4 °C for 1 h, and then, the unabsorbed virus was removed by washing three times with PBS. F and K, effect of rEv37 on viral replication. DENV-2 or HCV was incubated with cells for 2 or 4 h, and then rEv37 was added to the cells for 13 h after washing the unabsorbed virus with PBS. L, effect of rEv37 on the replication stage of HCV using the replicon. M, effect of rEv37 on HCV infectivity and the integrity of the HCV genome. HCV with or without rEv37 was incubated for 1 h at 37 °C, and then, the HCV genome was directly extracted using an extraction kit. HCV infectivity was analyzed as in B.
Next, we measured the effect of rEv37 at the stages of viral life cycle, including attachment, internalization (or fusion), and replication. The results showed that rEv37 mainly suppressed the entry process of DENV-2 (65%) (Fig. 3D) and HCV (71%) (Fig. 3I). To further investigate whether rEv37 had an effect on the attachment step of the DENV-2, DENV-2 or HCV particles were incubated with 10 μm rEv37 and Huh7 cells at 4 °C for 1 or 4 h, followed by washing the mixture and measuring by qRT-PCR. The results indicated that rEv37 had a slight effect on the attachment of DENV-2 (6%) (Fig. 3E) and HCV (10%) (Fig. 3J). To test the effect of rEv37 on DENV-2 and the HCV replication process, we conducted experiments as described under “Materials and methods.” The total intracellular RNA was extracted using TRIzol and measured by qRT-PCR, and rEv37 had little effect on the viral replication of DENV-2 and HCV (Fig. 3, F and K). Furthermore, to study replication only, we used the HCV replicon systems via in vitro transcription. First, we in vitro-transcribed the linearized template using a commercial T7 RNA polymerase kit according to the manufacturer's instructions. The purified RNA was transfected into cells by electrotransfection. The transfected cells were divided into three groups and were incubated with or without rEv37 and bafilomycin A1 (BAF) for 24 h to analyze the luciferase, which reflected replication. The results showed that Ev37 and BAF had no effect on the replication (Fig. 3L). As described above, rEv37 was slightly virucidal for HCV particles (Fig. 3G). To verify whether rEv37 could damage the HCV genome, HCV particles were incubated with 10 μm rEv37 for 1 h at 37 °C, and we extracted the RNA of HCV using an extraction kit. The RNA was analyzed via qRT-PCR, and the results showed that rEv37 was slightly virucidal for HCV particles but not for the HCV genome (Fig. 3M). Therefore, rEv37 mainly inhibit viral infection via affecting the internalization (fusion) stage of the DENV-2 and HCV life cycles.
rEv37 restricts the release of the DENV-2 genome into the cytoplasm from endosomes
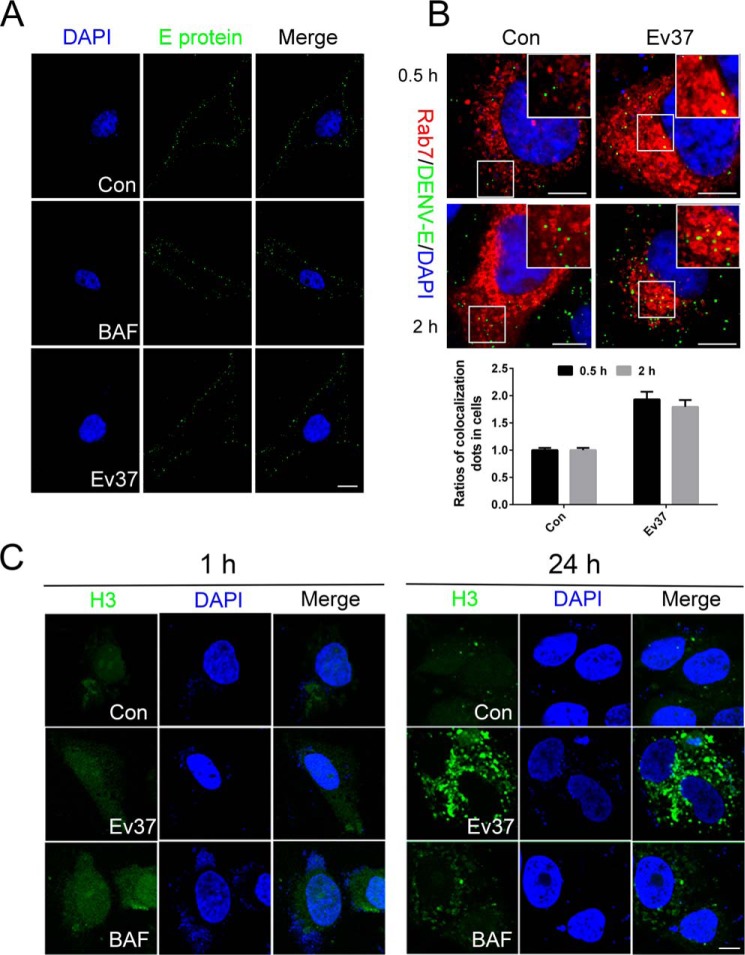
As we showed above, rEv37 vastly reduced DENV-2 infection during the post-entry process. To further demonstrate that rEv37 inhibited DENV-2 infection during the post-entry process and not during attachment, more experiments were carried out. First, we determined that rEv37 had few effects on DENV-2 attachment by using immunofluorescence (Fig. 4A). DENV-2 can attach to the cell surface of Huh7 cells but cannot enter at 4 °C. DENV-2 particles (m.o.i. = 20) were incubated with Huh7 cells with rEv37 at 4 °C for 4 h for attachment. We washed the unbound DENV-2 particles with cold PBS, treated the confocal dishes as described under “Materials and methods,” and detected cells using confocal microscopy.
Figure 4.
Restriction of rEv37 on DENV-2 release from the endosome into the cytoplasm. A, effect of rEv37 on attachment was detected by confocal microscopy. Cells were pretreated with 10 μm rEv37 or 100 nm BAF for 24 h and then incubated with DENV-2 at 4 °C for 4 h for viral attachment. B, colocalization of DENV-2 particles and the late endosome. The pt-Dimer-Rab7 plasmid was transfected into Huh7 cells for 24 h, followed by treatment with 10 μm rEv37 for 24 h. The cells were transferred to 37 °C for 0.5 or 2 h after incubation with DENV-2 at 4 °C for 2 h. DENV-2 E protein/Rab7 colocalization was quantified and described as a percentage of total examined DENV-2 particles. Red, late endosomes; green, DENV-2 particles; and blue, cell nucleus. C, influence of rEv37 on H3 distribution. FITC-H3 cells were treated with 10 μm rEv37 or 100 nm BAF for 1 and 24 h, stained with DAPI, and observed under a confocal microscopy. H3 is a derived peptide of Ctry2459 characterized from C/ tryznai, and it enters the cell by low pH-dependent endocytosis. Scale bar, 10 μm.
DENV-2 enters the cells by clathrin-mediated endocytosis. After internalization, the particles are delivered to Rab5-positive early endosomes and mature into Rab7-positive late endosomes, where membrane fusion in a low pH–dependent manner occurs (18, 19). The pt-Dimer-Rab7 construct was transiently transfected into Huh7 cells for 24 h, and then we detached the cells using trypsin and seeded the cells in confocal dishes. After 12 h, DENV-2 (m.o.i. = 20) was added to Huh7 cells at 4 °C for 4 h to allow viral attachment to the cells. After 4 h, the unabsorbed virus particles were washed with cold PBS, and cells were treated with rEv37 (10 μm) for 0.5 or 2 h at 37 °C to determine the entry stage affected. Finally, the results were determined using confocal microscopy. If fusion was suppressed, then the colocalization of DENV-2 and Rab7-positive late endosomes would be increased (20). DENV-2 E protein and Rab7 colocalization were quantified and expressed as a percentage of total examined DENV-2 particles. Counting the number of colocalizations, the colocalization percentage of DENV-2 increased about two times when treated with rEv37 (Fig. 4B).
BAF is a recognized inhibitor of organelle acidification, and we hypothesized that rEv37 probably alkalized organelles in the cells so that the fusion process between DENV-2 and late endosomes was suppressed. To verify our assumption, we used the FITC-labeled H3 peptide, which enters cells by endocytosis and is released into the cytoplasm in a low pH–dependent manner, as reported previously (9). Huh7 cells were seeded in the confocal dishes overnight; the adherent cells were treated with 10 μm rEv37 or 100 nm BAF for 1 or 24 h, and the cells were studied under confocal microscopy. Compared with the negative control (no drugs), rEv37 and BAF caused the peptide H3 to accumulate in Huh7 cells, which was more obvious at 24 h (Fig. 4C).
rEv37 inhibits viral infection with pH-dependent endocytosis entry
As reported previously, DENV-2, HCV, and ZIKV enter the host cells by clathrin-mediated endocytosis and a low pH–dependent pathway. The three viruses are positive single–stranded RNA viruses. To detect whether rEv37 only inhibited a class of viruses such as DENV-2, we used the DNA virus HSV-1 for the following experiments. Vero cells were incubated with HSV-1 and different concentrations of rEv37 for 1 h. We then washed the unbound virus with PBS. We added 1 ml/well MEM (no phenol red) containing 2% fetal bovine serum (FBS) and 1.5% carboxymethylcellulose to the cells, and the plate was cultured for 4 days. The covering layer was removed by washing three times with PBS, followed by adding a 1-ml mixture, including 1% crystal violet solution and 10% methanol in water for immobilization and staining. After 30 min, the plaques were counted after washing the mixture with water. The virus particle number was described as pfu/ml. The experimental data indicated that rEv37 could inhibit HSV-1 infection in a concentration-dependent manner (Fig. 5, A and B). Although HSV-1, a double-strand DNA virus, is different from DENV-2, it shares similar entry mechanisms into host cells as DENV-2 (21). The inhibitory activity of the peptide Ev37 against HSV-1 is correlated with the entry pathway of HSV-1.
Figure 5.
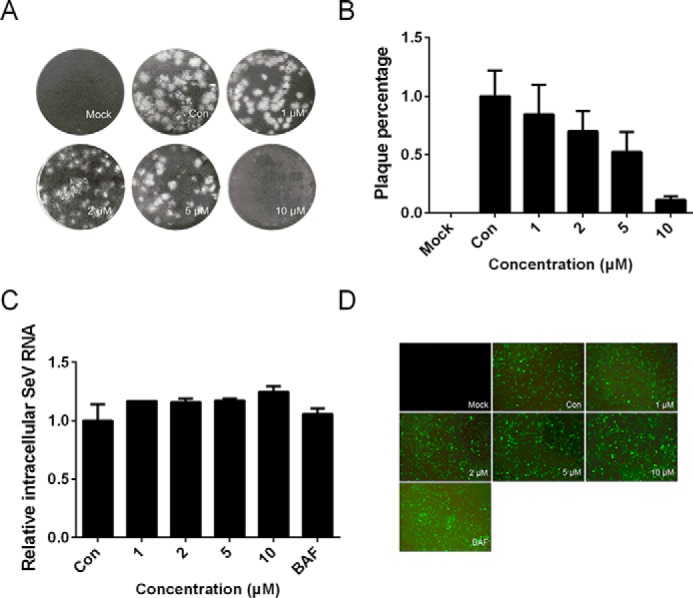
Effects of the rEv37 peptide on HSV-1, SeV, and AdV in vitro. A and B, pfu assay of HSV-1 in Vero cells. Vero cells were seeded for 24 h, and virus and rEv37 were coincubated in cells for 1 h followed by covering with a cover layer as described under “Materials and methods.” The plaque was exhibited (A), and the statistical analysis of the results was described as a percent (B). C, effect of rEv37 on SeV. The SeV-treated 293T cells were incubated for 24 h with rEv37 (0–10 μm) and BAF (10 nm), and the mRNA of SeV was assessed by qRT-PCR. D, adenovirus-treated 293A cells were incubated for 24 h with rEv37 (0–10 μm) and BAF (10 nm), and fluorescence was detected using fluorescence microscopy.
Next, we determined whether rEv37 inhibited other viruses that enter the host cell using different pathways from DENV-2. SeV is a negative single-stranded RNA virus that enters the host cell by direct fusion with cell membranes (22). SeV and different concentrations of rEv37 were incubated with 293T cells for 1 h. The cells were washed three times with PBS and sequentially cultured for 24 h with rEv37. The intracellular mRNA was measured by qRT-PCR using the SeV primer. The results showed that rEv37 could not inhibit SeV infection in vitro at a concentration of 10 μm (Fig. 5C).
Adenovirus (AdV) is a nonenveloped, double-strand DNA virus that causes upper respiratory tract, gastrointestinal tract, and ocular infections (23). AdV enters the host cell by clathrin-mediated endocytosis trigged by the interactions between the adenovirus penton base and αv integrins (24). Different from DENV-2, which releases its genome through a low pH–dependent pathway, the AdV partial capsid uncoats in the endosomes to release protein VI, which ruptures the endosomal membrane to allow partially disassembled virions to enter the cytoplasm (25). To determine whether rEv37 affected AdV infection, 293A cells were incubated with AdV and rEv37 for 48 h and then studied under a fluorescence microscope. There was no significant difference of fluorescence between the control group and rEv37 treatment group. As expected, BAF had no effect on AdV infection (Fig. 5D). Thus, rEv37 can inhibit the infections of viruses that enter the host cells by endocytosis and fusion in a low pH–dependent manner.
rEv37 alkalizes acidic organelles to inhibit the DENV-2 genome release into the cytoplasm from endosomes
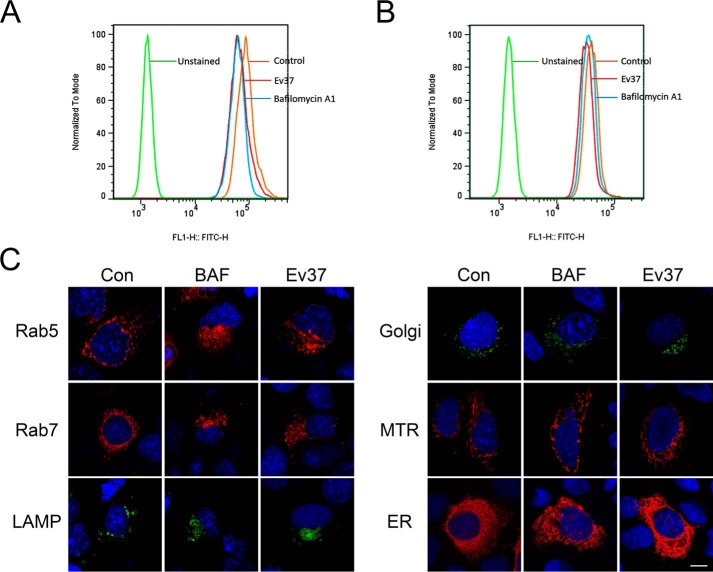
rEv37 inhibited viral entry to the host cells by the low pH–dependent endocytosis pathway, which is used by the positive single–strand RNA viruses DENV-2, HCV, and ZIKV and the double-strand DNA virus HSV-1, as described above. To detect whether rEv37 alkalizes the acidic organelles to affect their pH, the following experiments were carried out. BAF is a recognized inhibitor of acidification, so it was a positive control in the following experiments. First, an acid-sensitive indicator LysoSensorTM Green DND-189 was used to determine whether rEv37 impacted the acidic organelles' pH values (26). The samples with different treatments were measured by a flow cytometer using a 488-nm laser for excitation. The red (rEv37) and blue (BAF) curves (Fig. 6, A and B) in the treatment group shifted to the left relative to the control (orange curve). rEv37 and BAF inhibited organelle acidification (Fig. 6, A and B).
Figure 6.
Inhibition of the peptide rEv37 of acidic organelle acidification. A and B, measurement of organelle acidification in Huh7 cells (A) and 293T cells (B) by flow cytometry (FCM). Huh7 cells and 293T cells were pretreated with 10 μm rEv37 or 100 nm BAF for 24 h. Then, the cells were incubated with 1 μm LysoSensor DND-189 at 37 °C for 20 min. The intensity of FITC staining was determined on a Cytoflex flow cytometer. C, detection of organelle acidification after treatment with rEv37 and BAF, using confocal microscopy. The plasmids of acidic organelles (pt-Dimer-Rab5, pt-Dimer-Rab7, pEGFP-Golgi, and pEGFP-LAMP1) and nonacidic organelles (pRFP-mitochondria, pRFP-ER) were transfected into Huh7 cells for 24 h, and then the cells were seeded into confocal dishes and treated with 10 μm rEv37 or 100 nm BAF for 24 h, followed by staining with DAPI, and observation by confocal microscopy. Scale bar, 10 μm.
DENV-2 entry contains multiple steps, and the endocytosis pathway needs a series of cellular components, such as the early endosome, late endosome, and lysosome (27). To further determine whether rEv37 impacted the pH of acidic organelles, Huh7 cells were studied under a confocal microscope. As expected, the acidic organelles of Rab5 (early endosome), Rab7 (late endosome), and LAMP1 (lysosome) were enlarged and aggregated, and the Golgi was partially enlarged and aggregated (Fig. 6C). rEv37 had no effect on nonacidic organelles, such as RFP mitochondria and pRFP-ER (endoplasmic reticulum) (Fig. 6C). Therefore, rEV37 alkalifies and enlarges the acidic organelles but not the nonacidic organelles to suppress endocytosis.
Discussion
In the past decades, antiviral peptide drugs and small molecule drugs have been discovered. A peptide named EB, derived from the signal sequence of fibroblast growth factor 4, was determined to inhibit influenza viral infection by targeting viral attachment to cells (28). NP-1, a rabbit α-defensin, can suppress the entry and intercellular spread of HSV-2 (29). Eval418 and its derived peptides could suppress HSV-1 infection by blocking the initial step. The scorpion-derived defensin BmKDfsin4 could inhibit HBV replication in vitro (30). Chloroquine (CQ) is an antimalarial drug and can prevent DENV-2 infection by alkalizing the pH value of intracellular organelles, such as the endosome, lysosome, and vesicles of the Golgi complex, as described previously (31).
In this study, we expressed and purified a scorpion venom-derived peptide, Ev37, which contains 78 amino acid residues and is a scorpine-like peptide, including a CSα/β motif. rEv37 has six cysteines that can form three pairs of disulfide bonds at the C terminus. DENV-2, HCV, and ZIKV are enveloped, positive, single-strand RNA viruses and belong to Flaviviridae, and they enter the host cell through clathrin-mediated endocytosis and low pH–dependent fusion. rEv37 cannot only inhibit DENV-2 infection but can also suppress HCV and ZIKV infections in a concentration-dependent manner at noncytotoxic concentrations. There are four dengue virus serotypes (DENV1–4) that are clinically indistinguishable (2). The four serotypes of dengue virus share a common morphology, genomic structure and replication cycle. Especially, DENV serotypes 1, 3, and 4 have a similar entry pathway as DENV-2 (18). As reported, combined effects of ribavirin and compound A can inhibit DENV production infected with all DENV serotypes 1–4 (32). So, we proposed that rEv37 may have a similar antiviral effect on DENV-1, 3, and 4 serotypes, but it still needs to be further confirmed in future work. In contrast, rEv37 has no impact on the Sendai virus, which enters the host cell when the viral membrane directly fuses with the plasma membrane. Similarly, rEv37 has no effect on adenovirus, which enters the host cells by clathrin-mediated endocytosis but not low pH–dependent fusion, releasing viral genome into the cytoplasm.
As described above, we found that rEv37 may be a broad-spectrum antiviral peptide. Moreover, rEv37 had little effect on viral particles (DENV-2 and HCV) and was not a virucidal peptide, which was completely different from other scorpine-like peptides (Scorpine and Hge36) from scorpion venoms. Thus, rEv37 can enrich our knowledge about antiviral peptides from scorpion venoms and give us a better understanding of the scorpine-like peptide family. rEv37 has a specific antiviral mechanism in that it can inhibit DENV-2 infection by blocking the viral genome release from the endosome into the cytoplasm via alkalizing the pH of endosomes. In addition, we also discovered that rEv37 can enlarge the volume of acidic organelles and make them accumulate. Similarly, some chemicals or ion channels modulating organelle acidification were also reported to be related to viral infections. For example, BAF and CQ can block viral infection by alkalizing the pH of acidic organelles (33). The sodium–proton exchanger for K+/Na+ exchange (34) and two-pore channels, such as endosomal Ca2+ channels, are required for Ebola virus entry (35). High endosomal K+ plays a critical role in Bunyamwera virus infection (36). Vacuolar-type H+-ATPase (v-ATPase) is primarily responsible for proton secretion and is important for acidic organelle acidification to affect viral infection (37).
Although we found that rEv37 could alkalize the pH of acidic organelles, we still needed to investigate how rEv37 alkalized the pH of acidic organelles. We have studied that Ev37 can enter the cells alone (data not shown), so we can postulate that rEv37 inhibits the acidification of acidic organelles involved in two aspects of the viral life cycle. On the one hand, because rEv37 had a similar effect on acidic organelles as BAF, a known inhibitor for v-ATPase, we proposed that rEv37 may interact with v-ATPase or other ion channels to modulate the pH of acidic organelles. On the other hand, rEv37 contains 17 basic amino acid residues, close to 22% (17/78), and is a typical basic peptide with a pI of 9.22. Therefore, rEv37 may alkalize the pH of acidic organelles by its own basic properties.
Conclusion
In summary, we found that a scorpion venom peptide, Ev37, can inhibit DENV-2 and other viruses, such as HCV, ZIKV, and HSV-1, whose entry processes are similar to DENV-2. However, rEv37 has no effect on SeV and AdV, which enter host cells without low pH–dependent fusion. rEv37 mainly inhibited the late-entry stage of viral membrane–endosomal membrane fusion with low pH dependence by alkalizing organelles, but it had no effect on viral attachment and replication. Thus, rEv37 is a broad-spectrum antiviral peptide with specific molecular mechanisms that are different from scorpion venom and animal venom peptides. It is worth researching it further as an antiviral drug candidate.
Materials and methods
Peptide expression, purification, and characterization
Ev37 precursor contains 97 residues, which is composed of a signal peptide of 19 residues and a mature peptide of 78 residues (14).The cDNA sequence of the mature peptide of Ev37 was isolated from the cDNA library and was inserted into expression plasmid pET-32a with restriction enzymes KpnI and XhoI and five codons encoding an enterokinase cleavage site (DDDDK). Then, the recombinant vector was transformed into expression bacteria E. coli BL21(DE3).
E. coli BL21(DE3) containing recombinant vector was cultured in LB medium, including 100 μg/ml ampicillin at 37 °C, and added 0.8 mm isopropyl β-d-thiogalactoside (IPTG) to induce the synthesis of fusion protein at 25 °C for 8 h when the A600 = 0.8 of the cell density. Then, the cells were broken by ultrasonication at 400 Hz for 90 cycles, and the extract was clarified by centrifugation at 12,000 rpm for 15 min. The fusion protein in supernatant was purified by nickel-chelating affinity chromatography and then desalted by dialysis with 1 × enterokinase buffer for 3 h. To obtain rEv37, the fusion protein was cleaved by enterokinase (Morebio, China) for 16 h at 23 °C, and separated by RP-HPLC on a C18 column (Elite HPLC, China, 10 × 250 mm, 5 μm). Target peptide was collected with detection at a wavelength of 230 nm and freeze-dried to save.
To identify the characteristics of rEv37, we used MALDI-TOF MS to confirm its molecular mass and CD spectrum assay to determine its secondary structure.
MALDI-TOF MS
To measure the molecular weight of rEv37, we loaded the rEv37 purified by RP-HPLC to the instrument (Institute of Hydrobiology, Chinese Academy of Sciences). Its actual molecular weight was confirmed by MALDI-TOF MS. The peptide (1 μl) was spotted onto the plate along with an equal volume of matrix solution (10 mg/ml α-cyano-4-hydroxycinnamic acid, 50% acetonitrile, and 0.1% TFA). The mixture was left to dry at room temperature. The reflection mode and the accelerating voltage of 25 kV were chosen for work. Mass spectrometry was performed by the FlexControl software.
Cell culture
Cell lines Huh7, Huh7.5.1, BHK21, HEK293T, HEK293A, and Vero were cultured in Dulbecco's modified Eagle's medium (Gibco-Invitrogen) supplemented with 10% FBS (Gibco-Invitrogen) and 1% penicillin/streptomycin at 37 °C in a humidified 5% CO2 incubator. C6/36 cells were cultured at 28 °C without 5% CO2 in MEM (Gibco-Invitrogen) supplemented with 10% FBS and 1% penicillin/streptomycin.
Virus strains
Dengue virus serotype 2 TSV01 strain (DENV-2 TSV01) was kindly provided by Dr. Bo Zhang from Wuhan Institute of Virology, Chinese Academy of Sciences. HCV genotype 2a strain JFH-1 was kindly provided by Takaji Wakita from the National Institute of Infectious Diseases, Tokyo, Japan. Zika virus Puerto Rico strain (PRVABC59) cDNA plasmid was kindly provided by Dr. Ren Sun and Dr. Danyang Gong at UCLA. Sendai virus and adenovirus were kindly provided by Dr. Mingxiong Guo and Dr. Zan Huang from College of Life Sciences in Wuhan University, respectively.
Plasmids and transfection
pt-Dimer-Rab5, pt-Dimer-Rab7, and pRFP-ER were constructed by our laboratory. DNA sequences that were searched in the National Center of Biotechnology Information (NCBI) were inserted in vector plasmids. pEGFP-Golgi, pEGFP-LAMP1, and pRFP mitochondria were kindly provided by Dr. Hongbing Shu and Dr. Zhiyin Song from College of Life Sciences, Wuhan University. When covered about 80% of the culture flask, the cells were transfected with the DNA constructs using TurboFect transfection reagent (ThermoFisher Scientific, Waltham, MA) following the instructions.
Antibodies and agents
Dengue virus type 1–4 mouse mAb (D1-11(3)) (MA1-27093) was purchased from Invitrogen; mouse anti-HepC cAg (C7-50) antibody (sc-57800) was purchased from Santa Cruz Biotechnology; anti-flavivirus group antigen antibody and clone D1-4G2-4-15 (MAB10216) were purchased from Merck. Zika virus envelope protein antibody (GTX133314, GeneTex) was kindly provided by Dr. Jianguo Wu from College of Life Sciences, Wuhan University. Anti-GAPDH mouse antibody (60004-1-Ig) was from Proteintech Group. Goat HRP-conjugated anti-mouse IgG (H+L) (GM01H) was purchased from NovoGene, and Alexa Fluor 488 Affinipure donkey anti-mouse IgG (H+L) (34106ES60) was purchased from Yeasen. Bafilomycin A1 (ab120497) was ordered from Abcam; LysoSensorTM Green DND-189 (L7535)) and ThermoFisher Scientific PageRuler Prestained Protein Ladder (26617) were from ThermoFisher Scientific; Bestar® SYBR Green qPCR master mix reagent was purchased from DBI® Bioscience; 5× loading sample buffer (BL502A) was purchased from Biosharp; and M5 Superlow range prestained protein ladder (1.7–40 kDa) (MF215-T)) was purchased from Mei5 Biotechnology.
Time of addition assay
To determine rEv37's mechanism of action, the peptide was added to the virus or cells at different times. Cells were treated with the following conditions (38). (i) For co-inoculation, cells were incubated with DENV-2 (or HCV and ZIKV) and different concentrations of rEv37 (0–10 μm) in a 37 °C incubator for 1 h (DENV-2, ZIKV, and SeV) or 4 h (HCV), followed by washing off the unabsorbed virus three times with phosphate-buffered solution (PBS) and extracting total RNA after culturing for 48 h (DENV-2, ZIKV, and SeV) or 72 h (HCV). (ii) For free viral particles, a tube containing virus particles and 10 μm rEv37 mixture was incubated for 1 h at 37 °C, then the mixture was diluted and added to the cells for 1 h (DENV-2, ZIKV, SeV) or 4 h (HCV), and the following step was the same as step i. (iii) For pre-treating cells, the Huh7 cells were incubated with Ev37 at 37 °C for 1 h, and then the cells were incubated with virus for 1 h after removing redundant peptide; the following step is the same as step i. (iv) For entry, the cells and virus were incubated with rEv37 for 1 h (DENV-2) or 4 h (HCV) at 37 °C, followed by washing off the unabsorbed virus, and the cells were cultured for 24 h. (v) For attachment, cells, DENV-2 (or HCV), and 10 μm rEv37 were incubated at 4 °C for 1 h (or 4 h), and then the unabsorbed virus was removed, and total RNA were extracted and detected by qRT-PCR. (vi) For replication, 10 μm rEv37 was added to cells after they were infected with virus at 37 °C for 2 h, and the cells continued to grow for 13 h (in one life cycle). The m.o.i. in all treatments was 1 in addition assay.
qRT-PCR
To quantify the inhibitory effect of peptides on virus infection, the intracellular total RNA was extracted from the cells peptide-treated or untreated using TRIzol reagent (Takara), and the first-strand cDNA was reversed-transcribed by using the RevertAid first strand cDNA synthesis kit (ThermoFisher Scientific). The cDNA was quantitated by real-time qRT-PCR with DENV-2, HCV, ZIKV, and SeV primers using the Bestar® SYBR Green qPCR master mix reagent (DBI® Bioscience). DENV-2 primers were 5′-GGCCTCGACTTCAATGAGATGG-3′ (sense) and 5′-CCTGTTTCTTCGCATGGGGAT-3′ (antisense). HCV primers were 5′-TCTGCGGAACCGGTGAGTA-3′ (sense) and 5′-TCAGGCAGTACCACAAGGC-3′ (antisense). ZIKV primers were 5′-TTGTGGAAGGTATGTCAGGTG-3′ (sense) and 5′-ATCTTACCTCCGCCATGTTG-3′ (antisense). SeV primers were 5′-AAACGCATCACGTCTCTTCC-3′ (sense) and 5′-TTCTCAGCTCTGCTTAGGGG-3′ (antisense).
Plaque assay
To measure the virus particle number in the supernatant and DENV viral titration, we performed plaque assay using BHK21 cells (39). BHK21 cells were seeded in a 24-well (2 × 105 cells/well) plate with 10% FBS for 24 h at 37 °C. Serially diluted virus with 2% FBS was added to the monolayer cell after removing the medium. After 1 h, washing the unabsorbed virus and adding MEM containing 2% FBS and 1.5% carboxymethylcellulose to the cells for cover layer (1 ml/well), the plate then was cultured for 7 days. The covering layer was removed and washed three times with PBS, followed by adding 1 ml of mixture, including 1% crystal violet solution and 10% methanol in water for immobilization and staining. After 30 min, the plaques were counted after washing the mixture with water. The virus particle number was expressed as pfu/ml.
Western blotting
Cells with treatment or untreatment were collected and lysed on ice for 30 min by RIPA lysis buffer, including phenylmethanesulfonyl fluoride, a proteinase inhibitor. The lysates were centrifuged for 15 min at 12,000 rpm, and the supernatants containing total protein were measured using a BCA protein assay kit (ThermoFisher Scientific). The equal amounts (20 μg) of protein were separated by SDS-PAGE and transferred to nitrocellulose membranes (NC membrane, Millipore). The nonspecific proteins on an NC membrane were blocked by 5% milk for 2 h at room temperature on a table concentrator. Then the NC membrane was incubated with primary antibody of anti-DENV E protein (Invitrogen, MA1-27093), anti-HCV core protein (Santa Cruz Biotechnology, sc-57800), anti-Zika E protein (GeneTex, GTX133314), and GAPDH as a loading control for quantification normalization, respectively. The membranes were exposed to HRP-conjugated anti-mouse or anti-rabbit secondary antibodies at room temperature for 2 h, and then the results were visualized by enhanced chemiluminescence kit with FuJi medical X-ray film.
Confocal microscopy
To detect the attachment of DENV-2 in Huh7 cells, cells seeded in confocal dishes were precooled on ice and incubated with DENV-2 virus with or without rEv37 and bafilomycin A1 (Abcam, ab120497) at 4 °C for 1 h to make virus particles attach to the cell surface but not to enter the cell. The cells were washed three times with cold PBS to remove the unattached virus, and then immobilized with cold 4% paraformaldehyde and next permeabilized with 0.2% Triton X-100. Then, the cells were blocked by 5% BSA (BioFoxx) for 1 h at room temperature and were incubated with anti-DENV-2 E protein (Invitrogen, MA1–27093) in 1% BSA at 4 °C overnight. Subsequently, the cells were incubated with Alexa Fluor 488 donkey anti-mouse second antibody for 1 h at room temperature followed by staining the nucleus with DAPI (ANT10072) and being observed under a confocal microscope.
To detect whether rEv37 suppressed DENV-2 by releasing its genome to the cytoplasm from endosomes, we measured the colocalization of DENV-2 and Rab7 (late endosome). Huh7 cells were transfected with pt-Dimer-Rab7 for 24 h, then cells were seeded in the confocal dish and treated with 10 μm rEv37 or 100 nm BAF for 24 h, followed by adding with DENV-2 (m.o.i. = 20) at 4 °C for 2 h and then at 37 °C for 30 min or 2 h.
To observe the effect of rEv37 on acid organelles (early endosome, late endosome, lysosome, and Golgi) and nonacidic organelles (mitochondria and endoplasmic reticulum), pt-Dimer-Rab5, pt-Dimer-Rab7, pEGFP-Lamp1, pEGFP-Golgi, pRFP-mito, and pRFP-ER were transfected into Huh7 cells. Cells were seeded into confocal dishes after transfecting for 24 h and treated with rEv37 and BAF for the indicated times, then immobilized with cold 4% paraformaldehyde, and permeabilized with 0.2% Triton X-100, and nucleus was stained with DAPI.
H3 is a modified scorpion peptide of Ctry2459 separated from Chaerilus tryznai, and it depends on the endocytosis pathway and endocytic acidification to enter cells. To test whether rEv37 could alkalize the pH of acidic organelles, 1 μg/ml H3 was added into cells seeded in confocal dishes with 10 μm rEv37 or 100 nm BAF for 1 and 24 h, respectively. Then, the cells were immobilized, permeabilized, and stained as above.
Flow cytometry
To measure the effect of rEv37 on acid organelles by flow cytometry, we pretreated Huh7 cells and 293T cells for 24 h with 10 μm rEv37 or 100 nm BAF. Then, the cells were incubated with 1 μm LysoSensor DND-189 at 37 °C for 20 min and obtained by generating a single cell suspension, passing it through nylon net after digesting with 0.25% trypsin. Flow cytometry measuring the intensity of FITC was performed on a Cytoflex Flow cytometer. If rEv37 and BAF alkalize acidic organelles, the peak of FITC would shift to the left relative to the control.
Statistics analysis
Graphical representation and statistical analysis were performed using Adobe Photoshop CS6 (Adobe Systems Inc.) and Graphpad Prism6. Confocal statistics data are presented as the mean ± S.D. from a population of cells (n). Other data are presented as the mean ± S.D. from at least three separate experiments.
Author contributions
F. L. and Y. L. data curation; F. L., Y. L., Z. J., Y. C., Z. Chen, and Y. W. formal analysis; F. L. investigation; F. L., Y. L., Z. J., Z. X., G. L., F. S., and M. G. methodology; F. L. writing-original draft; Y. H. and Y. Z. software; Y. W., W. L., and Z. Cao conceptualization; W. L. project administration; Z. Cao funding acquisition.
This work was supported by National Science Fund of China Grants 31572289, 31872239, and 81630091; International S&T Cooperation Program of China Grant S2016G3110; Hubei Science Funds 2015CFA042 and 2016CFA018; China-Kazakhstan Cooperation Program Grant CK-07-09; and Fundamental Research Funds for the Central Universities in China 2042017kf0242 and 2042017kf0199. The authors declare that they have no conflicts of interest with the contents of this article.
- HCV
- hepatitis C virus
- DENV-2
- dengue virus type 2
- SeV
- Sendai virus
- AdV
- adenovirus
- ZIKV
- Zika virus
- HBV
- hepatitis B virus
- MEM
- minimum Eagle's medium
- FBS
- fetal bovine serum
- qPCR
- quantitative PCR
- DAPI
- 4′,6-diamidino-2-phenylindole
- m.o.i.
- multiplicity of infection
- pfu
- plaque-forming unit
- IPTG
- isopropyl β-d-thiogalactoside
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- HRP
- horseradish peroxidase
- NC
- nitrocellulose
- MTT
- 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide
- CQ
- chloroquine
- ER
- endoplasmic reticulum
- RFP
- red fluorescent protein
- RP-HPLC
- reversed-phase HPLC
- BAF
- bafilomycin A1.
References
- 1. Ravindran M. S., Bagchi P., Cunningham C. N., and Tsai B. (2016) Opportunistic intruders: how viruses orchestrate ER functions to infect cells. Nat. Rev. Microbiol. 14, 407–420 10.1038/nrmicro.2016.60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yacoub S., Mongkolsapaya J., and Screaton G. (2016) Recent advances in understanding dengue. F1000 Res. 5, F1000 10.12688/f1000research.6233.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Low J. G., Sung C., Wijaya L., Wei Y., Rathore A. P. S., Watanabe S., Tan B. H., Toh L., Chua L. T., Hou Y., Chow A., Howe S., Chan W. K., Tan K. H., Chung J. S., et al. (2014) Efficacy and safety of celgosivir in patients with dengue fever (CELADEN): a phase 1b, randomised, double-blind, placebo-controlled, proof-of-concept trial. Lancet Infect. Dis. 14, 706–715 10.1016/S1473-3099(14)70730-3 [DOI] [PubMed] [Google Scholar]
- 4. Shan C., Xie X., Zou J., Züst R., Zhang B., Ambrose R., Mackenzie J., Fink K., and Shi P. (2018) Using a virion assembly-defective dengue virus as a vaccine approach. J. Virol. 2018, JVI.01002-18 10.1128/JVI.01002-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Paul D., Madan V., and Bartenschlager R. (2014) Hepatitis C virus RNA replication and assembly: living on the fat of the land. Cell Host Microbe 16, 569–579 10.1016/j.chom.2014.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Song H., Qi J., Haywood J., Shi Y., and Gao G. F. (2016) Zika virus NS1 structure reveals diversity of electrostatic surfaces among flaviviruses. Nat. Struct. Mol. Biol. 23, 456–458 10.1038/nsmb.3213 [DOI] [PubMed] [Google Scholar]
- 7. Li Z., Xu X., Meng L., Zhang Q., Cao L., Li W., Wu Y., and Cao Z. (2014) Hp1404, a new antimicrobial peptide from the scorpion Heterometrus petersii. PLoS One 9, e97539 10.1371/journal.pone.0097539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zeng X. C., Wang S., Nie Y., Zhang L., and Luo X. (2012) Characterization of BmKbpp, a multifunctional peptide from the Chinese scorpion Mesobuthus martensii Karsch: gaining insight into a new mechanism for the functional diversification of scorpion venom peptides. Peptides 33, 44–51 10.1016/j.peptides.2011.11.012 [DOI] [PubMed] [Google Scholar]
- 9. Hong W., Zhang R., Di Z., He Y., Zhao Z., Hu J., Wu Y., Li W., and Cao Z. (2013) Design of histidine-rich peptides with enhanced bioavailability and inhibitory activity against hepatitis C virus. Biomaterials 34, 3511–3522 10.1016/j.biomaterials.2013.01.075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zeng Z., Zhang R., Hong W., Cheng Y., Wang H., Lang Y., Ji Z., Wu Y., Li W., Xie Y., and Cao Z. (2018) Histidine-rich modification of a scorpion-derived peptide improves bioavailability and inhibitory activity against HSV-1. Theranostics 8, 199–211 10.7150/thno.21425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhao Z., Hong W., Zeng Z., Wu Y., Hu K., Tian X., Li W., and Cao Z. (2012) Mucroporin-M1 inhibits hepatitis B virus replication by activating the mitogen-activated protein kinase (MAPK) pathway and down-regulating HNF4α in vitro and in vivo. J. Biol. Chem. 287, 30181–30190 10.1074/jbc.M112.370312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Carballar-Lejarazú R., Rodríguez M. H., de la Cruz Hernández-Hernández F., Ramos-Castañeda J., Possani L. D., Zurita-Ortega M., Reynaud-Garza E., Hernández-Rivas R., Loukeris T., Lycett G., and Lanz-Mendoza H. (2008) Recombinant scorpine: a multifunctional antimicrobial peptide with activity against different pathogens. Cell. Mol. Life Sci. 65, 3081–3092 10.1007/s00018-008-8250-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. El-Bitar A. M., Sarhan M. M., Aoki C., Takahara Y., Komoto M., Deng L., Moustafa M. A., and Hotta H. (2015) Virocidal activity of Egyptian scorpion venoms against hepatitis C virus. Virol. J. 12, 47 10.1186/s12985-015-0276-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Feng J., Yu C., Wang M., Li Z., Wu Y., Cao Z., Li W., He X., and Han S. (2013) Expression and characterization of a novel scorpine-like peptide Ev37, from the scorpion Euscorpiops validus. Protein Exp. Purif. 88, 127–133 10.1016/j.pep.2012.12.004 [DOI] [PubMed] [Google Scholar]
- 15. da Mata E. C., Mourao C. B., Rangel M., and Schwartz E. F. (2017) Antiviral activity of animal venom peptides and related compounds. J. Venom. Anim. Toxins Includ. Trop. Dis. 23, 3 10.1186/s40409-016-0089-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Conde R., Zamudio F. Z., Rodríguez M. H., and Possani L. D. (2000) Scorpine, an anti-malaria and anti-bacterial agent purified from scorpion venom. FEBS Lett. 471, 165–168 10.1016/S0014-5793(00)01384-3 [DOI] [PubMed] [Google Scholar]
- 17. Cheng G., Montero A., Gastaminza P., Whitten-Bauer C., Wieland S. F., Isogawa M., Fredericksen B., Selvarajah S., Gallay P. A., Ghadiri M. R., and Chisari F. V. (2008) A virocidal amphipathic α-helical peptide that inhibits hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. U.S.A. 105, 3088–3093 10.1073/pnas.0712380105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rodenhuis-Zybert I. A., Wilschut J., and Smit J. M. (2010) Dengue virus life cycle: viral and host factors modulating infectivity. Cell. Mol. Life Sci. 67, 2773–2786 10.1007/s00018-010-0357-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. van der Schaar H. M., Rust M. J., Chen C., van der Ende-Metselaar H., Wilschut J., Zhuang X., and Smit J. M. (2008) Dissecting the cell entry pathway of dengue virus by single-particle tracking in living cells. PLoS Pathog. 4, e1000244 10.1371/journal.ppat.1000244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Narayana S. K., Helbig K. J., McCartney E. M., Eyre N. S., Bull R. A., Eltahla A., Lloyd A. R., and Beard M. R. (2015) The interferon-induced transmembrane proteins, IFITM1, IFITM2, and IFITM3 inhibit hepatitis C virus entry. J. Biol. Chem. 290, 25946–25959 10.1074/jbc.M115.657346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nicola A. V. (2016) Herpesvirus entry into host cells mediated by endosomal low pH. Traffic 17, 965–975 10.1111/tra.12408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fan D. P., and Sefton B. M. (1978) The entry into host cells of Sindbis virus, vesicular stomatitis virus, and Sendai virus. Cell 15, 985–992 10.1016/0092-8674(78)90282-9 [DOI] [PubMed] [Google Scholar]
- 23. Marvin S. A., and Wiethoff C. M. (2012) Emerging roles for ubiquitin in adenovirus cell entry. Biol. Cell 104, 188–198 10.1111/boc.201100096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Huang S., Reddy V., Dasgupta N., and Nemerow G. R. (1999) A single amino acid in the adenovirus type 37 fiber confers binding to human conjunctival cells. J. Virol. 73, 2798–2802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wiethoff C. M., Wodrich H., Gerace L., and Nemerow G. R. (2005) Adenovirus protein VI mediates membrane disruption following capsid disassembly. J. Virol. 79, 1992–2000 10.1128/JVI.79.4.1992-2000.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bagh M. B., Peng S., Chandra G., Zhang Z., Singh S. P., Pattabiraman N., Liu A., and Mukherjee A. B. (2017) Misrouting of v-ATPase subunit V0a1 dysregulates lysosomal acidification in a neurodegenerative lysosomal storage disease model. Nat. Commun. 8, 14612 10.1038/ncomms14612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mercer J., Schelhaas M., and Helenius A. (2010) Virus entry by endocytosis. Annu. Rev. Biochem. 79, 803–833 10.1146/annurev-biochem-060208-104626 [DOI] [PubMed] [Google Scholar]
- 28. Jones J. C., Turpin E. A., Bultmann H., Brandt C. R., and Schultz-Cherry S. (2006) Inhibition of influenza virus infection by a novel antiviral peptide that targets viral attachment to cells. J. Virol. 80, 11960–11967 10.1128/JVI.01678-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sinha S., Cheshenko N., Lehrer R. I., and Herold B. C. (2003) NP-1, a rabbit α-defensin, prevents the entry and intercellular spread of herpes simplex virus type 2. Antimicrob. Agents Chemother. 47, 494–500 10.1128/AAC.47.2.494-500.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zeng Z., Zhang Q., Hong W., Xie Y., Liu Y., Li W., Wu Y., and Cao Z. (2016) A scorpion defensin bmKDfsin4 inhibits hepatitis B virus replication in vitro. Toxins 8, E124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kaptein S. J., and Neyts J. (2016) Towards antiviral therapies for treating dengue virus infections. Curr. Opin. Pharmacol. 30, 1–7 10.1016/j.coph.2016.06.002 [DOI] [PubMed] [Google Scholar]
- 32. Rattanaburee T., Junking M., Panya A., Sawasdee N., Songprakhon P., Suttitheptumrong A., Limjindaporn T., Haegeman G., and Yenchitsomanus P. T. (2015) Inhibition of dengue virus production and cytokine/chemokine expression by ribavirin and compound A. Antiviral Res. 124, 83–92 10.1016/j.antiviral.2015.10.005 [DOI] [PubMed] [Google Scholar]
- 33. Kang S., Shields A. R., Jupatanakul N., and Dimopoulos G. (2014) Suppressing dengue-2 infection by chemical inhibition of Aedes aegypti host factors. PLoS Negl. Trop. Dis. 8, e3084 10.1371/journal.pntd.0003084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Scott C. C., and Gruenberg J. (2011) Ion flux and the function of endosomes and lysosomes: pH is just the start: the flux of ions across endosomal membranes influences endosome function not only through regulation of the luminal pH. BioEssays 33, 103–110 10.1002/bies.201000108 [DOI] [PubMed] [Google Scholar]
- 35. Sakurai Y., Kolokoltsov A. A., Chen C. C., Tidwell M. W., Bauta W. E., Klugbauer N., Grimm C., Wahl-Schott C., Biel M., and Davey R. A. (2015) Ebola virus. Two-pore channels control Ebola virus host cell entry and are drug targets for disease treatment. Science 347, 995–998 10.1126/science.1258758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Whelan S. P., Hover S., Foster B., Fontana J., Kohl A., Goldstein S. A. N., Barr J. N., and Mankouri J. (2018) Bunyavirus requirement for endosomal K+ reveals new roles of cellular ion channels during infection. PLoS Pathog. 14, e1006845 10.1371/journal.ppat.1006845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yang D. Q., Feng S., Chen W., Zhao H., Paulson C., and Li Y. P. (2012) V-ATPase subunit ATP6AP1 (Ac45) regulates osteoclast differentiation, extracellular acidification, lysosomal trafficking, and protease exocytosis in osteoclast-mediated bone resorption. J. Bone Miner. Res. 27, 1695–1707 10.1002/jbmr.1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lin L. T., Chung C. Y., Hsu W. C., Chang S. P., Hung T. C., Shields J., Russell R. S., Lin C. C., Li C. F., Yen M. H., Tyrrell D. L., Lin C. C., and Richardson C. D. (2015) Saikosaponin b2 is a naturally occurring terpenoid that efficiently inhibits hepatitis C virus entry. J. Hepatol. 62, 541–548 10.1016/j.jhep.2014.10.040 [DOI] [PubMed] [Google Scholar]
- 39. Goh K. C., Tang C. K., Norton D. C., Gan E. S., Tan H. C., Sun B., Syenina A., Yousuf A., Ong X. M., Kamaraj U. S., Cheung Y. B., Gubler D. J., Davidson A., St John A. L., Sessions O. M., and Ooi E. E. (2016) Molecular determinants of plaque size as an indicator of dengue virus attenuation. Sci. Rep. 6, 26100 10.1038/srep26100 [DOI] [PMC free article] [PubMed] [Google Scholar]