Abstract
The unique nucleolar environment, the repetitive nature of ribosomal DNA (rDNA), and especially the possible involvement of RNA polymerase I (RNAPI) in transcription-coupled repair (TCR) have made the study of repair of rDNA both interesting and challenging. TCR, the transcription-dependent, preferential excision repair of the template strand compared with the nontranscribed (coding) strand has been clearly demonstrated in genes transcribed by RNAPII. Whether TCR occurs in rDNA is unresolved. In the present work, we have applied analytical methods to map repair events in rDNA using data generated by the newly developed XR-seq procedure, which measures excision repair genome-wide with single-nucleotide resolution. We find that in human and mouse cell lines, rDNA is not subject to TCR of damage caused by UV or by cisplatin.
Keywords: precursor ribosomal RNA (pre-rRNA), DNA repair, DNA damage, DNA transcription, bioinformatics, transcription-coupled repair, XR-seq
Introduction
Nucleotide excision repair (excision repair) is a universal repair mechanism that removes bulky DNA damage by concerted dual incisions bracketing the lesion (1–3). This repair system eliminates UV-induced cyclobutane pyrimidine dimers (CPDs)3 and 6–4 pyrimidine–primidone photoproducts as well as the Pt-d(GpG) guanine diadduct induced by the anti-cancer drug cisplatin (2). The efficiency of repair is affected by multiple factors, such as DNA sequence context, DNA and histone modifications, transcription factor binding, and chromatin domains, as well as DNA dynamics, including replication, recombination, and transcription. Among these, the effect of transcription on repair is unique in terms of quantitative impact and the relatively well-defined mechanistic details. This phenomenon is called transcription-coupled repair (TCR), and it has been observed in organisms ranging from Escherichia coli to humans (3–5).
Transcription-coupled repair is characterized by 3–10-fold higher efficiency of repair of the transcribed (template) strand (TS) compared with the nontranscribed (coding) strand (NTS) or nontranscribed regions of the genome (global repair) (3, 4, 6). In addition to the core excision repair proteins, TCR also depends on the transcription–repair coupling factor encoded by the mfd gene in E. coli (7) and the CSB gene in humans (8). In humans, in addition to CSB, the CSA gene is also required for TCR, and, of special interest, the XPC protein, which is essential for global repair, is not required for TCR (9). These unique features of global and TCR have been quite useful in mechanistic understanding of excision repair and in defining the contributions of global and TCR to the repair of particular regions in the genome. In Escherichia coli, all RNAs are transcribed by the sole RNAP in the cell, and therefore all transcribed E. coli genomic regions are subject to TCR in an Mfd-dependent manner (7). In contrast, in eukaryotes, in general, and in mammalian organisms, in particular, three RNA polymerases are responsible for transcribing various types of RNAs, including mRNA, rRNA, tRNA, and 5S RNA. Of particular relevance, mRNAs are transcribed by RNA polymerase II (RNAPII), whereas rRNA-encoding genes (rDNA) are transcribed by RNA polymerase I (RNAPI). Extensive studies have shown that genes transcribed by RNAPII are subject to TCR. In contrast, attempts to study TCR of rDNA have given conflicting results.
The classical approach used to investigate TCR in RNAPII-transcribed genes has been applied to rDNA (4). This approach employs T4 endoV, which cleaves the DNA strand at a CPD, and employs Southern blotting of restricted genomic DNA using strand-specific probes to detect full-length strands of any gene region of interest. Each restricted DNA sample from UV-irradiated cells is aliquoted; half of each is digested with T4 endoV. Samples are then processed by Southern blotting, and the reduction in full-length fragment due to T4 endoV digestion compared with the undigested fragment is used to calculate the number of CPDs per strand; loss of CPDs with time after UV is measured as repair. Studies of rDNA using this assay have shown the following: following UV, initial CPD induction is similar whether cellular or naked DNA is irradiated (10); in WT cells, CPD induction is either the same or similar in the two strands of rDNA (10, 11), although 2-fold more damage in the NTS of CSB amd XPC mutant cells was observed (12); excision repair is slower in rDNA genes compared with the genome overall (10, 11, 13, 14); no TCR is detected in rDNA (10–12, 14), even at varied transcription levels (10, 11); and repair of rDNA is absent in XPC mutant cells, which lack global repair (12). Interestingly, rDNA repair was relatively slow in CSB mutant cells (12). Except for the latter findings in CSB mutant cells, the results are consistent with a lack of RNAPI-dependent TCR in mammalian cells. However, another study measured repair of rDNA indirectly as the resumption of rRNA transcription following UV, which initially suppresses transcription (15). In this case, XPC mutant cells recovered normally and CSB mutant cells failed to recover, which implicates TCR in rDNA repair. Furthermore, TCR has also been implicated in rDNA repair in yeast (16).
The availability of our recently generated, genome-wide, high-resolution repair-mapping procedure with single-nucleotide resolution led us to address rDNA repair using this more direct high-throughput assay (6, 17, 18). However, because rDNA is repetitive, it is not included or accurately annotated in many of the earlier and currently available releases of human and mouse genome assemblies. Hence, we designed a specific computational pipeline to map our recently generated repair data to human and mouse rDNA and thus directly measure repair in the TS and NTS of rDNA genes. Our data show that in human cell lines, there is no preferential repair of UV-induced CPDs in the TS relative to the NTS of rDNA. Whereas the two strands are repaired with comparable efficiencies in WT and CSB mutant cell lines, repair is abolished in both strands in an XPC mutant cell line. Similar analyses of repair of cisplatin-induced DNA damage repair in human cell lines and UV-induced DNA damage repair in a mouse cell line also show that rDNA is repaired by the global repair mechanism and not by TCR.
Results
The XR-seq procedure involves isolation, repair, PCR, and sequencing of the excision products predominantly 26–27 nt in size that are generated during repair. In vivo, the excision products are concurrently formed and degraded; consequently, assessments made at different time points following damage reflect a snapshot of repair occurring at each different repair time point. In this study, we have analyzed repair at relatively early time points, when TCR is prevalent, and we have examined repair of CPDs and Pt-d(GpG), which are readily repaired by TCR as they are relatively poorly repaired by transcription-independent global repair.
Mapping of excision products to the genome requires a reference genome for the species of interest. rDNA genes are present in the genome as tandem repeats with ∼100–200 copies in mice and 200–400 in humans scattered over five chromosomes (chromosomes 12, 15, 16, 18, and 19 in mice and chromosomes 13, 14, 15, 21, and 22 in humans). Unfortunately, due to their repetitive nature, rDNA sequences are either not accurately annotated or not available in the previous or current reference genomes in a form that allows unique mapping of repair reads. In this study, as described under “Experimental procedures,” we utilized a single human or mouse 45S pre-rRNA sequence as references for mapping, and we also used bioinformatic programs for alignment and postalignment processing suited to this approach. This included stringent quality control (QC) procedures to remove any mismatches and gaps (Fig. S1) in the aligned reads due to the short-read nature of XR-seq. Mapping to the control (single-copy DHFR and Dhfr genes) was by analogous procedures.
Repair of CPDs is illustrated in screenshots and bar graphs, such as in Fig. 1, as “normalized repair,” or repair reads per TT site per strand per 20 million total reads. Similarly, repair of Pt-d(GpG) was normalized as reads per GG site per strand per 20 million reads. Because the number of copies of rDNA genes per cell varies and is not known with certainty, comparisons of relative repair in rDNA with other regions, such as DHFR and Dhfr, and comparisons between cell lines are made on a semiquantitative basis.
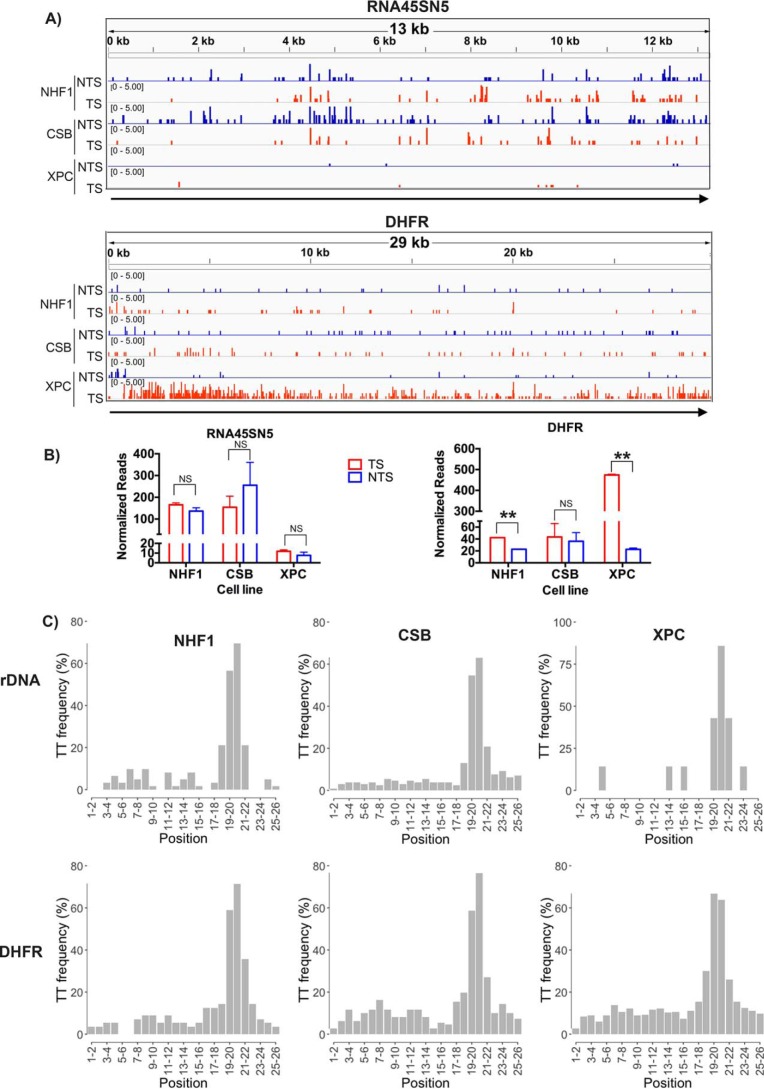
Figure 1.
Strand-specific excision repair of rDNA transcribed by RNA polymerase I and DHFR transcribed by RNA polymerase II in various genetic backgrounds. A, screenshots showing quantitative XR-seq repair reads from a representative experiment. Top, rDNA (RNA45SN5, NR_046235.3); bottom, DHFR. B, quantitative analysis of rDNA and DHFR XR-seq repair from two experiments. NS, not significant; **, p < 0.01, Student's t test. C, frequency of the relevant dinucleotide, T-T, at each position of the 26-nt XR-seq excision fragments mapped to rDNA (RNA45SN5) and DHFR for different cell lines, respectively.
Mapping CPD repair of rDNA in WT, CSB, and XPC mutant human cell lines
Two key properties of TCR in mammalian cells are its dependence on CSB translocase and independence of XPC damage recognition protein. Hence, to determine whether rDNA is subject to TCR, we analyzed the XR-seq data (6, 17, 18) for CPD repair in a normal human fibroblast (NHF1) cell line, in a CSB mutant cell line, which is defective in TCR but carries out normal global repair, and in an XPC mutant cell line, which is known to perform normal TCR but is defective in global repair. Fig. 1 shows the effects of these three genotypes on repair of rDNA genes, presented as screenshots (Fig. 1A, top) and in the form of bar graphs (Fig. 1B, left). As apparent from the figure, the relative levels of repair in the TS and NTS are comparable in WT and CSB mutant cell lines, indicating that rDNA is not subject to TCR. The strongest evidence, however, for this conclusion comes from the XPC mutant; in this cell line, there is no repair in either the TS or the NTS, indicating that the repair of rDNA is entirely dependent on the global repair pathway.
In contrast to the results with rDNA, when the XR-seq data were analyzed in the housekeeping gene DHFR, which is transcribed by RNAPII and has been traditionally used in TCR studies by conventional assays, the results shown in Fig. 1A (bottom) and Fig. 1B (right) were obtained. Specifically, in WT cells, the TS is repaired more efficiently than the NTS, and this preferential repair disappears in the CSB mutant, which cannot perform TCR and is greatly amplified in the XPC mutant because in this mutant there is virtually no repair in the NTS (or in either strand of the genomic regions that are not transcribed).
To ensure the quality of the data set used to generate Fig. 1 (A and B), two criteria were employed to ensure that the selected reads were products of excision repair. These criteria were 1) reads 26 nt in length and 2) a TT dinucleotide at 19–20 or 20–21 nt from the 5′ end. These criteria are based upon the incision sites made by the repair enzyme in vivo (15). Previous analyses of the data sets used for Fig. 1 illustrate an elevated proportion of genome-wide XR-seq reads 26 (and 27) nt in length (17). Similarly, Table 1 shows that the proportion of 26-nt reads mapping to rDNA and to DHFR was elevated, about 20% in all cases. The TT dinucleotide frequency at each position of the 26-nt reads mapped to rDNA or DHFR is shown in Fig. 1C and in Table 1. Thus, from Table 1, one can see that for mapping rDNA in Fig. 1 (A and B), from NHF1 cells, there were 31 oligonucleotides with T-T at 19–20 or 20–21 for each replicate, and from the CSB mutant cells, there were 53 and 77 oligonucleotides with T-T at 19–20 or 20–21 for two replicates; in contrast to these values, from XPC mutant cells, there were only three and four reads at these positions for two replicates. For mapping DHFR, from NHF1 and CSB mutant cells, there were similar reads compared with rDNA; however, from XPC mutant cells, there were more than 200 reads.
Table 1.
Number of XR-seq reads before and after gene alignment, size selection, and TT or GG site selection (quality-controlled mapped reads)
The number of reads for rDNA and DHFR are shown for each replicate across all queried cell lines. In the XR-seq assay, either anti-CPD-DNA or anti-cisplatin-DNA antibodies were used to purify excision products. Quality-controlled mapped reads are 26-nt reads with a TT dinucleotide at 19–20 or 20–21 nt from the 5′ end in the case of CPD repair or, in the case of Pt-(GpG) repair, 26-nt reads with GG dinucleotide 5–6 nt from the 3′ end.
| Cell line | Antibody | Replicate | Total genomic reads | Total genic reads |
Total genic reads (26 nt) |
Quality-controlled mapped reads |
|||
|---|---|---|---|---|---|---|---|---|---|
| rDNA | DHFRa | rDNA | DHFRa | rDNA | DHFRa | ||||
| NHF1 | CPD | 1 | 16,546,883 | 251 | 263 | 46 | 51 | 31 | 29 |
| 2 | 18,254,399 | 256 | 254 | 44 | 52 | 31 | 27 | ||
| CSB | CPD | 1 | 14,871,106 | 326 | 381 | 66 | 88 | 53 | 51 |
| 2 | 30,560,429 | 533 | 429 | 112 | 91 | 77 | 58 | ||
| XPC | CPD | 1 | 22,561,097 | 19 | 1526 | 4 | 364 | 3 | 253 |
| 2 | 21,964,428 | 20 | 1383 | 4 | 322 | 4 | 216 | ||
| GM12878 | cisplatin | 1 | 77,145,576 | 6371 | 1827 | 1201 | 376 | 576 | 194 |
| 2 | 80,424,964 | 6702 | 1817 | 1312 | 345 | 614 | 185 | ||
| MSF | CPD | 1 | 52,129,034 | 1651 | 1548 | 331 | 286 | 89 | 178 |
| 2 | 27,524,063 | 315 | 1504 | 79 | 296 | 51 | 213 | ||
a Dhfr in the mouse.
Repair of cisplatin damage in rDNA
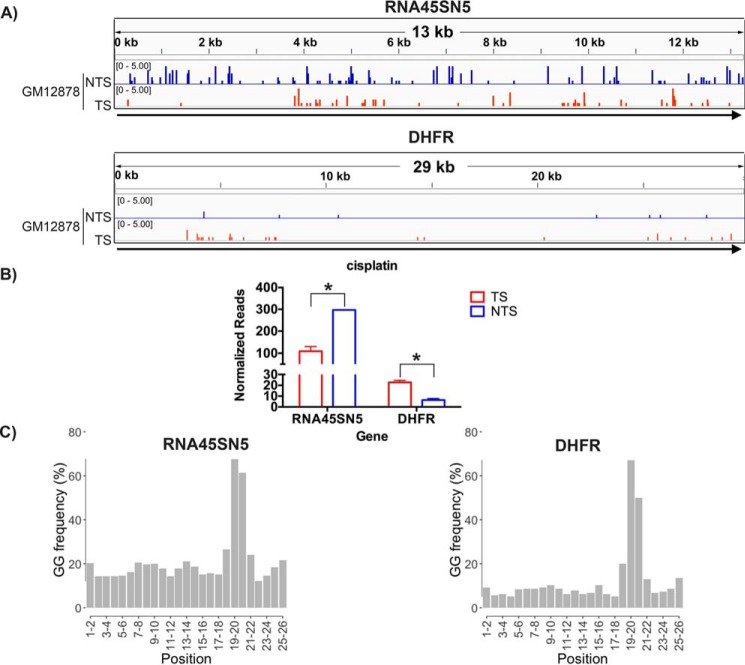
Using XR-seq, we previously reported that in the human lymphocyte cell line GM12878, cisplatin-induced Pt-d(GpG) damage is repaired by TCR of RNAPII-transcribed genes in a manner comparable with CPD repair in WT human cell lines (19). In this study, we mapped the cisplatin XR-seq data from Hu et al. (19) to rDNA. Fig. 2 (A and B) suggests that, in fact, the Pt-d(GpG) damage in rDNA is repaired more efficiently in the NTS than in the TS of rDNA. In contrast, in the RNAPII-transcribed DHFR gene, cisplatin damage in the TS is repaired about 3-fold more efficiently than the NTS, in agreement with the genome-wide data for RNAPII-transcribed genes.
Figure 2.
Repair of cisplatin damage in rDNA and DHFR in the GM12878 human lymphocyte cell line. A, representative screen shots are shown in the top panel for rDNA (RNA45SN5, NR_046235.3) and in the bottom panel for DHFR. B, quantitative analyses of two experiments are plotted in the bar graph. NS, not significant; *, p < 0.05, Student's t test. C, frequency of the relevant dinucleotide, G-G, at each position of the 26-nt XR-seq excision fragments mapped to rDNA (RNA45SN5) and DHFR, respectively.
To ensure the quality of the data set used to generate Fig. 2 (A and B), we used the criteria 1) 26-nt length and 2) GG dinucleotide 5–6 nt from the 3′ end to select XR-seq reads for mapping, based upon the mechanism of excising Pt-(GpG) in vivo (19). The distribution of GG dinucleotide frequencies among the 26-nt mapped reads is illustrated in Fig. 2C, and the total number of genomic and genic reads mapped (with GG 5–6 nt from the 3′ end) is given in Table 1. There are more reads on rDNA compared with DHFR because human rDNA has more GG than DHFR (Fig. S2).
Repair of CPD in rDNA in mouse fibroblasts
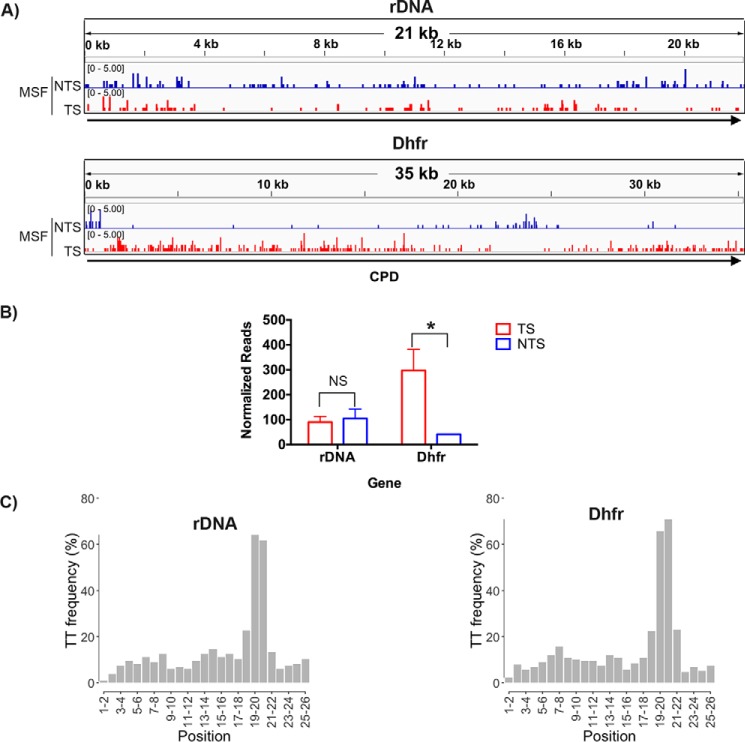
Mouse cell lines are known to exhibit more pronounced TCR compared with human cell lines. Therefore, we performed XR-seq with UV-irradiated mouse skin fibroblasts and analyzed CPD repair in rDNA and in Dhfr as sentinels for TCR in RNAPI and RNAPII-transcribed genes, respectively. Fig. 3 shows that whereas in Dhfr, the TS is repaired ∼7-fold more efficiently than the NTS, in rDNA, both strands are repaired with moderate efficiency and at a comparable level. Thus, even in a rodent cell line in which TCR, when it exists, is amplified relative to human cell lines, there is no detectable TCR of rDNA.
Figure 3.
CPD repair in MSFs. A, representative screen shots are shown for rDNA (M. musculus 45S pre-rRNA gene, X82564.1) in the top panel and Dhfr in the bottom panel. B, quantitative analysis of two experiments are plotted in the bar graph. NS, not significant; *, p < 0.05, Student's t test. C, frequency of the relevant dinucleotide, T-T, at each position of the 26-nt XR-seq excision fragments mapped to rDNA (X82564.1) and Dhfr, respectively.
The criteria for selecting XR-seq reads for mapping the fibroblast data in Fig. 3 (A and B) were the same as described above for the UV repair data in Fig. 1. The TT dinucleotide frequency distribution among 26-nt reads for fibroblasts is shown in Fig. 3C, with numerical values given in Table 1.
Discussion
This study of RNAPI-mediated TCR was encouraged by several factors, including a recent report indicating some similarities in the structures of RNAPI and RNAPII stalled at a CPD (20); another recent report concluding that TCR occurs in mammalian rDNA (15); reports of TCR in yeast rDNA, which is independent of the yeast TCR factor (16); and our recent development of methods to map repair events to the genome (18). By applying novel analytical methods to our data, we find that 1) there is no TCR of rDNA in human or mouse cells; 2) rDNA is repaired in WT and CSB mutant human cells at comparable rates; and 3) rDNA is not repaired in the XPC mutant cell line, indicating that it has the same requirement as global genomic excision repair.
Our results support and complement the findings of studies that utilized the classical T4 endoV/Southern blotting approach (10–14). The XR-seq method is valuable in that it provides high resolution, sensitivity, and specificity. Individual repair events are detected, and detection is sensitive due to a baseline of essentially zero repair. The classical approach, in contrast, detects repair of entire rDNA-containing fragments as a single end point, and repair is measured as an oftentimes small difference in potentially large signals. The classical approach requires an ideal combination of UV dose and restriction fragment size (10), and cells expressing the target gene in multiple copies are often used to obtain a meaningful signal. In some studies of rDNA, in fact, essentially no repair could be detected (10, 11, 13). However, the classical approach, unlike XR-seq, is relatively well-suited to monitor initial levels of DNA damage formation and to compare repair levels in rDNA with other genomic regions.
A potential drawback to both the classical approach and XR-seq is that only about half of the rDNA is transcribed at any one time (21), which would be expected to dilute any TCR signal. However, because XPC mutant cells lack global repair, they provide a very sensitive avenue to detect TCR. Thus, in XPC mutant cells, there is no repair signal from nontranscribed rDNA that could dilute the potential TCR signal from transcribed rDNA, and TCR was not detected in rDNA of XPC mutant cells by either the conventional or the XR-seq repair assays.
The measurement of rDNA repair as recovery of rRNA synthesis following UV (15) is interesting especially in the requirement for CSB but not XPC, which suggests involvement of TCR and not global repair in rDNA repair. However, the end point is indirect and is subject to general responses of damaged cells that could also be influenced by CSB and XPC. As such, these findings are less reliable than findings from the above approaches.
Mechanistically, template but not coding strand lesions block RNAPII (22, 23), and the blocked polymerase is thought to serve as a signal for TCR, as is the case in E. coli (24). The translocases required for TCR, Mfd in E. coli and CSB in eukaryotes, are thought to bind to both the upstream “faces” of their respective stalled RNAP substrates and the template immediately upstream, and then, via translocase action, “push” the polymerase forward (24, 25). In E. coli, this results in dissociation of the polymerase, which remains tethered to the template via Mfd. Mfd, in this template-bound, opened conformation reveals a high-affinity UvrA binding site that targets the transcription-blocking lesion for accelerated repair by the Uvr proteins (24). In vitro, CSB does not dissociate blocked RNAP but “pushes” it forward, even in the presence of the “backtracking” factor TFIIS (26–28). This action is thought to position RNAPII so as to allow repair of the transcription-blocking lesion to occur in the presence of stalled RNAPII at a rate comparable with the repair rate of naked DNA, which is faster than repair of histone-bound chromosomal DNA (23, 28–31). In vivo, RNAPII is dissociated from the template following excision of the transcription-blocking damage (32, 33), perhaps by CSB, or during repair synthesis.
There are several possible reasons for the absence of TCR in rDNA. For one, whereas both RNAPI and RNAPII are blocked by a CPD in the template, significant structural differences in the stalled complexes may explain the inability to promote repair (20). Notably, RNAPI elongation is both blocked and stabilized by damaged TT–RNAPI active site interactions. These active-site interactions do not occur with RNAPII, which adds one more rNTP to the nascent transcript than RNAPI. In addition, it is conceivable that CSB may not interact productively with RNAPI. Also, RNAPI blocked by DNA damage may prevent repair factors from access to the lesion. The latter possibility is consistent with the resilience of rDNA to repair as observed using the conventional repair assay, although stably blocked RNAPI would be expected to specifically hinder repair of the template strand, and this result was not consistently seen. In addition, TCR in rDNA could be impeded by a trailing RNAPI interacting with RNAPI blocked at a lesion. Finally, restricted access of necessary factors to the nucleolar environment could hinder TCR, although repair has been reported to occur at the periphery of the nucleolus (15). Additional work will be needed to determine the cause for no TCR in mammalian rDNA as well as to gain insight into the apparent Rad26 coupling factor–independent TRC of rDNA in yeast.
Experimental procedures
Cell line and culture
WT mouse skin fibroblast cells (34) were grown in Dulbecco's modified Eagle's medium (Life Technologies, Inc., Gaithersburg, MD) supplemented with 10% fetal bovine serum (Gemini, Woodland, CA). Cells were maintained in an incubator at 37 °C under 5% CO2.
UV irradiation and XR-seq
UV irradiation was performed as described previously (17). In brief, cells were grown to about 80% confluence in 20 150-mm dishes. Medium was removed, cells were washed with PBS, and irradiated with 20 J/m2 of UV, and then warm, fresh medium was added, and cells were incubated for 3 h at 37 °C. Cells were then harvested and processed using the XR-seq procedure described previously (17, 18). Here, we used anti-CPD antibody to capture CPD excision products for performing XR-seq.
Sequence and alignment
The NHF1, CSB, and XPC raw data were from Hu et al. (17) and are available on the Gene Expression Omnibus (GEO), accession number GSE67941. The GM12878 (damaged by cisplatin) raw data are from Hu et al. (19) and are available with GEO accession number GSE82213. The GEO accession number of WT mouse skin fibroblast cell (MSF) raw data is GSE121042.
GM12878 and MSF raw data were randomly selected (https://pythonforbiologists.com/randomly-sampling-reads-from-a-fastq-file/)4 to obtain 25–40 million reads from raw fastq file and then were aligned to rDNA, DHFR, and Dhfr.
Reconstruction of canonical rDNA genes, DHFR, and Dhfr sequences for humans and mice
For reference genomes, we started with hg38 for human samples and mm10 for mouse samples. However, both human and mouse rDNA consist of multiple clusters present on human chromosomes 13, 14, 15, 21, and 22, and mouse chromosomes 12, 15, 16, 18, and 19. Furthermore, each cluster contains multiple 45S rDNA repeat units that have extremely low polymorphisms and vary in number among individuals and chromosomes. As such, human and mouse rDNA are either poorly annotated with chromosomal location unknown or not included in the current assemblies. Therefore, we rebuilt the reference for the human and mouse ribosomal genes, respectively, using the canonical sequences downloaded from NCBI Nucleotide Database. Specifically, we used a 13,357-bp-long reference sequence for the human 45S preribosomal N5 (RNA45SN5, accession number NR_046235) and a 22,118-bp-long reference sequence for the mouse 45S pre-rRNA gene (accession number X82564) to build the reference fasta files for humans and mice. As a control and sanity check, we also included the gene DHFR for humans and Dhfr for mice, downloaded from Ensembl, in the reconstructed reference genome as an independent molecule. For all subsequent analysis, the reconstructed reference genome was used.
Bioinformatic pipeline, data normalization, and visualization
Here we outline the bioinformatic pipeline and statistical analyses that were used in this paper for the analysis of DNA repair of ribosomal genes by XR-seq. The same bioinformatic preprocessing, quality control, data normalization, and data visualization procedures are simultaneously performed on all sequences (rDNA, DHFR, and Dhfr) across all samples. These two genes went through the same analysis pipeline, yielding the same QC metrics yet distinct results and patterns shown in Fig. 1.
For sequencing data generated by XR-seq, cutadapt (35) was used to trim reads with adaptor sequence TGGAATTCTCGGGTGCCAAGGAACTCCAGTNNNNNNACGATCTCGTATGCCGTCTTCTGCTTG at the 3′ end and discard untrimmed reads (17). BWA-backtrack (36) was used for single-end read alignment, followed by Picard tools (http://broadinstitute.github.io/picard/)4 for filtering, sorting, deduplication, and indexing. We used bamtools (37) to remove reads with any mismatches/gaps (Fig. S1), which were prevalent due to the short lengths of the excised oligonucleotides and thus the trimmed reads. The damage caused by cisplatin and UV treatment is repaired in the XR-seq protocol (17); therefore, reads with mismatches should be removed. Additional postalignment QC procedures were adopted using the R packages Rsamtools (http://bioconductor.org/packages/Rsamtools/)4 and Biostrings (http://bioconductor.org/packages/Biostrings/).4 Specifically, we kept only reads that (i) had mapping quality greater than 20, (ii) were of length 22 to 30 bp, and (iii) had guanine-guanine (GG) or thymine-thymine (TT) dinucleotide sequence 5–8 bp upstream from the 3′ end of the reads for XR-seq of cisplatin- and UV-induced damage, respectively.
For data normalization, we scaled the observed total reads for the ribosomal gene as well as DHFR by a sample-specific library size factor and a gene- and strand-specific number of sites with GG or TT dinucleotides. These strand-specific total number of reads after normalization were plotted in the bar plots in Figs. 1–3.
For data visualization, we scaled each read by a sample-specific library size factor and used the R package rtracklayer (http://bioconductor.org/packages/rtracklayer/)4 to generate wig files across all samples, which were further loaded into the Integrative Genomics Viewer (38) with results shown in the screenshots in Figs. 1–3. All scripts and codes for the bioinformatic and statistical analyses can be found at https://github.com/yuchaojiang/damage_repair/tree/master/ribo.4
Author contributions
Y. Y. and A. S. designed research; J. H., C. P. S., W. L., and A. Y. performed research; Y. Y. and Y. J. analyzed data; Y. J. built the bioinformatics pipeline; and Y. Y., C. P. S., Y. J., and A. S. wrote the paper.
Supplementary Material
Acknowledgment
We thank John B. Hogenesch (Cincinnati Children's Hospital Medical Center) for useful comments.
This work was supported by National Institutes of Health Grants GM118102 and ES027255 (to A. S.) and 5 P01 CA142538-08 (to Y. J.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1 and S2.
The array data have been deposited in the NCBI Gene Expression Omnibus and are accessible through GEO Series accession numbers GSE67941, GSE82213, and GSE121042.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- CPD
- cyclobutane pyrimidine dimer
- TCR
- transcription-coupled repair
- XR-seq
- excision repair sequencing
- DHFR
- dihydrofolate reductase
- TS
- transcribed (template) strand
- NTS
- nontranscribed (coding) strand
- rRNA and rDNA
- ribosomal RNA and DNA, respectively
- RNAP
- RNA polymerase
- nt
- nucleotide(s)
- QC
- quality control
- GEO
- Gene Expression Omnibus
- MSF
- mouse skin fibroblast cell.
References
- 1. Wood R. D. (1997) Nucleotide excision repair in mammalian cells. J. Biol. Chem. 272, 23465–23468 10.1074/jbc.272.38.23465 [DOI] [PubMed] [Google Scholar]
- 2. Sancar A. (1996) DNA excision repair. Annu. Rev. Biochem. 65, 43–81 10.1146/annurev.bi.65.070196.000355 [DOI] [PubMed] [Google Scholar]
- 3. Sancar A. (2016) Mechanisms of DNA Repair by Photolyase and Excision Nuclease (Nobel Lecture). Angew. Chem. Int. Ed. Engl. 55, 8502–8527 10.1002/anie.201601524 [DOI] [PubMed] [Google Scholar]
- 4. Hanawalt P. C., and Spivak G. (2008) Transcription-coupled DNA repair: two decades of progress and surprises. Nat. Rev. Mol. Cell Biol. 9, 958–970 10.1038/nrm2549 [DOI] [PubMed] [Google Scholar]
- 5. Adebali O., Sancar A., and Selby C. P. (2017) Mfd translocase is necessary and sufficient for transcription-coupled repair in Escherichia coli. J. Biol. Chem. 292, 18386–18391 10.1074/jbc.C117.818807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hu J., Selby C. P., Adar S., Adebali O., and Sancar A. (2017) Molecular mechanisms and genomic maps of DNA excision repair in Escherichia coli and humans. J. Biol. Chem. 292, 15588–15597 10.1074/jbc.R117.807453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Selby C. P., and Sancar A. (1993) Molecular mechanism of transcription-repair coupling. Science 260, 53–58 10.1126/science.8465200 [DOI] [PubMed] [Google Scholar]
- 8. Venema J., Mullenders L. H., Natarajan A. T., van Zeeland A. A., and Mayne L. V. (1990) The genetic defect in Cockayne syndrome is associated with a defect in repair of UV-induced DNA damage in transcriptionally active DNA. Proc. Natl. Acad. Sci. U.S.A. 87, 4707–4711 10.1073/pnas.87.12.4707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Venema J., van Hoffen A., Karcagi V., Natarajan A. T., van Zeeland A. A., and Mullenders L. H. (1991) Xeroderma pigmentosum complementation group C cells remove pyrimidine dimers selectively from the transcribed strand of active genes. Mol. Cell. Biol. 11, 4128–4134 10.1128/MCB.11.8.4128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Christians F. C., and Hanawalt P. C. (1993) Lack of transcription-coupled repair in mammalian ribosomal RNA genes. Biochemistry 32, 10512–10518 10.1021/bi00090a030 [DOI] [PubMed] [Google Scholar]
- 11. Fritz L. K., and Smerdon M. J. (1995) Repair of UV damage in actively transcribed ribosomal genes. Biochemistry 34, 13117–13124 10.1021/bi00040a024 [DOI] [PubMed] [Google Scholar]
- 12. Christians F. C., and Hanawalt P. C. (1994) Repair in ribosomal-RNA genes is deficient in xeroderma-pigmentosum group-C and in Cockaynes-syndrome cells. Mutat. Res. 323, 179–187 10.1016/0165-7992(94)90031-0 [DOI] [PubMed] [Google Scholar]
- 13. Stevnsner T., May A., Petersen L. N., Larminat F., Pirsel M., and Bohr V. A. (1993) Repair of ribosomal RNA genes in hamster cells after UV irradiation, or treatment with cisplatin or alkylating agents. Carcinogenesis 14, 1591–1596 10.1093/carcin/14.8.1591 [DOI] [PubMed] [Google Scholar]
- 14. Fritz L. K., Suquet C., and Smerdon M. J. (1996) Strand breaks are repaired efficiently in human ribosomal genes. J. Biol. Chem. 271, 12972–12976 10.1074/jbc.271.22.12972 [DOI] [PubMed] [Google Scholar]
- 15. Daniel L., Cerutti E., Donnio L. M., Nonnekens J., Carrat C., Zahova S., Mari P. O., and Giglia-Mari G. (2018) Mechanistic insights in transcription-coupled nucleotide excision repair of ribosomal DNA. Proc. Natl. Acad. Sci. U.S.A. 115, E6770–E6779 10.1073/pnas.1716581115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Charton R., Guintini L., Peyresaubes F., and Conconi A. (2015) Repair of UV induced DNA lesions in ribosomal gene chromatin and the role of “Odd” RNA polymerases (I and III). DNA Repair (Amst.) 36, 49–58 10.1016/j.dnarep.2015.09.007 [DOI] [PubMed] [Google Scholar]
- 17. Hu J., Adar S., Selby C. P., Lieb J. D., and Sancar A. (2015) Genome-wide analysis of human global and transcription-coupled excision repair of UV damage at single-nucleotide resolution. Genes Dev. 29, 948–960 10.1101/gad.261271.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hu J., Li W., Adebali O., Yang Y., Oztas O., Selby C. P., and Sancar A. (2019) Genome-wide mapping of nucleotide excision repair with XR-seq. Nat Protoc. 1, 248–282 10.1038/s41596-018-0093-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hu J., Lieb J. D., Sancar A., and Adar S. (2016) Cisplatin DNA damage and repair maps of the human genome at single-nucleotide resolution. Proc. Natl. Acad. Sci. U.S.A. 113, 11507–11512 10.1073/pnas.1614430113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sanz-Murillo M., Xu J., Belogurov G. A., Calvo O., Gil-Carton D., Moreno-Morcillo M., Wang D., and Fernández-Tornero C. (2018) Structural basis of RNA polymerase I stalling at UV light-induced DNA damage. Proc. Natl. Acad. Sci. U.S.A. 115, 8972–8977 10.1073/pnas.1802626115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Xu B., Li H., Perry J. M., Singh V. P., Unruh J., Yu Z., Zakari M., McDowell W., Li L., and Gerton J. L. (2017) Ribosomal DNA copy number loss and sequence variation in cancer. PLoS Genet. 13, e1006771 10.1371/journal.pgen.1006771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Donahue B. A., Yin S., Taylor J. S., Reines D., and Hanawalt P. C. (1994) Transcript cleavage by RNA polymerase II arrested by a cyclobutane pyrimidine dimer in the DNA template. Proc. Natl. Acad. Sci. U.S.A. 91, 8502–8506 10.1073/pnas.91.18.8502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Selby C. P., Drapkin R., Reinberg D., and Sancar A. (1997) RNA polymerase II stalled at a thymine dimer: footprint and effect on excision repair. Nucleic Acids Res. 25, 787–793 10.1093/nar/25.4.787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Selby C. P. (2017) Mfd protein and transcription-repair coupling in Escherichia coli. Photochem. Photobiol. 93, 280–295 10.1111/php.12675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Xu J., Lahiri I., Wang W., Wier A., Cianfrocco M. A., Chong J., Hare A. A., Dervan P. B., DiMaio F., Leschziner A. E., and Wang D. (2017) Structural basis for the initiation of eukaryotic transcription-coupled DNA repair. Nature 551, 653–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Selby C. P., and Sancar A. (1997) Human transcription-repair coupling factor CSB/ERCC6 is a DNA-stimulated ATPase but is not a helicase and does not disrupt the ternary transcription complex of stalled RNA polymerase II. J. Biol. Chem. 272, 1885–1890 10.1074/jbc.272.3.1885 [DOI] [PubMed] [Google Scholar]
- 27. Selby C. P., and Sancar A. (1997) Cockayne syndrome group B protein enhances elongation by RNA polymerase II. Proc. Natl. Acad. Sci. U.S.A. 94, 11205–11209 10.1073/pnas.94.21.11205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lainé J. P., and Egly J. M. (2006) Initiation of DNA repair mediated by a stalled RNA polymerase IIO. EMBO J. 25, 387–397 10.1038/sj.emboj.7600933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tremeau-Bravard A., Riedl T., Egly J. M., and Dahmus M. E. (2004) Fate of RNA polymerase II stalled at a cisplatin lesion. J. Biol. Chem. 279, 7751–7759 10.1074/jbc.M309853200 [DOI] [PubMed] [Google Scholar]
- 30. Hara R., Mo J., and Sancar A. (2000) DNA damage in the nucleosome core is refractory to repair by human excision nuclease. Mol. Cell. Biol. 20, 9173–9181 10.1128/MCB.20.24.9173-9181.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liu X. (2015) In vitro chromatin templates to study nucleotide excision repair. DNA Repair (Amst.) 36, 68–76 10.1016/j.dnarep.2015.09.026 [DOI] [PubMed] [Google Scholar]
- 32. Chiou Y. Y., Hu J., Sancar A., and Selby C. P. (2018) RNA polymerase II is released from the DNA template during transcription-coupled repair in mammalian cells. J. Biol. Chem. 293, 2476–2486 10.1074/jbc.RA117.000971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Andrade-Lima L. C., Veloso A., Paulsen M. T., Menck C. F., and Ljungman M. (2015) DNA repair and recovery of RNA synthesis following exposure to ultraviolet light are delayed in long genes. Nucleic Acids Res. 43, 2744–2756 10.1093/nar/gkv148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gauger M. A., and Sancar A. (2005) Cryptochrome, circadian cycle, cell cycle checkpoints, and cancer. Cancer Res. 65, 6828–6834 10.1158/0008-5472.CAN-05-1119 [DOI] [PubMed] [Google Scholar]
- 35. Martin M. (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- 36. Li H., and Durbin R. (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Barnett D. W., Garrison E. K., Quinlan A. R., Strömberg M. P., and Marth G. T. (2011) BamTools: a C++ API and toolkit for analyzing and managing BAM files. Bioinformatics 27, 1691–1692 10.1093/bioinformatics/btr174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Thorvaldsdóttir H., Robinson J. T., and Mesirov J. P. (2013) Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 14, 178–192 10.1093/bib/bbs017 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.