Abstract
Siderophores make iron accessible under iron-limited conditions and play a crucial role in the survival of microorganisms. Because of their remarkable metal-scavenging properties and ease in crossing cellular envelopes, siderophores hold great potential in biotechnological applications, raising the need for a deeper knowledge of the molecular mechanisms underpinning the siderophore pathway. Here, we report the structural and functional characterization of a siderophore-interacting protein from the marine bacterium Shewanella frigidimarina NCIBM400 (SfSIP). SfSIP is a flavin-containing ferric-siderophore reductase with FAD- and NAD(P)H-binding domains that have high homology with other characterized SIPs. However, we found here that it mechanistically departs from what has been described for this family of proteins. Unlike other FAD-containing SIPs, SfSIP did not discriminate between NADH and NADPH. Furthermore, SfSIP required the presence of the Fe2+-scavenger, ferrozine, to use NAD(P)H to drive the reduction of Shewanella-produced hydroxamate ferric-siderophores. Additionally, this is the first SIP reported that also uses a ferredoxin as electron donor, and in contrast to NAD(P)H, its utilization did not require the mediation of ferrozine, and electron transfer occurred at fast rates. Finally, FAD oxidation was thermodynamically coupled to deprotonation at physiological pH values, enhancing the solubility of ferrous iron. On the basis of these results and the location of the SfSIP gene downstream of a sequence for putative binding of aerobic respiration control protein A (ArcA), we propose that SfSIP contributes an additional layer of regulation that maintains cellular iron homeostasis according to environmental cues of oxygen availability and cellular iron demand.
Keywords: flavoprotein, iron, nuclear magnetic resonance (NMR), X-ray crystallography, enzyme kinetics, electron transfer, ferredoxin, ferrozine, Shewanella, siderophore
Introduction
The metabolic versatility of the Shewanella genus enables these bacteria to thrive in a wide variety of iron-limited environments ranging from the ocean floor all the way to the infection of human hosts (1, 2). Shewanella are well-known spoilers of food products, notorious fish pathogens, and Shewanella oneidensis has been extensively studied for its remarkable ability of performing extracellular electron transfer (1, 2). Overall, this environmental versatility of Shewanella is highly dependent on the ability of these organisms to produce an abundance of iron-rich proteins, mainly multiheme c-type cytochromes (3). Shewanella frigidimarina, is a psychrophilic bacterial species isolated from Antarctic costal marine environments, in particular, from biofilms (4, 5). The multiheme c-type cytochromes of S. frigidimarina have been extensively characterized. However, being a marine microorganism, access to iron poses a permanent challenge (6–8).
Iron demand together with scarcity in the ocean limits microbial life, and all the respective biogeochemical processes essential for ocean productivity (9, 10). This high need for iron requires robust strategies for iron uptake while avoiding an uncontrolled oversupply that would be dangerous to organisms that live in oxic–anoxic transition zones (11). However, very limited knowledge exists regarding the iron-uptake strategies used by Shewanella in general, and by S. frigidimarina in particular. Over a decade ago, various Shewanella strains were identified as capable producers of soluble or surface-associated metal chelating compounds designated as siderophores (from the Greek “iron carrier”). This is a strategy of iron uptake that is widespread across the tree of life, but poorly explored in the Shewanella genus (12, 13).
Siderophores are low molecular weight compounds with high affinity for ferric-iron (13). These compounds play an essential role in the growth and virulence of many microorganisms. Since their discovery, over 500 distinct siderophores have been identified (14). This diversity reflects a veritable “arms race” to efficiently capture iron from the environment. Indeed, the combination of siderophores produced by an organism is crucial for bacterial competition as well as for the outcome of pathogenesis (15). The siderophore pathway involves the production and export of apo-siderophores (no iron), and their reuptake as ferric-siderophores (iron-loaded) so that iron is released for utilization in the cell metabolism (13). Within the siderophore pathway, the molecular mechanisms for iron release from the ferric-siderophore complexes remain poorly characterized, but ferric-siderophore reduction appears to be the more widespread mechanism.
In bacteria, ferric-siderophore reduction can occur in the cytosol and involves the superfamily of siderophore-interacting proteins composed of two distinct families: the ferric-siderophore reductase (FSRs)2 family, and the siderophore-interacting protein (SIPs) family (16, 17). SIPs can be distinguished in two broad groups: one that generates cytosolic reduced free flavins that subsequently reduce the ferric-siderophores, and a second group of SIPs that contain stably attached flavins and use NADH and/or NADPH as reducing agents. The latter includes the first reported SIP, the ViuB from Vibrio cholerae, and also other examples such as Escherichia coli YqjH and FscN from the Gram-positive bacterium Thermobifida fusca (16–20). Additionally, this group of SIPs can be further divided into two subgroups: subgroup I, which utilize NADH as electron donor and subgroup II, which utilize NADPH (16). This division is based upon the differences found in the C-terminal and N-terminal sequences: subgroup I proteins (e.g. ViuB, FscN) contain prominent C-terminal α-helix elements, whereas subgroup II contain N-terminal sequence extensions and insertions (e.g. YqhJ) (16, 17).
The SIP superfamily, which includes both SIPs and FSRs is widespread across several bacterial genera, however, very few of these proteins have been thoroughly characterized (YqhJ from E. coli, FscN from T. fusca, and Iruo from Staphylococcus aureus) and thus, the specific intracellular fate of the iron-loaded siderophores is among the least understood steps of the siderophore pathway at the molecular level (16, 17, 21). Across the different genomes in the Shewanella genus, some species contain genes that code for SIPs only (e.g. S. frigidimarina and Shewanella morhuae), others code for FSRs only (e.g. Shewanella oneidensis and Shewanella algae), whereas a few species contain genes that code for both SIPs and FSRs (e.g. Shewanella putrefaciens and Shewanella baltica). However, no knowledge exists on how these proteins are utilized, their importance and role for iron uptake and regulation. Based on sequence similarity with ViuB from V. cholerae, and with the structurally characterized S. putrefaciens homologue (SpSIP PBD code 2GPJ), a SIP was identified in S. frigidimarina NCIM400 (SfSIP) (22, 23). This work reports its characterization.
Results
SfSIP belongs to the ViuB subgroup: The proposed NADH-utilizing proteins
The SFRI_RS12295 gene from S. frigidimarina NCIM400 was inserted into the pETBlue-1 vector (Novagen) and SfSIP was overexpressed and purified by ion-exchange chromatography, as described elsewhere (23). The SDS-PAGE analysis revealed that the purified yellow protein migrated as a single band at 30 kDa, as expected from theoretical calculations based on amino acid and co-factor composition. N-terminal sequence analysis of the purified protein confirmed the identity of SfSIP and the UV-visible spectrum presented the typical features of an oxidized flavoprotein with absorption peaks at 387 and 470 nm (23).
The two subgroups of SIPs can be distinguished based on the length of their C terminus (16, 17). Subgroup I SIPs includes ViuB and FscN from T. fusca, and Subgroup II SIPs includes YqjH from E. coli. Comparative sequence analysis of SfSIP with its homologues (Fig. S1), revealed that the SfSIP contains a “long” C terminus with prominent α-helix elements. For this reason, it is possible to assign SfSIP to Subgroup I SIPs, which utilize NADH as an electron donor to reduce ferric-siderophores (16, 17). These results were further investigated by analyzing the three-dimensional structure of SfSIP.
Structural characterization of SfSIP
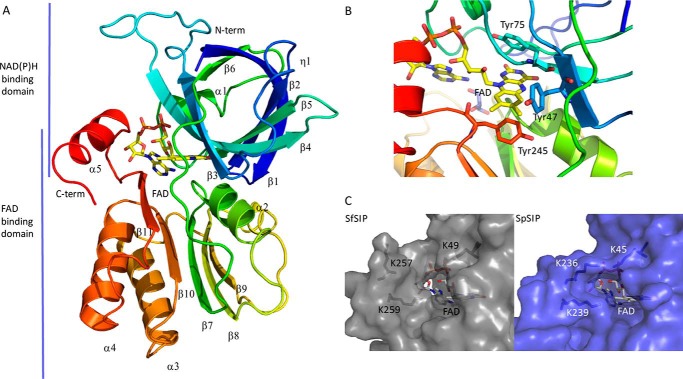
The crystal structure of SfSIP was determined to 1.15 Å resolution and deposited in PDB with accession number 6GEH. The structure reveals two domains, the FAD- and NAD(P)H-binding domains (Fig. 1A), as observed for previously determined structures of SIPs from S. putrefaciens (PDB code 2GPJ) and T. fusca (PDB code 4YHB), which share 32 and 28% sequence identity with SfSIP, respectively (17). The electron density is well-defined from residues Ser-9 to Leu-264, except for residues Pro-120 to Met-123, which are facing the solvent and are in the loop that connects the FAD- and NAD(P)H-binding domains, between strands β6 and β7 (Fig. 1A). The FAD-binding domain is composed of five antiparallel β-strands (β7–β11) with two short α-helices (α2 and η3) connecting the long loops. The FAD shows a planar conformation, and thus is oxidized as expected from the color and the spectrum of the sample used for crystallization. The FAD cofactor is well-conserved compared with both structures of YqhJ and FscN and this is stabilized through aromatic stacking interactions of the isoalloxazine ring by Tyr-47, Tyr-75, and Tyr-245 and hydrogen bonds with residues Asp-90 (backbone O), Val-92 (backbone N), and Thr-76 (backbone N). The negatively charged phosphate groups from FAD are also stabilized through hydrogen bonds with Arg-73 (NH2 and NE), His-96 (side chain NE2), Ala-100 (backbone N), Thr-101 (backbone N), and Ala-252 (N backbone). In addition, the ribose ring forms hydrogen bonds with the backbone of the protein, namely with Thr-250 (N and O backbone), Asn-247 (N backbone), and Lys-247 (O backbone), and with two water molecules (Fig. S2) (24). The NAD(P)H-binding domain consists of six antiparallel β-strands (β1–β6) and two short α-helices (α1 and η1). This has a β1–α1–β2 architecture consistent with the Rossmann-fold. The C-terminal of SfSIP contains an α-helix as predicted, confirming that SfSIP belongs to the subgroup I of SIPs. Furthermore, it has been proposed that a triad of basic amino acid residues (Lys-45, Lys-236, and Lys-239 based in the primary sequence of S. putrefaciens) forms a ferric-siderophore–binding pocket. In SfSIP, both comparative sequence analysis (Fig. S1) and structural determination of SfSIP revealed that it possesses a lysine residue in position 49, and two lysine residues in positions 257 and 259 (Fig. 1C). These could serve a similar role as predicted for the potential ferric-siderophore–binding pocket formed by this triad of basic amino acid residues (16). However, Lys-257 is placed in a different position compared with the corresponding lysine in SpSIP (Lys-236), further away from the FAD ring. This further opening of the pocket together with the absence of other surface locations that provide access to the FAD cofactor suggest that this pocket is also the location of NAD(P)H binding. So far no crystal structure has been reported for SIPs in complex with ferric-siderophores or NAD(P)H. In this work the same situation occurred, even though the crystallization was performed in the presence of both molecules. Therefore, the binding site of these substrates in SfSIP remains open to speculation.
Figure 1.
Structural characterization of SIP. A, overall structure of SIP; B, close-up of the FAD pocket and stacking residues (Tyr-47, Tyr-75, and Tyr-245); C, putative NAD(P)H-binding pocket: comparison of the C-terminal helix of SIPs from S. frigidimarina (SfSIP) and S. putrefaciens (SpSIP). Lys-236 from SpSIP seems to make a bridge across the pocket, whereas Lys-257 in SfSIP is more distant from the isoalloxazine ring and pointing outwards from the pocket making more space available for substrate binding (ferric-siderophore and/or electron donors).
SfSIP performs proton-coupled electron transfer
Proteins with a FAD as redox center can couple one- and two-electron transfer reactions with proton transfer. In SfSIP, this ability was investigated using protein film voltammetry (PFV), resulting in well-defined voltammetric responses (Fig. S3A). The width at half-height and midpoint potentials of the voltammetric signals are reported in Table 1 (25).
Table 1.
Redox properties of SfSIP at various pH values
| pH | Width at half-height peak | E(0) |
|---|---|---|
| mV | ||
| 5.4 | 86 ± 6 | −130 |
| 6.3 | 96 ± 1 | −201 |
| 7.0 | 94 ± 6 | −228 |
| 7.8 | 102 ± 6 | −286 |
| 8.5 | 110 ± 3 | −320 |
The data reveal that SfSIP presents a redox–Bohr effect (Fig. S3B and Table 1). The simulation of pH dependence of the reduction potentials (Fig. S3B) shows that the pKa values must be lower than 4.5 for pKaox and higher than 10 for pKared, outside of the pH range that could be tested (26). These results mean that in the physiological range of S. frigidimarina the release of electrons by SfSIP is accompanied by the release of protons. Because both electrons and protons originate from the isoalloxazine ring, this coupling provides a localized pH drop that enhances iron solubility. The reduction potential of SfSIP changes by 57 ± 2 mV for every pH unit and the half-height width of the anodic and cathodic signals was close to the theoretical value of 90 mV (Table 1) predicted for a Nernstian equilibrium for a proton coupled-one electron transfer (3.53 RT/nF). Trumpet plots were fitted (Fig. S4) using the Butler-Volmer model for 1 electron transfer, and proton transfer is not the limiting process at any pH value tested (27, 28). Furthermore, at pH 7 the reduction of SfSIP by NADH and NADPH (−320 mV versus SHE) should be feasible, however, both of these nucleotides perform a 2-electron step reduction, and this was not observed in PFV measurements.
SfSIP, a ferric siderophore reductase
To test SfSIP as a ferric siderophore reductase, siderophores, the putative substrates of SfSIP were produced and characterized (29). Three siderophores produced by Shewanella were obtained: a 4.3 mg of pure sample of bisucaberin, a 5.2 mg sample of of avaroferrin containing small traces of bisucaberin, and a sample of putrebactin containing small traces of the previous two siderophores. The addition of iron to the apo-siderophores resulted in an immediate transition from colorless to an end point that ranged from dark red to bright orange depending on the pH. These ferric-siderophores (BIS, ferric-bisucaberin; PUT, ferric-putrebactin; AVA, ferric-avaroferrin) native to Shewanella and commercially acquired ferric-ferrioxamine E (FER) presented a similar UV-visible spectrum with maximum absorption at ∼430 nm, in agreement with the literature (30).
Cyclic voltammograms of the three ferric-siderophores from Shewanella (Fig. S5) are complex and dominated by a pair of reduction and oxidation peaks. These peaks portray the speciation found in tetradentate siderophores, where two species are more prevalent in solution for each ferric-siderophore at neutral pH, one with lower reduction potential, and another with a higher reduction potential (Table 2) (31, 32). Of the measured reduction potentials, the species with positive potential can be spontaneously reduced by SfSIP, whereas the reduction of the species with negative potential is thermodynamically an uphill process.
Table 2.
Midpoint reduction potentials of ferric-siderophores used in this study
| Siderophore | pH, temperature | E1/2 (mV vs SHE) | Reference |
|---|---|---|---|
| Avaroferrin | 7.25, 25 °C | −295 and +25 | This study |
| Bisucaberin | 7.30, 25 °C | −311 and +35 | This study |
| Ferrioxamine E | 10.5, NDa | −478 | 30 |
| Putrebactin | 7.27, 25 °C | −309 and +33 | This study |
a ND, not determined.
SfSIP reduces hydroxamate siderophores produced by Shewanella
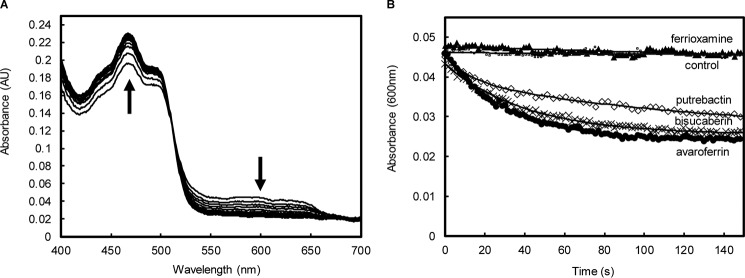
Having shown that SfSIP has adequate thermodynamic properties to reduce ferric-siderophores, its capability to perform the reaction was tested by stopped-flow kinetic assays. SfSIP in the semiquinone state (SfSIPsemi) was prepared using sodium dithionite (Fig. S6). Upon mixing of SfSIPsemi with ferric-siderophores (AVA, BIS, and PUT) absorption spectra changes showed an increase at 470 nm and a decrease at 600 nm (Fig. 2A). This is consistent with the transfer of one electron from the semiquinone flavin to the ferric-siderophore yielding fully oxidized SfSIP and Fe2+. Ferric-siderophore reduction was observed for all ferric-siderophores except for FER (Fig. 2B). The rate constant of ferric-siderophore reduction was determined (Table 3) from the kinetic trace at 600 nm to avoid signal interference from the ferric-siderophores. All kinetic traces were monophasic except for PUT, which was best fit by a biphasic exponential. This might be a consequence of the presence of small amounts of the other two siderophores (AVA and BIS) in this sample, adding extra minor features to the kinetic trace. All siderophores presented rate constants of similar magnitude in agreement with their reduction potentials (Tables 2 and 3).
Figure 2.
SfSIP-mediated reduction of ferric-siderophores. A, absorption spectra changes during 150 s after mixing SfSIPsemi with ferric-siderophore avaroferrin. The up arrow indicates the increase in absorbance at 470 nm and the down arrow indicates a decrease in the absorbance at 600 nm. B, time-resolved traces of SfSIP-mediated reduction of ferric-siderophores (600 nm): ▴, ferrioxamine E; ●, avaroferrin; ♢, putrebactin; ×, bisucaberin; □, control (apo-avaroferrin) and the respective fittings (solid lines).
Table 3.
Rate constants of ferric-siderophore reduction by SfSIP in the semiquinone state
| Siderophore | Rate constant (Kobs) |
|---|---|
| Avaroferrin | 0.033 ± 0.001 s−1 |
| Bisucaberin | 0.025 ± 0.001 s−1 |
| Ferrioxamine E | |
| Putrebactin | 0.08 ± 0.001 and 0.007 ± 0.005 s−1 |
Exploring the electron donors of SfSIP
Once the function of SfSIP was confirmed as a ferric-siderophore reductase, the interaction of SfSIP with the putative electron donors was tested.
The interaction of SfSIP with NAD(P)H was explored via 31P NMR spectroscopy. In solution, the two pyrophosphate phosphorus atoms of NADH and NADPH have identical chemical environments giving rise to only one resonance at −11.25 ppm in the 31P NMR spectra. Upon addition of increasing amounts of SfSIP to a solution of either NADH or NADPH, a pattern change was observed in the 31P NMR spectrum where the chemical shifts of the two pyrophosphate phosphorus atoms split from a singlet (−11.25 ppm) to two doublets centered at −11.33 and −11.65 ppm indicating that the local environment of the two phosphate groups becomes different upon interaction with the protein (Fig. 3). Furthermore, given that signals for the free and bound states coexist in the spectra, binding occurs in the slow-exchange regime. Using the relative peak intensities, the concentrations of free and bound species were calculated, allowing the determination of the dissociation constant. NADH and NADPH presented a KD of 20 ± 5 and 21 ± 6 μm, respectively. KD values in the micromolar range imply a relatively weak binding, characteristic of electron transfer reactions (33).
Figure 3.
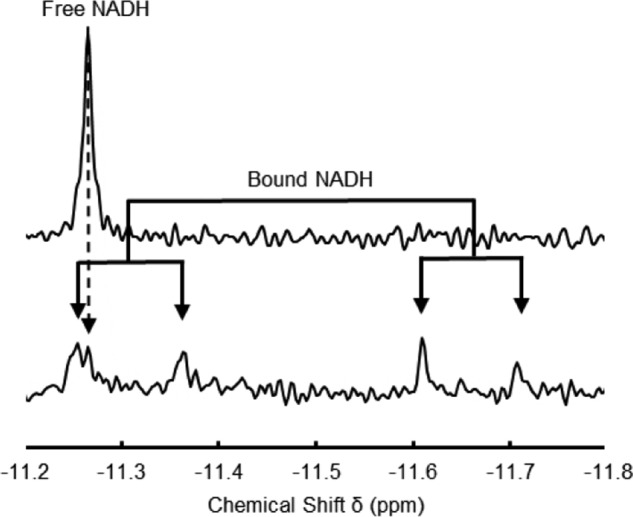
Proton-decoupled 31P NMR spectra. Top spectrum corresponds to 100 μm NADH and bottom spectrum corresponds to a NADH/SfSIP ratio of 0.58.
NADPH presented an extra resonance at 3.47 ppm, which corresponds to the 2′ phosphorous atom. This signal is unperturbed by binding, indicating that the chemical environment of the 2′ phosphorous atom in the bound form remains similar to what is found in the free form. This result strongly suggests that binding of NADPH to SfSIP occurs through the nicotinamide side of the molecule.
Despite the clear evidence for the interaction of NAD(P)H with SfSIP, no direct reduction of the enzyme by these compounds was observed, nor consumption of NAD(P)H (Fig. S7). This excludes the possibility of a ping-pong mechanism in SfSIP, in contrast to what was proposed for the YqjH protein in E. coli and the FscN protein from T. fusca (16, 17). In this context, other biologically relevant electron donors for SfSIP were considered. It is reported in the literature that ferric-reductases of the FSR family such as FchR from Bacillus halodurans can utilize other electron donors such as ferredoxin (34). The genome of S. frigidimarina codes for several small ferredoxins. A ferredoxin from Clostridium tetanomorphum, which presents a high sequence identity (39%) with a 4Fe-4S ferredoxin encoded in the genome of S. frigidimarina (sequence ID:WP_059746450.1) was used to explore the hypothesis of ferredoxin serving as electron donor for SfSIP.
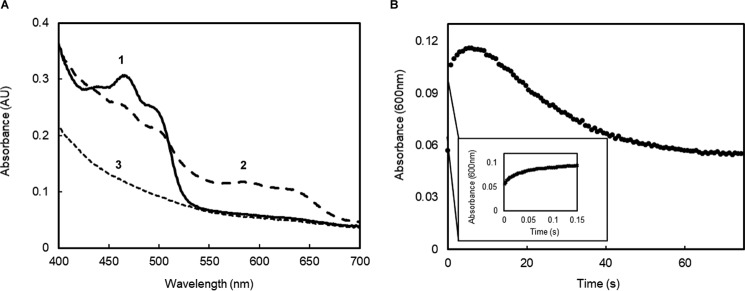
The reduction of SfSIP by ferredoxin revealed a two-step process characterized by an initial fast decrease in absorbance at 470 nm followed by a slower decrease (Fig. 4). Concomitantly, spectral changes at 600 nm revealed a fast increase in absorbance followed by a slow decrease. Altogether, these changes represent the formation of the semiquinone state followed by the transition to the fully reduced state of the flavin (FADH2). The fact that SfSIP can be fully reduced by ferredoxin shows that it is not restricted to be reduced to a FAD-semiquinone intermediate as proposed for the reduction of IruO from S. aureus by NADPH (21).
Figure 4.
Reduction of SfSIP using a ferredoxin. A, absorption spectral changes after mixing ferredoxin with SfSIP. Numbers in the figure indicate significant spectral features: 1) initial spectrum showing the typical features of oxidized SfSIP; 2) spectrum after 2 s, showing the typical features of the formation of the semi-quinone state of SfSIP; 3) final spectrum at 75 s, representing fully reduced SfSIP. B, time-resolved kinetic trace of SfSIP reduction followed at 600 nm.
Measuring SfSIP-mediated ferrous iron production
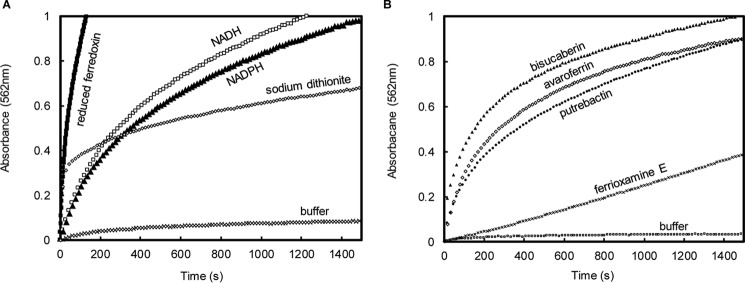
To assess the production of ferrous iron by SfSIP using different electron donors and different ferric-siderophores, ferric reductase assays were performed in the presence of the ferrous iron chelator ferrozine. Absorption spectra changes show a gradual increase in absorbance at 562 nm indicating the production of ferrous-ferrozine (Fig. 5). In the absence of SfSIP, no production of ferrous iron was observed (Fig. 5A). Ferredoxin and dithionite were able to drive the reduction of ferric-siderophores via SfSIP with ferredoxin presenting the fastest rates for the formation of ferrous iron. Surprisingly, the presence of ferrozine enabled NADH and NADPH to also drive the ferric reductase activity of SfSIP. Furthermore, NADH and NADPH show similar rates of ferrous iron production in agreement with their similar binding constants. The ferrous iron production activity of SfSIP using ferric-avaroferrin was compared for the different electron donors. Using ferredoxin as electron donor the value obtained (55 nmol of Fe2+ min− 1 mg−1) compares well with the reduction of fuscachelin by FscN of T. fusca using NADH (40 nmol of Fe2+ min−1 mg−1). By contrast, the values of the ferrous iron production activity of SfSIP using sodium dithionite, NADH, and NADPH (18, 15, and 14 nmol of Fe2+ min−1 mg−1, respectively) are more similar to the values reported for the ferrous iron production activity of YqhJ using NADPH (22 nmol of Fe2+ min−1 mg−1).
Figure 5.
Time-resolved formation of the Fe(II)-ferrozine complex by SfSIP. A, experiments performed using ferric-avaroferrin as iron-source and different electron donors. B, experiments performed using NADPH as electron donor with different ferric siderophore.
Additionally, in the presence of ferrozine, SfSIP could reduce all ferric-siderophores tested, including ferrioxamine E, which has a more negative reduction potential (Fig. 5B). The formation of ferrous iron was biphasic for all ferric-siderophores except ferrioxamine E, which is monophasic. These data tie with the voltammetric results, suggesting the existence of two iron-coordinated species in solution (Table 2), and reveal that the presence of a Fe2+-scavenger enables SfSIP to reduce iron forms that would otherwise be inaccessible.
Discussion
The characterization of SfSIP revealed several novelties relative to what was previously reported in the literature regarding this class of enzymes. First, it revealed that the length of the C terminus per se is not a good proxy for deducing the electron donor of a SIP. Indeed, although sequence comparison and structural analysis placed SfSIP in the SIP family subgroup I that uses NADH, this is the first reported case of a FAD-containing SIP where discrimination between these two electron donors does not occur. The structure of this enzyme suggests that the unprecedented placement of Lys-257 away from the entrance of the NAD(P)H-binding pocket may be responsible for this behavior. No other site in the structure of SfSIP provides access to the FAD cofactor and the placement of Lys-257 makes the triad basic amino acid residue pocket larger compared with the other SIPs from the same subgroup. Second, the observation that SfSIP can use ferredoxin as electron donor is also a first for the SIP family of enzymes. These observations provided two new insights on the reactivity of SIPs. On one hand, the electron donor repertoire of SfSIP is larger than previously reported for other flavin containing SIPs. On the other hand, ferredoxin can directly reduce SfSIP in marked contrast with NAD(P)H, which require the presence of an Fe2+-scavenger. Ferredoxins have two major differences with respect to NAD(P)H, both of which could contribute to different reactivity. Ferredoxins have more negative reduction potentials, and thus a stronger driving force (35). Furthermore, NAD(P)H operate as obligatory two-electron donors, unlike ferredoxins where the 4Fe-4S clusters operate as single electron donors.
Interestingly, the presence of ferrozine in the reaction mixture is sufficient to enable NAD(P)H to function as electron donors. This observation is reminiscent of the redox facilitated ligand exchange mechanism proposed for the reductive release of Fe2+ from low potential siderophores in the presence of a seemingly overwhelming unfavorable driving force (14). Ferrozine is known to have a “pulling” effect on reactions involving the formation of Fe2+ and caveats for the interpretation of results have been widely disseminated in the literature (36). Nonetheless, in the context of the data reported here, the presence of ferrozine may have provided a glimpse on the role and regulation of SfSIP in the context of iron uptake by Shewanella. Although ferrozine is not a physiologically relevant ligand, it provides an in vitro mimetic of naturally occurring ferrous iron ligands such as porphyrins or apo-proteins that promote concurrent ferric-siderophore reduction and ferrous iron uptake (36, 37). Shewanella are notorious for maintaining a high level of intracellular iron content but the process is by no means unregulated. This situation would be extraordinarily dangerous for these organisms that are found in environmental niches in oxic–anoxic transition zones, due to the participation of Fe2+ in the production of reactive species through the Haber–Weiss cycle (14). Proteins of the siderophore pathway such as the archetypal SIP, ViuB, are typically under the control of the Fur regulon (22). However, in S. frigidimarina, the Arc (aerobic respiration control) signal transduction system appears to be the best candidate for this role, given the fact that the SfSIP gene is found downstream of a putative ArcA-binding sequence (38, 39). This genetic context together with the functional characterization of SfSIP allow us to formulate a hypothesis for the overall regulation of its activity in vivo. Expression of SfSIP is likely controlled by the Arc transcriptional regulator, based on environmental cues indicating changes of oxygen availability. When oxygen levels are extremely low, cellular iron demand is increased and expression of SfSIP is up-regulated contributing to iron uptake without competing for its use because it is a flavoprotein. Furthermore, when cells find themselves in a reducing environment where synthesis of numerous iron containing proteins is stimulated in Shewanella, the pool of reduced ferredoxins is increased because of the favorable redox environment (39, 40). Reduced ferredoxins donate electrons to SfSIP, boosting the availability of intracellular iron given that they do not require the presence of Fe2+-acceptors and enable SfSIP to reduce ferric siderophore species of lower reduction potential. In contrast, when oxygen levels increase, the redox conditions are less favorable to the maintenance of a pool of reduced ferredoxins and the risk of producing reactive species is higher. Therefore, NAD(P)H can drive the reduction of ferric-siderophores only when a downstream acceptor is ready to receive the ferrous iron. This ensures that metabolic reducing power is only diverted to this activity when it is required without compromising cell integrity.
In conclusion, the novel aspects of SfSIP's reactivity revealed in this work offer opportunities to interfere with the iron metabolism of Shewanella at the level of SfSIP and its homologues. However, further knowledge is necessary to understand if the function and regulation of SIPs is conserved among the different species of Shewanella. Shewanella are notorious for having multiple redundant metabolic pathways that may hinder the identification of a clear-cut phenotype for SfSIP and its homologues (41, 42). However, studies using animal models have shown that knock-out of SIP genes greatly diminishes the virulence of pathogenic bacteria, revealing the importance of these proteins for survival fitness (43). This makes ferric reductases such as SfSIP, bona fide targets to disturb iron homeostasis in a wide diversity of microbial pathogens, contributing in this way, to avert losses derived from food spoilage and infections caused by Shewanella (44).
Experimental procedures
Expression and purification of SfSIP
SFRI_RS12295 gene from S. frigidimarina NCIM400 was inserted into pETBlue-1 expression vector (Novagen). Transformed E. coli Tuner (DE3) pLacI cells were grown for the overexpression of SIP in Luria-Bertani (LB) medium, supplemented with autoinduction solutions and selective antibiotics as previously described (23). Cells were grown at 30 °C for 32 h before collection, and the supernatant was purified by ion exchange chromatography as previously reported (23). The purity of SfSIP was assessed using SDS-PAGE analysis and UV-visible spectroscopy, and the SfSIP's sequence was confirmed via N-terminal sequencing.
Structure determination of SfSIP
SfSIP was crystallized using 22% PEG 3350 and 0.22 m magnesium formate dihydrate as precipitant by the hanging drop vapor diffusion method, where 1 μl of precipitant was mixed with 1 μl of a 30 mg/ml of protein solution and equilibrated against a 500-μl reservoir. Crystals were harvested and flash-cooled in liquid nitrogen directly from the crystallization drop, and diffraction data were collected at 100 K to a resolution of 1.35 Å on Diamond Light Source (DLS) beamline I04 (Didcot, UK) (23). The images were processed with xia2 (45), which makes use of XDS (46) and the CCP4 suite (47) for integration and conversion of integrated intensities to structure factors. The structure was solved by molecular replacement using PHASER in the CCP4 suite (47) and a previously determined SIP crystal structure (PDB 4YHB) (17) as phasing model to a resolution of 1.4 Å. The model was corrected and completed with COOT (48) after an initial refinement using REFMAC5 in the CCP4 suite (49). However, the subsequent refinement led to unsatisfactory values of R and R-free of 22.3 and 22.9%, respectively, and it was decided to use data collected from another crystal for structure determination and refinement.
The second crystal was obtained under the same crystallization conditions as the first, with the difference that 1 mm ferric-siderophore (bisucaberin), dissolved in DMSO, was added to the protein solution prior to setting up the crystallization experiment, and it was soaked for 1 min in a mother liquor solution to which 200 μm NADPH (dissolved in 20 mm Tris-HCl, pH 7.6, 150 mm NaCl) was added prior to flash cooling in liquid nitrogen. Diffraction data at 100 K were collected to 1.15 Å resolution on DLS beamline I04 and the images were likewise processed with xia2. The data collection and processing statistics are listed in Table 4. The structure was solved using molecular replacement and the previously determined structure of SfSIP as a template. Structure refinement was performed using PHENIX (50). Throughout the refinement, the model was periodically checked and corrected with COOT against 2|Fo| − |Fc| and |Fo| − |Fc| electron-density maps. Solvent molecules were added automatically by PHENIX and validated by inspection of electron-density maps in COOT (48). In the final refinement cycles, hydrogen atoms were included in calculated positions with the PHENIX READYSET tool and anisotropic displacement parameters were refined for nonhydrogen protein atoms. The final values of R and R-free (51) were 14.5 and 17.4%, respectively, and the maximum likelihood estimate of the overall coordinate error was 0.1 Å. The model stereochemical quality was analyzed with MOLPROBITY (52) and there are no outliers in the Ramachandran (54) ψ,φ plot. The final refinement statistics are included in Table 4. The coordinates and structure factors have been submitted to the Protein Data Bank in Europe (53) with accession codes 6GEH and r6GEHsf. Pictures were produced using PyMOL.
Table 4.
Data collection and refinement statistics
| Native | |
|---|---|
| Beamline and detector | DLS I04, PILATUS 6 M |
| Space group | P 21 |
| Unit cell parameters (Å, °) | a = 38.10, b = 77.75, c = 44.92, β = 109.64 |
| Resolution (Å) | 28.62–1.15 (1.19–1.15) |
| Unique reflections | 86,403 (8,002) |
| Completeness (%) | 99.0 (91.6) |
| Multiplicity | 5.0 (3.6) |
| Rmerge (%)a | 3.63 (44.6) |
| Rmeas (%)b | 4.05 (48.6) |
| CC1/2 | 1 (0.795) |
| 〈I/σ(I)〉 | 18.4 (2.4) |
| Wilson plot B (Å2) | 12.0 |
| Refinement | |
| Resolution limits (Å) | 28.62–1.15 (1.19–1.15) |
| R-factor (%)c | 14.5 (24.7) |
| No. reflections | 86396 |
| Free R-factor (%)d | 17.4 (27.6) |
| Model composition | |
| Non-hydrogen protein atoms | 2,274 |
| Solvent molecules | 431 |
| FAD ligand | 105 |
| Model r.m.s. deviations from ideality | |
| Bond lengths (Å) | 0.009 |
| Bond angles (°) | 1.38 |
| Model completeness and validation | |
| Regions omitted | 1–8 and 20–23 |
| Mean B values (Å2)e | |
| Protein | 16.9 |
| Solvent molecules | 30.2 |
| FAD ligand | 18.6 |
| Ramachandran plot | |
| Most favored regions (%) | 100.0 |
aRmerge = merging R-factor, (Σhkl Σi |Ii(hkl) − 〈I(hkl)〉|)/(Σhkl ΣiI(hkl)) × 100%.
b Rmeas = redundancy independent R-factor, Σh [Nhkl/(Nhkl−1)]1/2 Σi|Ii(hkl) − 〈I(hkl) 〉|/Σhkl ΣiIi(hkl) × 100%, where I is the observed intensity, 〈I〉 is the average intensity of multiple observations from symmetry-related reflections, and Nhkl is their multiplicity (58).
c R-factor = Σhkl‖Fo|−|Fc‖/Σhkl|Fo|, where |Fo| and |Fc| are the observed and calculated structure factor amplitudes, respectively.
d Free R-factor is the cross-validation R-factor computed from a randomly chosen subset of 5% of the total number of reflections, which were not used during the refinement.
e Calculated from the equivalent isotropic B values.
PFV experiments
Experiments were performed on a pyrolytic graphite edge (PGE) electrode purchased from IJ Cambria Scientific at 25 °C using a three-electrode electrochemical cell configuration with the PGE (working electrode), a platinum wire (counter electrode), and an Ag/AgCl (3 m KCl) (reference electrode) inside a Coy anaerobic chamber using an electrochemical analyzer from CHI Instruments controlled by the manufacturer's software (version 10.12). Oxygen levels were kept stable at ∼8 ppm. The electrode was cleaned (3 m nitric acid and distilled water) and freshly polished before every experiment. SfSIP was immobilized in the freshly polished PGE electrode by pipetting 3 μl of SIP in 20 mm potassium phosphate buffer, 100 mm KCl, pH 7.6. To guarantee protein immobilization, the electrode was left to dry for ∼15 min. Excess and/or unattached protein was removed by rinsing the electrode with distilled water. The electrode was then immersed in 2 ml of a mixed buffer solution containing 5 mm HEPES, 5 mm CHES, and 5 mm MES at the following pH values: 5.5, 6, 7.8, and 8.5. For each pH, voltammograms were acquired at the following scan rates (mV s−1): 10, 13, 17, 21, 28, 36, 41, 48, 60, 78, 100, 130, 170, 210, 280, 360, 410, 480, 600, 780, 1,000, 1,300, 1,700, 2,100, 2,800, 3,600, 4,100, 4,800, 6,000, 7800, 9,100, and 10,000 to ensure uniform data point coverage in a logarithmic scale plot. After each experiment, the solution was removed, and the pH was measured. These experiments were performed in duplicate by cleaning the electrode and repeating the procedure. Using the program Qsoas, the background current was subtracted for every voltammogram and all data were analyzed with Excel. The trumpet plots were fitted using the Butler-Volmer model using Jellyfit with the following parameters: n = 1, potential step 1 × 10−6, and temperature 25 °C; for every data potential a standard deviation (S.D.) of 0.005 was assumed (25, 27). Potentials were reported in mV versus the SHE by addition of 210 mV to those measured (54).
Production of ferric-siderophores
A deep-sea sediment metagenomic library was constructed and the mbs gene cluster (mbsA-D) was cloned as previously reported (29). As it tested positive for siderophore activity with the chrome azurol S assay, the metagenome-derived mbs cluster clone was cultured for 3 days at 37 °C in LB medium supplemented with isopropyl 1-thio-β-d-galactopyranoside (100 μm) and chloramphenicol (30 μg/ml). Centrifuge-clarified supernatant was extracted using a C18 resin, then eluted with a stepwise aqueous-MeOH gradient system. Fractions containing siderophores were further purified by Sephadex LH-20 gel filtration. Chrome azurol S active fractions were combined and further purified by two steps of a C18 HPLC using aqueous-MeOH (containing 0.2% acetic acid) gradient system to afford purified bisucaberin, putrebactin, and avaroferrin (29). The purity and nature of siderophore-containing samples was assessed by LC-MS analysis and NMR spectroscopy.
Characterization of ferric siderophores avaroferrin, bisucaberin, putrebactin, and ferrioxamine E
FER (1,12,23-Trihydroxy-1,6,12,17,17,23,28-hexaazacyclotritriacontane-2,5, 13,16,24,27-hexone iron(III) complex) was purchased from Sigma. A 2 mm stock solution was prepared in 20 mm potassium phosphate, 100 mm KCl buffer, pH 7.6.
The apo-siderophores (AVA, BIS, and PUT) were dissolved in DMSO and iron-containing siderophores were prepared for every experiment in working stocks of 500 μm in 20 mm phosphate buffer, 100 mm KCl, pH 7.6. Each 500 μm ferric-siderophore solution was prepared from the starting stock plus equivalent amounts of FeCl3, resulting in an iron-siderophore molar ratio of 1:1.
Cyclic voltammetry experiments were performed at 25 °C using a three-electrode electrochemical cell configuration with the PGE (working electrode), a platinum wire (counter electrode), and an Ag/AgCl (3 m KCl) (reference electrode) inside a Coy anaerobic chamber using an electrochemical analyzer from CHI Instruments controlled by the manufacturer's software (version 10.12). Oxygen levels were kept stable at ∼8 ppm. In each experiment, the freshly polished PGE electrode was first immersed in buffer to draw a baseline of the capacitive current and then immersed in the 500 μm ferric-siderophore–containing solution. Scans were acquired at 100 mV s−1 in triplicate. All data were analyzed using Excel and QSoas (25). Potentials were determined using the average of the oxidation and reduction peaks and reported in millivolts and with respect to the SHE by addition of 210 mV to those measured (54).
NMR interaction studies
NAD(P)H and SfSIP were prepared in 20 mm Tris-HCl, 100 mm KCl, pH 8, containing 10% D2O (99.9 atom %). Samples of 100 μm NAD(P)H were titrated against increasing concentrations of SfSIP. To determine the concentration of free and bound species triplicate experiments using a molar ratio of 0.58 (NAD(P)H:SIP) were performed. Using the standard Bruker pulse program “zgdc,” one-dimensional proton-decoupled 31P spectra were acquired with 1024 scans, d1 of 1.3 s at 25 °C on a Bruker Avance II 500 MHz equipped with a SEX probe for 31P detection.
Collected spectra were visualized and analyzed using TopSpin 3.5 (Bruker). The concentration of free and bound species was determined from the relative intensity of each peak and the dissociation constant (KD) was calculated using the following equation.
| (Eq. 1) |
Kinetic studies
The kinetic experiments were performed in HI-TECH Scientific Stopped-flow equipment (SF-61DX2) installed inside an anaerobic glove box (Mbraun MB150-GI). The temperature of the drive syringes and mixing chamber was maintained at 25 °C using a water bath, and the pH controlled with 20 mm potassium phosphate buffer, 100 mm KCl, pH 7.6. The time course of the reactions was monitored using a photodiode array. Solutions were prepared inside the anaerobic chamber with degassed water and all experiments were performed in triplicate. Data were analyzed using Excel and fits were performed and analyzed using Kinetics Studio version 2.32 (TgK Scientific).
Reduction of SfSIP with NAD(P)H, sodium dithionite, and ferredoxin
Experiments were performed at multiple time scales and at different concentrations for each reducing agent. The reduction of SfSIP with NAD(P)H was attempted using 0.5 mm NAD(P)H and also using a large excess of NAD(P)H with 10 μm SIP with spectra acquisition for a total 4000 s (67 min).
Reduction of SfSIP with sodium dithionite was performed using 50 μm sodium dithionite with 10 μm SIP. Reduction rates were obtained from experiments at the 6000-s time scale with a total of 200 spectra acquired.
The purity of the ferredoxin from C. tetanomorphum was confirmed by SDS-PAGE (15%). To avoid direct reduction of SIP with sodium dithionite the reduction of ferredoxin was carried out in a 1:1 molar ratio and the mixture was left to react for 10 min before initializing the experiments of reduced ferredoxin against SIP. The stock solutions of sodium dithionite were quantified by measuring the absorbance at 315 nm using the molar extinction coefficient of 8043 m−1 cm−1 (55). Reduction of 10 μm SfSIP was performed using 10 μm reduced ferredoxin.
Reduction of ferric siderophores
Reduction of ferric siderophores (AVA, BIS, PUT, and FER) by reduced SfSIP was performed after reducing SfSIP with sodium dithionite in a 1:1 molar ratio. The reduction of ferric-siderophores was performed using 10 μm SfSIP with 50 μm ferric-siderophores in the stopped-flow apparatus. The reduction rate constants were obtained from fitting the kinetic traces at 600 nm in a 150-s time scale (Table S1).
The ferrozine assay
The ferrozine assay monitored the formation of ferrous iron using an iron chelator with high affinity for Fe2+ and ferrozine, purchased from Fluka. The assay was performed with the following components: the Fe2+ chelator (0.25 mm ferrozine), the putative siderophore reductase (5 μm SIP), a reducing agent (0.25 mm NAD(P)H, 25 μm sodium dithionite, or 5 μm reduced ferredoxin), and 50 μm for each ferric-siderophore (AVA, BIS, PUT, and FER). The formation of Fe2+-ferrozine complexes was analyzed by measuring the absorbance at 562 nm (56, 57). Each experiment was performed in triplicate and controls were performed containing all the previously described components except for SIP.
Author contributions
I. B. T., J. M. S., B. M. F., T. C., P. M. M., E. M., and R. O. L. formal analysis; I. B. T., J. M. S., B. M. F., T. C., P. M. M., E. M., and R. O. L. investigation; I. B. T., P. M. M., E. M., and R. O. L. writing-original draft; I. B. T. and R. O. L. writing-review and editing; M. F. resources; R. O. L. conceptualization; R. O. L. supervision.
Supplementary Material
Acknowledgments
We thank Isabel Pacheco for help in the purification of SfSIP and Dr. Américo Duarte for providing ferredoxin. We also thank all members of the Inorganic Biochemistry and NMR Laboratory for discussions and comments and feedback regarding the preparation of the manuscript. The NMR experiments were performed at CERMAX (Centro de Ressonância Magnetica António Xavier) and was financially supported by Project LISBOA-01-0145-FEDER-007660 (Microbiologia Molecular, Estrutural e Celular) funded by FEDER funds through COMPETE2020-Programa Operacional Competitividade e Internacionalização (POCI), the European Community's Seventh Framework Program (FP7/2007–2013)) under Grant Agreement 283570 (BioStruct-X). Beamtime at I04 at the Diamond Light Source and assistance from the beamline staff during the synchrotron data collections are gratefully acknowledged.
This work was supported by the European Union's Horizon 2020 research and innovation program under Grant 810856, National Funds Grant ERA-MBT/0003/2014, Ph.D. fellowship PD/BD/135187/2017 (to I. B. T.), and postdoctoral fellowships SFRH/BPD/94050/2013 and SFRH/BPD/93164/2013 (to E. M. and B. F. M.), through the FCT-Fundação para a Ciência e a Tecnologia. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Table S1 and Figs. S1–S7.
The atomic coordinates and structure factors (code 6GEH) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- FSRs
- ferric siderophore reductase family
- SfSIP
- siderophore-interacting protein from S. frigidimarina
- SIP
- siderophore-interacting protein
- ArcA
- aerobic respiration control protein A
- ViuB
- vibriobactin utilization protein
- YqjH
- siderophore-interacting protein from E. coli
- FscN
- siderophore-interacting protein from the fuscachelin siderophore gene cluster
- PDB
- Protein Data Bank
- BIS
- ferric-bisucaberin
- AVA
- ferric-avaroferrin
- PUT
- ferric-putrebactin
- FER
- ferric-ferrioxamine E
- LB
- Luria-Bertani medium
- SHE
- standard hydrogen electrode
- FchR
- ferric citrate and hydroxamate reductase
- PFV
- protein film voltammetry
- DLS
- Diamond Light Source
- PGE
- pyrolytic graphite edge
- CHES
- 2-(cyclohexylamino)ethanesulfonic acid.
References
- 1. Janda J. M., and Abbott S. L. (2014) The genus Shewanella: from the briny depths below to human pathogen. Crit. Rev. Microbiol. 40, 293–312 [DOI] [PubMed] [Google Scholar]
- 2. Janda J. M. (2014) Shewanella: a marine pathogen as an emerging cause of human disease. Clin. Microbiol. Newsl. 36, 25–29 10.1016/j.clinmicnews.2014.01.006 [DOI] [Google Scholar]
- 3. Ghosal D., Omelchenko M. V., Gaidamakova E. K., Matrosova V. Y., Vasilenko A., Venkateswaran A., Zhai M., Kostandarithes H. M., Brim H., Makarova K. S., Wackett L. P., Fredrickson J. K., and Daly M. J. (2005) How radiation kills cells: survival of Deinococcus radiodurans and Shewanella oneidensis under oxidative stress. FEMS Microbiol. Rev. 29, 361–375 10.1016/j.femsre.2004.12.007 [DOI] [PubMed] [Google Scholar]
- 4. Bowman J. P., McCammon S. A., Nichols D. S., Skerratt J. H., Rea S. M., Nichols P. D., and McMeekin T. A. (1997) Shewanella gelidimarina sp. nov., and Shewanella frigidimarina sp. nov., novel Antarctic species with the ability to produce eicosapentaenoic acid (20:5 ω3) and grow anaerobically by dissimilatory Fe(III) reduction. Int. J. Syst. Bacteriol. 47, 1040–1047 10.1099/00207713-47-4-1040 [DOI] [PubMed] [Google Scholar]
- 5. Linares D., Jean N., Van Overtvelt P., Ouidir T., Hardouin J., Blache Y., and Molmeret M. (2016) The marine bacteria Shewanella frigidimarina NCIMB400 upregulates the type VI secretion system during early biofilm formation. Environ. Microbiol. Rep. 8, 110–121 10.1111/1758-2229.12358 [DOI] [PubMed] [Google Scholar]
- 6. Martin J. H., and Michael Gordon R. (1988) Northeast Pacific iron distributions in relation to phytoplankton productivity. Deep Sea Res. Part A Oceanogr. Res. Pap. 35, 177–196 10.1016/0198-0149(88)90035-0 [DOI] [Google Scholar]
- 7. Fonseca B. M., Paquete C. M., Salgueiro C. A., and Louro R. O. (2012) The role of intramolecular interactions in the functional control of multiheme cytochromes c. FEBS Lett. 586, 504–509 10.1016/j.febslet.2011.08.019 [DOI] [PubMed] [Google Scholar]
- 8. Martin J. H. (1990) Glacial-interglacial CO2 change: the iron hypothesis. Paleocenography 5, 1–13 10.1029/PA005i001p00001 [DOI] [Google Scholar]
- 9. Moore J. K., and Doney S. C. (2007) Iron availability limits the ocean nitrogen inventory stabilizing feedbacks between marine denitrification and nitrogen fixation. Global Biogeochem. Cycles 21, 1–12 [Google Scholar]
- 10. Moore C. M., Mills M. M., Arrigo K. R., Berman-Frank I., Bopp L., Boyd P. W., Galbraith E. D., Geider R. J., Guieu C., Jaccard S. L., Jickells T. D., La Roche J., Lenton T. M., Mahowald N. M., Marañón E., et al. (2013) Processes and patterns of oceanic nutrient limitation. Nat. Geosci. 6, 701–710 10.1038/ngeo1765 [DOI] [Google Scholar]
- 11. Yang Y., Harris D. P., Luo F., Xiong W., Joachimiak M., Wu L., Dehal P., Jacobsen J., Yang Z., Palumbo A. V., Arkin A. P., and Zhou J. (2009) Snapshot of iron response in Shewanella oneidensis by gene network reconstruction. BMC Genomics 10, 131 10.1186/1471-2164-10-131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Koebnik R. (2005) TonB-dependent trans-envelope signalling: the exception or the rule? Trends Microbiol. 13, 343–347 10.1016/j.tim.2005.06.005 [DOI] [PubMed] [Google Scholar]
- 13. Miethke M., and Marahiel M. A. (2007) Siderophore-based iron acquisition and pathogen control. Microbiol. Mol. Biol. Rev. 71, 413–451 10.1128/MMBR.00012-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Boukhalfa H., and Crumbliss A. L. (2002) Chemical aspects of siderophore mediated iron transport. BioMetals 15, 325–339 10.1023/A:1020218608266 [DOI] [PubMed] [Google Scholar]
- 15. Holden V. I., and Bachman M. A. (2015) Diverging roles of bacterial siderophores during infection. Metallomics 7, 986–995 10.1039/C4MT00333K [DOI] [PubMed] [Google Scholar]
- 16. Miethke M., Hou J., and Marahiel M. A. (2011) The siderophore-interacting protein YqjH acts as a ferric reductase in different iron assimilation pathways of Escherichia coli. Biochemistry 50, 10951–10964 10.1021/bi201517h [DOI] [PubMed] [Google Scholar]
- 17. Li K., Chen W. H., and Bruner S. D. (2015) Structure and mechanism of the siderophore-interacting protein from the fuscachelin gene cluster of Thermobifida fusca. Biochemistry 54, 3989–4000 10.1021/acs.biochem.5b00354 [DOI] [PubMed] [Google Scholar]
- 18. Sedlácek V., van Spanning R. J., and Kučcra I. (2009) Ferric reductase A is essential for effective iron acquisition in Paracoccus denitrificans. Microbiology 155, 1294–1301 10.1099/mic.0.022715-0 [DOI] [PubMed] [Google Scholar]
- 19. Fontecave M., Covès J., and Pierre J. L. (1994) Ferric reductases or flavin reductases? Biometals 7, 3–8 [DOI] [PubMed] [Google Scholar]
- 20. Coves J., and Fontecave M. (1993) Reduction and mobilization of iron by a NAD(P)H: flavin oxidoreductase from Escherichia coli. Eur. J. Biochem. 211, 635–641 10.1111/j.1432-1033.1993.tb17591.x [DOI] [PubMed] [Google Scholar]
- 21. Kobylarz M. J., Heieis G. A., Loutet S. A., and Murphy M. E. P. (2017) Iron uptake oxidoreductase (IruO) uses a flavin adenine dinucleotide semiquinone intermediate for iron-siderophore reduction. ACS Chem. Biol. 12, 1778–1786 10.1021/acschembio.7b00203 [DOI] [PubMed] [Google Scholar]
- 22. Butterton J. R., and Calderwood S. B. (1994) Identification, cloning, and sequencing of a gene required for ferric vibriobactin utilization by Vibrio cholerae. J. Bacteriol. 176, 5631–5638 10.1128/jb.176.18.5631-5638.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Trindade I. B., Fonseca B. M., Matias P. M., Louro R. O., and Moe E. (2016) A putative siderophore-interacting protein from the marine bacterium Shewanella frigidimarina NCIMB 400: cloning, expression, purification, crystallization and X-ray diffraction analysis. Acta Crystallogr. Sect. Struct. Biol. Commun. 72, 667–671 10.1107/S2053230X16011419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wallace A. C., Laskowski R. A., and Thornton J. M. (1995) Ligplot: a program to generate schematic diagrams of protein ligand interactions. Protein Eng. 8, 127–134 10.1093/protein/8.2.127 [DOI] [PubMed] [Google Scholar]
- 25. Fourmond V. (2016) QSoas: a versatile software for data analysis. Anal. Chem. 88, 5050–5052 10.1021/acs.analchem.6b00224 [DOI] [PubMed] [Google Scholar]
- 26. Louro R. O., Catarino T., Salgueiro C. A., LeGall J., and Xavier A. (1996) V Redox-Bohr effect in the tetrahaem cytochrome c3 from Desulfovibrio vulgaris: a model for energy transduction mechanisms. J. Biol. Inorg. Chem. 1, 34–38 10.1007/s007750050020 [DOI] [Google Scholar]
- 27. Jeuken L. J. C., McEvoy J. P., and Armstrong F. A. (2002) Insights into gated electron-transfer kinetics at the electrode-protein interface: a square wave voltammetry study of the blue copper protein azurin. J. Phys. Chem. B 106, 2304–2313 10.1021/jp0134291 [DOI] [Google Scholar]
- 28. Armstrong F. A., Heering H. A., Hirst J., Armstrong F. A., Heering H. A., and Hirst J. (1997) Reactions of complex metalloproteins studied by protein-film voltammetry. Chem. Soc. Rev. 26, 169–173 26528734 [Google Scholar]
- 29. Fujita M. J., and Sakai R. (2014) Production of avaroferrin and putrebactin by heterologous expression of a deep-sea metagenomic DNA. Mar. Drugs. 12, 4799–4809 10.3390/md12094799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rütschlin S., Gunesch S., and Böttcher T. (2017) One enzyme, three metabolites: Shewanella algae controls siderophore production via the cellular substrate pool. Cell Chem. Biol. 24, 598–604.e10 10.1016/j.chembiol.2017.03.017 [DOI] [PubMed] [Google Scholar]
- 31. Hou Z., Raymond K. N., O'Sullivan B., Esker T. W., and Nishio T. (1998) A preorganized siderophore: thermodynamic and structural characterization of alcaligin and bisucaberin, microbial macrocyclic dihydroxamate chelating agents. Inorg. Chem. 37, 6630–6637 10.1021/ic9810182 [DOI] [PubMed] [Google Scholar]
- 32. Spasojevic I., Armstrong S. K., Brickman T. J., and Crumbliss A. L. (1999) Electrochemical behavior of the Fe(III) complexes of the cyclic hydroxamate siderophores alcaligin and desferrioxamine E. Inorg. Chem. 38, 449–454 10.1021/ic980635n [DOI] [PubMed] [Google Scholar]
- 33. Díaz-quintana A., Cruz-gallardo I., Rosa M. A. De, and Díaz-moreno I. (2017) Diversity of interactions in redox systems: from short- to long-lived complexes. in Redox Proteins in Supercomplexes and Signalosomes (Louro R. O., and Díaz-moreno I., eds) pp. 1–48, CRC Press, Boca Raton, FL [Google Scholar]
- 34. Miethke M., Pierik A. J., Peuckert F., Seubert A., and Marahiel M. A. (2011) Identification and characterization of a novel-type ferric siderophore reductase from a Gram-positive extremophile. J. Biol. Chem. 286, 2245–2260 10.1074/jbc.M110.192468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Atkinson J. T., Campbell I., Bennett G. N., and Silberg J. J. (2016) Cellular assays for ferredoxins: a strategy for understanding electron flow through protein carriers that link metabolic pathways. Biochemistry 55, 7047–7064 10.1021/acs.biochem.6b00831 [DOI] [PubMed] [Google Scholar]
- 36. Pierre J. L., Fontecave M., and Crichton R. R. (2002) Chemistry for an essential biological process. BioMetals 15, 341–346 10.1023/A:1020259021641 [DOI] [PubMed] [Google Scholar]
- 37. Baysse C., Matthijs S., Schobert M., Layer G., Jahn D., and Cornelis P. (2003) Co-ordination of iron acquisition, iron porphyrin chelation and iron-protoporphyrin export via the cytochrome c biogenesis protein CcmC in Pseudomonas fluorescens. Microbiology 149, 3543–3552 10.1099/mic.0.26566-0 [DOI] [PubMed] [Google Scholar]
- 38. Gralnick J. A., Brown C. T., and Newman D. K. (2005) Anaerobic regulation by an atypical Arc system in Shewanella oneidensis. Mol. Microbiol. 56, 1347–1357 10.1111/j.1365-2958.2005.04628.x [DOI] [PubMed] [Google Scholar]
- 39. Gao H., Wang X., Yang Z. K., Palzkill T., and Zhou J. (2008) Probing regulon of ArcA in Shewanella oneidensis MR-1 by integrated genomic analyses. BMC Genomics 9, 42 10.1186/1471-2164-9-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Beliaev A. S., Klingeman D. M., Klappenbach J. A., Wu L., Romine M. F., Tiedje J. M., Nealson K. H., Fredrickson J. K., and Zhou J. (2005) Global transcriptome analysis of Shewanella oneidensis MR-1 exposed to different terminal electron acceptors. J. Bacteriol. 187, 7138–7145 10.1128/JB.187.20.7138-7145.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fonseca B. M., Paquete C. M., Neto S. E., Pacheco I., Soares C. M., and Louro R. O. (2013) Mind the gap: cytochrome interactions reveal electron pathways across the periplasm of Shewanella oneidensis MR-1. Biochem. J. 449, 101–108 10.1042/BJ20121467 [DOI] [PubMed] [Google Scholar]
- 42. Sturm G., Richter K., Doetsch A., Heide H., Louro R. O., and Gescher J. (2015) A dynamic periplasmic electron transfer network enables respiratory flexibility beyond a thermodynamic regulatory regime. ISME J. 9, 1802–1811 10.1038/ismej.2014.264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tu J., Lu F., Miao S., Ni X., Jiang P., Yu H., Xing L., Yu S., Ding C., and Hu Q. (2014) The siderophore-interacting protein is involved in iron acquisition and virulence of Riemerella anatipestifer strain CH3. Vet. Microbiol. 168, 395–402 10.1016/j.vetmic.2013.11.027 [DOI] [PubMed] [Google Scholar]
- 44. Xu N., Qian K., Dong Y., Chen Y., Yu Q., Zhang B., Xing L., and Li M. (2014) Novel role of the Candida albicans ferric reductase gene CFL1 in iron acquisition, oxidative stress tolerance, morphogenesis and virulence. Res. Microbiol. 165, 252–261 10.1016/j.resmic.2014.03.001 [DOI] [PubMed] [Google Scholar]
- 45. Winter G. (2010) xia2: an expert system for macromolecular crystallography data reduction. J. Appl. Crystallogr. 43, 186–190 10.1107/S0021889809045701 [DOI] [Google Scholar]
- 46. Kabsch W. (2010) XDS. Acta Crystallogr D Biol. Crystallogr. 66, 125–132 10.1107/S0907444909047337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., et al. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 10.1107/S0907444910045749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Murshudov G. N., Vagin A. A., and Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 10.1107/S0907444996012255 [DOI] [PubMed] [Google Scholar]
- 50. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr. 66, 213–221 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Brünger A. T. (1992) Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature 355, 472–475 10.1038/355472a0 [DOI] [PubMed] [Google Scholar]
- 52. Chen V. B., Arendall W. B. 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., and Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 10.1107/S0907444909042073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Velankar S., Best C., Beuth B., et al. (2010) PDBe: protein data bank in Europe. Nucleic Acids Res. 38, D308–D317 10.1093/nar/gkp916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Friis E. P., Andersen J. E. T., Madsen L. L., Bonander N., Moller P., and Ulstrup J. (1998) Dynamics of Pseudomonas aeruginosa azurin and its Cys3Ser mutant at single-crystal gold surfaces investigated by cyclic voltammetry and atomic force microscopy. Electrochim. Acta 43, 1114–1122 10.1016/S0013-4686(98)99006-5 [DOI] [Google Scholar]
- 55. Yousafzai F. K., and Eady R. R. (2002) Dithionite reduction kinetics of the dissimilatory copper-containing nitrite reductase of Alcalegenes xylosoxidans: the SO2˙̄ radical binds to the substrate binding type 2 copper site before the type 2 copper is reduced. J. Biol. Chem. 277, 34067–34073 10.1074/jbc.M204305200 [DOI] [PubMed] [Google Scholar]
- 56. Viollier E., Inglett P. W., Hunter K., Roychoudhury A. N., and Van Cappellen P. (2000) The ferrozine method revisited: Fe(II)/Fe(III) determination in natural waters. Appl. Geochem. 15, 785–790 10.1016/S0883-2927(99)00097-9 [DOI] [Google Scholar]
- 57. Jeitner T. M. (2014) Optimized ferrozine-based assay for dissolved iron. Anal. Biochem. 454, 36–37 10.1016/j.ab.2014.02.026 [DOI] [PubMed] [Google Scholar]
- 58. Diederichs K., and Karplus P. A. (1997) Improved R-factors for diffraction data analysis in macromolecular crystallography. Nat. Struct. Biol. 4, 269–275 10.1038/nsb0497-269 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.