Abstract
Lineage specification of the three germ layers occurs during early embryogenesis and is critical for normal development. The nucleosome remodeling and deacetylase (NuRD) complex is a repressive chromatin modifier that plays a role in lineage commitment. However, the role of chromodomain helicase DNA-binding protein 4 (CHD4), one of the core subunits of the NuRD complex, in neural lineage commitment is poorly understood. Here, we report that the CHD4/NuRD complex plays a critical role in neural differentiation of mouse embryonic stem cells (ESCs). We found that RNAi-mediated Chd4 knockdown suppresses neural differentiation, as did knockdown of methyl-CpG–binding domain protein Mbd3, another NuRD subunit. Chd4 and Mbd3 knockdowns similarly affected changes in global gene expression during neural differentiation and up-regulated several mesendodermal genes. However, inhibition of mesendodermal genes by knocking out the master regulators of mesendodermal lineages, Brachyury and Eomes, through a CRISPR/Cas9 approach could not restore the impaired neural differentiation caused by the Chd4 knockdown, suggesting that CHD4 controls neural differentiation by not repressing other lineage differentiation processes. Notably, Chd4 knockdown increased the acetylation levels of p53, resulting in increased protein levels of p53. Double knockdown of Chd4 and p53 restored the neural differentiation rate. Furthermore, overexpression of BCL2, a downstream factor of p53, partially rescued the impaired neural differentiation caused by the Chd4 knockdown. Our findings reveal that the CHD4/NuRD complex regulates neural differentiation of ESCs by down-regulating p53.
Keywords: embryonic stem cell, neurodifferentiation, nucleosome remodeling deacetylase (NuRD), p53, protein degradation, chromodomain helicase DNA-binding protein 4 (CHD4), lineage specification, methyl-CpG binding domain protein 3 (MBD3)
Introduction
Lineage specification of the three germ layers is an important event during early embryogenesis (1). In vitro differentiation of embryonic stem cells (ESCs)3 is a model system of early mammalian development. Neural lineage commitment of ESCs occurs in the absence of extrinsic cues, such as BMP4, which is called the default model (2). Previous studies have uncovered that the intrinsic programs mediated by transcription factors and epigenetic regulators play important roles in the default model of neural fate determination (3–7). Recent studies have shown that repressive chromatin modifiers, polycomb repressive complex 2 (PRC2) and Chromobox homolog 3, regulate lineage fidelity during neural differentiation of ESCs by enhancing neural gene expression and suppressing the genes specific to other cell lineages (8, 9). These results indicate the importance of repressive chromatin modifiers in neural lineage commitment.
The nucleosome remodeling and deacetylase (NuRD) complex, a repressive chromatin modifier, is involved in various biological processes, including development, DNA damage response, and cancer metastasis (10–13). The ATPase activity of the NuRD complex is provided by chromodomain helicase DNA-binding proteins (CHD3/4) and deacetylase activity of HDAC1 or HDAC2 (14–16). In addition, the NuRD complex contains methyl-CpG–binding domain proteins (MBD2/3), WD40 repeat proteins (RBBP4/7), metastasis-associated proteins (MTA1/2/3), and nuclear zinc-finger proteins (GATAD2a/b) (17).
CHD4, the largest component of the NuRD complex, has been shown to be important for cell fate in various developmental processes (18–22). In addition to its role as a component of the NuRD complex, CHD4 functions independently of the NuRD complex in some contexts (18, 20, 23, 24).
A recent study reported that Chd4 knockdown results in the promotion of endodermal differentiation of ESCs (25), leading to a different phenotype than that caused by Mbd3 knockdown or knockout (26), suggesting that CHD4 functions independently of the NuRD complex in this context. Although the involvement of CHD4 in ESC differentiation has been demonstrated, whether CHD4 regulates the neural lineage commitment of ESCs in a manner dependent on, or independent of, the NuRD complex remains unknown.
In this study, we found that the CHD4/NuRD complex plays an important role in neural differentiation of ESCs by regulating the p53 protein level.
Results
CHD4 is required for neural differentiation of ESCs
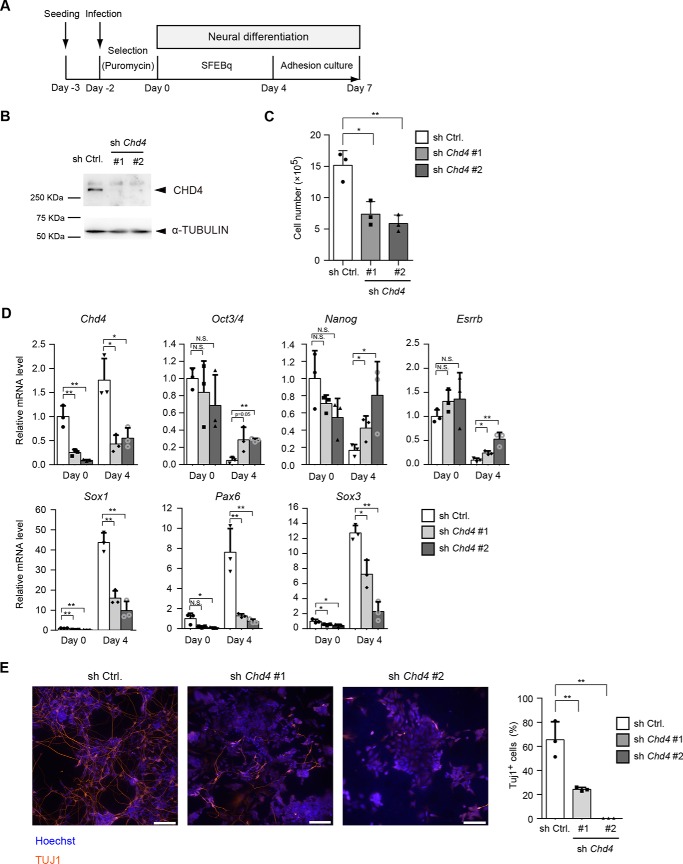
To study the role of the CHD4/NuRD complex in neural differentiation of mouse ESCs, we performed knockdown experiments. Short hairpin RNAs (shRNAs) against Chd4 were introduced into mouse embryonic stem cells (Fig. 1A). For early neural differentiation, we used a serum-free culture of embryoid body-like aggregates with quick reaggregation culture (SFEBq) that allowed highly-efficient neural differentiation (5). After 4 days of SFEBq culture, embryoid body-like aggregates were dissociated and subjected to adhesion culture for further differentiation into neurons. The protein levels of CHD4 were markedly reduced by each Chd4 shRNA in ESCs (day 0) (Fig. 1B). Chd4 knockdown decreased the number of ESCs, a finding that was consistent with that of a previous report (Fig. 1C) (25). Next, we examined the gene expression changes before (day 0) and after (day 4) the SFEBq culture. During neural differentiation, the pluripotent marker genes, Oct3/4, Nanog, and Esrrb, were markedly down-regulated in control cells (Fig. 1D). However, Chd4 knockdown did not alter the expression levels of pluripotent marker genes at day 0; Chd4 knockdown suppressed the down-regulation of pluripotent marker genes (Fig. 1D). In addition, Chd4 knockdown strongly suppressed the up-regulation of the early neural marker genes, Sox1, Pax6, and Sox3. Immunostaining also showed that Chd4 knockdown markedly decreased the number of TUJ1-positive neurons at day 7 (Fig. 1E).
Figure 1.
CHD4 is required for neural differentiation of ESCs. A, scheme for shRNA-mediated Chd4 knockdown and subsequent neural differentiation. ESCs were infected with a lentivirus encoding Chd4 shRNAs (sh Chd4 #1 or #2) or control shRNA (sh Ctrl.) and were cultured under selection with puromycin for 2 days. The SFEBq culture method was used for neural differentiation. SFEBq-cultured cells at day 4 were then subjected to adhesion culture for further neural differentiation. B, immunoblotting analysis for CHD4 and α-tubulin (as a loading control) in control shRNA- and Chd4 shRNA-expressing cells at day 0. C, effect of Chd4 shRNAs on the number of ESCs. One day before viral infection (day −3), 2.5 × 105 cells were plated, and cells were counted at day 0. D, qRT-PCR analysis of Chd4, pluripotency genes (Oct3/4, Nanog, and Esrrb), and neural genes (Sox1, Pax6, and Sox3) in control shRNA- and Chd4 shRNA-expressing cells at day 0 and day 4. Each mRNA level was normalized to the β-actin level, and the value of control shRNA-expressing cells at day 0 was set to 1. E, cells at day 7 were stained for TUJ1 (left). The scale bars represent 100 μm. The percentages of TUJ1-positive cells are shown (right). C–E, data are shown as the means ± S.D. (n = 3 independent experiments). *, p < 0.05, and **, p < 0.01. N.S., not significant. The p values were calculated using Student's unpaired two-tailed t tests compared with the control cells in the same day.
MBD3 is required for neural differentiation of ESCs
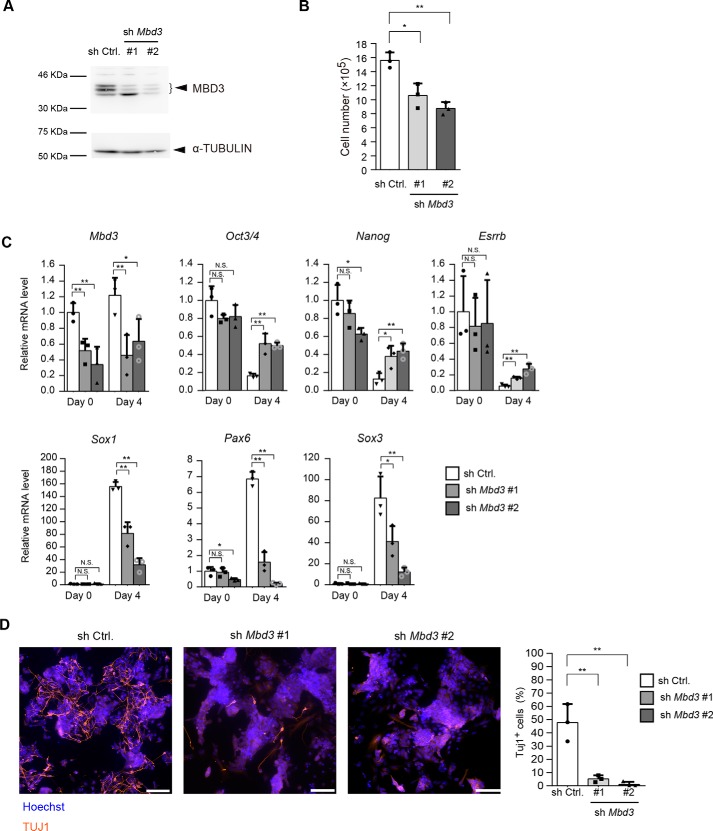
Next, we examined the effect of Mbd3 knockdown on the neural differentiation rate (Fig. 2A). Mbd3 knockdown also decreased the number of ESCs (Fig. 2B), a finding that was consistent with a previous finding that showed that Mbd3 knockout decreased the number of ESCs (26). During neural differentiation, Mbd3 knockdown suppressed the down-regulation of pluripotency marker genes and up-regulation of neural differentiation marker genes (Fig. 2C). Immunostaining also showed that Mbd3 knockdown markedly decreased the number of TUJ1-positive neurons at day 7 (Fig. 2D). These data indicate that the CHD4/NuRD plays a critical role in neural differentiation of ESCs.
Figure 2.
MBD3 is required for neural differentiation of ESCs. A, immunoblotting analysis for MBD3 and α-tubulin (as a loading control) in control shRNA-expressing cells and Mbd3 shRNA-expressing cells at day 0. B, effect of Mbd3 shRNAs on the number of ESCs. One day before viral infection (day −3), 2.5 × 105 cells were plated, and the cells were counted at day 0. C, qRT-PCR analysis. Each mRNA level was normalized to the β-actin level, and the value of control shRNA-expressing cells at day 0 was set to 1. D, cells at day 7 were stained for TUJ1 (left). The scale bars, 100 μm. The percentages of TUJ1-positive cells are shown (right). B–D, data are shown as the means ± S.D. (n = 3 independent experiments). *, p < 0.05 and **, p < 0.01. N.S., not significant. The p values were calculated using Student's unpaired two-tailed t tests compared with the control cells on the same day.
Changes in global gene expression upon Chd4 or Mbd3 knockdown in ESCs
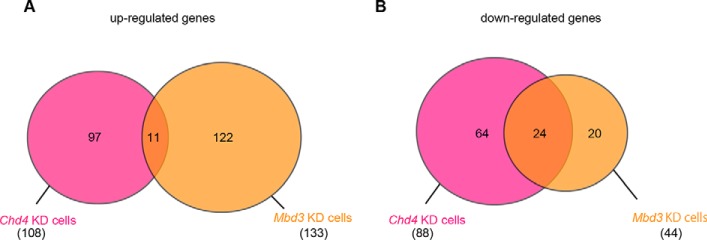
To investigate the molecular mechanisms by which the CHD4/NuRD complex regulates neural differentiation, we performed microarray experiments. We used six samples as follows: control cells at two time points (day 0 and day 4), Chd4-knockdown cells at two time points (day 0 and day 4), and Mbd3-knockdown cells at two time points (day 0 and day 4). First, we analyzed genes with expression levels that were changed upon Chd4 or Mbd3 knockdown in ESCs at day 0. In total, 108 genes and 133 genes were up-regulated in Chd4-knockdown cells and Mbd3-knockdown cells, respectively, compared with control ESCs (Fig. 3A and Table S1). Moreover, 88 genes and 44 genes were down-regulated in Chd4-knockdown cells and Mbd3-knockdown cells, respectively, compared with control ESCs (Fig. 3B and Table S2). However, few genes were commonly regulated by CHD4 and MBD3, indicating that CHD4 and MBD3 might have different functions in ESCs. Several pluripotency-related genes, Foxd3, Zscan4, and Myc, were down-regulated by both Chd4 and Mbd3 knockdown, indicating that these genes may be involved in the decreased number of ESCs caused by Chd4 and Mbd3 knockdown. There were no neural differentiation-related genes among the genes that were commonly regulated by CHD4 and MBD3 in ESCs.
Figure 3.
Changes in global gene expression upon Chd4 or Mbd3 knockdown in ESCs. A, Venn diagrams of genes whose expression level was up-regulated by more than 2-fold upon Chd4 or Mbd3 knockdown in ESCs (day 0). B, Venn diagrams of genes whose expression level was down-regulated by more than 2-fold upon Chd4 or Mbd3 knockdown in ESCs (day 0).
CHD4 and MBD3 commonly regulate global gene expression changes during neural differentiation
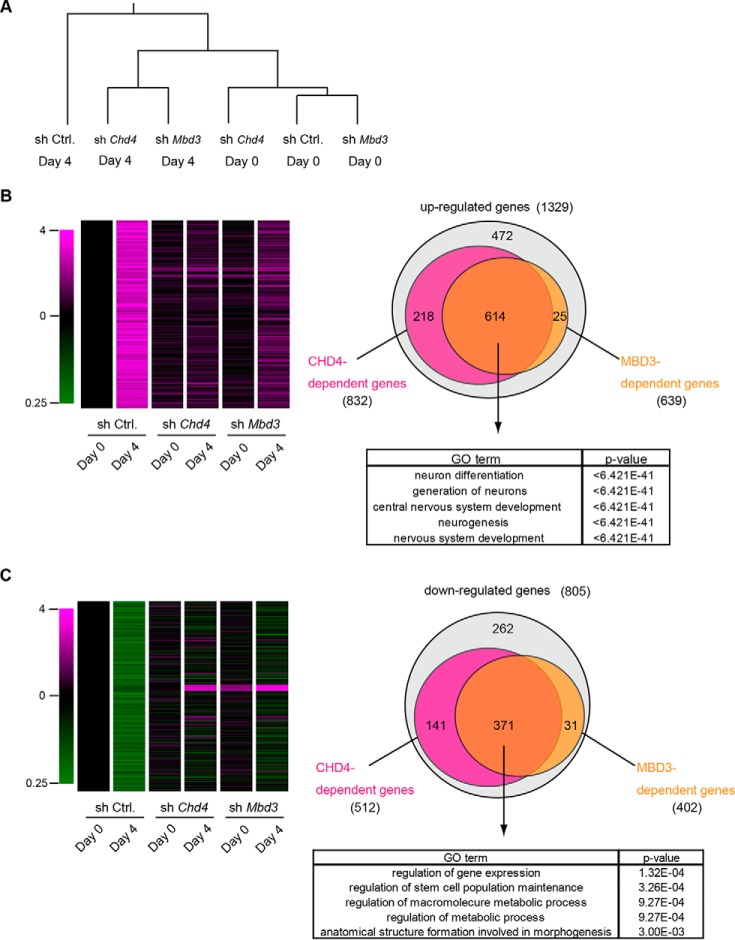
Remarkably, however, unsupervised hierarchical clustering analysis of gene expression in six samples showed that Chd4-knockdown cells and Mbd3-knockdown cells at day 4 were categorized into the same group, suggesting that CHD4 and MBD3 function in the same complex (Fig. 4A). Furthermore, this analysis showed that both Chd4-knockdown cells at day 4 and Mbd3-knockdown cells at day 4 were close to control cells at day 0 rather than control cells at day 4, suggesting that neither Chd4-knockdown cells nor Mbd3-knockdown cells were fully differentiated into the neural lineage and that both cell types remained in an ESC-like state. Additionally, 1329 genes were up-regulated by more than 2-fold during neural differentiation in control cells (Fig. 4B and Table S3). Among the 1329 genes, we identified 823 genes as CHD4-dependent up-regulated genes and 639 genes as MBD3-dependent up-regulated genes: their expression levels in control cells at day 4 were reduced by more than 50% in both Chd4-knockdown cells and Mbd3-knockdown cells. Remarkably, 614 genes were commonly regulated by CHD4 and MBD3 (Fig. 4B). GO analysis showed that genes related to neural differentiation were enriched in 614 genes. Zfp521, which functions upstream of early neural marker genes (5), was included among these 614 genes, suggesting that CHD4 and MBD3 function in the very early stage of neural differentiation.
Figure 4.
Microarray analysis of CHD4- and MBD3-dependent genes during neural differentiation of ESCs. A, unsupervised hierarchical cluster analysis of gene expression profiles. B, heatmap for the 1329 genes whose expression was up-regulated by more than 2-fold during neural differentiation (left). Venn diagrams of 1329 genes (right, upper). The magenta and yellow circles represent CHD4- and MBD3-dependent up-regulated genes, respectively. GO analysis of CHD4- and MBD3-dependent up-regulated genes (right, lower). C, heatmap for the 805 genes whose expression was down-regulated less than half during neural differentiation (left). Venn diagrams of 805 genes (right, upper). The magenta and yellow circles represent CHD4- and MBD3-dependent down-regulated genes, respectively. GO analysis of CHD4- and MBD3-dependent down-regulated genes (right, lower).
805 genes were down-regulated by less than half during neural differentiation in control cells (Fig. 4C and Table S4). Among these 805 genes, we identified 512 genes as Chd4-dependent down-regulated genes and 402 genes as Mbd3-dependent down-regulated genes: their expression levels in control cells at day 4 were increased by more than 2-fold in Chd4- and Mbd3-knockdown cells. Remarkably, 371 genes were commonly regulated by CHD4 and MBD3. Many pluripotency marker genes were included in the highly enriched GO terms “regulation of gene expression” and “regulation of stem cell population maintenance” (Fig. 4C). Collectively, our analysis indicated that the CHD4/NuRD complex regulated neural gene induction and stem cell gene suppression during neural differentiation.
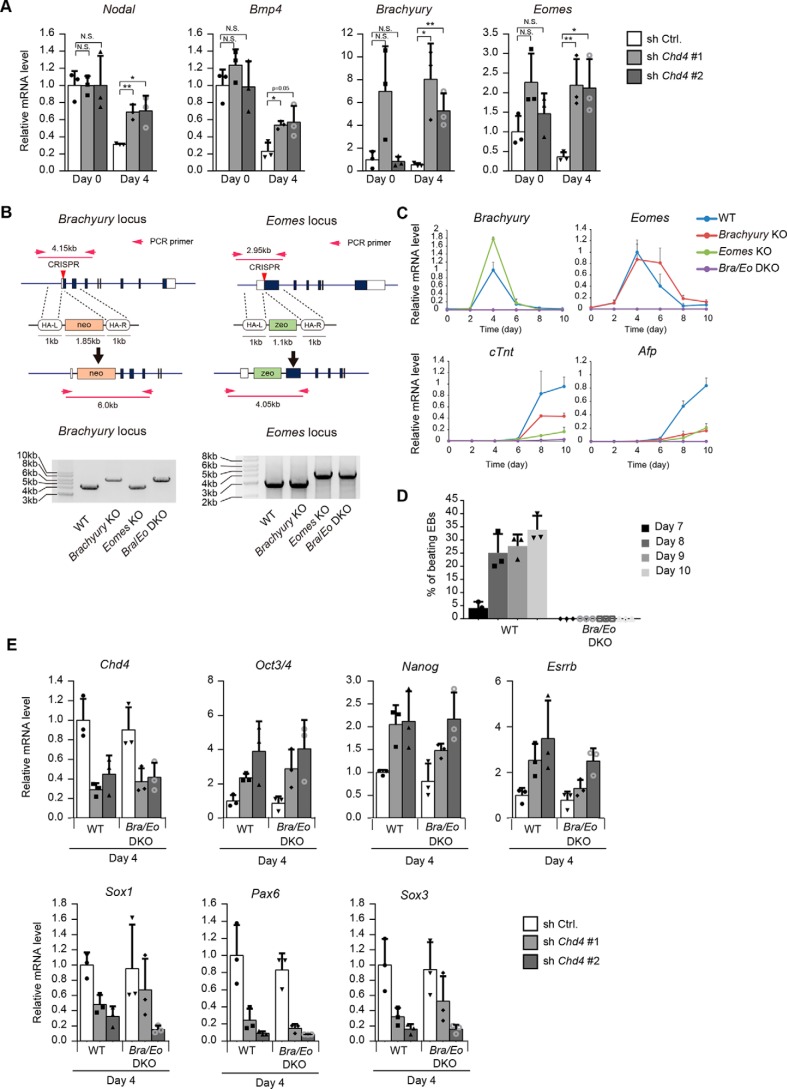
Inhibition of mesendoderm differentiation does not affect impaired neural differentiation caused by Chd4 knockdown
Because CHD4 represses the transcription of target genes, we focused on the CHD4-dependent down-regulated genes found in our microarray analysis as direct targets of the CHD4/NuRD complex. Notably, Nodal and Bmp4, which play important roles in mesendoderm differentiation of ESCs (27), were down-regulated during neural differentiation, and their down-regulation was suppressed by Chd4 knockdown, a finding that was also confirmed by qRT-PCR. Furthermore, qRT-PCR analysis showed that the expression levels of Brachyury and Eomes, which are T-box transcription factors important for mesendoderm differentiation (28–33), were increased by Chd4 knockdown at day 4 (Fig. 5A). Recent studies have shown that some repressive chromatin modifiers regulate lineage fidelity during neural differentiation of ESCs by enhancing neural gene expression and suppressing genes specific to other lineages (8, 9). Furthermore, a previous report has shown that the CHD4/NuRD complex establishes the identities of the two striated muscle types, cardiac and skeletal muscle, by suppressing the alternate lineage gene program (34). Therefore, we considered the possibility that the CHD4/NuRD complex may ensure the neural lineage determination of ESCs by inhibiting the mesoderm and endoderm gene programs.
Figure 5.
Inhibition of mesendodermal differentiation does not affect impaired neural differentiation caused by Chd4 knockdown. A, qRT-PCR analysis. Each mRNA level was normalized to the β-actin level, and the value of control shRNA-expressing cells at day 0 was set to 1. B, schematic representation of the strategy to generate Brachyury and Eomes knockout ESCs (left). Proper targeting of Brachuyury and Eomes loci is indicated by PCR genotyping (right). C, qRT-PCR analysis of Brachyury, Eomes, cTnt (mesodermal gene), and Afp (endodermal gene) at the indicated time points of embryoid body formation in WT, Brachyury single knockout, Eomes single knockout, and Bra/Eo DKO cells. The expression levels were normalized to that of β-actin. The y axis values represent the expression levels relative to the maximum expression levels in WT cells. D, percentages of embryoid bodies containing the beating area during the differentiation of WT and mutant cells. E, WT ESCs and Brachyury and Eomes double knockout (Bra/Eo DKO) ESCs were subjected to the experiments shown in Fig. 1A. The mRNA levels at day 4 were determined. Each mRNA level was normalized to the β-actin level, and the value of control shRNA-expressing WT ESCs was set to 1. A and C–E, data are shown as the means ± S.D. (n = 3 independent experiments). A, *, p < 0.05, and **, p < 0.01. N.S., not significant. The p values were calculated using Student's unpaired two-tailed t tests compared with cells on the same day.
To examine this possibility, we investigated whether inhibition of mesendoderm differentiation restored the impaired neural differentiation caused by Chd4 knockdown. To inhibit mesendoderm differentiation, we generated Brachyury-knockout ESCs (Brachyury-KO), Eomes-knockout ESCs (Eomes-KO), and Brachyury/Eomes double knockout (Bra/Eo DKO) ESCs by CRISPR/Cas9-mediated gene targeting (Fig. 5B). Proper targeting of the Brachuyury and Eomes loci was indicated by PCR genotyping. To examine the differentiation potential of Brachyury and/or Eomes KO ESCs, we induced their differentiation through embryoid body formation. The expression levels of a mesoderm marker, cTnt, and an endoderm marker, Afp, were up-regulated in WT cells, Brachyury-KO cells, and Eomes-KO cells, but they were not expressed in Bra/Eo DKO cells (Fig. 5C). In agreement with the qRT-PCR results, immunostaining showed that cTNT- and AFP-positive cells appeared in cultures of WT cells, Brachyury KO cells, and Eomes KO cells, but not in the culture of Bra/Eo DKO cells (data not shown). Double knockout of Brachyury and Eomes led to complete loss of beating embryoid bodies (Fig. 5D). Thus, we concluded that differentiation to mesoderm and endoderm could be almost completely lost when both Brachyury and Eomes were knocked out. Next, we examined whether the double knockout of Brachyury and Eomes restored the impaired neural differentiation caused by Chd4 knockdown. The obtained results demonstrated that the expression levels of pluripotency markers and neural differentiation markers were not altered by the double knockout of Brachyury and Eomes (Fig. 5E). Although Nodal, Bmp4, Brachyury, and Eomes were up-regulated by Chd4 knockdown, their expression levels might be too low to induce mesendoderm differentiation. Thus, our results suggest that CHD4 controls neural differentiation but not by repressing other lineage differentiation processes.
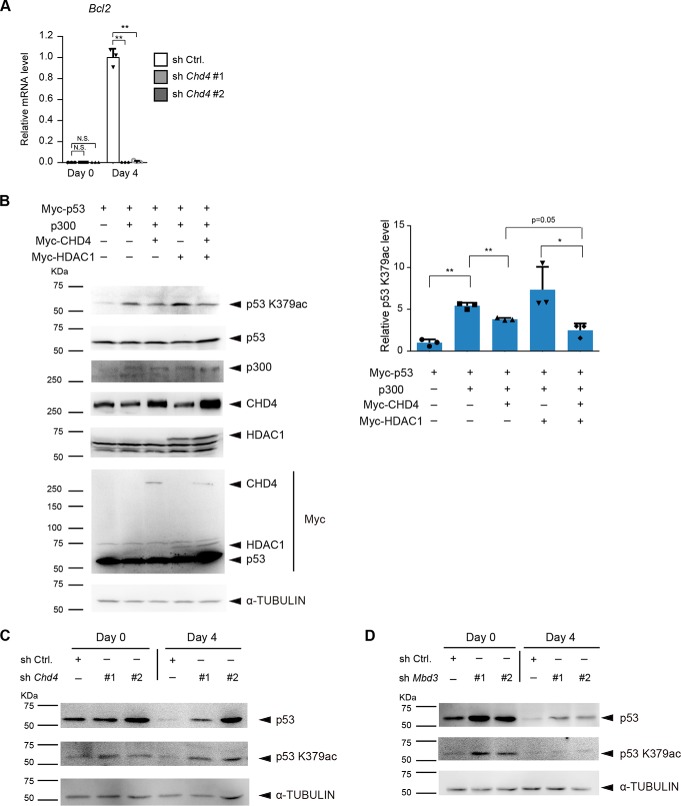
CHD4 and MBD3 regulate the acetylation and protein levels of p53
Next, we focused on CHD4-dependent up-regulated genes. Interestingly, up-regulation of Bcl2 was almost completely diminished by Chd4 knockdown (Fig. 6A). BCL2 is known for its negative role in apoptosis and plays an important role in the differentiation of ESCs (35). Previous reports have shown that p53 negatively regulates BCL2 activity by transcription-dependent and -independent mechanisms (36). Our results showed that one of the C-terminal lysines (Lys-379) of murine p53 (corresponding to Lys-382 of human p53) was acetylated by p300, and its acetylation was diminished by CHD4 and HDAC1 in HEK293T cells (Fig. 6B), which is consistent with the previous report using human cancer cells (37). Although it has been shown that deacetylation of C-terminal lysines of p53 leads to ubiquitination of the same lysines, resulting in p53 degradation (37, 38), total p53 expression levels were not changed, probably because the levels of the exogenously expressed p53 were too high in HEK293T cells. Next, we examined the effect of Chd4 and Mbd3 knockdown on the acetylation and protein levels of endogenous p53 during neural differentiation of ESCs. The protein levels of p53 were down-regulated during neural differentiation of control ESCs. Chd4 knockdown increased the acetylation levels of p53 and suppressed the down-regulation of the p53 protein levels during SFEBq culture (Fig. 6C). Mbd3 knockdown also increased the acetylation and protein levels of p53 (Fig. 6D).
Figure 6.
CHD4 and MBD3 regulate the acetylation and protein levels of p53. A, qRT-PCR analysis. Each mRNA level was normalized to the β-actin level, and the value of control shRNA-expressing cells at day 4 was set to 1. The data are shown as the means ± S.D. (n = 3 independent experiments). **, p < 0.01. The p values were calculated using Student's unpaired two-tailed t tests compared with control cells on the same day. B, immunoblotting analysis for acetyl-p53 (Lys-379), p53, p300, CHD4, HDAC1, Myc, and α-tubulin (as a loading control) in lysates from HEK293T cells transfected with the indicated combination of expression vectors (left). The relative acetyl-p53 (Lys-379) signal intensities are shown (right). The data are shown as the means ± S.D. (n = 3 independent experiments). *, p < 0.05, and **, p < 0.01. N.S., not significant. The p values were calculated using Student's unpaired two-tailed t tests. C, immunoblotting analysis for p53, acetyl-p53 (Lys-379), and α-tubulin (as a loading control) in control shRNA- and Chd4 shRNA-expressing ESCs at day 0 and day 4 of SFEBq culture. D, immunoblotting analysis for p53, acetyl-p53 (Lys-379), and α-tubulin (as a loading control) in control shRNA- and Mbd3 shRNA-expressing ESCs at day 0 and day 4 of SFEBq culture.
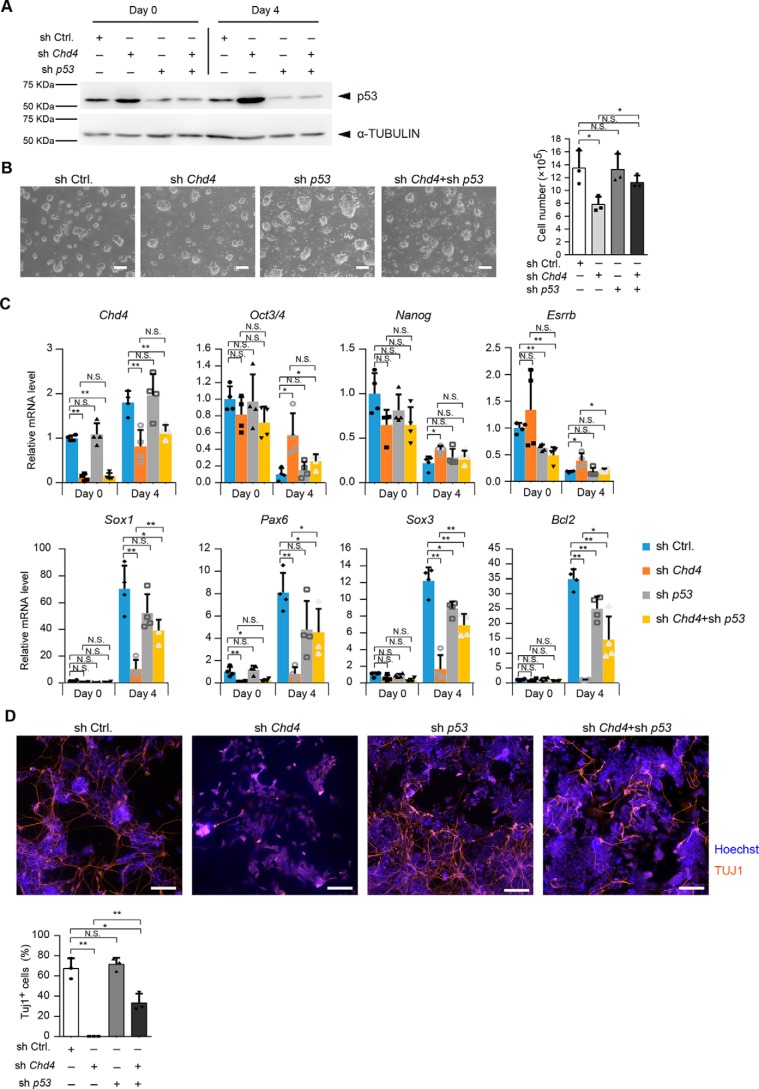
CHD4 regulates neural differentiation through p53 down-regulation
To examine whether the CHD4/NuRD complex regulates neural differentiation by suppressing p53 protein levels, we performed Chd4 and/or p53 knockdown experiments. RNAi against Chd4 and p53 in ESCs led to a reduction in the expression levels of CHD4 and p53, respectively, which was verified by immunoblotting analysis (Fig. 7A). In Chd4 and p53 double-knockdown ESCs, the number of cells was restored compared with that of the Chd4 single knockdown (Fig. 7B). During neural differentiation, the p53 single knockdown did not alter the expression levels of pluripotency marker genes compared with control cells. The expression levels of Nanog and Esrrb were down-regulated in Chd4 and p53 double-knockdown cells to the same extent as in control cells (Fig. 7C). Furthermore, the expression levels of neural differentiation markers were regained by the Chd4 and p53 double knockdown, compared with the Chd4 single knockdown (Fig. 7C). Immunoblotting analysis showed that TUJ1-positive cells were increased by the double knockdown of Chd4 and p53, compared with the Chd4 single knockdown (Fig. 7D). Thus, our results strongly suggest that the CHD4/NuRD complex regulates the neural differentiation of mESCs by suppressing the p53 proteins levels.
Figure 7.
CHD4 regulates neural differentiation through p53 down-regulation. A, immunoblotting analysis for p53 and α-tubulin (as a loading control) in Chd4 shRNA #2- and/or p53 shRNA-expressing cells at day 0 and day 4 of SFEBq culture. B, effect of Chd4 and/or p53 shRNAs on the number of ESCs. One day before viral infection (day −3), 2.5 × 105 cells were plated, and the cells were counted at day 0. C, qRT-PCR analysis. Each mRNA level was normalized to the β-actin level, and the value of control shRNA-expressing cells at the 0 was set to 1. D, SFEBq-cultured cells at day 4 were transferred to dishes for further neural differentiation. Three days later (day 7), cells were stained for TUJ1 (upper). The scale bars represent 100 μm. The percentages of TUJ1-positive cells are shown (lower). B–D, data are shown as the means ± S.D. (n = 4 independent experiments for C, and n = 3 independent experiments for B and D). *, p < 0.05, and **, p < 0.01. N.S., not significant. The p values were calculated using Student's unpaired two-tailed t tests compared with control cells on the same day. In addition, the p values were calculated using Student's unpaired two-tailed t tests between Chd4 shRNA-expressing cells and Chd4 and p53 shRNA-expressing cells on the same day.
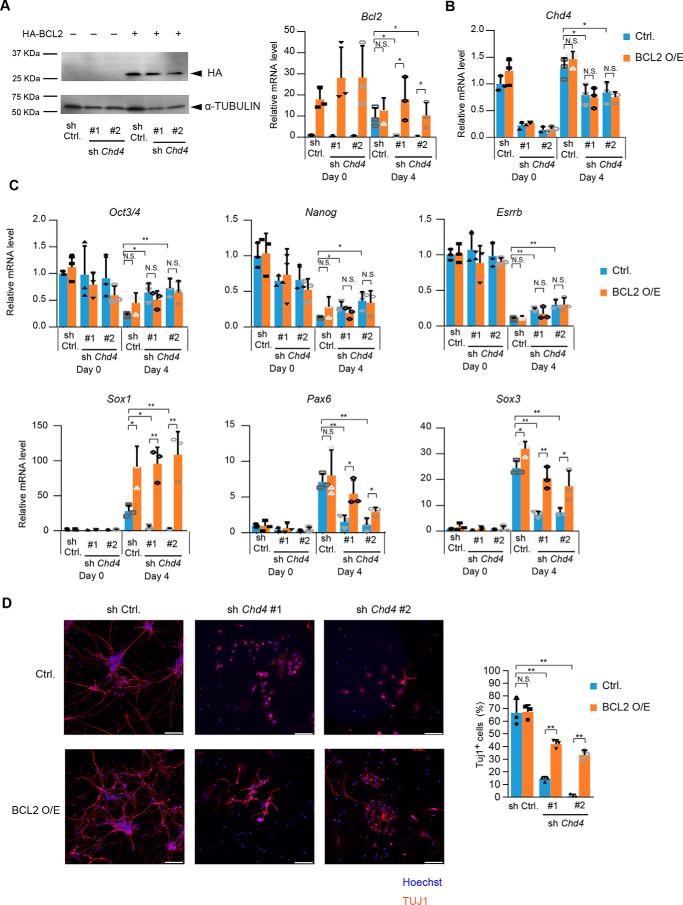
Overexpression of BCL2 partially rescues impaired neural differentiation caused by Chd4 knockdown
Next, we investigated the mechanisms by which the CHD4/NuRD–p53 axis regulates neural differentiation of ESCs. Our results showed that the up-regulation of Bcl2 was diminished by Chd4 knockdown, and Bcl2 expression levels were recovered by the Chd4 and p53 double knockdown. Because BCL2 is known as an important factor for neural commitment of ESCs (35), we examined whether BCL2 overexpression could restore the impaired neural differentiation caused by Chd4 knockdown. HA-BCL2 was introduced to ESCs, and its expression remained high in Chd4-knockdown cells at day 4 of neural differentiation (Fig. 8A). Chd4 expression was not altered by BCL2 overexpression (Fig. 8B). Overexpression of BCL2 did not affect the expression levels of pluripotency marker genes (Fig. 8C). Notably, the expression levels of neural genes were down-regulated by Chd4 knockdown and recovered by BCL2 overexpression. Immunostaining analysis also showed that the number of TUJ1-positive cells was increased by BCL2 overexpression in Chd4-knockdown cells (Fig. 8D). Thus, our results suggest that CHD4/NuRD–p53 axis regulates neural gene expression and neurite formation through BCL2 up-regulation.
Figure 8.
CHD4–p53 axis regulates neural differentiation through BCL2 up-regulation. A, immunoblotting analysis for HA and α-tubulin (as a loading control) in control shRNA-, Chd4 shRNA-, and/or HA-BCL2-expressing cells at day 0 (left). qRT-PCR analysis is shown at right. Each mRNA level was normalized to the β-actin level, and the value of control shRNA-expressing cells at the 0 was set to 1. B and C, qRT-PCR analysis. Each mRNA level was normalized to the β-actin level, and the value of control shRNA-expressing cells at the 0 was set to 1. D, SFEBq-cultured cells at day 4 were transferred to dishes for further neural differentiation. Three days later (day 7), cells were stained for TUJ1 (left). The scale bars represent 100 μm. The percentages of TUJ1-positive cells are shown (right). A–D, data are shown as the means ± S.D. (n = 3 independent experiments). *, p < 0.05, and **, p < 0.01. The p values were calculated using Student's unpaired two-tailed t tests compared with control cells. In addition, the p values were calculated using Student's unpaired two-tailed t tests between control vector-expressing cells (blue bars) and BCL2-expressing cells (orange bars).
Discussion
In this study, we demonstrated that the CHD4/NuRD complex plays an important role in the neural differentiation of mouse ESCs by controlling the p53 protein level. A previous report showed that CHD4 inhibits ESC differentiation into endoderm by repressing the expression of Tbx3 (25). Another report has shown that MBD3 is required for differentiation into all three lineages (26), suggesting the possibility that CHD4 inhibits endodermal differentiation independent of the classical NuRD complex. In this study, Chd4 knockdown and Mbd3 knockdown both resulted in a marked decrease in the neural differentiation efficiency, and it was shown that global gene expression changes are commonly regulated by CHD4 and MBD3, indicating that CHD4 regulates neural differentiation as a member of the NuRD complex. Because a previous report has shown that CHDs (CHD3, CHD4, and CHD5) are required to form the CHD/NuRD complex (22), Chd4 knockdown may result in the dissociation of the complex.
CHD4 and MBD3 have been shown to be important in regulating the proliferation of neural progenitors (22, 39). Our results showed that the expression levels of pluripotent marker genes remained high in SFEBq cultured Chd4- or Mbd3-knockdown cells, suggesting that the neural lineage commitment itself, rather than the proliferation of neural progenitors, is impaired by Chd4 or Mbd3 knockdown. The CHD4/NuRD complex may play various roles in the different stages of mammalian neural development.
It has been reported that CHD4 plays a role in cell cycle progression by regulating acetylation or altering the protein levels of p53 in human cancer cells (37). We found that the CHD4/NuRD complex suppresses the p53 acetylation levels and p53 protein levels in mouse ESCs. The number of ESCs was restored by Chd4 and p53 double knockdown compared with Chd4 single knockdown, suggesting that CHD4 regulates cell cycle progression through p53 down-regulation in ESCs.
Our results showed that impaired neural differentiation by Chd4 knockdown was partially rescued by inhibiting p53 expression. The induction of neural genes, such as Pax6, Sox3, and Bcl2, was partially inhibited by the single knockdown of p53, indicating that the loss of p53 may have a negative effect on neural differentiation independent of CHD4. Alternatively, CHD4 may also regulate neural differentiation via another mechanism. Although CHD4 is famous for its transcriptional repressive function, several studies have shown that CHD4 has the potential to promote gene expression; thus, CHD4 may transcriptionally activate neural genes.
Our results showed that impaired neural differentiation by Chd4 knockdown was rescued by BCL2 overexpression. BCL2 overexpression affected neural genes, not pluripotency genes, suggesting that CHD4/NuRD–p53–BCL2 axis is not involved in the suppression of pluripotency genes. A previous study has shown that the NuRD complex suppresses pluripotency gene expression by binding to their promoter regions (40). Collectively, both chromatin modification and post-translational regulation by the CHD4/NuRD complex are important for neural differentiation of ESCs. Because the CHD4/NuRD complex plays an important role not only in neural lineage commitment but also in mesendoderm lineage commitment (25, 26), whether CHD4/NuRD–p53 axis regulates mesendoderm differentiation should be examined in future studies.
Experimental procedures
Cell culture
EBRTcH3 ESCs (purchased from Riken Cell Bank, Tsukuba, Japan) (41) were cultured on 0.1% gelatin-coated dishes. The culture medium for EBRTcH3 included GMEM (Wako, Osaka, Japan, or Gibco, Waltham, MA), 10% FBS (Hyclone murine embryonic stem cell screened; ThermoFisher Scientific, Waltham, MA), 1 mm nonessential amino acids (Nacalai Tesque, Kyoto, Japan), 1 mm sodium pyruvate (Nacalai Tesque), 0.1 mm β-mercaptoethanol (Gibco), and 1000 units/ml LIF (Millipore, Billerica, MA). HEK293T cells were cultured in DMEM containing 10% FBS.
Viral production and transduction
Virus packaging vectors (PLKO.1, psPAX2, and pMD2.G) were obtained from Addgene (Addgene plasmids no. 8453). Lentiviral particles for shRNAs were produced in HEK293T cells. Packaging vectors (psPAX2 and pMD2.G), along with each pLKO.1 construct, were transfected into HEK293T cells using FuGENE HD (Promega, Madison, WI). The medium was changed 12 h later. Culture supernatants containing viruses were collected 48 h after transfection. ESCs were selected by puromycin at a final concentration of 1 μg/ml for 2 days. The shRNA sequences were as follows: control shRNA, CCTAAGGTTAAGTCGCCCTCG; Chd4 shRNA #1, CCATCTTGGGTTCTATTGACT; Chd4 shRNA #2, CGTAAACAGGTCAACTACAAT; Mbd3 shRNA #1, GAAAGATGTTGATGAACAAGA; Mbd3 shRNA #2, GGATTGAGTGCCTTTGACATT; and p53 shRNA, CCACTACAAGTACATGTGTAA.
In vitro differentiation of ESCs
Neural differentiation was performed using the SFEBq method as reported previously (5). Briefly, ESCs were re-aggregated using low-cell-adhesion 96-well plates (Primesurface, Sumitomo Bakelite, Tokyo, Japan) in GMEM-based differentiation medium supplemented with 10% KSR (5000 cells/150 μl/well). After 4 days of SFEBq culture, embryoid body-like aggregates were dissociated and plated onto poly-l-ornithine (Sigma)/fibronectin (Life Technologies, Inc.)–coated chambers and cultured in Neurobasal medium (Gibco) supplemented with B-27 (Gibco) for 3 days. For embryoid body formation-mediated differentiation into three germ layers, cells were plated in 100-mm bacterial dishes in EBRTcH3 maintenance medium without LIF. The embryoid bodies were transferred to 0.1% gelatin-coated dishes on day 6. The medium was replaced every 2 days.
CRISPR/Cas9-mediated gene targeting
pX330-U6-Chimeric_BB-CBh-hSpCas9 (pX330) (42) was a gift from Feng Zhang (Addgene plasmid no. 42230). The sgRNA sequences were inserted at the BbsI site. PGKneoF2L2DTA (43) was a gift from Philippe Soriano (Addgene plasmid no. 13445). The LoxP sites were removed by inserting a PCR-amplified drug resistance cassette without loxP sites into the vector digested with NotI and NheI. For the targeting vector that included the Zeocin resistance gene, the Zeocin resistance cassette was amplified from the CSIV–CMV–MCS–IRES2–Venus vector (a kindly gift from Dr. H. Miyoshi, Riken BRC, Japan) and was inserted into the PGKneoF2L2DTA vector instead of the neomycin resistance cassette. Next, 5′ and 3′ homology arms of ∼1 kb were amplified from the genome of ESCs and were inserted into the 5′ and 3′ sides of the drug-resistance cassette. The DTA expression cassette was removed at the time of insertion of the homology arms. ESCs were transfected with the pX330 vector and PGKneoF2L2DTA-derived targeting vector using Lipofectamine 2000 (Invitrogen). The transfected ESCs were selected by 500 μg/ml G418 or 100 μg/ml Zeocin for 4–6 days. After colonies were selected, recombination was checked by gel electrophoresis of the PCR-amplified genomic fragment.
Quantitative RT-PCR
Total RNA was isolated using the RNeasy mini kit (Invitrogen) or NucleoSpin RNA kit (TaKaRa, Shiga, Japan). During isolation, on-column DNase I digestion was performed. cDNA was synthesized using Moloney murine leukemia virus reverse transcriptase (Invitrogen) or ReverTra Ace (TOYOBO, Osaka, Japan). Real-time PCR analysis was performed using QuantiTect SYBR Green PCR kits (Qiagen, Hilden, Germany) or SYBR Premix Ex TaqII (TaKaRa) and the 7300 Real-Time PCR system or QuantStudio 3 (Applied Biosystems, Carlsbad, CA). The measured value was normalized to that of β-actin. The primers used for PCR analysis were as follows: β-actin, 5′-AGAGGGAAATCGTGCGTGAC-3′ and 5′-CAATAGTGATGACCTGGCCGT-3′; Chd4, 5′-GAAATTGCTGCGGCACCATTA-3′ and 5′-AGCCATCATTGTAGTTGACCTG-3′; Mbd3, 5′-GCTATGCCCACCTTATGTCCCT-3′ and 5′-AGGAAAGTGACTTCCTGGTGGG-3′; Oct3/4, 5′-GAAGGGCAAAAGATCAAGTATTGAG-3′ and 5′-GCCCCCCCTGGGAAAG-3′; Nanog, 5′-TCTCTCAGGCCCAGCTGTGT-3′ and 5′-GCTGGAGGCTGAGGTACTTCTG-3′; Esrrb, 5′-CCTGCTGTTTCTCCTTCTCCTC-3′ and 5′-TCTCTGCTATCCTACACCCCAAC-3′; Sox1, 5′-CCTCGGATCTCTGGTCAAGT-3′ and 5′-GCAGGTACATGCTGATCATCTC-3′; Pax6, 5′-TGGAGAAGAGAAGAGAAACTGAGGA-3′ and 5′-CTGTGGGATTGGCTGGTAGA-3′; Sox3, 5′-CGCGTTTAGGCTGTTGAGTTT-3′ and 5′-AATAACCCCTTCCCCACCAC-3′; Nodal, 5′-TCAAGCCTGTTGGGCTCTACT-3′ and 5′-GTCAAACGTGAAAGTCCAGTTCT-3′; Bmp4, 5′-ATTCCTGGTAACCGAATGCTG-3′ and 5′-CCGGTCTCAGGTATCAAACTAGC-3′; Brachyury, 5′-ACCTCTAATGTCCTCCCTTGTTG-3′ and 5′-GACGGTTCAGTTACAATCTTCTGG-3′; Eomes, 5′-GATTCGCTAAAGCATGCAGT-3′ and 5′-GAGCACTCTCCAGCGAGTAGAA-3′. The Brachyury coding sequences (for detection of expression from inducible cassette) were as follows: 5′-GCTTCAAGGAGCTAACTAACGAG-3′ and 5′-CCAGCAAGAAAGAGTACATGGC-3′. The Eomes coding sequences (for detection of expression from inducible cassette) were as follows: 5′-TGTTTTCGTGGAAGTGGTTCTGGC-3′ and 5′-AGGTCTGAGTCTTGGAAGGTTCATTC-3′; Bcl2, 5′-GCTACCGTCGTGACTTCGC-3′ and 5′-CCCCACCGAACTCAAAGAAGG-3′; and p53, 5′-GCCCCAGGATGTTGAGGAGTTT-3′ and 5′-AGGGACAAAAGATGACAGGGGC-3′.
Immunoblotting
Cells were washed in PBS and lysed in lysis buffer (20 mm Tris-Cl (pH 7.5), 2 mm EGTA, 1.5 mm MgCl2, 150 mm NaCl, 10 mm NaF, 12.5 mm β-glycerophosphate, 1 mm phenylmethylsulfonyl fluoride, and 1% Triton X-100). Proteins from whole-cell lysates were separated by 10 or 6% SDS-PAGE and were analyzed by immunoblotting. The antibodies used were as follows: anti-CHD4 (1:1000; Abcam, Cambridge, MA; ab70469); anti-MBD3 (1:1000; Bethyl, Montgomery, TX; A302-528A); anti-p53 (1:1000; Cell Signaling Technology, Danvers, MA; 1C12); anti-α-tubulin (1:10,000; Sigma; T6199); anti-acetyl-p53 (Lys-379) (1:1000; Cell Signaling Technology; 2570); anti-HA (1:1000, Covance, Princeton, NJ; 16B12); anti-Myc (1:2000, Santa Cruz Biotechnology; 9E10); anti-HDAC1 (1:1000, Merck, Rahway, NJ; 06-720); and anti-p300 (1:1000, Merck; 05-527).
Immunofluorescence
The cells were fixed with 4% paraformaldehyde in PBS for 15 min at room temperature. Then the cells were permeabilized with 0.5% Triton X-100 and blocked with 3% BSA in PBS. Next, the cells were incubated with primary antibodies overnight at 4 °C. The cells were then incubated with fluorescently labeled secondary antibodies (Molecular Probes) for 1 h at room temperature. Cells were finally mounted in a medium containing Mowiol 4-88 (Calbiochem; 475904). Images were acquired using the inverted microscope IX83 (Olympus, Tokyo, Japan). The primary antibodies used were anti-TUJ1 (1:250; Sigma), anti-cTNT (1:200; ThermoFisher Scientific; MS-295), and anti-AFP (1:400; DAKO, Glostrup, Denmark, or Santa Clara, CA; A000829).
Microarray analysis
Total RNA was isolated from control cells and knockdown cells using NucleoSpin RNA kit (TaKaRa). Biotinylated sense-strand DNA was synthesized using a GeneChip WT PLUS reagent kit (Affymetrix, Santa Clara, CA), hybridized to the Mouse Gene 2.0 ST Array (Affymetrix), and was analyzed using the GeneChip Scanner 7G System (Affymetrix). The data were analyzed by the GeneSpringGX12.6.1 software (Agilent Technologies, Santa Clara, CA). Hierarchical clustering analysis and GO analysis were performed using GeneSpring GX. Note that we refer to probe sets or entities as “genes” in the text. The microarray data have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO) under the accession number GSE114389.
Transfection
Mouse Hdac1, Chd4, p300, p53, and Bcl2 were amplified using PCR and cloned into Myc-pcDNA3 or HA-pcDNA3 expression vectors. The expression vectors were transfected into HEK293T cells or ESCs using FuGENE HD (Promega) or Lipofectamine 2000 (Invitrogen), respectively.
Author contributions
A. H. and M. N.-K. conceptualization; A. H., M. N.-K., and Y. A. data curation; A. H., M. N.-K., and Y. A. formal analysis; A. H. and M. N.-K. investigation; A. H. and M. N.-K. writing-original draft; E. N. supervision; E. N. funding acquisition; E. N. project administration; E. N. writing-review and editing.
Supplementary Material
This work was supported by grants from the Advanced Research and Development Programs for Medical Innovation from the Japan Agency for Medical Research and Development (AMED) (to E. N.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Tables S1–S4.
The microarray data have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO) under the accession number GSE114389.
- ESC
- embryonic stem cell
- NuRD
- nucleosome remodeling and deacetylase
- CHD4
- chromodomain helicase DNA-binding protein 4
- qRT
- quantitative RT
- CpG
- cytosine-phosphate-guanine
- GO
- gene ontology
- shRNA
- short hairpin RNA
- AFP
- α-fetoprotein
- GMEM
- Glasgow minimum essential medium
- LIF
- leukemia inhibitory factor.
References
- 1. Arnold S. J., and Robertson E. J. (2009) Making a commitment: cell lineage allocation and axis patterning in the early mouse embryo. Nat. Rev. Mol. Cell Biol. 10, 91–103 10.1038/nrm2618 [DOI] [PubMed] [Google Scholar]
- 2. Muñoz-Sanjuán I., and Brivanlou A. H. (2002) Neural induction, the default model and embryonic stem cells. Nat. Rev. Neurosci. 3, 271–280 10.1038/nrn786 [DOI] [PubMed] [Google Scholar]
- 3. Bylund M., Andersson E., Novitch B. G., and Muhr J. (2003) Vertebrate neurogenesis is counteracted by Sox1–3 activity. Nat. Neurosci. 6, 1162–1168 10.1038/nn1131 [DOI] [PubMed] [Google Scholar]
- 4. Iwafuchi-Doi M., Matsuda K., Murakami K., Niwa H., Tesar P. J., Aruga J., Matsuo I., and Kondoh H. (2012) Transcriptional regulatory networks in epiblast cells and during anterior neural plate development as modeled in epiblast stem cells. Development 139, 3926–3937 10.1242/dev.085936 [DOI] [PubMed] [Google Scholar]
- 5. Kamiya D., Banno S., Sasai N., Ohgushi M., Inomata H., Watanabe K., Kawada M., Yakura R., Kiyonari H., Nakao K., Jakt L. M., Nishikawa S., and Sasai Y. (2011) Intrinsic transition of embryonic stem-cell differentiation into neural progenitors. Nature 470, 503–509 10.1038/nature09726 [DOI] [PubMed] [Google Scholar]
- 6. Huang C., Xiang Y., Wang Y., Li X., Xu L., Zhu Z., Zhang T., Zhu Q., Zhang K., Jing N., and Chen C. D. (2010) Dual-specificity histone demethylase KIAA1718 (KDM7A) regulates neural differentiation through FGF4. Cell Res. 20, 154–165 10.1038/cr.2010.5 [DOI] [PubMed] [Google Scholar]
- 7. Aloia L., Di Stefano B., Sessa A., Morey L., Santanach A., Gutierrez A., Cozzuto L., Benitah S. A., Graf T., Broccoli V., and Di Croce L. (2014) Zrf1 is required to establish and maintain neural progenitor identity. Genes Dev. 28, 182–197 10.1101/gad.228510.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Thornton S. R., Butty V. L., Levine S. S., and Boyer L. A. (2014) Polycomb repressive complex 2 regulates lineage fidelity during embryonic stem cell differentiation. PLoS ONE 9, e110498 10.1371/journal.pone.0110498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Huang C., Su T., Xue Y., Cheng C., Lay F. D., McKee R. A., Li M., Vashisht A., Wohlschlegel J., Novitch B. G., Plath K., Kurdistani S. K., and Carey M. (2017) Cbx3 maintains lineage specificity during neural differentiation. Genes Dev. 31, 241–246 10.1101/gad.292169.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Baubec T., Ivánek R., Lienert F., and Schübeler D. (2013) Methylation-dependent and -independent genomic targeting principles of the MBD protein family. Cell 153, 480–492 10.1016/j.cell.2013.03.011 [DOI] [PubMed] [Google Scholar]
- 11. Le Guezennec X., Vermeulen M., Brinkman A. B., Hoeijmakers W. A., Cohen A., Lasonder E., and Stunnenberg H. G. (2006) MBD2/NuRD and MBD3/NuRD, two distinct complexes with different biochemical and functional properties. Mol. Cell. Biol. 26, 843–851 10.1128/MCB.26.3.843-851.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Spruijt C. G., Bartels S. J., Brinkman A. B., Tjeertes J. V., Poser I., Stunnenberg H. G., and Vermeulen M. (2010) CDK2AP1/DOC-1 is a bona fide subunit of the Mi-2/NuRD complex. Mol. Biosyst. 6, 1700–1706 10.1039/c004108d [DOI] [PubMed] [Google Scholar]
- 13. Yildirim O., Li R., Hung J.-H., Chen P. B., Dong X., Ee L.-S., Weng Z., Rando O. J., and Fazzio T. G. (2011) Mbd3/NURD complex regulates expression of 5-hydroxymethylcytosine marked genes in embryonic stem cells. Cell 147, 1498–1510 10.1016/j.cell.2011.11.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hassig C. A., Tong J. K., Fleischer T. C., Owa T., Grable P. G., Ayer D. E., and Schreiber S. L. (1998) A role for histone deacetylase activity in HDAC1-mediated transcriptional repression. Proc. Natl. Acad. Sci. U.S.A. 95, 3519–3524 10.1073/pnas.95.7.3519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Xue Y., Wong J., Moreno G. T., Young M. K., Côté J., and Wang W. (1998) NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol. Cell 2, 851–861 10.1016/S1097-2765(00)80299-3 [DOI] [PubMed] [Google Scholar]
- 16. Zhang Y., LeRoy G., Seelig H. P., Lane W. S., and Reinberg D. (1998) The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell 95, 279–289 10.1016/S0092-8674(00)81758-4 [DOI] [PubMed] [Google Scholar]
- 17. Allen H. F., Wade P. A., and Kutateladze T. G. (2013) The NuRD architecture. Cell. Mol. Life Sci. 70, 3513–3524 10.1007/s00018-012-1256-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hosokawa H., Tanaka T., Suzuki Y., Iwamura C., Ohkubo S., Endoh K., Kato M., Endo Y., Onodera A., Tumes D. J., Kanai A., Sugano S., and Nakayama T. (2013) Functionally distinct Gata3/Chd4 complexes coordinately establish T helper 2 (Th2) cell identity. Proc. Natl. Acad. Sci. U.S.A. 110, 4691–4696 10.1073/pnas.1220865110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kashiwagi M., Morgan B. A., and Georgopoulos K. (2007) The chromatin remodeler Mi-2 is required for establishment of the basal epidermis and normal differentiation of its progeny. Development 134, 1571–1582 10.1242/dev.001750 [DOI] [PubMed] [Google Scholar]
- 20. Williams C. J., Naito T., Arco P. G., Seavitt J. R., Cashman S. M., De Souza B., Qi X., Keables P., Von Andrian U. H., and Georgopoulos K. (2004) The chromatin remodeler Mi-2β is required for CD4 expression and T cell development. Immunity 20, 719–733 10.1016/j.immuni.2004.05.005 [DOI] [PubMed] [Google Scholar]
- 21. Yoshida T., Hazan I., Zhang J., Ng S. Y., Naito T., Snippert H. J., Heller E. J., Qi X., Lawton L. N., Williams C. J., and Georgopoulos K. (2008) The role of the chromatin remodeler Mi-2 in hematopoietic stem cell self-renewal and multilineage differentiation. Genes Dev. 22, 1174–1189 10.1101/gad.1642808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nitarska J., Smith J. G., Sherlock W. T., Hillege M. M., Nott A., Barshop W. D., Vashisht A. A., Wohlschlegel J. A., Mitter R., and Riccio A. (2016) A functional switch of NuRD chromatin remodeling complex subunits regulates mouse cortical development. Cell Rep. 17, 1683–1698 10.1016/j.celrep.2016.10.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Amaya M., Desai M., Gnanapragasam M. N., Wang S. Z., Zu Zhu S., Williams D. C. Jr., and Ginder G. D. (2013) Mi2-mediated silencing of the fetal-globin gene in adult erythroid cells. Blood 121, 3493–3501 10.1182/blood-2012-11-466227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. O'Shaughnessy-Kirwan A., Signolet J., Costello I., Gharbi S., and Hendrich B. (2015) Constraint of gene expression by the chromatin remodelling protein CHD4 facilitates lineage specification. Development 142, 2586–2597 10.1242/dev.125450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhao H., Han Z., Liu X., Gu J., Tang F., Wei G., and Jin Y. (2017) The chromatin remodeler Chd4 maintains embryonic stem cell identity by controlling pluripotency- and differentiation-associated genes. J. Biol. Chem. 292, 8507–8519 10.1074/jbc.M116.770248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kaji K., Caballero I. M., MacLeod R., Nichols J., Wilson V. A., and Hendrich B. (2006) The NuRD component Mbd3 is required for pluripotency of embryonic stem cells. Nat. Cell Biol. 8, 285–292 10.1038/ncb1372 [DOI] [PubMed] [Google Scholar]
- 27. Wilson S. I., and Edlund T. (2001) Neural induction: toward a unifying mechanism. Nat. Neurosci. 4, 1161–1168 10.1038/nn747 [DOI] [PubMed] [Google Scholar]
- 28. Faial T., Bernardo A. S., Mendjan S., Diamanti E., Ortmann D., Gentsch G. E., Mascetti V. L., Trotter M. W., Smith J. C., and Pedersen R. A. (2015) Brachyury and SMAD signalling collaboratively orchestrate distinct mesoderm and endoderm gene regulatory networks in differentiating human embryonic stem cells. Development 142, 2121–2135 10.1242/dev.117838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kalisz M., Winzi M., Bisgaard H. C., and Serup P. (2012) EVEN-SKIPPED HOMEOBOX 1 controls human ES cell differentiation by directly repressing GOOSECOID expression. Dev. Biol. 362, 94–103 10.1016/j.ydbio.2011.11.017 [DOI] [PubMed] [Google Scholar]
- 30. Lolas M., Valenzuela P. D., Tjian R., and Liu Z. (2014) Charting Brachyury-mediated developmental pathways during early mouse embryogenesis. Proc. Natl. Acad. Sci. U.S.A. 111, 4478–4483 10.1073/pnas.1402612111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Costello I., Pimeisl I. M., Dräger S., Bikoff E. K., Robertson E. J., and Arnold S. J. (2011) The T-box transcription factor Eomesodermin acts upstream of Mesp1 to specify cardiac mesoderm during mouse gastrulation. Nat. Cell Biol. 13, 1084–1091 10.1038/ncb2304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Izumi N., Era T., Akimaru H., Yasunaga M., and Nishikawa S. (2007) Dissecting the molecular hierarchy for mesendoderm differentiation through a combination of embryonic stem cell culture and RNA interference. Stem Cells 25, 1664–1674 10.1634/stemcells.2006-0681 [DOI] [PubMed] [Google Scholar]
- 33. Teo A. K., Arnold S. J., Trotter M. W., Brown S., Ang L. T., Chng Z., Robertson E. J., Dunn N. R., and Vallier L. (2011) Pluripotency factors regulate definitive endoderm specification through eomesodermin. Genes Dev. 25, 238–250 10.1101/gad.607311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gómez-Del Arco P., Perdiguero E., Yunes-Leites P. S., Acín-Pérez R., Zeini M., Garcia-Gomez A., Sreenivasan K., Jiménez-Alcázar M., Segalés J., López-Maderuelo D., Ornés B., Jiménez-Borreguero L. J., D'Amato G., Enshell-Seijffers D., Morgan B., et al. (2016) The chromatin remodeling complex Chd4/NuRD controls striated muscle identity and metabolic homeostasis. Cell Metab. 23, 881–892 10.1016/j.cmet.2016.04.008 [DOI] [PubMed] [Google Scholar]
- 35. Trouillas M., Saucourt C., Duval D., Gauthereau X., Thibault C., Dembele D., Feraud O., Menager J., Rallu M., Pradier L., and Boeuf H. (2008) Bcl2, a transcriptional target of p38α, is critical for neuronal commitment of mouse embryonic stem cells. Cell Death Differ. 15, 1450–1459 10.1038/cdd.2008.63 [DOI] [PubMed] [Google Scholar]
- 36. Hemann M. T., and Lowe S. W. (2006) The p53-Bcl-2 connection. Cell Death Differ. 13, 1256–1259 10.1038/sj.cdd.4401962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Polo S. E., Kaidi A., Baskcomb L., Galanty Y., and Jackson S. P. (2010) Regulation of DNA-damage responses and cell-cycle progression by the chromatin remodelling factor CHD4. EMBO J. 29, 3130–3139 10.1038/emboj.2010.188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ito A., Kawaguchi Y., Lai C.-H., Kovacs J. J., Higashimoto Y., Appella E., and Yao T.-P. (2002) MDM2-HDAC1-mediated deacetylation of p53 is required for its degradation. EMBO J. 21, 6236–6245 10.1093/emboj/cdf616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Knock E., Pereira J., Lombard P. D., Dimond A., Leaford D., Livesey F. J., and Hendrich B. (2015) The methyl binding domain 3/nucleosome remodelling and deacetylase complex regulates neural cell fate determination and terminal differentiation in the cerebral cortex. Neural Dev. 10, 13 10.1186/s13064-015-0040-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Reynolds N., Latos P., Hynes-Allen A., Loos R., Leaford D., O'Shaughnessy A., Mosaku O., Signolet J., Brennecke P., Kalkan T., Costello I., Humphreys P., Mansfield W., Nakagawa K., Strouboulis J., et al. (2012) NuRD suppresses pluripotency gene expression to promote transcriptional heterogeneity and lineage commitment. Cell Stem Cell 10, 583–594 10.1016/j.stem.2012.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Masui S., Shimosato D., Toyooka Y., Yagi R., Takahashi K., and Niwa H. (2005) An efficient system to establish multiple embryonic stem cell lines carrying an inducible expression unit. Nucleic Acids Res. 33, e43 10.1093/nar/gni043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cong L., Ran F. A., Cox D., Lin S., Barretto R., Habib N., Hsu P. D., Wu X., Jiang W., Marraffini L. A., and Zhang F. (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hoch R. V., and Soriano P. (2006) Context-specific requirements for Fgfr1 signaling through Frs2 and Frs3 during mouse development. Development 133, 663–673 10.1242/dev.02242 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.