Abstract
Iron is the most common transition metal cofactor across biological systems. As the earth transitioned from an anaerobic to aerobic environment, cellular mechanisms evolved to protect against iron-mediated oxidative damage, but the molecular details of these protective strategies remain unclear. In this report, the Lindahl group has combined spectroscopic, biochemical, and genetic approaches to inventory iron in Escherichia coli as a function of bacterial oxygen metabolism. Their results suggest that ferrous iron functions as an oxygen sink that is modulated by a “respiratory shield” of electron flux in the bacterial plasma membrane.
Introduction
The earth's ecosphere converted from reducing to oxidative ∼2.5 billion years ago, the boundary between the Archean and Proterozoic eons (1). This change meant a shift in iron's redox equilibrium from soluble FeII to FeIII, which is subsequently rendered insoluble by hydrolysis to [FeO(OH)]n (2). Organisms at this boundary—archaeal and bacterial anaerobes—were totally dependent on the ferrous iron pool in which they evolved. For example, the terminal electron transport pathway that fueled ATP synthesis at their plasma membranes was an enzyme cohort largely assembled using nutrient ferrous iron. In regard to energy metabolism, what distinguished nascent aerobes was a terminal dioxygen reductase coupled to this pathway, cytochrome c oxidase (3). Wofford et al. (4) provide a combination of 57Fe Mössbauer and iron EPR spectroscopy along with genetic manipulation and biochemical fractionation to suggest that this terminal respiration, in addition to using the free energy derived from reduction of dioxygen, assisted also in keeping the cytoplasm microaerobic and thus “protected” the cell's essential ferrous iron. When this protection is lost through run-down of terminal respiration in the bacterial inner membrane, oxygen tension increases in the cytoplasm, oxidizing a fraction of what is generally referred to as the labile iron pool of ferrous iron (5). This autoxidation results in the accumulation of a cytoplasmic pool of ferric iron nanoparticles. The Wofford et al. (4) study thus tells a compelling story about evolutionary aerobiosis and provides a compelling example of the power of precise spectroscopy in cell biology research.
Previous investigations of iron speciation in cells have utilized a variety of methods, including Mössbauer spectroscopy. Mössbauer is likely not the most familiar of spectroscopies, and yet it is a remarkably straightforward method. Briefly, it is γ-photon absorption spectroscopy. In iron Mössbauer, the photon comes from the first excited nuclear state of 57Fe in the source and is captured (absorbed) by the 57Fe in the sample (natural abundance 2.1%) resulting in a nuclear quantum transition that can be visualized as a loss of signal (6). This transition is called an isomer shift, but can be thought of as functionally equivalent to a chemical shift in NMR, where the iron species in the sample will have a specific readout. This isomer shift, δ, is therefore informative about the redox and electronic spin state of the metal. Broadly speaking, δ increases with spin state and with the increase in d-electron spin density in FeII compared with FeIII. Moreover, as in NMR, additional spectroscopic details can reveal the nuclear quantum state (via quadrupole splitting, Δ) or the electronic environment (via hyperfine splitting) (6). For example, quadrupole splitting occurs when the angular momentum quantum number, I, is >1/2 and is in a nonspherical electric field typical of an asymmetric ligand environment. Wofford et al. (4) use this splitting as an important diagnostic in their survey of cellular iron species. These elements of Mössbauer spectra are illustrated by some of their most significant data in Fig. 1A. Finally, Mössbauer has exceptional resolution but low sensitivity, meaning that spectral data accumulation takes hours not minutes; it is a credit to the authors that they have made the commitment to fully exploit this technique in the elucidation of cellular iron metabolism.
Figure 1.
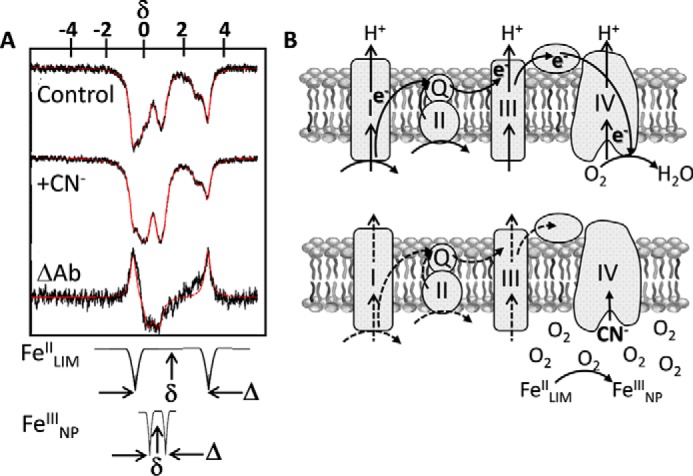
Using Mössbauer to track conversion of labile ferrous iron into ferric iron nanoparticles. A, illustrating how Wofford et al. use Mössbauer to catalogue cell iron species. Respiratory cyanide poisoning leads to conversion of FeII in a low-molecular-mass pool (FeIILIM) into a pool of FeIII nanoparticles FeIIINP. These two components of the experimental traces are idealized at the bottom of the panel. Note the typically larger δ for FeII compared with FeIII; the relatively large quadrupolar splitting, Δ, indicates that this FeII is high-spin. The trace labeled “ΔAb” is functionally the difference spectrum between the +CN− and control spectra in which a fraction of the FeIILIM in the control is replaced by an equivalent fraction of FeIIINP. These spectra are taken from Fig. 7 in Wofford et al. (4). B, the “respiratory shield” hypothesis illustrating how CN− inhibition of terminal reduction of O2 results in an increase in the cytoplasmic oxygen tension, the subsequent autoxidation of the LIM pool of ferrous iron, and the accumulation of ferric iron nanoparticles.
There are several unique take-home messages provided by the data presented by Wofford et al. (4). Previous work had provided evidence both for and against the storage of large amounts of iron as FeIII in ferritin (i.e. bacterioferritin), which could be used to protect cellular iron and avoid oxygen toxicity (7). In their system, using E. coli without overexpression of any proteins, Wofford et al. observed only a small amount of iron that displayed ferritin-like features, helping to resolve this controversy. Another novel observation is that the nonheme, high-spin FeII spectral signature of iron coordinated by O and N ligands is contributed by two biochemically separable species. Importantly, when cellular respiration in the form of electron flux to O2 via complex IV was inhibited by the addition of CN−, one of these species, designated FeIILMM, decreased (Fig. 1A). A significant fraction of this iron was converted to what the authors have characterized as magnetically ordered ferric oxyhydroxide nanoparticles, importantly differing from previous assignments of this species as ferritin iron. Based on biochemical fractionation, the authors further linked this FeIILMM species to what is commonly referred to as the cellular labile iron pool (4). The demonstration that this iron pool is sensitive to autoxidation and deposited as inert iron nanoparticles is a novel finding. Their model of this respiratory shield is illustrated in Fig. 1B.
The Lindahl group has previously demonstrated a similar link between diminished respiratory electron flux and iron nanoparticle deposition in mitochondria (7). Their conclusion is that whether in the bacterial inner membrane or in the mitochondrial matrix, aside from providing the H+-gradient that supports ATP synthesis, respiration maintains a microaerobic environment that stabilizes the ferrous iron required in the assembly of heme and FeS clusters. In this context, it is noteworthy that mitochondrial “anaerobiosis” also supports the assembly of an active nitrogenase, further evidence of this “respiratory shield” (8). After all, as Lynn Margulis (Sagan) pointed out over a half-century ago, mitochondria are simply repurposed bacteria (9). Wofford et al. (4) bring the story of evolutionary aerobiosis and the appearance of the eukaryotic cell full circle. It is a paper worth reading.
Footnotes
Research in the Kosman Lab is supported by Grant RO1NS102337 from the National Institute of Neurologic Diseases and Stroke of the Department of Health and Human Services (to D. J. K.). The author declares that he has no conflicts of interest with the contents of this article. The content of this article is solely the responsibility of the author and does not necessarily represent the official views of the National Institutes of Health.
References
- 1. Crichton R. R., and Pierre J. L. (2001) Old iron, young copper: From Mars to Venus. Biometals 14, 99–112 10.1023/A:1016710810701 [DOI] [PubMed] [Google Scholar]
- 2. Kosman D. J. (2013) Iron metabolism in aerobes: Managing ferric iron hydrolysis and ferrous iron autoxidation. Coord. Chem. Rev. 257, 210–217 10.1016/j.ccr.2012.06.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brochier-Armanet C., Talla E., and Gribaldo S. (2009) The multiple evolutionary histories of dioxygen reductases: Implications for the origin and evolution of aerobic respiration. Mol. Biol. Evol. 26, 285–297 10.1093/molbev/msn246 [DOI] [PubMed] [Google Scholar]
- 4. Wofford J. D., Bolaji N., Dziuba N., Outten F. W., and Lindahl P. A. (2018) Evidence that a respiratory shield in Escherichia coli protects a low-molecular-mass FeII pool from O2-dependent oxidation. J. Biol. Chem. 294, 50–62 10.1074/jbc.RA118.005233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lv H., and Shang P. (2018) The significance, trafficking and determination of labile iron in cytosol, mitochondria and lysosomes. Metallomics 10, 899–916 10.1039/C8MT00048D [DOI] [PubMed] [Google Scholar]
- 6. Pandelia M. E., Lanz N. D., Booker S. J., and Krebs C. (2015) Mössbauer spectroscopy of Fe/S proteins. Biochim. Biophys. Acta 1853, 1395–1405 10.1016/j.bbamcr.2014.12.005 [DOI] [PubMed] [Google Scholar]
- 7. Jhurry N. D., Chakrabarti M., McCormick S. P., Holmes-Hampton G. P., and Lindahl P. A. (2012) Biophysical investigation of the ironome of human Jurkat cells and mitochondria. Biochemistry 51, 5276–5284 10.1021/bi300382d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. López-Torrejón G., Jiménez-Vicente E., Buesa J. M., Hernandez J. A., Verma H. K., and Rubio L. M. (2016) Expression of a functional oxygen-labile nitrogenase component in the mitochondrial matrix of aerobically grown yeast. Nat. Commun. 7, 11426 10.1038/ncomms11426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sagan L. (1967) On the origin of mitosing cells. J. Theor. Biol. 14, 255–274 [DOI] [PubMed] [Google Scholar]


