Abstract
Explants of lymphoid tissue provide a rare opportunity to assess the organization of the immune system in a living, dynamic environment. Traditionally, ex vivo immunostaining is conducted in fixed tissue sections, while live tissues are analyzed using genetically engineered fluorescent reporters or adoptively transferred, pre-labelled cell populations. Here, we validated a protocol for immunostaining and imaging in live, thick slices of lymph node tissue, thus providing a spatial “map” of the lymph node while maintaining the viability and functionality of the slices. Using anti-B220/CD45R (B cell) as a prototype antibody, the procedure for immunostaining was tested for sufficient signal to noise with respect to staining time, temperature, and wash time, and the specificity was verified in comparison to isotype controls. Immunostaining signal in live tissue slices was detectable to least 120 μm deep for both whole antibodies and F(ab’)2 fragments using the staining procedure. This procedure revealed the expected changes in B cell organization in lymph nodes from immunized mice. Cell surface staining with most antibodies did not induce cytokine secretion, and cytokine secretion in response to T cell stimulation was unaffected by immunostaining. Staining with known a mitogenic antibody (anti-CD3) simultaneously labelled the cells and activated the tissue, confirming that reagents for live immunostaining must be selected judiciously. As a proof of concept, this method was used to reveal the dynamic distribution of CD69, a T cell activation marker, in lymph node slices before and after ex vivo stimulation.
Keywords: live imaging, fluorescent microscopy, immunocytochemistry, vaccination, tissue culture
1. Introduction
Live tissue explants have been used for decades as a unique experimental system that retains the complexity of intact tissue while making it experimentally accessible.1 A key advantage of tissue explants compared to in vitro cultures is that only the former preserve the spatial organization of cells, proteins, and matrix structures found in vivo. In the field of immunology, ex vivo slices of murine and human lymphoid tissue have been used to study thymocyte development and adaptive immune responses.2–7
While tissue slices could potentially accommodate spatially resolved analysis of dynamic immune responses, most current analysis techniques preempt such study by either dissociating or fixing the tissue. Dissociation into cell suspensions or lysates prior to gene expression analysis or flow cytometry means that these methods can provide no spatial information on the activated cell populations. On the other hand, spatially resolved analysis by protein immunostaining or RNA in situ hybridization is normally performed on fixed tissue sections, removing the opportunity to observe responses in living tissue after staining.
A method is needed to detect the organization of living lymphoid tissue samples without disrupting the function of the cells. One clever approach is to take advantage of differential uptake of vital dyes;8,9 this method best identifies divergent cell types and may not distinguish between resting vs activated cells or subtle polarization states as are expected in the lymph node. In addition, some vital dyes, e.g. DNA-binding molecules, may interfere with normal cellular function during on-going culture. Alternatively, immunofluorescent labelling has been reported recently for live lung slices and prostate explants.9,10 Immunofluorescence would be particularly useful to identify the organization of lymphoid tissues, where many cell types are well-described by surface proteins and fluorescently labelled antibodies are readily available. Immunostaining of live tissue is especially useful to study tissue organization in samples that are not readily labelled genetically, such as human or non-human primate tissue, as well as in readily available mouse models, bypassing the need to breed or acquire specialty transgenic strains that express specific fluorescent reporters. Here we report an optimized protocol to label cell surface markers and tissue structures in slices of live lymph node tissue using antibody-based reagents, while leaving the tissue viable and ready for further study.
2. Materials and Methods
2.1. Animal work
Female C57BL/6 mice were purchased from Jackson Laboratory or Taconic (USA) and housed in the University of Virginia vivarium with food and water ad libitum. All animal work was approved by the Institutional Animal Care and Use Committee at the University of Virginia under protocol #4042, and was conducted in compliance with guidelines the Office of Laboratory Animal Welfare at the National Institutes of Health (United States). Where noted, mice were vaccinated with chicken egg ovalbumin in complete Freund’s adjuvant prior to collection of lymph nodes (see Supporting Methods).
2.2. Slice preparation
On the day of the experiment, the animal was anesthetized with isoflurane and euthanized by cervical dislocation. The inguinal, brachial, and axillary lymph nodes were gently removed, carefully stripped of fat and connective tissue, and immediately placed into ice-cold DPBS without calcium or magnesium (Lonza, Walkersville MD, #17-512F) with 2% heat-inactivated FBS (Gibco, Fisher Scientific, Waltham, MA). Where noted, the popliteal node was collected as well. Nodes were embedded in 6% low melting point agarose (Lonza), prepared in 1x phosphate buffered saline (PBS), and allowed to chill on ice. Nodes were oriented flat side down to obtain the widest possible cross-section when sliced. A 10 mm tissue punch was used to obtain a column of agarose for each node. The LN tissue was sliced at a thickness of 300 μm in ice-cold PBS using a vibratome (Leica VT1000S, Bannockburn, IL) set to speed: 0.17 mm/s; freq: 30 Hz. Immediately after slicing, sections were transferred into a 12-well plate containing 2 mL complete media. Complete media consisted of RPMI (Lonza RPMI 1640 without L-glutamine, #12-167F) supplemented with 10% FBS, 1% L-glutamine, and 1% Pen/Strep, 50 μM beta-mercaptoethanol, 1 mM pyruvate, 1% non-essential amino acids, and 20 mM HEPES (Fisher Scientific). Immediately after slicing, the plate was transferred to a humidified sterile incubator at 37 °C with 5% CO2 for 1-2 hours for recovery. For sterile slicing, the procedure differed in the following ways: The agarose was sterilized by autoclaving prior to embedding tissue, 1% Pen-Strep was added to PBS buffer during slicing, the vibratome was placed in a biosafety cabinet to prevent contamination, and the vibratome frequency was reduced to 10 Hz.
2.3. Immunostaining and imaging of live lymph node slices
A 6-well plate was prepared by lining the wells with paraffin film that was sterilized in advance with 70% ethanol and dried. The paraffin film provided a hydrophobic surface to prevent the antibody solution from wicking across the plate under the tissue. Slices were transferred into the plate using a paintbrush and allowed to sit flat on the surface. To conserve antibodies, an A2 stainless steel flat washer (10 mm outer diameter, 5.3 mm inner; Grainger, USA) was placed on top of the slice, creating a 1-mm-deep well over each tissue sample. Slices were Fc-blocked by applying a 20-μL droplet of 25-μg/mL purified anti-mouse CD16/32 antibody (BioLegend, San Diego, CA) in 1x PBS with 2% heat-inactivated FBS (Gibco, Fisher Scientific) to the washer and incubating for 30 minutes in a humidified sterile incubator at 37 °C with 5% CO2. To stain, the blocking solution was left in place, an additional 10 μL of antibody cocktail was added (total volume: 30 μL), and the slice was incubated 1 hour or as noted. Antibodies are listed in Table S1, and preparation of fluorescently labelled antibodies and fragments is described in Supporting Methods. Antibodies were used at a concentration of 20 μg/mL (0.2 μg per tissue slice); preliminary tests showed little difference from 10 – 30 μg/mL. To remove unbound antibodies, the washer was removed, and the wells were filled with 10 mL of PBS and incubated 30 minutes at 37 °C or as noted, refreshing the PBS every 10-15 minutes. Slices were finally transferred into a 24-well plate in 0.5 mL PBS for immediate imaging on a Zeiss AxioZoom macroscope (Carl Zeiss Microscopy, Germany) with a Zeiss Axiocam 506 mono camera. Filters used were Zeiss Filter Set 38 HE (Ex: 470/40, Em: 525/50), 43 HE (Ex: 550/25, Em: 605/70); 64 HE (Ex: 587/25, Em: 647/70); and 50 (Ex: 640/30, Em: 690/50). Care was taken to image the slices from the side on which the antibodies had been applied, as they may not penetrate through the entire depth of the tissue.
2.4. Depth of antibody signal in tissue slices
Lymph node slices were double-stained with anti-B220 (CD45R) antibody and a fragment thereof (whole IgG–Alexa Fluor 647 and F(ab’)2–Alexa Fluor 555), using the staining protocol as described in section 2.3. Tissues were imaged on a Nikon A1Rsi upright confocal microscope (Nikon Instruments Inc., Melville NY) using GaAsP detectors and 40X/0.45NA Plan Apo NIR WD objective, in increments of 10 μm z-steps. Because B220+ cells are not uniformly distributed throughout the tissue, regions were selected for imaging based on having bright B220 staining near the surface of the tissue.
2.4. Viability of stained lymph node slices
To assess the viability of immunostained LN slices, LN slices were stained with anti-B220 as per method described above. After staining, a 24-well plate was filled with 2 μM Calcein AM (Fisher Scientific) in PBS, using 0.5 mL/well. Slices were immediately transferred into the wells and incubated for 20 minutes at room temperature, protected from light. Following incubation, wells were washed 3 times with PBS. Calcein intensity was averaged over the slice. Negative controls were prepared by treating a subset of slices with 70% ethanol for 10 minutes, and positive controls were prepared by keeping another subset of slices (unstained: no anti-B220) in a humidified sterile incubator at 37 °C with 5% CO2; these were Calcein-stained simultaneously with the experimental slices. Calcein intensity was imaged on a Zeiss AxioZoom macroscope.
2.5. IFN-γ secretion assay
Lymph node slices were obtained from 4 naive female C57BL/6 mice as described in section 2.2. Slices obtained were randomly assigned to each of 6 conditions: B220 + Lyve-1 staining or unstained for each PHA-L stimulated, CD3 stimulated, and unstimulated. Slices were stained as described in section 2.3 with anti-B220 FITC and anti-Lyve1 eFluor 660. Unstained control condition slices were left in media at 37 °C during this time. After staining, slices were placed in 500 μL of complete media and cultured for 20 hours at 37 °C with or without 25 μg/mL PHA-L (Fisher Scientific) or 1.25 μg/mL anti-CD3ε: (BioLegend). After 20 hours, supernatant was collected and analyzed by sandwich ELISA for IFNγ and TNFα (Supporting Methods).
2.6. Visualization of CD69 upregulation
Lymph node slices were obtained using sterile technique from 4 naive female C57BL/6 mice as described in section 2.2. The slices from each mouse were randomly assigned to each of 4 conditions: 1.) PreStain + Stimulation, 2.) PreStain + No Stimulation, 3.) No PreStain + Stimulation, or 4.) No PreStain + No Stimulation. Slices in the PreStain groups were stained as described in section 2.3 with anti-B220 FITC, anti-Lyve1 eFluor 570, and anti-CD69 Alexa Fluor 647 (Table S1). Unstained control slices were left in media at 37 °C during this time (no prestain). After prestaining, all slices were imaged on Zeiss AxioZoom macroscope. The slices were then placed in 500 μL of complete media in a 24-well plate and cultured for 16 hours at 37 °C, with or without 1.25 pg/mL anti-CD3ε. After incubation, slices were imaged again to quantify loss of signal from the prestain, stained once more using the same reagents and procedure, and imaged again.
2.7. Image analysis
Images were analyzed in ImageJ v1.48. To measure fluorescent intensity, a region of interest was drawn and the average signal was measured. For image display, brightness and contrast were adjusted uniformly across all compared slices unless otherwise specified.
3. Results and Discussion
3.1. Development of live immunostaining procedure
A naïve LN has a distinct cellular organization: B cells are concentrated in follicles around the periphery of the node, T cells are primarily in deep paracortex (center of the organ), and DCs are scattered throughout (Figure 1).11,12 These cells are organized within lobules surrounded by Lyve-1-positive lymphatic sinuses. In a LN undergoing an immune response, B cell follicles expand and/or multiply to maximize interaction with T cells and B-T cell region interfaces may be disrupted.12,13 Therefore, staining for B cells and lymphatics is often used to provide a first-order map of the gross structure of fixed sections of lymph node.
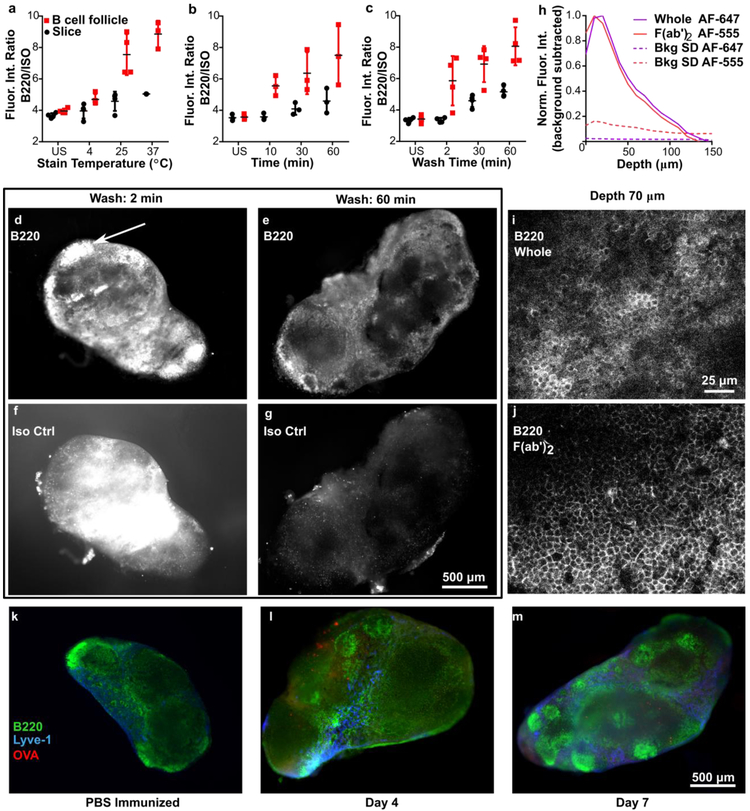
Figure 1.
Immunostaining procedure yields bright and specific staining at least 70 μm deep and reveals structures in lymph node, (a-c) Testing for sufficient stain time, temperature, and wash time. The fluorescent intensity ratio (FITC-anti-B220/ Alexa Fluor 594-IsoCtrl) was averaged over the entire slice or over a selected B cell follicular area, for stained and unstained (US) slices. Conditions for a: 1-hr stain, 30-min wash; b: 37 °C, 30-min wash; c: 37 °C, 1-hr stain. Each dot represents a single slice. Data from single experiment. Error bars denote standard deviation, (d-g) Representative images of 300-μm-thick lymph node slices stained with anti-B220 and its isotype control, after washing for 2 or 60 minutes. White arrow in (e) indicates a representative B cell follicle. Brightness and contrast were adjusted uniformly within each channel. (h) Fluorescent signal of anti-B220 IgG and F(ab’)2 fragment in a 300-μm-thick slice as measured by confocal microscopy. Dotted lines indicate one standard deviation (SD) of autofluorescence background as a function of depth, and solid colored lines indicate the average background-subtracted intensity (averaged across 20 positions). Data compiled from 2 replicate experiments, N = 20 z-stacks per condition. (i-j) Confocal images of antibody and fragment staining at a depth of 70 μm, both labeled with Alexa Fluor 647. Brightness and contrast were adjusted uniformly between the images, (k-m) Immunostaining of slices at different timepoints can reveal changes in substructure. After vaccination with rhodamine-OVA in CFA or with saline control, axillary LN slices were stained live immunostained with FITC-anti-B220 and eFluor 660-anti-Lyve-1. The images chosen were representative of the most common features observed in the majority of slices. Brightness and contrast were uniformly adjusted between images.
Here we tested procedures for fluorescent immunostaining of thick, live slices of lymph node. Briefly, immunofluorescence staining was performed by incubating the tissue first with CD16/32 (Fc receptor block) and then adding a cocktail of fluorophore-conjugated antibodies to label cells or structures of interest. Directly conjugated primary antibodies were used to avoid the additional processing time that would be needed to apply a secondary antibody. Only small-molecule fluorophores were used (e.g. Alexa Fluors), rather than the protein-based fluorophores commonly used for flow cytometry, to minimize the barrier to diffusion. After staining, the slices were washed with PBS to remove unbound reagents.
The staining procedure was performed using FITC-labelled anti-B220 as a prototype antibody, seeking to sufficiently maximize the ratio of fluorescent signals from anti-B220 and its Rat IgG2a isotype control (Iso Ctrl) in dual-stained slices. The signal ratio was tested as a function of incubation time, temperature, and wash time (Figure 1). We observed that the B220/Iso Ctrl ratio increased significantly with incubation temperature from 4 – 37 °C (Figure 1a) and time from 1 – 60 minutes (Figure 1b). For wash time, we observed that nonspecific binding of the isotype control was reduced to insignificant levels after a 30 - 60-minute wash in PBS at 37 °C (Figure 1c, d-g), thus increasing B220/Iso Ctrl signal. The best observed conditions were a 60-minute stain time at 37 °C followed by a 60-minute wash, but for practical purposes we find that 30-minute staining and washing is often sufficient. We now use this procedure routinely for other antibodies as well, including anti-Lyve-1 for lymphatics and CD69 (see section 3.3).
Despite the inclusion of Fc-blocking antibody in the cocktail, the IgG antibodies bound non-specifically to Fc receptors, leading to some undesirable off-target staining particularly in the sinus and medulla of the tissue (Figure 1g).14 We found that for this clone of anti-B220, the degree of off-target staining from the intact antibody was acceptably low, but for some other antibody isotypes it exceeded the on-target staining (data not shown), necessitating the use of antibody fragments instead.
As cells at the cut faces of the slice are expected to be damaged, it is useful for labelling reagents to penetrate at least several cell layers deep into the tissue. Using confocal microscopy, we observed that anti-B220 whole IgG and F(ab’)2 fragments labelled with Alexa Fluor 647 and Alexa Fluor 555, respectively, were both detectable above the autofluorescence signal to ~ 120 μm deep after a 60-minute stain time (Figure 1h and Figure S1). This is consistent with the expected diffusion coefficients for antibodies in intact tissues.15,16 Similar results were obtained when the whole IgG and F(ab’)2 fragment were labelled with DyLight 488 and Alexa Fluor 647, respectively (data not shown), suggesting that the observed depth of penetration was not influenced significantly by choice of fluorophore. However, we were unable to quantify the exact penetration of antibodies or fragments into the tissue with this method, due to limitations of light scattering in the tissue, and images collected deeper than 70-80 μm showed significant drop-off in intensity, quality, and clarity (Movie S1 and S2). When labelled with matching fluorophores, F(ab’)2 fragments tended to have marginally better image quality than whole IgG at the same depth (Figure 1i-j), as expected.
As an example of the types of images that can be collected using live immunostaining, we performed a simple vaccination, collected draining lymph nodes after varying time points, and performed live immunostaining with anti-B220 and anti-Lyve-1 (lymphatics). Consistent with prior studies,13 we observed that the LN structure was visibly altered in the vaccinated mice, with an increased variability in B cell follicle size and position throughout the LN (representative images shown in Figure 1 k-m).
One feature of tissue slices from is that the composition of the slice varies substantially with the position or depth in the lymph node from which the slice was collected. Therefore, structures can be highly variable between slices (see additional images provided in Figure S2), and a large number of slices must be examined to determine general trends or, in the future, conduct meaningful quantification of changes. Spatial heterogeneity would be expected in other structured organs as well, including tumors and brain tissue. This variability highlights the benefit of labelling live slices ex vivo to identify phenotypic regions prior to experiments intended to stimulate specific regions with drugs or immunotherapies.4,7,9
3.2. Slices remained viable and functional after live immunostaining
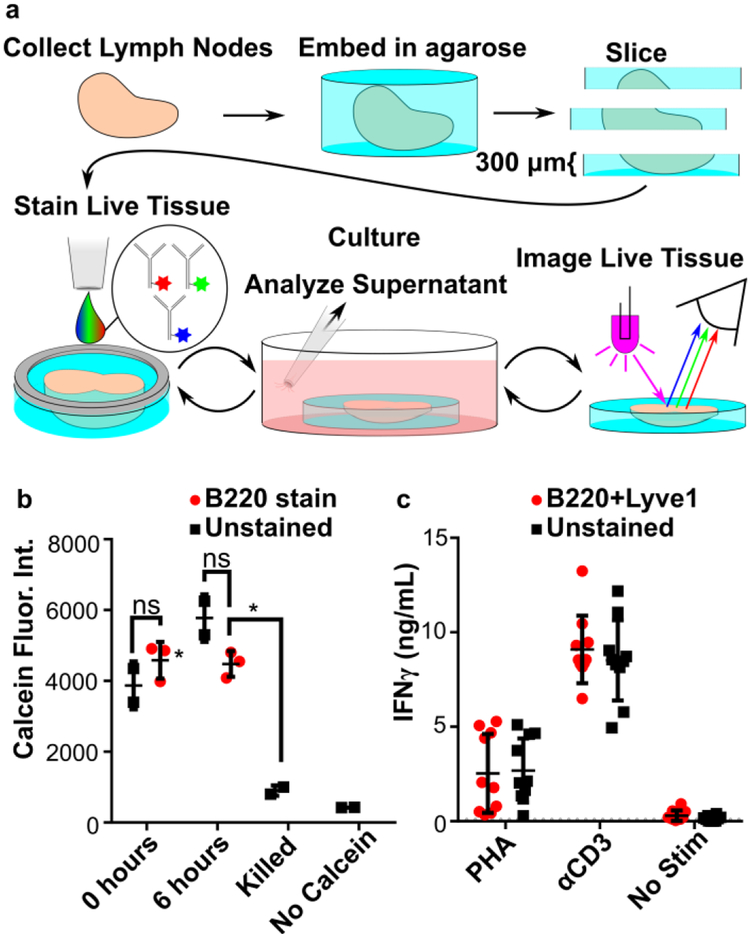
As we intended to use live immunostaining on tissue prior to short-term culture (Figure 2a), we tested the extent to which immunostaining altered the viability of the tissue or affected its ability to respond to stimuli. Slice viability was measured after immunostaining by using Calcein AM to assess cell membrane integrity.4 Compared to slices that were never immunostained, viability of immunostained slices was uncompromised (Figure 2b), both immediately after staining and 6 hours after staining.
Figure 2.
Live lymph tissue slices can be stained and cultured without impacting viability and function. (a) Overview of procedure for live immunostaining. During the culture period, stimuli can be added to slices. (b) Slices showed no difference in viability after immunostaining. Calcein fluorescent intensity was measured over whole slice at 0 and 6 hours after immunostaining with anti-B220. One-way ANOVA with Bonferroni’s multiple comparisons, data representative of 2 experiments, N=3 slices per condition. (c) Immunostaining neither inhibited nor promoted an immune response in slices. Slices dual-stained for B220 and Lyve-1 were incubated for 20 hours with or without activation by PHA or anti-CD3ε. IFN-γ was quantified in the supernatant by ELISA. LOD was 93 pg/mL (dotted line). Data compiled from 2 replicate experiments, N = 10 slices per condition. Error bars denote standard deviation.
Tissue function was assessed by measuring the secretion of IFN-γ after stimulation with phytohemagglutinin-L (PHA), a polyclonal T cell activator (Figure 2b). Slices that were dual-stained with anti-B220 and anti-Lyve-1 secreted similar quantities of IFN-γ as unstained slices after stimulation, indicating that immunostaining did not affect T cell responses. Furthermore, in the absence of stimulation, these antibodies did not induce IFN-γ secretion. TNF-α secretion was below the limit of detection (123 pg/mL) for all conditions.
In contrast to the results with anti-B220 and anti-Lyve-1, immunostaining with anti-mouse CD3ε antibody (clone: 145-201) activated T cells significantly more than PHA stimulation (p < 0.0001, two-way ANOVA with Sidak’s multiple comparisons). This was true for both labeled and unlabeled anti-CD3ε. Antibody-mediated crosslinking of cell surface CD3 is a standard strategy for T cell activation in cell cultures,4,5,6 and this result indicates that T cells in intact tissue can be activated in the same manner. Thus, when immunostaining for cell-surface antigens in live lymph node slices prior to subsequent experiments, care must be taken to avoid antigens such as CD3, whose cross-linking leads to cellular activation.
Here, we tested tissue function by a single metric. As for any protocol for live cells and tissues, live immunostaining must be validated for each specific application, depending on the cells or behavior of interest (e.g. to test the impact on function of B cells and lymphatic endothelial cells). Furthermore, while these experiments provided a relative assessment of viability and T cell function with respect to the immunostaining procedure, further work is needed to quantify the impact of slicing and extended culture on the viability and function of thick tissue slices. Both physical damage from the blade and depletion of oxygen or nutrient supply may play a role in slice physiology.
3.3. Live immunostaining revealed responses to ex vivo stimulation
Live immunostaining of thick tissue slices offers the unique advantage that the tissue can be imaged and then cultured ex vivo, stimulated, and analyzed repeatedly over time to detect changes in organization or function. Repeated analysis of the same tissue is not possible when using flow cytometry or conventional imaging of fixed tissue sections. As a proof of principle, live immunostaining was used to visualize immune-driven changes in the upregulation of CD69 (Figure 3). CD69 is a surface marker that is expressed in activated T Cells and is highly upregulated within several hours of T Cell receptor/CD3 ligation. In addition to testing CD69 upregulation by CD3 ligation, we also tested whether pre-staining the tissue had an impact on the observed response.
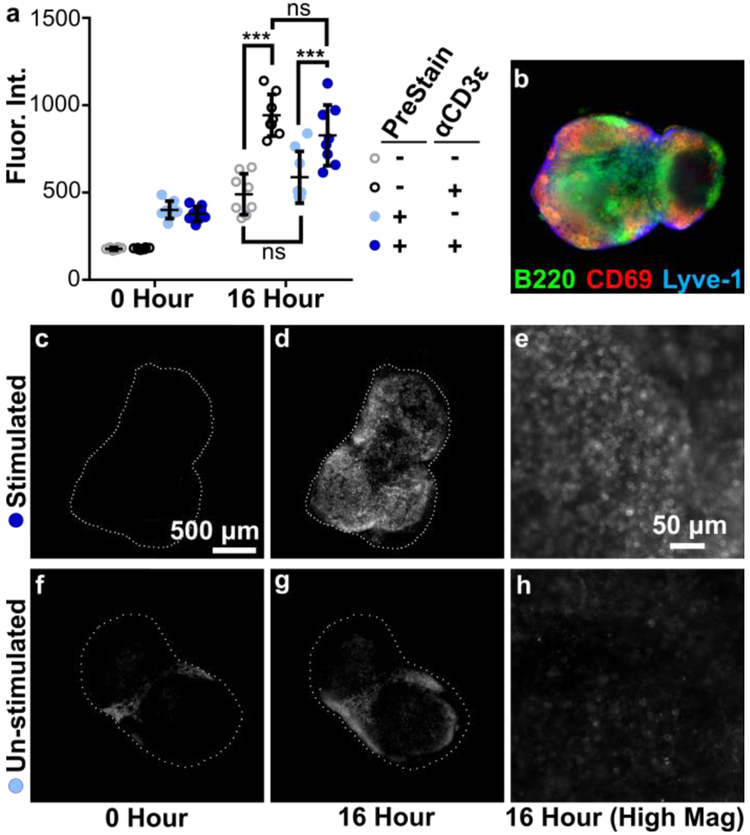
Figure 3.
Live immunostaining of LN slices before and after CD3 stimulation revealed increase of CD69 surface markers. Live LN’s were sliced and stained before stimulation (PreStain) to observe baseline in CD69 presentation. Slices were cultured and stimulated with anti-CD3ε and then stained again. (a) Mean fluorescent intensity of CD69 signal, averaged across the entire tissue, imaged before (0 Hour) and after stimulation (16 Hour). Data compiled from 2 replicate experiments, N=8 slices per condition, 2-way ANOVA analysis with Sidak multiple comparison. *** indicates p < .001. Error bars denote standard deviation, (b) A composite image of a representative slice after anti CD3ε stimulation, showing overlaid data for B220, Lyve-1, and CD69.(c-h) Representative single-channel images of Alexa Fluor 647 anti-CD69 data: PreStained + α.CD3ε (c) before and (d) after culture, and (e) high magnification after culture in the same slice; PreStained - α.CD3ε (f) before and (g) after culture, and (h) high magnification after culture in the same slice. Brightness and contrast uniformly adjusted for images (c,d,f, and g) and uniformly adjusted for (e and h). Dashed white lines show the outline of the tissue slice. Scalebar in (c) applies to (b-d,f, and g).
To observe baseline surface marker expression prior to stimulation, slices from naive LNs were stained with a cocktail of FITC anit-B220, eFluor570 anti-Lyve-1, and Alexa Fluor 647 anti-CD69, or left unstained, and imaged. After overnight culture with or without anti-CD3ε, all slices were imaged again, then stained and imaged a third time. Prior to restaining, overnight culture decreased the fluorescent intensity of all three markers by half (Figure S3), suggesting that the reagents slowly unbound and diffused out of the tissue over time. Thus, re-staining the same tissue slices after 1 – 2 days can reveal the organization of the tissue at that time, with only minimal interference from the previous staining step.
After culture, anti-CD3ε stimulated slices showed a drastic increase in CD69 and statistical analysis showed a significant difference between stimulated and unstimulated slices. Interestingly, un-stimulated slices also showed a slight increase in CD69 expression. In addition, pre-staining the tissue made no difference in overall fluorescent intensity, indicating that the immunostaining method did not, for this readout, significantly affect the behavior of the tissue. Furthermore, the fluorescent intensity of B220 and Lyve-1 did not significantly change before or after stimulation (Figure S3), confirming that the increase seen in CD69 signal was the result of a change in state of the tissue rather than an artifact of the procedure.
4. Conclusions
In conclusion, we describe a validated protocol for immunostaining live 300-μm-thick LN slices with fluorescent antibodies or antibody fragments. The live immunostaining protocol provides an organizational map of the LN slices while maintaining viability, allowing continued tissue culture, and preserving cytokine responses. The method was suitable to detect immune-driven changes not only in slices obtained from mice at different timepoints after vaccination (in-vivo structural changes) but also in the same slice at different timepoints during culture and stimulation (ex-vivo functional change). Because this method is similar to traditional immunocytochemistry and does not depend on genetic reporters, in principle it can be used to investigate other soft live tissue samples, including human tonsil tissue and murine thymus.2,3 It has been used previously to provide guidance for local stimulation of specific regions of LN tissue,4,7 and may prove useful to pre-screen tissue samples for regions of interest prior to experimentation.
Supplementary Material
Highlights.
Immunostaining was optimized for live slices of lymph node (LN) tissue.
The staining protocol reveals the organization of LN explants.
The protocol preserves the viability and cytokine secretion of LN tissue slices.
The protocol preserved activation marker expression after ex vivo stimulation.
Acknowledgements
This work was supported by The Hartwell Foundation (2015 Individual Biomedical Research Award) and the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (award number R01AI131723). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We would also like to thank Maura Belanger for assistance with lymph node collection, slicing, and for helpful discussions. Ben Groff was supported in part by a Double Hoo Award from the Center for Undergraduate Excellence at the University of Virginia. Jacob Woodroof was supported in part by a NanoSTAR Undergraduate Summer Research Award.
Footnotes
Competing Interests Statement:The authors have no competing interests to declare.
Supporting Information
Supporting information includes supplemental methods and SI Figures and Tables.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bach PH et al. The use of tissue slices for pharmacotoxicology studies: The report and recommendations of ecvam workshop 20. ATLA Altern. Lab. Anim. 24, 893–923 (1996). [Google Scholar]
- 2.Giger B et al. Human tonsillar tissue block cultures differ from autologous tonsillar cell suspension cultures in lymphocyte subset activation and cytokine gene expression. J. Immunol. Methods 289, 179–190 (2004). [DOI] [PubMed] [Google Scholar]
- 3.Ross J, Melichar H, Halkias J & Robey E Studying T Cell Development in Thymic Slices in T-Cell Development (eds. Bosselut R, Vacchio S, M. & Vacchio MS,) 131–140 (Springer; New York, 2016). doi: 10.1007/978-1-4939-2809-5_ll [DOI] [PubMed] [Google Scholar]
- 4.Ross AE, Belanger MC, Woodroof JF & Pompano RR Spatially resolved microfluidic stimulation of lymphoid tissue ex vivo. Analyst 142, 649–659 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoffmann P, Skibinski G & James K Organ culture of human lymphoid tissue. I. Characteristics of the system. J. Immunol Methods 179, 37–49 (1995). [DOI] [PubMed] [Google Scholar]
- 6.Katakai T, Habiro K & Kinashi T Dendritic Cells Regulate High-Speed Interstitial T Cell Migration in the Lymph Node via LFA-1/ICAM-1. J. Immunol 191, 1188–1199 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Ross AE & Pompano RR Diffusion of cytokines in live lymph node tissue using microfluidic integrated optical imaging. Anal. Chim. Acta 1000, 205–213 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Johnson S & Rabinovitch P Ex vivo imaging of excised tissue using vital dyes and confocal microscopy. Curr. Protoc. Cytom. Chapter 9, Unit 9.39 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson BP et al. Vital ex vivo tissue labeling and pathology-guided micropunching to characterize cellular heterogeneity in the tissue microenvironment. BioTechniqnes 64, 13–19 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thornton EE, Krummel MF & Looney MR Live imaging of the lung. Curr. Protoc. Cytom. Chapter 12, Unitl2.28 (2012). [DOI] [PubMed] [Google Scholar]
- 11.Germain RN et al. Making friends in out-of-the-way places: how cells of the immune system get together and how they conduct their business as revealed by intravital imaging. Immunol. Rev. 221, 163–181 (2008). [DOI] [PubMed] [Google Scholar]
- 12.Willard-Mack CL Normal Structure, Function, and Histology of Lymph Nodes. Toxicol. Pathol 34, 409–424 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Katakai T, Hara T, Sugai M, Gonda H & Shimizu A Lymph Node Fibroblastic Reticular Cells Construct the Stromal Reticulum via Contact with Lymphocytes. J. Exp. Med 200, 783–795 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kilarski WW et al. Intravital Immunofluorescence for Visualizing the Microcirculatory and Immune Microenvironments in the Mouse Ear Dermis. PLOS ONE 8, e57135 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li J, Czajkowsky DM, Li X & Shao Z Fast immuno-labeling by electrophoretically driven infiltration for intact tissue imaging. Sci. Rep. 5, 10640 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chung K et al. Structural and molecular interrogation of intact biological systems. Nature 497, 332–337 (2013).. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.