Abstract
An effective prophylactic vaccine against human immunodeficiency virus (HIV) will likely require a potent antibody response that can neutralize the virus at the mucosal portal of entry. The elicitation of potent broadly-neutralizing anti-sera will be an iterative process, optimizing candidates that only block a fraction of potential viral strains. This effect, termed “sieving”, is evidence of a partially efficacious vaccine. Understanding the mechanisms of resistance of the breakthrough viruses is important for improving vaccines. We developed a high-throughput assay that can be used on vaccine-elicited antisera or monoclonal antibodies. Using the SIVsmE660 swarm stock and sera from a large NHP vaccine/challenge study, our in vitro sieving assay identified the same viral subspecies as in the animal study—those with a canonical C1 amino acid variants conferring global neutralization resistance to antibodies. Using a genetically divergent swarm stock, we identified five other amino acid variants that confer global resistance; the C1 mutations in this stock were not selected, also in agreement with in vivo challenge studies. Thus, the in vitro sieving assay can be used with genetically diverse challenge stocks to predict the coverage of a vaccine-elicited sera and possibly inform candidate vaccine development efforts.
Keywords: Antibodies, vaccines, SIV, nonhuman primates
1. Introduction
An effective prophylactic vaccine against human immunodeficiency virus (HIV) will likely require a potent antibody response that can neutralize the virus at the mucosal portal of entry. Both HIV and its nonhuman primate (NHP) analog, simian immunodeficiency virus (SIV), can be neutralized by monoclonal antibodies and polyclonal sera in vitro (Mason et al., 2016). Traditional vaccines to date have failed to elicit the breadth and potency likely required to impact the HIV epidemic, leading to efforts directed at engendering broadly neutralizing antibodies (bNAbs) or delivering such bNAbs as preventive drugs.
Both HIV and SIV have an extraordinary range of virus sequence diversity because of their propensity to rapidly mutate and escape immune pressure (Allen et al., 2000; Borrow et al., 1997). Therefore, any potential vaccine candidates being developed for clinical trials must demonstrate protection by selectively blocking infection by the widest range of viral isolates. However, as yet, assays that can predict such selection have not been proven in clinical settings. As such, the NHP model has been a key part of this effort.
There have been a number of NHP vaccine/challenge studies demonstrating a range of vaccine efficacies (Bomsel et al., 2011; Hansen et al., 2011; Letvin et al., 2011; Santra et al., 2010; Vaccari et al., 2016). We carried out and published a large study as a model of a then-ongoing clinical trial which showed partial efficacy against SIVsmE660 virus swarm challenge (Roederer et al., 2014). By sequencing transmitted/founder (T/F) viruses, we demonstrated “sieving” – that is, the vaccine was particularly effective against viruses that carried a canonical amino acid signature in the Env gene, and it was ineffective against viruses with variants at these positions. Indeed, we found that the introduction of just two amino acid variants could dramatically alter the neutralization phenotype of E660-derived viruses in vitro.
However, a subsequent vaccine/challenge study using a related SIVsmE660 swarm did not identify the same sieving effect (Smith et al., 2016). In order to understand these apparently discrepant results, we developed an in vitro sieving assay presented here. This assay can also be adapted to test a wide range of HIV variants to identify the potential efficacy of vaccine elicited sera and/or bNAbs to inhibit infection.
2. Materials and methods
1.1. Virus expansion
PBMC was isolated from blood of rhesus macaques using Ficoll-Paque PLUS (GE Heathcare). For virus expansion, we followed the protocol developed by George Shaw (Li et al., 2016). Briefly, CD4 T cells isolated from human or rhesus PBMC using non-human primate CD4+ T Cell Isolation Kit (Miltenyi Biotec) were stimulated with T Cell Activation/Expansion Kit (Miltenyi Biotec). Four days after stimulation, cells were infected with viruses in the presence of 30 μg/ml of DEAE-Dextran (Sigma) for 4 hours with gentle mixing every 30 min. SIVsmE660 (2008) were expanded once on human CD4 T cells and SIVsmE660 (2015) were expanded once on rhesus CD4 T cells (matching the original innocula). After infection, cells were washed with medium three times and resuspended in RPMI medium (Thermo Fisher Scientific) supplemented with 15% Fetal Bovine Serum (FBS) (Thermo Fisher Scientific) and Recombinant Human IL-2 Protein (R&D SYSTEMS) at 30 IU/ml. Viruses were harvested every three days and p27 concentration was measured by p27 Antigen Capture Assay (ABL, Inc).
1.2. Virus titration for sieving assay
PBMC isolated from rhesus blood were stimulated with Concanavalin A from Canavalia ensiformis (Jack bean) (SIGMA-ALDLICH) at 25 μg/ml and IL-2 at 20 IU/ml in RPMI medium supplemented with 10% FBS (R-10). At 4 days post stimulation, viruses were diluted with R-10 (IL-2: 20 IU/ml) at 1:4 for 10 serial dilutions. 100 μl of virus diluents were transferred to a round bottom Corning® 96 Well TC-Treated Microplate (SIGMA-ALDLICH) in four replicates and 2 × 105 cells in 100ul of R-10 containing IL-2 at 20 IU/ml were added to each well. Culture supernatants were collected every week up to 2 weeks post infection (p.i.). p27 concentrations in the culture supernatant were measured by p27 Antigen Capture Assay and TCID50 values were calculated based on Spearman-Karber method(Kärber, 1931; Spearman, 1903).
1.3. Preparation of target cells
CD8+ cells were depleted from rhesus PBMC using non-human primate CD8 Microbeads (Miltenyi Biotec). CD8-depleted PBMC were stimulated with ConA at 25 μg/ml and IL-2 at 20 IU/ml in R-10. The medium was replaced with R-10 containing IL-2 at 20 IU/ml the next day. The assays were set up 4 days after stimulation in round bottom Corning® 96 Well TC-Treated Microplates (SIGMA-ALDLICH). 40 μl of viruses (ranging from 0.0006 to 0.02 M.O.I.) were mixed with 10 μl of VRC332 sera diluted at 1:2. For SIV mAb sieving assay, antibodies were diluted at 50 μg/ml and 10 μl of antibodies were mixed with 40 μl of virus. The mixtures were incubated at 37C, for 30 min. After incubation, 1.5 × 105 cells in 20 μl of R-10 containing IL-2: 20 IU/ml and 1mM indinavir (AIDS reagent program) were added to each well.
1.4. Cell harvest and amplification of env region by PCR
At 24 hours after infection, cells were spun and genomic DNA was extracted using QIAamp 96 DNA Blood Kit (QIAGEN). Genomic DNA was used as template for PCR and SIVsmE660 env region was amplified using KAPA HiFi PCR Kit (supplier). PCR was carried out in MicroAmp 96 well reaction plates (Thermo Fisher Scientific) with the following PCR parameters: 1 cycle of 95C for 3min, 30 cycles of a denaturing step of 98C for 20 sec, an annealing step of 62.5C for 15 sec and an extension step of 72C for 4 min followed by a final extension step of 72C for 4 min.
1.5. PCR product purification and preparation for MiSeq
PCR products were purified using Agencourt AMPure XP (Beckman Coulter). DNA concentrations were measured using Quant-iT™ PicoGreen™ dsDNA Assay Kit (Thermo Fisher Scientific). Purified PCR products were diluted to 0.1 ng/ul and Illumina libraries were generated using the Nextera XT DNA Library Preparation Kit (Illumina). Final products were measured on 2100 Bioanalyzer (Agilent) using High Sensitivity DNA kit (Agilent) to check the quality of the library.
1.6. Sequencing
Libraries were sequenced by 2 × 150 base paired-end reads an Illumina MiSeq sequencer. The final dataset comprised 1.7 × 107 read pairs in total and each sample corresponded to 1.9 × 105 read pairs on average.
1.7. Sequence analysis
Trimmomatic (version 0.22) was used to remove adapters and low quality bases. The trimmed paired-end reads were mapped to the reference SIV genome (“SIVsmE660”) using blastn program (default parameter except “-outfmt 6”) in BLAST+ software package (“BLAST+ 2.2.31”). We then wrote a Perl program “blast2vcf.pl” to extract all variations based on the blast result and save them in “vcf” format. In this process, only reads with alignment percentage >80% (at least 80% of trimmed reads mapped to the SIV reference genome) and identity score >80% were considered. We wrote another Perl program “vcfAnnot.pl” to annotate all the variations identified based on the detailed annotation of “SivsmE660.gbk” file. Our interest was to find the significant variations but not those rare ones supported by very few reads. Here, we define the significant variation as those supported by ≥ 10 reads and had frequency ≥0.1 in at least one sample.
1.8. Pseudovirus production
One day before transfection, 1.8 × 106 293T cells in 20 ml of DMEM–Dulbecco’s Modified Eagle Medium (Thermo Fisher Scientific) supplemented with 10% FBS (D-10) 100 IU/mL of penicillin-streptomycin (cDMEM) were seeded in T-75 SIGMA Corning® cell culture flasks (SIGMA-ALDLICH). The next day, 10 μg of SG3 delta env plasmid and 3 μg of pcDNA3.1 expressing SIVsmE660.CR54 mutant env were mixed and transfected to 293T cells using X-tremeGENE™ Transfection Reagents (SIGMA-ALDLICH). At 24 hours post transfection, the culture medium was replaced with fresh cDMEM. At 48 hours after transfection, the culture supernatants were harvested and filtered with Steriflip-HV, 0.45 µm, PVDF, radio-sterilized (Millipore). Harvested viruses were aliquoted and stored at −80C.
1.9. Virus titration for neutralization assay
Neutralization assays were performed as previously described (Li et al., 2005). Briefly, viruses were made in 2-fold dilution, 10 times with D-10. 10ul of D-10 were dispensed to all wells in Greiner CELLSTAR® 96 well plates black polystyrene wells flat bottom (SIGMA-ALDLICH). 40ul of diluted viruses or D-10 were transferred to the plate followed by the addition of trypsinized TZM-bl cells diluted at 1000 cells/20ul in D10. Next day, 80ul/well of D-10 was added to all wells. At 48 hours post infection, cells were harvested with 50ul/well of Steady-Glo® Luciferase Assay System (Promega) with shake on the shaker for 20 min. Fluorescence was measured by SpectraMax L Microplate Reader (MOLECULAR DEVICES). Titer was calculated based on Reed and Muench method. For neutralization assay, virus titer that reaches 50000 RLU was calculated.
1.10. Pseudovirus neutralization assay
Neutralization assays were performed as previously described (Li et al., 2005). In brief, viruses were diluted at a titer determined to result in 50,000 relative light units (RLU) in a luciferase assay. For SIV-specific monoclonal antibodies (mAb) (Mason et al., 2016), 5-fold serial dilutions starting at 50 μg/ml were made in duplicate and 10 μl/well of diluted mAb and 40 μl/well of viruses were mixed in black, flat bottom polystyrene CELLSTAR® 96 well plates and incubated at 37C for 30 min in a humidified 5% CO2–95% air environment. Freshly trypsinized TZM-bl (10,000 cells in 20 μl cDMEM containing 75 μg/ml DEAE-dextran) were added to the mAb-virus mixture and incubated at 37C in a humidified 5% CO2–95% air environment. The next day, 80 μl cDMEM was added to each well and 48 hours after infection, cells were harvested by adding 80 μl Steadylite plus Reporter Gene Assay System (Perkin Elmer) to each well. After a 2-min incubation at room temperature to allow cell lysis, luminescence was measured using Perkin Elmer luminometer.
3. Results
3.1. Comparability of sieve effects in vivo and in vitro
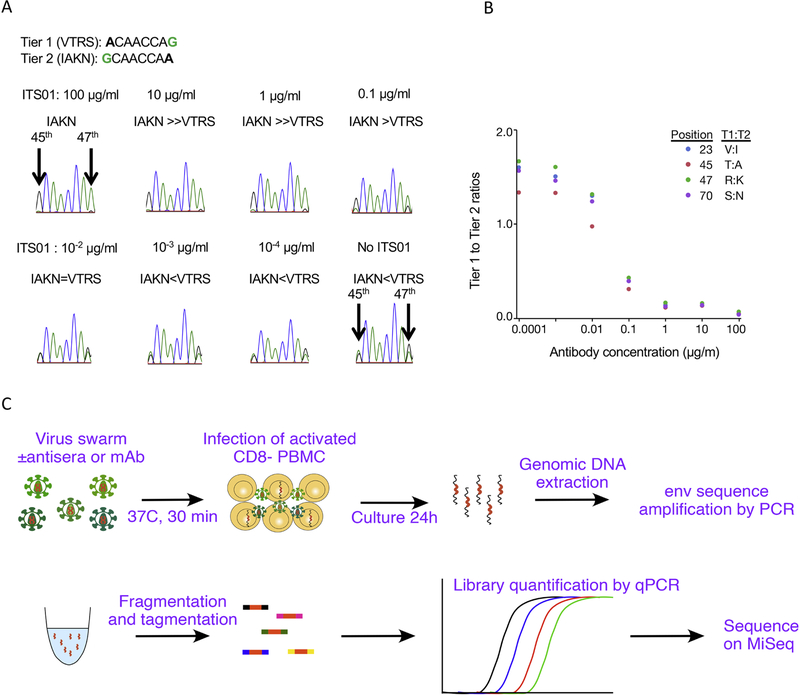
Sieving effects in a preclinical or clinical vaccine trial are indicative of partial vaccine efficacy. Defining mechanisms underlying differential transmission of variant sequences from the available infectious clones may help with vaccine development. As a prototype, we tested in vitro sieving assay using a mixture of two infectious molecular clones (IMC) with distinct neutralization profiles, SIVsmE660-FL14 and SIVsmE660-FL14.AK. These two IMC differ only at positions 23, 45, 46, and 70 in the C1 portion of Env (Roederer et al., 2014). The two viruses were mixed and used as an artificial swarm to infect target cells in the presence of serially diluted CD4bs-specific SIVmAb, ITS01. Based on Sanger sequencing, we observed the expected selection of resistant viruses in the presence of ITS01. (Figure 1A, B).
Fig. 1.
In vitro sieving assay using artificial mixtures of molecular clones, and schema of the assay using deep sequence. (A) in vitro sieving assay using mixture of tier 1 (VTRS) and tier 2 (IAKN) SIVsmE660 IMC after infection in the presence of various concentrations of ITS01. Peaks in Sanger sequence at 45th and 47th residues in Env are shown. (B) The ratios of Tier 1:Tier 2 variants at the 4 positions in ENV are graphed for each condition. The proportions show an expected selection for (enrichment) neutralization-resistant, Tier 2 viruses, in the presence of higher amounts of ITS01. (C) Schema of in vitro sieving assay and deep sequence analysis.
Based on these promising results, we set out to determine if we could recapitulate in vivo sieving using a high throughput in vitro assay using sera from animals in a large vaccine/challenge study. The experimental workflow is shown in Fig. 1C. We selected sera from five Gag vaccinated animals (for which no protection was observed in vivo), and sera from five animals in four different Env vaccines (for which we found a selection for viruses containing an “IAKN” signature in the C1 region). Viral inocula were incubated with antisera at 37C for 30 min, followed by addition of CD8 depleted, activated rhesus PBMC (with indinavir to prevent spreading). 24hr post infection, genomic DNA was extracted, and the Env region was amplified by nested PCR. The amplification largely eliminates the overall quantitation of entry while maintaining the ratio of subspecies; i.e., by this assay, sera that uniformly block all virus subtypes to varying degrees could not be distinguished, although that data is readily available by standard neutralization assays.
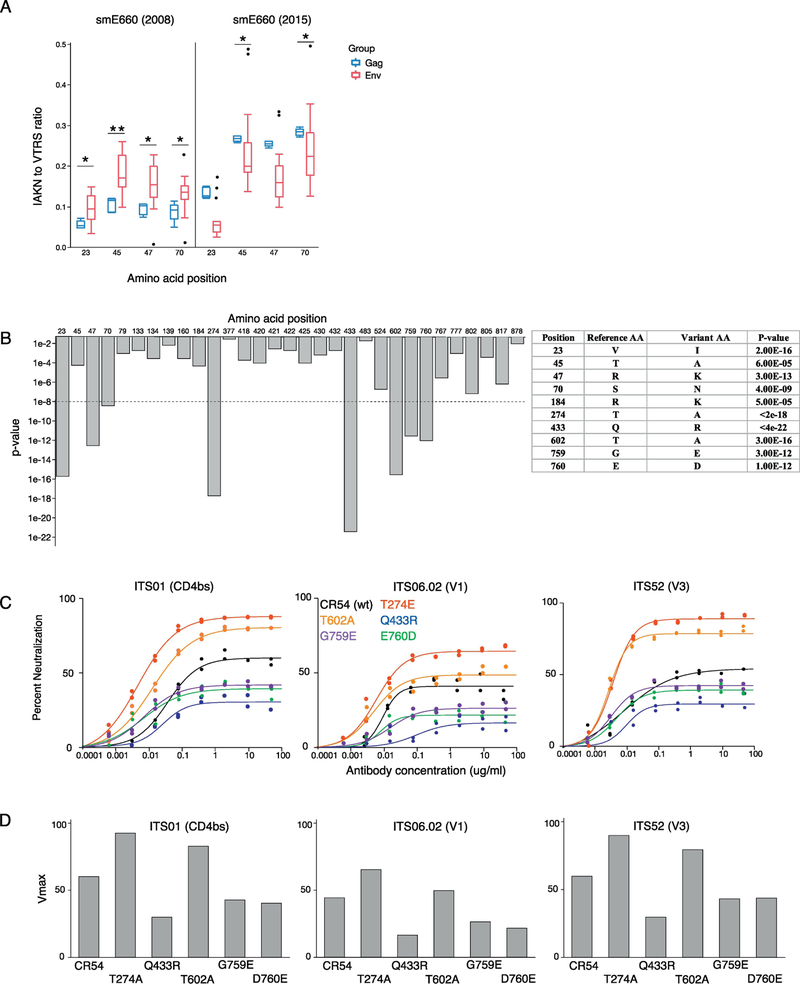
Sequences were mapped to SIVsmE660-FL14 (tier 1) Env(Wu et al., 2012), a clone derive from the smE660 swarm. Based on these reads, we quantified the frequency of the canonical IAKN sequence at residues 23, 45, 47 and 70 (neutralization-resistant genotypes) versus variant VTRS sequence at the same residues (neutralization-sensitive genotypes) in cell-associated DNA. These are from viruses able to infect cells in the presence of vaccine sera and reverse transcribe. As shown in Fig 2A (left), in the presence of sera from the partially efficacious Env vaccine, there was an over-representation of the IAKN sequences – mimicking the in vivo sieving effect we observed with this swarm.
Fig. 2.
In vitro sieving assay using VRC332 sera. (A) IAKN to VTRS ratios in SIVsmE660 (2008) and SIVsmE660 (2015) sieving assay. Blue boxes show ratios in Gag vaccinated sera and red boxes shows ratios in Env vaccinated sera. P-values: ** ≤ 0.01 < * ≤0.05 (Mann-Whitney). (B) The bar graph shows amino acid residues with p-values < .05 between Gag and Env vaccinated groups. The table is the list of amino acid variants with uncorrected p values < 6 × 10–5. Other than the first four, these variants were not found in the 2008 stock. (C) Neutralization curves of CR54 single mutants against ITS01 (CD4bs), ITS06.02 (V1) and ITS52 (V3). Black: CR54, Red: T274A, Blue: Q433R, Orange: T602A, Violet: G759E and Green: E760D (D) Vmax of single CR54 mutants calculated based on the neutralization curves.
The initial PCR amplification used primers outside the Env coding sequence in highly conserved regions. In the 2008 stock for which we have a deep sequence analysis, the forward primer encompassed 3 positions with 50–75% variation (in the middle of the sequence), with other positions highly conserved (<1% variation). In the reverse primer, only one position showed variation (5%) and all others were highly conserved. Thus, we did not expect a significant impact of PCR efficiency on amplicon representation in the assay. Note that PCR bias should not impact the results of this assay, since any systematic amplification bias would impact a variant in all wells equally. Importantly, we do not compare any given variant directly against another variant; we only compare a given variant’s prevalence under different conditions. If a given variant showed inefficient amplification, we presume that inefficiency would be the same in all wells, and any reduction in infection of that variant would be faithfully represented.
3.2. Identification of novel amino acid residues that modulate global neutralization profile
A later study using a different smE660 swarm stock did not reproduce this canonical sequence finding. To determine the potential mechanisms for this, we performed our sieving assay with the same sera and the 2015 viral inoculum. As shown in Fig 2A (right), we were able to reproduce the lack of selection for the IAKN signature; in fact, we found that the IAKN signature was selectively blocked in this viral swarm. Thus, we were able to recapitulate in vitro the apparently contradicting sieving effects observed in these studies.
We hypothesized that the different in sieving effect might be explained by the presence of other sequence variations within Env. We expanded our in vitro sieving assay using 80 sera from four vaccine groups, comparing the sequences selected by three Env vaccine regimens to those selected by the Gag vaccine (n = 20 animals each). Here, again, we used the 2015 stock of SIVsmE660 swarm. Across every position in Env, we defined the amino acid sequence variants observed in animals following Env or Gag vaccination and determined if differences were significant (Fig 2B). With this larger sample size, we confirmed the selection of VTRS/IAKN variants at position 23, 45, 47, and 70 in Envv.
We also identified a significant sieving effect at a number of other residues by our in vitro sieving assay which were not identified by T/F analysis of infected animals. We chose five new residues that showed the strongest sieving effect, to characterize their impact on virus neutralization. These included positions 274 (between V2 and V3), 433 (V4), and 602, 756 and (all in gp41). We introduced these individually into the SIVsmE660.CR54 clone, which has a tier 2 phenotype and contains the IAKN neutralization-resistant signature present in the C1 region. Using pseudotyped virus neutralization assay on TZM-bl, we evaluated neutralization sensitivities of these mutant viruses to a variety of mAbs against SIV, including ITS01 (CD4-binding site), ITS06.02 (V1) and ITS52 (V3) (Fig 2C). All single mutations changed the “global” neutralization profile, altering the ability of all of these viruses to escape control.
Against multiple distinct mAbs, the T274A and T602A mutations increased sensitivity to neutralization by increasing the neutralization sensitive fraction by 21–32% and 5–22%, respectively (Figure 2D). In contrast, Q433R, G759E and E760D conferred increased neutralizationresistance, by decreasing the neutralization fractions by 30%, 17%, and 16–23%, respectively. These results demonstrate that there are a range of amino acid variants across SIV Env that can impact the global neutralization profile to a range of mAbs in an epitope-independent manner.
The variants we found to confer these effects in the 2015 stock were not found in the 2008 challenge stock. In addition, the prevalence of the AK variants was much higher in the 2015 inoculum. These results suggest that the later stock had evolved away from the earlier stock both by expanding the AK variants as well as selecting for novel variants.
3.3. Neutralization profile of Env mutants by monoclonal antibodies
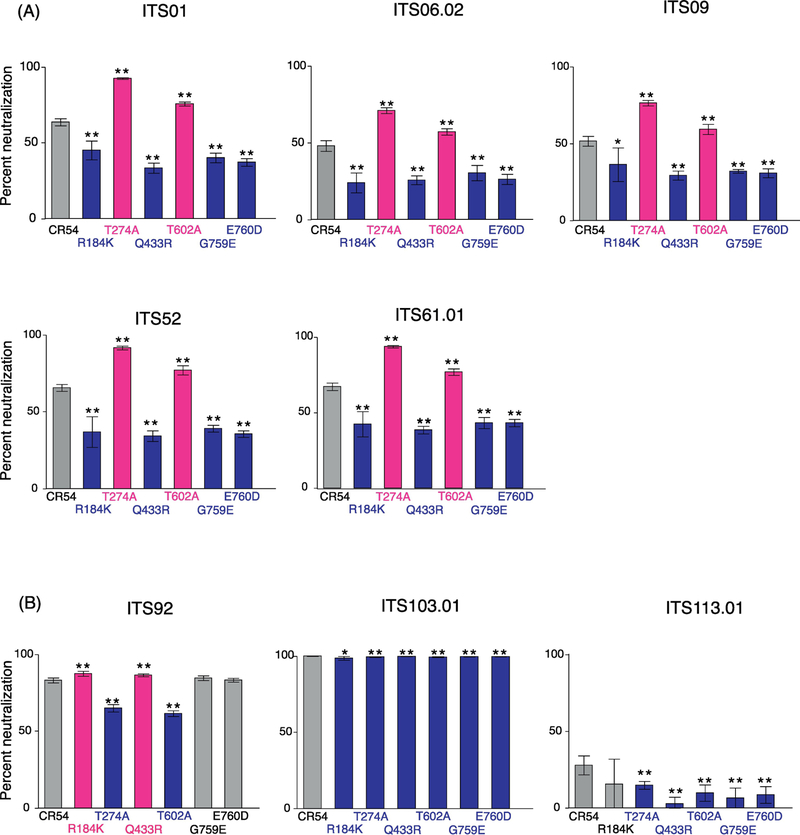
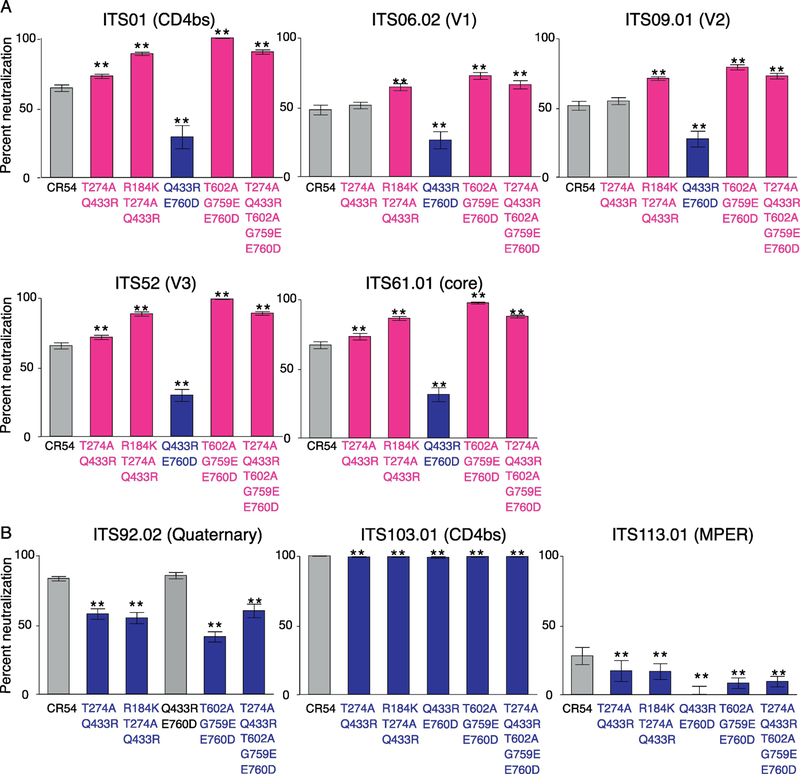
We investigated how each single mutation affects neutralization profiles against various SIVmAbs against a range of epitopes, including the CD4bs (ITS01, ITS103.01), gp120 core (ITS61.01), quaternary epitope (ITS92.02), MPER (ITS113.01) variable loops V1 (ITS06.02), V2 (ITS09.01) and V3. SIV mAbs ITS01, ITS06.02, ITS09.01, ITS52 and ITS61.01 are so-called “1st generation” mAbs because although they mediate nearly 100% neutralizing activity against Tier 1 SIV, they show incomplete neutralization of Tier 2 SIV and no neutralization of Tier 3 SIV. This is consistent with what has generally been observed for SIV vaccine-elicited sera. We have since isolated additional “2nd generation” or SIV broadly neutralizing antibodies (bnAbs), such as ITS92.02, ITS103.01 and ITS113.01, which show complete neutralization against Tier 1, 2 and 3 SIV, including the highly neutralization-resistant SIVmac239.
The impact of each mutation on the 1st generation antibodies was highly concordant (Fig 3a). Irrespective of the targeted epitope, each mutation conferred the same neutralization phenotype on the virus : either increasing neutralization sensitivity (T274A, T602A) or resistance (R184K, Q433R, G759E, E670D). Indeed, even the magnitude of these effects was similar. On the other hand, the SIV bnAbs were differently impacted by the same mutations, in particular, for ITS92 (targeting an unmapped quaternary epitope), the effect was nearly opposite to that observed for 1st generation SIV mAbs. By comparison, ITS103.01 (CD4bs) was able to mediate complete neutralization all mutants while all of these mutants increased resistance to ITS113.01 (MPER). That these mAbs are so distinct in their responses to these same mutations speaks to the unusual paths taken to elicit these mAbs, much like the case in humans with HIV, where only a minority generate bnAbs, and usually only after a several years post-infection.
Fig. 3.
One-point neutralization assay of SIVsmE660.CR54 single mutants by various SIVmAbs. Percentage of neutralization was assessed at 10 μg/ml of SIVmAbs in eight replicates. (A) Neutralization by 1st generation SIVmAbs. (B) Neutralization by 2nd generation SIVmAbs. Statistical differences in percent neutralization against original SIVsmE660.CR54 were analyzed by Mann-Whitney U test. P-value: ** ≤ 0.01 < * ≤ 0.05. Mutations that increased sensitivities to NAb are shown in magenta and mutations that made virus more resistant to NAb are shown in blue.
3.4. Correspondence of Sieved sequences by vaccine sera and monoclonal antibodies
We then tested whether our assay could identify antibody-mediated sieving effects on the SIVsmE660 swarm by setting up the in vitro sieving assay using SIV mAb with different specificities, including ITS01 (CD4bs), ITS06.02 (V1), ITS09.01 (V2), ITS52 (V3), ITS61.01 (gp120 core) and ITS113.01 (MPER). All mAbs were tested at 10 ug/ml in eight replicates, and the sieve effect compared against cultures with no mAb. As a comparison of sieving effect with polyclonal vaccine sera, we used 20 sera from the vaccine study, and compared Gag- (control, n = 5) to Env-vaccinated (n = 15) sera.
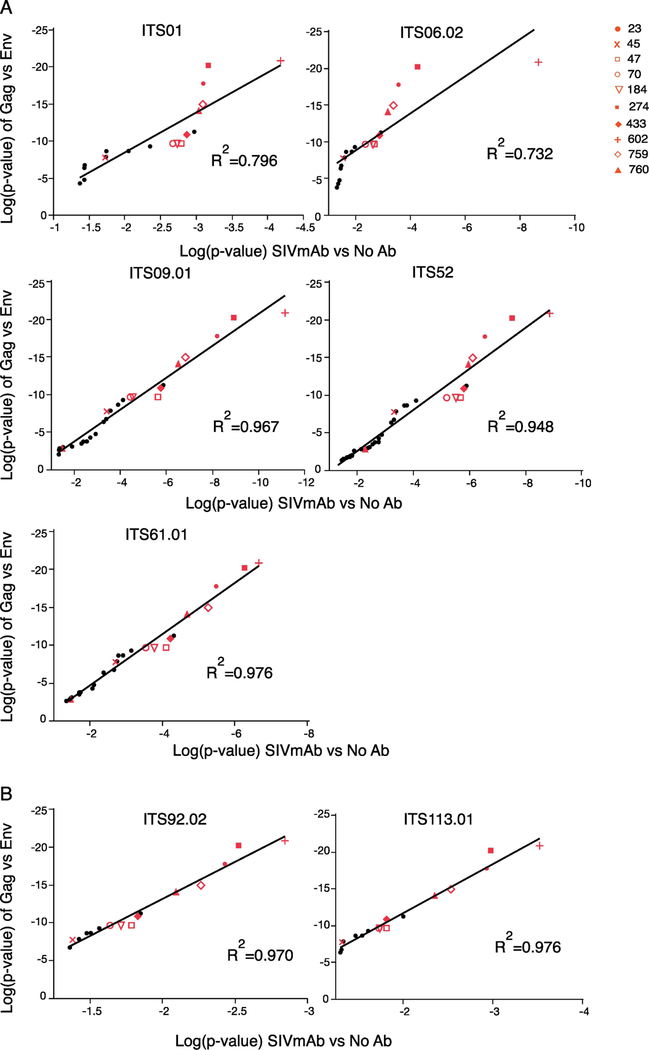
This approach would be expected to identify epitope-specific mutations that confer resistance, as demonstrated previously by Dingens et al (Dingens et al., 2017), using artificial mixtures of point mutants in HIV Env. However, we observed the same “global” sieve effect in response to the different SIV mAbs as well as the polyclonal vaccine sera. Indeed, as shown in Fig 4, there was a high degree of correlation between the p values (used here as an estimate of the selection) between a range of our 1st generation SIV mAbs and the vaccine-elicited sera. As was proposed by Georgiev et al. (Doria-Rose et al., 2017), the different neutralization profiles of the antisera and monoclonals might be used to identify the dominant neutralization response types of the sera.
Fig. 4.
SIVsmE660 (2015) sieving assay using SIVmAb and VRC332 sera. Ratios of Env variants were statistically analyzed between Gag and Env vaccinated animals or SIVmAb and no antibody samples by Mann-Whitney U test. Variants selected with significant p-value were converted into log and plotted on graphs. R-squared is shown on the graph. Red plots demonstrate variants that were selected with significant p-value in sieving assay using VRC332 sera. (A) Correlation of log(p-value) between Env and 1st generation SIVmAb (B) Correlation between log(p-value) of Env and 2nd generation SIVmAb.
3.5. Interaction of Env mutations
Finally, we asked whether the mutations in SIV Env could be combined to elicit a more pronounced effect on the viral neutralization phenotype. As shown in Fig 5, combinations of mutations can indeed alter the phenotype of the viruses where single mutations had minimal effect. In general, a mutation that increases “global” resistance is silenced by a mutation that increases susceptibility – that is, the “global” sensitivity mutations are dominant. Thus, the explanation for the lack of selection of “IAKN” variants in the E660 challenge studies using a different swarm can be explained by an, as yet unidentified, difference in the Env sequence that confers dominance over these variants (Fig 5A).
Fig. 5.
Neutralization profiles against combination mutants. Single mutations were combined in the CR54 backbone and neutralization sensitivities were evaluated by SIVmAbs. Statistical differences in percent neutralization against original SIVsmE660.CR54 were analyzed by Mann-Whitney U test. P-value: ** ≤ 0.01 < * ≤0.05. (A) Neutralization profile against 1st generation SIVmAbs. (B) Neutralization profile against 2nd generation SIVmAbs. Sensitizing mutations are shown in magenta and resistant mutations are shown in blue.
Interestingly, the broadly-neutralizing SIV mAbs do not necessarily follow these patterns, illustrating that their dependence on alternative Env structures is mechanistically different. ITS103.01, a CD4bs-directed mAb that completely neutralizes nearly all SIVs, was essentially unaffected by the mutations (Fig 5B) – although statistically significant, the loss in neutralization sensitivity averaged only 2%. In contrast, ITS92.02, an unmapped quaternary antibody, and ITS113.01, an MPER mAb, were nearly always much less potent against most combinations of mutations.
4. Discussion
We developed an in vitro “sieving” assay that can recapitulate in vivo sieving. Using a tissue-culture-expanded stock of the same SIVsmE660 “swarm” challenge virus used in our vaccine/challenge study (ref 9), together with sera from vaccinated animals, we identified several of the same amino acid mutations in SIV Env identified by T/F analysis of vaccinated and infected animals that correlated with neutralization sensitivity. We propose that this assay may be used in the future to characterize the potential potency of clinical vaccines and monoclonal antibodies, by rapidly assessing the neutralization resistance of a wide range of sequence variants simultaneously.
In this assay, we focused on comparing proviral sequence between control and vaccinated groups to have the best chance of identifying neutralization variants. It is conceivable that this assay could be configured to identify mutations impacting infectivity (e.g., by comparing variant representation in cells to that in the swarm). This would be hampered by the presence of a large fraction of non-infectious particles (Moore et al., 2006). We note that in our original in vivo study, the variant frequencies in transmitted/founders infecting non-Env vaccinated animals were represented at equal proportions as in the challenge stock itself (Roederer et al., 2014, Fig 2d).
A related method was published for antibody epitope mapping using mixtures of amino acid mutations in a single HIV strain (Dingens et al., 2017). By comparison, our in vitro sieving assay can simultaneously detect all possible amino acid variants within a swarm virus in response to immune selection pressure exerted by mAbs or polyclonal vaccine sera. Importantly, we observed the same mutations selected by 1st generation SIV mAbs as we previously observed in response to vaccine elicited sera. That our in vitro sieving assay reliably recapitulates the in vivo sieving effect on swarm virus highlights the utility of this assay in a clinical setting where single or multiple mAb combinations could be assessed for their in vitro efficacy against a diverse SIV swarm to predict in vivo efficacy against primary HIV infection. Alternatively, sieving effect against a large mixture of multiple HIV (or HIV pseudo-typed virus) strains could be evaluated (deCamp et al., 2014; Hraber et al., 2017). This assay is well-suited to high-throughput analysis, and potentially enables simultaneous testing of multiple serum samples against numerous viruses or mixtures of viruses. Given sera from recipients of different vaccine regimens (e.g. phase I safety trial or preclinical testing), our in vitro assay might be used to rank the potential protective effect in a more physiological setting, ie., exposure to a range of viruses rather than a clonal challenge.
We used our in vitro sieving assay to explore the partial or incomplete neutralization phenotype of SIV. Although less pronounced than for SIV, incomplete neutralization of HIV by glycan - targeting bnAbs is well documented (Bonsignori et al., 2011; Cale et al., 2017; Doria-Rose et al., 2014; Mason et al., 2016). However, incomplete neutralization by glycan-targeting mAbs may be best described as a “local” neutralization effect, in that this particular effect is only observed for viruses by glycan-targeting mAbs but not mAbs directed to other epitopes. On the other hand, particular SIV strains, unlike HIV, often show a “global” incomplete neutralization phenotype that impacts a wide range of antibodies across many epitope specificities. This phenomenon was first demonstrated using 1st generation SIV mAbs as well as vaccine-elicited sera from our NHP vaccine efficacy trial in a SIV Env pseudotype-based neutralization assay (ref 80 animal study) and then recapitulated in vitro here using the same SIVsmE660 swarm challenge stock used in the vaccine efficacy trial. Using our in vitro sieving assay, we identified the same 4 amino acid positions mediating “global” neutralization resistance (IAKN) or sensitivity (VTRS) in the SIVsmE660 strain.
However, our sieving assay revealed that this is not a universal neutralization signature amongst all SIV strains. All SIVmac251 lineage viruses have the “IAKN” neutralization-resistant signature, irrespective of how neutralization sensitive they are (Kilgore et al., 2015). In addition, more recent challenge studies also did not find this signature amongst the vaccinated animals (Burton et al., 2015; Kilgore et al., 2015; Smith et al., 2016). Indeed, when we tested the SIVsmE660 challenge stock in our in vitro sieving assay using NHP vaccine efficacy trial animal sera (ref 80 animal study), we also did not reproduce the same sequence signatures observed from the in vivo T/F analysis; indeed, we found an opposite dependence, that “VTRS” variants were the more resistant genotype. In addition, we identified multiple additional variant sequences conferring global neutralization resistance.
To better understand the effect of each virus variant on neutralization sensitivity, we constructed pseudotyped viruses for each variant. We found that there are multiple postions in SIV Env where mutation can have a significant impact on the “global” virus neutralization phenotype, and importantly, that combinations of mutations can either enhance or diminish the impact on “global” virus neutralization. However, we also observed that sensitizing mutations are often dominant over resistance mutations. We hypothesize that the SIVsmE660 swarms used in challenge studies subsequent to ours have a predominance of these mutations in trans, that silences the neutralization-resistant phenotype of the “IAKN” signature with the C1 region of SIV Env. Interestingly, these sites were still selected in our assay when we used the 2015 challenge stock – but in that stock, in vitro, they increased sensitivity. Thus, the structure alterations that the SIV Env undergoes to achieve this altered phenotype must rely on these sites as a fulcrum – and the result is either increased or decreased resistance to 1st generation and vaccine-elicited antibodies depending on the context of other amino acid mutations.
In summary, we have designed a high throughput in vitro sieving assay that can recapitulate the in vivo sieving in preclinical animal models. We used these data to identify a new set of amino acids signatures in SIV Env that confer the structural alterations underlying the “global” neutralization resistance phenotype. Furthermore, we determined that the impact of any single amino acid is dependent on the backbone context of the virus – i.e., mutation at a given amino acid position in the context of a particular viral sequence can lead to increased neutralization resistance but the same mutation can have the opposite effect in the context of another viral sequence. Overall, our in vitro sieving assay will be useful for screening antibody responses to understand the interactions between HIV/SIV Env and antibodies and to assess vaccine- and antibody-mediated protection against HIV.
Highlights.
A high throughput in vitro “sieving” assay can predict breakthrough sequences from a diverse swarm of challenge viruses
This assay is rapid, relies on deep-sequencing, and identifies viral variants that escape neutralization to infect
SIV Env variants escape vaccine-mediated control through an epitope-independent “global” neutralization resistance
Complex interactions between Env mutations can confer resistance or sensitivity depending on the sequence context.
Acknowledgements
We thank the members of the ImmunoTechnology Section at the VRC for discussion and advice.
Funding: This work was supported by the Intramural Research Program of the Vaccine Research Center, NIAID, NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Allen TM, O’Connor DH, Jing P, Dzuris JL, Mothe BR, Vogel TU, Dunphy E, Liebl ME, Emerson C, Wilson N, Kunstman KJ, Wang X, Allison DB, Hughes AL, Desrosiers RC, Altman JD, Wolinsky SM, Sette A, Watkins DI, 2000. Tat-specific cytotoxic T lymphocytes select for SIV escape variants during resolution of primary viraemia. Nature 407, 386–390. [DOI] [PubMed] [Google Scholar]
- Bomsel M, Tudor D, Drillet AS, Alfsen A, Ganor Y, Roger MG, Mouz N, Amacker M, Chalifour A, Diomede L, Devillier G, Cong Z, Wei Q, Gao H, Qin C, Yang GB, Zurbriggen R, Lopalco L, Fleury S, 2011. Immunization with HIV-1 gp41 subunit virosomes induces mucosal antibodies protecting nonhuman primates against vaginal SHIV challenges. Immunity 34, 269–280. [DOI] [PubMed] [Google Scholar]
- Bonsignori M, Hwang KK, Chen X, Tsao CY, Morris L, Gray E, Marshall DJ, Crump JA, Kapiga SH, Sam NE, Sinangil F, Pancera M, Yongping Y, Zhang B, Zhu J, Kwong PD, O’Dell S, Mascola JR, Wu L, Nabel GJ, Phogat S, Seaman MS, Whitesides JF, Moody MA, Kelsoe G, Yang X, Sodroski J, Shaw GM, Montefiori DC, Kepler TB, Tomaras GD, Alam SM, Liao HX, Haynes BF, 2011. Analysis of a clonal lineage of HIV-1 envelope V2/V3 conformational epitope-specific broadly neutralizing antibodies and their inferred unmutated common ancestors. J Virol 85, 9998–10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrow P, Lewicki H, Wei X, Horwitz MS, Peffer N, Meyers H, Nelson JA, Gairin JE, Hahn BH, Oldstone MB, Shaw GM, 1997. Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat Med 3, 205–211. [DOI] [PubMed] [Google Scholar]
- Burton SL, Kilgore KM, Smith SA, Reddy S, Hunter E, Robinson HL, Silvestri G, Amara RR, Derdeyn CA, 2015. Breakthrough of SIV strain smE660 challenge in SIV strain mac239-vaccinated rhesus macaques despite potent autologous neutralizing antibody responses. Proc Natl Acad Sci U S A 112, 10780–10785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cale EM, Gorman J, Radakovich NA, Crooks ET, Osawa K, Tong T, Li J, Nagarajan R, Ozorowski G, Ambrozak DR, Asokan M, Bailer RT, Bennici AK, Chen X, Doria-Rose NA, Druz A, Feng Y, Joyce MG, Louder MK, O’Dell S, Oliver C, Pancera M, Connors M, Hope TJ, Kepler TB, Wyatt RT, Ward AB, Georgiev IS, Kwong PD, Mascola JR, Binley JM, 2017. Virus-like Particles Identify an HIV V1V2 Apex-Binding Neutralizing Antibody that Lacks a Protruding Loop. Immunity 46, 777–791 e710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- deCamp A, Hraber P, Bailer RT, Seaman MS, Ochsenbauer C, Kappes J, Gottardo R, Edlefsen P, Self S, Tang H, Greene K, Gao H, Daniell X, Sarzotti-Kelsoe M, Gorny MK, Zolla-Pazner S, LaBranche CC, Mascola JR, Korber BT, Montefiori DC, 2014. Global panel of HIV-1 Env reference strains for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol 88, 2489–2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingens AS, Haddox HK, Overbaugh J, Bloom JD, 2017. Comprehensive Mapping of HIV-1 Escape from a Broadly Neutralizing Antibody. Cell Host Microbe 21, 777–787 e774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doria-Rose NA, Altae-Tran HR, Roark RS, Schmidt SD, Sutton MS, Louder MK, Chuang GY, Bailer RT, Cortez V, Kong R, McKee K, O’Dell S, Wang F, Abdool Karim SS, Binley JM, Connors M, Haynes BF, Martin MA, Montefiori DC, Morris L, Overbaugh J, Kwong PD, Mascola JR, Georgiev IS, 2017. Mapping Polyclonal HIV-1 Antibody Responses via Next-Generation Neutralization Fingerprinting. PLoS Pathog 13, e1006148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doria-Rose NA, Schramm CA, Gorman J, Moore PL, Bhiman JN, DeKosky BJ, Ernandes MJ, Georgiev IS, Kim HJ, Pancera M, Staupe RP, Altae-Tran HR, Bailer RT, Crooks ET, Cupo A, Druz A, Garrett NJ, Hoi KH, Kong R, Louder MK, Longo NS, McKee K, Nonyane M, O’Dell S, Roark RS, Rudicell RS, Schmidt SD, Sheward DJ, Soto C, Wibmer CK, Yang Y, Zhang Z, Program NCS, Mullikin JC, Binley JM, Sanders RW, Wilson IA, Moore JP, Ward AB, Georgiou G, Williamson C, Abdool Karim SS, Morris L, Kwong PD, Shapiro L, Mascola JR, 2014. Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies. Nature 509, 55–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen SG, Ford JC, Lewis MS, Ventura AB, Hughes CM, Coyne-Johnson L, Whizin N, Oswald K, Shoemaker R, Swanson T, Legasse AW, Chiuchiolo MJ, Parks CL, Axthelm MK, Nelson JA, Jarvis MA, Piatak M Jr., Lifson JD, Picker LJ, 2011. Profound early control of highly pathogenic SIV by an effector memory T-cell vaccine. Nature 473, 523–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hraber P, Rademeyer C, Williamson C, Seaman MS, Gottardo R, Tang H, Greene K, Gao H, LaBranche C, Mascola JR, Morris L, Montefiori DC, Korber B, 2017. Panels of HIV-1 Subtype C Env Reference Strains for Standardized Neutralization Assessments. J Virol 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kärber G, 1931. Beitrag zur kollektiven Behandlung pharmakologischer Reihenversuche. Naunyn-Schmiedebergs Archiv für experimentelle Pathologie und Pharmakologie 162, 480–483. [Google Scholar]
- Kilgore KM, Murphy MK, Burton SL, Wetzel KS, Smith SA, Xiao P, Reddy S, Francella N, Sodora DL, Silvestri G, Cole KS, Villinger F, Robinson JE, Pulendran B, Hunter E, Collman RG, Amara RR, Derdeyn CA, 2015. Characterization and Implementation of a Diverse Simian Immunodeficiency Virus SIVsm Envelope Panel in the Assessment of Neutralizing Antibody Breadth Elicited in Rhesus Macaques by Multimodal Vaccines Expressing the SIVmac239 Envelope. J Virol 89, 8130–8151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letvin NL, Rao SS, Montefiori DC, Seaman MS, Sun Y, Lim SY, Yeh WW, Asmal M, Gelman RS, Shen L, Whitney JB, Seoighe C, Lacerda M, Keating S, Norris PJ, Hudgens MG, Gilbert PB, Buzby AP, Mach LV, Zhang J, Balachandran H, Shaw GM, Schmidt SD, Todd JP, Dodson A, Mascola JR, Nabel GJ, 2011. Immune and Genetic Correlates of Vaccine Protection Against Mucosal Infection by SIV in Monkeys. Sci Transl Med 3, 81ra36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Wang S, Kong R, Ding W, Lee FH, Parker Z, Kim E, Learn GH, Hahn P, Policicchio B, Brocca-Cofano E, Deleage C, Hao X, Chuang GY, Gorman J, Gardner M, Lewis MG, Hatziioannou T, Santra S, Apetrei C, Pandrea I, Alam SM, Liao HX, Shen X, Tomaras GD, Farzan M, Chertova E, Keele BF, Estes JD, Lifson JD, Doms RW, Montefiori DC, Haynes BF, Sodroski JG, Kwong PD, Hahn BH, Shaw GM, 2016. Envelope residue 375 substitutions in simian-human immunodeficiency viruses enhance CD4 binding and replication in rhesus macaques. Proc Natl Acad Sci U S A 113, E3413–3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Gao F, Mascola JR, Stamatatos L, Polonis VR, Koutsoukos M, Voss G, Goepfert P, Gilbert P, Greene KM, Bilska M, Kothe DL, Salazar-Gonzalez JF, Wei X, Decker JM, Hahn BH, Montefiori DC, 2005. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol 79, 10108–10125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason RD, Welles HC, Adams C, Chakrabarti BK, Gorman J, Zhou T, Nguyen R, O’Dell S, Lusvarghi S, Bewley CA, Li H, Shaw GM, Sheng Z, Shapiro L, Wyatt R, Kwong PD, Mascola JR, Roederer M, 2016. Targeted Isolation of Antibodies Directed against Major Sites of SIV Env Vulnerability. PLoS Pathog 12, e1005537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore PL, Crooks ET, Porter L, Zhu P, Cayanan CS, Grise H, Corcoran P, Zwick MB, Franti M, Morris L, Roux KH, Burton DR, Binley JM, 2006. Nature of nonfunctional envelope proteins on the surface of human immunodeficiency virus type 1. J Virol 80, 2515–2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roederer M, Keele BF, Schmidt SD, Mason RD, Welles HC, Fischer W, Labranche C, Foulds KE, Louder MK, Yang ZY, Todd JP, Buzby AP, Mach LV, Shen L, Seaton KE, Ward BM, Bailer RT, Gottardo R, Gu W, Ferrari G, Alam SM, Denny TN, Montefiori DC, Tomaras GD, Korber BT, Nason MC, Seder RA, Koup RA, Letvin NL, Rao SS, Nabel GJ, Mascola JR, 2014. Immunological and virological mechanisms of vaccine-mediated protection against SIV and HIV. Nature 505, 502–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santra S, Liao HX, Zhang R, Muldoon M, Watson S, Fischer W, Theiler J, Szinger J, Balachandran H, Buzby A, Quinn D, Parks RJ, Tsao CY, Carville A, Mansfield KG, Pavlakis GN, Felber BK, Haynes BF, Korber BT, Letvin NL, 2010. Mosaic vaccines elicit CD8+ T lymphocyte responses that confer enhanced immune coverage of diverse HIV strains in monkeys. Nat Med 16, 324–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SA, Kilgore KM, Kasturi SP, Pulendran B, Hunter E, Amara RR, Derdeyn CA, 2016. Signatures in Simian Immunodeficiency Virus SIVsmE660 Envelope gp120 Are Associated with Mucosal Transmission but Not Vaccination Breakthrough in Rhesus Macaques. J Virol 90, 1880–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spearman C, 1903. The Method of “Right and Wrong Cases’ (Constant Stimuli) without Gauss’s Formulae. British Journal of Psychology 2, 227–242. [Google Scholar]
- Vaccari M, Gordon SN, Fourati S, Schifanella L, Liyanage NP, Cameron M, Keele BF, Shen X, Tomaras GD, Billings E, Rao M, Chung AW, Dowell KG, Bailey-Kellogg C, Brown EP, Ackerman ME, Vargas-Inchaustegui DA, Whitney S, Doster MN, Binello N, Pegu P, Montefiori DC, Foulds K, Quinn DS, Donaldson M, Liang F, Lore K, Roederer M, Koup RA, McDermott A, Ma ZM, Miller CJ, Phan TB, Forthal DN, Blackburn M, Caccuri F, Bissa M, Ferrari G, Kalyanaraman V, Ferrari MG, Thompson D, Robert-Guroff M, Ratto-Kim S, Kim JH, Michael NL, Phogat S, Barnett SW, Tartaglia J, Venzon D, Stablein DM, Alter G, Sekaly RP, Franchini G, 2016. Adjuvant-dependent innate and adaptive immune signatures of risk of SIVmac251 acquisition. Nat Med 22, 762–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F, Ourmanov I, Kuwata T, Goeken R, Brown CR, Buckler-White A, Iyengar R, Plishka R, Aoki ST, Hirsch VM, 2012. Sequential evolution and escape from neutralization of simian immunodeficiency virus SIVsmE660 clones in rhesus macaques. J Virol 86, 8835–8847. [DOI] [PMC free article] [PubMed] [Google Scholar]