Abstract
Purpose
Manual delineation of organs‐at‐risk (OARs) in radiotherapy is both time‐consuming and subjective. Automated and more accurate segmentation is of the utmost importance in clinical application. The purpose of this study is to further improve the segmentation accuracy and efficiency with a novel network named convolutional neural networks (CNN) Cascades.
Methods
CNN Cascades was a two‐step, coarse‐to‐fine approach that consisted of a simple region detector (SRD) and a fine segmentation unit (FSU). The SRD first used a relative shallow network to define the region of interest (ROI) where the organ was located, and then, the FSU took the smaller ROI as input and adopted a deep network for fine segmentation. The imaging data (14,651 slices) of 100 head‐and‐neck patients with segmentations were used for this study. The performance was compared with the state‐of‐the‐art single CNN in terms of accuracy with metrics of Dice similarity coefficient (DSC) and Hausdorff distance (HD) values.
Results
The proposed CNN Cascades outperformed the single CNN on accuracy for each OAR. Similarly, for the average of all OARs, it was also the best with mean DSC of 0.90 (SRD: 0.86, FSU: 0.87, and U‐Net: 0.85) and the mean HD of 3.0 mm (SRD: 4.0, FSU: 3.6, and U‐Net: 4.4). Meanwhile, the CNN Cascades reduced the mean segmentation time per patient by 48% (FSU) and 5% (U‐Net), respectively.
Conclusions
The proposed two‐step network demonstrated superior performance by reducing the input region. This potentially can be an effective segmentation method that provides accurate and consistent delineation with reduced clinician interventions for clinical applications as well as for quality assurance of a multicenter clinical trial.
Keywords: automated segmentation, CNN Cascades, deep learning, radiotherapy
1. Introduction
Modern radiotherapy techniques, such as intensity‐modulated radiotherapy (IMRT), volumetric‐modulated radiotherapy (VMAT), and TOMO therapy, have the ability to create highly conformal dose distributions to the tumor target and therefore better spare individual organs‐at‐risk (OARs) to reduce radiation‐induced toxicities. The increasing precision of radiotherapy treatment planning requires accurate definition of OARs in computed tomography (CT) images to fully realize the benefits afforded by these technological advances. However, this procedure is usually carried out manually by physicians, which is not only time‐consuming but may also need to be repeated several times during a treatment course due to significant anatomic changes (such as from tumor response). The delineation accuracy also depends on the physicians’ experience, and considerable inter‐ and intraobserver variations in the delineation of OARs have been noted in multiple disease sites including head‐and‐neck (H&N) cancer.1, 2, 3
A fully automated method of OARs delineation for radiotherapy is helpful to relieve physicians from this demanding process and to increase accuracy and consistency. Artificial intelligence (AI), especially the Convolutional Neural Networks (CNN),4, 5, 6, 7 is a potential tool for solving this problem. AI has the potential to change the landscape of medical physics research and practice8, 9, 10 and the utility of CNN in segmentation is a general trend. CNN consists of several convolutional and pooling layers. Multiple level visual features are extracted and predictions are made automatically. There has been increasing interest in applying CNN to radiation therapy.11, 12, 13, 14, 15 The group (Ibragimov and Xing) pioneered the introduction of CNN into radiotherapy contouring11 and achieved better or similar results in H&N site compared with state‐of‐the‐art algorithms. Soon, some varietal network models12, 13 succeeded in other anatomical sites and qualified for clinical use.14, 15 These methods improved contouring consistency and saved physicians’ time to some extent. However, improved accuracy and efficiency are highly desirable for widespread adoption.
One of the main drawbacks of CNN is its poor scalability with large input image size that is common for medical images. When performing segmentation of an isolated organ, the background is often uncorrelated and acts as a distraction to the primary task. The CNN is therefore burdened by relatively large background datasets, which affect the segmentation performance, especially for the smaller organs. Our work is inspired by the method by which physicians perform organ segmentation tasks. For an individual organ (e.g., the spinal cord) in a large image, they usually first focus on a relatively smaller region of interest (ROI) (e.g., the vertebral column) and then delineate the individual organ within the ROI. Here, we proposed CNN Cascades for segmentation of OARs in a similar fashion. It applied two cascaded networks of which the first for location and the second for precision segmentation. By filtering out the distractors in the big image, the proposed method could focus processing power on the specific discerning features of the organs, while simultaneously reducing time required for segmentation. There have been similar mechanisms named “attention models” for many computer vision tasks,16, 17 and recently, He et al.18 proposed a Mask R‐CNN that added a branch for predicting an object mask in parallel with the existing branch for bounding box recognition detection. This study has three main new contributions as compared with existing methods. First, the proposed method does not need a large number of additional manual annotations of the bounding boxes as required for the instance segmentation methods (e.g., the Mask R‐CNN). It has the benefit that it adopts a self‐attention mechanism to focus on the ROI and only requires the ground truth of the contours. This will greatly reduce the complexity of data preparation for training. Second, we trained two deep CNN separately, that is, the SRD to predict segmentation mask to shrink the input region and the FSU to achieve fine segmentation. It allows the usage of any existing method in each component of the process, and each model could be fine‐tuned for more accurate final segmentation. Finally, the proposed method can rapidly locate the OAR region in the form of a rectangular box which includes useful information around the OAR. Only the small region is used for fine segmentation which has greatly improved efficiency.
2. Methods and materials
2.A. Data and preprocessing
The imaging dataset used for this study is publically available via the cancer imaging archive (TCIA).19 It consists of 100 head‐and‐neck squamous cell carcinoma (HNSCC) patients’ images and DICOM RT data.20, 21 Simulation CT was scanned with the patient in the supine position. CT images were reconstructed with a matrix size of 512 × 512 and slice thickness of 2.5 or 3.0 mm. In total, there were 14,651 two‐dimensional (2D) CT slices. The pixel size was 0.88–1.27 mm with a median value of 1.07 mm. The radiotherapy contours were directly drawn on the CT by expert radiation oncologists and thereafter used for treatment planning.20 The relevant OARs studied in this research were the brainstem, spinal cord, left eye, right eye, left parotid, right parotid, and mandible.
The image data were preprocessed in MATLAB R2017b (MathWorks, Inc., Natick, Massachusetts, USA). The original CT data read from Dicom image were of 16‐bit. It was converted to an intensity image in the range 0–1 with the function “mat2gray” and then multiplied with 255 to create the 8‐bit data. A contrast‐limited adaptive histogram equalization (CLAHE) algorithm22 was followed to enhance the contrast. The final data used for CNN were the 2D CT slices and the corresponding contour labels. These processes were fully automated.
2.B. CNN Cascades for segmentation
In this study, we introduced an automated segmentation method for OARs delineation using ROI‐based serially connected CNN. Figure 1 depicts the flowchart of the CNN Cascades. It was an end‐to‐end segmentation framework that could predict pixel class labels in CT images. Different from the current single CNN methods, we used two cascaded networks to improve the accuracy and efficiency, including a simple region detector (SRD) and a fine segmentation unit (FSU). The SRD and FSU used the deep dilated CNN12 and the very deep dilated residual network (DD‐ResNet),13 respectively. Both were segmentation nets using CNN to classify every pixel of the object in the image into a given category. The SRD used a relative shallow network to identify the ROI where the organ was located, and the ROI image was then used by the FSU with a very deep network to fine segment the organs.
Figure 1.
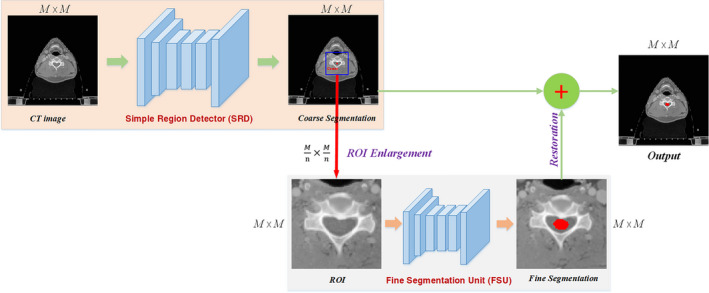
The overall framework of the CNN Cascades. [Color figure can be viewed at wileyonlinelibrary.com]
Specifically, the input of SRD was the 2D CT image (CT with size: M × M) and the output was the course segmentation (size: M ×M) of one OAR. Then, the center (C) of the segmented OAR was calculated and located in each CT slices. Taking the point C as the center, a square ROI with a dimension of (n = 2 for big organs, 4 for small organs) encompassing the OAR was selected in the CT image. Next, enlarging the ROI by n times to the original size M × M rendered an enlarged CT image (CTROI), which was used as the input to FSU for fine segmentation. The final result was restored from the output of FSU in the original image. The SRD may have generated either false‐negative or false‐positive ROIs. The slices containing false‐positive ROIs will introduce more images to the second step of the fine segmentation, which does not affect the final performance; however, the slices containing the false‐negative ROIs will be lost. According to our experience, the maximum number of slices at the boundary that may be ignored by our coarse segmentation was 3. In order to avoid losing such information, we took five more slices into consideration at the superior and inferior borders, respectively. The center of the segmented OAR was estimated with linear extrapolation method in these CT slices. With the ROI selection and fine segmentation operations, CNN Cascades could learn to focus processing and discriminatory power on the section of the image that is relevant for the specific organ.
2.C. Experiments
The performance of CNN Cascades was evaluated with fivefold cross‐validation. The dataset was randomly divided into five equal‐sized subsets. For each loop of validation, 80% of the data were used as the training set to “tune” the parameters of the segmentation model, and the remaining 20% cases were used as the test set to evaluate the performance of the model.
For data augmentation, we adopt some most popular and effective methods, such as random resize between 0.5 and 1.5 (scaling factors: 0.5, 0.75, 1, 1.25, and 1.5), random cropping (crop size: 417 × 417), and random rotation (between −10° and 10°) for training dataset. This comprehensive scheme greatly enlarged the existing training dataset and made the network resist overfitting.
The two nets, SRD and FSU, were trained independently and were combined only during the inference stage. The model parameters for each network were initialized using the weights from the corresponding model trained on ImageNet and were then “fine‐tuned” using training data. We used a batch size of 12 for SRD with shallow network and 1 for FSU with deep network due to memory limitations. The input images and their corresponding segmentation labels were used to train the network with the Stochastic Gradient Descent implementation of Caffe23 We used the “poly” learning rate policy with initial learning rate of 0.0001, learning rate decay factor of 0.0005, and momentum of 0.9, respectively. Both SRD and FSU models were fine‐tuned for 80 K iterations.
2.D. Quantitative evaluation
The cross‐validation set was used to assess the performance of the model. All the 2D CT slices of the validation set were segmented one by one. The input was the 2D CT image and the final output was pixel‐wise classification (1 for segmented target and 0 for background). The boundary of the segmented target was extracted as the contour. Manual segmentations (MS) generated by the experienced physicians were defined as the reference segmentations. The segmentation accuracy was quantified using two metrics: the Dice similarity coefficient (DSC)24 and the Hausdorff distance (HD).25 Both of them measure the degree of mismatch between the automated segmentation (A) and the manual segmentation (B). The DSC is calculated as DSC = 2TP/(2TP + FP + FN) using the definition of true positive (TP), false positive (FP), and false negative (FN). It ranges from 0, indicating no spatial overlap between the two segmentations, to 1, indicating complete overlap. The HD is the greatest of all the distances from a point in A to the closest point in B. Smaller value usually represents better segmentation accuracy.
In addition, the performance of our CNN Cascades was compared with the state‐of‐the‐art CNN methods (U‐Net7 and FSU) in medical segmentation. We also evaluated the accuracy of the coarse segmentation with SRD. The DSC and HD values for each OAR with the four methods were analyzed and compared. All values are presented as mean ± SD. A multigroup comparison of means was first carried out by one‐way analysis of variance (ANOVA) test. If it was significant, then a post hoc test by least significant difference (LSD) test was performed to detect whether a significant difference lies between the proposed method and each of the other methods. All analyses were performed with a P < 0.05.
3. Results
3.A. Accuracy
The detailed results of are shown in Tables 1 and 2. It can be seen from the quantitative evaluation metrics that the proposed CNN Cascades approach gave the best accuracy compared with other methods. The advantage over U‐Net and SRD was significantly for all the OARs (P < 0.05). Although the LSD test showed some of the metrics (DSC of left eye and Mandible, and HD of left eye, right eye, and left parotid) between CNN Cascades and FSU were not so significant, CNN Cascades had the highest mean DSC values and the lowest mean HD values for each OAR.
Table 1.
DSC values of the OARs segmentation
| OARs | Methods | P‐value | ||||||
|---|---|---|---|---|---|---|---|---|
| ANOVA | LSD test | |||||||
| 1. SRD | 2. FSU | 3. U‐Net | 4.CNN Cascades | 4 vs 1 | 4 vs 2 | 4 vs 3 | ||
| Brainstem | 0.86 ± 0.03 | 0.87 ± 0.02 | 0.84 ± 0.04 | 0.90 ± 0.02 | <0.001 | =0.001 | =0.017 | <0.001 |
| Spinal cord | 0.86 ± 0.02 | 0.86 ± 0.02 | 0.85 ± 0.03 | 0.91 ± 0.01 | <0.001 | <0.001 | <0.001 | <0.001 |
| Left eye | 0.89 ± 0.03 | 0.91 ± 0.02 | 0.88 ± 0.04 | 0.93 ± 0.01 | <0.001 | <0.001 | =0.060 | <0.001 |
| Right eye | 0.89 ± 0.02 | 0.90 ± 0.02 | 0.89 ± 0.02 | 0.92 ± 0.02 | =0.002 | <0.001 | =0.037 | =0.001 |
| Left parotid | 0.82 ± 0.03 | 0.83 ± 0.04 | 0.81 ± 0.03 | 0.86 ± 0.03 | <0.001 | <0.001 | =0.010 | <0.001 |
| Right parotid | 0.81 ± 0.06 | 0.82 ± 0.04 | 0.80 ± 0.05 | 0.86 ± 0.03 | =0.001 | =0.001 | =0.013 | <0.001 |
| Mandible | 0.89 ± 0.02 | 0.90 ± 0.02 | 0.87 ± 0.04 | 0.92 ± 0.02 | <0.001 | =0.005 | =0.159 | <0.001 |
Table 2.
Hausdorff distance (mm) of the OARs segmentation
| OARs | Methods | P‐value | ||||||
|---|---|---|---|---|---|---|---|---|
| ANOVA | LSD test | |||||||
| 1. SRD | 2. FSU | 3. U‐Net | 4. CNN Cascades | 4 vs 1 | 4 vs 2 | 4 vs 3 | ||
| Brainstem | 3.9 ± 0.5 | 3.4 ± 0.3 | 4.3 ± 0.7 | 2.9 ± 0.3 | <0.001 | <0.001 | =0.017 | <0.001 |
| Spinal Cord | 2.3 ± 0.3 | 2.2 ± 0.3 | 2.4 ± 0.4 | 1.7 ± 0.2 | <0.001 | <0.001 | <0.001 | <0.001 |
| Left Eye | 2.3 ± 0.4 | 2.0 ± 0.4 | 2.6 ± 0.6 | 1.7 ± 0.3 | <0.001 | <0.001 | =0.062 | <0.001 |
| Right Eye | 2.5 ± 0.5 | 2.1 ± 0.4 | 2.7 ± 0.6 | 1.8 ± 0.3 | =0.001 | =0.004 | =0.152 | <0.001 |
| Left Parotid | 6.4 ± 1.4 | 5.9 ± 1.4 | 7.0 ± 1.6 | 5.1 ± 1.1 | =0.002 | =0.008 | =0.130 | <0.001 |
| Right Parotid | 7.1 ± 1.5 | 6.5 ± 1.4 | 7.6 ± 1.5 | 5.4 ± 1.1 | =0.001 | =0.002 | =0.045 | <0.001 |
| Mandible | 3.2 ± 0.4 | 3.0 ± 0.4 | 4.0 ± 1.0 | 2.4 ± 0.4 | <0.001 | =0.001 | =0.022 | <0.001 |
The mean values of evaluation metrics for all the OARs with the different methods were evaluated. CNN Cascades was also the best with mean DSC of 0.90 (SRD: 0.86, FSU: 0.87, and U‐Net: 0.85) and the mean HD of 3.0 mm (SRD: 4.0, FSU: 3.6, and U‐Net: 4.4).
Figure 2 shows the visualization organ segmentation in axial cross sections. The autosegmented contours with all the methods were in good agreement with the reference contours. However, the single CNN (U‐Net and FSU) missed some contours for mandible and parotids, especially at the superior and inferior border and small regions. At the same time, U‐Net produced some wrong scattered points for mandible.
Figure 2.
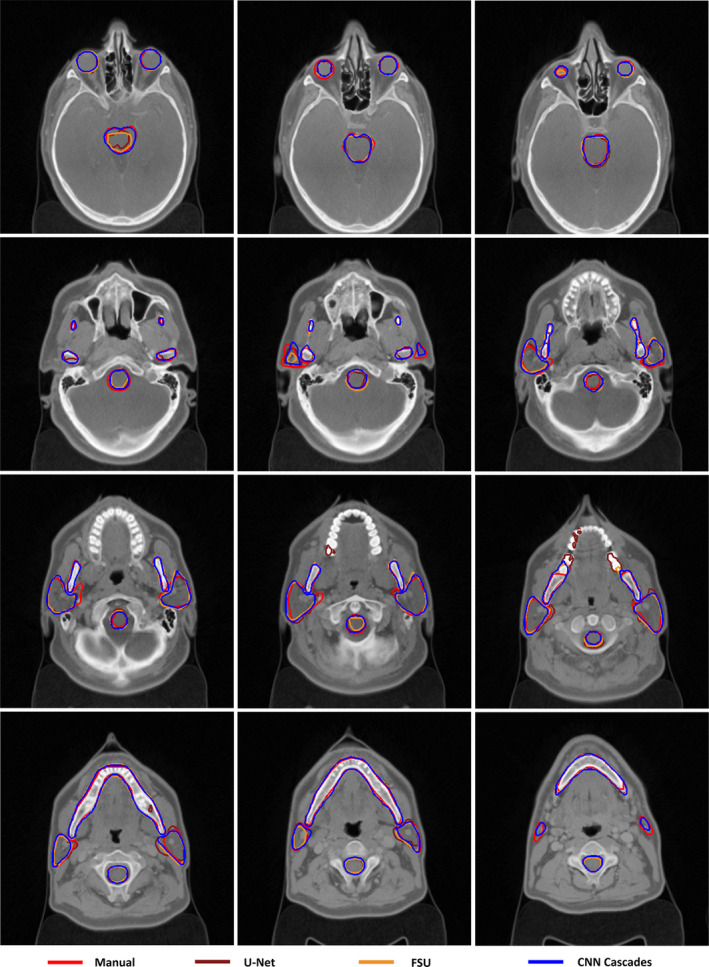
Segmentation results of CNN Cascades. [Color figure can be viewed at wileyonlinelibrary.com]
3.B. Time cost
The mean time for automated segmentation with FSU, U‐Net, and CNN Cascades was about 10.6 (SD ± 0.8) min, 5.8 (SD ± 0.4) min, and 5.5 (SD ± 0.3) min per patient, respectively, using Amazon Elastic Compute Cloud with NVIDIA K80 GPU. The proposed CNN Cascades significantly reduced the mean segmentation time by 48% (FSU, P < 0.05) and 5% (U‐Net, P < 0.05), respectively.
4. Discussion
This study proposed a two‐step CNN Cascades model to improve the segmentation accuracy of OARs in radiotherapy. For all the OARs, CNN Cascades performed well with good agreement to the delineations contoured manually by clinical experts. It can be seen from Table 1 that the CNN Cascades outperformed the current state‐of‐the‐art nets (U‐Net and ResNet) significantly. The reasons for the better performance of the proposed method can be explained as follows: The segmentation using CNN achieves pixel‐wise prediction based on the features extracted from images with a set of convolutional filters. As for the CT image for radiotherapy, the image is big and there is no organ to be segmented in some slices. CNN needs to classify the pixel into two regions (organ and background) using lots of different features for all the slices. Some features might be more relevant to the organ in the image while others might be more relevant to the background. Each filter extracts a different feature; however, the number of filters is fixed in a certain CNN. The first net in out method could predict segmentation mask to shrink the input region. In this way, it can ignore the big background and focus on optimizing the parameters of the filters used for segmentation. This means using more parameters to solve the simplified problem, which is bound to improve accuracy.
We compared the DSC value that is the most common evaluation index reported in the literature with other studies (Table 3) and found our results to of similar or improved accuracy. Our segmentation was done on CT images; however, magnetic resonance imaging (MRI) may be recommended for a better delineation of the low‐contrast region36 due to its superior soft‐tissue visualization compared with CT for some organs. Further studies combining CT with MRI could improve the segmentation accuracy further.
Table 3.
Comparison of DSC values with other studies
| OARs | DSC | |
|---|---|---|
| Other studies | Ours | |
| Brainstem | 0.76,26 0.77,27 0.78,28 0.76,29 0.86,30 0.83,31 0.9132 | 0.90 |
| Spinal cord | 0.76,26 0.76,27 0.85,28 0.76,33 0.75,34 0.8711 | 0.91 |
| Left eye | 0.83,26 0.80,33 0.8811 | 0.93 |
| Right eye | 0.83,26 0.8033 , 0.8711 | 0.92 |
| Left parotid | 0.71,27 0.78,35 0.76,29 0.84,30 0.81,31 0.8332 , 0.7711 | 0.86 |
| Right parotid | 0.72,27 0.79,35 0.76,29 0.81,30 0.79,31 0.83,32 0.7811 | 0.86 |
| Mandible | 0.78,27 0.86,35 0.87,31 0.93,32 0.9011 | 0.92 |
We quantitatively evaluated the time efficiency of the proposed two‐step framework. The proposed method had one more step in the process than single CNN; however, the segmentation time was less. The reason may be that the first network is very shallow and can define the region where the OAR located fairly quickly, while the second deep network only used reduced CT images for the fine prediction, saving inference time for 2D CNN.
One limitation of this study is the lack of an independent test set. The reason is that the data available are limited and separating an independent test set will drastically reduce the number of samples for training a robust model. The general approach in medical research to work around this limitation is a procedure called k‐fold cross‐validation. The dataset is split into k smaller sets. A model is trained using k − 1 of the folds as training data and tested on the remaining part of the data. This step is repeated until k models are trained. The average performance is then used as the evaluation index of the studied method. This approach can be computationally expensive, but it takes full advantage of the entire dataset, especially when the number of samples is very small. This approach can also demonstrate how the trained model is generalizable to unseen data to avoid deliberately choosing data with superior results for testing.
The proposed method achieved more accurate segmentation with a relatively smaller input region than the state‐of‐the‐art networks used in the field of radiotherapy for automated contouring. Efficiency and accuracy are highly desirable for radiotherapy segmentation. Unlike the networks which require the annotation of the bounding boxes, our method features a self‐attention mechanism to focus on the ROI only with the labeling of contour. Moreover, the previous two‐step networks usually need to be trained together; it is, therefore, more difficult to fine‐tune the two networks independently. In contrast, we are able to fine‐tuned the two networks separately and optimize each. Since the proposed model is flexible, effective, and efficient, we hope that it is a promising solution to further improve automated contouring in radiotherapy.
5. Conclusions
The proposed CNN Cascades with ROI identification and fine segmentation with very deep network from reduced image regions demonstrated superior performance in both accuracy and efficiency. It has the potential for implementations into radiotherapy clinical workflow as well as for quality assurance needs of multicenter clinical trials.
Conflicts of interest
The authors have no conflicts to disclose.
Acknowledgments
This project was supported by the funds from the National Cancer Institute (NCI): U24CA180803 (IROC) and CA223358.
References
- 1. Li XA, Tai A, Arthur DW, et al. Variability of target and normal structure delineation for breast‐cancer radiotherapy: a RTOG multi‐institutional and multi‐observer study. Int J Radiat Oncol Biol Phys. 2009;73:944–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nelms BE, Tomé WA, Robinson G, et al. Variations in the contouring of organs at risk: test case from a patient with oropharyngeal cancer. Int J Radiat Oncol Biol Phys. 2012;82:368–378. [DOI] [PubMed] [Google Scholar]
- 3. Feng MU, Demiroz C, Vineberg KA, et al. Intra‐observer variability of organs at risk for head and neck cancer: geometric and dosimetric consequences. Int J Radiat Oncol Biol Phys. 2010;78:S444–S445. [Google Scholar]
- 4. Long J, Shelhamer E, Darrell T. Fully convolutional networks for semantic segmentation. In: Proceedings of the IEEE conference on computer vision and pattern recognition; 2015:3431–3440. [Google Scholar]
- 5. Simonyan K, Zisserman A. Very deep convolutional networks for large‐scale image recognition. arXiv preprint arXiv:1409.1556, 2014.
- 6. He K, Zhang X, Ren S, et al. Deep residual learning for image recognition. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition; 2016:770–778.
- 7. Ronneberger O, Fischer P, Brox T. U‐net: convolutional networks for biomedical image segmentation. In: International Conference on Medical Image Computing and Computer‐Assisted Intervention. Cham: Springer; 2015:234–241. [Google Scholar]
- 8. Xing L, Krupinski EA, Cai J. Artificial intelligence will soon change the landscape of medical physics research and practice. Med Phys. 2018;45:1791–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ibragimov B, Toesca D, Chang D, et al. Development of deep neural network for individualized hepatobiliary toxicity prediction after liver SBRT. Med Phys. 2018;45:4763–4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bibault JE, Giraud P, Burgun A. Big data and machine learning in radiation oncology: state of the art and future prospects. Cancer Lett. 2016;382:110–117. [DOI] [PubMed] [Google Scholar]
- 11. Ibragimov B, Xing L. Segmentation of organs‐at‐risks in head and neck CT images using convolutional neural networks. Med Phys. 2017;44:547–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Men K, Dai J, Li Y. Automatic segmentation of the clinical target volume and organs at risk in the planning CT for rectal cancer using deep dilated convolutional neural networks. Med Phys. 2017;44:6377–6389. [DOI] [PubMed] [Google Scholar]
- 13. Men K, Zhang T, Chen X, et al. Fully automatic and robust segmentation of the clinical target volume for radiotherapy of breast cancer using big data and deep learning. Phys Med. 2018;50:13–19. [DOI] [PubMed] [Google Scholar]
- 14. Ibragimov B, Toesca D, Chang D, et al. Combining deep learning with anatomical analysis for segmentation of the portal vein for liver SBRT planning. Phys Med Biol. 2017;62:8943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lustberg T, Van JS, Gooding M, et al. Clinical evaluation of atlas and deep learning based automatic contouring for lung cancer. Radiother Oncol. 2018;126:321–317. [DOI] [PubMed] [Google Scholar]
- 16. Li S, Bak S, Carr P, et al. Diversity regularized spatiotemporal attention for video‐based person re‐identification. In: Proceedings of IEEE Conference on Computer Vision and Pattern Recognition; 2018:369–378. [Google Scholar]
- 17. Eppel S. Setting an attention region for convolutional neural networks using region selective features, for recognition of materials within glass vessels. arXiv preprint arXiv:1708.08711, 2017.
- 18. He K, Gkioxari G, Dollár P, et al. Mask r‐cnn[C]//Computer Vision (ICCV), 2017. In: IEEE International Conference, 22–29 October 2017. Venice, Italy: IEEE; 2017:2980–2988. [Google Scholar]
- 19. Clark K, Vendt B, Smith K, et al. The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. J Digit Imaging. 2013;26:1045–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vallières M, Kayrivest E, Perrin LJ, et al. Radiomics strategies for risk assessment of tumour failure in head‐and‐neck cancer. Sci Rep. 2017;7:10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vallières M, Kayrivest E, Perrin LJ, et al. Data from head‐neck‐PET‐CT. The Cancer Imaging Archive; 2017, 10.7937/k9/tcia.2017.8oje5q00 [DOI]
- 22. Reza AM. Realization of the contrast limited adaptive histogram equalization (CLAHE) for real‐time image enhancement. J VLSI Signal Process Syst Signal Image Video Technol. 2004;38:35–44. [Google Scholar]
- 23. Jia Y, Shelhamer E, Donahue J, et al. Caffe: convolutional architecture for fast feature embedding; 2014.
- 24. Crum WR, Camara O, Hill DLG. Generalized overlap measures for evaluation and validation in medical image analysis. IEEE Trans Med Imaging. 2006;25:1451–1461. [DOI] [PubMed] [Google Scholar]
- 25. Huttenlocher DP, Klanderman GA, Rucklidge WJ. Comparing images using the Hausdorff distance. IEEE Trans Pattern Anal Mach Intell. 1993;15:850–863. [Google Scholar]
- 26. Isambert A, Dhermain F, Bidault F, et al. Evaluation of an atlas‐based automatic segmentation software for the delineation of brain organs at risk in a radiation therapy clinical context. Radiother Oncol. 2008;87:93–99. [DOI] [PubMed] [Google Scholar]
- 27. Tsuji SY, Hwang A, Weinberg V, et al. Dosimetric evaluation of automatic segmentation for adaptive IMRT for head‐and‐neck cancer. Int J Radiat Oncol Biol Phys. 2010;77:707–713. [DOI] [PubMed] [Google Scholar]
- 28. Fortunati V, Verhaart RF, Fedde VDL, et al. Tissue segmentation of head and neck CT images for treatment planning: a multiatlas approach combined with intensity modeling. Med Phys. 2013;40:071905. [DOI] [PubMed] [Google Scholar]
- 29. Jean‐François D, Andreas B. Atlas‐based automatic segmentation of head and neck organs at risk and nodal target volumes: a clinical validation. Radiat Oncol. 2013;8:154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fritscher KD, Peroni M, Zaffino P, et al. Automatic segmentation of head and neck CT images for radiotherapy treatment planning using multiple atlases, statistical appearance models, and geodesic active contours. Med Phys. 2014;41:051910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zaffino P, Limardi D, Scaramuzzino S, et al. Feature based atlas selection strategy for segmentation of organs at risk in head and neck district. In: Proceedings of Imaging and Computer Assistance in Radiation Therapy Workshop, MICCAI; 2015:34–41.
- 32. Qazi AA, Pekar V, Kim J, et al. Auto‐segmentation of normal and target structures in head and neck CT images: a feature‐driven model‐based approach. Med Phys. 2011;38:6160–6170. [DOI] [PubMed] [Google Scholar]
- 33. Verhaart RF, Fortunati V, Verduijn GM, et al. CT‐based patient modeling for head and neck hyperthermia treatment planning: manual versus automatic normal‐tissue‐segmentation. Radiother Oncol. 2014;111:158–163. [DOI] [PubMed] [Google Scholar]
- 34. Duc AKH, Eminowicz G, Mendes R, et al. Validation of clinical acceptability of an atlas‐based segmentation algorithm for the delineation of organs at risk in head and neck cancer. Med Phys. 2015;42:5027–5034. [DOI] [PubMed] [Google Scholar]
- 35. Peroni M, Ciardo D, Spadea MF, et al. Automatic segmentation and online virtual CT in head‐and‐neck adaptive radiation therapy. Int J Radiat Oncol Biol Phys. 2012;84:e427–e433. [DOI] [PubMed] [Google Scholar]
- 36. Brouwer CL, Steenbakkers RJ, Bourhis J, et al. CT‐based delineation of organs at risk in the head and neck region: DAHANCA, EORTC, GORTEC, HKNPCSG, NCIC CTG, NCRI, NRG oncology and TROG consensus guidelines. Radiother Oncol. 2015;117:83–90. [DOI] [PubMed] [Google Scholar]


