Abstract
Loss-of-function and hypomorphic ECHS1 variants are associated with mitochondrial short-chain enoyl-CoA hydratase deficiency, an inborn error of valine metabolism. We report an 8-year-old boy with developmental delay, ataxia, hemiplegia, and hearing loss with abnormalities in the basal ganglia. Biochemical studies were essentially normal except for a persistent mildly elevated CSF alanine. This patient demonstrates an intermediate phenotype between a Leigh-like, early-onset presentation and paroxysmal exercise-induced dyskinesia. Two novel ECHS1 variants (c.79T>G; p.Phe27Val and c.789_790del; p.Phe263fs) were identified via exome sequencing in the proband, and pathogenicity was confirmed by enzyme assay performed on patient fibroblasts. Neither of the ECHS1 variants detected in the child were present in the mother. However, due to nearby polymorphisms, it was possible to determine that p.Phe263fs occurred de novo on the maternal chromosome and that p.Phe27Val likely derived from the paternal chromosome. Nearby polymorphisms can help set phase of variants when only a single parent is available for testing or when an identified variant occurs de novo.
Keywords: ECHS1, Exome interpretation, Mitochondrial short-chain enoyl-CoA hydratase deficiency, Phase determination
Introduction
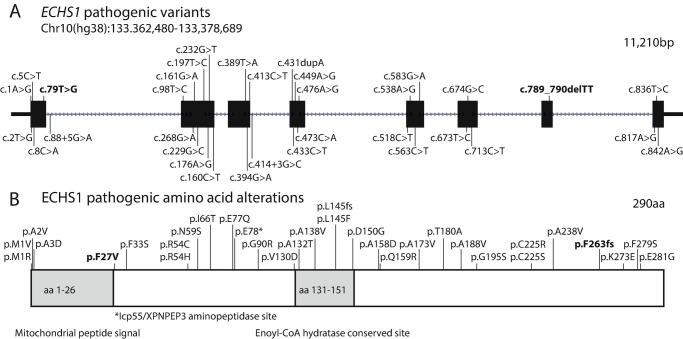
Deficiency of short-chain enoyl-CoA hydratase (MIM: 616277) is an inborn error of valine metabolism (Peters et al. 2014). To date, 43 patients from 34 families have been reported (Peters et al. 2014; Sakai et al. 2015; Haack et al. 2015; Ferdinandusse et al. 2015; Tetreault et al. 2015; Yamada et al. 2015; Ganetzky et al. 2016; Stark et al. 2016; Olgiati et al. 2016; Nair et al. 2016; Al Mutairi et al. 2017; Mahajan et al. 2017; Bedoyan et al. 2017; Balasubramaniam et al. 2017; Ogawa et al. 2017; Huffnagel et al. 2017; Fitzsimons et al. 2018). Thirty-two pathogenic ECHS1 variants were detected in these families, including 28 missense variants with no obvious mutational hotspots (the most recurrent variant c.476A>G; p.Gln159Arg has been identified in 7 unrelated patients), although a few variants have been identified in multiple families (Fig. 1).
Fig. 1.
Mutational spectrum of ECHS1. (a) A gene diagram illustrating the 32 previously identified pathogenic ECHS1 variants (28 missense, 2 splicing, 1 frameshift, and 1 nonsense) as well as the novel pathogenic variants detected in this patient (in bold). (b) Protein diagram of pathogenic amino acid alterations (changes associated with splicing not shown, and the patient’s amino acid changes are in bold). Protein diagram illustrates that p.Phe27Val affects the first amino acid following the mitochondrial peptide signal and that Phe27 is predicted to be recognized and cleaved by the Icp55/XPNPEP3 aminopeptidase in order to stabilize the N-terminus of ECHS1 in mitochondria
ECHS1 deficiency can cause severe, intermediate, or mild disease (termed paroxysmal exercise-induced dystonia), although there is substantial phenotypic overlap. Most cases in the literature are severe and present at birth or in early infancy with a Leigh-like syndrome including often fatal lactic acidosis (Haack et al. 2015; Ferdinandusse et al. 2015). Recurrent findings include encephalopathy, microcephaly, epilepsy, hearing loss, optic nerve atrophy, and cardiomyopathy (Sakai et al. 2015; Haack et al. 2015; Ferdinandusse et al. 2015; Tetreault et al. 2015; Stark et al. 2016; Huffnagel et al. 2017). A recent report also described cutis laxa in a 4.5-year-old patient with soft dysmorphic features (Balasubramaniam et al. 2017).
There is some evidence of genotype-phenotype correlation with higher ECHS1 activity associated with increased lifespan (Haack et al. 2015). Intermediate ECHS1 deficiency patients typically presented with developmental delay, hearing and/or vision loss, and bilateral T2 signals in the globi pallidi on brain MRI (Table 1). Age at diagnosis in these patients ranged from age 8 to 31 years.
Table 1.
Clinical/laboratory findings in patients with intermediate ECHS1 deficiency phenotype
| This report | Haack et al. (2015) | Tetreault et al. (2015) | Huffnagel et al. (2017) | |||||
|---|---|---|---|---|---|---|---|---|
| F8, II:1 | F9, II:2 | F10, II:1 | P2 | P3 | P4 | |||
| ECHS1 pathogenic variants | c.79T>G; p.Phe27Val c.789_790del; p.Phe263fs |
c.161G>A; p.Arg54His c.394G>A; p.Ala132Thr |
c.161G>A; p.Arg54His c.431dup; p.Leu145fs |
c.229G>C; p.Glu77Gln c.476A>G; p.Gln159Arg |
c.538A>G; p.Thr180Ala c.713C>T; p.Ala238Val |
c.538A>G; p.Thr180Ala c.713C>T; p.Ala238Val |
c.476A>G; p.Gln159Arg c.538A>G; p.Thr180Ala |
c.229G>C; p.Glu77Gln c.563C>T; p.Ala188Val |
| Gender | M | F | F | F | M | M | F | F |
| Age at presentation | 1 year | 1 year | Birth | 11 months | 2.9 years | 10 months | 6 months | 1.5 months |
| Reported at age | 9 years | 8 years | 16 years | 31 years | 18 years | 13 years | 12 years | 26 years |
| Hypotonia | + | + | − | − | − | − | + | Nd |
| Regression/developmental delay | + | + | + | + | + | + | + | + |
| Hearing loss | + | + | + | + | + | + | + | − |
| Optic atrophy | − | − | + | + | + | + | + | + |
| Epilepsy | − | – (One seizure) | − | + | Nd | Nd | Nd | − |
| Dystonia | − | − | + | + | + | − | + | + |
| MRI: basal ganglia T2 hyperintensity | + | Nd | + | + | + | + | + | + |
| MRS: lactate | + | Nd | + | − | − | − | − | − |
| Elevated plasma lactate | − | + | + | + | Normal-mild increase | Normal-mild increase | − | + (Normalized with age) |
| Acylcarnitines | Normal | Normal | Normal | Normal | Normal | Normal | Normal | Normal |
| Urine organic acids | Normal | Nd | Nd | Normal | Normal (especially note that 2-methyl-2,3-dihydroxybutyrate levels were normal) | Normal | Ketosis and hyper-lactaturia (resolved with treatment) | 3-Methylglutaconic aciduria (normalized with age) Elevated cysteine metabolites (S-(2-carboxypropyl)-cysteamine, S-(2-carboxypropyl)cysteine and N-acetyl-S-(2-carboxypropyl)cysteine) |
| Additional information | Hemiparesis, syringomyelia, partial mtDNA depletion | Truncal ataxia, gastroschisis | Uses voice computer | Spastic tetraparesis, wheelchair from 9.5 years | Failure to thrive, microcephaly, nystagmus | Failure to thrive, microcephaly, nystagmus | Failure to thrive, nystagmus | Spastic tetraparesis, contractures, nystagmus |
nd Not determined
Three older patients (including a sibling pair) presented with paroxysmal exercise-induced dyskinesia (Olgiati et al. 2016; Mahajan et al. 2017). Although these patients had bilateral T2 hyperintensities in the globus pallidus, they lacked other finding characteristic of ECHS1 deficiency such as psychomotor delays. In all three cases, the same hypomorphic allele (p.Ala173Val) was detected. One patient responded well to treatment with a mitochondrial cocktail (thiamine, riboflavin, carnitine, coenzyme Q, vitamin B6, and vitamin C), reporting less frequent and less severe attacks (Mahajan et al. 2017).
Here, we present an intermediate ECHS1 deficiency patient, who exhibits developmental delay, hypotonia, ataxia, hemiplegia, and hearing loss. Only a maternal sample was available for testing, and extrapolation from nearby polymorphisms was required to help set phase for the two identified novel ECHS1 variants.
Materials and Methods
Genomic DNA from peripheral blood was extracted using a Gentra PureGene Blood Kit (Qiagen, Germantown, MD) and enriched for targeted regions using SureSelect Clinical Research Exome Kit (Agilent Technologies, Santa Clara, CA). Exome sequencing was performed on a HiSeq2500 instrument (Illumina, San Diego, CA). Sequencing reads were aligned to the human genome reference (GRch37) using Burrows-Wheeler Aligner (BWA 0.5.9) (Li and Durbin 2009). Variant calling was performed with Genome Analysis Toolkit (GATK v1.3) (McKenna et al. 2010). Sanger sequencing was performed in the proband and mother to confirm variants and to assist in phasing (FinchTV 1.4.0, Geospiza, Seattle, WA). Family relationships were adjudicated using short tandem repeat (STR) markers.
Results
Clinical Phenotype
This 8-year-old boy is a twin of Afro-Caucasian ancestry who was conceived by in vitro fertilization. The fraternal twin sister is unaffected. At 1 year of age, the child was noted to be delayed in development and to be clumsy. An MRI of his spine and brain indicated the presence of syringohydromyelia and bilateral lesions of the globi pallidi (more advanced on the left side). These lesions had low N-acetylaspartate suggesting neuronal/myelin loss. At 22 months of age, the patient experienced regression of language skills and new-onset, right-sided weakness with a febrile illness. A repeat brain MRI confirmed alterations in the basal ganglia, and MRS identified a prominent lactate peak in the left globus pallidus. He also has hematuria (due to a known diagnosis of Alport syndrome), with normal kidneys on ultrasound. With time, the child learned to walk using a walker and at 8 years of age has short sentences. He has significant truncal hypotonia, ataxia of the lower limbs, right hemiplegia, hearing loss (requiring bilateral hearing aids), and global developmental delays. The child is fed via a G-tube and has normal growth. Repeated brain MRIs indicate gliosis of bilateral globi pallidi, right caudate head, and left cerebral peduncle with no new brain lesions. Despite extensive biochemical studies, the only notable finding was a persistent, mild elevation of CSF alanine.
Previous normal genetic and biochemical testing includes a cytogenomic microarray (CGH 1-megabase), karyotype, PANK2 sequencing and del/dup, comprehensive metabolic panel, CoQ10 and electron transport chain functional enzyme assays, CDG transferrin, plasma lactate, CSF pyruvic acid, urine organic acids, plasma free and total carnitine, plasma acylcarnitine profile, very long-chain fatty acids, plasma amino acids (by liquid chromatography, mass spectrometry, and ion exchange chromatography), heavy metals, uric acid, prolactin, ceruloplasmin, copper, urine purines and pyrimidines, serum thymidine, ammonia, CSF neurotransmitters, folate, pyridoxal 5-phosphate, succinyladenosine, lysosomal enzymes, pyruvate dehydrogenase, and carboxylase in fibroblasts. Mitochondrial DNA sequencing performed by an outside laboratory was nondiagnostic. The patient has a partial deletion of exon 31 of the COL4A5 gene causing Alport syndrome (maternally inherited). A muscle biopsy had normal histochemistry but showed partial depletion of mitochondrial DNA (but not to the degree seen in mtDNA depletion syndromes).
Molecular Genetic Analysis
Clinical exome sequencing of the proband detected two rare variants, c.79T>G; p.Phe27Val and c.789_790del; p.Phe263fs, in the ECHS1 gene (NM_004092.3), neither of which were present in the unaffected mother (nor in two unaffected siblings). There was substantial phenotypic overlap of the patient’s presentation with previous descriptions of patients with ECHS1 deficiency, and autosomal recessive conditions may be caused by a combination of inherited and de novo variants occurring in trans. Unfortunately, the two variants were in different exons and therefore not close enough to determine phase directly from NGS data.
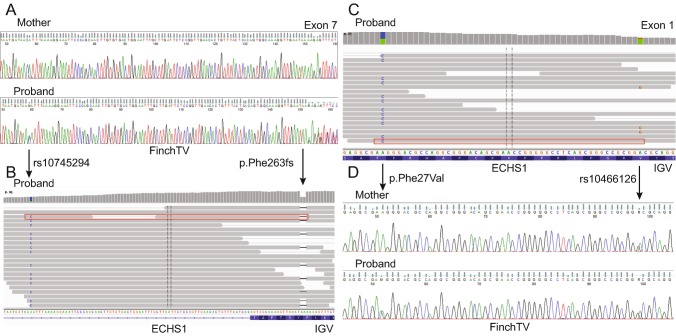
We noted that the patient’s Caucasian mother was homozygous for an intronic ECHS1 polymorphism (rs10745294) [the most common allele in non-Finnish Europeans with an allele frequency of 99.5% in the genome aggregation database (gnomAD)] (Lek et al. 2016), while the Afro-Caucasian patient was heterozygous (allele frequency is 66.6% in Africans) (Fig. 2a). In Integrative Genomics Viewer (IGV), p.Phe263fs was in cis with this polymorphism (Fig. 2b), and therefore was a de novo change that occurred on the maternal chromosome (Robinson et al. 2011; Thorvaldsdóttir et al. 2013). A second polymorphism, p.Val11Ala (rs10466126), was in trans from p.Phe27Val in IGV (Fig. 2c). The p.Val11Ala polymorphism (with an allele frequency of 72.9% in non-Finnish Europeans versus only 30.9% in Africans) was likely maternally inherited (Fig. 2d), implying that the p.Phe27Val derived from the paternal chromosome. The most parsimonious explanation is that p.Phe263fs occurred de novo on the maternal chromosome, while p.Phe27Val was paternally inherited.
Fig. 2.
Next-generation sequencing (NGS) suggests that the ECHS1 variants identified in the patient are in trans. (a) The patient’s Caucasian mother is homozygous for the common exon 7 polymorphism rs10745295 (99.5% allele frequency in Caucasians), while the Afro-Caucasian patient is heterozygous for the polymorphism (66.6% allele frequency in Africans). (b) NGS reads demonstrate that the rs10745295 polymorphism is in cis with p.Phe263fs (example read in red box). Thus, p.Phe263fs occurred de novo on the maternal chromosome. (c) NGS data demonstrate that p.Phe27Val (0.0924% allele frequency in Africans) is in trans from the exon 1 rs10466126 polymorphism (example read in red box). (d) Both the patient’s mother and the patient are heterozygous for the rs10466126 polymorphism (allele frequency of 72.9% in non-Finnish Europeans but only 30.9% in Africans), suggesting that p.Phe27Val is likely paternally inherited and in trans from p.Phe263fs
The de novo frameshift variant, c.789_790del, is absent from population databases and predicted to introduce a premature termination codon seven codons downstream (p.Phe263Leufs*7). The frameshift occurs in the penultimate exon potentially targeting the transcript for nonsense-mediated decay, and loss-of-function ECHS1 variants are a well-established cause of ECHS1 deficiency. The c.79T>G variant has an allele frequency of 0.0924% in Africans (8 out of 8,654 alleles) and is absent from more than 18,000 European alleles in the gnomAD browser, further supporting that p.Phe27Val was likely paternally inherited. The ECHS1 precursor protein contains a mitochondrial peptide signal (amino acids 1–26) that is cleaved after import by a mitochondrial processing peptidase (Fig. 1b). The Icp55 aminopeptidase in yeast and plants stabilizes mitochondrial proteins by specifically cleaving a single destabilizing amino acid on the modified N-terminus, and the XPNPEP3 protein is thought to play the same role in humans (Singh et al. 2017). The absence of such processing has a severe effect on the half-lives of substrate proteins (Vögtle et al. 2009). In the case of ECHS1, the destabilizing phenylalanine (tyrosine, leucine, phenylalanine, and tryptophan are the primary destabilizing amino acids) at position 27 would be predicted to be cleaved revealing the stabilizing amino acid alanine (serine, alanine, and threonine are the amino acids that form the N-terminus after processing in nearly all substrates) at position 28. The p.Phe27Val amino acid alteration would disrupt the normal cleavage process, potentially resulting in a destabilized protein (Fukasawa et al. 2015).
Enzyme Assay
Activity of short-chain enoyl-CoA hydratase was measured in the patient’s fibroblasts and was reduced to <31 nmol/min per mg protein with a reference range of 179–616 nmol/min per mg protein. This result provides biochemical confirmation of ECHS1 deficiency.
Discussion
We describe a new case of ECHS1 deficiency presenting with developmental delay, hypotonia, ataxia, hemiplegia, and hearing loss. The longer survival and less severe presentation with the absence of urine metabolites, compared to many of the classical ECHS1 deficiency cases, may be attributable to the presence of residual enzyme activity (Haack et al. 2015) or an as of yet unidentified compensatory mechanism. ECHS1 deficiency likely constitutes a continuous spectrum of disease severity affected both by genotype (correlating with ECHS1 activity) and environmental events that may increase metabolic demand such as illness or trauma. The delineation of an intermediate ECHS1 deficiency phenotype (Table 1) is therefore somewhat arbitrary but may help in counseling the families of patients diagnosed later in childhood or in adulthood.
Certain missense variants in a subset of ECHS1 deficiency patients with prolonged survival (Table 1) suggest that these could be hypomorphic alleles (particularly p.Arg54His, p.Glu77Gln, and p.Ala238Val). However, the most recurrent amino acid alteration in intermediate cases, p.Thr180Ala, has also been observed in a homozygous state in three siblings who died at the ages of 21 months, 28 months, and 13 months (Fitzsimons et al. 2018). Similarly, unrelated patients homozygous for the variant encoding p.Gln159Arg died at 28 months and age 4, respectively (Haack et al. 2015; Fitzsimons et al. 2018). Notably, an ECHS1 deficiency patient who was compound heterozygous for variants encoding p.Gln159Arg and p.Thr180Ala was alive at age 12 (Tetreault et al. 2015), so there may be considerable variability in patient survival (possibly related to access to supportive care) even with variants that have been associated with a severe phenotype in other ECHS1 deficiency patients. Alternatively, it is possible that p.Gln159Arg and p.Thr180Ala contribute to ECHS1 deficiency in different ways and are able to partially compensate for one another in a compound heterozygous individual compared to in individuals homozygous for either. The description here of p.Phe27Val, a novel, likely hypomorphic allele that appears to be associated with African ancestry, may help in the diagnosis of future ECHS1 deficiency patients with an intermediate phenotype.
Biochemical testing, especially in cases of patients with ECHS1 deficiency surviving into adulthood, is complex, and metabolic abnormalities are often more subtle and non-specific than in cases with early lethality (Sharpe and McKenzie 2018). Mildly elevated butyrylcarnitine in blood has been reported in two severe cases, and elevated methylmalonyl/succinyl-CoA and decreased hydroxybutyryl-CoA were reported in frozen liver from one of these patients (Ganetzky et al. 2016). However, in our patient, elevated butyrylcarnitine was not observed. For several patients with early-onset lactic acidosis, both pyruvate and lactate were elevated, often with a normal pyruvate-lactate ratio (Peters et al. 2014; Ferdinandusse et al. 2015; Ganetzky et al. 2016; Bedoyan et al. 2017), while individuals with more variable phenotypes have inconsistently elevated lactate, in some cases only during episodes of deterioration (Sakai et al. 2015; Haack et al. 2015; Ferdinandusse et al. 2015; Tetreault et al. 2015; Yamada et al. 2015; Nair et al. 2016; Huffnagel et al. 2017). Elevations of plasma lactate or plasma alanine were not observed in our patient; however, mildly elevated CSF alanine was observed on multiple occasions, suggesting lactic acidosis. Elevated 2,3-dihydroxy-2-methylbutyric acid is considered a biomarker of ECHS1 deficiency, though it is also not consistently present nor is its presence in urine unique to ECHS1 deficiency (Peters et al. 2015; Fitzsimons et al. 2018). 2,3-Dihydroxy-2-methylbutyric acid was not found in our patient, nor was it detected in excess in any of the patients presenting with an intermediate phenotype (Table 1), although it was observed in a patient with a more severe form of ECHS1 deficiency (Ferdinandusse et al. 2015). For some longer-surviving patients, increased S-(2-carboxypropyl)cysteamine, S-(2-carboxypropyl)cysteine, and N-acetyl-S-(2-carboxypropyl) cysteine have been useful diagnostic markers in urine (Peters et al. 2014; Ferdinandusse et al. 2015; Yamada et al. 2015; Olgiati et al. 2016; Huffnagel et al. 2017), and 3-methylglutaconic aciduria may also be observed later or transiently in patients with an intermediate phenotype compared to those who are more severely affected (Ferdinandusse et al. 2015; Huffnagel et al. 2017; Fitzsimons et al. 2018). However, as the presentation and evolution of the phenotype are so variable in ECHS1 deficiency patients, the metabolic abnormalities may reflect changing metabolic demands placed upon these biochemical pathways at any given time.
This case highlights the important issue of how to proceed with sequencing analysis when only a single parent is available for testing. Under these circumstances it may be difficult to determine whether variants are in trans. Polymorphisms may be informative during dyad analysis or when determining parent of origin for a de novo variant associated with an autosomal recessive condition or an imprinting disorder. Particularly with NGS-based testing, it may be possible to determine on which parental allele a variant resides, even in the case of suspected de novo variants.
Acknowledgments
We are grateful to the family for their willingness to share this case with the medical community. We also would like to thank the members of Genomics Lab and Genetic Sequencing Lab at ARUP laboratories for performing testing on the individuals included in this study.
Author Contributions
Colleen M. Carlston: exome data interpretation, study design, and writing manuscript.
Dr. Ferdinandusse: metabolite analysis (ECHS1 testing), data analysis, and interpretation.
Dr. Hobert: metabolite analysis (previous metabolic testing), data analysis and interpretation, and critical revision of manuscript.
Dr. Mao: exome lab director, exome data interpretation, and STR interpretation.
Dr. Longo: study concept, design and supervision, patient care, and critical revision of manuscript.
Compliance with Ethics Guidelines
Colleen Carlston, Sacha Ferdinandusse, Judith Hobert, Rong Mao, and Nicola Longo declare that they have no conflicts of interest.
All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1975, as revised in 2000. Informed consent was obtained from the family included in the study and is available upon request.
Take-Home Message
During exome analysis performed without both biological parents and/or when a de novo variant is suspected, the use of nearby polymorphisms can help set phase of variants as in this case of a boy with biochemically confirmed ECHS1 deficiency caused by two novel ECHS1 variants.
References
- Al Mutairi F, Shamseldin HE, Alfadhel M, et al. A lethal neonatal phenotype of mitochondrial short-chain enoyl-CoA hydratase-1 deficiency. Clin Genet. 2017;91:629–633. doi: 10.1111/cge.12891. [DOI] [PubMed] [Google Scholar]
- Balasubramaniam S., Riley L. G., Bratkovic D., Ketteridge D., Manton N., Cowley M. J., Gayevskiy V., Roscioli T., Mohamed M., Gardeitchik T., Morava E., Christodoulou J. Unique presentation of cutis laxa with Leigh-like syndrome due to ECHS1 deficiency. Journal of Inherited Metabolic Disease. 2017;40(5):745–747. doi: 10.1007/s10545-017-0036-4. [DOI] [PubMed] [Google Scholar]
- Bedoyan JK, Yang SP, Ferdinandusse S, et al. Lethal neonatal case and review of primary short-chain enoyl-CoA hydratase (SCEH) deficiency associated with secondary lymphocyte pyruvate dehydrogenase complex (PDC) deficiency. Mol Genet Metab. 2017;120:342–349. doi: 10.1016/j.ymgme.2017.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferdinandusse S, Friederich MW, Burlina A, et al. Clinical and biochemical characterization of four patients with mutations in ECHS1. Orphanet J Rare Dis. 2015;10:79. doi: 10.1186/s13023-015-0290-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzsimons PE, Alston CL, Bonnen PE, et al. Clinical, biochemical, and genetic features of four patients with short-chain enoyl-CoA hydratase (ECHS1) deficiency. Am J Med Genet A. 2018;176:1115–1127. doi: 10.1002/ajmg.a.38658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukasawa Y, Tsuji J, Fu SC, et al. MitoFates: improved prediction of mitochondrial targeting sequences and their cleavage sites. Mol Cell Proteomics. 2015;14(4):1113–1126. doi: 10.1074/mcp.M114.043083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganetzky RD, Bloom K, Ahrens-Nicklas R, et al. ECHS1 deficiency as a cause of severe neonatal lactic acidosis. JIMD Rep. 2016;30:33–37. doi: 10.1007/8904_2016_538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haack TB, Jackson CB, Murayama K, et al. Deficiency of ECHS1 causes mitochondrial encephalopathy with cardiac involvement. Ann Clin Transl Neurol. 2015;2:492–509. doi: 10.1002/acn3.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffnagel Irene C., Redeker Egbert J. W., Reneman Liesbeth, Vaz Frédéric M., Ferdinandusse Sacha, Poll-The Bwee Tien. JIMD Reports. Berlin, Heidelberg: Springer Berlin Heidelberg; 2017. Mitochondrial Encephalopathy and Transient 3-Methylglutaconic Aciduria in ECHS1 Deficiency: Long-Term Follow-Up; pp. 83–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan A, Constantinou J, Sidiropoulos C. ECHS1 deficiency-associated paroxysmal exercise-induced dyskinesias: case presentation and initial benefit of intervention. J Neurol. 2017;264:185–187. doi: 10.1007/s00415-016-8381-z. [DOI] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair P, Hamzeh AR, Mohamed M, et al. Novel ECHS1 mutation in an Emirati neonate with severe metabolic acidosis. Metab Brain Dis. 2016;31:1189–1192. doi: 10.1007/s11011-016-9842-x. [DOI] [PubMed] [Google Scholar]
- Ogawa E, Shimura M, Fushimi T, et al. Clinical calidity of biochemical and molecular analysis in diagnosing Leigh syndrome: a study of 106 Japanese patients. J Inherit Metab Dis. 2017;40:685–693. doi: 10.1007/s10545-017-0042-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olgiati S, Skorvanek M, Quadri M, et al. Paroxysmal exercise-induced dystonia within the phenotypic spectrum of ECHS1 deficiency. Mov Disord. 2016;31:1041–1048. doi: 10.1002/mds.26610. [DOI] [PubMed] [Google Scholar]
- Peters H, Buck N, Wanders R, et al. ECHS1 mutations in Leigh disease: a new inborn error of metabolism affecting valine metabolism. Brain. 2014;137:2903–2908. doi: 10.1093/brain/awu216. [DOI] [PubMed] [Google Scholar]
- Peters H, Ferdinandusse S, Ruiter JP, et al. Metabolite studies in HIBCH and ECHS1 defects: implications for screening. Mol Genet Metab. 2015;115:168–173. doi: 10.1016/j.ymgme.2015.06.008. [DOI] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai C, Yamaguchi S, Sasaki M, et al. ECHS1 mutations cause combined respiratory chain deficiency resulting in Leigh syndrome. Hum Mutat. 2015;36:232–239. doi: 10.1002/humu.22730. [DOI] [PubMed] [Google Scholar]
- Sharpe AJ, McKenzie M. Mitochondrial fatty acid oxidation disorders associated with short-chain enoyl-CoA hydratase (ECHS1) deficiency. Cell. 2018;7(6):46. doi: 10.3390/cells7060046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh Rahul, Jamdar Sahayog N., Goyal Venuka Durani, Kumar Ashwani, Ghosh Biplab, Makde Ravindra D. Structure of the human aminopeptidase XPNPEP3 and comparison of itsin vitroactivity with Icp55 orthologs: Insights into diverse cellular processes. Journal of Biological Chemistry. 2017;292(24):10035–10047. doi: 10.1074/jbc.M117.783357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark Z, Tan TY, Chong B, et al. A prospective evaluation of whole-exome sequencing as a first-tier molecular test in infants with suspected monogenic disorders. Genet Med. 2016;18:1090–1096. doi: 10.1038/gim.2016.1. [DOI] [PubMed] [Google Scholar]
- Tetreault M, Fahiminiya S, Antonicka H, et al. Whole-exome sequencing identifies novel ECHS1 mutations in Leigh syndrome. Hum Genet. 2015;134:981–991. doi: 10.1007/s00439-015-1577-y. [DOI] [PubMed] [Google Scholar]
- Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative genomics viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vögtle F-N, Wortelkamp S, Zahedi RP, et al. Global analysis of the mitochondrial N-proteome identifies a processing peptidase critical for protein stability. Cell. 2009;139:428–439. doi: 10.1016/j.cell.2009.07.045. [DOI] [PubMed] [Google Scholar]
- Yamada K, Aiba K, Kitaura Y, et al. Clinical, biochemical and metabolic characterisation of a mild form of human short-chain enoyl-CoA hydratase deficiency: significance of increased N-acetyl-S-(2-carboxypropyl)cysteine excretion. J Med Genet. 2015;52:691–698. doi: 10.1136/jmedgenet-2015-103231. [DOI] [PubMed] [Google Scholar]