Abstract
Postmenopausal osteoporosis (PMOP) is a major public health concern worldwide. The present study aimed to provide evidence to assist in the development of specific novel biomarkers for PMOP. Differentially expressed genes (DEGs) were identified between PMOP and normal controls by integrated microarray analyses of the Gene Expression Omnibus (GEO) database, and the optimal diagnostic gene biomarkers for PMOP were identified with LASSO and Boruta algorithms. Classification models, including support vector machine (SVM), decision tree and random forests models, were established to test the diagnostic value of identified gene biomarkers for PMOP. Functional annotations and protein-protein interaction (PPI) network constructions were also conducted. Integrated microarray analyses (GSE56815, GSE13850 and GSE7429) of the GEO database were employed, and 1,320 DEGs were identified between PMOP and normal controls. An 11-gene combination was also identified as an optimal biomarker for PMOP by feature selection and classification methods using SVM, decision tree and random forest models. This combination was comprised of the following genes: Dehydrogenase E1 and transketolase domain containing 1 (DHTKD1), osteoclast stimulating factor 1 (OSTF1), G protein-coupled receptor 116 (GPR116), BCL2 interacting killer, adrenoceptor β1 (ADRB1), neogenin 1 (NEO1), RB binding protein 4 (RBBP4), GPR87, cylicin 2, EF-hand calcium binding domain 1 and DEAH-box helicase 35. RBBP4 (degree=12) was revealed to be the hub gene of this PMOP-specific PPI network. Among these 11 genes, three genes (OSTF1, ADRB1 and NEO1) were speculated to serve roles in PMOP by regulating the balance between bone formation and bone resorption, while two genes (GPR87 and GPR116) may be involved in PMOP by regulating the nuclear factor-κB signaling pathway. Furthermore, DHTKD1 and RBBP4 may be involved in PMOP by regulating mitochondrial dysfunction and interacting with ESR1, respectively. In conclusion, the findings of the current study provided an insight for exploring the mechanism and developing novel biomarkers for PMOP. Further studies are required to test the diagnostic value for PMOP prior to use in a clinical setting.
Keywords: postmenopausal osteoporosis, differentially expressed genes, biomarker, Gene Expression Omnibus
Introduction
Osteoporosis is a bone metabolic disorder, characterized by low bone mineral density (BMD) and microarchitectural deterioration with increased bone fragility and subsequent susceptibility to fractures (1). It has been reported that osteoporosis is induced by an imbalance between bone resorption by osteoclasts and bone deposition by osteoblasts (2). Postmenopausal osteoporosis (PMOP) is a major public health concern worldwide that frequently presents in postmenopausal women due to the estrogen deficiency and continuous calcium loss that occurs with aging (3). A proactive approach that identifies patients at high risk of developing PMOP is recommended to prevent bone loss (4).
With the advancement of high-throughput technologies, gene microarray analysis has become an effective method for identifying differentially expressed genes (DEGs) and, therefore, potential biomarkers in various diseases. Multiple studies (5–7) have utilized gene microarray analysis to identify key genes in the pathogenesis of PMOP. Integrated multiple gene microarray analysis may contribute to the identification of more accurate gene biomarkers.
The present study aimed to develop accurate biomarkers and provide clues for exploring the underlying mechanism of PMOP. By integrating multiple microarray analysis in this present study, DEGs between PMOP patients and normal controls were identified. Based on these DEGs, the optimal gene combination with the greatest diagnostic value for PMOP was determined. Functional annotation and protein-protein interaction (PPI) network constructions were also performed to explore the biological functions of DEGs. These findings will help elucidate the mechanism underlying PMOP development and uncover novel diagnostic biomarkers.
Materials and methods
Microarray expression profiling
Microarray datasets of PMOP and normal controls were downloaded from the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo). All datasets that contained whole-genome mRNA expression profiles between PMOP patient and control blood samples were enrolled in the current study. The datasets were scale normalized.
Identification of DEGs between PMOP patients and normal controls
MetaMA is an R package-implementing meta-analysis approach for microarray data (8). Data from multiple microarray analyses were combined by metaMA (inverse normal method), and individual P-values were obtained. In the integrated analysis performed in the present study, DEGs between PMOP patients and normal controls were identified at a P-value of <0.05. Hierarchical clustering analyses of mRNAs were conducted with ‘pheatmap’ package in R (version 3.3.3; www.r-project.org).
Identification of optimal diagnostic gene biomarkers for PMOP
The LASSO algorithm was applied with the glmnet package (https://cran.r-project.org/web/packages/glmnet/) in order to reduce the dimensions of the data (9). The Boruta algorithm (https://cran.r-project.org/web/packages/Boruta/) employs a wrapper approach, built around a random forest classifier (10). This algorithm is used to compare the relevance of the features to those of the random probes (11). The scale-standardized datasets were merged, the DEGs between PMOP patients and normal controls were retained for feature selection, and gene biomarkers for PMOP were identified with the LASSO and Boruta algorithms. Furthermore, the optimal gene biomarkers for PMOP were identified by overlapping biomarkers derived from these two algorithms. Hierarchical clustering analysis of these shared gene biomarkers, obtained by LASSO and Boruta algorithms, was conducted with the R package ‘pheatmap’ (R version 3.3.3).
Based on these optimal gene biomarkers, several classification models, including support vector machine (SVM), decision tree and random forest models, were established to further identify the diagnostic value of these biomarkers in PMOP. An SVM model was established with an ‘e1071’ package (https://cran.r-project.org/web/packages/e1071/index.html). A decision tree model was established with the ‘rpart’ package (https://cran.r-project.org/web/packages/rpart/). A random forest model was established with the ‘randomForests’ package (https://cran.r-project.org/web/packages/randomForest/). These three classification models were compared by the average misjudgment rates of their 10-fold cross validations. The diagnostic ability of the three classification models was assessed by calculating the receiver operating characteristic curve, and measuring the area under the curve (AUC), accuracy, sensitivity and specificity.
PMOP-specific PPI network
To further investigate the biological functions of these optimal gene biomarkers, a PPI network was constructed with the BioGRID (also known as Biological General Repository for Interaction Datasets; http://thebiogrid.org/) and Cytoscape (http://www.cytoscape.org). Nodes and edges in the PPI network represented the proteins and the interactions between two proteins, respectively.
Functional annotation
Based on the PMOP-specific PPI network, the proteins that integrated with proteins encoded by the optimal gene biomarkers were identified. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses of all gene biomarkers and other DEGs that encode proteins within the PMOP-specific PPI network were conducted with the online software GeneCodis (http://genecodis.cnb.csic.es/analysis). Differences (using the Benjamini and Hochberg method) were defined as statistically significant when the false discovery rate (FDR) was <0.05.
Results
DEGs between PMOP patients and normal controls
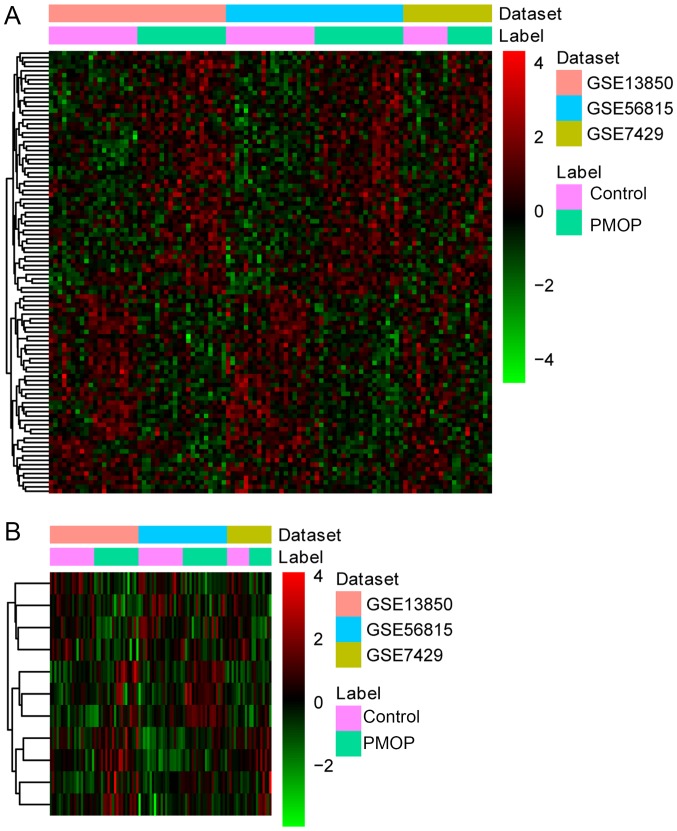
Three datasets, including GSE56815, GSE13850 and GSE7429, were downloaded from the GEO database [(Table I) (12)]. Based on these three datasets, 1,320 DEGs (710 upregulated DEGs and 613 downregulated DEGs) with FDR<0.05 were identified between PMOP patients and normal controls. Hierarchical clustering analysis of the top 100 DEGs between PMOP patients and normal controls is presented in Fig. 1.
Table I.
Datasets used in the present study.
| GEO ID | Sample | Country | Year | First author | PMOP to control ratio |
|---|---|---|---|---|---|
| GSE56815 | Blood | USA | 2016 | Liu | 20:20 |
| GSE13850 | Blood | USA | 2009 | Xiao | 20:20 |
| GSE7429 | Blood | USA | 2008 | Xiao | 10:10 |
The platform used for all datasets was GPL96 [HG-U133A] Affymetrix Human Genome U133A Array. GEO, Gene Expression Omnibus. PMOP, postmenopausal osteoporosis.
Figure 1.
Hierarchical clustering analysis of DEGs between PMOP patients and normal controls. (A) Analysis of top 100 DEGs between PMOP patients and normal controls. (B) Analysis of 11 shared gene biomarkers for PMOP obtained by both the LASSO and Boruta algorithms. Rows and columns represent DEGs and samples, respectively. The color scale represents the expression levels. DEGs, differentially expressed genes; PMOP, postmenopausal osteoporosis. BMD, bone mineral density; DEGs, ESR1, estrogen receptor 1; GEO, Gene Expression Omnibus; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; NE, norepinephrine; PPI, protein-protein interaction; RANKL, receptor activator of NF-κB ligand; SVM, support vector machine.
Identification of optimal diagnostic gene biomarkers for PMOP
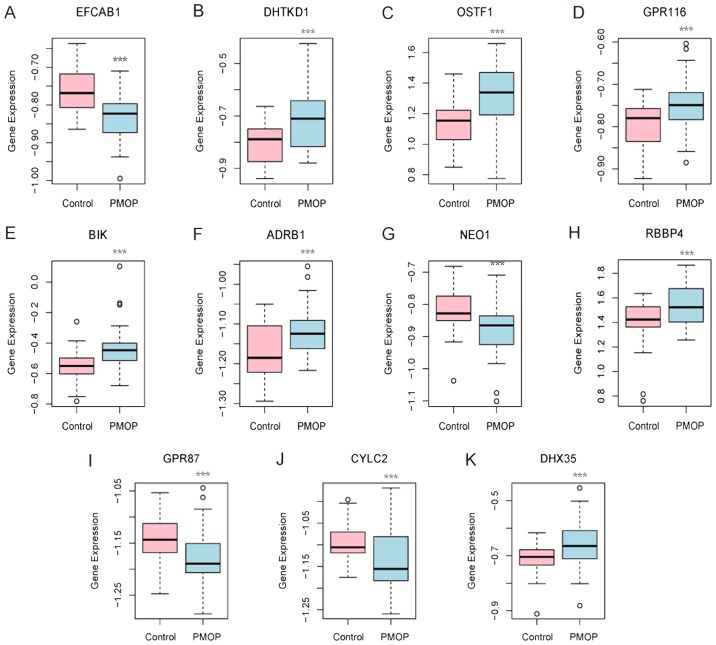
A total of 31 and 32 gene biomarkers were identified with the LASSO and Boruta algorithms, respectively. Furthermore, 11 shared mRNA biomarkers for PMOP were identified by overlapping the biomarkers derived from these two algorithms (Table II and Fig. 2). These 11 mRNA biomarkers included dehydrogenase E1 and transketolase domain containing 1 (DHTKD1), osteoclast stimulating factor 1 (OSTF1), G protein-coupled receptor 87 (GPR87), GPR116 (also known as adhesion G protein-coupled receptor F5), BCL2 interacting killer (BIK), adrenoceptor β1 (ADRB1), neogenin 1 (NEO1), RB binding protein 4 (RBBP4), cylicin 2 (CYLC2), EF-hand calcium binding domain 1 (EFCAB1) and DEAH-box helicase 35 (DHX35). Hierarchical clustering analysis of these 11 mRNA biomarkers was performed.
Table II.
A total of 11 shared gene biomarkers for postmenopausal osteoporosis, obtained using the LASSO and Boruta algorithms.
| Gene ID | Gene symbol | Combined ES | P-value | FDR | Regulation |
|---|---|---|---|---|---|
| 79645 | EFCAB1 | −1.16 | 4.51×10−8 | 2.80×10−4 | Down |
| 55526 | DHTKD1 | 1.07 | 4.61×10−7 | 1.45×10−3 | Up |
| 26578 | OSTF1 | 1.05 | 1.99×10−6 | 3.38×10−3 | Up |
| 221395 | GPR116 | 0.98 | 2.58×10−6 | 3.38×10−3 | Up |
| 638 | BIK | 0.92 | 9.30×10−6 | 8.69×10−3 | Up |
| 153 | ADRB1 | 0.89 | 1.97×10−5 | 1.27×10−2 | Up |
| 4756 | NEO1 | −0.85 | 5.61×10−5 | 2.09×10−2 | Down |
| 5928 | RBBP4 | 0.81 | 7.70×10−5 | 2.32×10−2 | Up |
| 53836 | GPR87 | −0.80 | 1.20×10−4 | 2.61×10−2 | Down |
| 1539 | CYLC2 | −0.74 | 3.42×10−4 | 4.12×10−2 | Down |
| 60625 | DHX35 | 0.73 | 5.64×10−4 | 5.08×10−2 | Up |
ES, effective size; FDR, false discovery rate; EFCAB1, EF-hand calcium binding domain 1; DHTKD1, dehydrogenase E1 and transketolase domain containing 1; OSTF1, osteoclast stimulating factor 1; GPR, G protein-coupled receptor; BIK, BCL2 interacting killer; ADRB1, adrenoceptor β1; NEO1, neogenin 1; RBBP4, RB binding protein 4; CYLC2, cylicin 2; DHX35, DEAH-box helicase 35.
Figure 2.
Expression levels of 11 gene biomarkers in the blood samples of postmenopausal osteoporosis patients and normal controls. Gene expression levels of (A) EFCAB1, (B) DHTKD1, (C) OSTF1, (D) GPR116, (E) BIK, (F) ADRB1, (G) NEO1, (H) RBBP4, (I) GPR87, (J) CYLC2 and (K) DHX35 are shown. ***P<0.001 vs. the normal control group. EFCAB1, EF-hand calcium binding domain 1; DHTKD1, dehydrogenase E1 and transketolase domain containing 1; OSTF1, osteoclast stimulating factor 1; GPR, G protein-coupled receptor; BIK, BCL2 interacting killer; ADRB1, adrenoceptor β1; NEO1, neogenin 1; RBBP4, RB binding protein 4; CYLC2, cylicin 2; DHX35, DEAH-box helicase 35.
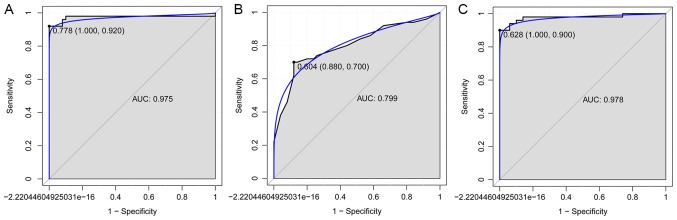
The SVM, decision tree and random forest models were established with these 11 mRNA biomarkers, and the accuracy of these three models was 93, 78 and 94%, respectively. The AUC of SVM, decision tree and random forest models was 0.975, 0.799 and 0.975, respectively. In addition, the sensitivity and specificity of the SVM model were 92 and 100%, respectively (Fig. 3A). The sensitivity and specificity of the decision tree model were 70 and 88%, respectively (Fig. 3B). Finally, the sensitivity and specificity of the random forest model were 90 and 100%, respectively (Fig. 3C).
Figure 3.
Receiver operating characteristic curves of the combination of 11 gene biomarkers between patients with postmenopausal osteoporosis and normal controls, based on three classification models. The results of the (A) Support vector machine, (B) decision tree and (C) random forest models are displayed. The X-axis represents the 1-specificity and Y-axis represents the sensitivity. AUC, area under the curve.
PMOP-specific PPI network
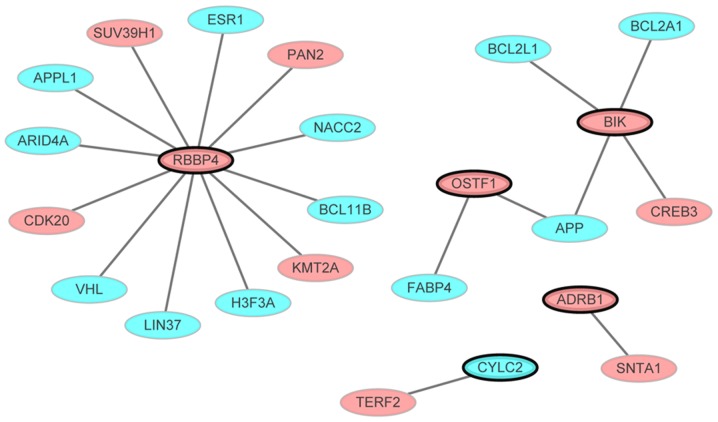
The PMOP-specific PPI network consisted of 24 nodes and 20 edges (Fig. 4). Out of the 11 genes not all interacted with other differentially expressed genes. Only the gene that interacted with other differentially expressed genes (even though one gene) in the PPI network were presented. Nodes and edges represented the proteins and the interactions between two proteins, respectively. The red and blue ellipses represented the proteins encoded by up- and downregulated DEGs between PMOP patients and normal controls, respectively. RBBP4 (degree=12) was the hub gene of this PMOP-specific PPI network.
Figure 4.
PMOP-specific protein-protein interaction network, comprised of 24 nodes and 20 edges. Nodes and edges represented proteins and interactions between two proteins, respectively. Red and blue ellipses represented the proteins encoded by up- and downregulated DEGs, respectively. Ellipses with black borders were DEGs derived from the combination of 11 gene biomarkers. DEGs, differentially expressed genes; PMOP, postmenopausal osteoporosis.
Functional annotation
Based on the functional annotations of these 11 biomarkers and DEGs that encode proteins of the PMOP-specific PPI network, prostate epithelial cord elongation (FDR<0.05), estrogen response element binding (FDR<0.05) and nucleosome remodeling deacetylase complex (FDR<0.05) were the most significant GO terms. Furthermore, endocrine and other factor-regulated calcium reabsorption (FDR=2.23×10−5), was a significantly enriched pathway in PMOP; Estrogen receptor 1 (ESR1) was revealed to be upregulated within this pathway (Table III).
Table III.
Top 10 significantly GO terms and KEGG pathways in postmenopausal osteoporosis.
| A, GO terms | |||
|---|---|---|---|
| ID | Term | FDR | Genes |
| Biological process | |||
| GO:0060523 | Prostate epithelial cord elongation | <0.05 | ESR1 |
| GO:0031649 | Heat generation | <0.05 | ADRB1 |
| GO:0045986 | Negative regulation of smooth muscle contraction | <0.05 | ADRB1 |
| GO:0002025 | Vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure | <0.05 | ADRB1 |
| GO:0031077 | Post-embryonic camera-type eye development | <0.05 | BCL11B |
| GO:0048386 | Positive regulation of retinoic acid receptor signaling pathway | <0.05 | ESR1 |
| GO:0003382 | Epithelial cell morphogenesis | <0.05 | BCL11B |
| GO:0051124 | Synaptic growth at neuromuscular junction | <0.05 | APP |
| GO:0060750 | Epithelial cell proliferation involved in mammary gland duct elongation | <0.05 | ESR1 |
| GO:0046878 | Positive regulation of saliva secretion | <0.05 | ADRB1 |
| Molecular function | |||
| GO:0034056 | Estrogen response element binding | <0.05 | ESR1 |
| GO:0051400 | BH domain binding | <0.05 | BIK |
| GO:0051380 | Norepinephrine binding | <0.05 | ADRB1 |
| GO:0004535 | Poly(A)-specific ribonuclease activity | <0.05 | PAN2 |
| GO:0051434 | BH3 domain binding | <0.05 | BCL2L1 |
| GO:0051425 | PTB domain binding | <0.05 | APP |
| GO:0004939 | β-adrenergic receptor activity | <0.05 | ADRB1 |
| GO:0004591 | Oxoglutarate dehydrogenase (succinyl-transferring) activity | <0.05 | DHTKD1 |
| GO:0004940 | β1-adrenergic receptor activity | <0.05 | ADRB1 |
| GO:0031798 | Type 1 metabotropic glutamate receptor binding | <0.05 | ESR1 |
| Cellular component | |||
| GO:0016581 | NuRD complex | <0.05 | APPL1, RBBP4 |
| GO:0097136 | Bcl-2 family protein complex | <0.05 | BCL2L1 |
| GO:0033150 | Cytoskeletal calyx | <0.05 | CYLC2 |
| GO:0033553 | rDNA heterochromatin | <0.05 | SUV39H1 |
| GO:0033186 | CAF-1 complex | <0.05 | RBBP4 |
| GO:0044429 | Mitochondrial part | <0.05 | BCL2L1, ESR1 |
| GO:0030870 | Mre11 complex | <0.05 | TERF2 |
| GO:0030891 | VCB complex | <0.05 | VHL |
| GO:0001740 | Barr body | <0.05 | H3F3A |
| GO:0016589 | NURF complex | <0.05 | RBBP4 |
| B, KEGG pathways | |||
| ID | Pathway | FDR | Genes |
| KEGG:05200 | Pathways in cancer | <0.05 | BCL2L1, VHL, APPL1 |
| KEGG:04962 | Vasopressin-regulated water reabsorption | <0.05 | CREB3 |
| KEGG:04961 | Endocrine and other factor-regulated calcium reabsorption | <0.05 | ESR1 |
| KEGG:00310 | Lysine degradation | <0.05 | SUV39H1 |
| KEGG:05014 | Amyotrophic lateral sclerosis (ALS) | <0.05 | BCL2L1 |
| KEGG:05210 | Colorectal cancer | <0.05 | APPL1 |
| KEGG:03018 | RNA degradation | <0.05 | PAN2 |
| KEGG:03320 | PPAR signaling pathway | <0.05 | FABP4 |
| KEGG:05211 | Renal cell carcinoma | <0.05 | VHL |
| KEGG:05212 | Pancreatic cancer | <0.05 | BCL2L1 |
GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; FDR, false discovery rate.
Discussion
PMOP increases the risk of fragility fractures in postmenopausal women, and imposes a significant burden on patients' families and society. Previous studies have indicated that identification of patients at high risk of developing PMOP can contribute to the prevention of bone loss (4,13).
In the present study, 1,320 DEGs between PMOP patients and normal controls were identified with integrated microarray analysis. The gene biomarkers for PMOP were further identified with the LASSO and Boruta algorithms. An 11-gene combination (EFCAB1, DHTKD1, OSTF1, GPR116, BIK, ADRB1, NEO1, RBBP4, GPR87, CYLC2 and DHX35) was revealed as an optimal biomarker for PMOP with feature selection and classification procedures using SVM, decision tree and random forest models. Based on the random forest model, the 11-gene combination achieved a 94% prediction accuracy in distinguishing patients with PMOP from normal controls, with 90% sensitivity and 100% specificity. The results obtained using the other two models (SVM and decision tree) further supported this finding.
Among these 11 genes, CYLC2 has previously been shown to be upregulated in B cells from postmenopausal women with low BMD compared with that in postmenopausal women with high BMD (14). In addition, CYLC2 is involved in the structural component of the cytoskeleton. A previous study indicated that the structural component of the cytoskeleton is associated with PMOP, which may suggest a potential role of CYLC2 in PMOP (14).
Three DEGs identified in the present study, namely OSTF1, ADRB1 and NEO1, have previously been reported to be associated with the balance between bone formation and bone resorption (15–19). OSTF1 is an intracellular protein that is highly expressed in osteoclasts, which indirectly enhances osteoclast formation and bone resorption (15). ADRB1 belongs to the family of guanine nucleotide-binding regulatory protein-coupled receptors, which regulate the physiological effects of the hormone epinephrine and the neurotransmitter norepinephrine (NE) (16). The β-adrenergic system is also involved in leptin-dependent central regulation of bone turnover (17,18). Intraosseous sympathetic nerve fibers can be activated and release NE via leptin stimulation (17). Adrenergic receptors expressed on osteoblasts bind to the released NE and result in suppression of bone formation. In addition, β-adrenergic-stimulated production of the receptor activator of nuclear factor (NF)-κB ligand by osteoblasts may contribute to a negative bone mineral balance (19). In the present study, OSTF1 and ADRB1 were upregulated in the blood of patients with PMOP compared with that of normal controls. Furthermore, NEO1 encodes a cell surface protein that belongs to the immunoglobulin superfamily, and has been speculated to serve roles in cell growth and differentiation and in cell-cell adhesion. A previous study reported abnormal chondrocyte maturation and endochondral bone growth in NEO1 knockout mice (20). Additionally, the association between NEO1 and bone mass was identified by high-throughput screening of mouse gene knockouts (21). To the best of our knowledge, the present study is the first to reveal a downregulation of NEO1 in the blood of patients with PMOP.
Estrogen deficiency is a pivotal cause of postmenopausal bone loss (22). RBBP4 is an estrogen-associated gene, which was included in the 11-gene combination described in the present study. It is also a chromatin remodeling factor that encodes a ubiquitously expressed nuclear protein that belongs to a highly conserved subfamily of WD-repeat proteins (23). RBBP4 has been reported to be involved in the chromatin remodeling and transcriptional repression associated with histone deacetylation (24). An upregulation in the expression of RBBP4 was detected in the tibia callus of estrogen-deficient rats (25). To the best of our knowledge, the present study is the first to reveal an upregulation of RBBP4 in the blood of patients with PMOP. Furthermore, as the hub protein of the PMOP-specific PPI network, RBBP4 was integrated with ESR1, a well-known PMOP-associated gene, which was revealed to be enriched in the endocrine and other factor-regulated calcium reabsorption pathway (KEGG ID: 04961). ESR1 is expressed on cells that contribute to bone formation, such as osteoblasts, osteocytes and osteoclasts. It also increases the formation and function of osteoblasts and reduces bone resorption activities (26). Therefore, the RBBP4-ESR1 interaction may serve a key role in PMOP.
To the best of our knowledge, no previous study has reported the association between PMOP and the six other genes described in the current study, including DHTKD1, GPR87, GPR116, BIK, EFCAB1 and DHX35. DHTKD1 is a nuclear gene that is involved in mitochondrial lysine metabolism and adenosine triphosphate production (27,28). DHTKD1 has also been demonstrated to link mitochondrial dysfunction and eosinophilic esophagitis (29). Kim and Lee (30) indicated that mitochondrial dysfunction may be a potential pathophysiological mechanism of PMOP, which suggested that DHTKD1 may regulate mitochondrial dysfunction in PMOP. In addition, GPR87 is a cell surface G protein-coupled receptor that has been reported to be overexpressed in various types of cancer (31,32), and it serves a critical oncogenic role in pancreatic cancer progression by activating the NF-κB signaling pathway (33). GPR116 is a member of the G protein-coupled receptor family predominantly expressed in the alveolar type II epithelial cells of the lung. Since NF-κB signaling pathway is an important mediator in osteoblast differentiation, it can be speculated that both GPR87 and GPR116 may serve a role in PMOP by regulating the NF-κB signaling pathway. Another identified gene, BIK, is a member of the BH3-only Bcl-2 family of pro-apoptotic proteins, which is suppressed in various types of cancer (34). Methylated BIK was identified in the bone marrow of patients with multiple myeloma, and dysregulated BIK expression was observed in hematopoietic cell fractions of patients with myelodysplastic syndrome, highlighting the importance of BIK in bone disease (34,35). Furthermore, the gene DHX35 is a putative RNA helicase, and its variants have been reported to be involved with facial morphology, thyroid cancer and colorectal cancer (36–38) Finally, DNA methylation of EFCAB1 was demonstrated to be involved in multi-organ carcinogenesis (39). However, further research is required to explore the roles of DHX35 and EFCAB1 in PMOP.
In conclusion, the present study identified 11 genes that were significantly associated with PMOP and provided clues for exploring the molecular mechanism of PMOP. Three of the identified genes (OSTF1, ADRB1 and NEO1) were speculated to be involved in PMOP by regulating the balance between bone formation and bone resorption, while two genes (GPR87 and GPR116) may regulate the NF-κB signaling pathway. RBBP4 and DHTKD1 may also be potential regulators of PMOP via interacting with ESR1 and regulating mitochondrial dysfunction, respectively. Furthermore, the constituents of this 11-gene combination may serve as potential biomarkers for PMOP. However, biological investigations and validation with a larger sample size are lacking, and are considered to be limitations of the present study. Further investigations are required to validate the diagnostic abilities of this gene combination for PMOP prior to its clinical application.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- BMD
bone mineral density
- DEGs
differentially expressed genes
- ESR1
estrogen receptor 1
- GEO
Gene Expression Omnibus
- GO
Gene Ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- NE
norepinephrine
- PMOP
postmenopausal osteoporosis
- PPI
protein-protein interaction
- RANKL
receptor activator of NF-κB ligand
- SVM
support vector machine
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
CY and XS made substantial contributions to the study conception and design. CY, JR, BL, CJ, CM, CC and YS collected and analyzed the data. CY, JR, BL and CJ interpreted the data. All authors were involved in drafting the manuscript and provided final approval of this manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Kanis JA, McCloskey EV, Johansson H, Oden A, Melton LJ III, Khaltaev N. A reference standard for the description of osteoporosis. Bone. 2008;42:467–475. doi: 10.1016/j.bone.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 2.Paschalis EP, Gamsjaeger S, Hassler N, Fahrleitner-Pammer A, Dobnig H, Stepan JJ, Pavo I, Eriksen EF, Klaushofer K. Vitamin D and calcium supplementation for three years in postmenopausal osteoporosis significantly alters bone mineral and organic matrix quality. Bone. 2017;95:41–46. doi: 10.1016/j.bone.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 3.Meng J, Zhang D, Pan N, Sun N, Wang Q, Fan J, Zhou P, Zhu W, Jiang L. Identification of mir-194-5p as a potential biomarker for postmenopausal osteoporosis. PeerJ. 2015;3:e971. doi: 10.7717/peerj.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sanders S, Geraci SA. Osteoporosis in postmenopausal women: Considerations in prevention and treatment: (women's health series) Southern Med J. 2013;106:698–706. doi: 10.1097/SMJ.0b013e3182a0df8b. [DOI] [PubMed] [Google Scholar]
- 5.Yan B, Li J, Zhang L. Identification of B cells participated in the mechanism of postmenopausal women osteoporosis using microarray analysis. Int J Clin Exp Med. 2015;8:1027–1034. [PMC free article] [PubMed] [Google Scholar]
- 6.Liu Y, Wang Y, Yang N, Wu S, Lv Y, Xu L. In silico analysis of the molecular mechanism of postmenopausal osteoporosis. Mol Med Rep. 2015;12:6584–6590. doi: 10.3892/mmr.2015.4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma M, Luo S, Zhou W, Lu L, Cai J, Yuan F, Yin F. Bioinformatics analysis of gene expression profiles in B cells of postmenopausal osteoporosis patients. Taiwan J Obstet Gynecol. 2017;56:165–170. doi: 10.1016/j.tjog.2016.04.038. [DOI] [PubMed] [Google Scholar]
- 8.Marot G, Foulley JL, Mayer CD, Jaffrézic F. Moderated effect size and P-value combinations for microarray meta-analyses. Bioinformatics. 2009;25:2692–2699. doi: 10.1093/bioinformatics/btp444. [DOI] [PubMed] [Google Scholar]
- 9.Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw. 2010;33:1–22. doi: 10.18637/jss.v033.i01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Breiman L. Random forests. Machine Learning. 2001;45:5–32. doi: 10.1023/A:1010933404324. [DOI] [Google Scholar]
- 11.Stoppiglia H, Dreyfus G, Dubois R, Oussar Y. Ranking a random feature for variable and feature selection. J Mach Learn Res. 2003;3:1399–1414. [Google Scholar]
- 12.Xiao P, Chen Y, Jiang H, Liu YZ, Pan F, Yang TL, Tang ZH, Larsen JA, Lappe JM, Recker RR, Deng HW. In vivo genome-wide expression study on human circulating B cells suggests a novel esr1 and mapk3 network for postmenopausal osteoporosis. J Bone Miner Res. 2008;23:644–654. doi: 10.1359/jbmr.080105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tella SH, Gallagher JC. Prevention and treatment of postmenopausal osteoporosis. J Steroid Biochem Mol Biol. 2014:155–170. doi: 10.1016/j.jsbmb.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma M, Chen X, Lu L, Yuan F, Zeng W, Luo S, Yin F, Cai J. Identification of crucial genes related to postmenopausal osteoporosis using gene expression profiling. Aging Clin Exp Res. 2016;28:1067–1074. doi: 10.1007/s40520-015-0509-y. [DOI] [PubMed] [Google Scholar]
- 15.Vermeren M, Lyraki R, Wani S, Airik R, Albagha O, Mort R, Hildebrandt F, Hurd T. Osteoclast stimulation factor 1 (ostf1) knockout increases trabecular bone mass in mice. Mamm Genome. 2017;28:498–514. doi: 10.1007/s00335-017-9718-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y, Lin Y, Zhao H, Guo Q, Yan C, Lin N. Revealing the effects of the herbal pair of euphorbia kansui and glycyrrhiza on hepatocellular carcinoma ascites with integrating network target analysis and experimental validation. Int J Biol Sci. 2016;12:594–606. doi: 10.7150/ijbs.14151. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Takeda S, Elefteriou F, Levasseur R, Liu X, Zhao L, Parker KL, Armstrong D, Ducy P, Karsenty G. Leptin regulates bone formation via the sympathetic nervous system. Cell. 2002;111:305–317. doi: 10.1016/S0092-8674(02)01049-8. [DOI] [PubMed] [Google Scholar]
- 18.Karsenty G. Leptin controls bone formation through a hypothalamic relay. Recent Prog Horm Res. 2001;56:401–415. doi: 10.1210/rp.56.1.401. [DOI] [PubMed] [Google Scholar]
- 19.Elefteriou F, Ahn JD, Takeda S, Starbuck M, Yang X, Liu X, Kondo H, Richards WG, Bannon TW, Noda M, et al. Leptin regulation of bone resorption by the sympathetic nervous system and cart. Nature. 2005;434:514–520. doi: 10.1038/nature03398. [DOI] [PubMed] [Google Scholar]
- 20.Zhou Z, Xie J, Lee D, Liu Y, Jung J, Zhou L, Xiong S, Mei L, Xiong WC. Neogenin regulation of bmp-induced canonical smad signaling and endochondral bone formation. Dev cell. 2010;19:90–102. doi: 10.1016/j.devcel.2010.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brommage R, Liu J, Hansen GM, Kirkpatrick LL, Potter DG, Sands AT, Zambrowicz B, Powell DR, Vogel P. High-throughput screening of mouse gene knockouts identifies established and novel skeletal phenotypes. Bone Res. 2014;2:14034. doi: 10.1038/boneres.2014.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sharma D, Larriera AI, Palacio-Mancheno PE, Gatti V, Fritton JC, Bromage TG, Cardoso L, Doty SB, Fritton SP. The effects of estrogen deficiency on cortical bone microporosity and mineralization. Bone. 2018;110:1–10. doi: 10.1016/j.bone.2018.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang X, Yao F, Liang X, Zhu X, Zheng R, Jia B, Hou L, Zou X. Cloning and expression of retinoblastoma-binding protein 4 gene in embryo diapause termination and in response to salinity stress from brine shrimp artemia sinica. Gene. 2016;591:351–361. doi: 10.1016/j.gene.2016.06.010. [DOI] [PubMed] [Google Scholar]
- 24.Balboula AZ, Stein P, Schultz RM, Schindler K. Rbbp4 regulates histone deacetylation and bipolar spindle assembly during oocyte maturation in the mouse. Biol Reprod. 2015;92:105. doi: 10.1095/biolreprod.115.128298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saul D, Ninkovic M, Komrakova M, Wolff L, Simka P, Gasimov T, Menger B, Hoffmann DB, Rohde V, Sehmisch S. Effect of zileuton on osteoporotic bone and its healing, expression of bone, and brain genes in rats. J Appl Physiol. 2018;124:118–130. doi: 10.1152/japplphysiol.01126.2016. [DOI] [PubMed] [Google Scholar]
- 26.Shalan NA, Mustapha NM, Mohamed S. Noni leaf and black tea enhance bone regeneration in estrogen-deficient rats. Nutrition. 2017;33:42–51. doi: 10.1016/j.nut.2016.08.006. [DOI] [PubMed] [Google Scholar]
- 27.Xu W, Zhu H, Gu M, Luo Q, Ding J, Yao Y, Chen F, Wang Z. Dhtkd1 is essential for mitochondrial biogenesis and function maintenance. FEBS Lett. 2013;587:3587–3592. doi: 10.1016/j.febslet.2013.08.047. [DOI] [PubMed] [Google Scholar]
- 28.Danhauser K, Sauer S, Haack TB, Wieland T, Staufner C, Graf E, Zschocke J, Strom TM, Traub T, Okun JG, et al. Dhtkd1 mutations cause 2-aminoadipic and 2-oxoadipic aciduria. Am J Hum Genet. 2012;91:1082–1087. doi: 10.1016/j.ajhg.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sherrill JD, Kc K, Wang X, Wen T, Chamberlin A, Stucke EM, Collins MH, Abonia JP, Peng Y, Wu Q, et al. Whole-exome sequencing uncovers oxidoreductases dhtkd1 and ogdhl as linkers between mitochondrial dysfunction and eosinophilic esophagitis. JCI Insight. 2018;3 doi: 10.1172/jci.insight.99922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim JH, Lee DC. Mitochondrial DNA copy number in peripheral blood is associated with femoral neck bone mineral density in postmenopausal women. J Rheumatol. 2012;39:1465–1472. doi: 10.3899/jrheum.111444. [DOI] [PubMed] [Google Scholar]
- 31.Glatt S, Halbauer D, Heindl S, Wrnitznig A, Kozina D, Su KC, Puri C, Garin-Chesa P, Sommergruber W. Hgpr87 contributes to viability of human tumor cells. Int J Cancer. 2008;122:2008–2016. doi: 10.1002/ijc.23349. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Y, Scoumanne A, Chen X. G protein-coupled receptor 87: A promising opportunity for cancer drug discovery. Mol Cell Pharmacol. 2010;2:111–116. [PMC free article] [PubMed] [Google Scholar]
- 33.Wang L, Zhou W, Zhong Y, Huo Y, Fan P, Zhan S, Xiao J, Jin X, Gou S, Yin T, et al. Overexpression of g protein-coupled receptor gpr87 promotes pancreatic cancer aggressiveness and activates nf-κb signaling pathway. Mol Cancer. 2017;16:61. doi: 10.1186/s12943-017-0627-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hatzimichael E, Dasoula A, Kounnis V, Benetatos L, Lo Nigro C, Lattanzio L, Papoudou-Bai A, Dranitsaris G, Briasoulis E, Crook T. Bcl2-interacting killer cpg methylation in multiple myeloma: A potential predictor of relapsed/refractory disease with therapeutic implications. Leuk Lymphoma. 2012;53:1709–1713. doi: 10.3109/10428194.2012.661854. [DOI] [PubMed] [Google Scholar]
- 35.Langemeijer SM, Mariani N, Knops R, Gilissen C, Woestenenk R, de Witte T, Huls G, van der Reijden BA, Jansen JH. Apoptosis-related gene expression profiling in hematopoietic cell fractions of MDS patients. PLoS One. 2016;11:e0165582. doi: 10.1371/journal.pone.0165582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoff AM, Johannessen B, Alagaratnam S, Zhao S, Nome T, Løvf M, Bakken AC, Hektoen M, Sveen A, Lothe RA, Skotheim RI. Novel rna variants in colorectal cancers. Oncotarget. 2015;6:36587–36602. doi: 10.18632/oncotarget.5500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Figlioli G, Kohler A, Chen B, Elisei R, Romei C, Cipollini M, Cristaudo A, Bambi F, Paolicchi E, Hoffmann P, et al. Novel genome-wide association study-based candidate loci for differentiated thyroid cancer risk. J Clin Endocrinol Metab. 2014;99:E2084–E2092. doi: 10.1210/jc.2014-1734. [DOI] [PubMed] [Google Scholar]
- 38.Cha S, Lim JE, Park AY, Do JH, Lee SW, Shin C, Cho NH, Kang JO, Nam JM, Kim JS, et al. Identification of five novel genetic loci related to facial morphology by genome-wide association studies. BMC Genomics. 2018;19:481. doi: 10.1186/s12864-018-4865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ohara K, Arai E, Takahashi Y, Ito N, Shibuya A, Tsuta K, Kushima R, Tsuda H, Ojima H, Fujimoto H, et al. Genes involved in development and differentiation are commonly methylated in cancers derived from multiple organs: A single-institutional methylome analysis using 1007 tissue specimens. Carcinogenesis. 2017;38:241–251. doi: 10.1093/carcin/bgw209. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.