Abstract
The present study aimed to examine the roles and mechanisms of metformin in the stemness and epithelial-mesenchymal transition (EMT) of colorectal cancer cells. The formation of spheroid cells, and the results of reverse transcription-quantitative polymerase chain reaction and western blot analyses showed that metformin suppressed the ability to form spheroid cells and the expression of stemness markers in HCT116 colorectal cancer cells. Additionally, metformin attenuated the EMT process, characterized by a decrease of mesenchymal marker Vimentin and an increase in the expression of an epithelial marker. Mechanistically, metformin inactivated the Wnt3a/β-catenin signaling pathway, and reactivation of Wnt3a/β-catenin signaling attenuated the inhibition of metformin on the stemness of HCT116 colorectal cancer cells and EMT. Finally, it was revealed that metformin re-sensitized HCT116 sphere cells to 5-fluorouracil resistance. These results suggest that metformin can attenuate stemness and EMT in colorectal cancer cells.
Keywords: metformin, epithelial-mesenchymal transition, stemness, colorectal cancer
Introduction
Metformin is a first-line drug used in the treatment of type 2 diabetes, and is positively correlated with increased risk of liver cancer, pancreatic cancer and colorectal cancer (1). A previous study showed that metformin decreased the risk and mortality rate of colorectal cancer (2), however, the detailed mechanisms underlying the effect of metformin remain to be fully elucidated.
The stemness of cancer cells is involved in tumor progression, recurrence and chemoresistance (3). Non-migratory tumorigenic intrinsic cancer stem cells (CSCs) ensure breast cancer metastasis by the generation of C-X-C chemokine receptor 4 migrating CSCs (4). Aspirin can attenuate chemoresistance in breast cancer by disrupting the nuclear factor-κB-interleukin-6 signaling axis, which is responsible for CSC generation (5). Previous studies have indicated that nemo-like kinase maintains the proliferation and stemness of non-small cell lung cancer (NSCLC) (6) and is a target of metformin, in which metformin inhibits the stemness of glioma cells and epithelial-mesenchymal transition (EMT) via suppressing the activity of yes-associated protein (7). However, whether metformin can attenuate the stemness of colorectal cancer cells remains to be elucidated.
A previous study demonstrated that activation of the WNT/β-catenin pathway is involved in vascular endothelial growth factor-A/neuropilin 1 axis-induced breast cancer stemness (8). The WNT/β-catenin pathway has been shown to direct the self-renewal symmetric cell division of human telomerase reverse transcriptase-high prostate CSCs (9). In addition, the downregulation of BORIS/CTCFL efficiently regulates cancer cells stemness and metastasis in the MYCN-amplified neuroblastoma cell line by modulating the Wnt/β-catenin signaling pathway (10). However, whether Wnt3a/β-catenin pathway is also engaged in colorectal cancer cell stemness remains to be elucidated.
In the present study, the results indicated that metformin attenuated the stemness and EMT process in colorectal cancer cells. Additionally, the Wnt3a/β-catenin signaling pathway was suppressed by metformin in colorectal cancer cells. Notably, re-activation of the Wnt3a/β-catenin pathway via its agonist or the overexpression of Wnt3a attenuated the metformin-mediated inhibition of stemness and EMT in colorectal cancer cells. Metformin enhanced the sensitivity of the colorectal cancer cells to 5-fluorouracil (5-FU). Therefore, it was suggested that metformin suppresses the stemness and enhances the chemotherapeutic sensitivity of colorectal cancer cells.
Materials and methods
Cell culture and reagents
The HCT116 human colorectal cell line was purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). The HCT116 colorectal cancer cells were cultured in 1640 medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) under a humidified atmosphere at 37°C with 5% CO2. The Wnt3a/β-catenin agonist (SKL2001, cat. no. S8320) and 5-FU (cat. no. S1209) were purchased from Selleck Chemicals (Shanghai, China). For cell spheroid formation assay, the concentration of SKL2001 was 10 nM, cell density was 1,000 cells/ml, duration was 10 days at 37°C with 5% CO2; and for other experiments the concentration of SKL2001 was 5 µM at cell density of 50% confluence, duration was 48 h at 37°C with 5% CO2.
Cell spheroid formation assay
The HCT116 colorectal cancer cells were cultured on ultra-low attachment 24-well plates (Corning Incorporated, Union City, CA, USA) at 500 cells/well, and maintained in MammoCult™ Human Medium (cat. no. 05620; StemCell Technologies, Inc., Vancouver, BC, Canada), and followed by treatment of various concentrations of metformin (1, 2 or 4 mM) or solvent (control) for 10 days at 37°C with 5% CO2. Cell spheroid formation efficiency was determined by counting the number of mammospheres and identifying those with a diameter >50 µm under a light microscope. All experiments were performed at least in triplicate.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis
Total RNA was extracted from the HCT116 colorectal cancer cells treated with or without metformin using TRIzol (cat. no. R0016; Beyotime Institute of Biotechnology, Beijing, China). cDNA was synthesized using a BeyoRT™ First Strand cDNA Synthesis kit (RNaseH minus; cat. no. D7166; Beyotime Institute of Biotechnology) following the manufacturer's protocols. The RT-qPCR analysis was performed on the StepOne Plus PCR system with BeyoFast™ SYBR-Green qPCR mix (2X, Low ROX; cat no. D7262; Beyotime Institute of Biotechnology) containing 1 µl BeyoFast™ TaqDNA Polymerase, 1 µl PCR Buffer, 0.5 µl dNTPs, 6.5 µl SYBR-Green I and 1 µl cDNA was used as templates in a 10 µl reaction volume. The denaturing process was 95°C for 5 min, the annealing process was 58°C for 30 sec and the elongation process was 72°C for 30 sec. A total of 35 cycles of this RT-qPCR was performed. All samples were analyzed in triplicate and repeated at least three times. The relative expression of genes was normalized to GAPDH. The 2−∆∆Cq method was used to determine the expression of each transcript (11). Primers sequences were: ALDH1, forward, 5′-CACGCCAGACTTACCTGTCCTACT-3′, reverse, 5′-TGTCAACATCCTCCTTATCTCCTT-3′; Nanog, forward, 5′-CACGCCAGACTTACCTGTCCTACT-3′, reverse, 5′-TGTCAACATCCTCCTTATCTCCTT-3′; EpCAM, forward, 5′-TCCAGAAAGAAGAGAATGGCAAAG-3′, reverse, 5′-ACAAGACTCAAGTAAATAGAAAGG-3′; CD44, forward, 5′-GCAGGTATGGGTTCATAGAAGGGC-3′, reverse, 5′-TGTGAGTGTCTGGTAGCAGGGATT-3′; E-cadherin, forward, 5′-ATGGCTTCCCTCTTTCATCTCCTG-3′, reverse, 5′-CATAGTTCCGCTCTGTCTTTGGCT-3′; Vimentin, forward, 5′-GCCAGATGCGTGAAATGGAAGAGA-3′, reverse, 5′-TCAGGGAGGAAAAGTTTGGAAGAG-3′; Wnt3a, forward, 5′-GCAGGAGGGCCCAGCGACGCCGCCG-3′, reverse, 5′-CGGCGGCGTCGCTGGGCCCTCCTGC-3′; β-catenin, forward, 5′-TTCGGGCTGGTGACAGGGAAGACA-3′, reverse, 5′-TTTGCGGGACAAAGGGCAAGATTT-3′.
Western blot analysis
The HCT166 cells exposed to the different treatments were harvested by ice-cold scraping and lysed with RIPA lysis buffer (cat. no. P0013B; Beyotime Institute of Biotechnology), and protein concentration was measured using a Bradford protein concentration assay kit (cat. no. P0009; Beyotime Institute of Biotechnology). Subsequently, 30 µg of total protein was uploaded and separated by 10% SDS-PAGE, and transferred onto a PVDF membrane (cat. no. 1620177; Bio-Rad Laboratories, Inc., Hercules, CA, USA). The membranes were blocked with 5% non-fat milk in Tris-buffered saline with 0.1% Tween-20 (TBST) for 1.5 h at room temperature and then incubated with primary antibodies overnight at 4°C. The primary antibodies were purchased from Abcam (Cambridge, MA, USA) and were: ALDH1 (cat. no. ab52492), Nanog (cat. no. ab109250), EpCAM (cat. no. ab32392), CD44 (cat. no. ab189524), E-cadherin (cat. no. ab1416), Vimentin (cat. no. ab8978), Wnt3a (cat. no. ab219412), β-catenin (cat. no. ab16051) and β-actin (cat. no. ab8227). The dilution was 1:3,000 for all primary antibodies. The membranes were then incubated with the following secondary antibodies: HRP-labeled goat anti-rabbit IgG(H+L) from Beyotime Institute of Biotechnology (cat. no. A0208) and HRP-labeled goat anti-mouse IgG(H+L) from Beyotime Institute of Biotechnology (cat. no. A0216) for 1 h at room temperature at a dilution of 1:500. The membranes were washed with TBST for 15 min three times. An enhanced chemiluminescence kit (BeyoECL Star, cat. no. P0018A; Beyotime Institute of Biotechnology) was used to develop images in the Tanon 5200 machine (Tanon Science and Technology Co., Ltd., Shanghai, China).
Plasmid and transfection
The Human Wnt3a expression plasmid (pcDNA-Wnt3a, cat. no. 35908) was purchased from Addgene, Inc. (Cambridge, MA, USA), which was transfected into the HCT116 colorectal cancer cells using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) in accordance with the manufacturer's protocol. A total of 2 µg plasmid was used for transfection.
Cell viability assay
To determine whether metformin enhances the sensitivity of HCT116 colorectal cancer cells to 5-FU. The HCT116 colorectal cancer cells were treated with 5-FU (25 µg/ml), with or without metformin. The density of cells was 30–50% at 37°C with 5% CO2. Cells that were left untreated served as the control group. After 24, 48 and 72 h, the HCT116 colorectal cancer cell viability was evaluated using a Cell Counting Kit-8 (cat no. C0037; Beyotime Institute of Biotechnology) according to the manufacturer's protocol. The experiments were repeated at least three times.
Statistical analysis
GraphPad Prism (version 5.01; GraphPad Software, Inc., La Jolla, CA, USA) was used for data analysis. All data were obtained from at least three independent experiments, and presented as the mean ± standard deviation. Datasets with only two groups were analyzed using Student's t-test. Differences between multiple groups were analyzed using one-way analysis of variance with the Tukey-Kramer post hoc test. P<0.05 was considered to indicate a statistically significant difference.
Results
Metformin attenuates stemness of HCT116 colorectal cancer cells in a concentration-dependent manner
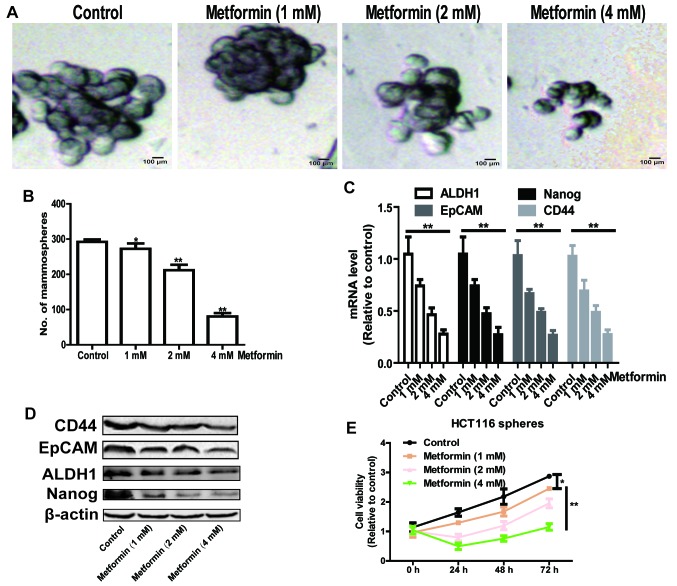
As CSCs are efficiently enriched in spheroid cells, and they have the capacity of self-renewal and multi-differentiation (12), the present study initially identified whether metformin was able to decrease the formation of cell spheroids in HCT116 colorectal cancer cells. As shown in Fig. 1A and B, the formation of cell spheroids was attenuated by metformin treatment in a concentration-dependent manner, which was characterized by a decrease in spheroid size and number. Additionally, the expression levels of stemness markers epithelial cell adhesion molecule (EpCAM), CD44, Nanog and aldehyde dehydrogenase 1 (ALDH1) were detected, and it was found that metformin also decreased the expression levels of these stemness markers in a concentration-dependent manner (Fig. 1C and D). Furthermore, cells digested from the spheres formed by HCT116 colorectal cancer cells were subjected to a cell viability assay following metformin treatment. As shown in Fig. 1E, metformin significantly decreased cell viability in a concentration-dependent manner. These results suggest that metformin attenuated the stemness of HCT116 colorectal cancer cells in a concentration-dependent manner.
Figure 1.
Metformin attenuates the stemness of HCT116 colorectal cancer cells in a concentration-dependent manner. The potential of HCT116 colorectal cancer cells to form spheroids was detected in HCT116 colorectal cancer cells treated with different concentrations of metformin by measuring sphere (A) size and (B) number. (C) mRNA and (D) protein expression levels of stemness markers (EpCAM, CD44, Nanog and ALDH1) were examined in cells in the different treatment groups. (E) Cell viability was examined in cells digested from by HCT116 colorectal cancer sphere cells treated with different concentrations of metformin. Data are presented as the mean ± standard deviation, *P<0.05 and **P<0.01 vs. control. EpCAM, epithelial cell adhesion molecule; ALDH1, aldehyde dehydrogenase 1.
Metformin suppresses the EMT process in HCT116 colorectal cancer cells
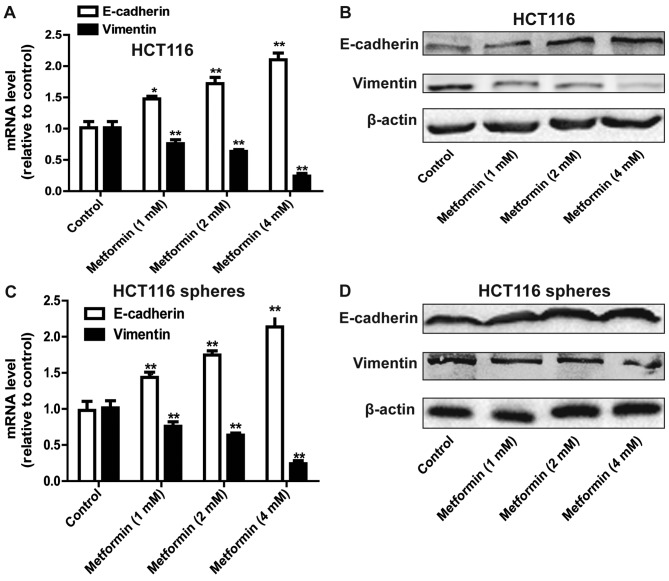
As the stemness of cells can confer mesenchymal traits (13), it was hypothesized that metformin can inhibit the EMT process in HCT116 colorectal cancer cells. As expected, metformin attenuated the EMT process in HCT116 colorectal cancer cells, characterized by a decrease in the expression of mesenchymal marker Vimentin, and an increase in the expression of epithelial marker E-cadherin (Fig. 2A and B). The EMT process in HCT116 colorectal cancer sphere cells was also suppressed by metformin treatment (Fig. 2C and D).
Figure 2.
Metformin suppresses the EMT process in HCT116 colorectal cancer cells. (A) mRNA and (B) protein levels of EMT markers (E-cadherin and Vimentin) were detected in HCT116 colorectal cancer cells treated with different concentrations of metformin. (C) mRNA and (D) protein levels of EMT markers (E-cadherin and Vimentin) were detected in HCT116 colorectal cancer sphere cells treated with treatment of different concentrations of metformin. Data are presented as the mean ± standard deviation, *P<0.05 and **P<0.01 vs. control. EMT, epithelial-mesenchymal transition.
Metformin inhibits the Wnt3a/β-catenin pathway in HCT116 colorectal cancer cells
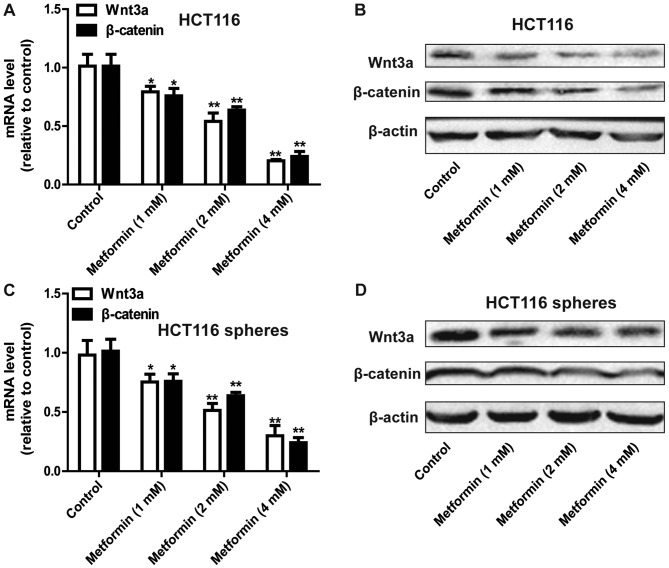
The present study further examined the mechanisms by which metformin inhibited the stemness of HCT116 colorectal cancer cells. The focus of this investigation was on the Wnt3a/β-catenin pathway owing to its critical promotive roles in cancer cell stemness (8,9). As shown in Fig. 3A and B, the expression levels of Wnt3a and β-catenin were decreased by metformin treatment in a concentration-dependent manner in the HCT116 colorectal cancer cells. Consistently, metformin decreased the expression of Wnt3a and β-catenin in the sphere cells formed by HCT116 colorectal cancer cells (Fig. 3C and D). Therefore, metformin inhibited the Wnt3a/β-catenin signaling pathway in HCT116 colorectal cancer cells.
Figure 3.
Metformin inhibits the Wnt3a/β-catenin pathway in HCT116 colorectal cancer cells. (A) mRNA and (B) protein levels of Wnt3a and β-catenin were detected in HCT116 colorectal cancer cells treated with different concentrations of metformin. (C) mRNA and (D) protein levels of Wnt3a and β-catenin were detected in HCT116 colorectal cancer sphere cells treated with different concentrations of metformin. Data are presented as the mean ± standard deviation, *P<0.05 and **P<0.01 vs. control.
Reactivation of the Wnt3a/β-catenin signaling pathway rescues metformin-mediated inhibition on stemness and EMT of HCT116 colorectal cancer cells
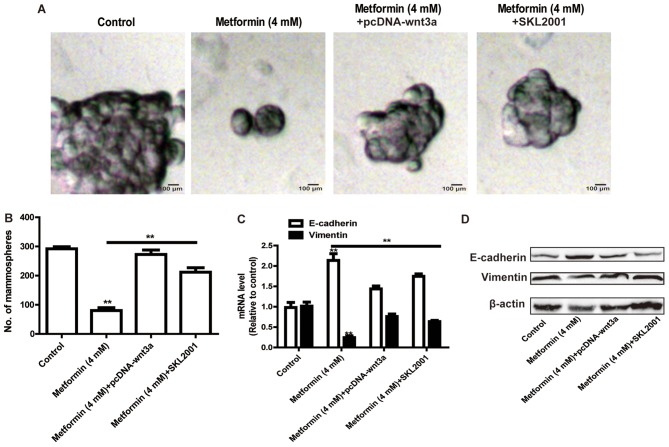
The present study further assessed whether metformin inhibited the stemness of HCT116 colorectal cancer cells and the EMT process via the Wnt3a/β-catenin signaling pathway. The HCT116 colorectal cancer cells were transfected with the pcDNA-Wnt3a plasmid or treated with the Wnt3a/β-catenin agonist (SKL2001), followed by metformin treatment. As shown in Fig. 4A and B, the overexpression of Wnt3a or treatment with SKL2001 attenuated the metformin-mediated inhibition of the size and number of sphere cells. Additionally, the metformin-induced inhibition of EMT was rescued by the overexpression of Wnt3a or treatment with SKL2001 (Fig. 4C and D).
Figure 4.
Reactivation of Wnt3a/β-catenin signaling rescues the metformin-mediated inhibition of stemness and EMT of HCT116 colorectal cancer cells. The potential of HCT116 colorectal cancer cells to form spheroids was detected in HCT116 colorectal cancer cells with Wnt3a overexpression or treatment with SKL2001, with or without metformin treatment, via measuring sphere (A) size and (B) number. (C) mRNA and (D) protein levels of EMT markers (E-cadherin and Vimentin) were determined in the cells in the different treatment groups. Data are presented as the mean ± standard deviation, **P<0.01 vs. control. EMT, epithelial-mesenchymal transition.
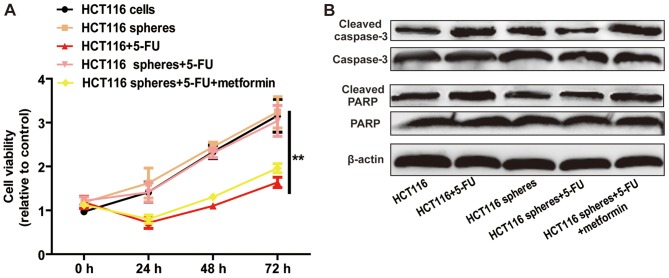
Metformin attenuates 5-FU resistance of HCT116 sphere cells
As CSCs contribute to chemoresistance, it was hypothesized that metformin can attenuate 5-FU resistance in HCT116 cell spheres. The results of the cell viability assay indicated that metformin enhanced 5-FU sensitivity, characterized by a decrease in cell viability (Fig. 5A) and increase in cell apoptosis (Fig. 5B). These results demonstrated that metformin attenuated 5-FU resistance in HCT116 sphere cells.
Figure 5.
Metformin attenuates 5-FU resistance of HCT116 sphere cells. (A) Cell viabilities of HCT116 colorectal cancer cells treated with 5-FU and of HCT116 colorectal cancer sphere cells treated with 5-FU with or without metformin treatment, were determined using a Cell Counting Kit-8 assay. (B) Expression levels of apoptosis executors (cleaved caspase-3 and cleaved PARP) were detected in the cells in the different treatment groups. Data are presented as the mean ± standard deviation; **P<0.01 vs. control. 5-FU, 5-fluorouracil; PARP, poly(ADP-ribose) polymerase.
Discussion
Metformin, a first-line drug for treating type 2 diabetes, has suppressive effects in various tumors (14,15). However, the application of metformin in clinical treatments for colorectal cancer has not been approved based on the fact its functions and mechanisms remain to be fully elucidated. Therefore, elucidation of the mechanisms underlying the role of metformin in colorectal cancer progression is urgently required. The present study focused on the roles and mechanisms of metformin on the stemness of colorectal cancer cells. To the best of our knowledge, this is the first study revealing the metformin-mediated inhibition of the stemness of colorectal cancer cells, which may facilitate the clinical evaluation of metformin in the treatment of colorectal cancer.
In the present study, it was shown that metformin inhibited the Wnt3a/β-catenin pathway in HCT116 colorectal cancer cells. The Wnt3a/β-catenin pathway promotes tumor progression by facilitating tumor metastasis, angiogenesis, EMT and CSC formation (8,9). Notably, reactivation of the Wnt3a/β-catenin pathway rescued the metformin-mediated inhibition of HCT116 colorectal cancer cell stemness and the EMT process, indicating that metformin exerted its effects at least partially via the Wnt3a/β-catenin pathway. The present study also confirmed that metformin enhanced 5-FU sensitivity in HCT116 sphere cells, which is consistent with previous studies showing that metformin mediated 5-FU sensitivity in hepatocellular carcinoma (16), pancreatic cancer cells (17) and NSCLC (18). However, it is noteworthy that the results presented here are based on in vitro experiments, and further in vivo experiments are required to confirm the inhibitory effects of metformin in colorectal cancer.
In conclusion, the results of the present study suggest that the interaction between metformin and the Wnt3a/β-catenin may be therapeutically targetable providing novel approaches to treat colorectal cancer and potentially other diseases in which Wnt3a/β-catenin signaling transactivation is aberrant.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
CZ and YW designed the study, analyzed the data, performed the experiments and wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Lee MS, Hsu CC, Wahlqvist ML, Tsai HN, Chang YH, Huang YC. Type 2 diabetes increases and metformin reduces total, colorectal, liver and pancreatic cancer incidences in Taiwanese: A representative population prospective cohort study of 800,000 individuals. BMC Cancer. 2011;11:20. doi: 10.1186/1471-2407-11-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee JH, Kim TI, Jeon SM, Hong SP, Cheon JH, Kim WH. The effects of metformin on the survival of colorectal cancer patients with diabetes mellitus. Int J Cancer. 2012;131:752–759. doi: 10.1002/ijc.26421. [DOI] [PubMed] [Google Scholar]
- 3.Wang N, Docherty F, Brown HK, Reeves K, Fowles A, Lawson M, Ottewell PD, Holen I, Croucher PI, Eaton CL. Mitotic quiescence, but not unique ‘stemness,’ marks the phenotype of bone metastasis-initiating cells in prostate cancer. FASEB J. 2015;29:3141–3150. doi: 10.1096/fj.14-266379. [DOI] [PubMed] [Google Scholar]
- 4.Mukherjee S, Manna A, Bhattacharjee P, Mazumdar M, Saha S, Chakraborty S, Guha D, Adhikary A, Jana D, Gorain M, et al. Non-migratory tumorigenic intrinsic cancer stem cells ensure breast cancer metastasis by generation of CXCR4(+) migrating cancer stem cells. Oncogene. 2016;35:4937–4948. doi: 10.1038/onc.2016.26. [DOI] [PubMed] [Google Scholar]
- 5.Saha S, Mukherjee S, Khan P, Kajal K, Mazumdar M, Manna A, Mukherjee S, De S, Jana D, Sarkar DK, Das T. Aspirin suppresses the acquisition of chemoresistance in breast cancer by disrupting an NFκB-IL6 signaling axis responsible for the generation of cancer stem cells. Cancer Res. 2016;76:2000–2012. doi: 10.1158/0008-5472.CAN-15-1360. [DOI] [PubMed] [Google Scholar]
- 6.Suwei D, Liang Z, Zhimin L, Ruilei L, Yingying Z, Zhen L, Chunlei G, Zhangchao L, Yuanbo X, Jinyan Y, et al. NLK functions to maintain proliferation and stemness of NSCLC and is a target of metformin. J Hematol Oncol. 2015;8:120. doi: 10.1186/s13045-015-0203-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yuan X, Wei W, Bao Q, Chen H, Jin P, Jiang W. Metformin inhibits glioma cells stemness and epithelial-mesenchymal transition via regulating YAP activity. Biomed Pharmacother. 2018;102:263–270. doi: 10.1016/j.biopha.2018.03.031. [DOI] [PubMed] [Google Scholar]
- 8.Zhang L, Wang H, Li C, Zhao Y, Wu L, Du X, Han Z. VEGF-A/neuropilin 1 pathway confers cancer stemness via activating Wnt/β-catenin axis in breast cancer cells. Cell Physiol Biochem. 2017;44:1251–1262. doi: 10.1159/000485455. [DOI] [PubMed] [Google Scholar]
- 9.Zhang K, Guo Y, Wang X, Zhao H, Ji Z, Cheng C, Li L, Fang Y, Xu D, Zhu HH, Gao WQ. WNT/β-catenin directs self-renewal symmetric cell division of hTERThigh prostate cancer stem cells. Cancer Res. 2017;77:2534–2547. doi: 10.1158/0008-5472.CAN-16-1887. [DOI] [PubMed] [Google Scholar]
- 10.Garikapati KR, Patel N, Makani VKK, Cilamkoti P, Bhadra U, Bhadra MP. Down-regulation of BORIS/CTCFL efficiently regulates cancer stemness and metastasis in MYCN amplified neuroblastoma cell line by modulating Wnt/β-catenin signaling pathway. Biochem Biophys Res Commun. 2017;484:93–99. doi: 10.1016/j.bbrc.2017.01.066. [DOI] [PubMed] [Google Scholar]
- 11.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔC(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 12.Kida K, Ishikawa T, Yamada A, Shimada K, Narui K, Sugae S, Shimizu D, Tanabe M, Sasaki T, Ichikawa Y, Endo I. Effect of ALDH1 on prognosis and chemoresistance by breast cancer subtype. Breast Cancer Res Treat. 2016;156:261–269. doi: 10.1007/s10549-016-3738-7. [DOI] [PubMed] [Google Scholar]
- 13.Ren D, Zhu X, Kong R, Zhao Z, Sheng J, Wang J, Xu X, Liu J, Cui K, Zhang XH, et al. Targeting brain-adaptive cancer stem cells prohibits brain metastatic colonization of triple-negative breast cancer. Cancer Res. 2018;78:2052–2064. doi: 10.1158/0008-5472.CAN-17-2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu S, Yang Z, Jin P, Yang X, Li X, Wei X, Wang Y, Long S, Zhang T, Chen G, et al. Metformin suppresses tumor progression by inactivating stromal fibroblasts in ovarian cancer. Mol Cancer Ther. 2018;17:1291–1302. doi: 10.1158/1535-7163.MCT-17-0927. [DOI] [PubMed] [Google Scholar]
- 15.Lee JE, Lim JH, Hong YK, Yang SH. High-dose metformin plus temozolomide shows increased anti-tumor effects in glioblastoma in vitro and in vivo compared with monotherapy. Cancer Res Treat. 2018;50:1331–1342. doi: 10.4143/crt.2017.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tian Y, Tang B, Wang C, Sun D, Zhang R, Luo N, Han Z, Liang R, Gao Z, Wang L. Metformin mediates resensitivity to 5-fluorouracil in hepatocellular carcinoma via the suppression of YAP. Oncotarget. 2016;7:46230–46241. doi: 10.18632/oncotarget.10079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng G, Lanza-Jacoby S. Metformin decreases growth of pancreatic cancer cells by decreasing reactive oxygen species: Role of NOX4. Biochem Biophys Res Commun. 2015;465:41–46. doi: 10.1016/j.bbrc.2015.07.118. [DOI] [PubMed] [Google Scholar]
- 18.Liu Y, He C, Huang X. Metformin partially reverses the carboplatin-resistance in NSCLC by inhibiting glucose metabolism. Oncotarget. 2017;8:75206–75216. doi: 10.18632/oncotarget.20663. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.