Abstract
Uncontrolled proliferation and defective apoptosis are two major factors responsible for maintaining the malignant properties of melanoma cells. Our previous study demonstrated that induced expression of four reprogramming factors remodeled the phenotype of B16-F10 mouse melanoma cells into melanoma stem cells. The present study was conducted to investigate the effect of the four Yamanaka reprogramming factors, namely Oct4, Sox2, Klf4 and c-Myc (OSKM), on the proliferation and apoptosis of melanoma cells, and to identify the responsible molecular signals. The results identified that expression of the four reprogramming factors was highly induced by doxycycline treatment in the stable melanoma cell clone that was transfected with a plasmid expressing these factors, driven by the Tet-On element. It was further confirmed that induced expression of these factors enhanced the proliferation and suppressed the apoptosis of the melanoma cells. In addition, induced OSKM expression increased cell proliferation, accelerated the progression of the cell cycle, and upregulated the mRNA expression levels of Janus kinase 2 (JAK2) and Cyclin-B1. Induced expression of these factors also decreased the apoptosis, as well as upregulated B-cell lymphoma 2 (BCL-2) and downregulated BCL-2-associated X (BAX) mRNA expression levels. Taken together, the results suggested that upregulated JAK2 and Cyclin-B1 may be responsible for the enhanced proliferation of melanoma cells, and that BCL-2 upregulation and BAX downregulation may account for the suppressed apoptosis of these cells.
Keywords: melanoma, reprogramming factors, proliferation, apoptosis, gene expression
Introduction
Malignant melanoma is a highly aggressive disease exhibiting drug-resistant behavior (1). Higher melanoma incidence is reported in children and adolescents, whose longer life expectancy than adult patients may be seriously affected (2). Melanoma treatments include conventional surgery, chemotherapy, radiotherapy and biotherapy; however, these are not always successful. Furthermore, certain of these treatment strategies are associated with adverse reactions and/or emergence of drug resistance (3,4), due to the involvement of relatively complicated cellular and molecular mechanisms.
Uncontrolled proliferation and defective apoptosis have been recognized as major factors responsible for the transformation of normal melanocytes into malignant melanoma cells (5). This has been further proven by studies reporting that benign nevi can be transformed into melanoma cells through uncontrolled proliferation and decreased apoptosis (6,7).
The Yamanaka transcription factors: Oct4, Sox2, Klf4, and c-Myc (OSKM) have been successfully used to induce the differentiation of osteosarcoma and breast cancer cells into osteosarcoma stem cells and breast cancer stem cells, respectively (8–11). Our previous study demonstrated that the plasmid expressing these four factors driven by the Tet-On element is transfected into melanoma F10-B16 cells and doxycycline (DOX) is used to induce the expression of these factors in stable transfected cell clones; thus the expression of four reprogramming factors OSKM were induced, remodeling the phenotype of B16-F10 mouse melanoma cells into melanoma stem cells (12). Therefore, the present study was conducted to seek evidence for the effect of induced expression of these four reprogramming factors on the proliferation and apoptosis of melanoma cells, as well as to identify the responsible molecular signals involved.
Materials and methods
Materials
High-glucose Dulbecco's modified Eagle's medium (H-DMEM), sucrose-based solution, SYBR Green PCR Master Mix, Lipofectamine® 2000 and Zeocin were obtained from Thermo Fisher Scientific, Inc. (Waltham, MA, USA). Fetal bovine serum (FBS) was purchased from GE Healthcare Life Science (Hyclone; Logan, UT, USA). Doxycycline (DOX) was from Sigma-Aldrich (Merck KGaA, Darmstadt, Germany). The Cell Counting kit-8 (CCK-8) assay kit was provided by Dojindo Molecular Technologies, Inc. (Tokyo, Japan). The Cell Cycle Detection kit was from Beyotime Institute of Biotechnology (Jiangsu, China). The Annexin V/PI Apoptosis Detection kit was purchased from BD Biosciences (San Jose, CA, USA). The RNAiso Plus reagent, polymerase chain reaction (PCR) primers and reverse transcription (RT) reaction kit were purchased from Takara Biotechnology Co., Ltd. (Dalian, China). The plasmids TetO-FUW-OSKM and FUW-M2rtTA were kindly provided by Professor Rudolf Jaenisch (Whitehead Institute for Biomedical Research, Cambridge, MA, USA; Addgene plasmid nos. 20321 and 20342, respectively).
Cell culture and transfection
B16-F10 mouse melanoma cells were obtained from the American Type Culture Collection (Manassas, VA, USA). These cells were maintained in H-DMEM supplemented with 10% FBS at 37°C in a humidified atmosphere with 5% CO2. Cell transfection with the plasmid TetO-FUW-OSKM or FUW-M2rtTA was performed using the Lipofectamine® 2000 reagent, according to the manufacturer's protocol. After 24 h, the transfection was terminated by washing away the media, following which fresh complete medium was added to the cells. On day 3, we seeded the cells in a 10-cm plate, and then Zeocin (400 µg/ml) was added at day 5 to start the selection. The cells were checked daily, and the medium was changed when necessary in order to remove dead cells. When the clones were clearly visible and isolated from one another, they were selected, transferred to 96-well plates (one clone/well) and then expanded.
Cell proliferation assay
The transfected cells were seeded into 96-well plates at a density of 1,000 cells/well and incubated at 37°C overnight to allow for cell attachment. Then, the expression of OSKM was induced via a Tet-On inducer, DOX (2 µg/ml,). After 0, 24, 48, 72 and 96 h of incubation with the control group that absence of DOX, the CCK-8 assay kit was used to respectively evaluate the cell proliferation, according to the manufacturer's protocols. Briefly, following incubation, the cells were incubated with CCK-8 solution, and then the absorbance was determined at a wavelength of 450 nm with a microplate reader. The absorbance values determined the cell number based on a standard curve of cell numbers against the absorbance value, which resulted from a series of samples analyses in which the cell numbers were known.
Cell cycle progression assay
The B16-F10 cells were plated in 6-cm diameter dishes (0.5×106 cells/dish) and allowed to grow for 3 days. A Cell Cycle Detection kit was then used to determine the cell cycle status, according to the manufacturer's protocol. Briefly, the cells were washed with cooled PBS, fixed with 70% alcohol at for >2 h. Next, cells were treat ed with RNase A (included in kit) and stained with PI solution. Flow cytometry was then conducted at an excitation wavelength of 488 nm and analyzed. The percentage of S-phase cells was calculated according to the number of cells at each phase, as follows: SPF(%)=S/(G0/G1+S+G2M). In addition, the proliferation index was calculated according to the following formula: PI(%)=(S+G2M)/(G0/G1+S+G2M).
Cell apoptosis assay
The cells were plated in 6-cm diameter dishes (0.5×106 cells/dish) and allowed to grow for 3 days. Flow cytometry with Annexin V-FITC/PI dual staining was conducted to detect the apoptotic cells, according to the protocol suggested by the manufacturer. Briefly, subsequent to staining with Annexin V-FITC and PI, the cells were analyzed with a FACSCalibur flow cytometer (BD Biosciences). The emitted green fluorescence of Annexin V (FL1) and red fluorescence of PI (FL3) were detected at excitation wavelengths of 488 and 546 nm, respectively, and at emission wavelengths of 525 and 647 nm, respectively. The degrees of early and late apoptosis were determined based on the percentages of Annexin V+/PI− and Annexin V+/PI+ cells, respectively.
RNA purification and RT-quantitative PCR (qPCR)
Total RNA purification and RT-qPCR were performed according to the detailed procedure described in a previous study (13). The primer sets used in the PCR amplification are listed in Table I. Following amplification, a melting curve was generated, and data analysis was performed using Dissociation Curve software, version 1.0 (Applied Biosystems; Thermo Fisher Scientific, Inc.). The normalized value was given by the ratio of the target gene mRNA to the reference gene (β-actin) mRNA in each sample.
Table I.
Primer sets used in reverse transcription-quantitative polymerase chain reaction.
| Genes | Primer | Sequence | GenBank no. |
|---|---|---|---|
| Oct4 | Forward | 5′-CAGCCAGACCACCATCTGTC-3′ | NM_013633.3 |
| Reverse | 5′-GTCTCCGATTTGCATATCTCCTG-3′ | ||
| Sox2 | Forward | 5′-GCTCGCAGACCTACATGAAC-3′ | NM_011443.4 |
| Reverse | 5′-GCCTCGGACTTGACCACAG-3′ | ||
| Klf4 | Forward | 5′-CTTCAGCTATCCGATCCGGG-3′ | NM_010637.3 |
| Reverse | 5′-GAGGGGCTCACGTCATTGAT-3′ | ||
| c-Myc | Forward | 5′-TCTCCATCCTATGTTGCGGTC-3′ | NM_010849.4 |
| Reverse | 5′-TCCAAGTAACTCGGTCATCATCT-3′ | ||
| JAK2 | Forward | 5′- CTGGCGAGGTGGTCGCTGTG −3 | NM_146145.2 |
| Reverse | 5′- GCCGACCCGCACTGTAGCAC −3′ | ||
| Cyclin-B1 | Forward | 5′- GGCGAGCCTCAAAAGCCGGA −3′ | NM_008655.1 |
| Reverse | 5′- GCGGCTGCCACCTGAGAAGG −3′ | ||
| BCL2 | Forward | 5′- ATGGCGCAAGCCGGGAGAAC −3′ | NM_009741.5 |
| Reverse | 5′- CGCGTCCGCATCTCCAGCAT −3′ | ||
| BAX | Forward | 5′- TCCGGGGAGCAGCTTGGGAG −3′ | NM_007527.3 |
| Reverse | 5′- GGCGGCTGCTCCAAGGTCAG −3′ | ||
| CDK1 | Forward | 5′- GTTGCTGGGCTCGGCTCGTT −3′ | NM_007658.3 |
| Reverse | 5′- GCGGCTTCTTGGTGGCCAGT −3′ | ||
| SKP2 | Forward | 5′- TGCCCCAACCTCATCCGCCT −3′ | NM_013787.3 |
| Reverse | 5′- ACCGGCTGAGCGAGAGGTGT −3′ | ||
| Caspase 3 | Forward | 5′- TCATTCAGGCCTGCCGGGGT −3′ | NM_009810.3 |
| Reverse | 5′- TGGATGAACCACGACCCGTCC −3′ | ||
| Caspase 9 | Forward | 5′- AGGGTGCGCCTAGTGAGCGA −3′ | NM_015733.5 |
| Reverse | 5′- CCTGATCCCGCCGAGACCCA −3′ | ||
| P53 | Reverse | 5′-GGACGATCTGTTGCTGCCCCGAGA −3′ | NM_011640.3 |
| Forward | 5′- TGACAGGGGCCATGGAGTGGCT −3′ | ||
| β-actin | Reverse | 5′-CATGTACGTTGCTATCCAGGC-3′ | NM_001101 |
| Reverse | 5′-CTCCTTAATGTCACGCACGAT-3′ |
JAK2, Janus kinase 2; CDK1, cyclin-dependent kinase 1; SKP2, S-phase kinase-associated protein 2; BCL-2, B-cell lymphoma 2; BAX, BCL-2-associated X; Caspase, cysteinyl aspartate specific proteinase.
Statistical analysis
Statistical analyses were performed using GraphPad Prism software, version 5.0 (San Diego, CA, USA). Student's t-test was used to analyze the statistical significance of any differences between two groups. P<0.05 was considered to indicare a statistically significant difference.
Results
Induction of OSKM expression by DOX treatment
In our previous study, B16-F10 mouse melanoma cells were transfected with a plasmid expressing the four Yamanaka transcription factors, known as OSKM, driven by the Tet-On element. The stable cell clones were then used to study the role of induced expression of these factors. The present study further attempted to ensure the induction of OSKM expression by a Tet-On inducer, DOX (2 µg/ml). The mRNA levels of the OSKM factors were analyzed by RT-qPCR in a selected stable cell clone, cultured in the presence or absence of DOX. As shown in Fig. 1, the expression of the four factors at the mRNAs level was significantly induced by the DOX treatment, when compared with that in cells without DOX exposure. The induction of elevated protein levels has been proven in our previous study (12). These results indicated that high expression of these factors was obtained by treating the melanoma cells with DOX.
Figure 1.
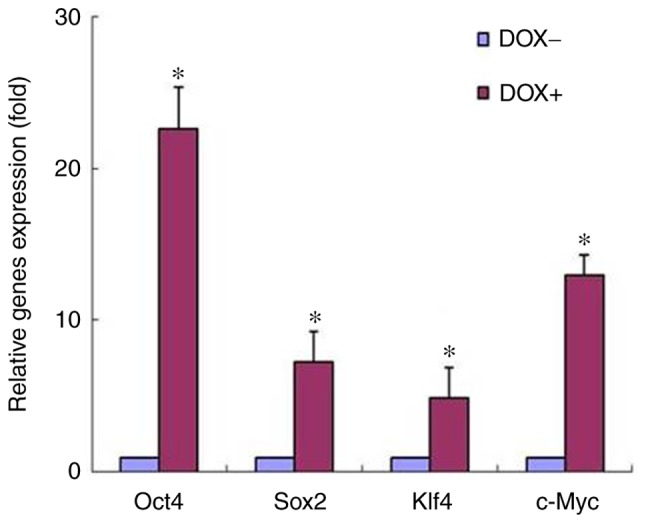
Induction of OSKM expression by DOX treatment. B16-F10 mouse melanoma cells, stably-transfected with TetO-FUW-OSKM, were cultured in the presence or absence of DOX. The OSKM mRNA levels were analyzed using reverse transcription-quantitative polymerase chain reaction and normalized to β-actin mRNA. The relative fold activation was obtained based on the ratio of the normalized values of the DOX-induced cells to that of the cells without DOX exposure. *P<0.01 vs. cells without exposure to DOX. OSKM, Oct4, Sox2, Klf4 and c-Myc; DOX, doxycycline.
Induced expression of OSKM enhances the proliferation of melanoma cells
Uncontrolled proliferation is a crucial factor that characterizes the malignancy of melanoma cells (6). To examine whether induction of OSKM expression was able to enhance the proliferation of melanoma cells, a CCK-8 cell proliferation assay was used in cells cultured in the presence or absence of DOX. As shown in Fig. 2, cells in the two culture conditions proliferated in a time-dependent manner; however, the cell proliferation was significantly increased following exposure to DOX as compared with that of cells without DOX exposure. These findings indicated that induction of OSKM expression enhanced the proliferation of melanoma cells.
Figure 2.
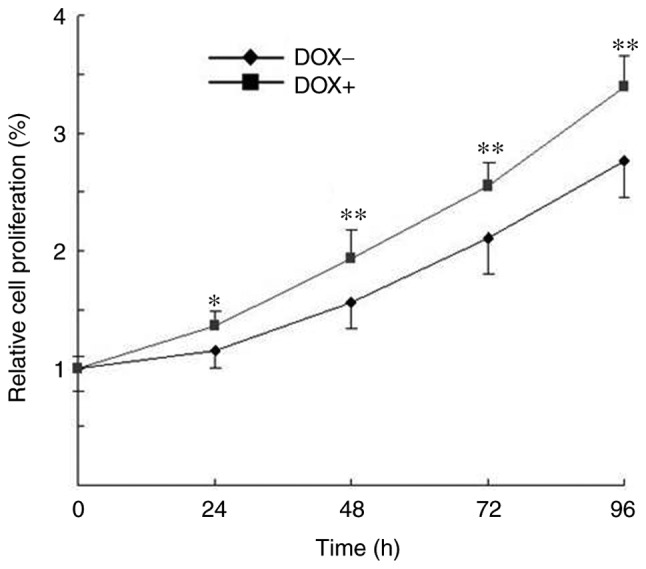
Induced expression of OSKM promoted the proliferation of melanoma cells, as examined using the Cell Counting kit-8 assay. Relative cell proliferation was calculated based on the ratio of the number of cells at a specific time point over the total number of cells at 0 h. *P<0.05 and **P<0.01, vs. cells without exposure to DOX. OSKM, Oct4, Sox2, Klf4 and c-Myc; DOX, doxycycline.
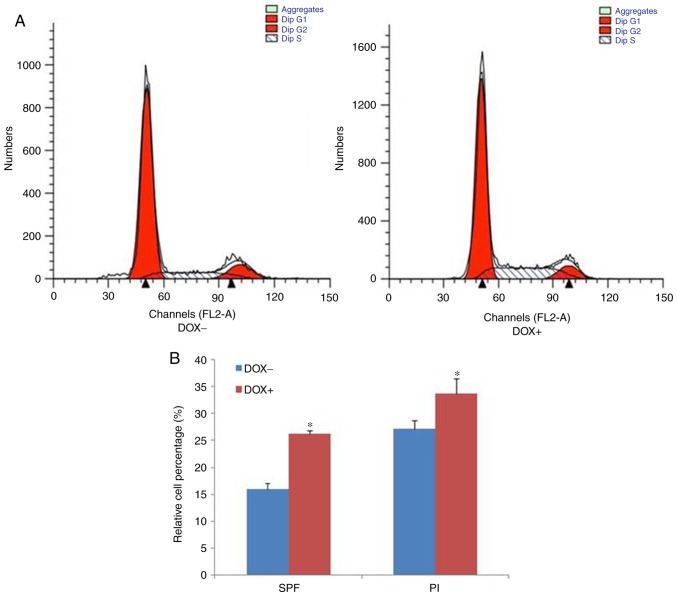
Induced expression of OSKM accelerates the cell cycle progression
The cell proliferation rate is determined by the progression of the cell cycle (14). Based on the earlier finding that the proliferation of melanoma cells is enhanced by the induction of OSKM expression, the present study next analyzed the percentage of melanoma cells at each cell division phase following culture in the absence or presence of DOX. The representative histograms of the flow cytometric assay are shown in Fig. 3A, while statistically analyzed data are presented in Fig. 3B. The percentage of S-phase cells and the proliferation index significantly increased in the cells cultured in the presence of DOX, when compared with those in cells cultured in the absence of DOX. Taken together, these results indicated that the induced expression of OSKM enhanced cell proliferation by accelerating cell cycle progression.
Figure 3.
Induced expression of OSKM accelerated the procession of the cell cycle. (A) Representative histograms of the flow cytometric assay, and (B) quantified results of the relative cell percentage are shown. *P<0.05 vs. cells without exposure to DOX. OSKM, Oct4, Sox2, Klf4 and c-Myc; SPF, S-phase fraction; PI, proliferation index; DOX, doxycycline.
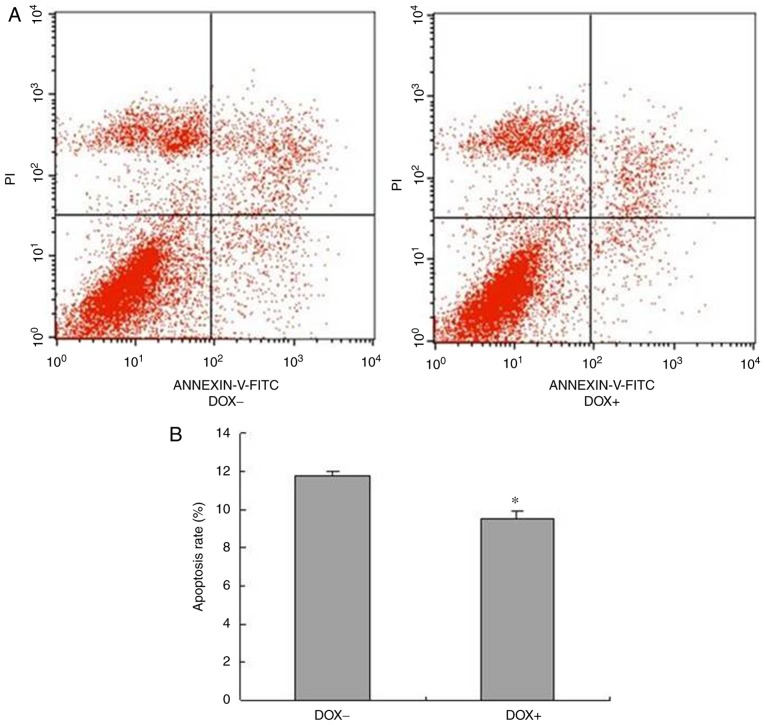
Induced expression of OSKM inhibits the apoptosis of melanoma cells
Defective apoptosis is another major factor deciding the malignancy of melanoma cells (7,14). To determine whether induced OSKM expression was able to decrease the apoptosis of melanoma cells, the proportion of apoptotic cells was analyzed through a flow cytometric assay. The representative histograms of the flow cytometric assay are shown in Fig. 4A, while statistically analyzed results are shown in Fig. 4B. As shown in Fig. 4, the percentage of apoptotic cells significantly decreased following exposure to DOX as compared with that in cells without DOX exposure. These results indicated that induction of OSKM expression suppressed the apoptosis of melanoma cells.
Figure 4.
Induced expression of OSKM suppressed the apoptosis of melanoma cells. (A) Representative histograms of the flow cytometric assay, and (B) quantified results of apoptotic cells are shown. Cells at the early apoptosis (Annexin V+/PI−) and late apoptosis (Annexin V+/PI+) stages were used in the statistical analysis. *P<0.05 vs. cells without exposure to DOX. OSKM, Oct4, Sox2, Klf4 and c-Myc; DOX, doxycycline.
Induced expression of OSKM changes the expression of associated genes
To explore the underlying mechanism by which OSKM factors enhance the proliferation and suppress the apoptosis of melanoma cells, the mRNA levels of certain associated genes were analyzed with RT-qPCR. These genes included Janus kinase 2 (JAK2), Cyclin-B1, cyclin-dependent kinase 1 (CDK1) and S-phase kinase-associated protein 2 (SKP2), whose products serve crucial roles in regulating the cell cycle progression and cell proliferation (15–17). In addition, B-cell lymphoma 2 (BCL-2), BCL-2-associated X (BAX), cysteinyl aspartate specific proteinase 3 (Caspase 3), Caspase 9 and P53 (also known as transformation-related protein 53) were detected, whose products are important in controlling cell apoptosis (18–21).
As shown in Table II, among the analyzed genes that are associated with cell proliferation, the mRNA expression levels of JAK2 and Cyclin-B1 were significantly increased when the cells were exposed to DOX, as compared with those in cells not exposed to DOX. By contrast, the mRNA expression levels of CDK1 and SKP2 were unchanged in the DOX-induced cells. Among the analyzed genes associated with cell apoptosis, the mRNA level of BCL-2 was increased, while the mRNA of BAX was decreased, when the cells were exposed to DOX. However, the expression of other genes associated with cell apoptosis was unchanged in the DOX-induced cells. The increased expression of JAK2 and Cyclin-B1 mRNAs supported the observation that induced expression of the four OSKM factors enhanced cell proliferation, suggesting that upregulation of JAK2 and Cyclin-B1 may be responsible for the enhanced proliferation. Furthermore, the upregulated BCL-2 and downregulated BAX mRNAs verified the conclusion that induced expression of the OSKM factors was able to suppress cell apoptosis, suggesting that the altered expression levels of BCL-2 and BAX may be involved in the suppression of apoptosis.
Table II.
Induction of OSKM expression by DOX treatment altered the expression levels of associated genes.
| Genes | Fold change (DOX+/DOX-) |
|---|---|
| JAK2 | 7.9±0.81a |
| Cyclin-B1 | 1.8±0.10b |
| CDK1 | 0.90±0.07 |
| SKP2 | 1.22±0.05 |
| BCL-2 | 2.4±0.18b |
| BAX | 0.56±0.02a |
| Caspase 3 | 1.4±0.10 |
| Caspase 9 | 1.3±0.08 |
| P53 | 1.39±0.17 |
P<0.01
P<0.05, vs. group without DOX exposure. mRNA levels were analyzed by reverse transcription-quantitative polymerase chain reaction and normalized to β-actin mRNA. Relative fold activation was calculated based on the ratio of the normalized values of the cells induced by DOX to that of the cells without DOX exposure. DOX, doxycycline; JAK2, Janus kinase 2; CDK1, cyclin-dependent kinase 1; SKP2, S-phase kinase-associated protein 2; BCL-2, B-cell lymphoma 2; BAX, BCL-2-associated X; Caspase, cysteinyl aspartate specific proteinase.
Discussion
The effects of induced expression of the four reprogramming factors on the proliferation and apoptosis of melanoma cells, which are the two major factors affecting the malignancy of the cells, has not been extensively investigated (22). In our previous study, stable B16-F10 mouse melanoma cell clones that were transfected with a plasmid expressing the Yamanaka reprogramming factors, namely OSKM, driven by the Tet-On element, had been used to study the role of the induced expression of these factors in remodeling the phenotype of melanoma cells. In the present study, the effects of these factors on the proliferation and apoptosis of melanoma cells, as well as the involved mechanisms, were investigated. Initially, the mRNA expression of these four factors in a stable cell clone cultured in the presence or absence of DOX was analyzed to ensure that the expression of OSKM was successfully induced. As expected, the mRNA levels of the OSKM factors were significantly enhanced when the cells were exposed to DOX, indicating that high expression of these factors was achieved by DOX exposure. This had also been reported in our earlier study (12), in which high induction of the protein levels of these factors had been observed. Furthermore, this was also supported by the knowledge that the plasmid TetO-FUW-OSKM is a single polycistronic vector encoding the four transcription factors, driven by the Tet-On element (23,24). These results provided the preliminary basis for the next phase of the present study.
The malignant growth of tumors is a complex process, involving activation of sustained growth signals and inhibition of cellular death signals (22). In the present study, it was observed that induction of the expression of the four reprogramming OSKM factors in B16-F10 melanoma cells significantly increased the proliferation and suppressed the apoptosis of melanoma cells. These results indicated that the reprogramming factors were able to elevate the malignancy of B16-F10 melanoma cells, based on the crucial role of uncontrolled cell proliferation and defective cell apoptosis in the development of melanoma and the development of the malignant phenotype of other various tumors (6,25,26). This observation was also supported by other studies reporting that induction of the reprogramming factors increased the cell proliferation rate of mature epithelial cells and Kaposi's sarcoma tumorigenesis (27,28).
To explore the molecular mechanism by which the induced expression of the four reprogramming factors affected the proliferation and apoptosis of melanoma cells, the mRNA levels of the associated genes was analyzed. Special attention was paid to the genes whose functions are involved in cell cycle progression, proliferation and apoptosis. The results revealed that among all analyzed mRNAs associated with cell cycle progression and proliferation, only the mRNA expression levels of JAK2 and Cyclin-B1 were significantly increased, while the levels of other associated genes remained unchanged. These results indicated that JAK2 and Cyclin-B1 upregulation may be responsible for the increased proliferation of melanoma cells, which was induced by the reprogramming factors. This finding was supported by other studies reporting that Cyclin-B1 controlled the cell cycle progression and JAK2 upregulated Cyclin-B1 (29,30). Furthermore, the current study demonstrated that among all the analyzed mRNAs that control cell apoptosis, the mRNA expression of BCL-2 was significantly increased and that of BAX was significantly decreased by the induced expression of the reprogramming factors, while all other apoptosis-associated genes were unchanged. These results indicated that the increased expression of BCL-2 and decreased expression of BAX may be responsible for the suppressed apoptosis of melanoma cells, induced by the reprogramming factors. This finding was also supported by previous studies reporting that cell apoptosis was controlled by the upregulation of BCL-2 and downregulation of BAX (18,19).
In conclusion, the present study observed that induced expression of four reprogramming factors enhanced the proliferation and suppressed the apoptosis of B16-F10 mouse melanoma cells. The enhanced proliferation, indicated by the increase in cell proliferation and accelerated progression of the cell cycle, was found to be associated with upregulation of JAK2 and Cyclin-B1 expression levels. In addition, the suppression of apoptosis was supported by the upregulation of BCL-2 and downregulation of BAX mRNA expression levels. Taken together, the results revealed that upregulated JAK2 and Cyclin-B1 may be responsible for enhanced proliferation, while upregulated BCL-2 and downregulated BAX may be responsible for apoptosis suppression in melanoma cells with DOX-induced OSKM expression.
Acknowledgements
The authors would like to express their appreciation to Professor F. William Orr from the University of Manitoba (Manitoba, Canada) for his great help in revising the manuscript and to Professor Rudolf Jaenisch from the Whitehead Institute for Biomedical Research (Cambridge, MA, USA) for supplying the plasmids FUW-M2rtTA and TetO-FUW-OCT4 of Addgene.
Glossary
Abbreviations
- OSKM
Oct4, Sox2, Klf4 and c-Myc
- SPF
S-phase fraction
- PI
proliferation index
- DOX
doxycycline
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
YW provided the related materials and methods from previous studies, and made substantial contributions to the study conception and design. H-JS analyzed and interpreted the data. R-GL and X-MW wwere involved in the data collection and disposal, the manuscript drafting and critical revision. Z-QC and NL conceived and designed parts of the study and agreed to be accountable for all aspects of the work in ensuring that questions related to the accuracy and integrity of any part of the work are appropriately investigated and resolved. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.DeSantis CE, Lin CC, Mariotto AB, Siegel RL, Stein KD, Kramer JL, Alteri R, Robbins AS, Jemal A. Cancer treatment and survivorship statistics, 2014. CA Cancer J Clin. 2014;64:252–271. doi: 10.3322/caac.21235. [DOI] [PubMed] [Google Scholar]
- 2.Hoang MT, Eichenfield LF. The rising incidence of melanoma in children and adolescents. Dermatol Nurs. 2000;12:188–189. [PubMed] [Google Scholar]
- 3.Dudek-Peric AM, Ferreira GB, Muchowicz A, Wouters J, Prada N, Martin S, Kiviluoto S, Winiarska M, Boon L, Mathieu C, et al. Antitumor immunity triggered by melphalan is potentiated by melanoma cell surface-associated calreticulin. Cancer Res. 2015;75:1603–1614. doi: 10.1158/0008-5472.CAN-14-2089. [DOI] [PubMed] [Google Scholar]
- 4.Ribas A, Hamid O, Daud A, Hodi FS, Wolchok JD, Kefford R, Joshua AM, Patnaik A, Hwu WJ, Weber JS, et al. ssociation of pembrolizumab with tumor response and survival among patients with advanced melanoma. JAMA. 2016;315:1600–1609. doi: 10.1001/jama.2016.4059. [DOI] [PubMed] [Google Scholar]
- 5.Bandarchi B, Jabbari CA, Vedadi A, Navab R. Molecular biology of normal melanocytes and melanoma cells. J Clin Pathol. 2013;66:644–648. doi: 10.1136/jclinpath-2013-201471. [DOI] [PubMed] [Google Scholar]
- 6.Croce CM. Oncogenes and cancer. N Engl J Med. 2008;358:502–511. doi: 10.1056/NEJMra072367. [DOI] [PubMed] [Google Scholar]
- 7.Zhao H, Tang W, Chen X, Wang S, Wang X, Xu H, Li L. The NAMPT/E2F2/SIRT1 axis promotes proliferation and inhibits p53-dependent apoptosis in human melanoma cells. Biochem Biophys Res Commun. 2017;493:77–84. doi: 10.1016/j.cbpb.2016.11.011. [DOI] [PubMed] [Google Scholar]
- 8.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 9.Levings PP, McGarry SV, Currie TP, Nickerson DM, McClellan S, Ghivizzani SC, Steindler DA, Gibbs CP. Expression of an exogenous human Oct-4 promoter identifies tumor-initiating cells in osteosarcoma. Cancer Res. 2009;69:5648–5655. doi: 10.1158/0008-5472.CAN-08-3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beltran AS, Rivenbark AG, Richardson BT, Yuan X, Quian H, Hunt JP, Zimmerman E, Graves LM, Blancafort P. Generation of tumor-initiating cells by exogenous delivery of OCT4 transcription factor. Breast Cancer Res. 2011;13:94. doi: 10.1186/bcr3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corominas-Faja B, Cufi S, Oliveras-Ferraros C, Cuyàs E, López-Bonet E, Lupu R, Alarcón T, Vellon L, Iglesias JM, Leis O, et al. Nuclear reprogramming of luminal-like breast cancer cells generates Sox2-overexpressing cancer stem-like cellular states harboring transcriptional activation of the mTOR pathway. Cell cycle. 2013;12:3109–31024. doi: 10.4161/cc.26173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Y, Mou Y, Zhang H, Wang X, Li R, Cheng Z, Liu X. Reprogramming factors remodel melanoma cell phenotype by changing Stat3 expression. Int J Med Sci. 2017;14:1402–1409. doi: 10.7150/ijms.21952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y, Wei Y, Zhang H, Shi Y, Li Y, Li R. Arsenic trioxide induces apoptosis of p53 null osteosarcoma MG63 cells through the inhibition of catalase. Med Oncol. 2012;29:1328–1334. doi: 10.1007/s12032-011-9848-5. [DOI] [PubMed] [Google Scholar]
- 14.Sherr CJ. Principles of tumor suppression. Cell. 2004;116:235–246. doi: 10.1016/S0092-8674(03)01075-4. [DOI] [PubMed] [Google Scholar]
- 15.Pan XW, Chen L, Hong Y, Xu DF, Liu X, Li L, Huang Y, Cui LM, Gan SS, Yang QW, et al. EIF3D silencing suppresses renal cell carcinoma tumorigenesis via inducing G2/M arrest through downregulation of Cyclin B1/CDK1 signaling. Int J Oncol. 2016;48:2580–2590. doi: 10.3892/ijo.2016.3459. [DOI] [PubMed] [Google Scholar]
- 16.Kralovics R, Passamonti F, Buser AS, Teo SS, Tiedt R, Passweg JR, Tichelli A, Cazzola M, Skoda RC. A gain-of-function mutation of JAK2 in myeloproliferative disorders. N Engl J Med. 2005;352:1779–1790. doi: 10.1056/NEJMoa051113. [DOI] [PubMed] [Google Scholar]
- 17.Xu J, Zhou W, Yang F, Chen G, Li H, Zhao Y, Liu P, Li H, Tan M, Xiong X, Sun Y. The beta-TrCP-FBXW2-SKP2 axis regulates lung cancer cell growth with FBXW2 acting as a tumour suppressor. Nat commun. 2017;8:14002. doi: 10.1038/ncomms14002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ashkenazi A, Fairbrother WJ, Leverson JD, Souers AJ. From basic apoptosis discoveries to advanced selective BCL-2 family inhibitors. Nat Rev Drug Discov. 2017;16:273–284. doi: 10.1038/nrd.2016.253. [DOI] [PubMed] [Google Scholar]
- 19.Carrington EM, Tarlinton DM, Gray DH, Huntington ND, Zhan Y, Lew AM. The life and death of immune cell types: The role of BCL-2 anti-apoptotic molecules. Immunol Cell Biol. 2017;95:870–877. doi: 10.1038/icb.2017.72. [DOI] [PubMed] [Google Scholar]
- 20.Herr I, Debatin KM. Cellular stress response and apoptosis in cancer therapy. Blood. 2001;98:2603–2614. doi: 10.1182/blood.V98.9.2603. [DOI] [PubMed] [Google Scholar]
- 21.Ferraz da Costa DC, Fialho E, Silva JL. Cancer chemoprevention by resveratrol: The p53 tumor suppressor protein as a promising molecular target. Molecules. 2017;22 doi: 10.3390/molecules22061014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hainaut P, Plymoth A. Targeting the hallmarks of cancer: Towards a rational approach to next-generation cancer therapy. Curr Opin Oncol. 2013;25:50–51. doi: 10.1097/CCO.0b013e32835b651e. [DOI] [PubMed] [Google Scholar]
- 23.Hockemeyer D, Soldner F, Cook EG, Gao Q, Mitalipova M, Jaenisch R. A drug-inducible system for direct reprogramming of human somatic cells to pluripotency. Cell Stem Cell. 2008;3:346–353. doi: 10.1016/j.stem.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carey BW, Markoulaki S, Hanna J, Saha K, Gao Q, Mitalipova M, Jaenisch R. Reprogramming of murine and human somatic cells using a single polycistronic vector. Proc Nati Acad Sci USA. 2009;106:157–162. doi: 10.1073/pnas.0811426106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Imran A, Qamar HY, Ali Q, Naeem H, Riaz M, Amin S, Kanwal N, Ali F, Sabar MF, Nasir IA. Role of molecular biology in cancer treatment: A review article. Iran J Public Health. 2017;46:1475–1485. [PMC free article] [PubMed] [Google Scholar]
- 26.Celia-Terrassa T, Kang Y. Distinctive properties of metastasis-initiating cells. Genes Dev. 2016;30:892–908. doi: 10.1101/gad.277681.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo L, Karoubi G, Duchesneau P, Shutova MV, Sung HK, Tonge P, Bear C, Rogers I, Nagy A, Waddell TK. Generation of induced progenitor-like cells from mature epithelial cells using interrupted reprogramming. Stem Cell Reports. 2017;9:1780–1795. doi: 10.1016/j.stemcr.2017.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gramolelli S, Ojala PM. Kaposi's sarcoma herpesvirus-induced endothelial cell reprogramming supports viral persistence and contributes to Kaposi's sarcoma tumorigenesis. Curr Opin Virol. 2017;26:156–162. doi: 10.1016/j.coviro.2017.09.002. [DOI] [PubMed] [Google Scholar]
- 29.Qian X, Tan C, Yang B, Wang F, Ge Y, Guan Z, Cai J. Astaxanthin increases radiosensitivity in esophageal squamous cell carcinoma through inducing apoptosis and G2/M arrest. Dis Esophagus. 2017;30:1–7. doi: 10.1093/dote/dox027. [DOI] [PubMed] [Google Scholar]
- 30.Toyoshima-Morimoto F, Taniguchi E, Shinya N, Iwamatsu A, Nishida E. Polo-like kinase 1 phosphorylates cyclin B1 and targets it to the nucleus during prophase. Nature. 2001;410:215–220. doi: 10.1038/35065617. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.