Abstract
The importance of microRNAs (miRNAs) in cancer development has been widely recognized in recent decades. In the present study, the function and mechanism of miRNA-363-3p (miR-363-3p), formerly characterized as a tumor suppressor, in the hepatocarcinogenesis of liver cancer cells was investigated. Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) was applied to detect the expression of miR-363-3p in liver cancer tissues. Cell proliferation, survival and migration capacities were determined by MTT, colony formation and wound-healing assays, respectively. The targeting of high mobility group AT-hook 2 (HMGA2) mRNA by miR-363-3p was confirmed by bioinformatics analysis, and RT-qPCR, luciferase reporter and western blot assays. The correlation between the expression levels of HMGA2 and miR-363-3p was analyzed. The RT-qPCR results revealed that the levels of miR-363-3p were downregulated in liver cancer tissues. Cellular assays validated that miR-363-3p exerted tumor suppressing functions, including the inhibition of cell proliferation, survival and migration abilities in two liver cancer cell lines. Bioinformatics prediction and subsequent experiments demonstrated that HMGA2 was a direct target of miR-363-3p. Restoration of the expression of HMGA2 in miR-363-3p mimic-transfected cells reversed the tumor suppressing effects caused by miR-363-3p. Finally, there was a significant negative correlation between the expression levels of HMGA2 and miR-363-3p in liver cancer tissues. miR-363-3p was identified as an important tumor suppressor in liver cancer via targeting HMGA2, which may have potential benefits in liver cancer therapy.
Keywords: liver cancer, microRNA-363-3p, tumor suppressor, hepatocarcinogenesis, high mobility group AT-hook 2
Introduction
Liver cancer is one of the most common types of human cancer and the third most frequent cause of cancer-associated mortality worldwide (1). Currently, the primary method for treating liver cancer is surgical resection. Although important progress has been made in liver cancer treatment via surgery, chemotherapy and biological agents, a cure is not achieved in the majority of patients, particularly for those with metastasis (2). Liver cancer is associated with a poor prognosis due to the high frequency of intrahepatic and extrahepatic metastasis (3). Hepatocarcinogenesis is a multi-step process beginning with specific genetic alterations, which ultimately cause the malignant transformation of hepatocytes (4). However, the mechanisms underlying hepatocarcinogenesis remain to be fully elucidated. Addressing the pathological mechanisms underlying the development and progression of liver cancer is of critical importance for developing effective therapeutic agents to treat this disease.
MicroRNAs (miRNAs) are defined as a class of small (~20 nucleotides in length), endogenous, non-coding RNAs. They have been investigated substantially in the last two decades and are known to exert crucial regulatory roles by targeting mRNAs for cleavage or translational repression (5). Emerging evidence has revealed that miRNAs are involved in almost all cellular processes and human diseases, particularly in cancer (6–11). miRNAs can be characterized as either oncogenes or tumor suppressors, and the same miRNA can have opposing functions in different types of cancer (12–14). Among these, miRNA-363-3p (miR-363-3p) functions as a tumor suppressor in various types of cancer, including liver cancer (15–17). For example, miR-363-3p inhibits tumor growth, epithelial-to-mesenchymal transition and metastasis in lung and colorectal cancer (18,19). One study demonstrated that miR-363-3p was downregulated in liver cancer, and suppressed cell proliferation, migration and invasion by targeting specificity protein 1 (SP1) (17). However, bioinformatics analyses suggest that one miRNA can simultaneously target several protein-coding mRNAs (5). Identifying more targets for miR-363-3p increases the understanding of the role of miR-363-3p in liver cancer.
The present study focused on the functions of miR-363-3p in liver cancer. The expression, functions and mechanisms of miR-363-3p in liver cancer were characterized. The results revealed that miR-363-3p can reduce the proliferation, survival and migration of liver cancer cells by regulating the expression of high mobility group AT-hook 2 (HMGA2). Therefore, miR-363-3p acts as a tumor suppressor in liver cancer by targeting HMGA2. The findings provide valuable insight into the molecular mechanisms of hepatocarcinogenesis, and highlight the potential of targeting the miR-363-3p/HMGA2 axis in liver cancer therapy in the future.
Materials and methods
Tissues and patients
Tissue samples were obtained between January 2016 and December 2017 at Tianjin Medical University General Hospital Airport Site (Tianjin, China). A summary of patients and the relevant clinical characteristic is in Table I. Informed consent was obtained from all 50 patients in advance. The tissues samples were obtained during surgery and immediately frozen in liquid nitrogen and stored at −80°C prior to use.
Cell culture
Human liver cancer cell lines (HepG2 and Huh-7) were provided by the Cell Bank of Chinese Academy of Sciences (Shanghai, China). All cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco) with 10% fetal bovine serum (FBS; both Thermo Fisher Scientific, Inc., Waltham, MA, USA) at 37°C in a humidified atmosphere with 5% CO2.
miRNA mimics, small interfering RNAs (siRNAs) and transfection
The miR-363-3p mimics and control mimics were purchased from Shanghai GenePharma Co., Ltd. (Shanghai, China), and transfected into Huh-7 or HepG2 cells using Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. The HMGA2 siRNAs and negative control siRNA were also designed and purchased from Shanghai GenePharma Co., Ltd. The sequences were: Negative control siRNA: 5′-UUCUCCGAACGUGUCACGUTT-3′; HMGA2 siRNA 1#: 5′-GGCAUUCAUAUAGGAAGAACGTT-3′; HMGA2 siRNA 2#: 5′-GCUGCUAUACACAAGCAAUGCTT-3′. These were used to transfect Huh-7 or HepG2 cells using Lipofectamine® 2000 according to the manufacturer's protocol.
Reverse transcription-quantitative-polymerase chain reaction (RT-qPCR) analysis
Total RNA from the liver cancer tissues, and cells with or without the indicated treatments was extracted using TRIzol reagent (Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. RT-qPCR using the SYBR-Green Realtime PCR Master mix (Toyobo Life Science, Osaka, Japan) was performed to quantify the levels of miR-363-3p in the liver cancer tissues and cells transfected with miRNA mimics, under the following conditions: 95°C for 30 sec, followed by 40 cycles at 95°C for 15 sec, and 60°C for 30 sec, and 72°C for 30 sec. For qPCR system, 2 µl diluted RT products were combined with 10 µl of 2X mix, 1 µl forward and reverse primers (10 µM) and 6 µl ddH2O in a final volume of 20 µl following the manufacturer's protocols. U6 was used as the internal control. RT-qPCR analysis was also performed to quantify the levels of HMGA2 in the liver cancer tissues, cells transfected with miRNA mimics or HMGA2 siRNAs alone, and cells co-transfected with miRNA mimics plus HMGA2 plasmid. β-actin was used as the internal control. The fold changes of miRNA and mRNA levels were calculated using the 2−ΔΔCq method (20). The primers for qPCR were as follows: miR-363-3p forward (F), 5′-CGGCGAATTGCACGGTATCCA-3′; U6 forward (F), 5′-CGCAAGGATGACACGCAAATTCG-3′; miR-363-3p/U6 reverse (R), 5′-CAGTGCAGGGTCCGAGGT-3′; HMGA2 F, 5′-AAAGCAGAAGCCACTGGAGAAA-3′; HMGA2 R, 5′-TTCCTCCTGAGCAGGCTTCTT-3′; β-actin F, 5′-GATCATTGCTCCTCCTGAGC-3′; β-actin R, 5′-ACTCCTGCTTGCTGATCCAC-3′.
Colony formation assay
The Huh-7 or HepG2 cells transfected with miRNA mimics were resuspended in DMEM with 10% FBS, seeded in 6-well plates (5×103 cells per well) and incubated for 7 days. The colonies were fixed with 4% formaldehyde solution and stained with 0.25% crystal violet in PBS for 30 min. Images of the colonies were captured using a digital camera (Canon PowerShot A2000 IS, Canon, Inc., Tokyo, Japan). The experiments were repeated in triplicate.
MTT assay
The Huh-7 or HepG2 cells transfected with miRNA mimics or siRNAs alone, or co-transfected with miRNA mimic and HMGA2 plasmid were seeded into 96-well plates at a density of 2.5×103 cells per well (six replicates). Cell viability was assessed using an MTT assay (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). Following incubation for 1, 2, 3 and 4 days, 10 µl MTT (0.5 mg/ml) was added into each well for an additional 3 h. Subsequently, 250 µl dimethyl sulfoxide was added into each well and the absorbance at 490 nm was measured using a microplate reader (Molecular Devices, LLC, Sunnyvale, CA, USA).
Wound-healing assay
For the wound-healing experiments, the Huh-7 or HepG2 cells transfected with miRNA mimics or siRNAs alone, or co-transfected with miRNA mimic plus HMGA2 plasmid were seeded into 6-well plates and cultured to confluency. Following transfection, the cells were serum-starved and scraped using a P200 tip (0 h). Images were captured using an inverted optical microscope 48 h later.
Dual luciferase reporter assay
The Huh-7 or HepG2 cells were seeded in 24-well plates and co-transfected with luciferase reporters plus miRNA mimics. The transfection efficiency was normalized using a Renilla luciferase reporter (pRL-CMV; Promega Corporation, Madison, WI, USA). The reporter activity was measured using a Luciferase Assay Detection kit (Promega Corporation) according to the manufacturer's protocol.
Western blot analysis
Protein samples from the Huh-7 or HepG2 cells transfected with miRNA mimics were extracted using radioimmunoprecipitation assay reagent supplemented with protease inhibitors (both Beyotime Institute of Biotechnology, Haimen, China). The protein was quantified by using BCA Protein Assay kit (Beyotime Institute of Biotechnology). Denatured protein (40 µg) was separated on 8% SDS-PAGE gels and transferred onto nitrocellulose membranes (EMD Millipore, Billerica, MA, USA). The membranes were blocked with a buffer containing 5% skimmed milk in PBS with 0.05% Tween-20 for 1 h at room temperature. The nitrocellulose membranes were incubated with primary antibody (anti-HMGA2, 1:1,000, 7777; and anti-β-actin, 1:6,000, 4970; both Cell Signaling Technology, Inc., Danvers, MA, USA) overnight at 4°C. Then the membranes were incubated with peroxidase-conjugated secondary antibodies (1:3,000; A0208; Beyotime Institute of Biotechnology). Subsequently, the membranes were incubated with an enhanced chemiluminescence detection system (EMD Millipore).
Target Prediction
Targets of miR-363-3p were searched on TargetScan Release 3.1 (http://www.targetscan.org/mamm_31/) and the results suggested that HMGA2 was a potential target of miR-363-3p. To further confirm that HMGA2 is directly targeted by miR-363-3p, more information about the 3′UTR of HMGA2 mRNAs was obtained on TargetScan.
Statistical analysis
Statistical analysis was performed using GraphPad Prism v6.0 (GraphPad Software, Inc., La Jolla, CA, USA). Student's two-tailed t-test was used to evaluate the significance of the differences between two groups. The correlation between the expression of miR-363-3p and HMGA2 in tissues was calculated by Spearman's rank correlation analysis. Data are presented as the mean ± standard deviation from three independent experiments. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-363-3p is downregulated in liver cancer and is associated with tumor grade
The RT-qPCR analysis demonstrated that miR-363-3p was downregulated in the 50 liver cancer samples compared with the paired adjacent non-cancerous liver tissue samples (Fig. 1A). Specifically, downregulation of miR-363-3p (≤2-fold change) was observed in 50% (25/50) of all the examined liver cancer samples (Fig. 1B). The expression of miR-363-3p in liver cancer tissues was also analyzed with samples grouped by different tumor grades. Lower levels of miR-363-3p were expressed in tumors with higher grades (Fig. 1C), indicating the potential use of miR-363-3p in liver cancer diagnosis. These results suggest a vital tumor-suppressing role for miR-363-3p in liver cancer.
Figure 1.
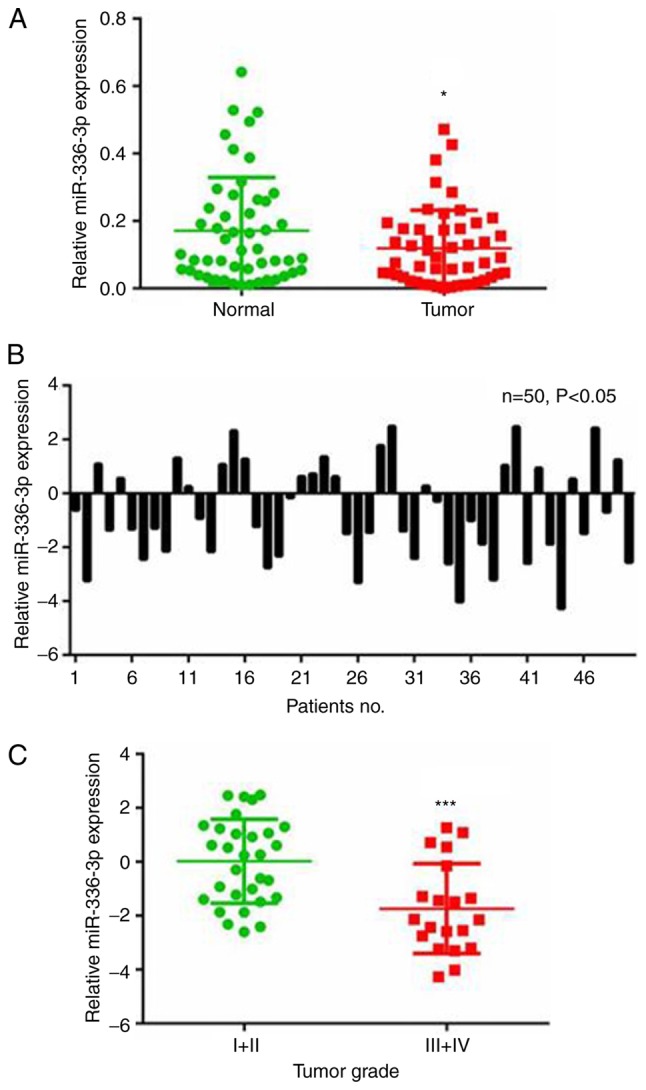
Levels of miR-363-3p are downregulated in liver cancer tissues. (A) Expression of miR-363-3p was assessed by RT-qPCR in 50 paired liver cancer (Tumor) and adjacent non-cancerous tissues (Normal). (B) RT-qPCR analysis of the expression of miR-363-3p in 50 paired liver cancer tissues normalized to that in matched non-cancerous tissues. (C) miR-363-3p was decreased in liver cancer tissues with a higher tumor grade (Grade III+IV, vs. Grade I+II). *P<0.05; ***P<0.001. miR, microRNA; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
miR-363-3p inhibits hepatocarcinogenesis in Huh-7 and HepG2 cells
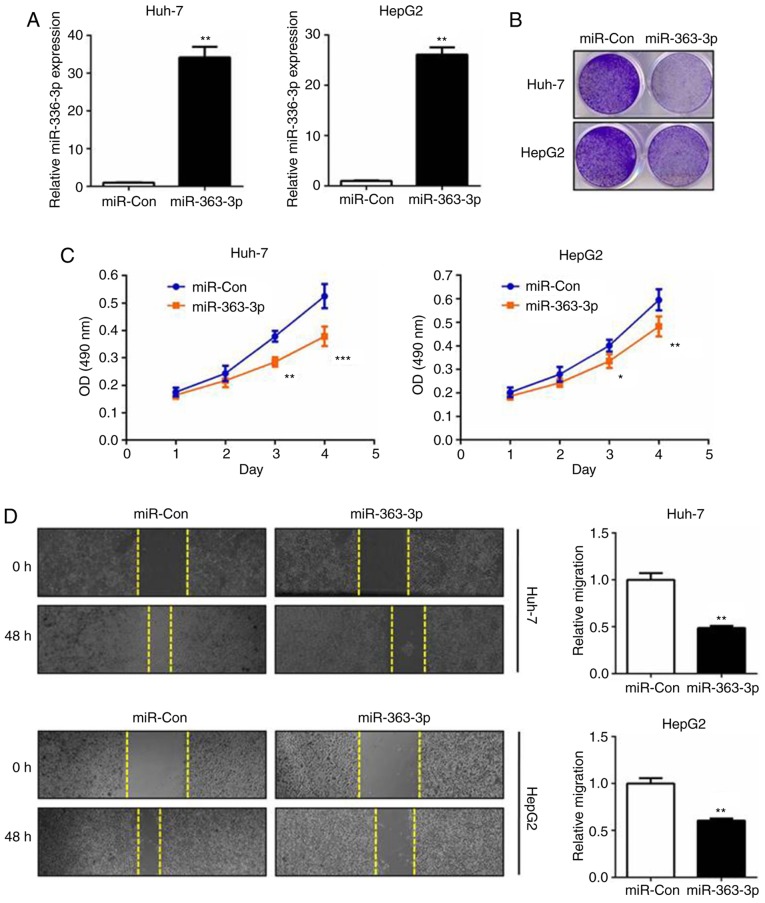
Prompted by the expression results, the role of miR-363-3p in the cell proliferation and survival of liver cancer was subsequently investigated. miR-363-3p mimics were transfected into cells and RT-qPCR analysis was performed (Fig. 2A). The MTT assay demonstrated that miR-363-3p effectively inhibited cell proliferation, and the colony formation assay validated that miR-363-3p significantly impaired clonogenic survival in the two liver cancer cell lines (Fig. 2B and C). In the wound-healing experiments, miR-363-3p exerted potent anti-migration effects on the liver cancer cells (Fig. 2D). Collectively, these results validated that the tumor-suppressing effect of miR-363-3p in liver cancer was mediated by inhibition of the proliferation, survival and migration of liver cancer cells in vitro.
Figure 2.
miR-363-3p inhibits hepatocarcinogenesis in liver cancer cells. (A) miR-363-3p mimics or miR-Con were transfected into Huh-7 and HepG2 cells. After 48 h, cells were harvested for reverse transcription-quantitative polymerase chain reaction analysis. (B) Colony formation assay in the miR-363-3p- and miR-Con-transfected Huh-7 and HepG2 cells. (C) MTT assay in the miR-363-3p- and miR-Con-transfected Huh-7 and HepG2 cells. (D) Wound-healing assay in miR-363-3p- and miR-Con-transfected Huh-7 and HepG2 cells. Magnification, ×40. *P<0.05; **P<0.01; ***P<0.001. miR, microRNA; miR-Con, control mimics.
HMGA2 is a direct target of miR-363-3p in liver cancer
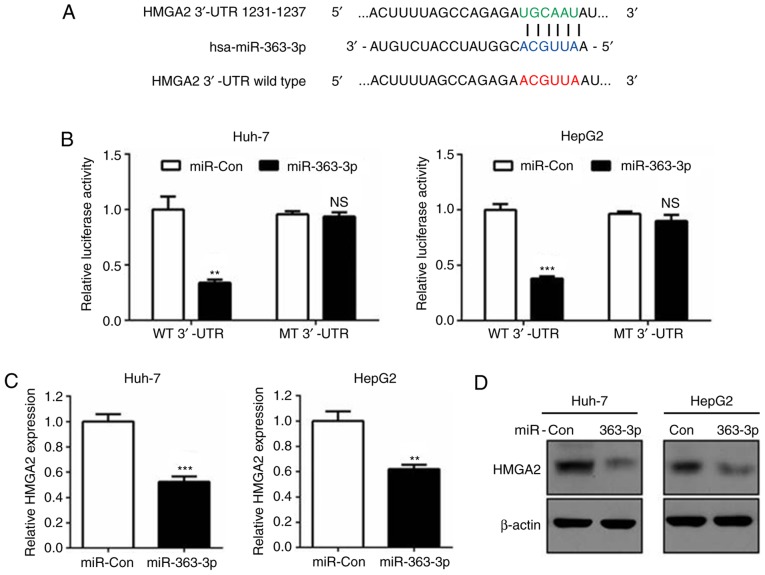
Bioinformatics prediction revealed that the 3′untranslated region (3′UTR) of the HMGA2 gene contained a putative miR-363-3p target sequence (Fig. 3A). Dual luciferase reporter assays further confirmed this prediction and demonstrated that HMGA2 was a direct target of miR-363-3p, as reporters with deletions in the miR-363-3p target bases of the 3′-UTR eliminated the silencing of HMGA2 mRNA transcripts (Fig. 3A and B). Additionally, RT-qPCR and western blot analysis verified that miR-363-3p effectively reduced the mRNA and protein levels of HMGA2 compared with those in the control group (Fig. 3C and D). Therefore, the findings demonstrated that miR-363-3p directly targeted the HMGA2 3′-UTR and induced its mRNA degradation in liver cancer cells.
Figure 3.
HMGA2 is a direct target of miR-363-3p in liver cancer. (A) Putative miR-363-3p binding sites in the HMGA2 3′-UTR. WT binding sequences are highlighted in green. MT binding sequences are highlighted in red. (B) Firefly luciferase activity normalized to Renilla luciferase activity in Huh-7 and HepG2 cells co-transfected with luciferase reporters of WT or MT 3′-UTR of HMGA2 plus miR-363-3p mimics or miR-Con. (C) Expression levels of HMGA2 were detected by reverse transcription-quantitative polymerase chain reaction analysis following transfection with miR-363-3p mimics or miR-Con in Huh-7 and HepG2 cells. (D) Expression levels of HMGA2 were detected by western blotting following transfection with miR-363-3p mimics or miR-Con in Huh-7 and HepG2 cells. **P<0.01; ***P<0.001. miR, microRNA; HMGA2, high mobility group AT-hook 2; 3′-UTR, 3′-untranslated region; WT, wild-type; MT, mutant; miR-Con, control mimics; NS, not significant.
miR-363-3p inhibits cell proliferation and migration by targeting HMGA2
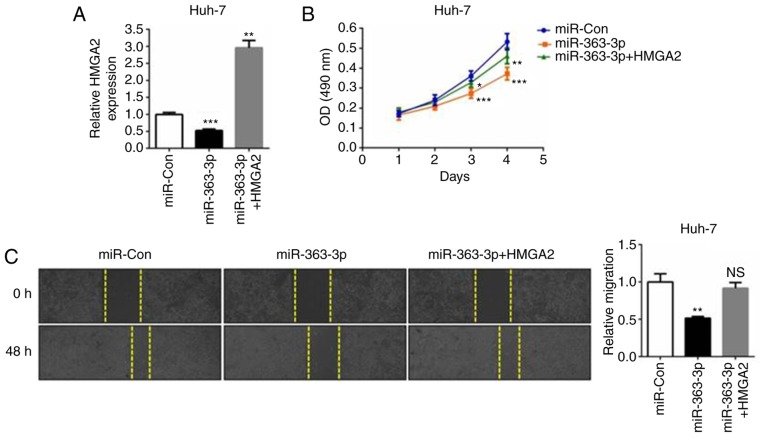
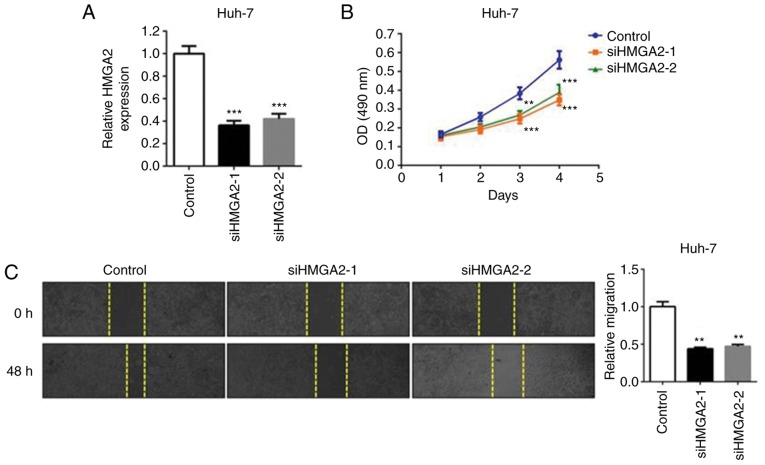
HMGA2 is known to promote proliferation and metastasis in liver cancer and other types of cancer (21,22). To better understand the association between HMGA2 and the tumor-suppressing role of miR-363-3p in liver cancer cells, miR-363-3p mimics and the HMGA2 plasmid were co-transfected into Huh-7 cells (Fig. 4A). The MTT assay revealed that liver cancer cell proliferation was suppressed in the miR-363-3p-overexpressing cells, whereas the ectopic expression of HMGA2 abrogated this inhibitory effect (Fig. 4B). The same effect was observed in the cell migration assay; that is, the overexpression of HMGA2 reversed the anti-migration effects caused by miR-363-3p mimics (Fig. 4C). Additionally, the transient knockdown of endogenous HMGA2 (si-HMGA2-1 or si-HMGA2-2; Fig. 5A) simulated the suppressive effect of miR-363-3p mimics on the cell proliferation and migration of liver cancer cells (Fig. 5B and C). Therefore, the findings demonstrated that HMGA2 is a functional target of miR-363-3p in liver cancer cells, and the overexpression of HMGA effectively enhances cell proliferation and migration.
Figure 4.
miR-363-3p inhibits liver cancer cell proliferation and migration by targeting HMGA2. (A) miR-Con or miR-363-3p mimics, with or without the HMGA2-overexpressing plasmid were co-transfected into Huh-7 cells. After 48 h, cells were harvested for reverse transcription-quantitative polymerase chain reaction analysis. (B) MTT assay of Huh-7 cells transfected as above. (C) Wound-healing assay of Huh-7 cells transfected as above. Magnification, ×40. *P<0.05; **P<0.01; ***P<0.001. miR, microRNA; HMGA2, high mobility group AT-hook 2; miR-Con, control mimics; NS, not significant.
Figure 5.
Knockdown of endogenous HMGA2 suppresses cell proliferation and migration in liver cancer cells. (A) HMGA2 siRNAs (siHMGA2-1, siHMGA2-2) or negative control siRNA were transfected into Huh-7 cells. After 48 h, cells were harvested for reverse transcription-quantitative polymerase chain reaction analysis. (B) MTT assay of Huh-7 cells transfected as above. (C) Wound-healing assay of Huh-7 cells transfected as above. **P<0.01; ***P<0.001. miR, microRNA; HMGA2, high mobility group AT-hook 2; siRNA, small interfering RNA; NS, not significant.
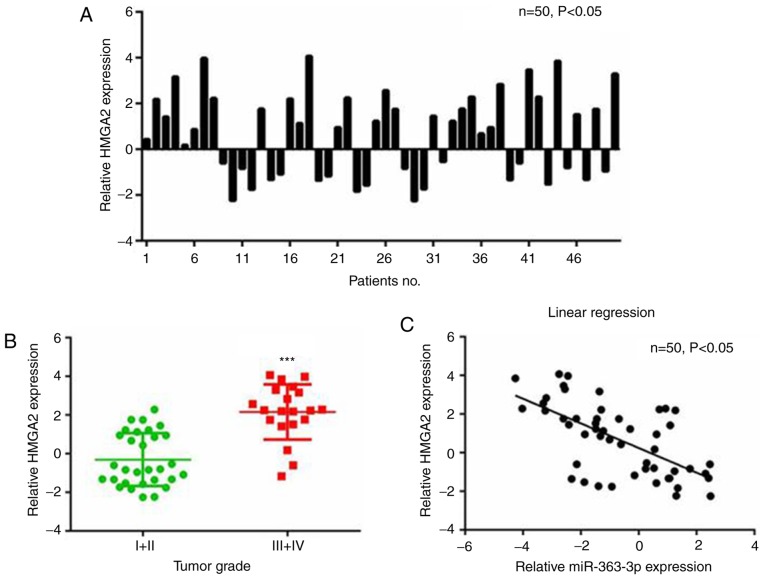
Upregulated HMGA2 is correlated with downregulated miR-363-3p in patients with liver cancer
The correlation between expression levels of HMGA2 and miR-363-3p in liver cancer tissues was analyzed. The RT-qPCR results revealed that the mRNA levels of HMGA2 were higher in liver cancer than in paired non-cancerous tissues (Fig. 6A), and continued to rise with increasing tumor grade (Fig. 6B). Furthermore, the expression of HMGA2 in liver cancer tissues was negatively correlated with the expression of miR-363-3p (Fig. 6C). Therefore, there was a significant negative correlation between the expression of HMGA2 and miR-363-3p in liver cancer tissues.
Figure 6.
HMGA2 is negatively correlated with miR-363-3p in liver cancer tissues. (A) Reverse transcription-quantitative polymerase chain reaction analysis of the expression of HMGA2 in 50 paired liver cancer tissues normalized to that in matched non-cancerous tissues. (B) HMGA2 was increased in liver cancer tissues with higher tumor grade (Grade III+IV, vs. Grade I+II). (C) Linear correlation between the expression of miR-363-3p and HMGA2 in paired liver cancer tissues (n=50). Statistical analysis was performed using Pearson's correlation analysis. ***P<0.001. HMGA2, high mobility group AT-hook 2; miR, microRNA.
Discussion
In the present study, miR-363-3p was revealed to be downregulated in liver cancer tissues. As miR-363-3p is decreased in other types of cancer, including lung and colorectal cancer (18,19), this finding of its downregulation in liver cancer reinforces the evidence for its important potential tumor suppressor role. It has been previously reported that miR-363-3p was decreased in liver cancer and suppressed cell proliferation, migration and invasion by targeting SP1 (17). However, the identification of HMGA2 as another target for miR-363-3p in the present study increases current understanding of the role of miR-363-3p in liver cancer. miR-363-3p was demonstrated to have tumor suppressor activity via the direct targeting of HMGA2, resulting in inhibition of cell proliferation, survival and migration in two liver cancer cell lines, which suggests a novel mechanism of hepatocarcinogenesis.
Regarding the transcriptional control of miR-363-3p in liver cancer, Han et al (21) reported that c-Myc represses the transcription of miR-363-3p by binding to the conserved region of the miR-363-3p promoter. As c-Myc is pathologically activated in liver cancer, the downregulation of miR-363-3p may be largely due to the transcriptional repression caused by c-Myc.
Bioinformatics prediction and in vitro experiments demonstrated that HMGA2 is a direct target of miR-363-3p. HMGA2 is known to promote cancer cell proliferation and metastasis in liver cancer and other cancer types (22,23). HMGA proteins function as architectural factors required for chromosome structure and are essential components of the enhanceosome. The literature indicates that the overexpression of HMGA in tumors is associated with a highly malignant phenotype, resulting in poor patient prognosis (24). The findings of the present study confirmed the overexpression of HMGA2 in liver cancer, which was increased in parallel with the increasing tumor grade. A significant negative correlation between the expression levels of HMGA2 and miR-363-3p in liver cancer tissues was also established, which provided important insight into the association between HMGA2 and miR-363-3p.
Functionally, restoring HMGA2 in miR-363-3p-overexpressing cells significantly reversed the tumor-suppressing effects caused by miR-363-3p mimics. Notably, although the overexpression of HMGA2 was marked in the co-transfection group (miR-363-3p + HMGA2), growth and migration were not elevated in this group compared with the control group. This may be explained by the functions of other miR-363-3p target genes; miR-363-3p also targets SP1 in liver cancer cells (17). However, the results of the present study adequately demonstrated that HMGA2 is an important target of miR-363-3p in liver cancer cells. Additionally, the knockdown of endogenous HMGA2 simulated the suppressive roles of miR-363-3p on the proliferation and migration of liver cancer cells. These findings were in accordance with the conclusion from a previous study that HMGA2 is a potential oncogene and tumor biomarker in human cancer (24); however, the detailed mechanisms remain to be fully elucidated. Structurally, HMGA2 protein contains DNA-binding domains and may act as a transcriptional regulatory factor. High-throughput sequencing methods, including RNA-Seq and chromatin immunoprecipitation-Seq, may provide more information on the downstream gene regulation networks that HMGA2 is involved in.
In conclusion, the findings of the present study revealed that miR-363-3p inhibited liver cancer cell proliferation, survival and migration by directly targeting HMGA2. Abnormal decreases in miR-363-3p and increases of its target, HMGA2, are important factors that may be involved in hepatocarcinogenesis and liver cancer progression. This novel miR-363-3p/HMGA2 axis provides insight into the mechanisms of liver cancer onset and may facilitate drug development against liver cancer. Additionally, miR-363-3p and HMGA2 may be effective biomarkers used for liver cancer grading and prognosis prediction.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
JW and HL designed the study; HG and XG performed data collection; DG performed the data analysis and YY collected the clinical data, prepared the manuscript and approved the publication of the final version.
Ethics approval and consent to participate
This study was approved by the Medical Ethics Committee of Tianjin Medical University General Hospital Airport Site.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet. 2012;379:1245–55. doi: 10.1016/S0140-6736(11)61347-0. [DOI] [PubMed] [Google Scholar]
- 2.Blum HE. Hepatocellular carcinoma: Therapy and prevention. World J Gastroenterol. 2005;11:7391–7400. doi: 10.3748/wjg.v11.i47.7391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.El-Serag HB, Rudolph KL. Hepatocellular carcinoma: Epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–2576. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 4.Thorgeirsson SS, Grisham JW. Molecular pathogenesis of human hepatocellular carcinoma. Nat Genet. 2002;31:339–346. doi: 10.1038/ng0802-339. [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 6.He L, Hannon GJ. MicroRNAs: Small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 7.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 8.Rapado-González Ó, Majem B, Muinelo-Romay L, Álvarez-Castro A, Santamaría A, Gil-Moreno A, López-López R, Suárez-Cunqueiro MM. Human salivary microRNAs in Cancer. J Cancer. 2018;9:638–649. doi: 10.7150/jca.21180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hosseinahli N, Aghapour M, Duijf PHG, Baradaran B. Treating cancer with microRNA replacement therapy: A literature review. J Cell Physiol. 2018;233:5574–5588. doi: 10.1002/jcp.26514. [DOI] [PubMed] [Google Scholar]
- 10.Ma J, Li X. MicroRNAs are involved in the toxicity of microcystins. Toxin Reviews. 2017;36:165–175. doi: 10.1080/15569543.2016.1258713. [DOI] [Google Scholar]
- 11.Ma J, Li Y, Yao L, Li X. Analysis of microRNA expression profiling involved in MC-LR-induced cytotoxicity by high-throughput sequencing. Toxins (Basel) 2017;9(pii):E23. doi: 10.3390/toxins9010023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chakraborty C, Sharma AR, Sharma G, Sarkar BK, Lee SS. The novel strategies for next-generation cancer treatment: miRNA combined with chemotherapeutic agents for the treatment of cancer. Oncotarget. 2018;9:10164–10174. doi: 10.18632/oncotarget.24309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vannini I, Fanini F, Fabbri M. Emerging roles of microRNAs in cancer. Curr Opin Genet Dev. 2018;48:128–133. doi: 10.1016/j.gde.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lou W, Liu J, Gao Y, Zhong G, Chen D, Shen J, Bao C, Xu L, Pan J, Cheng J, et al. MicroRNAs in cancer metastasis and angiogenesis. Oncotarget. 2017;8:115787–115802. doi: 10.18632/oncotarget.23115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin Y, Xu T, Zhou S, Cui M. MicroRNA-363 inhibits ovarian cancer progression by inhibiting NOB1. Oncotarget. 2017;8:101649–101658. doi: 10.18632/oncotarget.21417. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Song B, Yan J, Liu C, Zhou H, Zheng Y. Tumor Suppressor Role of miR-363-3p in Gastric Cancer. Med Sci Monit. 2015;21:4074–4080. doi: 10.12659/MSM.896556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ying J, Yu X, Ma C, Zhang Y, Dong J. MicroRNA-363-3p is downregulated in hepatocellular carcinoma and inhibits tumorigenesis by directly targeting specificity protein 1. Mol Med Rep. 2017;16:1603–1611. doi: 10.3892/mmr.2017.6759. [DOI] [PubMed] [Google Scholar]
- 18.Wang Y, Chen T, Huang H, Jiang Y, Yang L, Lin Z, He H, Liu T, Wu B, Chen J, et al. miR-363-3p inhibits tumor growth by targeting PCNA in lung adenocarcinoma. Oncotarget. 2017;8:20133–20144. doi: 10.18632/oncotarget.15448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu F, Min J, Cao X, Liu L, Ge Z, Hu J, Li X. MiR-363-3p inhibits the epithelial-to-mesenchymal transition and suppresses metastasis in colorectal cancer by targeting Sox4. Biochem Biophys Res Commun. 2016;474:35–42. doi: 10.1016/j.bbrc.2016.04.055. [DOI] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Han H, Sun D, Li W, Shen H, Zhu Y, Li C, Chen Y, Lu L, Li W, Zhang J, et al. A c-Myc-MicroRNA functional feedback loop affects hepatocarcinogenesis. Hepatology. 2013;57:2378–2389. doi: 10.1002/hep.26302. [DOI] [PubMed] [Google Scholar]
- 22.Gao X, Dai M, Li Q, Wang Z, Lu Y, Song Z. HMGA2 regulates lung cancer proliferation and metastasis. Thorac Cancer. 2017;8:501–510. doi: 10.1111/1759-7714.12476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang W, Li J, Guo X, Zhao Y, Yuan X. miR-663a inhibits hepatocellular carcinoma cell proliferation and invasion by targeting HMGA2. Biomed Pharmacother. 2016;81:431–438. doi: 10.1016/j.biopha.2016.04.034. [DOI] [PubMed] [Google Scholar]
- 24.Pallante P, Sepe R, Puca F, Fusco A. High mobility group a proteins as tumor markers. Front Med (Lausanne) 2015;2:15. doi: 10.3389/fmed.2015.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.