Abstract
Ribosomal protein S15A (RPS15A), a member of the ribosomal protein gene family, was demonstrated to be closely associated with tumorigenesis in multiple human malignancies. Nevertheless, the role of RPS15A in the progression of renal cell carcinoma (RCC) remains unknown. In the present study, by comparing the publicly available data from RCC tissues and reverse transcription-quantitative polymerase chain reaction results, it was identified that RPS15A was upregulated in RCC tissues and cell lines (P<0.001). Notably, knockdown of RPS15A suppressed 786-O cell proliferation (P<0.001) and promoted its apoptosis/necrotic (P=0.0001) in vitro. Additionally, tumour formation and growth of transfected 786-O cells were observed to be restrained in a mouse model (P<0.05). Subsequent to analysing the microarray data, 747 genes were differentially expressed in the RPS15A-knockdown 786-O cells. The enriched canonical pathways, diseases and functions of differentially expressed genes, and the interactive network of RPS15A in RCC were successfully constructed by ingenuity pathway analysis. Overall, the present results provided a preliminary experimental basis for RPS15A as a novel oncogene and potential therapeutic target in RCC.
Keywords: renal cell carcinoma, ribosomal protein S15A, tumour progression, interactive network, apoptosis
Introduction
Ribosomal proteins (RPs) are a family of RNA-binding proteins that have primary roles in ribosome biogenesis and protein translation (1). Previous data suggested a close association of the extraribosomal functions of RPs with cell growth and proliferation (2,3), apoptosis (4,5), DNA repair (6), cellular development (7) and differentiation (8,9). Ribosomal protein S15A (RPS15A), a member of the RP gene family, was highly expressed in hepatic cancer (10), glioblastoma (11,12), non-small cell lung (13) and colorectal cancer (14).
In colorectal cancer, RPS15A depletion contributed to cell cycle arrest and cell growth suppression via p21 upregulation and cyclin-dependent kinase (CDK)1 downregulation (14). The p53 pathway, well documented for its anticancer functions (15), was significantly activated in RPS15A-specific short hairpin RNA (shRNA)-expressing lentivirus (Lv)-shRPS15A-infected A549 cells (16). In another previous study, the knockdown of RPS15A induced lung cancer cell apoptosis (13). RPS15A may additionally contribute to glioblastoma growth, proliferation and migration via the protein kinase B pathway, and the knockdown of RPS15A may significantly inhibit B cell lymphoma 2 and activate caspase-3 and poly (ADP-ribose) polymerase (11,12). Overall, the majority of the previous studies indicated that the dysfunction of RPS15A is significantly associated with tumorigenesis in human malignancies. However, the role of RPS15A in the development of renal cell carcinoma (RCC) remains unknown.
RCC was reported to be the seventh most common cancer, with >350,000 people diagnosed worldwide in 2013 (17). Despite numerous advances in the systemic treatment of RCC over the past years, including targeted therapies, the life expectancy of patients remains generally unsatisfactory due to the side effects and tolerance of these drugs. Based on the increasing number of patients with RCC and prevalence of resistance to currently available drugs (18), the identification of novel target pathways remains an active area of RCC (17).
The aim of the present study was to investigate the expression levels and functions of RPS15A in RCC. Through analysing data from multiple patient cohorts, and validating its expression in RCC cell lines, the results revealed that RPS15A was overexpressed in RCC. Additionally, the function of RPS15A in RCC was further examined in vitro and in vivo using RCC cells transfected with Lv-shRPS15A. A regulatory network of RPS15A was also constructed. Thus, the present study provided evidence that RPS15A may serve a potential role in the tumorigenesis of RCC.
Materials and methods
Data collection and analysis
The gene expression data of RPS15A in kidney cancer tissues were collected from The Cancer Genome Atlas (TCGA; cancergenome.nih.gov) and included 507 cancer samples and 72 adjacent normal tissues. Differential analysis was performed and visualized using starBase V2.0 (19). In the validation tests, data from the Oncomine database (www.oncomine.org) were collected (20), with a cut-off value of a fold change >1.5 and P<0.05.
Cell culture
Normal human renal cells (HK-2), 293T and the RCC cell lines 786-O and Caki-1 were obtained from The Shanghai Biological Institute (Shanghai, China). The 786-O, 293T and HK-2 cells were cultured in RPMI 1640 medium with 10% foetal bovine serum, and Caki-1 cells were grown as a monolayer to a subconfluent state in Falcon tissue culture dishes in McCoy's 5A medium (all Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% FBS. All cells were cultured in a 37°C humidified incubator with 95% air and 5% CO2. The cultured cells were washed briefly with PBS, harvested with a rubber policeman, frozen in liquid nitrogen and stored at −80°C until further use.
RNA extraction and reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis
Total RNA was isolated from the cells using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.), and RT of 2.0 µg total RNA was performed using the M-MLV kit (Promega Corporation, Madison, WI, USA), both according to the manufacturer's protocol. The primers used for qPCR were as follows: RPS15A forward, 5′-CTCCAAAGTCATCGTCCGGTT-3′ and reverse, 5′-TGAGTTGCACGTCAAATCTGG-3′; GAPDH forward, 5′-TGACTTCAACAGCGACACCCA-3′ and reverse, 5′-CACCCTGTTGCTGTAGCCAAA-3′. GAPDH was used as an endogenous control. qPCR was performed using SYBR-Green Real-Time PCR Master Mix (Agilent Technologies, Inc., Santa Clara, CA, USA) and measured using a CFX96 Real-Time PCR system (Bio-Rad Laboratories, Inc., Hercules, CA, USA) to quantify RPS15A expression levels. The thermo cycling conditions were as follows: 30 sec at 95°C, followed by 45 cycles of 5 sec at 95°C and 30 sec at 60°C. Following amplification, melting curve analysis was performed to calculate the product melting temperature. Relative gene expression levels were calculated using the 2−ΔΔCq method and normalized to GAPDH (21).
Lentiviral vector construction and cell infection
To knockdown RPS15A expression, an shRNA sequence targeting the human RPS15A gene (shRPS15A; NM_001019) was designed: Sense, 5′-CCGGGTGCAACTCAAAGACCTGGAATTCAAGAGATTCCAGGTCTTTGAGTTGCACTTTTTG-3′, antisense, 3′-CACGTTGAGTTTCTGGACCTTAAGTTCTCTAAGGTCCAGAAACTCAACGTGAAAAACTTAA-5′. A non-targeting shRNA was designed as the control: 5′-GCGGAGGGTTTGAAAGAATATCTCGAGATATTCTTTCAAACCCTCCGCTTTTTT-3′. The stem-loop-stem oligos were subsequently synthesized, annealed and inserted into the linearized vector GV115 (Shanghai GeneChem Co., Ltd., Shanghai, China) to generate the reconstructed vector. Recombinant lentiviral vectors and packaging vectors (1.8×109 TU/ml) were subsequently co-transfected into 293T cells (>1×106 cells/ml) at 37°C for 48–72 h using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol for the generation of recombinant lentiviruses Lv-shRPS15A and negative control Lv-shCtrl. Following centrifugation (50,000 × g; 4°C; 2 h) and purification, recombinant lentiviruses were collected, and the viral titre was counted according to the percentage of green fluorescent protein (GFP)-positive cells, observed under a fluorescence microscope (magnification, ×100). 786-O cells at 30–45% confluency were transfected with the Lv-shRPS15A and Lv-shCtrl (8 µg/ml) to obtain cell lines stably expressing the shRPS15A. At 72 h following transfection, cells were observed under fluorescence microscope to confirm successful establishment and were used in subsequent experiments. The target gene knockdown efficiency in 786-O cells was verified by RT-qPCR, and the expression levels of RPS15A protein was detected by western blot analysis.
Western blot analysis
Lv-shRNA-transduced cells were washed twice with ice-cold PBS and lysed in 2X lysis buffer (100 mM Tris-HCl, pH 6.8; 2% mercaptoethanol; 20% glycerinum; 4% SDS). The lysates were centrifuged at 12,000 × g for 15 min at 4°C, and the supernatant was collected and stored at −80°C prior to use. BCA Protein Quantitation kit used for protein determination. Proteins were loaded (30 µg each well) and separated by 10% SDS-PAGE and transferred onto polyvinylidene membranes (Merck KGaA, Darmstadt, Germany). The membranes were blocked for 1 h at room temperature with 5% non-fat milk. Subsequently, the membranes were incubated with the following primary antibodies overnight at 4°C: Mouse anti-Flag (1:2,000; cat. no. F1804; Sigma-Aldrich; Merck KGaA), rabbit anti-RPS15A (1:1,000; cat. no. AP4804a; Abgent, Inc., San Diego, CA, USA) and mouse anti-GAPDH (1:2,000; cat. no. sc-32233; Santa Cruz Biotechnology, Inc., Dallas, TX, USA). On the second day, the membranes were washed using TBS + 0.1% Tween-20 and incubated with the following horseradish peroxidase-conjugated secondary antibodies: Goat anti-rabbit immunoglobulin (Ig)G (1:2,000; cat. no. sc-2004) and rabbit anti-mouse IgG (1:2,000; cat. no. sc-2005; both Santa Cruz Biotechnology, Inc.) for 2 h at room temperature. Proteins were visualized using the Enhanced Chemiluminescence-PLUS/kit (cat. no. RPN2132; Amersham; GE Healthcare, Chicago, IL, USA) according to the manufacturer's protocol. GAPDH served as the loading control.
Cell proliferation and apoptosis/necrosis detection
A Celigo Fluorescent Scanner (Nexcelom Bioscience, Lawrence, MA, USA) was used to detect the number of lentivirus-transduced 786-O cells expressing GFP for 5 consecutive days, and growth curves of cells and colonies were constructed. For the MTT proliferation assay, lentivirus-transduced 786-O cells were seeded into 96-well plates with 2,000 cells/well. MTT (20 µl; 5 mg/ml; Gen-View Scientific, Inc., El Monte, CA, USA) was used according to the manufacturer's protocol, and DMSO was used to dissolve the purple formazan crystals. The absorbance of each well was measured at 490 nm using a microplate reader, and cell proliferation curves were plotted according to the optical density values.
For the apoptosis/necrosis assays, the collected cells were washed with binding buffer at room temperature and subsequently stained with Annexin V-APC (cat. no. 88-8007; eBioscience; Thermo Fisher Scientific, Inc.) in the dark at room temperature for 15 min prior to flow cytometric analysis. Apoptosis/necrosis was measured using the FACSAria III flow cytometer (BD Biosciences, San Jose, CA, USA).
Xenograft model
For tumorigenesis evaluation in vivo, female BALB/c nude mice (age, 4 weeks; weight, 20–22 g) were purchased from Shanghai Linchang Biotechnology (Shanghai, China). Mice were raised in a specific pathogen-free environment at 25°C, 40–70% humidity with a 12-h light/dark cycle; the mice were allowed to move freely and had free access to food and water. The mice were divided randomly into two groups (n=8/group), including the shRPS15A group and the Lv-shCtrl group. In total, ~2×107 shRPS15A and shCtrl cells were inoculated into the right armpit of mouse in each group. The body weights of the mice and bi-dimensional tumour measurements were taken once each week for 7 weeks, and the tumour size was estimated using the standard formula π/6 × L × W × W; where L represents length and W represents width. The mice were sacrificed following seven measurement periods, and the tumour tissues were measured and weighed. All experimental procedures and animal work were conducted under the principles and procedures by The National Institutes of Health (Bethesda, MA, USA) and approved by the Animal Ethics Committee of West China Hospital (Chengdu, China).
Gene chip and bioinformatic analyses
Total RNA was isolated from shRPS15A- and shCtrl-transduced 786-O as aforementioned. The quality of total RNA was evaluated by its concentration and A260/A280 ratio, and the integrity was evaluated using the Agilent RNA 6000 Nano kit (Affymetrix; Thermo Fisher Scientific, Inc.). Gene expression profiling was performed using the GeneChip® PrimeView™ Human Gene Expression Array (cat. no. 901838; Affymetrix; Thermo Fisher Scientific, Inc.) covering >36,000 transcripts and variants. Subsequent to preparing a total RNA/Poly-A RNA control mixture, 100 ng total RNA was reverse transcribed (2 h at 42°C, followed by 1 h at 16°C and 10 min at 65°C), labelled, purified and fragmented using the GeneChip 3′IVT PLUS kit (Affymetrix; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. Hybridization, washing and staining were performed using the GeneChip Hybridization Wash and Stain kit (Affymetrix; Thermo Fisher Scientific, Inc.) in a GeneChip Hybridization Oven 645 and a GeneChip Fluidics Station 450. The arrays were subsequently scanned using a GeneChip Scanner 3000 7G (Thermo Fisher Scientific, Inc.). The signal histogram, relative signal box plot and Pearson's correlation (signal) methods were used to control the quality of the gene chip analysis. All the data analyses were first performed in R (v3.2.2; www.r-project.org) using Bioconductor (www.bioconductor.org) to verify the differential expression of genes. The probe number and fold change (FC) information (ratio of the expression amounts of the treatment and control groups) of the gene chip was analysed using Ingenuity Pathway Analysis (IPA; Ingenuity Systems; Qiagen, Inc., Valencia, CA, USA). Interaction networks were constructed using the model of network creation algorithm in IPA with a cut-off of |FC| >1.5 and P<0.05.
Statistical analysis
All values are presented as the mean ± standard deviation, and experiments were repeated in triplicate. The data of two groups were compared with an unpaired standard Student's t-test using SPSS 16.0 (SPSS, Inc., Chicago, IL, USA). For comparisons that involved >2 groups, one-way analysis of variance with the Tukey-Kramer post-hoc test was used. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of RPS15A in RCC samples and cell lines
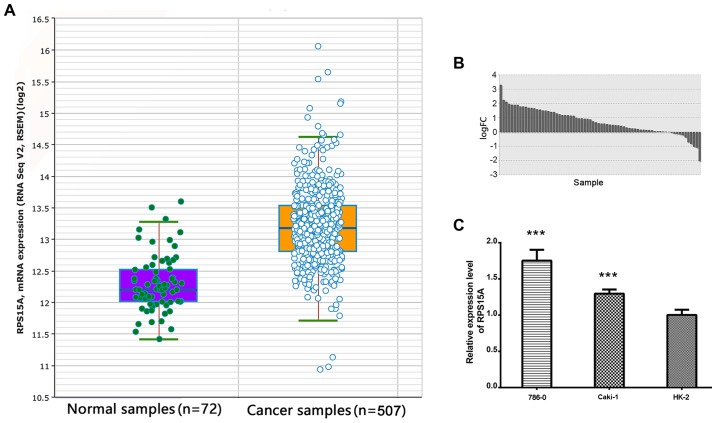
The RNA-seq data from TCGA was analysed, and the expression of RPS15A was identified to be significantly higher in RCC compared with the normal group (FC=1.89; P<0.001; Fig. 1A). In the 72 paired RCC tissues, RPS15A expression was further identified to be higher in RCC compared with the adjacent normal tissues (FC=1.73; P<0.001; Fig. 1B). Subsequently, the overexpression of RPS15A was validated in multiple RCC cohorts from the Oncomine database (P<0.05; Table I). These data suggested that RPS15A was highly expressed in RCC tissues.
Figure 1.
Expression levels of RPS15A in human RCC cells and tissues. (A) RPS15A mRNA was significantly higher in the RCC group (507 samples) compared with the normal group (72 samples); FC=1.89; P<0.001. (B) Expression levels of RPS15A in RCC tissues was higher compared with adjacent normal tissues; FC=1.73; P<0.001. (C) Expression of RPS15A mRNA in RCC cell lines. ***P<0.001. FC, fold change; RCC, renal cell carcinoma; RPS15A, ribosomal protein S15A.
Table I.
Ribosomal protein S15A differential transcript expression in renal cell carcinoma extracted from multiple studies in the Oncomine databases.
| Author, year | Comparison between groups | Fold change | P-value | (Refs.) |
|---|---|---|---|---|
| Jones et al, 2005 | Papillary renal cell carcinoma (n=11) vs. normal (n=23) | 2.123 | 9.33×10−16 | (36) |
| Jones et al, 2005 | Clear cell renal cell carcinoma (n=23) vs. normal (n=23) | 1.840 | 4.93×10−12 | (36) |
| Higgins et al, 2003 | Clear cell renal cell carcinoma (n=24) vs. normal (n=3) | 1.788 | 3.0×10−2 | (37) |
| Yusenko et al, 2009 | Chromophobe renal cell carcinoma (n=4) vs. normal (n=5) | 1.514 | 2.6×10−2 | (38) |
To determine whether the high expression of RPS15A transcripts exists in RCC cell lines, RT-qPCR was performed to detect the RPS15A mRNA expression levels in 786-O, Caki-1 and HK-2 cells. Among them, the 786-O and Caki-1 cell lines highly expressed RPS15A mRNA compared with the HK-2 cells (P<0.001; Fig. 1C).
Lv-shRPS15A-mediated knockdown of RPS15A inhibits the growth and proliferation of 786-O cells
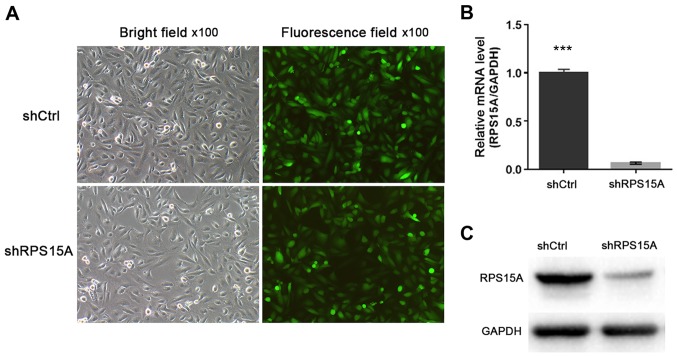
To study the function of RPS15A in RCC, RPS15A expression in 786-O cells was knocked down using stable Lv-shRPS15A. The 786-O cells were infected with Lv-shRPS15A or Lv-shCtrl. Subsequently, the infection efficiency was calculated to be >80% according to the ratio of GFP-expressing cells to total cells under a fluorescence microscope at 72 h post-transduction (Fig. 2A). RT-qPCR and western blot assays were used to examine the targeted gene silencing effects of Lv-shRPS15A in 786-O cells (Fig. 2B and C).
Figure 2.
Efficacy of Lv-shRPS15A in 786-O cells. (A) Green fluorescent protein-expressing cells were observed under a fluorescence microscope at 72 h post-transfection. (B) Expression levels of RPS15A mRNA in the Lv-shRPS15A- and Lv-shCtrl-transduced 786-O cells were analysed by reverse transcription-quantitative polymerase chain reaction. The results are represented as the mean ± standard deviation of three independent experiments; ***P<0.001. (C) Western blotting indicating a notable reduction of RPS15A protein expression in the transfected 786-O cells. Ctrl, control Lv, lentivirus; RPS15A, ribosomal protein S15A; sh, short hairpin RNA.
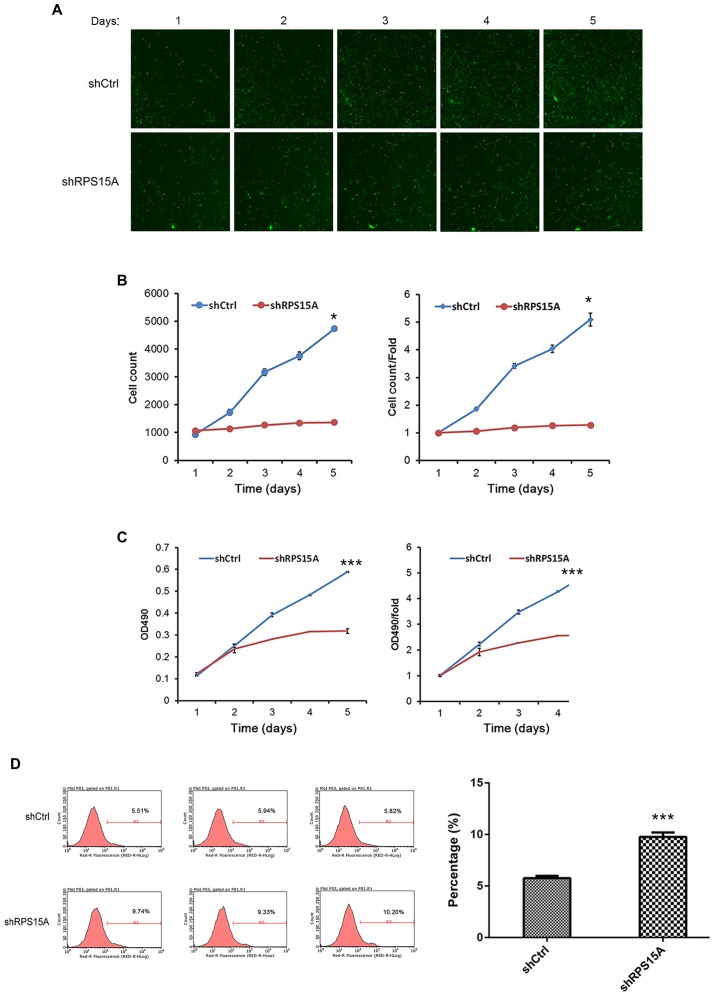
Subsequently, the growth and proliferative abilities of Lv-shRPS15A-infected and Lv-shCtrl-infected 786-O cells were examined by Celigo counting and MTT assay, respectively. From the cell fluorescence images and cell growth curve diagrams, the cell growth rate in the Lv-shRPS15A group was significantly inhibited compared with the Lv-shCtrl group (P<0.05; Fig. 3A and B). Additionally, the MTT assay was performed, and the proliferation of Lv-shRPS15A-infected cells was decreased compared with the Lv-shCtrl-infected cells at the time points of 2, 3, 4 and 5 days (P<0.001; Fig. 3C). The results suggested that the depletion of RPS15A significantly inhibited the growth and proliferative abilities of 786-O cells in vitro.
Figure 3.
Lv-shRNA-mediated knockdown of RPS15A regulates the growth, proliferation and apoptosis/necrosis of 786-O cells. (A) Following 3 days of incubation with Lv-shRNA or Lv-Ctrl, 786-O cell growth was recorded in cell fluorescence; magnification, ×200. (B) Celigo counting and (C) the MTT assay on 5 consecutive days. (D) Apoptotic rates of transfected 786-O cells at day 5. The results demonstrated that the percentage of apoptotic and necrotic cells was significantly higher shRPS15A compared with the shCtrl group; *P<0.05, ***P<0.001. Ctrl, control; Lv, lentivirus; OD, optical density; RPS15A, ribosomal protein S15A; shRNA, short hairpin RNA.
Lv-shRPS15A-mediated knockdown induces the apoptosis and necrosis of 786-O cells
To examine the underlying mechanism of RPS15A in RCC cell apoptosis/necrosis, Annexin V-APC staining was performed, and the percentage of apoptotic/necrotic cells was assessed by flow cytometry. Compared with the Lv-shCtrl group, the proportion of apoptotic/necrotic cells was significantly higher in the Lv-shRPS15A group (9.75±0.44% vs. 5.76±0.22%; P=0.0001; Fig. 3D). This result suggested an association between RPS15A and 786-O cell apoptosis/necrosis; the knockdown of RPS15A may trigger apoptosis in RCC cells; however, this requires further investigation.
Lv-shRPS15A-mediated knockdown of RPS15A inhibits tumour formation and growth in vivo
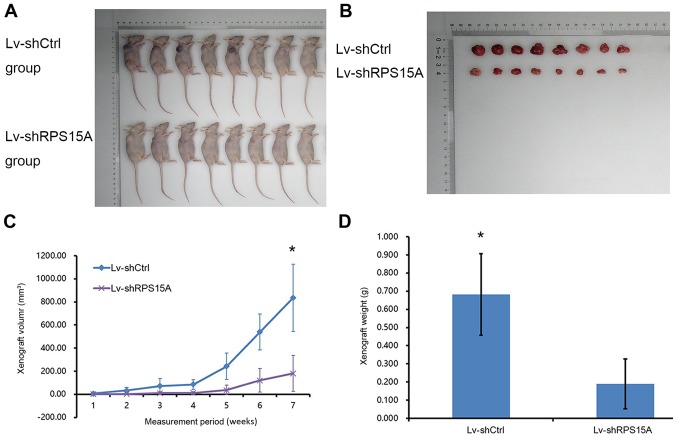
To test the potential role of RPS15A in RCC cell growth in vivo, a nude mouse xenograft model was established (Fig. 4A and B). From the curves of tumour formation, it was observed that the daily xenograft growth volume of the Lv-shRPS15A group was significantly decreased compared with the Lv-shCtrl group (P<0.05; Fig. 4C). Additionally, the average weight of xenografts from Lv-shRPS15A-injected mice was 0.189±0.137 g, which was decreased compared with the control group (0.683±0.225 g) at week 7 post-injection (P<0.05; Fig. 4D). These results demonstrated that the knockdown of RPS15A expression decreased the growth of tumours in size and weight, which suggested an important function of RPS15A in RCC tumour formation in vivo.
Figure 4.
Knockdown of RPS15A inhibits 786-O xenograft formation and growth in vivo. (A) Mice were divided into two groups: Group 1, The Lv-shRPS15A, and Group 2, Lv-shCtrl group. (B) Images of the tumours. (C) Mean estimated tumour volume of the Lv-shRPS15A group was significantly lower compared with the Lv-shCtrl group. (D) The average tumour weight of the xenografts from the Lv-shRPS15A group was significantly reduced compared with the Lv-shCtrl group in the seventh week. Data are presented as the mean ± standard deviation; *P<0.05. Ctrl, control; Lv, lentivirus; RPS15A, ribosomal protein S15A; sh, small hairpin RNA.
Identification of differentially expressed genes in lentiviral-transduced 786-O cells
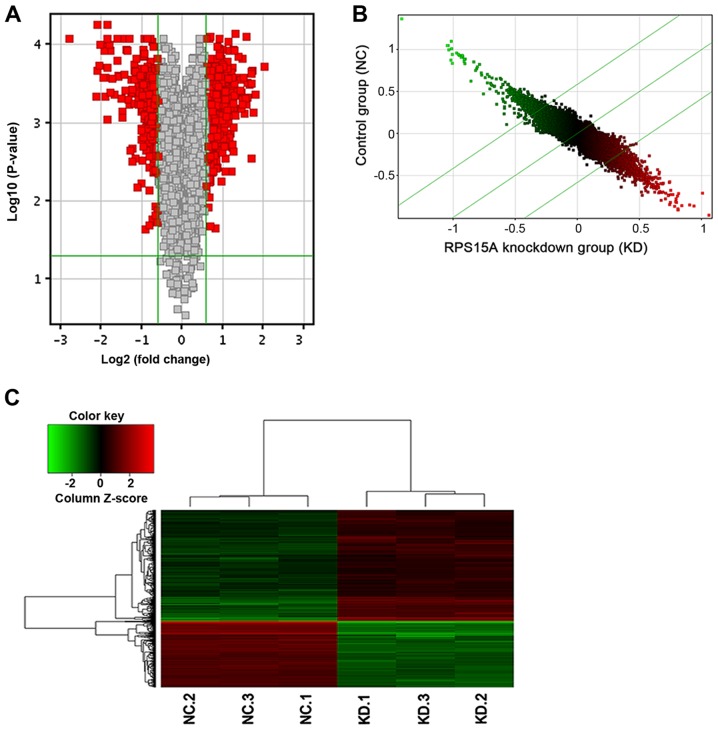
The Affymetrix GeneChip® PrimeView™ Human Gene Expression Arrays were used to determine differences in gene expression levels in total RNA samples between Lv-shRPS15A- and Lv-shCtrl-infected 786-O cells. In total, 747 genes were identified as differentially expressed. Among these, the expression levels of 469 genes were upregulated and 278 genes were downregulated (Fig. 5).
Figure 5.
(A) Volcano plot. The red point in the plot represents the differentially expressed mRNAs with statistical significance. (B) Scatter plot. The plot located in median (Y=X) represents a Fold Change value=1. X axial: KD group; Y axial: NC group. (C) Cluster map; red indicates upregulated and green indicates downregulated. The top cluster tree was generated according to the value of each sample; the left cluster tree was generated according to the value of each gene. RPS15A, ribosomal protein S15A. NC, negative control; KD, knockdown.
Pathway, functional enrichment and interaction network analyses of RPS15A
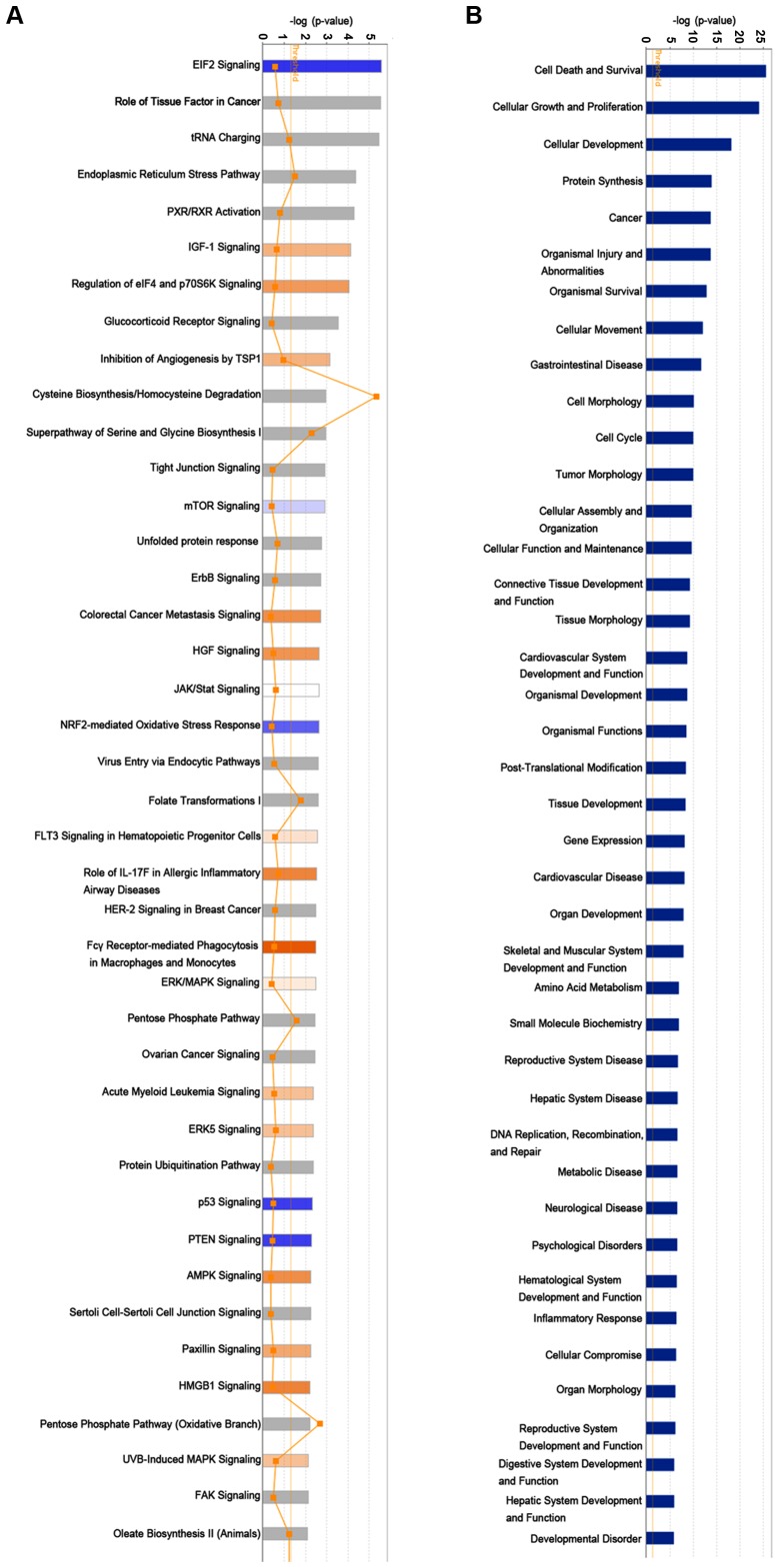
To further examine the associated signatures and functions of RPS15A in RCC, the microarray information was analysed by IPA. The data demonstrate the enrichment condition of differentially expressed genes in canonical signalling pathways (Fig. 6A), and it was identified that subsequent to silencing RPS15A, multiple signalling pathways were enriched, including the ‘Fcγ receptor-mediated phagocytosis in macrophages and monocytes’ and ‘EIF2 signaling’ pathways. Additionally, in the disease and functions enrichment analysis it was identified that the knockdown of RPS15A may induce the alteration of numerous biological functions, such as ‘cell death and survival’, ‘cellular growth and proliferation’ and ‘cellular development’ (Fig. 6B).
Figure 6.
Ingenuity Pathway Analysis. (A) Canonical signalling pathways. The signalling pathways with Z-score >0 were marked in orange; those with Z-score <0 were marked in blue. (B) Disease and function analysis. Ranking according to the value of -log (P-value).
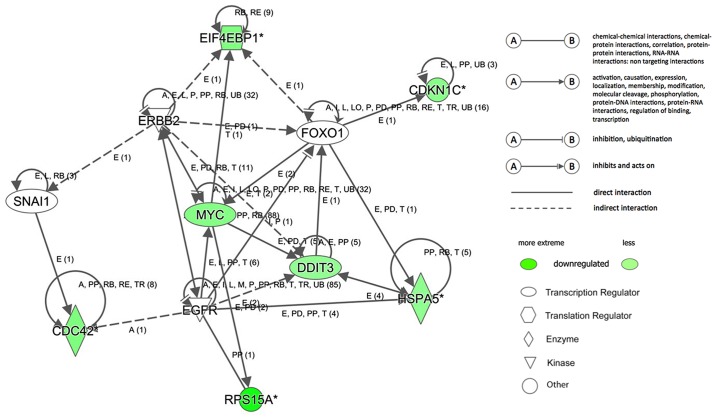
Subsequently, the interaction networks of genes were integrated based on microarray data and previous reports (Fig. 7). The results demonstrated a multiple gene network of RPS15A in RCC. In this network, no upregulated genes were noted. Among the downregulated genes, cell division cycle 42, cyclin dependent kinase inhibitor 1C (CDKN1C), DNA damage inducible transcript 3 (DDIT3), eukaryotic translation initiation factor 4E binding protein 1 (EIF4EBP1), heat shock protein family A (Hsp70) member 5 and MYC proto-oncogene, bHLH transcription factor (MYC) were predicted to be the upstream genes of RPS15A, but also detected to be downregulated following the silencing of RPS15A. These data further suggested an indirect and complicated association between RPS15A and these signatures in the regulation of proliferation and apoptosis in RCC cells.
Figure 7.
Networks and RPS15A downstream genes. Regulator effects analysis and interaction networks of genes. The predicted downregulated molecules are marked in green. CDKN1C, cyclin dependent kinase inhibitor 1C; DDIT3, DNA damage inducible transcript 3; EGFR, epidermal growth factor receptor 1; EIF4EBP1, eukaryotic translation initiation factor 4E binding protein 1; ERBB2, erb-b2 receptor tyrosine kinase 2; FOXO1, forkhead box O1; HSPA5, heat shock protein family A (Hsp70) member 5; MYC, MYC proto-oncogene, bHLH transcription factor; RPS15A, ribosomal protein S15A; SNAI1, snail family transcriptional repressor 1.
Discussion
RPs, initially thought to be ‘housekeeping’ genes, are now considered to be associated with tumorigenesis due to their diverse extraribosomal functions (9). The dysregulation of RPS15A was observed to serve a key role in multiple tumours (22). The present study aimed to examine the function of RPS15A in RCC, and detected abnormally high expression levels of RPS15A in RCC samples and cell lines. This high expression suggested a malignant function of RPS15A in RCC. Using an Lv-mediated RNA interference system, the suppression of RPS15A notably inhibited 786-O cell proliferation and induced its apoptosis and necrosis. Additionally, the transfected cells demonstrated a decreased ability of tumour formation and growth in vivo. Furthermore, it was identified that 469 genes were upregulated and 278 were downregulated in the Lv-shRPS15A group using gene chip analysis and a genetic interaction network of RPS15A in RCC was mapped using IPA analysis.
By summarizing the IPA outcomes, it was observed that subsequent to knocking down RPS15A in 786-O cells, the functional signalling pathway of ‘cell death and survival’ and ‘cellular growth and proliferation’ were altered. Furthermore, the possible signalling pathways involved were evaluated, such as ‘Fcγ receptor-mediated phagocytosis in macrophages and monocytes’. Fc receptor (FcR) activation was demonstrated to be tightly regulated to prevent immune responses, and cytokines associated with inflammation were able to increase FcR avidity (23). Additionally, in the inflammation environment, tumour-associated macrophages contribute to RCC progression and tumour angiogenesis (24). These data demonstrated that multiple regulatory signalling pathways are involved in the RPS15A network, which were important in RCC development.
Downstream of RPS15A, CDKN1C was recognized as a negative regulator of cell proliferation and tumour invasion (25), and DDIT3 was activated by endoplasmic reticulum stress, which was responsible for an anti-proliferative effect (26). These genes were downregulated, suggesting decreased activities. Additionally, MYC was identified as a hallmark gene in multiple cancer cells (27). The MYC/EIF4E axis was previously observed to induce mammalian target of rapamycin (mTOR) inhibition in small cell lung cancer, and EIF4EBP1, the upstream direct inhibitor of EIF4E, was observed to be unaltered in this axis (28). However, these genes were identified to be downregulated in RPS15A-knockdown 786-O cells and the findings provided preliminary evidence on the changes in signalling pathways and related molecules. MYC is a key inducer of the oncogenic pathway, regulating the antitumor immune response through cluster of differentiation 47 and programmed death-ligand 1, and its gene expression is closely associated with disease stage and an adverse prognosis (28–30). EIF4EBP1 encodes a member of the family of translation repressor proteins. The expression levels of phosphorylated EIF4EBP1 was demonstrated to be a prognostic predictor in patients with RCC (31,32), and it may serve as a funnel factor that converges the upstream proliferative oncogenic signals (33). Previous studies have indicated that eukaryotic translation initiation factor 4E-binding proteins (which have three family members: EIF4EBP1, 2 and 3) mediate the effect of mTORC1 to promote cell proliferation; however, not growth (thus regulating the number, but not the size), of mammalian cells (34,35).
There are some limitations for the present study. Only 786-O cells were used in the functional experiments owing to the potential tumour heterogeneity in different cell lines, such as metastatic RCC cell line, Caki-1 cells. However, the results provide new evidence that enriched the interaction relationships of RPS15A, in which the specific molecular mechanisms are still waiting to be explored.
In conclusion, the present study confirmed that RPS15A is highly expressed in RCC samples and cell lines. Suppression of RPS15A successfully inhibited 786-O cell growth and induced cell apoptosis and necrosis. The direct effects of RPS15A on promoting tumour progression through various potential intracellular signalling pathways indicated RPS15A as a potential therapeutic target in RCC.
Acknowledgements
The abstract of the present study was presented at the 38th congress of the Société Internationale d'Urologie Seoul Dragon City, October 4–7, 2018 and published in World Journal of Urology 36 (Suppl 1): 2018.
Funding
The present study was supported by The Science and Technology Foundation of Sichuan Province (grant no. 2014JY0183 to Yiping Lu and grant no. 2017SZ0123 to Zhihong Liu) and 1.3.5 Project for Disciplines of Excellence, West China Hospital, Sichuan University (Chengdu, China).
Availability of data and materials
The results from the present study are based partly on data generated by The Cancer Genome Atlas Research Network (cancergenome.nih.gov). The analysed data sets generated during the current study are available from the corresponding author on reasonable request.
Authors' contributions
JL, ZL and YL conceived and designed the present study. JL, ZL, ZZ, XW and CZ developed the methodology. JL, ZL, YT, FZ, KW and CZ analysed and interpreted the data. JL, ZL and YL wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
All data in the present study involving human participants were collected from a publicly available database. For this type of study, formal consent was not required. All experimental procedures and animal work were conducted under the principles and procedures by The National Institutes of Health (Bethesda, MD, USA) and approved by the Animal Ethics Committee of West China Hospital.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Warner JR, McIntosh KB. How common are extraribosomal functions of ribosomal proteins? Mol Cell. 2009;34:3–11. doi: 10.1016/j.molcel.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lindstrom MS, Zhang Y. Ribosomal protein S9 is a novel B23/NPM-binding protein required for normal cell proliferation. J Biol Chem. 2008;283:15568–15576. doi: 10.1074/jbc.M801151200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Volarevic S, Stewart MJ, Ledermann B, Zilberman F, Terracciano L, Montini E, Grompe M, Kozma SC, Thomas G. Proliferation, but not growth, blocked by conditional deletion of 40S ribosomal protein S6. Science. 2000;288:2045–2047. doi: 10.1126/science.288.5473.2045. [DOI] [PubMed] [Google Scholar]
- 4.He H, Sun Y. Ribosomal protein S27L is a direct p53 target that regulates apoptosis. Oncogene. 2007;26:2707–2716. doi: 10.1038/sj.onc.1210073. [DOI] [PubMed] [Google Scholar]
- 5.Jang CY, Lee JY, Kim J. RpS3, a DNA repair endonuclease and ribosomal protein, is involved in apoptosis. FBS Lett. 2004;560:81–85. doi: 10.1016/S0014-5793(04)00074-2. [DOI] [PubMed] [Google Scholar]
- 6.Hegde V, Wang M, Deutsch WA. Human ribosomal protein S3 interacts with DNA base excision repair proteins hAPE/Ref-1 and hOGG1. Biochemistry. 2004;43:14211–14217. doi: 10.1021/bi049234b. [DOI] [PubMed] [Google Scholar]
- 7.Anderson SJ, Lauritsen JP, Hartman MG, Foushee AM, Lefebvre JM, Shinton SA, Gerhardt B, Hardy RR, Oravecz T, Wiest DL. Ablation of ribosomal protein L22 selectively impairs αβ T cell development by activation of a p53-dependent checkpoint. Immunity. 2007;26:759–772. doi: 10.1016/j.immuni.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 8.Zhan Y, Melian NY, Pantoja M, Haines N, Ruohola-Baker H, Bourque CW, Rao Y, Carbonetto S. Dystroglycan and mitochondrial ribosomal protein L34 regulate differentiation in the Drosophila eye. PLoS One. 2010;5:e10488. doi: 10.1371/journal.pone.0010488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang W, Nag S, Zhang X, Wang MH, Wang H, Zhou J, Zhang R. Ribosomal proteins and human diseases: Pathogenesis, molecular mechanisms, and therapeutic implications. Med Res Rev. 2015;35:225–285. doi: 10.1002/med.21327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu M, Wang Y, Chen L, Pan B, Chen F, Fang Y, Yu Z, Chen G. Down-regulation of ribosomal protein S15A mRNA with a short hairpin RNA inhibits human hepatic cancer cell growth in vitro. Gene. 2014;536:84–89. doi: 10.1016/j.gene.2013.11.075. [DOI] [PubMed] [Google Scholar]
- 11.Yao Y, Liu Y, Lv X, Dong B, Wang F, Li J, Zhang Q, Xu R, Xu Y. Down-regulation of ribosomal protein S15A inhibits proliferation of human glioblastoma cells in vivo and in vitro via AKT pathway. Tumour Biol. 2016;37:4979–4990. doi: 10.1007/s13277-015-4323-0. [DOI] [PubMed] [Google Scholar]
- 12.Zhang C, Fu J, Xue F, Ryu B, Zhang T, Zhang S, Sun J, Xu X, Shen Z, Zheng L, Chen X. Knockdown of ribosomal protein S15A induces human glioblastoma cell apoptosis. World J Surg Oncol. 2016;14:129. doi: 10.1186/s12957-016-0891-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao X, Shen L, Feng Y, Yu H, Wu X, Chang J, Shen X, Qiao J, Wang J. Decreased expression of RPS15A suppresses proliferation of lung cancer cells. Tumour Biol. 2015;36:6733–6740. doi: 10.1007/s13277-015-3371-9. [DOI] [PubMed] [Google Scholar]
- 14.Chen J, Wei Y, Feng Q, Ren L, He G, Chang W, Zhu D, Yi T, Lin Q, Tang W, et al. Ribosomal protein S15A promotes malignant transformation and predicts poor outcome in colorectal cancer through misregulation of p53 signaling pathway. Int J Oncol. 2016;48:1628–1638. doi: 10.3892/ijo.2016.3366. [DOI] [PubMed] [Google Scholar]
- 15.Levine AJ, Oren M. The first 30 years of p53: Growing ever more complex. Nat Rev Cancer. 2009;9:749–758. doi: 10.1038/nrc2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y, Zhang G, Li X, Li B, Zhang X. The effect of ribosomal protein S15a in lung adenocarcinoma. Peer J. 2016;4:e1792. doi: 10.7717/peerj.1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Capitanio U, Montorsi F. Renal cancer. Lancet. 2016;387:894–906. doi: 10.1016/S0140-6736(15)00046-X. [DOI] [PubMed] [Google Scholar]
- 18.Abu Aboud O, Wettersten HI, Weiss RH. Inhibition of PPARα induces cell cycle arrest and apoptosis, and synergizes with glycolysis inhibition in kidney cancer cells. PLoS One. 2013;8:e71115. doi: 10.1371/journal.pone.0071115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92–D97. doi: 10.1093/nar/gkt1248. (Database Issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: A cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/S1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Xu X, Xiong X, Sun Y. The role of ribosomal proteins in the regulation of cell proliferation, tumorigenesis, and genomic integrity. Sci China Life Sci. 2016;59:656–672. doi: 10.1007/s11427-016-0018-0. [DOI] [PubMed] [Google Scholar]
- 23.Brandsma AM, Jacobino SR, Meyer S, ten Broeke T, Leusen JH. Fc receptor inside-out signaling and possible impact on antibody therapy. Immunol Rev. 2015;268:74–87. doi: 10.1111/imr.12332. [DOI] [PubMed] [Google Scholar]
- 24.Yang Z, Xie H, He D, Li L. Infiltrating macrophages increase RCC epithelial mesenchymal transition (EMT) and stem cell-like populations via AKT and mTOR signaling. Oncotarget. 2016;7:44478–44491. doi: 10.18632/oncotarget.9873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo H, Li Y, Tian T, Han L, Ruan Z, Liang X, Wang W, Nan K. The role of cytoplasmic p57 in invasion of hepatocellular carcinoma. BMC Gastroenterol. 2015;15:104. doi: 10.1186/s12876-015-0319-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shimizu T, Kamel WA, Yamaguchi-Iwai S, Fukuchi Y, Muto A, Saya H. Calcitriol exerts an anti-tumor effect in osteosarcoma by inducing the endoplasmic reticulum stress response. Cancer Sci. 2017;108:1793–1802. doi: 10.1111/cas.13304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Casey SC, Tong L, Li Y, Do R, Walz S, Fitzgerald KN, Gouw AM, Baylot V, Gütgemann I, Eilers M, Felsher DW. MYC regulates the antitumor immune response through CD47 and PD-L1. Science. 2016;352:227–231. doi: 10.1126/science.aac9935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsumoto M, Seike M, Noro R, Soeno C, Sugano T, Takeuchi S, Miyanaga A, Kitamura K, Kubota K, Gemma A. Control of the MYC-eIF4E axis plus mTOR inhibitor treatment in small cell lung cancer. BMC Cancer. 2015;15:241. doi: 10.1186/s12885-015-1202-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pelengaris S, Khan M, Evan G. c-MYC: More than just a matter of life and death. Nat Rev Cancer. 2002;2:764–776. doi: 10.1038/nrc904. [DOI] [PubMed] [Google Scholar]
- 30.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10:789–799. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 31.Nishikawa M, Miyake H, Harada K, Fujisawa M. Expression level of phosphorylated-4E-binding protein 1 in radical nephrectomy specimens as a prognostic predictor in patients with metastatic renal cell carcinoma treated with mammalian target of rapamycin inhibitors. Med Oncol. 2014;31:792. doi: 10.1007/s12032-013-0792-4. [DOI] [PubMed] [Google Scholar]
- 32.Nishikawa M, Miyake H, Harada K, Fujisawa M. Expression of molecular markers associated with the mammalian target of rapamycin pathway in nonmetastatic renal cell carcinoma: Effect on prognostic outcomes following radical nephrectomy. Urol Oncol. 2014;32:49.e15–e21. doi: 10.1016/j.urolonc.2013.07.014. [DOI] [PubMed] [Google Scholar]
- 33.Qu Y, Zhao R, Wang H, Chang K, Yang X, Zhou X, Dai B, Zhu Y, Shi G, Zhang H, Ye D. Phosphorylated 4EBP1 is associated with tumor progression and poor prognosis in Xp11.2 translocation renal cell carcinoma. Sci Rep. 2016;6:23594. doi: 10.1038/srep23594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu J, Chen J, Dong Z, Meyuhas O, Chen JK. Phosphorylation of ribosomal protein S6 mediates compensatory renal hypertrophy. Kidney Int. 2015;87:543–556. doi: 10.1038/ki.2014.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dowling RJ, Topisirovic I, Alain T, Bidinosti M, Fonseca BD, Petroulakis E, Wang X, Larsson O, Selvaraj A, Liu Y, et al. mTORC1-mediated cell proliferation, but not cell growth, controlled by the 4E-BPs. Science. 2010;328:1172–1176. doi: 10.1126/science.1187532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jones J, Otu H, Spentzos D, Kolia S, Inan M, Beecken WD, Fellbaum C, Gu X, Joseph M, Pantuck AJ, et al. Gene signatures of progression and metastasis in renal cell cancer. Clin Cancer Res. 2005;11:5730–5739. doi: 10.1158/1078-0432.CCR-04-2225. [DOI] [PubMed] [Google Scholar]
- 37.Higgins JP, Shinghal R, Gill H, Reese JH, Terris M, Cohen RJ, Fero M, Pollack JR, van de Rijn M, Brooks JD. Gene expression patterns in renal cell carcinoma assessed by complementary DNA microarray. Am J Pathol. 2003;162:925–932. doi: 10.1016/S0002-9440(10)63887-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yusenko MV, Kuiper RP, Boethe T, Ljungberg B, van Kessel AG, Kovacs G. High-resolution DNA copy number and gene expression analyses distinguish chromophobe renal cell carcinomas and renal oncocytomas. BMC Cancer. 2009;9:152. doi: 10.1186/1471-2407-9-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The results from the present study are based partly on data generated by The Cancer Genome Atlas Research Network (cancergenome.nih.gov). The analysed data sets generated during the current study are available from the corresponding author on reasonable request.