Abstract
Breast cancer is one of the most frequently diagnosed cancers among females worldwide. Long noncoding RNAs (lncRNAs) have been revealed to serve significant roles in diagnosis and treatment of breast cancer. In the present study, the novel lncRNA RUSC1-AS-N was demonstrated to promote cell viability and metastasis. A total of 100 patients with breast cancer were recruited for this study and it was revealed that RUSC1-AS-N was upregulated in tumor tissues compared with in adjacent non-cancerous counterparts. In addition, using several breast cancer cell lines, it was demonstrated that the mRNA levels of RUSC1-AS-N were highest in the notably metastatic cell lines MDA-MB-231 and MDA-MB-468. Knockdown of RUSC1-AS-N in breast cancer cells inhibited cell proliferation in the colony formation and cell proliferation assays. Furthermore, depletion of RUSC1-AS-N suppressed cell metastasis, as revealed by wound-healing and western blot assays. In addition, the protein levels of Wnt1 and β-catenin were significantly decreased when RUSC1-AS-N was knocked down. However, Wnt signaling pathway activator Wnt agonist 1 reversed the effects of RUSC1-AS-N knockdown on cell proliferation and metastasis. The present study demonstrated that lncRNA RUSC1-AS-N promoted cell viability and metastasis via Wnt/β-catenin signaling in human breast cancer, which may indicate novel targets for the treatment of breast cancer in clinic.
Keywords: RUSC1-AS-N, proliferation, metastasis, Wnt/β-catenin signaling, breast cancer
Introduction
Breast cancer remains a great threat to female health worldwide. According to a cancer statistic (1,2), breast cancer is the most frequently diagnosed cancer and the leading cause of cancer-associated mortality among females in the United States, accounting for ~232,340 of the total cases of cancer and ~39,620 cases of cancer-associated mortality. The high incidence has made breast cancer the most common non-skin cancer in females (1,2). Current knowledge has attributed several factors, including histone modification (3,4), hormone disorder (5) and transcription factors (6), to the development and progression of breast cancer. Breast cancer development is thought to occur via the accumulation of genetic alterations or mutations, and is a multistep disease that involves the co-ordinal interaction of numerous genes, leading to molecular and morphologic changes within normal gland epithelium (7). Although treatments have improved over the last decades, the prognosis of breast cancer remains poor. Therefore, novel therapeutic strategies for the treatment of breast cancer are still required.
With the rapid development of genome and transcriptome sequencing technologies, as well as establishment of genomics consortiums, including Encyclopedia of DNA Elements and Functional Annotation of the Mammalian Genome, the classic view of the transcriptome landscape and its mRNA-centric paradigm for transcript annotation has undergone fundamental changes (8,9). It is well recognized that the vast majority of the genome serves as a template for the transcription of noncoding RNAs. Among these noncoding RNAs, long noncoding RNAs (lncRNAs) are a novel class of RNA that are >200 nucleotides in length and function by regulating various biological processes, including cell proliferation, apoptosis, differentiation, migration and invasion (10,11). Increasing evidence has also indicated the potential role of lncRNAs in numerous diseases, such as cancer (12–15). lncRNAs may have pro-oncogenic or suppressive roles in breast cancer development. A recent study used a Global run-on sequencing, and RNA-sequencing-integrated genomic and molecular approaches to identify and characterize growth-regulating lncRNAs in breast cancers (12), revealing novel insight into breast cancer diagnosis and treatments. For example, nuclear factor κ-light-chain-enhancer of activated B cells interacting lncRNA has been demonstrated to suppress breast cancer metastasis by blocking IκB phosphorylation (13). Furthermore, lncRNA HOXA11-antisense (AS) can promote cell proliferation and metastasis in human breast cancer (14), and six lncRNAs have been identified to be significantly altered in invasive ductal breast carcinoma with the use of sure independence screening procedures based on distance correlation (15).
lncRNA RUSC1-AS-N is a novel lncRNA, whose function remains largely unknown. RUSC1-AS-N was initially identified from microarray data (16), and has recently been identified to be upregulated in hepatocellular carcinoma (HCC) tissue. It was associated with poor prognosis in patients with HCC from GSE54238 and GSE40144 datasets (16), indicating the oncogenic role of RUSC1-AS-N in human tumorigenesis. However, the role of RUSC1-AS-N in breast cancer remains to be elucidated.
In the present study, systemic investigation into the role of RUSC1-AS-N in breast cancer was performed. Firstly, the expression levels of RUSC1-AS-N in breast cancer tissues and cell lines were examined. Subsequently, RUSC1-AS-N expression was knocked down in breast cancer cell lines to evaluate the role of RUSC1-AS-N in cell proliferation, migration and invasion. To the best of our knowledge, this is the first study of its kind to provide experimental evidence for the functional role of RUSC1-AS-N in human solid tumors. The data from the present study indicated that lncRNA RUSC1-AS-N as a therapeutics target may be valuable and promising for the treatment of breast cancer.
Materials and methods
Human samples
A total of 100 breast cancer tissues and their adjacent non-cancerous tissues were collected from female patients (age range, 45–70 years; mean age, 59 years) diagnosed with triple-negative breast infiltrating ductal carcinoma at The Third Department of Breast Cancer, Tianjin Medical University Cancer Institute and Hospital (Tianjin, China) between January 2014-January 2016. Patients did not receive radiotherapy or chemotherapy treatment prior to surgery. All tissues were frozen in liquid nitrogen as soon as they were collected via surgery. All patients provided informed consent to participate in the present study, which was approved by the Ethics Committee of Tianjin Medical University Cancer Institute and Hospital.
Cell culture and transfection
Human breast cancer cell lines T47D, MCF7, MDA-MB-231, MDA-MB-468 and SK-BR-3 were purchased from the Cell Bank of Type Culture Collection of Shanghai Biological Institute, Chinese Academy of Science (Shanghai, China). The breast epithelial cell line MCF10A was purchased from the American Type Culture Collection (Manassas, VA, USA) and used as control cells. Cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.). Cells were maintained at 37°C in a humidified atmosphere with 5% CO2.
Cell transfection was conducted using Lipofectamine® 3000 (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturers' protocols. The transfection was performed 48 h prior to the subsequent analysis. The siRNAs were provided by GenePharm Co. (Shanghai, China; cat. no. 17892). Wnt agonist 1 was purchased from Selleck Chemicals (Shanghai, China) and used at a final concentration of 10 µM at 37°C for 24 h. The culture medium was replaced every two days unless otherwise stated.
RNA isolation and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from human tissues and cultured cells using TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.). RNA was quantified using a NanoDrop™ 2000 spectrophotometer (NanoDrop Technologies; Thermo Fisher Scientific, Inc., Wilmington, DE, USA) and then immediately reversely transcribed into cDNA using PrimeScript™ RT Master Mix (Takara Biotechnology Co., Ltd., Dalian, China), according to the manufacturer's protocols. Subsequently, qPCR was performed with SYBR® Premix EX Taq™ II (Takara Biotechnology Co., Ltd.) on an ABI PRISM® 7900HT Sequence Detection system (Thermo Fisher Scientific, Inc.). The thermocycling protocol was as follows: Initial denaturation at 95°C for 5 min, followed by 45 cycles of 10 sec at 95°C (denaturation), 10 sec at 60°C (primer annealing) and 10 sec at 72°C (elongation), and a final extension step for 10 min at 72°C. The primer sequences used for qPCR were as follows: RUSC1-AS-N, forward, 5′-TCTTTCCCAGAAGTAGCAC-3′ and reverse, 5′-ATTTTATCAACGGAGACGC-3′; Wnt1, forward, 5′-CGATGGTGGGGTATTGTGAAC-3′ and reverse, 5′-CCGGATTTTGGCGTATCAGAC-3′; β-catenin, forward, 5′-AAAGCGGCTGTTAGTCACTGG-3′ and reverse, 5′-CGAGTCATTGCATACTGTCCAT-3′; and GAPDH, forward, 5′-GTGGACATCCGCAAAGAC-3′ and reverse, 5′-AAAGGGTGTAACGCAACTA-3′. GAPDH was used as the internal reference, and relative gene expression levels were calculated using the 2−ΔΔCq method (17). Each experiment was repeated at least three times.
Colony formation assay
To observe the effects of RUSC1-AS-N on cell proliferation, colony formation assays were performed. Briefly, MDA-MB-231 and MDA-MB-468 cells were seeded in 6-well plates at a density of 900 cells/well and were later transfected with scramble negative control siRNA (siNC) or specific siRNA against RUSC1-AS-N (siRUSC1-AS-N) when the cell confluence reached 10%. Transfected cells were cultured for 14 days to form natural colonies. Subsequently, cells were washed with PBS for three times, treated with (1%) crystal violet for 30 min at room temperature and washed twice with deionized water. The colonies were viewed under a light microscope (Nikon Corporation, Tokyo, Japan) with at least five fields randomly and the number of colonies was counted.
Cell viability analysis
Cell viability was determined using an MTT assay. MDA-MB-231 and MDA-MB-468 cells were trypsinized and seeded in triplicate in 96-well plates at a density of 4,000 cells/well. Cells were transfected with siNC and siRUSC1-AS-N at the presence or absence of RUSC1-AS-N knockdown. Cell viability was monitored over the course of 5 days. At each indicated time point (day 1, 2, 3, 4 and 5), 10 µl/well of 5 mg/ml MTT solution was added to the cell cultures. DMSO was used to dissolve the formazan. Following 2 h incubation at room temperature, the absorbance at 490 nm was recorded with a Tecan microplate reader. Cell viability was defined as the cell number ratio of experimental groups to control cells.
Wound-healing assay
MDA-MB-231 and MDA-MB-468 cells were transfected with siRUSC1-AS-N or siNC, seeded at a density of 5×105 cells/well in 6-well culture plates and allowed to reach 90% confluence overnight. Subsequently, the culture medium was replaced with serum-free DMEM, and a scratch wound was created using a 10-µl pipette tip and the cells were washed three times with PBS. Following incubation at 37°C for 12 h, the migrating cells were observed and images were captured using a light microscope (magnification, ×100; Nikon Corporation).
Western blot analysis
Cells were lysed in radioimmunoprecipitation assay buffer (Beyotime Institute of Biotechnology, Nantong, China) with a protease inhibitor cocktail (Thermo Fisher Scientific, Inc.). The total protein extracts were quantified using a Bicinchoninic Acid assay kit (Thermo Fisher Scientific, Inc.), according to the manufacturers' protocols. A total of 40 µg protein was separated by 12% SDS-PAGE and transferred to a polyvinylidene fluoride membrane (EMD Millipore, Billerica, MA, USA). After blocking in 5% milk for 1 h at room temperature, proteins were detected using specific primary antibodies (incubated at 4°C overnight) against Wnt1 (1:1,000; cat. no. ab15251; Abcam, Cambridge, UK), β-catenin (1:1,000; cat. no. ab16051; Abcam), E-cadherin (1:1,000; cat. no. sc-71009; Santa Cruz Biotechnology, Inc., Dallas, TX, USA), N-cadherin (1:1,000; cat. no. sc-53488; Santa Cruz Biotechnology, Inc.), cyclin B1 (1:1,000; cat. no. sc-70898; Santa Cruz Biotechnology, Inc.) and GAPDH (1:1,000; cat. no. sc-32233; Santa Cruz Biotechnology, Inc.). Goat anti-rabbit IgG-HRP secondary antibody and goat anti-mouse IgG-HRP secondary antibody were purchased from Santa Cruz Biotechnology (cat. nos. sc-2004 and sc-2005; 1:2,500) and incubated with the membrane at room temperature for 1 h. Proteins were visualized using an enhanced chemiluminescence (Immobilon Western HRP; Millipore) and the bands were quantified by densitometry using ImageJ software (V1.8.0; National Institutes of Health, Bethesda, MD, USA). All experiments were performed in triplicate.
Transwell assay
For cell migration assays, MDA-MB-231 and MDA-MB-468 cells were transfected with siRUSC1-AS-N or siNC for 48 h. Subsequently, cells were trypsinized and collected by low-speed centrifugation (1,000 × g, 4°C for 5 min). A total of 1×104 cells in 200 µl serum-free DMEM were seeded into the upper chamber of Transwell cell culture chambers (pore size, 8 µm; Corning Life Sciences, New York, NY, USA). The lower chamber was filled with 600 µl DMEM containing 10% FBS. The plate was incubated at 37°C and the cells were allowed to migrate for 24 h. Next, the membrane was fixed with pre-cooled methanol for 5 min at room temperature and stained with 1% crystal violet for 5 min at room temperature. Cell migration was assessed by counting the cells that had migrated through the membrane to the underside of the membrane. A total of 5 random fields of view were selected and images were captured using a light microscope (magnification, ×100; Nikon Corporation).
For cell invasion assays, the same protocol was followed; however, membrane was pre-coated with Matrigel (20%; Corning Incorporated, Corning, NY, USA) and cells were incubated for 6 h at 37°C.
Statistical analysis
GraphPad Prism version 5.0 (GraphPad Software, Inc., La Jolla, CA, USA) was used to perform statistical analyses. The data are presented as the mean ± standard deviation. A two-tailed Student's t-test was used for comparisons between two groups. Differences between tumor and adjacent normal control samples were analyzed using paired Student's t-test. For comparisons among multiple groups, one-way analysis of variance was applied followed by Least Significance Difference post hoc test. P<0.05 was considered to indicate a statistically significant difference. Each experiment was repeated three times.
Results
lncRNA RUSC1-AS-N is upregulated in human breast cancer
Firstly, the relative expression levels of RUSC1-AS-N in human breast cancer were examined in vivo and in vitro. A total of 100 patients with breast cancer were included in the study. As presented in Fig. 1A, patients had significantly higher transcript levels of RUSC1-AS-N in the tumor tissues compared with in the adjacent non-cancerous tissues. Next, five different breast cancer cell lines were used to compare expression levels of RUSC1-AS-N; normal MCF10A cells were included as a control. It was demonstrated that the transcription levels of RUSC1-AS-N were significantly upregulated in all breast cancer cell lines compared with the control (Fig. 1B), with MDA-MB-231 and MDA-MB-468 cells exhibiting the highest expression of RUSC1-AS-N. Therefore, MDA-MB-231 and MDA-MB-468 were selected for subsequent knockdown experiments. Interestingly, MDA-MB-231 and MDA-MB-468 cells are known to have high metastatic potential (7,14), indicating a potential association of RUSC1-AS-N with cell metastasis. The data suggested that the expression of RUSC1-AS-N was upregulated in human breast cancer both in vivo and in vitro.
Figure 1.
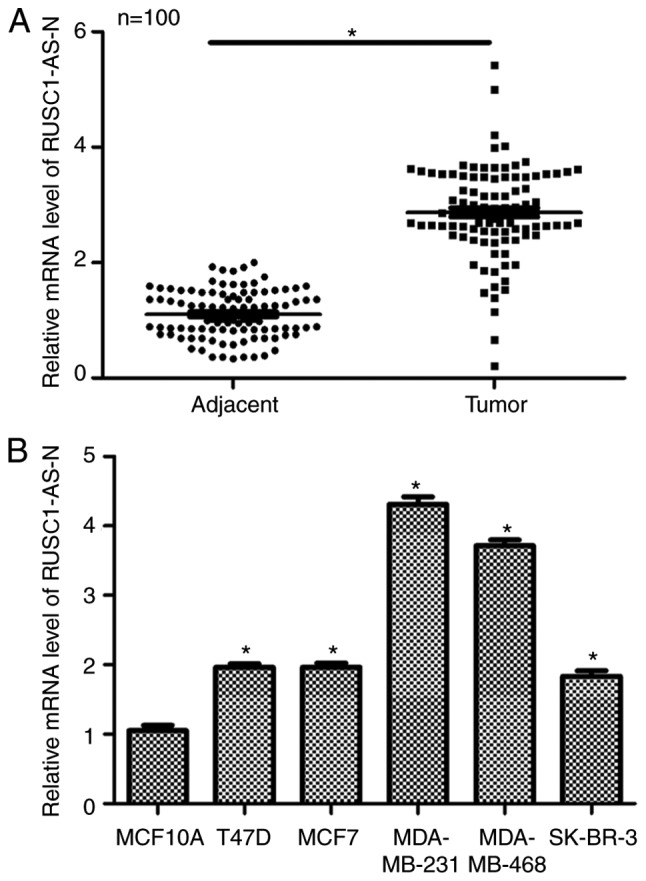
Long noncoding RNA RUSC1-AS-N is upregulated in human breast cancer. (A) A total of 100 patients with breast cancer were recruited, and reverse transcription-quantitative polymerase chain reaction was performed to examine the expression levels of RUSC1-AS-N in the tumor and adjacent non-cancerous tissue. The data was analyzed using a paired Student's t-test. *P<0.05. (B) Relative transcript levels of RUSC1-AS-N were detected in T47D, MCF7, MDA-MB-231, MDA-MB-468 and SK-BR-3 breast cancer cell lines, as well as the MCF10A normal human breast epithelial cell line. *P<0.05 vs. MCF10A. AS, antisense.
Knockdown of RUSC1-AS-N inhibits cell proliferation and viability in human breast cancer
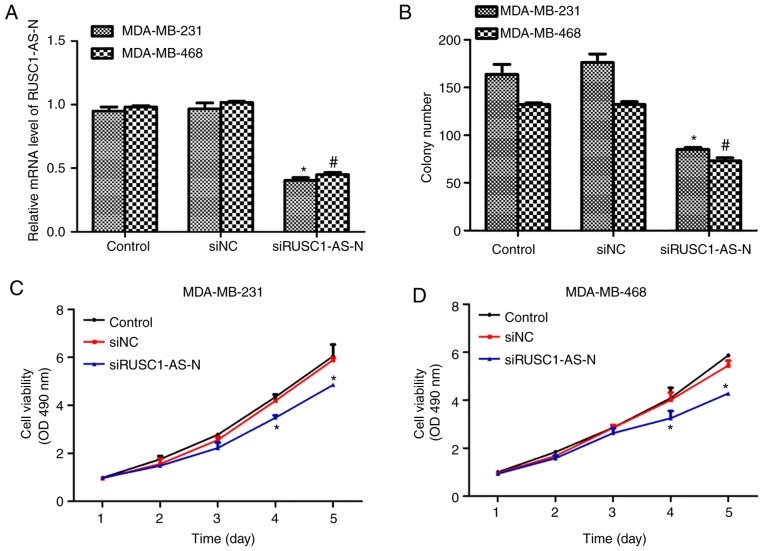
Next, the role of RUSC1- AS-N in human breast cancer cell proliferation was explored. A specific siRNA against RUSC1-AS-N, siRUSC1-AS-N, was transfected into MDA-MB-231 and MDA-MB-468 cells. It was revealed that the transcription levels of RUSC1-AS-N were decreased by ~50% in both cell lines following transfection with siRUSC1-AS-N (Fig. 2A). Colony formation assays were also performed, and it was demonstrated that knockdown of RUSC1-AS-N significantly decreased the number of colonies by 24 and 20% in MDA-MB-231 and MDA-MB-468 cells, respectively, compared with the control (Fig. 2B). Subsequently, cell viability was detected in both cell lines transfected with or without siRUSC1-AS-N. For the first 3 days following transfection, there was no notable difference amongst the three experimental groups; however, cell viability of MDA-MB-231 cells was suppressed by 16 and 22% on days 4 and 5, respectively (Fig. 2C). Similar results were observed in MDA-MB-468 cells (Fig. 2D), whereby significant suppression of cell proliferation was also recorded on day 4 and 5. These results revealed that knockdown of RUSC1-AS-N inhibited cell proliferation of human breast cancer cells in vitro.
Figure 2.
Knockdown of RUSC1-AS-N inhibits the cell viability of human breast cancer cells. (A) Relative transcript levels of RUSC1-AS-N were detected in MDA-MB-231 and MDA-MB-468 cells following siRUSC1-AS-N transfection using reverse transcription-quantitative polymerase chain reaction. (B) Colony formation assays were performed with MDA-MB-231 and MDA-MB-468 cells transfected with siRUSC1-AS-N. *P<0.05 vs. MDA-MB-231 Control. #P<0.05 vs. MDA-MB-468 Control. Cell viability of (C) MDA-MB-231 and (D) MDA-MB-468 cells transfected with siRUSC1-AS-N was examined over a 5-day period using MTT assay. *P<0.05 vs. Control. siNC, scramble negative control siRNA; siRNA, small interfering RNA; siRUSC1-AS-N, siRNA against RUSC1-antisense-N.
Knockdown of RUSC1-AS-N suppresses cell migration in human breast cancer
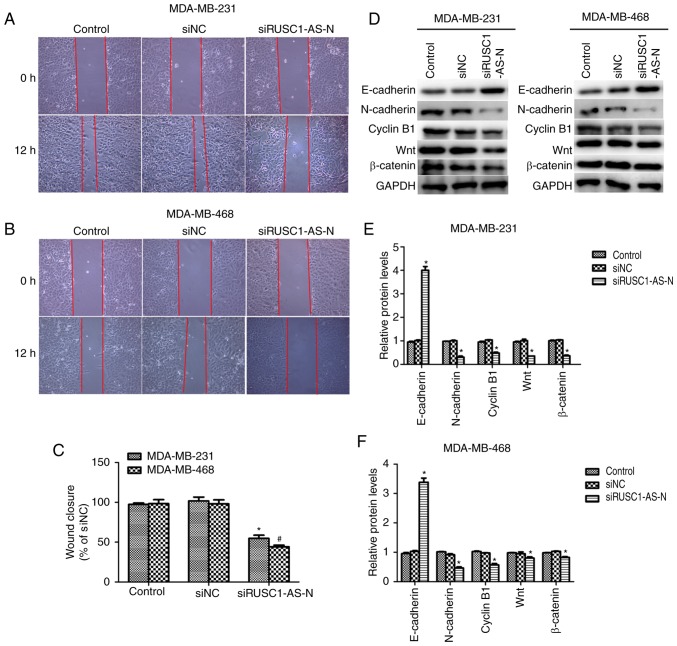
Cell proliferation and migration are the two main manifestations of cancer; therefore, the role of RUSC1-AS-N in cell migration was investigated. Wound healing assays were performed and as presented in Fig. 3A and B, transfection with specific siRNA against RUSC1-AS-N decreased the wound closure rate in both cell lines. Quantification of wound gap area revealed that cell migration was significantly suppressed by ~50% in MDA-MB-231 and MDA-MB-468 cells (Fig. 3C). The protein expression levels of epithelial-to-mesenchymal transition (EMT) markers were analyzed using western blotting. It was demonstrated that when RUSC1-AS-N was knocked down, the protein levels of E-cadherin were significantly increased, while that of N-cadherin, cyclin B1, Wnt1 and β-catenin were significantly decreased in MDA-MB-231 and MDA-MB-468 cells (Fig. 3D-F) compared with the control. The results suggested that knockdown of RUSC1-AS-N inhibited cell migration via the Wnt/β-catenin signaling pathway in human breast cancer.
Figure 3.
Knockdown of RUSC1-AS-N suppresses cell migration in human breast cancer. Representative images of cell migration by wound healing assay for (A) MDA-MB-231 and (B) MDA-MB-468 cells treated with siRUSC1-AS-N. Magnification, ×200. (C) Quantification of wound closure area relative to siNC at 0 h. *P<0.05 vs. MDA-MB-231 Control; #P<0.05 vs. MDA-MB-468 Control. (D) Western blot analysis of E-cadherin, N-cadherin, cyclin B1, Wnt1 and β-catenin in MDA-MB-231 and MDA-MB-468 cells transfected with siRUSC1-AS-N. Quantification of relative protein expression levels in (E) MDA-MB-231 and (F) MDA-MB-468 cells. *P<0.05 vs. Control. siNC, scramble negative control siRNA; siRNA, small interfering RNA; siRUSC1-AS-N, siRNA against RUSC1-antisense-N.
Activation of the Wnt/β-catenin signaling pathway reverses the inhibitory effects of siRUSC1-AS-N on cell proliferation in human breast cancer
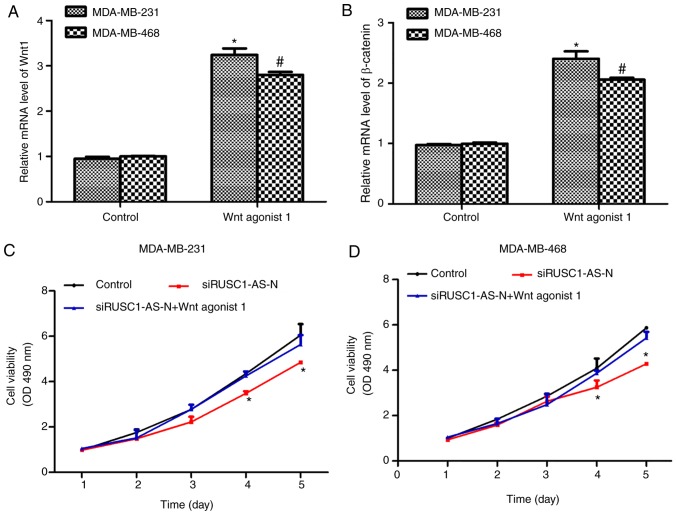
Next, the Wnt/β-catenin signaling pathway was activated using a specific activator, Wnt agonist 1, at a final concentration of 10 µM. As presented in Fig. 4A and B, treatment of MDA-MB-231 and MDA-MB-468 cells with Wnt agonist 1 significantly increased the mRNA levels of Wnt1 and β-catenin by ~3-fold. In addition, cell viability was assessed in cells treated with Wnt agonist 1 in the presence or absence of siRUSC1-AS-N. As presented in Fig. 4C, knockdown of RUSC1-AS-N in MDA-MB-231 cells significantly suppressed cell viability on days 4 and 5 following transfection compared with the control; however, treatment with Wnt agonist 1 reversed the inhibitory effects of siRUSC1-AS-N. Similarly, depletion of RUSC1-AS-N in MDA-MB-468 cells also decreased cell viability, whereas activation of the Wnt/β-catenin signaling pathway suppressed the inhibitory effects of siRUSC1-AS-N (Fig. 4D). These data suggested that RUSC1-AS-N promoted cell viability by regulating the Wnt/β-catenin signaling pathway in human breast cancer cells.
Figure 4.
Activation of Wnt/β-catenin signaling reverses the inhibitory effects of siRUSC1-AS-N on cell viability in human breast cancer. The mRNA levels of (A) Wnt1 and (B) β-catenin were examined in MDA-MB-231 and MDA-MB-468 cells treated with Wnt agonist 1, using reverse transcription-quantitative polymerase chain reaction. *P<0.05 vs. MDA-MB-231 Control; #P<0.05 vs. MDA-MB-468 Control. Cell viability was measured in siRUSC1-AS-N-transfected (C) MDA-MB-231 and (D) MDA-MB-468 cells in the presence or absence of Wnt agonist 1, over the course of 5 days. *P<0.05 vs. Control. OD, optical density; siRUSC1-AS-N, small interfering RNA against RUSC1-antisense-N.
Activation of Wnt/β-catenin signaling reverses the inhibitory effects of siRUSC1-AS-N on cell metastasis in human breast cancer
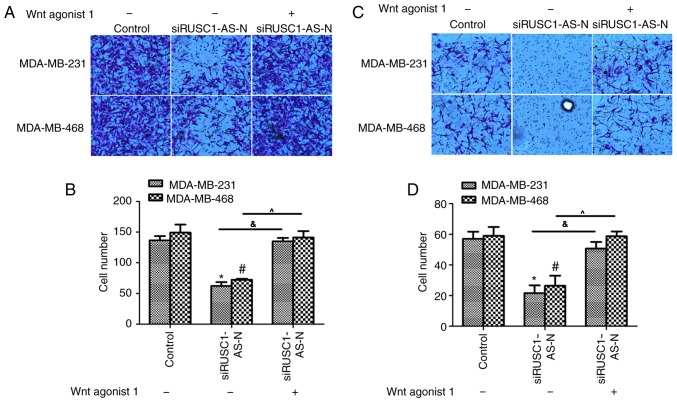
The effects of Wnt agonist 1 on human breast cancer cell lines were also examined using Transwell assays. The migration assays revealed that transfection with siRUSC1-AS-N significantly inhibited cell migration via the Transwell membrane for MDA-MB-231 and MDA-MB-468 cells, while co-treatment with Wnt agonist 1 reversed the inhibitory effect of siRUSC1-AS-N (Fig. 5A and B). The invasion assay demonstrated that cell metastasis was significantly inhibited when cells were transfected with siRUSC1-AS-N compared with the control; cell metastasis returned to normal levels when cells were co-treated with Wnt agonist 1 (Fig. 5C and D). These results indicated that RUSC1-AS-N promoted cell metastasis via regulation of the Wnt/β-catenin signaling pathway in human breast cancer cells.
Figure 5.
Activation of the Wnt/β-catenin signaling pathway reverses the inhibitory effects of siRUSC1-AS-N on cell metastasis in human breast cancer. (A) Representative images of Transwell cell migration assays with siRUSC1-AS-N-transfected MDA-MB-231 and MDA-MB-468 cells treated with or without Wnt agonist 1. Magnification, ×200. (B) Quantification of the number of migratory cells. (C) Representative images of Transwell cell invasion assays with siRUSC1-AS-N-transfected MDA-MB-231 and MDA-MB-468 cells treated with or without Wnt agonist 1. Magnification, ×200. (D) Quantification of the number of invasive cells. *P<0.05 vs. MDA-MB-231 Control; #P<0.05 vs. MDA-MB-468 Control; &P<0.05 vs. MDA-MB-231 siRUSC1-AS-n/Wnt agonist 1; ^P<0.05 vs. MDA-MB-468 siRUSC1-AS-n/Wnt agonist 1 siRUSC1-AS-N, small interfering RNA against RUSC1-antisense-N.
Discussion
Breast cancer is one of the most common type of cancer in females. According to an investigation in 2016, there are 3,053,450 females living with breast cancer, which significantly affects the quality of life of these patients (1,3,18). Although great efforts have been made to improve breast cancer diagnosis and treatment, most tumors are often clinically diagnosed at an advanced stage (19). Additionally, karyotyping suggests that breast cancer becomes increasingly aggressive via the stepwise accumulation of genetic alteration (6,20); therefore, novel therapeutic strategies for breast cancer are required.
Recent advances in gene sequencing have greatly broadened current knowledge of gene regulation. lncRNAs are a novel class of noncoding RNAs that have been implicated in multiple cellular processes, including proliferation (21), apoptosis (22), migration and invasion (14,23). lncRNA RUSC1-AS-N is a novel lncRNA that has unknown function. A previous study revealed that RUSC1-AS-N is upregulated in HCC tissues and its expression levels indicate poor prognosis in patients with HCC (16). To the best of our knowledge, the present study is the first to demonstrate that RUSC1-AS-N is also upregulated in breast cancer tissues compared with in non-cancerous tissues.
RUSC1-AS-N was also highly expressed in breast cancer cell lines MDA-MB-231 and MDA-MB-468; therefore, siRNA was used to deplete RUSC1-AS-N expression, and cell proliferation, migration and invasion were examined. Interestingly, it was identified that depletion of RUSC1-AS-N significant decreased cell viability over the course of 5 days. Furthermore, depletion of RUSC1-AS-N also decreased cell migration ability as breast cancer cells transfected with siRUSC1-AS-N exhibited significantly lower wound closure rates. EMT is a marker of cell invasiveness and has been widely recognized as a critical process that implicates distant metastases (24). Consistent with the wound healing assay results, knockdown of RUSC1-AS-N increased the expression of the epithelial marker E-cadherin, but decreased that of the mesenchymal marker N-cadherin, confirming that RUSC1-AS-N may promote the EMT processes during breast tumorigenesis. Knockdown of RUSC1-AS-N also decreased the protein levels of cyclin B1, a marker of cell cycle progression, reinforcing the idea that RUSC1 also promoted cell cycle progression, thereby promoting cell proliferation in breast cancer. The results of the present study collectively suggested that RUSC1-AS-N promoted cell proliferation and migration in breast cancer.
Interestingly, it was observed that the protein levels of Wnt and β-catenin, two pivotal proteins that execute the Wnt/β-catenin pathway cascade, were also decreased following knockdown of RUSC1-AS-N. With the use of Wnt agonist 1, which selectively activates the Wnt/β-catenin pathway, it was demonstrated that siRUSC1-AS-N-induced inhibition of cell proliferation, migration and invasion was reversed. Activation of the Wnt/β-catenin pathway by Wnt agonist 1 led to cell viabilities and invasion capacities that were comparable with control untreated cells. Previous studies have reported that the Wnt/β-catenin pathway is crucial for the development and progression of breast cancer (25,26). In addition, high levels of Wnt expression and aberrant activation of β-catenin have been detected in breast cancer tissues (27). However, downregulation of the Wnt/β-catenin pathway can inhibit EMT and suppress the spontaneous invasion of breast cancer cells (28). Taken together, the results in the present study suggested that RUSC1-AS-N positively regulated the Wnt/β-catenin pathway, thereby promoting the proliferation and metastasis of breast cancer cells.
In conclusion, the present study identified lncRNA RUSC1-AS-N as a critical mediator of breast cancer cell proliferation and metastasis. This is the first report that systemically investigated the functional roles of RUSC1-AS-N in solid tumors. The results suggested that RUSC1-AS-N promoted cell proliferation and metastasis via Wnt/β-catenin signaling in human breast cancer. The present study provided novel evidence that the therapeutic targeting of RUSC1-AS-N or Wnt/β-catenin may be a promising strategy for the treatment of breast cancer in clinic.
Acknowledgements
Not applicable.
Funding
The present study was financially supported by the National Natural Science Foundation of China (grant no. 81672623).
Availability of data and materials
All data and materials are available.
Authors' contributions
PZ performed most of the experiments, analyzed the data and prepared the manuscript, PL helped with certain of the experiments and revised the manuscript and JZ provided the funding, designed the study and revised the manuscript.
Ethics approval and consent to participate
This study was approved by the Ethics Committee of Tianjin Medical University Cancer Institute and Hospital (Tianjin, China). All patients gave their full consent to participate in the present study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.DeSantis C, Ma J, Bryan L, Jemal A. Breast cancer statistics, 2013. CA Cancer J Clin. 2014;64:52–62. doi: 10.3322/caac.21203. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 3.Li LL, Xue AM, Li BX, Shen YW, Li YH, Luo CL, Zhang MC, Jiang JQ, Xu ZD, Xie JH, Zhao ZQ. JMJD2A contributes to breast cancer progression through transcriptional repression of the tumor suppressor ARHI. Breast Cancer Res. 2014;16:R56. doi: 10.1186/bcr3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feng W, Lu Z, Luo RZ, Zhang X, Seto E, Liao WS, Yu Y. Multiple histone deacetylases repress tumor suppressor gene ARHI in breast cancer. Int J Cancer. 2007;120:1664–1668. doi: 10.1002/ijc.22474. [DOI] [PubMed] [Google Scholar]
- 5.Travis RC, Key TJ. Oestrogen exposure and breast cancer risk. Breast Cancer Res. 2003;5:239–247. doi: 10.1186/bcr628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li L, Gao P, Li Y, Shen Y, Xie J, Sun D, Xue A, Zhao Z, Xu Z, Zhang M, et al. JMJD2A-dependent silencing of Sp1 in advanced breast cancer promotes metastasis by downregulation of DIRAS3. Breast Cancer Res Treat. 2014;147:487–500. doi: 10.1007/s10549-014-3083-7. [DOI] [PubMed] [Google Scholar]
- 7.Zhang H, Stephens LC, Kumar R. Metastasis tumor antigen family proteins during breast cancer progression and metastasis in a reliable mouse model for human breast cancer. Clin Cancer Res. 2006;12:1479–1486. doi: 10.1158/1078-0432.CCR-05-1519. [DOI] [PubMed] [Google Scholar]
- 8.de Hoon M, Shin JW, Carninci P. Paradigm shifts in genomics through the FANTOM projects. Mamm Genome. 2015;26:391–402. doi: 10.1007/s00335-015-9593-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pennisi E. Genomics. ENCODE project writes eulogy for junk DNA. Science. 2012;337:1159–1161. doi: 10.1126/science.337.6099.1159. [DOI] [PubMed] [Google Scholar]
- 10.Chen L, Wang W, Cao L, Li Z, Wang X. Long non-coding RNA CCAT1 acts as a competing endogenous RNA to regulate cell growth and differentiation in acute myeloid leukemia. Mol Cells. 2016;39:330–336. doi: 10.14348/molcells.2016.2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo X, Hua Y. CCAT1: An oncogenic long noncoding RNA in human cancers. J Cancer Res Clin Oncol. 2017;143:555–562. doi: 10.1007/s00432-016-2268-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun M, Gadad SS, Kim DS, Kraus WL. Discovery, annotation, and functional analysis of long noncoding RNAs controlling cell-cycle gene expression and proliferation in breast cancer cells. Mol Cell. 2015;59:698–711. doi: 10.1016/j.molcel.2015.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu B, Sun L, Liu Q, Gong C, Yao Y, Lv X, Lin L, Yao H, Su F, Li D, et al. A cytoplasmic NF-κB interacting long noncoding RNA blocks IκB phosphorylation and suppresses breast cancer metastasis. Cancer Cell. 2015;27:370–381. doi: 10.1016/j.ccell.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 14.Su JC, Hu XF. Long non-coding RNA HOXA11-AS promotes cell proliferation and metastasis in human breast cancer. Mol Med Rep. 2017;16:4887–4894. doi: 10.3892/mmr.2017.7163. [DOI] [PubMed] [Google Scholar]
- 15.Chen X, Yang J, Qian L, Cao T. Aberrantly expressed mRNAs and long non-coding RNAs in patients with invasive ductal breast carcinoma: A pilot study. Mol Med Rep. 2015;11:2185–2190. doi: 10.3892/mmr.2014.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang R, Wu JC, Zheng LM, Li ZR, Zhou KL, Zhang ZS, Xu DF, Chen C. Long noncoding RNA RUSC1-AS-N indicates poor prognosis and increases cell viability in hepatocellular carcinoma. Eur Rev Med Pharmacol Sci. 2018;22:388–396. doi: 10.26355/eurrev_201801_14185. [DOI] [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 18.Banas T, Juszczyk G, Pitynski K, Nieweglowska D, Ludwin A, Czerw A. Incidence and mortality rates in breast, corpus uteri, and ovarian cancers in Poland (1980–2013): An analysis of population-based data in relation to socioeconomic changes. Onco Targets Ther. 2016;9:5521–5530. doi: 10.2147/OTT.S112187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meissner HI, Klabunde CN, Han PK, Benard VB, Breen N. Breast cancer screening beliefs, recommendations and practices: Primary care physicians in the United States. Cancer. 2011;117:3101–3111. doi: 10.1002/cncr.25873. [DOI] [PubMed] [Google Scholar]
- 20.Lee EY, Muller WJ. Oncogenes and tumor suppressor genes. Cold Spring Harb Perspect Biol. 2010;2:a003236. doi: 10.1101/cshperspect.a003236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun H, Wang G, Peng Y, Zeng Y, Zhu QN, Li TL, Cai JQ, Zhou HH, Zhu YS. H19 lncRNA mediates 17β-estradiol-induced cell proliferation in MCF-7 breast cancer cells. Oncol Rep. 2015;33:3045–3052. doi: 10.3892/or.2015.3899. [DOI] [PubMed] [Google Scholar]
- 22.Arase M, Horiguchi K, Ehata S, Morikawa M, Tsutsumi S, Aburatani H, Miyazono K, Koinuma D. Transforming growth factor-β-induced lncRNA-Smad7 inhibits apoptosis of mouse breast cancer JygMC(A) cells. Cancer Sci. 2014;105:974–982. doi: 10.1111/cas.12454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu P, Chu J, Wu Y, Sun L, Lv X, Zhu Y, Li J, Guo Q, Gong C, Liu B, Su S. NBAT1 suppresses breast cancer metastasis by regulating DKK1 via PRC2. Oncotarget. 2015;6:32410–32425. doi: 10.18632/oncotarget.5609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nieto MA, Huang RY, Jackson RA, Thiery JP. EMT: 2016. Cell. 2016;166:21–45. doi: 10.1016/j.cell.2016.06.028. [DOI] [PubMed] [Google Scholar]
- 25.Wu Y, Ginther C, Kim J, Mosher N, Chung S, Slamon D, Vadgama JV. Expression of Wnt3 activates Wnt/β-catenin pathway and promotes EMT-like phenotype in trastuzumab-resistant HER2-overexpressing breast cancer cells. Mol Cancer Res. 2012;10:1597–1606. doi: 10.1158/1541-7786.MCR-12-0155-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao Z, Lu P, Zhang H, Xu H, Gao N, Li M, Liu C. Nestin positively regulates the Wnt/β-catenin pathway and the proliferation, survival and invasiveness of breast cancer stem cells. Breast Cancer Res. 2014;16:408. doi: 10.1186/s13058-014-0408-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu Z, Xie L. LHX6 inhibits breast cancer cell proliferation and invasion via repression of the Wnt/β-catenin signaling pathway. Mol Med Rep. 2015;12:4634–4639. doi: 10.3892/mmr.2015.4269. [DOI] [PubMed] [Google Scholar]
- 28.Proffitt KD, Madan B, Ke Z, Pendharkar V, Ding L, Lee MA, Hannoush RN, Virshup DM. Pharmacological inhibition of the Wnt acyltransferase PORCN prevents growth of WNT-driven mammary cancer. Cancer Res. 2013;73:502–507. doi: 10.1158/0008-5472.CAN-12-2258. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data and materials are available.