Abstract
Background
Ginsenosides are known as the principal pharmacological active constituents in Panax medicinal plants such as Asian ginseng, American ginseng, and Notoginseng. Some ginsenosides, especially the 20(R) isomers, are found in trace amounts in natural sources and are difficult to chemically synthesize. The present study provides an approach to produce such trace ginsenosides applying biotransformation through Escherichia coli modified with relevant genes.
Methods
Seven uridine diphosphate glycosyltransferase (UGT) genes originating from Panax notoginseng, Medicago sativa, and Bacillus subtilis were synthesized or cloned and constructed into pETM6, an ePathBrick vector, which were then introduced into E. coli BL21star (DE3) separately. 20(R)-Protopanaxadiol (PPD), 20(R)-protopanaxatriol (PPT), and 20(R)-type ginsenosides were used as substrates for biotransformation with recombinant E. coli modified with those UGT genes.
Results
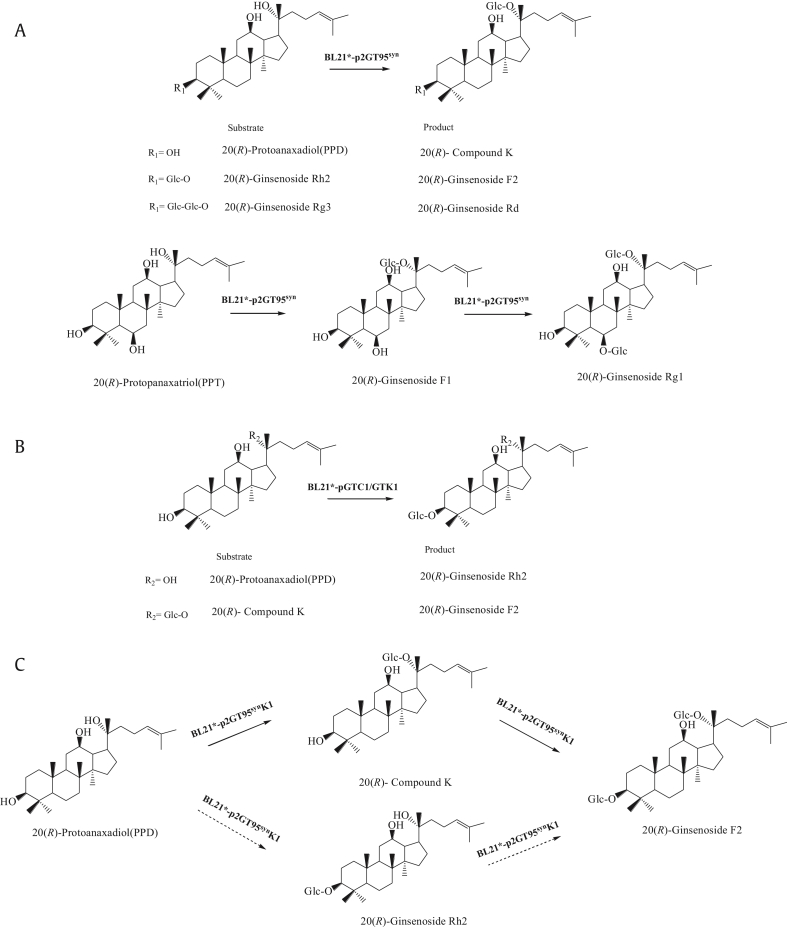
E. coli engineered with GT95syn selectively transfers a glucose moiety to the C20 hydroxyl of 20(R)-PPD and 20(R)-PPT to produce 20(R)-CK and 20(R)-F1, respectively. GTK1- and GTC1-modified E. coli glycosylated the C3—OH of 20(R)-PPD to form 20(R)-Rh2. Moreover, E. coli containing p2GT95synK1, a recreated two-step glycosylation pathway via the ePathBrich, implemented the successive glycosylation at C20—OH and C3—OH of 20(R)-PPD and yielded 20(R)-F2 in the biotransformation broth.
Conclusion
This study demonstrates that rare 20(R)-ginsenosides can be produced through E. coli engineered with UTG genes.
Keywords: biosynthesis, 20(R)-ginsenosides, ginsenoside, UDP-glycosyltransferase
1. Introduction
Ginsenosides are the main pharmacological active dammarane-type triterpene saponins distributed in the genus Panax, such as Panax ginseng Meyer, Panax quinquefolius L., and Panax notoginseng (Burk.) F. H. Chen [1], [2], [3]. There are nearly 300 saponins reported from Panax species [4]. Those saponins can be classified into six types: protopanaxadiol (PPD), protopanaxatriol (PPT), octillol (OT), oleanolic acid (OA), and C17 side-chain varied and miscellaneous types. Some PPD-type saponins with one to three monosaccharides are limited in plants and are regarded as rare ginsenosides, such as ginsenosides Rh1, Rh2, compound (CK), Rg3, and notoginsenosides ST-4 and Ft1, which showed potent biological activities [5], [6], [7], [8], [9].
According to the chemical configuration at C20, the dammarane-type triterpenoids could also be classified into the 20(S)-type and the 20(R)-type groups. Most of the saponins in Panax plants are in 20(S) type, yet a few of them exist in both 20(S) and 20(R) types, such as ginsenosides Rg3, Rh2, Rh1, Rg2, R2, and notoginsenosides ST-4/Ft1 [4].
Ginsenosides, especially the rare ginsenosides, possess a broad spectrum of bioactivities [10], [11]. For instance, ginsenoside CK was proved to protect against myocardial infarction [12] and inhibits angiogenesis [13]. Ginsenoside F2 showed antitumor [14] and antiobesity [15] activities.
Previous research focused on 20(S)-type ginsenosides, and little attention was paid to 20(R)-type ginsenosides because only a few 20(R)-type ginsenosides were available. In fact, ginsenosides with different chemical configurations often showed inequable biological responses. For example, ginsenoside Rg3 was stereospecific in stimulating the immune response, and 20(R)-Rg3 showed more potent adjuvant activity than 20(S)-Rg3 [16]. Conversely, 20(R)-Rg2 showed a stronger effect on improving cortical neuron cell vitality against oxygen-glucose deprivation/reperfusion (OGD/R)-induced injury than 20(S)-Rg2 [17].
To obtain more active rare ginsenosides, some rich ginsenosides have been used as substrates for conversion via various methods such as heating, mild acid hydrolysis, alkali treatment, as well as microbial and enzymatic biotransformation [18]. As reported, CK was transformed from Rb1 with β-glucosidase [19]. Ginsenosides Rb1, Rb2, and Rc were also selected for use as substrates to prepare CK through biotransformation with microorganisms such as intestinal bacteria [20], fungus [21], and food microorganisms [22]. In our recent studies, the rare notoginsenoside ST-4 [20 (S) type] and Ft1 [20 (R) type] through enzymatic transformation and acid hydrolyzing strategy, respectively [23], [24]. Ginsenoside CK has also been produced using PPD as substrate through microbial metabolic engineering by heterologous expressing uridine diphosphate glycosyltransferase (UGT) in yeast [25]. Although aglycone PPD is an easily available and ideal substrate, there is no report on the biotransformation of the 20(R)-type ginsenosides.
Here, we report the biotransformation of 20(R)-CK, 20(R)-Rh2, and 20(R)-F2 from 20(R)-PPD and 20(R)-F1, 20(R)-Rg1 from 20(R)-PPT through Escherichia coli expressing relevant UGT genes.
2. Materials and methods
2.1. Bacterial strains, vectors, and substrates
E. coli DH5α was utilized to propagate all the plasmids. BL21star (DE3) was used as a host cell for ginsenoside production. The pETM6, an ePathBrick vector and a kind gift from Professor Koffas (Rensselaer Polytechnic Institute, Troy, NY, USA), was used to create the expression constructs. The substrates used in this study were purchased from Chengdu Biopurify Phytochemicals Ltd. (Sichuan, China), and their structures were confirmed by NMR analyses (Fig. S1).
2.2. Gene selection, codon optimization, and synthesis
Seven UGT genes (Table 1) originating from different species were selected according to the functional property of enzymes and chemical profiles of the species [26], [27]. Five (1–5) of them (Table 1) were codon-optimized and synthesized by Life Technologies (Shanghai, China): GT71syn, GT73syn, GT74syn, GT82syn, GT95syn. The other two genes, GTK1 and GTC1, were cloned from Bacillus subtilis using PlantDirect polymerase chain reaction (PCR) kit (HeroGen Biotech, Shanghai, China) with corresponding primers (Table S1). PCR mixtures contained 2.5 μL of DNA sample, 10 μmol/L of each primer, and 25 μL of 2 × Direct PCR mix in 50 μL. Amplification was performed under the following program: 94°C for 5 min; 30 cycles of 94°C for 30 s, 59°C for 30 s, and 72°C for 45 s; and a hold at 72°C for 10 min.
Table 1.
Genes used in the study
| No. | UGT | Origin | GenBank Accession number of the original sequence |
|---|---|---|---|
| 1 | GT71syn1) | Medicago sativa | AY747627 |
| 2 | GT73syn1) | Medicago sativa | AY747626 |
| 3 | GT74syn1) | Vaccaria v. wolf | DQ915168 |
| 4 | GT82syn1) | Panax notoginseng | GU997661 |
| 5 | GT95syn1) | Panax notoginseng | GU997660 |
| 6 | GTK11) | Bacillus subtilis 1.1470 | JX982975 |
| 7 | GTC11) | Bacillus subtilis 1.1470 | JX982974 |
UGT, uridine diphosphate glycosyltransferase.
A number was given to a certain gene for standing for the gene in this study; e.g., GT71 stands for the UGT gene from Medicago sativa.
2.3. Ginsenoside pathway construction with ePathBrick vector
All plasmids were constructed using standard molecular cloning protocols. Six basic (Table 2, 2–8) and one double gene (9) ginsenoside ePathBrick plasmid (Table 2) were constructed according to the literature [28]. Plasmid with an additional copy of GT95syn (p2GT95syn) was also constructed accordingly [29].
Table 2.
Strains and vectors used in this study
| Number | Strain or vector | Relevant properties | Reference |
|---|---|---|---|
| S1 | Escherichia coli DH5α | F−, φ80d lacZΔM15, Δ(lacZYA-argF)U169, recA1, endA1, hsdR17(rk−, mk+), phoA,supE44λ−, thi−1, gyrA96, relA1 | [28] |
| S2 | E. coli BL21 Star (DE3) | F−ompT gal dcm rne131 lon hsdSB (rB_mB_) λ(DE3) | [28] |
| 1 | pETM6 | ColE1 ori, AmpR | [28] |
| 2 | BL21*-pGT71syn | BL21*-pETM6 carrying GT71syn | This study |
| 3 | BL21*-pGT73syn | BL21*-pETM6 carrying GT73syn | This study |
| 4 | BL21*-pGT74syn | BL21*-pETM6 carrying GT74syn | This study |
| 5 | BL21*-pGT82syn | BL21*-pETM6 carrying GT82syn | This study |
| 6 | BL21*-pGT95syn BL21*-p2GT95syn |
BL21*-pETM6 carrying GT95syn BL21*-pETM6 carrying GT95syn-GT95syn |
This study |
| 7 | BL21*-pGTK1 | BL21*-pETM6 carrying GTK1 | This study |
| 8 | BL21*-pGTC1 | BL21*-pETM6 carrying GTC1 | This study |
| 9 | BL21*-p2GT95synK1 | BL21*-pETM6 carrying GT95syn-GT95syn-GTK1 | This study |
2.4. Heterologous expression of the UGT genes in E. coli BL21star (DE3)
The heterologous expression of individual UGT genes in E. coli was analyzed with 8% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). Recombinant E. coli BL21* strains were grown in 1 × M9 medium at 37°C to an absorbance at 600 nm (OD600) of 0.6. Then, exogenous proteins were induced to express with isopropyl-β-d-thioglactopyranoside (IPTG) at a final concentration of 1mM for 3 h at 30°C prior to centrifugation (4,800 g for 15 min). Cells were harvested and resuspended in lysis buffer, and proteins were extracted through ultrasonication. Finally, the cell lysis was centrifuged at 13,000 g for 15 min. The supernatant and cell debris was used for SDS-PAGE analysis as water-soluble and dissoluble proteins, respectively.
2.5. Biotransformation procedure of recombinant E. coli BL21star (DE3)
The recombinant E. coli BL21* strain containing individual construct was used to do the biotransformation according to Zhao et al. [29]. Briefly, a single colony of E. coli was inoculated in Luria-Bertani (LB) liquid medium with 100 μg/mL ampicillin and cultured at 37°C, 215 g for 14–16 h. Then, it was subcultured into 1 × M9 medium and grew at 37°C, 215 g in a horizontal shaker until the OD600 reached 0.6–0.8. Then, 1mM IPTG was added and cultured at 30°C, 215 g for another 3 h. The cell culture was concentrated by centrifugation and resuspended as a high-density culture with an OD600 of about 15 in 1 × M9 medium containing 1mM IPTG, 100 mg/L ampicillin, and 0.3 mg substrate and cultured at 30°C, 215 g for 2 d. To obtain the yield of the products in the medium, the biotransformation broth was extracted with n-butyl alcohol at a 1:1 ratio three times, and the n-butyl alcohol layer was collected and evaporated for UPLC-electrospray ionization(ESI)-MS analysis. E. coli BL21* with pETM6 vector served as negative control.
2.6. Concentration calculation and UPLC-ESI-MS analysis
The ginsenosides used as substrates were dissolved in dimethyl sulfoxide as stock solution of 10 mg/mL, and 30 μL of the substrate solution was added in 1 mL medium as a supersaturated solution for cell cultures. To calculate the concentration of substrates, the medium was then centrifuged at 13,000 g for 15 min, and the supernatant was used for UPLC-ESI-MS analysis. The conversion rate of each substrate with the corresponding enzyme was calculated according to the following formula:
| Conversion rate = wp/ws × 100% | (1) |
where wp is the amount of product and ws is the amount of substrate dissolved in the medium.
Biotransformation broth was analyzed with an Agilent 1290 series UPLC and an Agilent 6410 Triple Quadrupole mass spectrometer equipped with an ESI source (Agilent Technologies, Waldbronn, Germany). Chemical identification and concentration assay were performed with an ACQUITY UPLC BEH C18 column (50 mm × 2.1 mm i.d., 1.7 μm; Waters, Massachusetts, USA) at 55°C using a mobile phase of 0.1% formic acid (A) and acetonitrile (B) at a flow rate of 0.4 mL/min. The gradient elution (B) was as follows: 0–1 min (20–25%), 1–4 min (25–34%), 4–6 min (34–52%), 6–6.1 min (52–95%), and 6.1–8 min (95%). As reported, C-18 column was practicable to separate the (R) and (S) configuration of ginsenosides [30]. To do the chirality assay of related compounds, an optimized condition with Ultimate UHPLC XB-C18 column [100 mm × 2.1 mm (i.d.), 1.8 μm; Welch, Massachusetts, USA] was established using 0.1% formic acid (A) and acetonitrile (B) at a flow rate of 0.3 mL/min at 45°C. The gradient elution (B) was as follows: 0–2 min (2–8%), 2–5 min (8–18%), 5–9 min (18–28%), 9–14 min (28–60%), 14–19 min (60–90%), and 19–25 min 90%. Mass spectrometric analysis was performed in the negative ion multiple reaction monitoring mode with 3.4-kV capillary voltage for all experiments except for PPD analysis, which uses the positive ion multiple reaction monitoring mode. The transitions were set at m/z 425.4 → 217 for PPD, at m/z 475.4 → 391.4 for PPT, at m/z 621.3 → 161.1 for CK and Rh2, at m/z 829.3 → 621.4 for F2, at m/z 637.5 → 475.5 for F1, at m/z 845.5 → 637.4 for Rg1, and at m/z 945.5 → 783.3 for Rd, respectively.
3. Results and discussion
3.1. Glycosylation of the C20 hydroxyl of PPD/PPT- and PPD/PPT-type ginsenosides
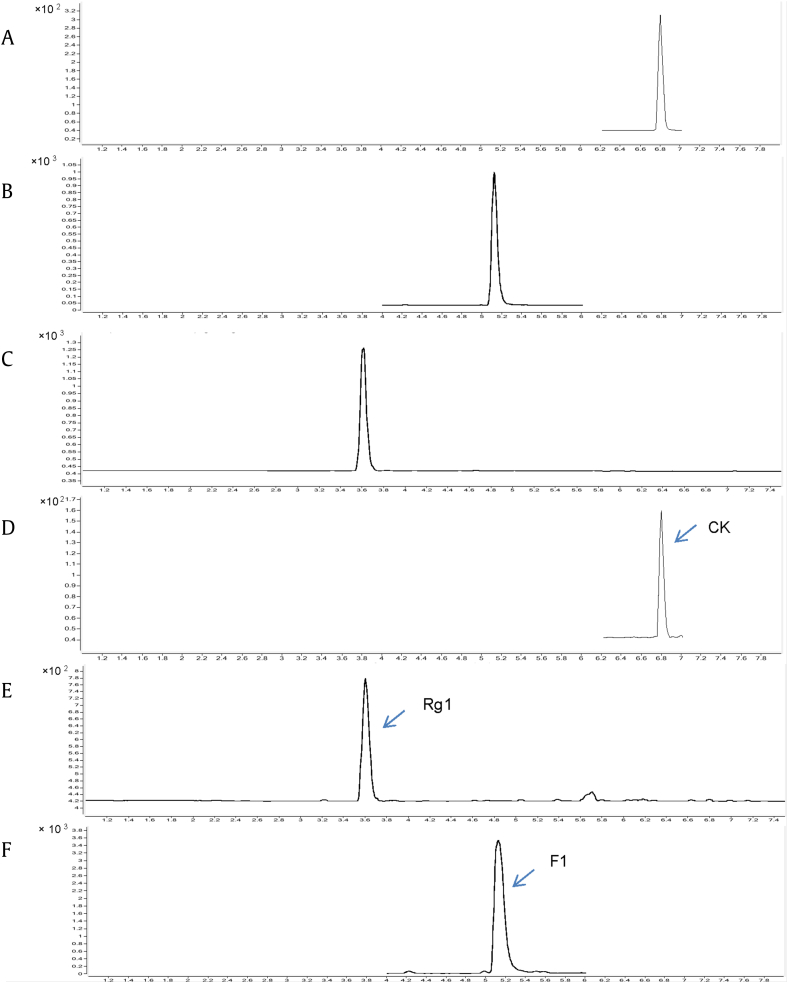
Among the seven UGTs, GT95syn showed the highest activity in glycosylation of 20(R)-PPD and PPD-type ginsenosides with a free C20 hydroxyl in E. coli. When 20(R)-PPD was used as substrate, BL21*-pGT95syn produced 20(R)-CK as monitored by UPLC-ESI-MS (Fig. 1). 20(R)-CK was accumulated in the biotransformation broth at 0.49 mg/L. The concentration of 20(R)-CK increased to 0.57 mg/L when using E. coli containing construct with additional copies of GT95syn(BL21*-p2GT95syn). Thus, BL21*-p2GT95syn was selected for further studies. BL21*-p2GT95syn could also convert 20(R)-PPT into 20(R)-F1 and a small amount of 20(R)-Rg1 (Fig. 1) when using 20(R)-PPT as substrate.
Fig. 1.
Typical UPLC-MS chromatograms. Reference substance: (A) ginsenoside CK; (B) ginsenoside F1; (C) ginsenoside Rg1; (D) BL21*-p2GT95syn converts 20(R)-PPD into 20(R)-CK; (E) BL21*-p2GT95syn converts 20(R)-PPT into 20(R)-Rg1; and (F) 20(R)-F1. CK, compound K; PPD, protopanaxadiol; PPT, protopanaxatriol.
Current reports about UGTs mostly focus on the glycosylation of 20(S)-PPD, and some UGTs can only catalyze 20(S)-PPD-type substrates [25], [31], [32]. This study demonstrated that GT95syn glycosylated both 20(S)- and 20(R)-PPD. It could also utilize 20(S)-PPT, the stereoisomer of 20(R)-PPT, as substrate and formed 20(S)-F1.
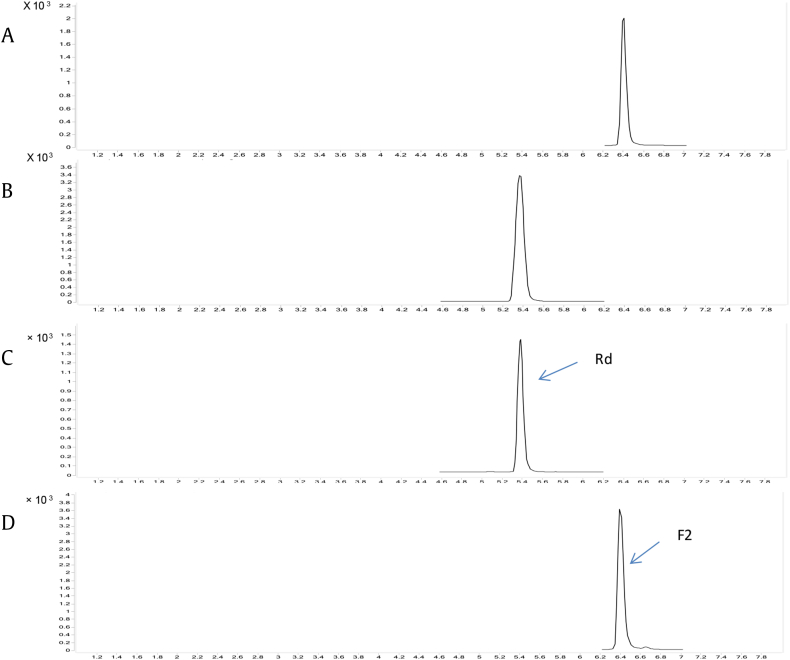
To investigate whether GT95syn had the site-specific selectivity to the free hydroxyl, we performed biotransformation with four different structure ginsenosides—20(S)-Rg3, 20(S)-Rh2, 20(S)-CK, and 20(S)-F2—as substrates. All four compounds are PPD-type ginsenosides but with different groups at C20 and C3 positions. Both 20(S)-Rg3 and 20(S)-Rh2 have a free hydroxyl at C20 site, and 20(S)-Rg3 possesses two glucosyl groups at C3 site whereas 20(S)-Rh2 has one at C3 site. The C20 sites of both 20(S)-CK and 20(S)-F2 are occupied by a glucosyl group. 20(S)-CK has a free hydroxyl at C3 site and 20(S)-F2 has a glucosyl group at C3 site. When incubation of BL21*-p2GT95syn with these compounds individually, only 20(S)-Rg3 and 20(S)-Rh2 were transformed into their corresponding glycosylated products, 20(S)-Rd and 20(S)-F2 (Fig. 2).
Fig. 2.
Typical UPLC-MS chromatograms. Reference substance: (A) ginsenoside F2; (B) ginsenoside Rd; (C) BL21*-p2GT95syn converts 20(S)-Rg3 into 20(S)-Rd; and (D) BL21*-p2GT95syn converts 20(S)-Rh2 into 20(S)-F2.
Taken together, these results indicate that GT95syn can specifically glycosylate the 20(S) and 20(R)-stereo configuration PPD- and PPD-type ginsenosides with a free hydroxyl at C20 site, and the glycosylation reaction is not affected by the glucosyl groups at C3 site, even though the water-soluble protein of this enzyme was not obvious in the SDS-PAGE profiles (Fig. S2). When the C20 site is already linked with glucosyl groups, GT95syn could not glycosylate the free hydroxyl at the C3 site instead or assemble additional glucosyl group into the glucosyl groups already linked with the PPD core structure.
Two other UGTs from Panax ginseng, UGTPg1 and UGT71A27, were reported to have a similar catalytic property to GT95syn. UGT71A27, which has 94.74% identity with GT95syn at the amino acid level (Fig. S3B), could only convert 20(S)-PPD into 20(S)-CK [32], and UGTPg1 had a specificity to glycosylate the hydroxyl at C20 site of 20(S)-PPD but not 20(R)-PPD [25]. In addition, UGTPg1 only glycosylated the C20—OH, but not the C6—OH of 20(S)-PPT. As shown above, GT95syn glycosylated the C20—OH of PPD and both the C6- and C20—OH of PPT. UGTPg1 had high homology (98.11% identity) with GT95syn (Fig. S3A). We attempted to construct the molecular models of UGTPg1 and GT95syn using the SWISS-MODEL homology modeling software (Swiss Institute of Bioinformatics, Basel, Switzerland; https://www.swissmodel.expasy.org) to explain the formation of stereospecific ginsenosides. Yet, the homology modeling report showed that the templates of the two UGTs were the same one, namely, triterpene uridine diphosphate (UDP)-glucosyl transferase UGT71G1 (PDB: 2ACV, Fig. S4). Therefore, the primary structures of UGTPg1 and GT95syn were compared with each other to explain the difference between the two enzymes. As displayed in Fig. S3A, only eight amino acid residues were different between them, which were T71, Q110, S281, K324, S355, K410, D411, N438, and A455 in UGTPg1 corresponding to S71, R110, T281, E324, A355, N410, E411, K438, and L455 in GT95, respectively. It is possible that these eight amino acids cooperated together and determined the substrate stereospecific and free hydroxyl site-specific properties of these UTGs.
The E. coli engineered with constructs containing the other individual UGT genes—GT71syn, GT73syn, GT74syn, and GT82syn—did not produce any glycosylated products with either PPD or PPT as substrates. As shown in typical SDS-PAGE profiles (Fig. S5), all five genes were expressed at high level as inclusion bodies (Fig. S5B), whereas only GT71 and GT73 were also expressed as water-soluble proteins and the water-soluble proteins of GT74, GT82, and GT95 were undetectable with SDS-PAGE (Fig. S5A); even all the sequences had been codon-optimized. Despite this, GT95 still exhibited the highest glycosylation activity toward PPD and PPT in E. coli. The amino acid alignment of the UGTs used in this study is listed in Fig. S6. The different amino acid residues may contribute to the divergent expression property and catalytic activity in E. coli host cells.
3.2. Glycosylation of the C3 hydroxyl of PPD- and PPD-type ginsenosides
UGTs were reported to exist widely in nature, and two UGTs (YojK1 and YjiC1) from B. subtilis were identified to have catalytic activity toward 20(S)-Rh1, which is a PPT-type ginsenoside. The PPD-type ginsenosides, another primary group of saponins in P. notoginseng, were not studied in that work [27]. To explore the catalytic activity toward PPD and PPD-type ginsenosides, two recombinant E. coli strains containing either YojK1 (GTK1) or YjiC1 (GTC1) genes were constructed to do the biotransformation.
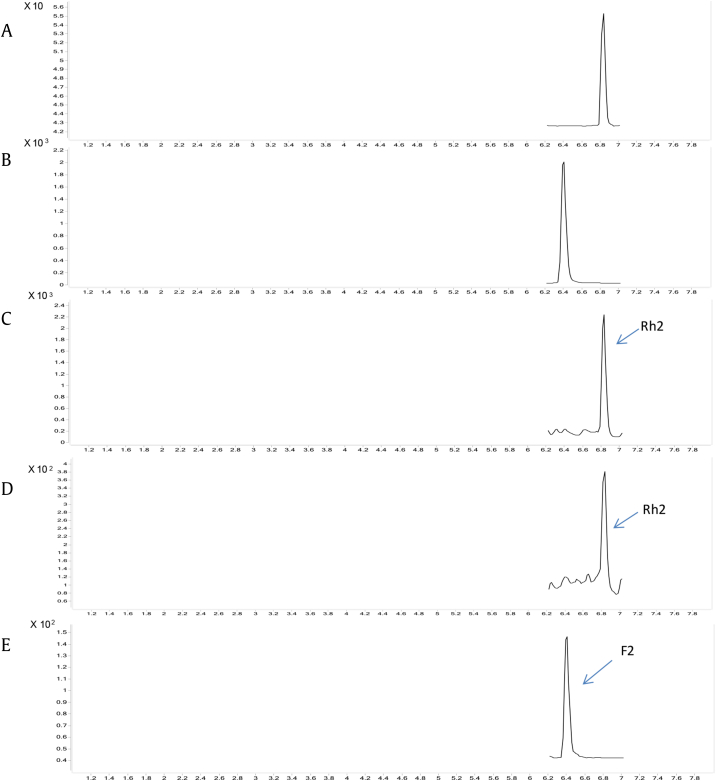
As monitored by UPLC-ESI-MS, both GTK1 and GTC1 glycosylated the C3—OH of 20(R)-PPD to 20(R)-Rh2 (Fig. 3). However, neither of them could catalyze the free C3—OH of 20(R)-PPT, which could be regarded as a C6 hydroxylated product of 20(R)-PPD. It appeared that the hydroxyl at C6 site of 20(R)-PPT affected either the catalytic activity or the substrate affinity of GTK1 and GTC1 to some extent. Moreover, these two UGTs also catalyzed 20(S)-PPD to form 20(S)-Rh2. The site-specific selectivity of GTK1 and GTC1 to the free hydroxyl was also studied. Out of the four different ginsenosides tested, PPD, CK, Rh2, and F2, BL21*-pGTK1, and BL21*-pGTC1 converted PPD and CK to Rh2 and F2, respectively, but neither could further utilize Rh2 or F2 as substrates.
Fig. 3.
Typical UPLC-MS chromatograms. Reference substance: (A) 20(R)-ginsenoside Rh2; (B) 20(S)-ginsenoside F2; (C) BL21*-pGTK1 converts 20(R)-PPD into 20(R)-Rh2; (D) BL21*-pGTC1 converts 20(R)-PPD into 20(R)-Rh2; and (E) BL21*-p2GT95synK1 converts 20(R)-PPD into 20(R)-F2. PPD, protopanaxadiol; PPT, protopanaxatriol.
As depicted above, these ginsenosides have different groups at C20 and C3 positions. These results suggested that the two UGTs could also catalyze PPD- and PPD-type ginsenosides, and only specifically glycosylate the C3—OH, but not the C20—OH, of the substrates. UPLC-ESI-MS analysis also showed that GTK1 had higher conversion efficiency compared with GTC1, as listed in Table 3. Taking these factors together, we used GTK1 for the next studies.
Table 3.
Production of ginsenosides in four genetically engineered strains
| Substrate (concentration in medium, mg/L) | Products (yield, mg/L and conversion, w/w%) |
|||
|---|---|---|---|---|
| BL21*-p2GT95 | BL21*-pGTK1 | BL21*-pGTC1 | BL21*-p2GT95K1 | |
| 20(R)-PPD (3.64 ± 0.32) | 20(R)-CK (0.57 ± 0.04, 15.7%) | 20(R)-Rh2 (0.13 ± 0.007, 3.6%) | 20(R)-Rh2 (0.06 ± 0.01, 1.6%) | 20(R)-CK (0.44 ± 0.04, 12.1%) 20(R)-F2 (0.06 ± 0.01, 1.6%) |
| 20(R)-Rh23) | 20(R)-F2 (1.08 ± 0.21) | −1) | – | −2) |
| 20(R)-Rg34) | 20(R)-Rd (0.01) | – | – | — |
| 20(R)-PPT (19.53 ± 0.68) | 20(R)-F1 (6.48 ± 0.03, 33.2%) 20(R)-Rg1 (0.2 ± 0.01, 1.0%) |
– | – | – |
| 20(S)-PPD (3.64 ± 0.29) | 20(S)-CK (0.36 ± 0.07, 9.9%) | 20(S)-Rh2 (0.77 ± 0.02, 21.2%) | 20(S)-Rh2 (0.3 ± 0.03, 8.2%) | 20(S)-CK (0.61 ± 0.06, 16.8%) 20(S)-F2 (0.36 ± 0.01, 9.9%) |
| 20(S)-Rh25) | 20(S)-F2 (1.46 ± 0.12) | – | – | — |
| 20(S)-Rg3 (6.71 ± 0.11) | 20(S)-Rd (0.36 ± 0.04, 5.4%) | – | – | — |
| 20(S)-PPT (23.31 ± 0.43) | 20(S)-F1 (6.34 ± 0.18, 27.2%) 20(S)-Rg1 (0.23 ± 0.01, 1.0%) |
– | – | — |
| 20(S)-CK (5.47 ± 0.20) | – | 20(S)-F2 (0.62 ± 0.12, 11.3%) | 20(S)-F2 (0.36 ± 0.03, 6.6%) | — |
CK, compound K; ESI, electrospray ionization; PPD, protopanaxadiol; PPT, protopanaxatriol; UPLC, ultra performance liquid chromatography.
1) –, not produced.
2) —, the experiment was not implemented.
3)–5) These substrates had low solubility in water, which was not detected by (UPLC-ESI-MS).
3.3. Glycosylation of both C3 and C20 hydroxyl of PPD
Considering that GT95syn could convert PPD to form CK and GTK1 could catalyze CK to form F2, we constructed p2GT95synK1, an ePathBrick expression vector harboring two copies of GT95syn and one copy of GTK1 gene, with the aim of recreating a two-step glycosylation pathway from 20(R)-PPD to CK then to 20(R)-F2, or from 20(R)-PPD to 20(R)-Rh2 then to 20(R)-F2 (Fig. 4). As determined with UPLC-ESI-MS, E. coli BL21* engineered with p2GT95synK1 (BL21*-p2GT95synK1) yielded 20(R)-F2 when using 20(R)-PPD as substrate (Fig. 3). However, only CK was detected as the intermediate product but not Rh2. This result demonstrated that this recreated two-step pathway worked as expected, simultaneously glycosylating 20(R)-PPD to CK then to 20(R)-F2 in the engineered E. coli. Similar to BL21*-p2GT95syn and BL21*-pGTK1, BL21*-p2GT95synK1 also exhibited the ability to convert 20(S)-PPD to 20(S)-F2 (Table 3).
Fig. 4.
The supposed biosynthetic pathway of 20(R)-type ginsenosides in UGT-modified Escherichia coli. Plain arrows indicate the pathway that really happened in the biotransformation. Dash arrows represent the deduced pathway that theoretically existed, but no 20(R)-ginsenoside Rh2 was detected in the broth. UGT, uridine diphosphate glycosyltransferase.
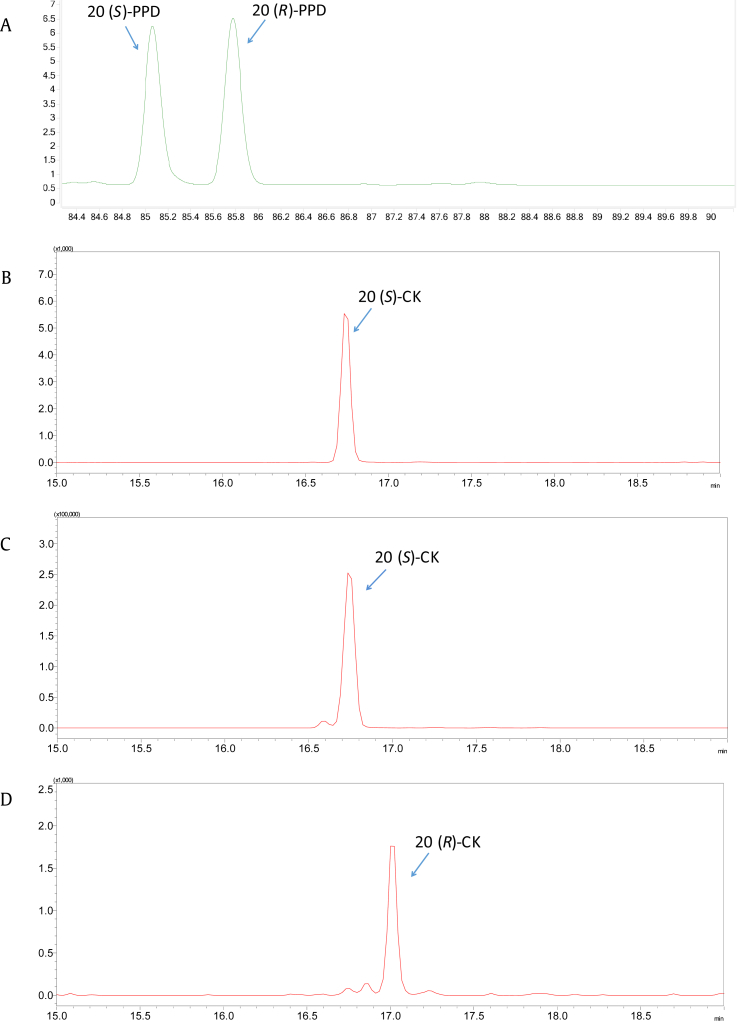
To investigate whether GT95syn would change the chirality at C-20 site of the ginsenosides, the biotransformation products of GT95syn were analyzed with UPLC-ESI-MS under an optimized condition separately using 20(S)- and 20(R)-PPD as substrates. As shown (Fig. 5), GT95syn converted 20(S)-PPD into 20(S)-CK and 20(R)-PPD into 20(R)-CK, respectively, demonstrating that GT95syn did not change the chirality of C-20 site. Similar results were observed in GTK1. When 20(R)-PPD was used as a substrate, GTK1 converted 20(R)-PPD to 20(R)-Rh2 (Fig. S7), proving the CTK1 also did not change the chirality of C-20 of ginsenosides.
Fig. 5.
Typical UPLC-MS chromatograms under the optimized condition. Reference substance: (A) 20(S)-PPD, 20(R)-PPD; (B) 20(S)-CK; (C) BL21*-p2GT95syn converts 20(S)-PPD into 20(S)-CK; and (D) BL21*-p2GT95syn converts 20(R)-PPD into 20(R)-CK. CK, compound K; PPD, protopanaxadiol; PPT, protopanaxatriol.
At present, most studies on UGTs are concentrated on the catalyzing capability using purified recombinant proteins with the much more expensive active UDP-glucose as substrate in vitro [23], [28]. By taking advantage of the host E. coli cells to synthesize the active UDP-glucose with its own UDP and supplemented glucose, PPD or PPT could be converted into the corresponding glycosylated products with UGT-modified E. coli in vivo, which provides a much more convenient and economical method to explore the activity of UGTs.
Biotransformation optimization experiments had been conducted to improve the yield of products, including the concentration of IPTG, the induction temperature, pH value, and fermentation time, which showed that biotransformation with 1mM IPTG, pH value ranging from 6.5 to 7.0 at 30 °C was optimal except that the production of 20(S)-CK from 3 d was slightly better than that from 2 d (Fig. S8). Taking these factors together, we carried out the biotransformation of other substrates and enzymes with 1mM IPTG, pH 7.0 at 30 °C for 2 d. The poor water solubility of these substrates may lead to the relatively low production of ginsenosides. To improve the solubility of these ginsenosides, several efforts have been attempted such as adding 0.1–2% Tween-80 in the medium and by applying cyclodextrin camplexation techniques for PPD. Yet, no positive results were obtained (data not shown). Moreover, excess substrates were added in the medium in order to promote the biotransformation reaction, and the yield of product increased with the increasing amount of the substrate fed in the medium (data not shown). Structural modifications for these substrates to increase the water solubility may be helpful to increase the production of ginsenosides.
In addition, the specific catalytic activities of UGTs reported here provided candidate genes to produce specific ginsenosides through engineering of ginsenoside biosynthetic pathway when combined with other biosynthetic genes in microorganisms, plants, or other organisms, as shown in this study (i.e., when GT95syn and GTK1 were combined, a 2-step pathway worked as expected).
4. Conclusion
In general, the present study demonstrated that GT95syn could glycosylate the C20—OH of PPD, and the C20—OH and C6—OH of PPT in E. coli host cells. We also proved that GTK1 could glycosylate the C3—OH of PPD, and the recreated two-step glycosylation pathway via combination of GT95syn and GTK1 implemented the biosynthesis of F2 from PPD in E. coli. Both GT95syn and GTK1 could catalyze 20(S)- and 20(R)-configuration substrates to the corresponding rare 20(S)- and 20(R)-ginsenosides.
These results implied that rare ginsenosides could be produced through E. coli engineered with recreated exogenous biosynthetic pathway with ordinary substrates such as PPD and PPT.
Acknowledgments
This work was financially supported by National Natural Science Foundation of China (81603229, U1032604) and China Postdoctoral Science Foundation (2016T90382, 2015M580349, 2015M581654).
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.jgr.2017.09.005.
Contributor Information
Shujuan Zhao, Email: zhaoshujuan@126.com, zhaoshujuan@shutcm.edu.cn.
Zhengtao Wang, Email: ztwang@shutcm.edu.cn.
Conflicts of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Nicol R.W., Traquair J.A., Bernards M.A. Ginsenosides as host resistance factors in American ginseng (Panax quinquefolius) Can J Bot. 2011;80:557–562. [Google Scholar]
- 2.Cho W.C.S., Chung W.S., Lee S.K.W., Leung A.W.N., Cheng C.H.K. Ginsenoside Re of Panax ginseng possesses significant antioxidant and antihyperlipidemic efficacies in streptozotocin-induced diabetic rats. Eur J Pharmacol. 2006;550:173–179. doi: 10.1016/j.ejphar.2006.08.056. [DOI] [PubMed] [Google Scholar]
- 3.Wang T., Guo R.X., Zhou G.H., Zhou X.D., Kou Z.Z., Sui F., Li C., Tang L.Y., Wang Z.J. Traditional uses, botany, phytochemistry, pharmacology and toxicology of Panax notoginseng (Burk.) F.H. Chen: a review. J Ethnopharmacol. 2016;188:234–258. doi: 10.1016/j.jep.2016.05.005. [DOI] [PubMed] [Google Scholar]
- 4.Yang W.Z., Hu Y., Wu W.Y., Ye M., Guo D.A. Saponins in the genus Panax L. (Araliaceae): a systematic review of their chemical diversity. Phytochemistry. 2014;106:7–24. doi: 10.1016/j.phytochem.2014.07.012. [DOI] [PubMed] [Google Scholar]
- 5.Gao B., Huang L.F., Liu H.S., Wu H., Zhang E.Y., Yang L., Wu X.J., Wang Z.T. Platelet P2Y12 receptor involved in the hemostatic effect of notoginsenoside Ft1, a saponin isolated from Panax notoginseng. Br J Pharmacol. 2014;171:214–223. doi: 10.1111/bph.12435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao B., Shi H.L., Li X., Qiu S.P., Wu H., Zhang B.B., Wu X.J., Wang Z.T. p38 MAPK and ERK1/2 pathways are involved in the pro-apoptotic effect of notoginsenoside Ft1 on human neuroblastoma SH-SY5Y cells. Life Sci. 2014;108:63–70. doi: 10.1016/j.lfs.2014.05.010. [DOI] [PubMed] [Google Scholar]
- 7.Shen K.K., Ji L.L., Gong C.Y., Ma Y.B., Yang L., Fan Y., Hou M.Q., Wang Z.T. Notoginsenoside Ft1 promotes angiogenesis via HIF-1α mediated VEGF secretion and the regulation of PI3K/AKT and Raf/MEK/ERK signaling pathways. Biochem Pharmacol. 2012;84:784–792. doi: 10.1016/j.bcp.2012.05.024. [DOI] [PubMed] [Google Scholar]
- 8.Shen K.K., Leung S.W.S., Ji L.L., Huang Y., Hou M.Q., Xu A.M., Wang Z.T., Vanhoutte P.M. Notoginsenoside Ft1 activates both glucocorticoid and estrogen receptors to induce endothelium-dependent, nitric oxide-mediated relaxations in rat mesenteric arteries. Biochem Pharmacol. 2014;88:66–74. doi: 10.1016/j.bcp.2014.01.007. [DOI] [PubMed] [Google Scholar]
- 9.Zhang J.J., Ding L.L., Wang B.C., Ren G.Y., Sun A.N., Deng C., Wei X.H., Mani S., Wang Z.T., Dou W. Notoginsenoside R1 attenuates experimental inflammatory bowel disease via pregnane X receptor activation. J Pharmacol Exp Ther. 2015;352:1–10. doi: 10.1124/jpet.114.218750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deng J., Jiang Y.X., Ying-Li C., Chen X.Q. Effects of ginsenoside CK on gastric cancer SGC-7901 cell line and endogenous VEGF secreted by tumor cells. Cancer Res Prev Treat. 2011;38:17–20. [Google Scholar]
- 11.Oh M., Choi Y.H., Choi S., Chung H., Kim K. Anti-proliferating effects of ginsenoside Rh2 on MCF-7 human breast cancer cells. Int J Oncol. 1999;14:869–875. doi: 10.3892/ijo.14.5.869. [DOI] [PubMed] [Google Scholar]
- 12.Tsutsumi Y.M., Tsutsumi R., Mawatari K., Nakaya Y., Kinoshita M., Tanaka K., Oshita S., Compound K. A metabolite of ginsenosides, induces cardiac protection mediated nitric oxide via Akt/PI3K pathway. Life Sci. 2011;88:725–729. doi: 10.1016/j.lfs.2011.02.011. [DOI] [PubMed] [Google Scholar]
- 13.Shin K.O., Seo C.H., Cho H.H. Ginsenoside compound K inhibits angiogenesis via regulation of sphingosine kinase-1 in human umbilical vein endothelial cells. Arch Pharmacol Res. 2014;37:1183–1192. doi: 10.1007/s12272-014-0340-6. [DOI] [PubMed] [Google Scholar]
- 14.Mai T.T., Moon J.Y., Song Y.W. Ginsenoside F2 induces apoptosis accompanied by protective autophagy in breast cancer stem cells. Cancer Lett. 2012;321:144–153. doi: 10.1016/j.canlet.2012.01.045. [DOI] [PubMed] [Google Scholar]
- 15.Siraj F.M., SathishKumar N., Kim Y.J. Ginsenoside F2 possesses anti-obesity activity via binding with PPAR and inhibiting adipocyte differentiation in the 3T3-L1 cell line. J Enzym Inhib Med Ch. 2014;30:9–14. doi: 10.3109/14756366.2013.871006. [DOI] [PubMed] [Google Scholar]
- 16.Wei X.J., Chen J., Su F., Su X.Y., Hu T.J., Hu S.H. Stereospecificity of ginsenoside Rg3 in promotion of the immune response to ovalbumin in mice. Int Immunol. 2012;24:465–471. doi: 10.1093/intimm/dxs043. [DOI] [PubMed] [Google Scholar]
- 17.Pi M.S., Ru Q., Gong X.K., Wu R.H., Tian X., Xiong Q., Li C.Y. Effect of ginsenoside Rg2 and its stereoisomers on oxygen-glucose deprivation and reperfusion induced cortical neuronal injury model. Chin J Integr Tradit West Med. 2016;36:333–338. [PubMed] [Google Scholar]
- 18.Yang X.D., Yang Y.Y., Ouyang D.S., Yang G.P. A review of biotransformation and pharmacology of ginsenoside compound K. Fitoterapia. 2015;100:208–220. doi: 10.1016/j.fitote.2014.11.019. [DOI] [PubMed] [Google Scholar]
- 19.Yan Q., Zhou X.W., Zhou W., Li X.W., Feng M.Q., Zhou P. Purification and properties of a novel beta-glucosidase, hydrolyzing ginsenoside Rb1 to CK, from Paecilomyces bainier. J Microbiol Biotechnol. 2008;18:1081–1089. [PubMed] [Google Scholar]
- 20.Bae E.A., Choo M.K., Park E.K., Park S.Y., Shin H.Y., Kim D.H. Metabolism of ginsenoside R(c) by human intestinal bacteria and its related antiallergic activity. Biol Pharm Bull. 2002;25:743–747. doi: 10.1248/bpb.25.743. [DOI] [PubMed] [Google Scholar]
- 21.Zhou W., Feng M.Q., Li J.Y., Zhou P. Studies on the preparation, crystal structure and bioactivity of ginsenoside compound K. J Asian Nat Prod Res. 2006;8:519–527. doi: 10.1080/10286020500208600. [DOI] [PubMed] [Google Scholar]
- 22.Quan L.H., Kim Y.J., Li G.H., Choi K.T., Yang D.C. Microbial transformation of ginsenoside Rb1 to compound K by Lactobacillus paralimentarius. World J Microbiol Biotechnol. 2013;29:1001–1007. doi: 10.1007/s11274-013-1260-1. [DOI] [PubMed] [Google Scholar]
- 23.Wang R.F., Zheng M.M., Cao Y.D., Li H., Li C.X., Xu J.H., Wang Z.T. Enzymatic transformation of vina-ginsenoside R7 to rare notoginsenoside ST-4 using a new recombinant gylcoside hydrolase from Herpetosiphon aurantiacus. Appl Microbiol Biotechnol. 2015;99:3433–3442. doi: 10.1007/s00253-015-6446-z. [DOI] [PubMed] [Google Scholar]
- 24.Wang R.F., Li J., Hu H.J., Li J., Yang Y.B., Yang L., Wang Z.T. Chemical transformation and target preparation of saponins in stems and leaves of Panax notoginseng. J Ginseng Res. 2018;42:270–276. doi: 10.1016/j.jgr.2016.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yan X., Fan Y., Wei W., Wang P.P., Liu Q.F., Wei Y.J., Zhang L., Zhao G.P., Yue J.M., Zhou Z.H. Production of bioactive ginsenoside compound K in metabolically engineered yeast. Cell Res. 2014;24:770–773. doi: 10.1038/cr.2014.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu M.H., Yang B.R., Cheung W.F., Yang K.Y., Zhou H.F., Kwok J.S., Liu G.C., Li X.F., Zhong S., Lee S.M. Transcriptome analysis of leaves, roots and flowers of Panax notoginseng identifies genes involved in ginsenoside and alkaloid biosynthsis. BMC Genomics. 2015;16:1–12. doi: 10.1186/s12864-015-1477-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo S.L., Dang L.Z., Zhang K.Q., Liang L.M., Li G.H. Cloning and heterologous expression of UDP-glycosyltransferase genes from Bacillus subtilis and its application in the glycosylation of ginsenoside Rh1. Lett Appl Microbiol. 2014;60:72–78. doi: 10.1111/lam.12339. [DOI] [PubMed] [Google Scholar]
- 28.Xu P., Vansiri A., Bhan N., Koffas M.A.G. ePathBrick: a synthetic biology platform for engineering metabolic pathways in E. coli. ACS Synth Biol. 2012;1:256–266. doi: 10.1021/sb300016b. [DOI] [PubMed] [Google Scholar]
- 29.Zhao S.J., Jones J.A., Lachance D.M., Bhan N., Khalidi O., Venkataraman S., Wang Z.T., Koffas M.A. Improvement of catechin production in Escherichia coli through combinatorial metabolic engineering. Metab Eng. 2015;28:43–53. doi: 10.1016/j.ymben.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 30.Liu L., Zhu X.M., Wang Q.J., Zhang D.L., Fang Z.M., Wang C.Y., Wang Z., Sun B.S., Wu H., Sung C.K. Enzymatic preparation of 20(S, R)-protopanaxadiol by transformation of 20(S,R)-Rg3 from black ginseng. Phytochemistry. 2010;71:1514–1520. doi: 10.1016/j.phytochem.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 31.Wang P.P., Wei Y.J., Fan Y., Liu Q.F., Wei W., Yang C.S., Zhang L., Zhao G.P., Yue J.M., Yan X. Production of bioactive ginsenosides Rh2 and Rg3 by metabolically engineered yeasts. Metab Eng. 2015;29:97–105. doi: 10.1016/j.ymben.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 32.Jung S.C., Kim W., Park S.C., Jeong J., Park M.K., Lim S., Lee Y., Im W.T., Lee J.H., Choi G. Two ginseng UDP-glycosyltransferases synthesize ginsenoside Rg3 and Rd. Plant Cell Physiol. 2014;55:2177–2188. doi: 10.1093/pcp/pcu147. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.