Abstract
Accumulation of unfolded proteins in the endoplasmic reticulum (ER) initiates a stress response mechanism to clear out the unfolded proteins by either facilitating their re-folding or inducing their degradation. When this fails, an apoptotic cascade is initiated so that the affected cell is eliminated. IRE1α is a critical sensor of the unfolded-protein response, essential for initiating the apoptotic signaling. Here, we report an infantile neurodegenerative disorder associated with enhanced activation of IRE1α and increased apoptosis. Three unrelated affected individuals with congenital microcephaly, infantile epileptic encephalopathy, and profound developmental delay were found to carry heterozygous variants (c.932T>C [p.Leu311Ser] or c.935T>C [p.Leu312Pro]) in RNF13, which codes for an IRE1α-interacting protein. Structural modeling predicted that the variants, located on the surface of the protein, would not alter overall protein folding. Accordingly, the abundance of RNF13 and IRE1α was not altered in affected individuals’ cells. However, both IRE1α-mediated stress signaling and stress-induced apoptosis were increased in affected individuals’ cells. These results indicate that the RNF13 variants confer gain of function to the encoded protein and thereby lead to altered signaling of the ER stress response associated with severe neurodegeneration in infancy.
Keywords: RNF13, microcephaly, epilepsy, XBP1 splicing, ER stress
Main Text
Apoptosis is a cellular response to stress conditions such as the accumulation of unfolded proteins in the endoplasmic reticulum (ER). Under this stress, an intracellular unfolded-protein response (UPR) is activated, triggering a cascade of signaling events resulting in attenuation of protein synthesis and transcriptional upregulation of genes encoding ER chaperones, folding enzymes, and ER-associated degradation (ERAD) components. When UPR fails, an apoptotic cascade is triggered, leading to cell death.1 Mammalian UPR is initiated by the activation of three ER stress sensors: protein kinase RNA-like ER kinase (PERK), activating transcription factor 6 (ATF6), and inositol-requiring transmembrane kinase/endonuclease (IRE1α).2 We now report on an infantile neurodegenerative disorder associated with enhanced activation of the ER stress sensor IRE1α and increased apoptosis.
Of the three affected individuals, all of whom were males, two are alive at 2 and 8 years of age, respectively, whereas the third died of sepsis at 33 months. The affected individuals originated from three unrelated families; their clinical and radiologic data are summarized in Table 1, and an illustrative clinical description is included in the Supplemental Note. Abnormally high α-fetoprotein in maternal blood was noted in the only pregnancy tested (individual 1). Pregnancies were otherwise uneventful for all three affected individuals. The affected individuals were born at term; although birth weight varied, the head circumference was invariably low (−2 SD to −3.1 SD). The perinatal course was characterized by feeding difficulties, restlessness, and abnormally increased muscle tone. Tonic, clonic, and myoclonic seizures were first documented at 7 weeks to 7 months of age and were resistant to multiple drugs. An electroencephalogram (EEG) and video EEG revealed a slowing of background activity, with interictal bilateral nonsynchronous spikes and sharp waves, as well as focal ictal discharges correlating with tonic and clonic seizures that progressed to secondary generalization. All affected individuals had cortical visual impairment accompanied by roving eye movements and a pupillary response to light but failure to fixate or track moving objects; fundoscopy was essentially normal. All affected individuals had bilateral hearing loss. An auditory brainstem response test in affected individuals 1 and 3 indicated profound sensorineural deafness with prolonged latencies and slow conductance. Affected individual 1 was treated with cochlear implants, but the subsequent response to noise was inconsistent. None of the affected individuals achieved any developmental milestones, and all three showed no voluntary movements and no communication. Increased muscle tone, limb contractures, and scoliosis (Figure 1) were invariably present. The affected individuals were fed via gastrostomy but showed failure to thrive and microcephaly; head circumference ranged from −5.5 SD (affected individual 2) to −2 SD (affected individual 3) according to the latest measurements.
Table 1.
Clinical and Radiologic Features and RNF13 Variant in the Three Affected Individuals
| Pt/Sex/Age | Head Circumference at Birth (cm) (Percentile) | Presenting Symptoms | Age at First Seizure/ Types | Intellectual Disability | Hearing | Other Clinical Findings | Brain MRI | Hetero-zygous Variant in RNF13 |
|---|---|---|---|---|---|---|---|---|
| 1/M/died at 33 months | 31.5 (3rd percentile) | feeding difficulties, irritability, increased tone |
7 weeks/multiple | profound | deaf bilaterally | contractures, FTT, scoliosis | thin CC | p.Leu311Ser |
| 2/M/8 years | 30.0 (−2.6 SD) | respiratory distress, irritability, low central tone, arching, feeding difficulties | 7 months/ infantile spasms | profound | normal | dysmorphic features,a FTT, Inguinal hernia, contractures, spastic extremities, scoliosis, hip dysplasia, delayed bone age, idiopathic high B12 levelsb | delayed myelination, thin CC, subsequent volume loss | p.Leu312Pro |
| 3/M/21 months | 29.5 (−2.8 SD) | seizures | 2 months/GTCS | profound | deaf bilaterally | contractures, cataract, FTT, inguinal hernia | N.A. | p.Leu312Pro |
Abbreviations are as follows: FTT, failure to thrive; CC, corpus callosum; N.A., not available; GTCS, general tonic clonic; ahypodontia, dysplastic nails, tapered fingers, edema and puffiness of hands and feet, very hypermobile fingers and wrists, and short toes; b >2000 pg/mL.
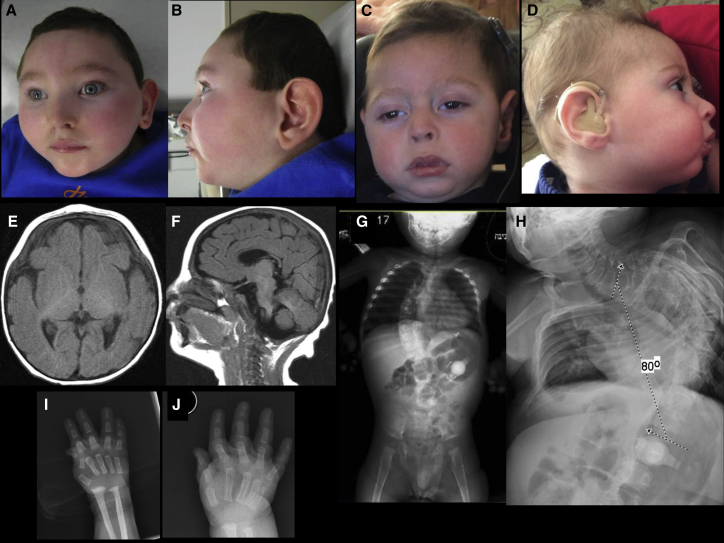
Figure 1.
Clinical and Radiographic Findings
(A–D) Craniofacial features of affected individual 2 at age 3 years (A and B) and affected individual 1 at age 22 months (C) and 18 months (D). Images show microcephaly with midface hypoplasia, a narrowed forehead, a short nose, a small chin, and a narrowed nasal bridge and base.
(E and F) A brain MRI of affected individual 2 at age 10 months reveals delayed myelination with thin corpus callosum (E: T1 axial, F: T1 sagittal).
(G and H) A scoliosis X-ray of affected individual 1 at age 21 months (G) and affected individual 2 at age 8 years (H) shows 80° levoscoliosis.
(I and J) A hand X-ray of affected individual 2 at 3 months (I) and 17 months of age (J) shows initial flexion deformity of metacarpophalangeal joint and joint hypermobility as well as markedly delayed bone age (i.e., absent epiphyses and ossification of carpal bones).
Brain MRI of affected individual 1 at age 3 months showed a thin corpus callosum, whereas a brain MRI of affected individual 2 showed thin corpus callosum and delayed myelination at 1 year of age. Subsequent imaging revealed supra- and infratentorial volume loss.
Laboratory investigations, including blood lactate, ammonia, very long chain fatty acids, amino acids, biotinidase activity, isoelectrofocusing of transferrins, and acylcarnitine levels, were normal, as were cerebrospinal-fluid amino acids and glucose; urinary organic acid analysis repeatedly displayed increased lactate excretion in one of the three affected individuals. Chromosomal microarray was normal in all three affected individuals, and mitochondrial DNA sequencing performed in affected individual 2 disclosed no pathogenic variants.
Whole-exome analysis (WES) for affected individual 1 and his parents was performed on exon targets captured with the SureSelect Human All Exon 50 Mb Kit V4 (Agilent Technologies). Sequences were determined by HiSeq2500 (Illumina). The full sequencing methodology and variant interpretation protocol were previously described.3 For individuals 2 and 3 and their parents, WES was performed at GeneDX on exon targets isolated by capture with the Clinical Research Exome kit (Agilent Technologies). The full sequencing methodology and variant interpretation protocol has been previously described.4 The general assertion criteria for variant classification are publicly available on the GeneDx ClinVar submission page (see Web Resources below). All procedures were performed in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national); proper informed consent was obtained from all guardians.
In the WES of the affected individuals, average depth of coverage was 81×, 351×, and 30×, with 95.6%, 85.1%, and 85.3% of the target covered at least 20x for affected individuals 1, 2, and 3, respectively. Following read alignment, variant calling, and filtration, 334 heterozygous variants remained; all but one were inherited from the parents. For affected individual 1, the non-inherited variant was GRCh37/Hg19 Chr3: 149678677T>C (GenBank: NM_007282.4) (c.932T>C[ p.Leu311Ser]) in RING finger protein 13 (RNF13, MIM: 609247), whereas for affected individuals 2 and 3, this was GRCh37/Hg19 Chr3: 149678680T>C (GenBank: NM_007282.4) (c.935T>C [p.Leu312Pro]) in RNF13. We verified the findings by Sanger sequencing in the affected individuals and confirmed their absence from the DNA of all the parents and the healthy siblings. The variants were not carried by any of the ∼123,000 individuals whose exome analyses were deposited at gnomAD (see Web Resources below), nor were they found at the Hadassah and GeneDx WES databases. The three affected individuals became known to each other’s physicians through the GeneMatcher website.26
RNF13 encodes a 381 amino acid protein, which belongs to the PA (protease associated)-TM (transmembrane)-RING (really interesting new gene) protein family. RNF13 was shown to associate with IRE1α and promote its activation.5 IRE1α is the main ER stress sensor and functions as a transcription factor of ER chaperones.6 Activated IRE1α cleaves the mRNA of X-box-binding protein 1 (XBP1).7 It also interacts with tumor-necrosis-factor-receptor-associated factor-2 (TRAF2), leading to the activation of apoptosis-signal-regulating kinase 1 (ASK1) and c-Jun NH2-terminal kinase (JNK).8 In turn, phosphorylated JNK activates the cytochrome-c-mediated apoptotic pathway by phosphorylating specific members of the BCL-2 family of proteins.9, 10, 11 Thus, RNF13 is considered a crucial mediator of ER stress-induced JNK activation and apoptosis through its interaction with and activation of IRE1α. Because heterozygotes for RNF13 loss-of-function variants are present at the expected rate (relative to protein length, pLI = 0.00) in the ExAC cohort of ∼60,000 healthy individuals (see Web Resources below), we explored the possibility that the c.932T>C (p.Leu311Ser) or c.935T>C (p.Leu312Pro) variant in RNF13 confers a gain of function, i.e., increased ER stress-induced apoptosis relative to that in unrelated, sex-matched control cells from a young, healthy adult.
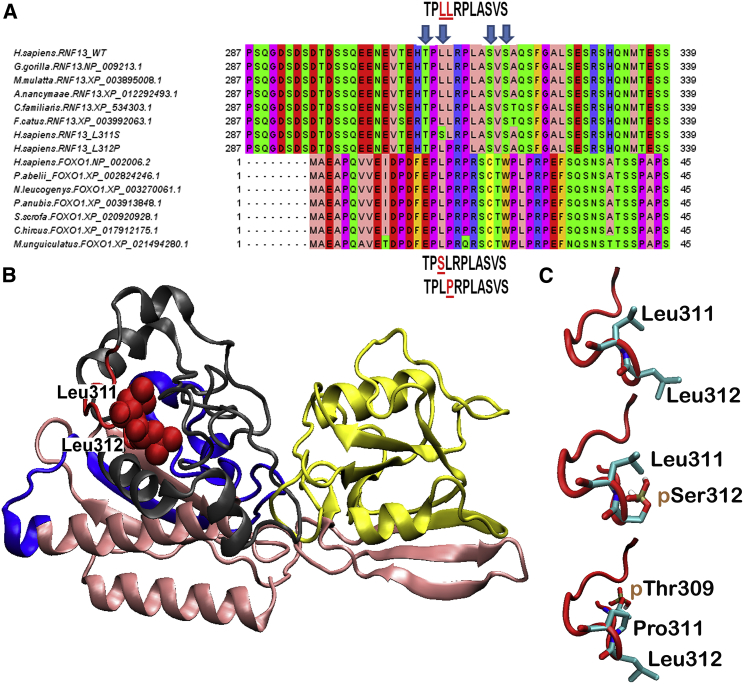
The two RNF13 variants are localized in a highly conserved sequence (Figure 2A) that includes a set of charged, hydrophilic residues, 309-TPLLRPLASVS-319 (see PhosphoSite Plus in Web Resources below), which is the target of post-translational modification (PTM) by phosphorylation at Thr309 and Ser319 (Figure S1). No structures of RNF13 or its close paralogs are currently available. Thus, to gain insights into the impact of these mutations, we created a computational 3D comparative model of RNF13 structure (for methods of 3D model generation, see Supplemental Data). The 3D model revealed that the di-leucine motif, together with the entire 309-TPLLRPLASVS-319 sequence, is located on the protein surface and forms a loop between the PEST and the Ser-rich sequence motifs (Figure 2B; Figure S2). In silico mutagenesis simulations showed that the two mutations result in only slightly different orientations of side chains in adjacent amino acids (Figure 2C), thus ruling out a significant perturbation of the local tertiary structure (i.e., the global structural arrangement of PEST and Ser-rich motifs containing the mutated loop). Furthermore, the new variants containing motifs 309-TPSLRPLASVS-319 and 309-TPLPRPLASVS-319 are highly similar to the sequence 24-PLPRPRSCT-32, hosting phosphorylation at Thr32 in FOXO111 (Figure 2A), suggesting that these mutations might alter PTMs of RNF13. This is further corroborated by the results of the NetPhos 3.1 PTM prediction server,12 which reported a novel high-probability phosphorylation site at Ser311 (p = 0.91), in the case of variant c.932T>C (p.Leu311Ser), and an increased probability of Thr309 phosphorylation (from p = 0.56 to p = 0.64), in the case of c.935T>C( p.Leu312Pro) compared to wild-type (Figure 2C). Altogether, these observations led us to explore the possibility of a gain-of-function effect of these variants.
Figure 2.
3D Modeling of RNF13 Protein Structure
(A) RNF13 orthologous sequences from various mammalian species. The dileucine motif in position 311-312, altered in the affected individuals reported here, is highly conserved. FOXO1 sequences are reported for comparative purposes.
(B) The 3D model of RNF13. The PA_C_RZF-like domain located at the N-terminal of RNF13 (residues 55-171) is shown in yellow. The RING_H2_RNF167 domain (residues 233-283) is shown in blue. Residues 309-319, including the discussed 309-TPLLRPLASVS-319 sequence, are presented in red. The putative PEST motif and a serine-rich sequence (See alignment panel), located close to the 309-TPLLRPLASVS-319 sequence at the end of the C-terminal domain (residues 284-381), are shown in gray. Finally, the N-terminal domain (residues 1-54) and the domain including residues 172-232 are presented in pink.
(C) The 309-TPLLRPLASVS-319 loop and the resulting two variants, 309-TPSLRPLASVS-319 and 309-TPLPRPLASVS-319, are shown in a red cartoon representation. High-probability phosphorylated residues are indicated.
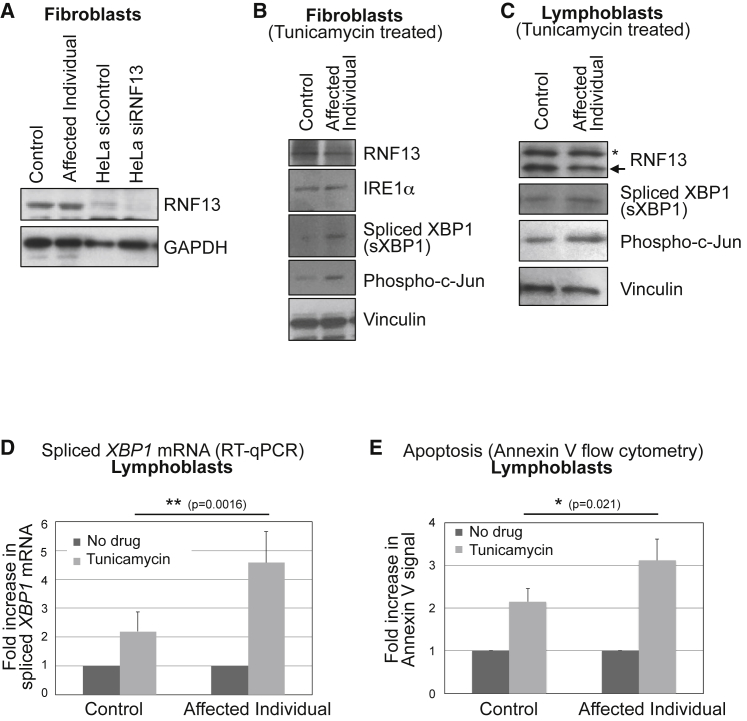
We therefore studied the signaling of ER stress in fibroblast and lymphoblast cells derived from affected individual 1 and an unrelated control (detailed materials and methods are described in the Supplemental Data). We first investigated RNF13 abundance in these cells. Immunoblot experiments indicated no major difference between control cells and those from the affected individual (Figure 3A–3C). Treatment of HeLa cells with RNF13-targeting siRNA resulted in its depletion, confirming that the observed band corresponds to RNF13 (Figure 3A). Given the role of RNF13 in the modulation of signaling of ER stress, such as that induced by tunicamycin,5 we treated control cells and cells from affected individual 1 with 1 μg/mL tunicamycin for 48 hr. Immunoblot experiments showed that, although IRE1α abundance was unchanged, both XBP1 splicing and c-Jun phosphorylation increased in fibroblast (Figure 3B) and lymphoblast (Figure 3C) cells derived from the affected individual. Moreover, quantitative determination of spliced XBP1 mRNA by RT-qPCR13 confirmed increased levels in cells from the affected individual upon tunicamycin treatment (Figure 3D, Figure S3A). Because ER stress signaling mediated by RNF13 had previously been shown to be associated with induction of apoptosis,5 we investigated apoptosis levels under increased ER stress. To this end, we measured annexin V positivity in control and affected individual lymphoblasts after 1 μg/mL tunicamycin treatment for 48 hr. Compared to control cells, cells from the affected individual showed significantly higher tunicamycin-induced annexin V signal, suggesting increased apoptosis (Figure 3E, Figure S3B). These results indicate that the c.932T>C (p.Leu311Ser) RNF13 mutation identified in individual 1 has a gain-of-function effect, resulting in enhanced signaling of ER stress and increased ER stress-induced apoptosis in mutated relative to control cells.
Figure 3.
Increased ER Stress-Induced Apoptosis in RNF13-Mutant Cells
(A) Control fibroblasts and fibroblasts derived from affected individual 1 were analyzed for RNF13 protein abundance by immunoblot. To confirm that the indicated band corresponds to RNF13, we treated HeLa cells with RNF13-targeting siRNA and analyzed cells in parallel.
(B and C) Increased signaling of ER stress in fibroblasts (B) and lymphoblasts (C) derived from affected individual 1. Cells were treated with 1 μg/mL tunicamycin for 48 hr, and lysates were analyzed for XBP1 splicing (2.4- and 2.5-fold increases in the affected individual’s fibroblasts and lymphoblasts, respectively, when results were normalized against those for vinculin) and Jun phosphorylation at Ser73 (3.2- and 3.7-fold increases in the affected individual’s fibroblasts and lymphoblasts, respectively). The asterisk indicates a crossreactive band.
(D) Increased tunicamycin-induced XBP1 splicing in the affected individual’s lymphoblasts, quantified by qRT-PCR. Cells were treated with 1 μg/mL tunicamycin for 48 hr. The average of six experiments, with standard deviations indicated as error bars, is shown. Tunicamycin-treated samples were normalized to the “no drug” condition for each cell type. The statistical difference between tunicamycin-treated control cells and those from the affected individual (TTEST, two-tailed, unequal variance) is indicated. Baseline (no drug treatment) amounts of spliced XBP1 mRNA are not significantly different in the affected individual (Figure S3A).
(E) Increased tunicamycin-induced apoptosis in the affected individual’s lymphoblasts. Cells were treated with 1 μg/mL tunicamycin for 48 hr and analyzed for Annexin V labeling by flow cytometry. The average of four experiments, for which standard deviations are indicated as error bars, is shown. Tunicamycin-treated samples were normalized to the “no drug” condition for each cell type. The statistical difference between tunicamycin-treated control cells and the affected individual’s cells (TTEST, two-tailed, unequal variance) is indicated. Baseline (no drug treatment) levels of apoptosis are not significantly different in the affected individual (Figure S3B).
Apoptosis, the activation of the caspase cascade leading to cell death, is an essential feature of cortical brain development. During neurogenesis, about 70% of progenitor cells in the ventricular and subventricular zone undergo apoptosis; this number is similar to the number of postmitotic cells that undergo apoptosis, leading to the suggestion that apoptosis controls the stem-cell population.14 Regulation of apoptosis is necessary for normal brain development; an increase in neuronal apoptosis, with the resultant depletion of the intermediate neuron-progenitor population, is observed in the microcephalic Mos+/− mice. This strain is haploinsufficient for Magoh, the exon-junction-complex component that regulates neural stem-cell division.15 Conversely, apoptosis reduction due to Caspase 9 knockout is associated with a markedly enlarged and malformed brain in mice.16 Collectively, these data support the hypothesis that mitigation of endoplasmic reticulum (ER) stress and precision in spatio-temporal regulation of apoptosis is critical for normal brain development.
RNF13 was previously identified as a critical regulator of staurosporine-mediated apoptosis. RNF13-knockdown cells showed a markedly reduced response to the triggering of ER stress-mediated apoptosis, whereas overexpression of RNF13 induced an enhanced ER stress response and caspase-dependent apoptosis.5 RNF13 is synthesized as an endosomal integral membrane protein17 and was shown to localize with markers of the late endosomal-lysosomal system.18 RNF13 expression is prominent in embryonic and adult brain tissues, and RNF13 was shown to be involved in neuronal development. Brain RNF13 mRNA is upregulated after initiation of neurite outgrowth.17 Genetic disruption of Rnf13 leads to a learning and memory defect in mice,19 whereas its ectopic expression promotes spontaneous neurite outgrowth of PC12 cells in vitro.20, 21 Our work suggests that these neuro-cognitive phenotypes might be attributed to altered levels of apoptosis.
Here we show that heterozygous missense mutations in the Leu 311-Leu 312 dileucine motif of RNF13 are associated with gain of function of its apoptosis-related activity without affecting RNF13 abundance. Abnormally increased apoptosis was previously reported in neurological syndromes in humans; individuals suffering from the renal-neurological disease Galloway-Mowat syndrome [MIM: 251300], characterized by microcephaly, brain anomalies, and developmental delay, accompanied by early-onset nephrotic syndrome, were shown to carry recessive mutations in subunits of the KEOPS complex. This complex is involved, among its other activities, in tRNA modification; defects in the KEOPS complex would thus result in translational errors that can impair protein folding and thereby activate a UPR. In agreement, knocking down KEOPS subunits resulted in an increase in phosphorylated IRE1α, spliced XBP-1, phosphorylated eIF2α, and ATF4, indicating activation of the UPR.22 Another example of the association between increased apoptosis and neurological syndromes is seen in affected individuals with bi-allelic mutations in IER3IP1 (immediate early response 3 interacting protein 1 [MIM: 609382]); these mutations manifest as a simplified gyral pattern in combination with severe infantile epileptic encephalopathy and early-onset permanent diabetes. Although the role of IER3IP1 in apoptosis induction is unknown, increased apoptosis is observed in the affected individual’s cortex, as well as in control cells treated with IER3IP1 siRNA after dithiothreitol-induced ER stress.23
RNF13 was previously assumed to function as an E3 ubiquitin-protein ligase.18, 19 Mutations in another E3 ubiquitin ligase, encoding Malin (NHLRC1 [MIM: 608072], were shown to cause the Lafora progressive myoclonus epilepsy.24 Indeed, from a clinical point of view, RNF13-associated disease can be partly regarded as early-onset progressive myoclonus epilepsy.
In summary, we propose that the neurodegenerative disorder affecting the individuals described here is attributed to the heterozygous RNF13 variants c.932T>C( p.Leu311Ser) or c.935T>C( p.Leu312Pro), which occurred de novo in the three individuals. The variants were absent from large cohorts of healthy individuals, and the three affected individuals share a distinctive clinical phenotype. Furthermore, the variants are adjacent and reside in an amino acid stretch that is highly conserved throughout evolution, and we found them to be associated with RNF13 gain of function manifesting as abnormally increased ER-stress-induced apoptosis, which in itself is reported to cause neurological disorder in humans. We hypothesize that the presence of a new PTM site or the modification of the amino acid sequence flanking a native PTM target site might be responsible for altering RNF13 function, perhaps by changing its half-life or its interactions with other proteins, such as IRE1α, that are involved in apoptosis regulation.25
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
We would like to thank the Penn State College of Medicine Flow Cytometry core for technical support.
Published: December 27, 2018
Footnotes
Supplemental Data include Supplemental Material and Methods, a case report, and three figures and can be found with this article online at https://doi.org/10.1016/j.ajhg.2018.11.018.
Contributor Information
Orly Elpeleg, Email: elpeleg@hadassah.org.il.
George-Lucian Moldovan, Email: gmoldovan@pennstatehealth.psu.edu.
Web Resources
ExAC for RNF13, http://exac.broadinstitute.org/gene/ENSG00000082996
GeneDx ClinVar Submission Page, http://www.ncbi.nlm.nih.gov/clinvar/submitters/26957
GeneMatcher, https://www.genematcher.org
gnomAD, http://gnomad.broadinstitute.org
NetPhos 3.1, http://www.cbs.dtu.dk/services/NetPhos/
Online Mendelian Inheritance in Man, http://www.omim.org
PhosphoSite Plus, https://www.phosphosite.org/proteinAction.action?id=3664450&showAllSites=true
Supplemental Data
References
- 1.Merksamer P.I., Papa F.R. The UPR and cell fate at a glance. J. Cell Sci. 2010;123:1003–1006. doi: 10.1242/jcs.035832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Malhotra J.D., Kaufman R.J. The endoplasmic reticulum and the unfolded protein response. Semin. Cell Dev. Biol. 2007;18:716–731. doi: 10.1016/j.semcdb.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ta-Shma A., Zhang K., Salimova E., Zernecke A., Sieiro-Mosti D., Stegner D., Furtado M., Shaag A., Perles Z., Nieswandt B. Congenital valvular defects associated with deleterious mutations in the PLD1 gene. J. Med. Genet. 2017;54:278–286. doi: 10.1136/jmedgenet-2016-104259. [DOI] [PubMed] [Google Scholar]
- 4.Tanaka A.J., Cho M.T., Millan F., Juusola J., Retterer K., Joshi C., Niyazov D., Garnica A., Gratz E., Deardorff M. Mutations in SPATA5 are associated with microcephaly, intellectual disability, seizures, and hearing loss. Am. J. Hum. Genet. 2015;97:457–464. doi: 10.1016/j.ajhg.2015.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arshad M., Ye Z., Gu X., Wong C.K., Liu Y., Li D., Zhou L., Zhang Y., Bay W.P., Yu V.C., Li P. RNF13, a RING finger protein, mediates endoplasmic reticulum stress-induced apoptosis through the inositol-requiring enzyme (IRE1α)/c-Jun NH2-terminal kinase pathway. J. Biol. Chem. 2013;288:8726–8736. doi: 10.1074/jbc.M112.368829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ye J., Rawson R.B., Komuro R., Chen X., Davé U.P., Prywes R., Brown M.S., Goldstein J.L. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol. Cell. 2000;6:1355–1364. doi: 10.1016/s1097-2765(00)00133-7. [DOI] [PubMed] [Google Scholar]
- 7.Hetz C., Martinon F., Rodriguez D., Glimcher L.H. The unfolded protein response: integrating stress signals through the stress sensor IRE1α. Physiol. Rev. 2011;91:1219–1243. doi: 10.1152/physrev.00001.2011. [DOI] [PubMed] [Google Scholar]
- 8.Urano F., Wang X., Bertolotti A., Zhang Y., Chung P., Harding H.P., Ron D. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science. 2000;287:664–666. doi: 10.1126/science.287.5453.664. [DOI] [PubMed] [Google Scholar]
- 9.Lei K., Davis R.J. JNK phosphorylation of Bim-related members of the Bcl2 family induces Bax-dependent apoptosis. Proc. Natl. Acad. Sci. USA. 2003;100:2432–2437. doi: 10.1073/pnas.0438011100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tournier C., Hess P., Yang D.D., Xu J., Turner T.K., Nimnual A., Bar-Sagi D., Jones S.N., Flavell R.A., Davis R.J. Requirement of JNK for stress-induced activation of the cytochrome c-mediated death pathway. Science. 2000;288:870–874. doi: 10.1126/science.288.5467.870. [DOI] [PubMed] [Google Scholar]
- 11.Scheid M.P., Woodgett J.R. PKB/AKT: Functional insights from genetic models. Nat. Rev. Mol. Cell Biol. 2001;2:760–768. doi: 10.1038/35096067. [DOI] [PubMed] [Google Scholar]
- 12.Blom N., Sicheritz-Pontén T., Gupta R., Gammeltoft S., Brunak S. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics. 2004;4:1633–1649. doi: 10.1002/pmic.200300771. [DOI] [PubMed] [Google Scholar]
- 13.van Schadewijk A., van’t Wout E.F., Stolk J., Hiemstra P.S. A quantitative method for detection of spliced X-box binding protein-1 (XBP1) mRNA as a measure of endoplasmic reticulum (ER) stress. Cell Stress Chaperones. 2012;17:275–279. doi: 10.1007/s12192-011-0306-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanes D.H., Reh T.A., Harris W.A., editors. Development of the nervous system. Second Edition. Elsevier Academic Press; Oxford: 2006. pp. 173–206. [Google Scholar]
- 15.Silver D.L., Watkins-Chow D.E., Schreck K.C., Pierfelice T.J., Larson D.M., Burnetti A.J., Liaw H.J., Myung K., Walsh C.A., Gaiano N., Pavan W.J. The exon junction complex component Magoh controls brain size by regulating neural stem cell division. Nat. Neurosci. 2010;13:551–558. doi: 10.1038/nn.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuida K., Haydar T.F., Kuan C.Y., Gu Y., Taya C., Karasuyama H., Su M.S., Rakic P., Flavell R.A. Reduced apoptosis and cytochrome c-mediated caspase activation in mice lacking caspase 9. Cell. 1998;94:325–337. doi: 10.1016/s0092-8674(00)81476-2. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Q., Meng Y., Zhang L., Chen J., Zhu D. RNF13: A novel RING-type ubiquitin ligase over-expressed in pancreatic cancer. Cell Res. 2009;19:348–357. doi: 10.1038/cr.2008.285. [DOI] [PubMed] [Google Scholar]
- 18.Bocock J.P., Carmicle S., Chhotani S., Ruffolo M.R., Chu H., Erickson A.H. The PA-TM-RING protein RING finger protein 13 is an endosomal integral membrane E3 ubiquitin ligase whose RING finger domain is released to the cytoplasm by proteolysis. FEBS J. 2009;276:1860–1877. doi: 10.1111/j.1742-4658.2009.06913.x. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Q., Li Y., Zhang L., Yang N., Meng J., Zuo P., Zhang Y., Chen J., Wang L., Gao X., Zhu D. E3 ubiquitin ligase RNF13 involves spatial learning and assembly of the SNARE complex. Cell. Mol. Life Sci. 2013;70:153–165. doi: 10.1007/s00018-012-1103-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saito S., Honma K., Kita-Matsuo H., Ochiya T., Kato K. Gene expression profiling of cerebellar development with high-throughput functional analysis. Physiol. Genomics. 2005;22:8–13. doi: 10.1152/physiolgenomics.00142.2004. [DOI] [PubMed] [Google Scholar]
- 21.Tranque P., Crossin K.L., Cirelli C., Edelman G.M., Mauro V.P. Identification and characterization of a RING zinc finger gene (C-RZF) expressed in chicken embryo cells. Proc. Natl. Acad. Sci. USA. 1996;93:3105–3109. doi: 10.1073/pnas.93.7.3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Braun D.A., Rao J., Mollet G., Schapiro D., Daugeron M.C., Tan W., Gribouval O., Boyer O., Revy P., Jobst-Schwan T. Mutations in KEOPS-complex genes cause nephrotic syndrome with primary microcephaly. Nat. Genet. 2017;49:1529–1538. doi: 10.1038/ng.3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poulton C.J., Schot R., Kia S.K., Jones M., Verheijen F.W., Venselaar H., de Wit M.C., de Graaff E., Bertoli-Avella A.M., Mancini G.M. Microcephaly with simplified gyration, epilepsy, and infantile diabetes linked to inappropriate apoptosis of neural progenitors. Am. J. Hum. Genet. 2011;89:265–276. doi: 10.1016/j.ajhg.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chan E.M., Young E.J., Ianzano L., Munteanu I., Zhao X., Christopoulos C.C., Avanzini G., Elia M., Ackerley C.A., Jovic N.J. Mutations in NHLRC1 cause progressive myoclonus epilepsy. Nat. Genet. 2003;35:125–127. doi: 10.1038/ng1238. [DOI] [PubMed] [Google Scholar]
- 25.Karve T.M., Cheema A.K. Small changes huge impact: the role of protein posttranslational modifications in cellular homeostasis and disease. J. Amino Acids. 2011;2011:207691. doi: 10.4061/2011/207691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.